Abstract
Medical image segmentation is a challenging task due to the high variation in shape, size and position of infections or lesions in medical images. It is necessary to construct multi-scale representations to capture image contents from different scales. However, it is still challenging for U-Net with a simple skip connection to model the global multi-scale context. To overcome it, we proposed a dense skip-connection with cross co-attention in U-Net to solve the semantic gaps for an accurate automatic medical image segmentation. We name our method MCA-UNet, which enjoys two benefits: (1) it has a strong ability to model the multi-scale features, and (2) it jointly explores the spatial and channel attentions. The experimental results on the COVID-19 and IDRiD datasets suggest that our MCA-UNet produces more precise segmentation performance for the consolidation, ground-glass opacity (GGO), microaneurysms (MA) and hard exudates (EX). The source code of this work will be released via https://github.com/McGregorWwww/MCA-UNet/.
Keywords: Medical image segmentation, U-Net, Attention, Multi-scale feature fusion
Introduction
Medical image segmentation [1–5] of target objects provides valuable information for the analysis of pathologies. However, the high variation in shape, size and position of infections or lesions is one of the key challenges in medical image segmentation. As observed in Fig. 1, the size and shape with irregular and blurred appearances in CT between consolidation and ground-glass opacity (GGO) lesions vary significantly. The microaneurysms and hard exudates in fundus photography are tiny/small and dispersedly distributed, which easily results in the false-negative detection.
Fig. 1.
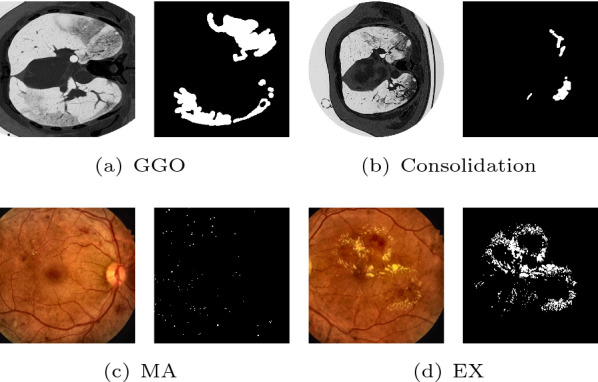
The major challenges in medical segmentation: the various shape and size of COVID-19 lesions in CT images (a, b), and small lesion scattered in fundus images (c, d). The right subfigures are the segmentation results of the left original images. ‘MA’ denotes microaneurysms and ‘EX’ denotes hard exudates
Recently, deep learning has shown its strong power of feature learning in image segmentation area. For medical image segmentation, U-Net-like encoder-decoder architectures have shown their power in medical image segmentation applications [6]. Although U-shaped networks have achieved good performances in many medical image segmentation applications [7–9], they still have several key limitations. (1) Insufficient capability of extracting context information for reconstructing the fine-grained segmentation map. The global context information is generally captured by deeper layers of the encoder and is gradually transmitted to shallower layers, which may be progressively diluted. (2) Although skip connection can help recover the spatial information which gets lost through the pooling layers, it is unnecessarily restrictive due to demanding the feature maps fusion of the encoder and decoder of the same level without considering the semantic gap [10, 11]. Therefore, it raises an important question to the U-Net methods: can we solve the limitation and develop a new framework that can improve over the restrictive skip connections in U-Net that requires fusion of only same-scale feature maps with simply concatenating?
To this end, we propose a U-shaped architecture with a more flexible multi-scale cross co-attention skip connection enabling flexible feature fusion in decoders for automatic segmentation. With the proposed dense connectivity, each node in a decoder is connected with the aggregation of all feature maps from the encoder by relaxing the unnecessarily restrictive skip connections where only the feature maps with the same scale are connected. It is different from UNet++ which fuses only the encoder features from the deeper layers without considering the fusion of the shallower layers (please refer to Fig. 2b). On the other hand, we design an attention mechanism [12, 13] from both the perspectives of channel-wise and spatial-wise to reduce the semantic gap between the encoder and decoder, termed co-attention mechanism. The co-attention mechanism can not only eliminate the semantic gap in feature fusion but also highlight salient features that are passed through the skip connections. Due to the reuse of feature maps, no extra computations and parameters are required, compared with UNet++ and MultiResUNet which solve the semantic gap by combining a series of convolution blocks. To facilitate the learning of the multi-scale feature fusion with cross co-attention connections, we employ deep supervision to facilitate the feature learning in different stages of the decoder. Our experimental results indicate that the deep supervision mechanism is effective in improving the segmentation performances of U-shaped networks, especially in the cases that the target objects have multiple scales. The performance of deep supervision highly depends on appropriate corresponding task weights. Therefore, we regard it as a multi-task learning task and make the weights learnable through a balanced multi-task dynamic weight (BMTD) optimization algorithm. The contribution of this work are three-folds:
Fig. 2.
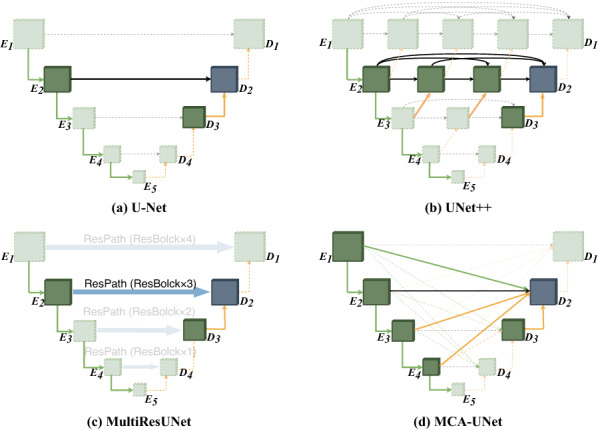
The comparison of the different skip connection settings
We dissect the skip connections in U-Net and empirically demonstrate appropriate connections are important for segmentation. We propose a multi-scale cross skip connection to boost semantic segmentation by bridging the semantic gaps between low-level and high-level features by an effective feature fusion scheme. Compared with the plain skip connections, the multi-scale cross skip connection improve the receptive field of U-Net by jointly considering the multi-scale features and hence able to extract multi-scale features of the target object and incorporate larger context.
While encoders have been studied rigorously, relatively few studies focus on the decoder side. The proposed bi-decoder module differs from the original decoder in three ways: (1) cross co-attention, which bridges the semantic gap between encoder and decoder feature maps by highlighting regions that present a significant interest for the diseases. (2) dual upsampling, which improves the upsampling performance by exploiting the finer spatial recovery in the decoder. (3) deep supervision, which further facilitates the multi-scale features fusion with a direct supervision for each level. Based on a U-shape network, the proposed decoder module can be easily embedded in the frameworks in the medical image segmentation tasks.
The proposed MCA-UNet is evaluated on four lesion segmentation tasks of two different datasets with difficulties including large variations of shape/size, blurred boundaries and small lesions, and it is shown that it achieves better performance than the related UNet-based architectures.
