Abstract
FLASH radiotherapy is a novel technique that has been shown in numerous preclinical in vivo studies to have the potential to be the next important improvement in cancer treatment. However, the biological mechanisms responsible for the selective FLASH sparing effect of normal tissues are not yet known. An optimal translation of FLASH radiotherapy into the clinic would require a good understanding of the specific beam parameters that induces a FLASH effect, environmental conditions affecting the response, and the radiobiological mechanisms involved. Even though the FLASH effect has generally been considered as an in vivo effect, studies finding these answers would be difficult and ethically challenging to carry out solely in animals. Hence, suitable in vitro studies aimed towards finding these answers are needed. In this review, we describe and summarise several in vitro assays that have been used or could be used to finally elucidate the mechanisms behind the FLASH effect.
Keywords: Cell models, cells, dose rate, radiation, radiotherapy, ultra-high
Introduction
Recent preclinical studies have shown that FLASH irradiation, which is radiation delivered in a fraction of a second, reduces incidence and severity of radiation side effects compared to conventional dose rate (CONV) irradiation used in clinical practice (Refs 1–16). However, the treatment effect on tumours is not reduced (Refs 1, 5, 11, 12, 17, 18). This has been called the ‘FLASH effect’. The FLASH sparing effect has mainly been observed using relatively large single doses in vivo, though a few studies have shown an effect also in vitro (Refs 7, 8, 19–22). The benefit of FLASH radiotherapy (RT) has been further shown in veterinary clinical studies and in the first treatment of a human (Refs 6, 10, 23). FLASH-RT is delivered with irradiation systems with a high radiation output, capable of generating the ultra-high dose rates and short delivery times required for producing an observable FLASH sparing effect, which permit treatments to be delivered in fractions of a second, compared to several minutes for conventional treatments (Refs 4, 24–26).
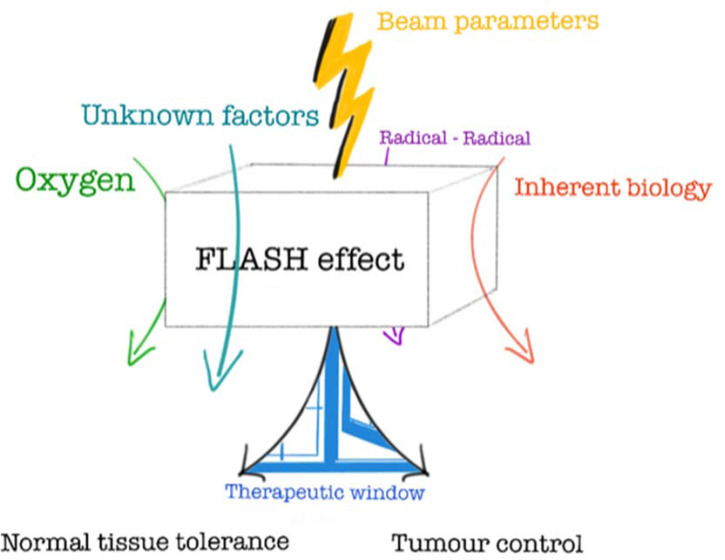
The short treatment times used in FLASH-RT, often less than 0.1 s, have the added value of minimising treatment delivery uncertainties caused by patient motion during delivery, for example, reduced risk of missing a lung tumour due to the breathing motion. The potential to ‘freeze’ physiological motion could allow for the use of smaller motion-related target margins, and thereby smaller volumes of normal tissue being unnecessarily irradiated. Due to these advantages, FLASH-RT has the potential to be an important (r)evolutionary step in cancer treatment (Refs 16, 27). However, the radiobiological mechanisms responsible for this differential FLASH sparing effect observed between normal tissue and tumour tissue are not yet known, though several hypotheses have been proposed (Fig. 1) (Ref. 16), e.g. radiochemical depletion of oxygen leading to transient hypoxia (Refs 28–31), radical-radical interaction (Refs 32, 33), and a modified immune response following FLASH relative to CONV irradiation (Refs 34, 35).
Fig. 1.
‘The FLASH box’ - Illustrating what we know and what is yet to be discovered about the radiobiological mechanisms behind the highly beneficial ‘FLASH effect’ © Gabriel Adrian.
For an optimal translation of FLASH-RT into clinical trials, it is essential to know what specific beam parameters induces a FLASH effect and what the radiobiological mechanisms involved are. The FLASH effect is currently considered by many as an effect only seen in vivo. However, elucidating the mechanisms underpinning the FLASH sparing effect solely utilising in vivo models may prove to be difficult and ethically unviable. Hence, suitable in vitro studies are needed, aimed towards finding optimal beam parameters and pin-pointing the biological mechanisms. Here, we describe several in vitro assays that have been used in preclinical FLASH studies as well as other assays that could be used to fully understand and exploit the benefits associated with FLASH-RT.
