Abstract
Our previous studies showed that Blastomyces dermatitidis yeast activates the human complement system, leading to deposition of opsonic complement fragments onto the yeast surface. This report examines the influence of altered surface expression of glucan or BAD1 protein (formerly WI-1) on the yeast's ability to activate and bind C3. Compared to the wild type, a glucan-deficient mutant yeast delayed initiation of C3 deposition and reduced C3-binding capacity by 50%. Linkage of baker's-yeast β-glucan to the glucan-deficient yeast restored initial C3 deposition kinetics to the wild-type level and partially restored C3-binding capacity, suggesting that β-glucan is an initiator of complement activation and a C3 acceptor. The role of BAD1 in B. dermatitidis yeast-complement interaction was also assessed. BAD1 knockout yeast initiated faster C3 deposition and increased C3-binding capacity compared to the wild-type yeast or a BAD1-reconstituted yeast, suggesting either a lack of an intrinsic ability in BAD1 or an inhibitory role of BAD1 in complement activation and binding. However, both complement activation and the capacity for C3 binding by the wild-type yeast were enhanced in normal human serum supplemented with an anti-BAD1 monoclonal antibody (MAb) or in immune sera from blastomycosis patients. Microscopic analysis revealed that more initial C3-binding sites were formed on yeast in the presence of both naturally occurring complement initiators and exogenous anti-BAD1 MAb, suggesting that anti-BAD1 antibody enhanced the ability of B. dermatitidis yeast to interact with the host complement system. Thus, glucan and BAD1 have distinctly different regulatory effects on complement activation by B. dermatitidis.
Blastomyces dermatitidis is the etiological agent of blastomycosis, which is one of the principal endemic systemic mycoses of humans and other mammals (22). Host defense mechanisms against B. dermatitidis infection include the complement system. Complement promotes attachment of human phagocytes to B. dermatitidis and is required for neutrophil-mediated killing of B. dermatitidis yeast (5). Our previous studies showed that B. dermatitidis yeast cells activate the human complement system via both the classical and alternative pathways, leading to deposition of C3b and iC3b complement fragments on the yeast surface (26). Furthermore, we found that naturally occurring anti-β-glucan antibodies promote activation of the classical complement pathway by the yeast (26). How yeast cell surface structures modulate the yeast's ability to interact with the human complement system remains unknown.
The cell surface of B. dermatitidis yeast is associated with a dynamic array of proteins and carbohydrates. Prominent among them are glucan and surface protein BAD1, formerly termed WI-1 (21), both of which have been implicated as virulence factors (2, 10). About 95% of the glucan components are α-glucan, and the remainder are β-glucan (8). BAD1 is a 120-kDa immunodominant antigen; it is also an adhesin that binds the yeast to CD14 and complement receptors (13, 19). Both human patients with blastomycosis and mice vaccinated with purified BAD1 develop strong humoral and cell-mediated immune responses to BAD1 (13, 15, 25). Vaccinated mice are more resistant than unvaccinated controls to experimental pulmonary blastomycosis (25).
In this study, we investigated the roles of glucan and BAD1 in activation of the human complement system by B. dermatitidis yeast. The goals of our study were (i) to assess the effect of surface β-glucan on the kinetics of C3 deposition, (ii) to determine the role of surface BAD1 in activation of the complement system, and (iii) to determine the effect of anti-BAD1 antibody on the kinetics of C3 deposition. Our findings demonstrate that surface glucan and BAD1 have distinctly different regulatory roles. β-Glucan supports complement activation, whereas BAD1 is not required for activation of either the classical or the alternative complement pathway and appears to retard complement activation. However, anti-BAD1 antibody markedly enhances the ability of the yeast to activate the human complement system.
MATERIALS AND METHODS
B. dermatitidis strains.
American Type Culture Collection (Manassas, Va.) strains 26199 and 60916 were used in this study. Strain 26199 was originally isolated from a human patient, and strain 60916 was derived from strain 26199 through serial passages (3). Yeast cells of strain 26199 express both glucan and BAD1, whereas yeast cells of strain 60916 express undetectable amounts of α-glucan (7), much less β-glucan (M. X. Zhang and B. S. Klein, unpublished data), and 5 to 10 times more BAD1 than do those of the parental strain (11). B. dermatitidis 55 was derived from strain 26199 by targeted disruption of the BAD1 gene, and B. dermatitidis strain 4-55 was derived from strain 55 by reconstitution of BAD1 through gene transfer (2). All isolates were grown in the yeast form on Middlebrook 7H10 agar medium containing oleic acid-albumin complex (Sigma Chemical Co., St. Louis, Mo.) at 37°C for 72 h. Yeast cells were then collected, washed in phosphate-buffered saline (PBS; 1.9 mM NaH2PO4, 8.1 mM Na2HPO4, 154 mM NaCl, pH 7.2), heat killed at 60°C, and stored in PBS–0.05% azide at 4°C (26).