Related works
Recently, deep learning has shown their strong power of feature learning in image segmentation applications, for example, brain lesion segmentation [14], organ segmentation [15], electron microscopy image segmentation [6]. U-shaped architectures offer the advantages in medical image segmentation applications [6]. However, they still have some limitations such as lack of ability of modeling multi-scale global context and the semantic gap between the encoder and the decoder. To solve the issue, some methods with different skip connections for more flexible feature fusion are proposed, as illustrated in Fig. 2. Zhou et al. [10] propose a nested U-shaped framework, UNet++, with nested dense skip pathways which replace the restrictive skip connections fusing only the same-scale feature maps in U-Net. Ibtehaz et al. [11] propose MultiResUNet to incorporate some residual convolutional layers along the skip connections. The study hypothesizes that the features propagating from the encoder stage may balance the possible semantic gaps. Attention-UNet [13] is proposed to reduce the semantic gap between the encoder and decoder by a spatial attention mechanism. The advantage of the methods is that they improve the segmentation performance by alleviating the semantic gap and incorporating extra convolution layers or attention mechanism. Despite achieving good performance, the works above are still incapable of effectively exploring sufficient information from full scales due to the designs of the skip connections which ignore the correlation of multiple scale encoder features.
Methods
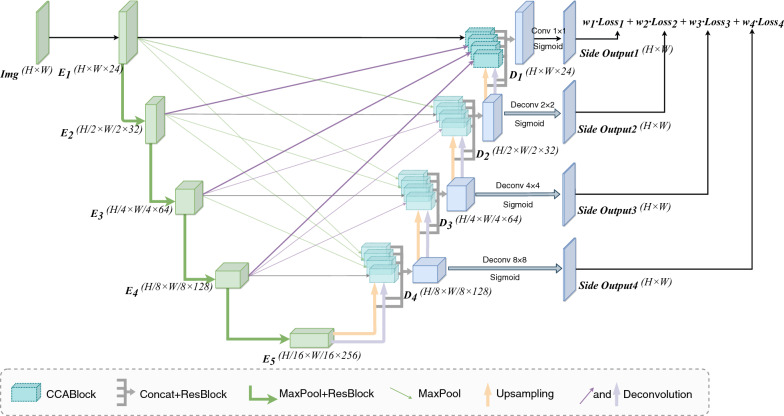
The overall framework of MCA-UNet
Our network consists of three parts: the encoder, the Multi-scale Cross Skip Connection and the Bidirectional Decoder (Bi-decoder) which consists of Dual Upsampling, Cross Co-Attention (CCA) and Deep Supervision. We also employ a BMTD algorithm to optimize the multi-task loss from the deep supervised decoder layers. Figure 3 illustrates the architecture of our proposed MCA-UNet network. To improve the representation capacity of the segmentation network, we replace the original two-layer convolution block with a Residual Block [16]. To better fuse features of inconsistent semantics and scales, we propose a cross co-attention guided multi-scale fusion scheme, which addresses the issues that arise when fusing features given at different scales. To effectively fuse the multi-scale features from different encoder levels to produce the final segmentation mask, we proposed a bi-decoder module which is directly enhanced by multi-scale context extracted from the contracting path. The bi-decoder module also involves a dual upsampling process that improves the upsampling performance and a deep supervision scheme to facilitate back-propagation and convergence. We provide details for each step in the following sections.
Fig. 3.
The architecture of the proposed MCA-UNet
Encoder
The encoder of the original U-Net consists of four doubled convolution layers with an activation function, which is insufficient for feature extraction and representation. Thus, we replace each convolution with a Residual Block [16] which has been proven to be useful for increasing the ability of learning richer representations and mitigating the degradation problem. The details can be seen in Table. 1.
Table 1.
The architecture of our segmentation network
| Stage encoder | Input | Kernel | Output | Stage | Input | Kernel | Output |
|---|---|---|---|---|---|---|---|
| Encoder | Decoder | ||||||
| Image | – | – | 6402 × 3 | 402 × 376 | ResBlock × 3 + DU | 802 × 128 | |
| 6402 × 3 | ResBlock[16] × 2 | 6402 × 24 | 802 × 248 | ResBlock × 2 + DU | 1602 × 64 | ||
| 6402 × 24 | ResBlock × 2 + MP | 3202 × 32 | 1602 × 184 | ResBlock × 2 + DU | 3202 × 32 | ||
| 3202 × 32 | ResBlock × 2 + MP | 1602 × 64 | 3202 × 168 | ResBlock × 2 + DU | 6402 × 24 | ||
| 1602 × 64 | ResBlock × 3 + MP | 802 × 128 | SO1 | 6402 × 24 | Conv,1 × 1 | 6402 × 1 | |
| 802 × 128 | ResBlock × 4 + MP | 402 × 256 | SO2 | 3202 × 32 | Deconv,× 2 | 6402 × 1 | |
| – | – | – | – | SO3 | 1602 × 64 | Deconv,4 × 4 | 6402 × 1 |
| – | – | – | – | SO4 | 802 × 128 | Deconv,8 × 8 | 6402 × 1 |
The input (output) shapes are represented by (size2 × channel). ‘MP’ denotes the MaxPooling operation, ‘DU’ denotes the proposed dual upsampling module which is a concatenated result of deconvolution and upsampling and ‘SOi’ denotes ith Side Output. For simplicity, we omit the upsampling operations in skip connections and the detail of CCA module which can be seen in Fig. 4
Multi-scale cross skip connection
Skip connection was first proposed in U-Net, which transmits the low-level information (textures, shapes, etc.) in the shallower encoder stages to the corresponding stages of the decoder. However, each stage of the decoder can only get feature from one scale through the original skip connection, which may harm the decoder features due to the semantic gaps and lacks the ability of capturing multi-scale context information which has been proven essential for lesion segmentation tasks [17, 18]. To solve these problems, we replace the original skip connection scheme with a multi-scale cross skip connection scheme. The proposed scheme transmits the resized (using up-samples or max-pooling) features from all the four encoder stages to each decoder stage, then combines them with a Bi-decoder block which will be introduced through the next section. The cross skip paths between the encoder and the decoder can aggregate features generated by multiple scales thus leads to better segmentation prediction.
Bidirectional decoder, Bi-decoder
The bi-decoder block is designed as a gating operation of the skip connection based on a learned attention map given to multiple feature maps from encoder. Unlike the traditional decoder, the proposed decoder has two inputs and two outputs. Each decoder block is connected with all encoder blocks via attentional skip connections as in the U-Net architecture. The inputs of bi-decoder involves two parts: multi-scale features from the encoder, and a complementary dual upsampled information from the deeper layers. The bi-decoder processes the two inputs with two directions of horizontal and vertical paths, and then learns a more powerful representation and finer recovery by dealing with the feature learning in both directions. With the different scales inputs, the decoder further encode the feature maps as the inputs for extracting global contexts with attention mechanism to enhance finer details by recovering localized spatial information. The outputs are dual upsampled information to the shallower layers and the direct segmentation prediction with another upsampling to the original resolution.