Clonogenic assays
In radiobiological research, the clonogenic assay has been a reference method for in vitro studies since Puck & Markus introduced it in 1956 (Ref. 36). Radiation-induced cell death can occur in several ways, including mitotic catastrophe, apoptosis, necrosis, senescence, autophagy and ferroptosis (Ref. 37). The advantage of the clonogenic assay is its ability to capture cancer cells ability (or inability) of ‘endless’ division, i.e. including all kinds of cell deaths. Hence, the assay provides an in vitro-surrogate for the complete sterilisation of tumour cells in vivo. Although the assay may have limitations, such as being dependent on cell densities, it should be a reliable method for head-to-head comparisons in dose rate studies, provided all other experimental conditions are equal (Refs 38–40).
Interestingly, both early (from the 60's) and recent (here defined as in the 21st century) studies of ultra-high dose rate irradiations in mammalian cells using clonogenic assays have provided inconsistent results. In the early days of ultra-high dose rate irradiation, a ‘hockey-stick’-shaped survival curve for ultra-high dose rate irradiated samples was described. Typically, CONV and FLASH-curves were indistinguishable at lower doses, and then separated at higher doses, i.e. an increased survival fraction after FLASH irradiation was noted at higher irradiation doses (Refs 41, 42). Such survival curves were initially described for normoxic conditions (ambient air, 21%), though other experiments could not reproduce the findings (Refs 43, 44). Attention was then turned to the dependence of oxygen. It was found that the ‘break’ of the survival curve was influenced by the oxygen tension (Refs 43–46). Increasing the level of hypoxia in cells resulted in a lower total dose required to ‘break’ the survival curve. In addition to these results, similarly shaped survival curves were also described when irradiating bacteria at ultra-high dose rates, with breaks at doses around 60–70 Gy in normoxic conditions (Refs 47, 48). During the 90's, two independent studies could not detect any survival difference after FLASH and CONV irradiation in neither normoxic nor anoxic conditions (Refs 49, 50).
Recent investigations using clonogenic assays in normoxic conditions have also provided inconsistent results. Despite the fact that the FLASH sparing in vivo is found in the healthy tissue, few of the recent studies have used normal cell lines. Instead, most have investigated the potential sparing of cancer cell lines. An increased survival fraction was found for H454 murine glioblastoma cells after 20 Gy FLASH compared with CONV irradiation in normoxia (Ref. 8). Congruently, an increased survival fraction was found for 4/7 cell lines after FLASH compared with CONV in our recent investigations (Ref. 51). On the other hand, Venkatesulu et al. found opposing FLASH effects, hence a lower survival fraction after FLASH for two murine pancreas cancer cell lines in normoxia (Ref. 52). Other studies could not distinguish any differences in survival fraction in normoxia for IMR90 normal human lung fibroblasts, DU145 prostate cancer cells, or A549 lung cancer cells (Refs 19, 20, 22). In hypoxia, we have previously described an oxygen-dependent FLASH sparing for DU145 prostate cancer cells (Ref. 19). Similarly, A549 lung cancer cells irradiated as spheroids with naturally occurring hypoxic cores exhibited a FLASH sparing, in contrast to the results for cells irradiated as a normoxic monolayer (Ref. 22). Noteworthy, in the early publications, the dose required to break the survival curve under normoxia was found to be at 7–10 Gy with a distinct inflexion point (Refs 41, 42). Such inflexion points have since only been described for cells in hypoxia and/or for much higher (non-clinical) doses and seem to indicate the dose required to consume all available oxygen (Ref. 46). Later studies have instead successfully used the linear-quadratic (LQ) model to fit the data (Ref. 53).
The reasons for the inconsistent findings of FLASH versus CONV using clonogenic assays could be several (Table 1). Firstly, we would not expect to see significant oxygen depletion effects for FLASH versus CONV in fully anoxic cell cultures as there is no oxygen available to deplete, nor in normoxic cultures as there is too much oxygen to deplete for clinical doses to have a meaningful impact on the available oxygen. Furthermore, beam characteristics differ between laboratories. Single-pulse versus pulsed delivery, instantaneous dose rate, dose per pulse, pulse repetition frequency, average dose rate, total delivery time and type of irradiation (electron/photon versus ion) could possibly all affect the radiobiological response (Refs 5, 16). Experimental conditions vary between laboratories. Temperature during irradiation, time outside the incubator, type of cell medium used, the volume of cell medium per flask or dish might influence results (Ref. 54). The definition of survival is stated to be a single cell that has proliferated to form a colony of at least 50 cells (Ref. 36). Laboratories may use different approaches to determine the colony size, i.e. to decide which clones are to be counted as survivors. It has been shown that different clone-size cut-offs influences the results in clonogenic assays (Refs 55–57).