Linkage of glucan to the yeast surface.
Glucan purified from baker's yeast (Saccharomyces cerevisiae) (catalog no. G5011) was purchased from Sigma, St. Louis, Mo. Glucan was linked to B. dermatitidis 60916 by using the 1.82-nm cross-linker Sulfo-SANPAH (SPH; Pierce, Rockford, Ill.) in accordance with the manufacturer's instructions, with minor modifications. Briefly, 5 mg of glucan was solubilized in 450 μl of 1 N NaOH at 60°C for 30 min, cooled to room temperature, treated with 450 μl of 1 N HCl and 450 μl of 0.2 M sodium phosphate buffer (pH 7.3) at 4°C for 18 to 36 h, and centrifuged to remove insoluble material. Cross-linker SPH was conjugated to the yeast surface in a siliconized microcentrifuge tube containing 500 μl of 0.5 mM SPH and 5 × 105 heat-killed B. dermatitidis 60916 yeast cells that were rocked in the dark for 30 min at room temperature. Cross-linker-treated yeast cells were washed three times with PBS and resuspended in 600 μl of soluble glucan. Glucan molecules were conjugated to yeast-attached cross-linkers by use of UV irradiation at 365 nm for 20 min. In a flow cytometry analysis, rabbit anti-β-glucan serum (20) (a generous gift of D. M. Schmatz, Merck Laboratories, Rahway, N.J.) and fluorescein isothiocyanate (FITC)-conjugated goat anti-rabbit antibody detected an approximately 30% gain in mean channel fluorescence in glucan-treated 60916 yeast cells compared to that of untreated 60916 yeast cells or yeast cells treated with the cross-linker alone.
Serum and serum proteins.
C3 was isolated from frozen human plasma (Reno Blood Services, Reno, Nev.) (17, 23) and labeled with 125I by use of Iodogen reagent (Pierce) (6). Pooled normal human serum (NHS) was prepared from peripheral blood collected from at least 10 normal donors with no history of blastomycosis and was stored at −80°C. Yeast-absorbed serum was prepared by incubation of NHS twice, each time with 108 26199 yeast cells for 1 h on ice with frequent mixing, followed by centrifugation to separate the serum from the yeast. Yeast-absorbed serum was filtered through a 0.45-μm-pore-size filter and used immediately. The concentration of absorbed serum was corrected for the dilution that occurred during the absorption procedure. Sera from patients with blastomycosis were collected and stored, and the presence of serum anti-BAD1 antibodies was confirmed as previously described (13).
Quantitative analysis of C3 binding using 125I-labeled C3.
Binding of C3 to B. dermatitidis yeast cells was analyzed by the procedure of Kozel et al. (18). Briefly, each complement-binding medium contained (i) 40% nonabsorbed NHS, yeast-absorbed NHS, or sera of blastomycosis patients and (ii) 125I-labeled C3 sufficient to provide a specific activity of 50,000 cpm/μg of C3 for the mixture of labeled and unlabeled C3 in the serum (assuming that NHS contains 1,200 μg of C3/ml). To study classical pathway activation, the binding medium contained GVB2+ buffer (sodium Veronal [5 mM]-buffered saline [142 mM], pH 7.3, containing 0.1% gelatin, 1.5 mM CaCl2, and 1 mM MgCl2). To study alternative pathway activation, the medium contained GVB-Mg-EGTA (sodium Veronal [5 mM]-buffered saline [142 mM], pH 7.3, containing 0.1% gelatin, 5 mM EGTA, and 5 mM MgCl2); EGTA chelates calcium, which is needed for classical pathway activity. In some experiments, MAb DD5-CB4, specific for a 25-amino-acid tandem repeat contained in BAD1 (12), was added to the binding medium prior to addition of the yeast cells. The reaction medium was warmed to and kept at 37°C, and 106 yeast cells per ml of reaction medium were added to initiate C3 binding. To study C3 deposition kinetics, 50-μl samples were withdrawn in duplicate at various time intervals and added to 200 μl of a stop solution (PBS, 0.1% sodium dodecyl sulfate, 20 mM EDTA) in Millipore MABX-N12 filter plates fitted with BV 1.2-μm-pore-size filter membranes (Millipore, Bedford, Mass.). The particles were washed five times with PBS containing 0.1% sodium dodecyl sulfate. The membranes were removed, and the amount of radioactivity bound to the yeast cells collected on the membranes was determined with a Packard AUTO-GAMMA counter. Specific binding was determined by subtracting the amount of nonspecific binding observed in reaction medium that contained GVB-EDTA (sodium Veronal [5 mM]-buffered saline [142 mM], pH 7.3, 0.1% gelatin, 10 mM EDTA), as EDTA blocks activation of the complement system.