In summary, we introduce three enhancements to the conventional decoder module in our proposed bi-decoder: (1) Directly concatenating the feature maps from the encoder may cause redundancy, hence we proposed a co-correlation with channel- and spatial-wise attention module to guide the channel and spatial information filtration of the encoder feature maps through skip connections, allowing a fine spatial recovery in the decoder. (2) Both deconvolution and upsampling were added in the splicing process of the high-resolution features in the contraction path to leverage the complementarity between two different upsampling operations. (3) Finally, the incorporation of deep supervision can further facilitate the multi-scale features fusion.
Dual upsampling
The bi-decoder contains two upsampling components of nearest neighbor upsampling and deconvolution to recover resolution from the previous layers. We argue that the two processions are totally different from each other in terms of operation mode and can be complementary for the following cross correlation. Among the existing algorithms, the upsampling or deconvolution algorithm is seperately used in the decoder. In our work, the upsampling and deconvolution comprises a dual-path decoder. The combination of the upsampling and deconvolution can enhance the performance of the cross co-attention when the multi-feature encoder and decoder are fused.
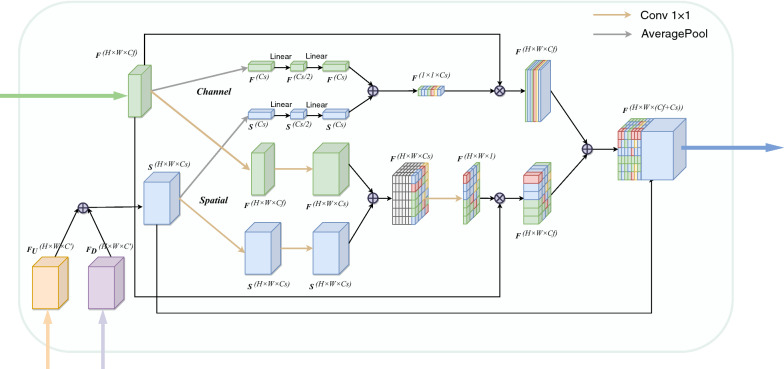
Cross co-attention (CCA)
Attention Mechanism for medical image segmentation have also been used recently [13, 19, 20], showing great potential in improving the segmentation performance. In our work, we hypothesize that the information from multi-scale encoder blocks are different. We are focusing on the cross correlation between the feature maps from encoder and decoder rather than a self-attention within a single feature map. Hence, to better fuse features of inconsistent semantics and scales, we propose a multi-scale channel-wise and spatial-wise attention module. The proposed module is incorporated into the bi-decoder to guide the channel and spatial information filtration of the encoder features through skip connections and eliminate the ambiguity with the decoder features as signals.
Specifically, instead of simply aggregating features from all levels, we propose to learn the attention in four parallel different level features. Unlike the previously proposed attention modules, most of which only explore channel- or spatial-wise attention, the proposed multi-scale cross co-attention module applies attention mechanism of channel- and spatial-wise for high-level and low-level features to exploit the complementary space and channel simultaneously. With the cross co-attention, the decoder can learn the importance of each feature channels which come from multi-level feature maps, and emphasize a meaningful feature selection in the spatial map to locate the critical structures.
Motivated by Squeeze-and-Excitation (SE) block, we extend the self attention mechanism to a cross co-attention in the multi-scale feature fusion to model the interactions of encoder-decoder with different scales for better feature representations. We introduce a cross co-attention module and the process is shown in Fig. 4. It involves channel and spatial attention branches. As illustrated in Fig. 4, the two branches are conducted simultaneously rather than sequentially, thus better feature representations for pixel-level prediction are obtained. It takes the concatenated results of two up-sampled features and as query feature , and the encoder features from different scales as key features , indicates the level of encoder which the feature is skip-connected from. For the th level encoder, each pair of feature maps (, ) are fed into the CCA module.
Fig. 4.
The structure of cross co-attention (CCA) module. The process of attention is similar to the self attention. The cross co-attention module takes three inputs, an upsampled tensor as well as a deconvoluted tensor as queries, and an encoder tensor as a key and a value at the same time
Mathematically, we consider the encoder feature maps and decoder feature maps as combinations of channels and , where W , H and C indicate width, height and channel dimension, respectively. Let and are the channel and spatial attention mask. A global average pooling layer is used for Spatial squeezing. This operation embeds the global spatial information in vector . This vector is transformed by
| 1 |
where , and being weights of three Linear layers and the ReLU operator .
This operation in Eq. (2) encodes the channel-wise dependencies. The resultant vector is used to recalibrate or excite as follow:
| 2 |
where the activation indicates the importance of the ith channel, which are rescaled.
The process of modeling the spatial relationship is similar to the channel attention. We consider it as an alternative slicing of the input feature maps and , where and correspond to the spatial location (i, j) with and . The spatial squeeze operation is achieved through a convolution
| 3 |
where is the weight of spatial squeeze convolution layer, and reduce the feature channels of and to the same number . Each of the projection represents the linearly combined representation for all channels C for a spatial location (i, j). This projection is passed through a sigmoid layer to rescale activations to [0, 1].
| 4 |
where each value corresponds to the relative importance of a spatial information (i, j) of a given feature map.
After computing the relevance between decoder and encoder during the fusion with the channel and spatial attention, next, we perform a tensor multiplication between the two attention tensor and the original encoder features. Third, we use an element-wise sum operation between the above tensor and original features to obtain the final representations reflecting effective fusion with skip connections for better segmentation. At last, we aggregate the features from these two attention modules, a cleaned up version is indicated as , which is the element-wise addition of the channel and spatial excited features, where is the element-wise multiplication. The final output feature is expressed by concatenating all the features: .
Deep supervision
To improve the back-propagation and make the decoder more stable, we introduce deep supervision [21] to the four stages of the decoder. Deep supervision is capable of guiding the feature learning of the hidden layers directly under the supervision of the loss and labels. We up-sample the features from the first three hidden stages to the size of the last prediction stage and add three more losses to supervise them. The final output of the decoder is then re-scaled to the original input size. The re-scaled output is further fed into a softmax layer to produce the class probability distribution. Note that the deep supervision does not work in the inference stage, we only use the last layer of decoder Side Output 1 for producing the segmentation prediction.
Training and inference
For the main idea of enhancing the decoder of U-Net, we add horizontal deep supervision in the four decoder levels. We choose deconvolution with kernel size 2 × 2, 4 × 4 and 8 × 8 to resize the output of every layer in decoder to meet the size of the ground truth. Then we compute the losses of those four layers, and use back propagation to update the weights of them, so we can deploy a direct guidance to the decoder and further improve the accuracy of the reconstruction operation.