Immunofluorescence analysis of C3 binding.
The pattern of C3 binding to the yeast cell surface was determined by immunofluorescence in the presence of various concentrations of anti-BAD1 MAb (27). Yeast cells were opsonized in the same manner as described above for the quantitative analysis, except that the reaction mixtures did not contain 125I-labeled C3. At various time intervals, 200-μl samples were transferred to 900 μl of ice-cold PBS containing 10 mM EDTA and unbound C3 was removed by three washes with PBS. The yeast-bound C3 was detected with FITC-conjugated goat anti-human C3 antibodies (Kent Laboratories, Indianapolis, Ind.). The FITC-conjugated goat anti-human C3 had been absorbed with B. dermatitidis 26199 yeast cells to remove nonspecific antibodies. The fluorescence images of C3 deposition on the yeast surface were acquired at 0.4-μm intervals through individual cells by use of an epifluorescence microscope equipped with a Photonic Science (East Sussex, United Kingdom) Color CoolView image acquisition system and were processed with the aid of image analysis software including Image-Pro Plus (Media Cybernetics, Silver Spring, Md.) and MicroTome (VayTeK, Fairfield, Iowa). Each stack of images acquired from a single cell was digitally deconvolved and projected onto a single plane. A minimum of 100 yeast cells were examined for C3-binding patterns at each dose of anti-BAD1 MAb, and reproducible observations from two independent experiments were obtained.
RESULTS
Surface-linked glucan accelerates initiation of C3 deposition.
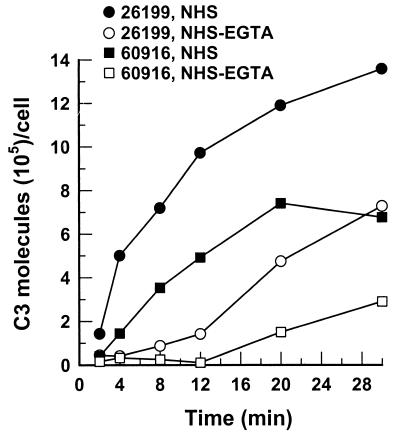
Our previous study showed that removal of natural antibody against β-glucan significantly delayed C3 deposition onto 26199 yeast cells, suggesting a requirement for β-glucan in complement activation by B. dermatitidis yeast. To test this hypothesis, we compared the kinetics of C3 deposition for yeast cells of glucan-deficient mutant strain 60916 and parental wild-type strain 26199. Yeast cells were incubated in pooled NHS with or without EGTA for up to 30 min. The number of bound C3 molecules was expressed either per unit of surface area or per yeast cell because the wild-type yeast differed from the mutant in shape. The kinetics of C3 deposition per unit of surface area were comparable to the kinetics of C3 deposition per cell, and only the latter are presented here. Although yeast cells of both strains initiated C3 deposition via either the classical pathway or the alternative pathway, glucan-deficient yeast exhibited a noticeably longer delay than the wild type in binding detectable amounts of C3 generated by either pathway (Fig. 1). This reduced ability of the mutant yeast to active the complement was also reflected in the rate of C3 deposition. The C3 deposition rate for the first 8 min of incubation in NHS was used as a measure of classical pathway activity because an approximately 8-min delay in accumulation of C3 was observed in EGTA-treated NHS where the classical pathway was blocked (Fig. 1 [see also Fig. 3], NHS-EGTA). During this 8-min interval, the rate of C3 deposition to the mutant yeast incubated in NHS was estimated to be 5 × 104 C3 molecules/min/cell, compared to 9 × 104 C3 molecules/min/cell for the wild-type yeast (Fig. 1). In addition to the slower rate of C3 deposition, the capacity of the mutant cells to bind C3 molecules was about 50% of that of the wild type (Fig. 1).
FIG. 1.
Kinetics of C3 deposition on B. dermatitidis yeast cells of wild-type (26199) and glucan-deficient mutant (60916) strains. Kinetics were assessed under conditions that permitted both the classical and alternative complement pathways (40% NHS; ●, 26199; ▪, 60916) or that limited C3-binding activity to the alternative complement pathway (40% EGTA-chelated NHS; ○, 26199; □, 60916). Data representative of duplicate experiments are shown.
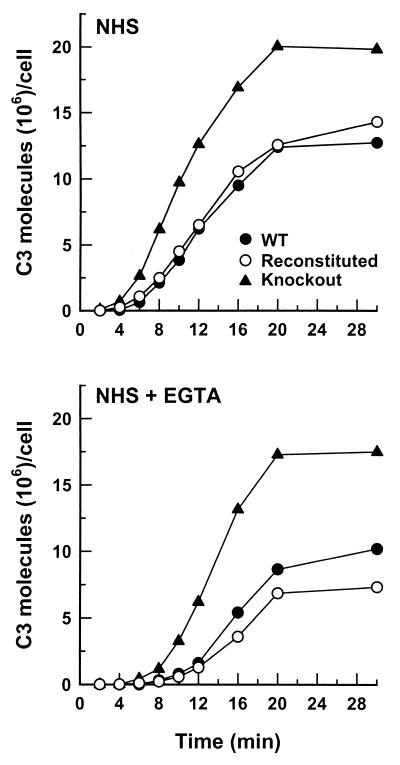
FIG. 3.
Effect of surface-protein BAD1 expression on kinetics of C3 deposition on B. dermatitidis yeast cells. Kinetics were assessed under conditions that permitted both the classical and alternative complement pathways (40% NHS) or only the alternative complement pathway (40% EGTA-chelated NHS) for the wild-type (WT) yeast (●), BAD1 knockout yeast (▴), or BAD1-reconstituted yeast (○). Data representative of duplicate experiments are shown.
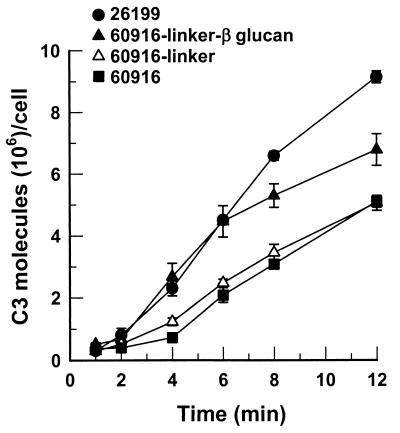
We next analyzed the effect of exogenous β-glucan on the ability of the mutant yeast to activate the classical complement pathway. Glucan-deficient yeast (60916) cells were conjugated with baker's yeast β-glucan via cross-linkers or with cross-linkers alone as a control. Kinetics of classical pathway-mediated C3 deposition onto the chemically modified mutant yeasts was compared to the kinetics of C3 deposition onto the wild-type yeast. Addition of β-glucan restored wild-type-like kinetics of early C3 deposition onto the glucan-deficient yeast, whereas addition of cross-linkers alone had no effect (Fig. 2). Surface-linked β-glucan also partially restored C3-binding capacity to the mutant (Fig. 2).
FIG. 2.
Effect of linked exogenous β-glucan on the kinetics of C3 deposition on B. dermatitidis yeast cells of glucan-deficient mutant strain 60916. The kinetics were assessed in an assay mixture containing 40% NHS that permitted both the classical and alternative complement pathways for wild-type strain 26199 (●), mutant yeast with a linker and β-glucan (▴), mutant yeast with the linker alone (▵), and untreated mutant yeast (▪). Results are expressed as the mean ± the standard error of the mean of three experiments.
BAD1 is not required for complement activation.
In addition to deficiency in glucan expression, the 60916 mutant yeast expresses 5 to 10 times more BAD1 than does the wild type (11). The slower initiation of complement activation by the glucan-deficient mutant cells prompted us to examine the role of BAD1 in complement activation and C3 deposition. We compared the kinetics of C3 deposition for yeast cells of isogenic strains that express or lack surface protein BAD1: wild-type 26199, BAD1 knockout 55, and BAD1-reconstituted 4-55. The BAD1 knockout yeast activated both the classical and alternative complement pathways, and C3 was deposited more rapidly on BAD1 knockout yeast than on wild-type yeast or BAD1-reconstituted yeast (Fig. 3). The rate of C3 deposition following 8 min of incubation in NHS was approximately 8 × 105 C3 molecules/min/cell for BAD1 knockout yeast and 3 × 105 C3 molecules/min/cell for either the wild-type or the BAD1-reconstituted yeast (Fig. 3). Yeast cells deficient in BAD1 also accumulated approximately twice as many C3 molecules as did wild-type or BAD1-reconstituted cells following a 30-min incubation in either NHS or EGTA-treated NHS (Fig. 3). These observations indicate that surface-expressed BAD1 is not required for complement activation by B. dermatitidis yeast cells and may, instead, retard complement opsonization of the yeast.
Anti-BAD1 antibody promotes C3 deposition onto yeast cells.
The assessment of the role of BAD1 in complement activation and binding as described above was conducted with pooled nonimmune human serum. BAD1 is immunogenic; strong humoral and cell-mediated immune responses against BAD1 have been observed both in people with blastomycosis (13, 15) and in an animal model of blastomycosis (25). Consequently, we analyzed the influence of anti-BAD1 antibodies on the ability of B. dermatitidis yeast cells to interact with the human complement system.