For each layer, we employ the combined binary cross entropy loss and dice loss as our loss function:
| 5 |
where and denote the ground truth labels and predicted probabilities in the batch, and denote the nth pixel of and , N indicates the number of pixels within one batch. We empirically set the weights of the two terms in Eq. (5) to the same. The overall loss function for MCA-UNet is then defined as the weighted summation of the combined loss from each level of decoder:
| 6 |
where i indexes the level of the decoder and is the weight of each loss.
The performance of deep supervision highly depends on an appropriate choice of weights among the different tasks. How to appropriately set the weights of different tasks is a key issue in the deep supervision. A naive approach is to assign each individual task with an equal weight. It is not appropriate because the multiple tasks to be optimized have different difficulty levels. In this work, we consider the deep supervision as a multi-task learning formulation and assign different weights for different tasks. We propose a dynamic task weighting algorithm, named BMTD, which helps the model to automatically achieve balanced training by dynamically tuning the weight of each task during the model optimization. The weight of each task changes every batch. Hence, we measure how well the model is trained by considering the loss ratio between the current loss and the initial loss for each task. The task which is not trained well has a larger loss ratio. Hence, the harder tasks are optimized with more priority than the easier tasks.
Experiment and results
Implementation details
The proposed architecture is listed in Table 1. We used Adam as the optimizer and set the learning rate and batch size to 5e−3 and 24. To avoid over-fitting, we used early stopping and set the patience as 50 epochs. The final number of training epochs is about 200. For all the compared methods, we used the same parameter settings.
Data and experimental setting
COVID-19 lung CT images segmentation
We used the public COVID-19 CT images collected by Italian Society of Medical and Interventional Radiology (SIRM) dataset1 that contains 100 training and 10 testing images. The ground-truth segmentation was done by a trained radiologist. Raw data are public available.2 We performed 5-fold cross validation and augmented the data by rotating and rescaling. To improve the computational efficiency of the model, we resized the image to 256256 pixels. Three evaluation metrics were adopted, including Dice coefficient (Dice), Precision and Recall.
Retinal microaneurysms segmentation
For this task, we used the Indian Diabetic Retinopathy Image Dataset (IDRiD) [22], which contains 81 images including 54 images for training and 27 images for testing. The ground-truth segmentation has precise pixel level annotation of abnormalities associated with DR. We chose microaneurysms (MA) and hard exudates (EX) as the target lesion in our experiment since both lesions are small and dispersedly distributed. We computed the area under the precision-recall curve (AUC-PR), the area under the receiver operating characteristic curve (AUC-ROC) and Dice coefficient (Dice) to quantitatively evaluate the segmentation results. We used online data augmentation including resize, random crop, random rotate and CLAHE.
The comparison on COVID-19 dataset
We carried out experiments on the COVID-19 dataset to evaluate the effectiveness of our method. Note that the comparable models have the same encoder-decoder framework as MCA-UNet, including the number of channels, network depth and training strategies. We chose U-Net with ResBlock as our backbone segmentation architecture. The average Dice, Precision, and Sensitivity of all the methods were listed in Table 2. As shown in Table 2, it shows that these enhancements lead to notable improvements on the two segmentation tasks. Our model yields the overall highest performance, with an increase of 3.66% Dice for GGO segmentation and 12.30% Dice for consolidations segmentation compared to the baseline U-Net. Particularly for Consolidation, the increase of performance is striking. Compared to U-Net, our MCA-UNet improves the performance remarkably. Compared with the U-Net with the residual blocks, the cross co-attention module brings 3.15% improvement. The attention information from different layers in the encoder has complementary features, which obviously improves the segmentation accuracy. Meanwhile, deep supervision module individually outperforms the baseline by 1.71%. Therefore, learning the feature representation with direct supervision in the deeper layers is important. When we integrated the deep supervision and MCA together, the performance further improves to 52.60%, which outperforms the individual component of DS and MCA. With the BMTD optimization algorithm, improvements of 0.47% and 1.27% are achieved in ground glass and consolidations, respectively. These observation shows the crucial role of BMTD optimization. Moreover, it also indicates that the side outputs cannot be simply used with the same weights.
Table 2.
The results of effectiveness of the proposed components
| Index | Base | Res | DS | MCA | BMTD | Ground glass(%) | Consolidations (%) | Average (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dice | Prec. | Sen. | Dice | Prec. | Sen. | Dice | Prec. | Sen. | ||||||
| 1 | ✓ | 61.15 | 59.00 | 68.01 | 38.69 | 38.21 | 46.73 | 49.92 | 48.61 | 57.37 | ||||
| 2 | ✓ | ✓ | 62.08 | 59.94 | 68.00 | 38.73 | 43.11 | 39.62 | 50.41 | 51.52 | 53.81 | |||
| 3 | ✓ | ✓ | ✓ | 62.36 | 61.49 | 67.00 | 40.19 | 41.53 | 45.56 | 51.27 | 51.51 | 56.28 | ||
| 4 | ✓ | ✓ | ✓ | 62.72 | 61.46 | 62.78 | 41.27 | 44.07 | 44.31 | 52.00 | 52.77 | 55.55 | ||
| 5 | ✓ | ✓ | ✓ | ✓ | 62.92 | 60.92 | 68.82 | 42.28 | 43.24 | 47.77 | 52.60 | 52.08 | 58.30 | |
| 6 | ✓ | ✓ | ✓ | ✓ | ✓ | 63.39 | 61.24 | 68.91 | 43.45 | 44.91 | 49.11 | 53.42 | 53.07 | 59.01 |
Best results are boldfaced
Res residual block, DS Deep supervision, MCA Multi-scale cross corelation Attention block, BMTD balanced multi-task dynamic weighting, ‘Prec.’ and ‘Sen.’ Precision and Sensitivity
To more comprehensively evaluate our model, we chose some typical methods for further comparison. For the Covid19 dataset, we compared the proposed MCA-UNet to UNet++ (Resblock) [10], MultiResUNet [11], and Attention-UNet [13]. All of the networks have an encoder-decoder based architecture. We also compared to the UNet++ with ResNet-101 as powerful encoder.