NHS was absorbed with B. dermatitidis 26199 yeast cells to remove natural initiators of complement activation (26). Kinetics of C3 deposition were determined for 26199 yeast cells that were incubated either in nonabsorbed NHS or in yeast-absorbed NHS supplemented with 0 to 100 μg of anti-BAD1 MAb/ml of assay mixture. In the yeast-absorbed serum, complement activation required the anti-BAD1 antibody and the rate of early C3 deposition was antibody dose dependent (Fig. 4A). At 25 or 100 μg/ml, the anti-BAD1 MAb enhanced the C3 molecule-binding capacity of the yeast (Fig. 4A).
FIG. 4.
Effect of anti-BAD1 antibodies on complement activation and C3 binding to B. dermatitidis yeast cells (26199). (A) Kinetics of C3 deposition were assessed in complement-binding medium containing either pooled NHS (●) or yeast-absorbed NHS that was supplemented, per milliliter of medium, with anti-BAD1 MAb DD5-CB4 at 0 μg (○), 1.6 μg (▴), 6.3 μg (▪), 25 μg (♦), or 100 μg (▾). (B) Capacity of C3 binding to strain 26199 yeast cells was determined following a 30-min incubation in nonimmune NHS or in sera from three blastomycosis patients. Anti-BAD1 endpoint titers for the patient sera were as follows: 7758, 1:10,240; 7740A, 1:5,000; 8637, 1:2,000. Data representative of duplicate experiments are shown.
Because the MAb described above was of mouse origin, anti-BAD1 antibody-mediated C3 deposition was further analyzed in the sera of three patients with blastomycosis. The presence of anti-BAD1 antibodies in these sera had been confirmed (13). The amount of C3 accumulation on 26199 yeast cells incubated in the patient sera for 30 min was at least twice that on the yeast incubated in pooled nonimmune serum and correlated with the endpoint titer of anti-BAD1 antibody in the immune serum (Fig. 4B). The latter observation is consistent with the contention that BAD1 is an immunodominant antigen displayed on B. dermatitidis yeast (13, 14).
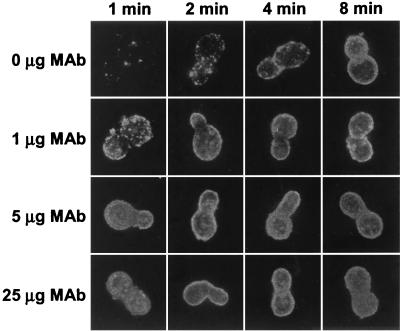
The above two observations indicated a quantitative enhancement of C3 deposition on B. dermatitidis yeast by anti-BAD1 antibodies. We then analyzed microscopic patterns of C3 deposition on strain 26199 yeast incubated in NHS with or without exogenous anti-BAD1 antibodies. In the absence of anti-BAD1 MAb, C3 deposition first appeared in a limited number of sites on yeast cells following 1 min of incubation in NHS but eventually occurred uniformly over the entire surface as incubation proceeded (Fig. 5). In the presence of the MAb, at 1 to 25 μg per ml of assay mixture, the appearance of C3 deposition sites was quantitatively dependent on the antibody dose (Fig. 5). Initiation sites were numerous yet distinguishable on yeast cells following 1 min of incubation in NHS containing 1 μg of MAb but became confluent and indistinguishable on yeast cells that were incubated in NHS containing 5 μg or more of the MAb (Fig. 5).
FIG. 5.
Effect of exogenous anti-BAD1 MAb on formation of C3 deposition sites. B. dermatitidis yeast cells (26199) were incubated for 1 to 8 min in a complement-binding medium that contained 40% NHS and 0, 1, 5, or 25 μg of anti-BAD1 MAb DD5-CB4 per ml of binding medium. Yeast cells were stained with FITC-labeled goat anti-human C3 antibodies.
DISCUSSION
Our previous studies demonstrated that B. dermatitidis yeast activates the human complement system, leading to deposition of opsonic complement fragments onto the yeast (26). The focus of the present study was to determine the structural requirements for complement activation by B. dermatitidis yeast cells. We analyzed the influence of alterations in display of glucan and surface protein BAD1 on the ability of B. dermatitidis yeast cells to interact with the human complement system. Our approach was to employ genetically related B. dermatitidis strains that differ in the expression of these two macromolecules.
First, we further established the role of β-glucan as an initiator of complement activation by B. dermatitidis yeast and as a potential C3 acceptor. We found that yeast cells of a glucan-deficient strain had both a reduced rate of initial C3 deposition and a 50% reduction in C3-binding capacity compared to yeast cells of the wild-type parental strain (Fig. 1). Importantly, linkage of baker's yeast β-glucan to the surface of the glucan-deficient mutant fully restored the early C3 deposition kinetics observed in the wild-type yeast and improved the C3-binding capacity (Fig. 2). These results confirmed and extended our previous observations that removal of naturally occurring antibodies against β-glucan inhibited complement activation by B. dermatitidis yeast and that the majority of yeast-bound C3 molecules were attached to acceptors through an ester linkage, suggesting an important contribution of surface carbohydrates to C3 binding (26). A requirement of glucan for complement activation has also been demonstrated with nonencapsulated Cryptococcus neoformans and zymosan particles (9, 24).
A failure to restore C3-binding capacity to the mutant yeast by exogenous β-glucan alone was not unexpected and may be attributed to a number of factors. First, cell wall components other than β-glucan may contribute additional C3-binding sites. Chemical analysis of the cell wall of B. dermatitidis yeast revealed that yeast glucan is composed of 95% α-glucan and 5% β-glucan (8). α-Glucan was undetected in the glucan-deficient mutant but present in the parental wild-type strain (7), and its absence may thus account for some of the reduced C3-binding capacity. Second, linked glucan molecules may not have formed an optimal surface for C3 binding, possibly due to the linker's length and/or density. The third possibility is that BAD1, which is expressed 5- to 10-fold more on the glucan-deficient mutant yeast than on the parental wild-type yeast, may have prevented efficient coating of the mutant cells with exogenous glucan. The overexpressed BAD1 on the mutant may have also masked available C3-binding sites contained in other cell wall components. These three factors, together and/or independently, may influence C3-binding capacity by altering the number of available binding sites on the yeast surface. They may also influence the cell surface topography that determines how deposition of additional C3 occurs following the initial binding of C3 molecules.
We analyzed the effect of surface protein BAD1 on the ability of B. dermatitidis yeast to activate complement and bind C3 by comparing C3 deposition kinetics in wild-type strain 26199 and isogenic BAD1 knockout strain 55. BAD1 could theoretically serve as an initiator of complement activation and/ or an acceptor of C3. However, we observed a noticeably faster rate of initial C3 deposition on the BAD1 knockout yeast than on the wild-type or the BAD1 reconstituted yeast (Fig. 3). This result was observed in either NHS, where both the classical and alternative complement pathways are active, or in EGTA-treated NHS, where only the alternative complement pathway is operative. This observation suggests that BAD1 may lack an intrinsic ability to activate complement or it may activate complement poorly. Alternatively, it may, instead, retard complement activation.
Strikingly, the C3 molecule-binding capacity was almost doubled in the BAD1 knockout yeast compared to that in the wild type (Fig. 3). The increased C3-binding capacity in the absence of BAD1 suggests that BAD1 molecules are poor C3 acceptors. This interpretation is supported by the dominant role of ester linkage in C3 attachment to B. dermatitidis yeast (26) and by the lack of carbohydrate in BAD1 molecules (14). It is also consistent with the observation of reduced C3-binding capacity in the glucan-deficient mutant that expresses 5 to 10 times more BAD1 than the parental wild type (Fig. 1 and 2). Alternatively, the increased C3-binding capacity in the absence of BAD1 suggests that BAD1 molecules block otherwise available C3-binding sites. A recent study demonstrated that BAD1 is associated with chitin through both covalent linkages and noncovalent interactions (1). Thus, the BAD1 knockout yeast may display more accessible chitin fibrils that support C3 deposition. However, the role of chitin in complement activation and C3 binding by B. dermatitidis yeast or other pathogenic fungi has largely been uncharacterized (16). Previously, we showed that absorption of NHS with purified chitin had no discernible effect on the ability of 26199 yeast cells to activate the classical complement pathway (26). Nonetheless, B. dermatitidis chitin fibrils could serve as C3 acceptors. The masking effect of surface-associated proteins on the ability of other cell surface structures to activate complement was recently reported with gram-negative bacteria. It was found that binding of serum amyloid P component to lipopolysaccharides of gram-negative bacteria prevented lipopolysaccharide-mediated activation of the classical complement pathway (4). How BAD1 retards C3 binding by B. dermatitidis yeast cells remains to be resolved.