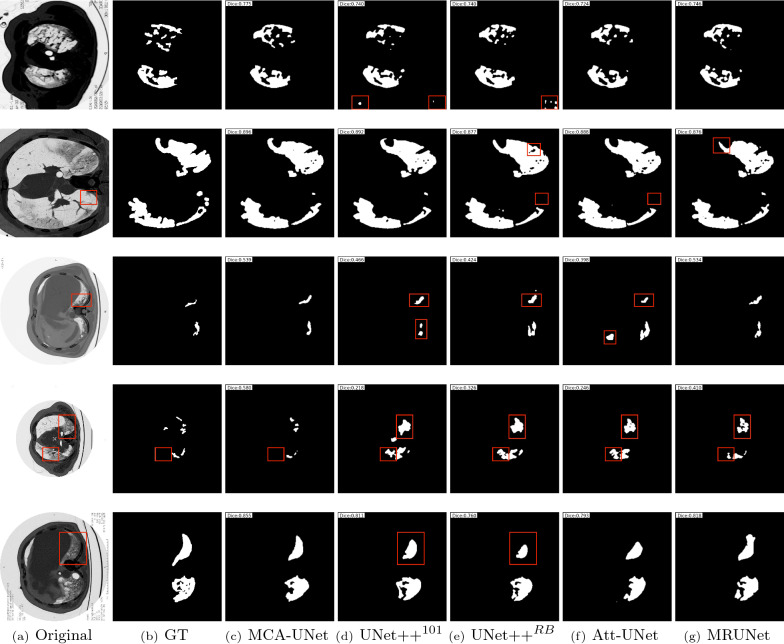
The experimental results obtained by several state-of-the-art segmentation networks are reported in Table 3. By comparing the results from Table 3, we can observe that the segmentation task achieves better performance in MCA-UNet. Compared to other networks that were proposed in the context of medical image segmentation: UNet++ (ResNet-101), MultiResUNet and Attention-UNet, our network achieves average improvements of 6.59%, 4.89% and 5.21% (in terms of Dice), 5.40%, 3.33% and 6.06% (in terms of Precision) and 5.09%, 4.76% and 1.88% (in terms of Sensitivity), respectively. Except for the sensitivity, our model also obtains improvements of 4.21% and 6.85% in terms of dice and precision compared with UNet++(ResBlock). Based on the above quantitative analysis, we can see that the cross skip connections guided by co-attention mechanisms are helpful for the refinement and fusion of complementary information between multi-scale features. Particularly, the proposed multi-scale guided attention network performs better results than Attention-UNet, which also integrates attention modules. Besides, we visualized the segmentation results of the comparable models in Fig. 5. The red boxes highlight regions where MCA-UNet performs better than the other methods by making better use of the multi-scale context fusion and attention scheme. It shows that our MCA-UNet generates better segmentation results, which are more similar to the ground truth than the results of the competing models. Through the empirical results, we summarize the following findings:
For the 1st and 2nd cases where the boundaries of GGO often have low contrast and blurred appearances, making them difficult to be identified. MCA-UNet predicts finer boundary information and maintain the object coherence, which demonstrates the effectiveness of modeling global context representations. It indicates that the multi-scale fusion help to discover more complete and accurate areas of classes of interest with low contrast.
2. Consolidations vary significantly in size and shape and have irregular and ambiguous boundaries. For the 3rd and 4th cases, the consolidations have a narrow shape. It can be seen that the predictions of MCA-UNet captures the boundary well. It is obvious that MCA-UNet keeps more details due to its multi-scale features from different encoder levels. For the 5th case where the lesions contain irregular boundaries, the segmentation results generated by our method are closer to the ground truths. Moreover, it also introduces fewer mislabeled pixels, which leads to better performance than other methods. These visual results indicate that our approach can successfully recover finer segmentation details while avoiding getting distracted in ambiguous regions. Nevertheless, the other networks produce smoother segmentations, resulting in a loss of fine grained details. As UNet++ and UNet++(ResBlock) also employed a multi-scale architecture, these differences suggest that the higher scale incorporation and effective cross co-attention can actually improve the performance of segmentation networks. It can be seen that both methods tend to have over-segmentation problems, which may be caused by the lack of higher resolution features.
Table 3.
Comparison of our method and the state-of-the-art methods on the COVID dataset
| Methods | Ground glass (%) | Consolidations (%) | Average (%) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Dice | Prec. | Sen. | Dice | Prec. | Sen. | Dice | Prec. | Sen. | |
| UNet(Baseline) | 61.15 | 59.00 | 68.01 | 38.69 | 38.21 | 46.73 | 49.92 | 48.61 | 57.37 |
| UNet++(Backbone:ResNet-101) | 61.50 | 60.32 | 66.46 | 39.20 | 40.14 | 45.83 | 50.35 | 50.23 | 56.15 |
| UNet++(ResBlock) | 62.07 | 57.86 | 71.56 | 40.94 | 41.48 | 47.39 | 51.50 | 49.67 | 59.48 |
| MultiResUNet | 61.46 | 59.94 | 66.99 | 40.88 | 42.78 | 45.66 | 51.17 | 51.36 | 56.33 |
| Attention-UNet | 62.18 | 60.90 | 68.25 | 39.84 | 39.17 | 47.59 | 51.01 | 50.04 | 57.92 |
| MCA-UNet | 63.39 | 61.24 | 68.91 | 43.95 | 44.91 | 49.11 | 53.67 | 53.07 | 59.01 |
Best results are boldfaced
Fig. 5.
The qualitative comparison of the segmentation results on COVID-19 dataset produced by the comparable models and the proposed MCA-UNet. The red boxes indicate the regions where our method exhibits better segmentation performance than the others
In summary, the previous approaches suffer from two main limitations in the segmentation of COVID-19: large variations of consolidation and blurred boundary of GGO. For large variations of consolidation in CT lead to inaccurate prediction for the baseline and the comparable methods due to the insufficient multi-scale feature which fails to deal with such variations. The blurred boundary of GGO leads to inaccurate prediction due to the lack of the high spatial information which is lost or distorted in the pooling and upsampling. Both the quantitative evaluation in Table 3 and qualitative comparison in Fig. 5 demonstrate the effectiveness of the proposed MCA-UNet for COVID19 segmentation.
The comparison on IDRiD dataset
For the IDRiD dataset, we compared MCA-UNet with SESV-DLab [26], SSCL [24], DRU-Net [23], and three top-ranking methods on the IDRiD challenge leaderboard [22]. DRU-UNet (Deep Recurrent U-Net) is a model which combines the deep residual model and recurrent convolutional operations into U-Net. SSCL is an advanced semi-supervised collaborative learning (SSCL) model. DeepLabv3+ is an extension of DeepLabv3, which introduces a decoder module to better recover the spatial resolutions and further refine the final segmentation masks. Different from the common methods for constructing a more accurate segmentation model, the aim of SESV-DLab is to predict the segmentation errors produced by an existing model and then correct them.
The performance of these methods is shown in Tables 4 and 5. The results show that our model achieves the highest AUC-PR and AUC-ROC, especially for the segmentation of MA in Table 4, our model beats the top-3 ranking methods by 3.77%, 5.15% and 9.83% in terms of AUC-PR, setting the new state of the art. It demonstrates again that our model is able to produce precise and reliable results for medical image segmentation.
Table 4.
The comparison with the state-of-the-art MA segmentation methods on the IDRiD dataset
| Methods | AUC-PR (%) | AUC-ROC (%) | Dice (%) |
|---|---|---|---|
| iFLYTEK-MIG (Rank #1) | 50.17 | N/A | N/A |
| VRT (Rank #2) | 49.51 | N/A | N/A |
| PATech (Rank #3) | 47.40 | N/A | N/A |
| DRU-Net [23] | N/A | 98.20 | N/A |
| SSCL [24] | 49.60 | 98.28 | N/A |
| DeepLabv3+ [25] | 48.65 | 98.91 | N/A |
| SESV-Dlab [26] | 50.99 | 99.13 | N/A |
| U-Net [6](Baseline) | 45.04 | 94.46 | 16.24 |
| Attention-UNet [13] | 49.01 | 98.89 | 25.49 |
| MultiResUNet [11] | 49.13 | 99.19 | 25.51 |
| UNet++ [10](ResNet-101) | 46.92 | 98.01 | 32.03 |
| UNet++(ResBlock) | 49.32 | 99.27 | 19.45 |
| MCA-UNet | 52.06 | 99.12 | 38.50 |
Best results are boldfaced
Table 5.
The comparison with the state-of-the-art EX segmentation methods on the IDRiD dataset
| Methods | AUC-PR (%) | AUC-ROC (%) | Dice (%) |
|---|---|---|---|
| U-Net(Baseline) | 75.52 | 99.34 | 60.46 |
| Attention-UNet | 75.33 | 99.16 | 62.18 |
| MultiResUNet | 70.76 | 98.60 | 62.03 |
| UNet++(ResNet-101) | 74.95 | 99.32 | 61.91 |
| UNet++(ResBlock) | 72.82 | 96.41 | 50.54 |
| MCA-UNet | 79.45 | 99.53 | 65.91 |
Best results are boldfaced
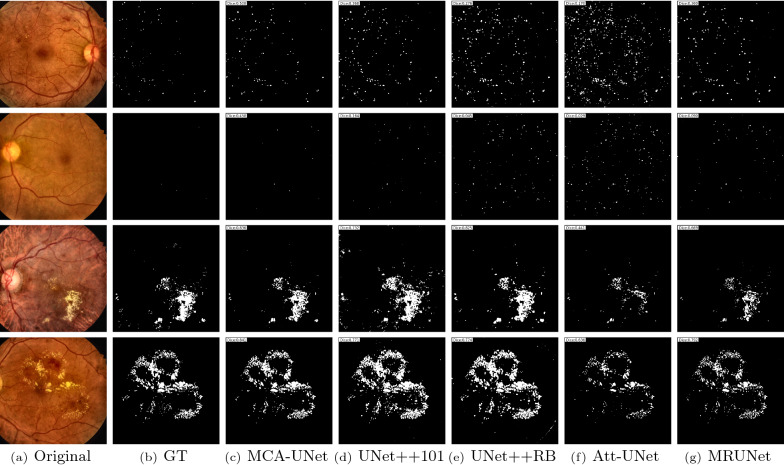
Most of the existing U-shaped methods perform well on the large object segmentation, but fail to the detection of the small objects, which are particularly prevalent in the eye diseases. Due to the downsampling and upsampling operations in U-Net, the feature maps in hidden layers are sparser than the original inputs, which causes a loss of image details and results in the comparable segmentation models yield inferior segmentation performance for the small lesions. Figure 6 shows some representative results and the comparable methods to exhibit the superiority of the proposed method on the segmentation of MA and EX. As illustrated in Fig. 6, from the top three examples, we can find that the comparable segmentation methods are limited in small lesion segmentation and produce amounts of false positives. From the bottom three examples, it can be observed that both UNet++(ResNet-101) and UNet++(ResBlock) have over-segmentation problems. On the contrary, the boundary of the EX is under-segmented by both Attention-UNet and MultiResUNet. All the comparable segmentation models are not capable of precise segmentation of the small lesions. In the medical image domain, the multi-scale information is required to be learned by the segmentation models which then facilitates the target segmentation. It shows that MCA-UNet can significantly reduce the false positives and correct some inaccurately segmented regions by the previous algorithms.
Fig. 6.
The qualitative comparison of the segmentation results on IDRiD dataset produced by the comparable models and the proposed MCA-UNet
Discussion
Discussion on the number of dense skip connections
Multi-scale dense connection and cross co-attention (CCA) are two vital modules in our segmentation model to achieve better segmentation performances. To further investigate the relative contribution of each component, we conduct a series of experiments on the EX segmentation, to investigate the individual contribution by varying the number of skip connections, skip connection schemes, and positions of skip connections. By varying the number of skip connections in the bi-decoder, we explored the influence of different skip connections on the EX segmentation performance. Moreover, to evaluate the segmentation performance of the CCA, we replace CCA in all the bi-decoders with a simple concatenation fusion used in the U-Net. The illustration of the competing models can be referred to Fig. 7. ‘w/o up’ or ‘w/o down’ means that the up-sampling or down-sampling operation in the skip connection is removed.
Fig. 7.
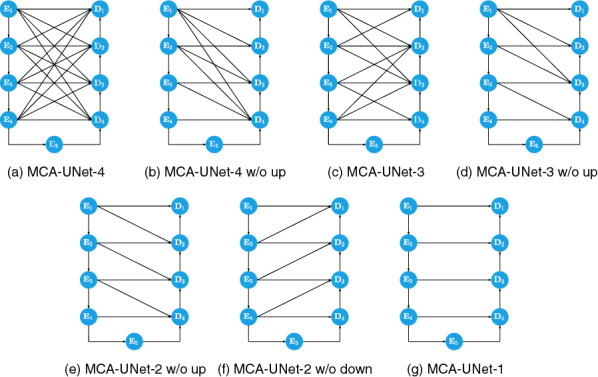
Comparison among the MCA-UNets with different number of the skip connections
As shown in Table 6, our proposed CCA is able to consistently achieve better performance compared with the simple concatenation fusion, which demonstrates its robustness and high flexibility for integrating information from the earlier feature maps. Moreover, it can be seen from Table 6 that the segmentation performance of the model improves with the increase of the number of skip connections. For the comparison between models with up-sampled connection remained and ones with up-sampled connection removed, the former is worse when the connection number is the same. For example, MCA-UNet-2 (w/o up) achieves an improvement by 1.05% compared with MCA-UNet-2(w/o down). which indicates that the higher resolution is important for the fine spatial recovery, whereas the connections from the encoders with lower resolution is not helpful for the decoders. Our findings show that the spatial information is more critical for the segmentation of the multi-scale lesion objects, especially for the small lesions. MCA-UNet-2 (w/o down) performs the worst, even worse than MCA-UNet-1. The skip connection scheme in MCA-UNet-2 (w/o down) is similar as the UNet++ where the decoders are connected with the lower resolution feature maps of encoders. Another interesting finding is that MCA-UNet-4 without CCA achieved a relatively poor performance compared to MCA-UNet-1 with CCA in terms of AUPR. The results once again validate that simply connecting the feature maps with same level from the encoder and the decoder is not an optimal solution.