The apparent inert or perhaps inhibitory properties of surface protein BAD1 in complement activation by B. dermatitidis could be markedly altered by anti-BAD1 antibodies in two ways. First, in the absence of naturally occurring activators of the classical complement pathway, a murine anti-BAD1 antibody alone could mediate a dose-dependent activation of the classical complement pathway (Fig. 4). In addition, more initial C3-binding sites were formed on yeast incubated in NHS plus anti-BAD1 MAb than on yeast incubated in NHS alone (Fig. 5). These observations demonstrate that either the natural initiators of complement activation, including anti-β-glucan antibodies, are limiting or the corresponding surface structures are limiting and that anti-BAD1 antibodies synergistically enhance the ability of B. dermatitidis yeast to interact with the human complement system. Second, the C3-binding capacity of B. dermatitidis yeast was markedly expanded in the presence of either murine anti-BAD1 MAb or human anti-BAD1 antibodies as supplied in blastomycosis-positive sera (Fig. 4).
Observations presented in this report demonstrate how two major surface components on B. dermatitidis yeast regulate interaction of the yeast with the host complement system. In each experiment, we focused on the independent effects of each component. Future studies might address the influence of the interplay of multiple yeast surface components on interactions between B. dermatitidis and the host complement system. These studies will require additional isogenic mutants simultaneously altered in more than one molecule, such as glucan and BAD1.
Our previous (26) and current studies allow us to propose a model of complement activation by B. dermatitidis yeast cells. In nonimmune serum, naturally occurring antibodies against β-glucan components initiate complement activation by B. dermatitidis yeast cells, and β- and α-glucans and other structures including chitin serve as major C3 acceptors. Surface protein BAD1 lacks an intrinsic ability to mediate complement activation by B. dermatitidis yeast cells and masks C3-binding sites on chitin and/or β- and α-glucans. However, anti-BAD1 antibodies mediate complement activation and enhance the C3-binding capacity of B. dermatitidis yeast by providing additional binding sites for C3 molecules and/or by modifying the surface topography for maximum binding of C3.
ACKNOWLEDGMENTS
We thank Robert Audet and Randall MacGill for technical assistance.
This work was supported in part by National Institutes of Health grants AI14209 and AI37194 (T.R.K.) and AI40996 and AI35681 (B.S.K.) and by a Burroughs Wellcome Fund Scholar Award in Molecular Mycology (B.S.K).
REFERENCES
- 1.Brandhorst T, Klein B. Cell wall biogenesis of Blastomyces dermatitidis. Evidence for a novel mechanism of cell surface localization of a virulence-associated adhesin via extracellular release and reassociation with cell wall chitin. J Biol Chem. 2000;275:7925–7934. doi: 10.1074/jbc.275.11.7925. [DOI] [PubMed] [Google Scholar]
- 2.Brandhorst T T, Wuthrich M, Warner T, Klein B. Targeted gene disruption reveals an adhesin indispensable for pathogenicity of Blastomyces dermatitidis. J Exp Med. 1999;189:1207–1216. doi: 10.1084/jem.189.8.1207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brass C, Volkmann C M, Philpott D E, Klein H P, Halde C J, Stevens D A. Spontaneous mutant of Blastomyces dermatitidis attenuated in virulence for mice. Sabouraudia. 1982;20:145–158. [PubMed] [Google Scholar]
- 4.de Haas C J, van Leeuwen E M, van Bommel T, Verhoef J, van Kessel K P, van Strijp J A. Serum amyloid P component bound to gram-negative bacteria prevents lipopolysaccharide-mediated classical pathway complement activation. Infect Immun. 2000;68:1753–1759. doi: 10.1128/iai.68.4.1753-1759.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Drutz D J, Frey C L. Intracellular and extracellular defenses of human phagocytes against Blastomyces dermatitidis conidia and yeasts. J Lab Clin Med. 1985;105:737–750. [PubMed] [Google Scholar]
- 6.Fraker P J, Speck J C., Jr Protein and cell membrane iodinations with a sparingly soluble chloroamide, 1,3,4,6-tetrachloro-3a,6a-diphenylglycoluril. Biochem Biophys Res Commun. 1978;80:849–857. doi: 10.1016/0006-291x(78)91322-0. [DOI] [PubMed] [Google Scholar]
- 7.Hogan L H, Klein B S. Altered expression of surface alpha-1,3-glucan in genetically related strains of Blastomyces dermatitidis that differ in virulence. Infect Immun. 1994;62:3543–3546. doi: 10.1128/iai.62.8.3543-3546.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kanetsuna F, Carbonell L M. Cell wall composition of the yeastlike and mycelial forms of Blastomyces dermatitidis. J Bacteriol. 1971;106:946–948. doi: 10.1128/jb.106.3.946-948.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Keller R G, Pfrommer G S, Kozel T R. Occurrences, specificities, and functions of ubiquitous antibodies in human serum that are reactive with the Cryptococcus neoformans cell wall. Infect Immun. 1994;62:215–220. doi: 10.1128/iai.62.1.215-220.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Klein B S. Role of cell surface molecules of Blastomyces dermatitidis in the pathogenesis and immunobiology of blastomycosis. Semin Respir Infect. 1997;12:198–205. [PubMed] [Google Scholar]