Table 6.
Different mappings from encoder to decoder
| Models with different Layer nums | Attention | w/o Attention | ||
|---|---|---|---|---|
| AUPR (%) | AUC (%) | AUPR (%) | AUC (%) | |
| MCA-UNet-4 | 79.45 | 99.53 | 77.14 | 99.27 |
| MCA-UNet-4(w/o up) | 78.37 | 99.09 | 77.05 | 99.15 |
| MCA-UNet-3 | 77.25 | 98.81 | 77.09 | 97.20 |
| MCA-UNet-3(w/o up) | 77.35 | 99.09 | 76.25 | 99.27 |
| MCA-UNet-2(w/o up) | 77.21 | 99.06 | 77.25 | 98.39 |
| MCA-UNet-2(w/o down) | 76.41 | 98.53 | 75.41 | 98.98 |
| MCA-UNet-1 | 77.78 | 99.15 | 77.56 | 99.31 |
Best results are boldfaced
Discussion on different attention mechanisms
Based on the skip connections for information fusion, we systematically conduct the experiment of different attention mechanisms. The result is shown in Table 7. We conduct a series of comparison including the spatial- and channel-wise CCA vs. the spatial-wise CCA, the self-attention (SA) of encoder or decoder vs. CCA and the sequential CCA vs. the concurrent CCA. The traditional self-attention mechanism is to capture the dependencies within the same feature map from the spatial- and channel-wise perspective. Our CCA is to capture the correlation between two feature maps from the encoder and decoder. It is apparent to see that, the proposed concurrent CCA method obtain improvements upon the traditional self attention methods in terms of Dice and precision. The channel maps help capture the context information for the feature fusion. When we integrate the spatial- and channel-wise together, the performance further improves to 62.48% with respect to Dice. Furthermore, when we compare the sequential and concurrent fashion for the encoder-decoder cross co-attention, the concurrent fashion improves the segmentation performance over the sequential model by 0.35% in terms of Dice.
Table 7.
The study on different fusion and attention mechanism
| Index | Attention structure | Ground glass (%) | ||
|---|---|---|---|---|
| Dice | Prec. | Sen. | ||
| 1 | S (SA in encoder) | 61.59 | 60.71 | 66.56 |
| 2 | S+C (SA in encoder) | 62.48 | 58.29 | 72.50 |
| 3 | S+C (SA in encoder and decoder) | 60.93 | 59.99 | 66.94 |
| 4 | S+C (sequential en-de CCA) | 62.54 | 59.43 | 69.42 |
| 5 | S+C (concurrent en-de CCA) | 62.76 | 60.93 | 66.98 |
Best results are boldfaced
S spatial-wise, C channel-wise, SA self-attention
Discussion on positions of the proposed dense skip connections
We performed a series of experiments with respect to the positions of the proposed skip connection in Table 8. Figure 8 shows the illustration of the settings. Let , where , indicates how the encoder features are connected to the decoders. For example, indicates that encoder is connected to the decoders of and . Although the proposed CCA module contributes to the performance improvement as shown in the previous results, it is interesting to investigate 1) which level of encoder is more important for the decoders; and 2) which layer of decoder is more beneficial for the same combination of multi-scale encoder features. Obviously, MCA-UNet with multiple dense connection leads to improved performance than the other models with the certain connections removed. It can be seen that obtains the best performance in terms of AUPR, which indicates that the low-level features with higher resolution is important. The can take full advantage of the rich spatial information, which can help refine the predicted boundary for the lesions with complex structure. On the contrary, shows the worst performance. The reason may be that the spatial information is lost in the contracting path and semantic gap is too large, resulting in poor fusion performance.
Table 8.
The performance of the proposed connections with different positions and side outputs
| Positions of encoder and decoder | AUPR (%) | AUC (%) | Side outputs for prediction | AUPR (%) | AUC (%) |
|---|---|---|---|---|---|
| 79.45 | 99.53 | 79.45 | 99.53 | ||
| 77.67 | 98.35 | 78.82 | 99.26 | ||
| 76.56 | 98.65 | 78.53 | 99.33 | ||
| 75.06 | 99.17 | 73.46 | 99.48 | ||
| 72.53 | 99.06 | 79.59 | 99.28 | ||
| 75.66 | 99.29 | 78.46 | 99.50 | ||
| 76.50 | 99.15 | 78.78 | 99.37 | ||
| 75.86 | 98.13 | – | – | – | |
| 72.90 | 99.26 | – | – | – |
Best results are boldfaced
Fig. 8.

The comparison among MCA-UNet with CCA on the different positions
Deep supervision
To test the effectiveness of the deep supervision scheme, we show the performance of each individual side output. From the Table 8, we observe small difference for the multiple predictions of side outputs except . Furthermore, we find the performance of are slightly better than and . We also try to employ an ensemble-based methods, where the multiple side outputs are combined to make a final prediction. We find the ensemble of achieves a slightly better performance than the individual performance.
Conclusion
In this work, we introduced a multi-scale Cross Co-Attentional Skip Connection U-Net architecture for the medical image segmentation. Our MCA-UNet utilized the multi-scale feature fusion strategy to combine semantic information at different levels and the cross co-attention module to aggregate relevant global dependencies. To validate our approach, we conducted experiments on three different segmentation tasks on the two different medical image datasets: consolidation, GGO, Microaneurysms and Hard Exudates, indicating that it can be broadly applied to the other medical images segmentation tasks. We provided extensive experiments to evaluate the impact of the individual components of the proposed architecture. Moreover, we will extend our 2D model to a 3D version for capturing the inter-slice continuity of the lesion in the future work.
Funding
This research was supported by the National Natural Science Foundation of China (No. 62076059), the Science Project of Liaoning province (2021-MS-105) and the National Natural Science Foundation of China (No. 61971118).
Availability of data and materials
Data are publicly available.
Declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Informed consent
Not applicable
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Peng Cao, Email: caopeng@mail.neu.edu.cn.
Jinzhu Yang, Email: yangjinzhu@cse.neu.edu.cn.
References
- 1.Litjens G, Kooi T, Bejnordi BE, Setio AA, Ciompi F, Ghafoorian M, Van Der Laak JA, Van Ginneken B, Sánchez CI. A survey on deep learning in medical image analysis. Med Image Anal. 2017;42:60–88. doi: 10.1016/j.media.2017.07.005. [DOI] [PubMed] [Google Scholar]
- 2.Pham DL, Chenyang X, Prince JL. Current methods in medical image segmentation. Ann Rev Biomed Eng. 2000;2(1):315–337. doi: 10.1146/annurev.bioeng.2.1.315. [DOI] [PubMed] [Google Scholar]
- 3.Tan W, Huang P, Li X, Ren G, Chen Y, Yang J. Analysis of segmentation of lung parenchyma based on deep learning methods. J X-Ray Sci Technol. 2021;29(6):945–959. doi: 10.3233/XST-210956. [DOI] [PubMed] [Google Scholar]
- 4.Tan W, Liu P, Li X, Shaoxun X, Chen Y, Yang J. Segmentation of lung airways based on deep learning methods. IET Image Process. 2022;16(5):1444–1456. doi: 10.1049/ipr2.12423. [DOI] [Google Scholar]
- 5.Wang L, Juan G, Chen Y, Liang Y, Zhang W, Jiantao P, Chen H. Automated segmentation of the optic disc from fundus images using an asymmetric deep learning network. Pattern Recognit. 2021;112:107810. doi: 10.1016/j.patcog.2020.107810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ronneberger O, Fischer P, Brox T. U-net: convolutional networks for biomedical image segmentation. In: Medical Image Computing and Computer-Assisted Intervention (MICCAI), volume 9351 of LNCS, 2015;234–241. Springer.
- 7.Isensee F, Kickingereder P, Wick W, Bendszus M, Maier-Hein KH. Brain Tumor Segmentation and Radiomics Survival Prediction: Contribution to the BRATS 2017 Challenge. In: Brainlesion: Glioma, Multiple Sclerosis, Stroke and Traumatic Brain Injuries, Lecture Notes in Computer Science, pp 287–297, Cham, 2018.
- 8.Falk T, Mai D, Bensch R, Ronneberger O. U-Net: deep learning for cell counting, detection, and morphometry. Nat Methods. 2019;16(1):67–70. doi: 10.1038/s41592-018-0261-2. [DOI] [PubMed] [Google Scholar]
- 9.Qian Y, Gao Y, Zheng Y, Zhu J, Dai Y, Shi Y. Crossover-Net: leveraging vertical-horizontal crossover relation for robust medical image segmentation. Pattern Recognit. 2021;113:107756. doi: 10.1016/j.patcog.2020.107756. [DOI] [Google Scholar]
- 10.Zongwei Zhou Md, Siddiquee MR, Tajbakhsh N, Liang J. UNet++: redesigning skip connections to exploit multiscale features in image segmentation. IEEE Trans Med Imaging. 2020;39(6):1856–1867. doi: 10.1109/TMI.2019.2959609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ibtehaz N, Sohel RM. MultiResUNet : rethinking the u-net architecture for multimodal biomedical image segmentation. Neural Netw. 2020;121:74–87. doi: 10.1016/j.neunet.2019.08.025. [DOI] [PubMed] [Google Scholar]
- 12.Vaswani A, Shazeer N, Parmar N, Uszkoreit J, Jones L, Gomez AN, Kaiser Ł, Polosukhin I. Attention is all you need. In: Advances in Neural Information Processing Systems, volume 30. Curran Associates, Inc., 2017.
- 13.Oktay O, Schlemper J, Folgoc LL, Lee M, Heinrich M, Misawa K, Mori K, McDonagh S, Hammerla NY, Kainz B, Glocker B, Rueckert D. Attention U-Net: Learning Where to Look for the Pancreas. arXiv:1804.03999 [cs], 2018.
- 14.ISLES 2015-A public evaluation benchmark for ischemic stroke lesion segmentation from multispectral MRI. [DOI] [PMC free article] [PubMed]
- 15.Li X, Chen H, Qi X, Dou Q, Chi-Wing F, Heng P-A. H-DenseUNet: hybrid densely connected UNet for liver and tumor segmentation From CT volumes. IEEE Trans Med Imaging. 2018;37(12):2663–2674. doi: 10.1109/TMI.2018.2845918. [DOI] [PubMed] [Google Scholar]
- 16.He K, Zhang X, Ren S, Sun J. Deep residual learning for image recognition. In: 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), pp 770–778, Las Vegas, NV, USA, 2016. IEEE.
- 17.Yang J, Bo W, Li L, Cao P, Zaiane O. MSDS-UNet: a multi-scale deeply supervised 3D U-Net for automatic segmentation of lung tumor in CT. Comput Med Imaging Graph. 2021;92:101957. doi: 10.1016/j.compmedimag.2021.101957. [DOI] [PubMed] [Google Scholar]
- 18.Kamnitsas K, Ledig C, Newcombe VF, Simpson JP, Kane AD, Menon DK, Rueckert D, Glocker B. Efficient multi-scale 3D CNN with fully connected CRF for accurate brain lesion segmentation. Med Image Anal. 2017;36:61–78. doi: 10.1016/j.media.2016.10.004. [DOI] [PubMed] [Google Scholar]
- 19.Li X, Xiaowei H, Lequan Y, Zhu L, Chi-Wing F, Heng P-A. CANet: cross-disease attention network for joint diabetic retinopathy and diabetic macular edema grading. IEEE Trans Med Imaging. 2020;39(5):1483–1493. doi: 10.1109/TMI.2019.2951844. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Wu B, Zhu P, Li P, Zuo W, Hu Q. ECA-Net: efficient channel attention for deep convolutional neural networks. p 9.
- 21.Lee C-Y, Xie S, Gallagher P, Zhang Z, Tu Z. Deeply-supervised nets. In: Guy Lebanon and S. V. N. Vishwanathan, (eds), In: Proceedings of the eighteenth international conference on artificial intelligence and statistics, volume 38 of Proceedings of Machine Learning Research, pp 562–570, San Diego, 2015. PMLR.
- 22.Porwal P, Pachade S, Kamble R, Kokare M, Deshmukh G, Sahasrabuddhe V, Meriaudeau F. Indian diabetic retinopathy image dataset (IDRiD): a database for diabetic retinopathy screening research. Data. 2018;3(3):25. doi: 10.3390/data3030025. [DOI] [Google Scholar]
- 23.Kou C, Li W, Liang W, Zekuan Y, Hao J. Microaneurysms segmentation with a U-Net based on recurrent residual convolutional neural network. J Med Imaging. 2019;6(02):1. doi: 10.1117/1.JMI.6.2.025008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhou Y, He X, Huang L, Liu L, Zhu F, Cui S, Shao L. Collaborative learning of semi-supervised segmentation and classification for medical images. In: Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, pp. 2079–2088, 2019.
- 25.Chen L-C, Papandreou G, Kokkinos I, Murphy K, Yuille AL. DeepLab: semantic image segmentation with deep convolutional nets, Atrous convolution, and fully connected CRFs. IEEE Trans Pattern Anal Mach Intell. 2018;40(4):834–848. doi: 10.1109/TPAMI.2017.2699184. [DOI] [PubMed] [Google Scholar]
- 26.Xie Y, Zhang J, Hao L, Shen C, Xia Y. SESV: accurate medical image segmentation by predicting and correcting errors. IEEE Trans Med Imaging. 2021;40(1):286–296. doi: 10.1109/TMI.2020.3025308. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Data are publicly available.