- 11.Klein B S, Chaturvedi S, Hogan L H, Jones J M, Newman S L. Altered expression of surface protein WI-1 in genetically related strains of Blastomyces dermatitidis that differ in virulence regulates recognition of yeasts by human macrophages. Infect Immun. 1994;62:3536–3542. doi: 10.1128/iai.62.8.3536-3542.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Klein B S, Hogan L H, Jones J M. Immunologic recognition of a 25-amino acid repeat arrayed in tandem on a major antigen of Blastomyces dermatitidis. J Clin Investig. 1993;92:330–337. doi: 10.1172/JCI116571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein B S, Jones J M. Isolation, purification, and radiolabeling of a novel 120-kD surface protein on Blastomyces dermatitidis yeasts to detect antibody in infected patients. J Clin Investig. 1990;85:152–161. doi: 10.1172/JCI114406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Klein B S, Jones J M. Purification and characterization of the major antigen WI-1 from Blastomyces dermatitidis yeasts and immunological comparison with A antigen. Infect Immun. 1994;62:3890–3900. doi: 10.1128/iai.62.9.3890-3900.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Klein B S, Sondel P M, Jones J M. WI-1, a novel 120-kilodalton surface protein on Blastomyces dermatitidis yeast cells, is a target antigen of cell-mediated immunity in human blastomycosis. Infect Immun. 1992;60:4291–4300. doi: 10.1128/iai.60.10.4291-4300.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kozel T R. Complement activation by pathogenic fungi. Res Immunol. 1998;149:309–320. doi: 10.1016/s0923-2494(98)80755-4. [DOI] [PubMed] [Google Scholar]
- 17.Kozel T R, Pfrommer G S. Activation of the complement system by Cryptococcus neoformans leads to binding of iC3b to the yeast. Infect Immun. 1986;52:1–5. doi: 10.1128/iai.52.1.1-5.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kozel T R, Weinhold L C, Lupan D M. Distinct characteristics of initiation of the classical and alternative complement pathways by Candida albicans. Infect Immun. 1996;64:3360–3368. doi: 10.1128/iai.64.8.3360-3368.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Newman S L, Chaturvedi S, Klein B S. The WI-1 antigen of Blastomyces dermatitidis yeasts mediates binding to human macrophage CD11b/CD18 (CR3) and CD14. J Immunol. 1995;154:753–761. [PubMed] [Google Scholar]
- 20.Nollstadt K H, Powles M A, Fujioka H, Aikawa M, Schmatz D M. Use of β-1,3-glucan-specific antibody to study the cyst wall of Pneumocystis carinii and effects of pneumocandin B0 analog L-733,560. Antimicrob Agents Chemother. 1994;38:2258–2265. doi: 10.1128/aac.38.10.2258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rooney P J, Sullivan T D, Klein B S. Selective expression of the virulence factor BAD1 upon morphogenesis to the pathogenic yeast form of Blastomyces dermatitidis: evidence for transcriptional regulation by a conserved mechanism. Mol Microbiol. 2001;39:875–889. doi: 10.1046/j.1365-2958.2001.02300.x. [DOI] [PubMed] [Google Scholar]
- 22.Sarosi G A, Davies S F. Blastomycosis. Am Rev Respir Dis. 1979;120:911–938. doi: 10.1164/arrd.1979.120.4.911. [DOI] [PubMed] [Google Scholar]
- 23.Tack B F, Janatova J, Thomas M L, Harrison R A, Hammer C H. The third, fourth, and fifth components of human complement: isolation and biochemical properties. Methods Enzymol. 1981;80:64–101. [Google Scholar]
- 24.Wilson M A, Kozel T R. Contribution of antibody in normal human serum to early deposition of C3 onto encapsulated and nonencapsulated Cryptococcus neoformans. Infect Immun. 1992;60:754–761. doi: 10.1128/iai.60.3.754-761.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wuthrich M, Chang W L, Klein B S. Immunogenicity and protective efficacy of the WI-1 adhesin of Blastomyces dermatitidis. Infect Immun. 1998;66:5443–5449. doi: 10.1128/iai.66.11.5443-5449.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang M X, Klein B S. Activation, binding, and processing of complement component 3 (C3) by Blastomyces dermatitidis. Infect Immun. 1997;65:1849–1855. doi: 10.1128/iai.65.5.1849-1855.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang M X, Lupan D M, Kozel T R. Mannan-specific immunoglobulin G antibodies in normal human serum mediate classical pathway initiation of C3 binding to Candida albicans. Infect Immun. 1997;65:3822–3827. doi: 10.1128/iai.65.9.3822-3827.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]