Table 1.
Published Clonogenic FLASH data using mammalian cells, separated by electron and other type of irradiation
| Publication | Year | Cell line | Plating method | Dose-range (Gy) | Average dose rate for a 10 Gy delivery (Gy/s) | Instantaneous dose rate (Gy/s) | Dose/pulse (Gy) | Pulse repetition frequency (Hz) | Total delivery time | Energy | Type of irradiation | FLASH sparing normoxia | FLASH sparing hypoxia |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Town (Ref. 42) | 1967 | HeLa S3 | post | 0–25 | 7 × 106 or 4 × 103 |
<3.5 × 107 | single or two pulse(s) | single pulse or 400 | 1.3 μs or 2.5 ms | 15 MeV | electron | yes | N/A |
| Nias et al. (Ref. 58) | 1969 | HeLa | post | 0–27 | 1 × 107 | 1 × 107 | single pulse | single pulse | 1 μs | 8–14 MeV | electron | N/A | probably, but no direct comparison |
| Nias et al. (Ref. 44) | 1970 | HeLa | post | 0–33 | 1 × 109 | ~109 | single pulse | single pulse | 10 ns | 10 MeV | electron | N/A | N/A |
| Epp et al. (Ref. 45) | 1972 | HeLa-S3 | pre | 0–35 | 3 × 109 | ~109 | single pulse | single pulse | 3 ns | 350 keV | electron | N/A | probably, but no direct comparison |
| Michaels et al. (Ref. 46) | 1978 | CHO | pre | 0–45 | 3 × 109 | ~109 | single pulse | single pulse | 3 ns | 600 keV | electron | no | probably, but no direct comparison |
| Zackrisson et al. (Ref. 50) | 1991 | V79–379-A | post | 0–40 | 380 | 2.7 × 105 | 1.6 | 200 | ~30 ms (10 Gy) | 50 MeV | electron | no | no (anoxic) |
| Cygler et al. (Ref. 49) | 1994 | U87-MG, HT-144 | post | 0–27 | 3 x106 | ~ 5 × 106 | single pulse | single pulse | 3.2 μs | 20 MeV | electron | no | no (anoxic) |
| Montay-Gruel et al. (Ref. 8) | 2019 | H454 | post | 20 | 1000 | 1.8 × 106 | 6.7 | 100 | 20 ms | 6 MeV | electron | yes | yes |
| Venkatesulu et al. (Ref. 52) | 2019 | KPC, Panc02 | pre | 0–8 | 35 | – | – | – | – | 20 MeV | electron | reversed | N/A |
| Adrian et al. (Ref. 19) | 2019 | DU145 | pre | 0–25 | 800 | 8.6 × 105 | 3 | 200 | ~10 ms (10 Gy) | 10 MeV | electron | no | yes |
| Adrian et al. (Ref. 51) | 2021 | MCF7, MRC-5, Lu-HNSCC4, MDA-MB-231, HeLa, WiDr | pre | 0–12 | 800 | 8.6 × 105 | 3 | 200 | ~10 ms (10 Gy) | 10 MeV | electron | yes | N/A |
| Khan et al. (Ref. 22) | 2021 | A549, MDA-MB-231, HT29 | post | 0–20 | 90 | 4.0 × 105 | 1 | 90 | ~100 ms (10 Gy) | 16 MeV | electron | no | yes |
| Berry et al. (Ref. 41) | 1969 | HeLa S3 | post | 0–15 | ~ 1 × 109 | ~109 | single pulse | single pulse | 7 ns | 2.0 MVp | photon | yes | N/A |
| Auer et al. (Ref. 59) | 2011 | HeLa-RIKEN | post | 3 | – | 109 | 3 | – | < 1 ns | 20–25 MeV | proton | no | no |
| Pommarel et al. (Ref. 60) | 2017 | HCT116-WT, HCT116-p53-/- | post | 0–10 | – | 9 × 107 | 1.15 | N/A | – | 5 MeV | laser-accelerated protons | no | no |
| Manti et al. (Ref. 61) | 2017 | HUVEC | – | 0–5 | – | – | N/A | N/A | – | – | laser-accelerated protons | no | N/A |
| Bayart et al. (Ref. 62) | 2019 | SF763, U87 | post | 0–10 | – | 1.5 × 108 | 0.72 | N/A | – | – | laser-accelerated protons | no | N/A |
| Buonanno et al. (Ref. 20) | 2019 | IMR90 | post | 0–10 | 1000 | – | – | – | – | 5.5 MV | protons | no | N/A |
| Tessonnier et al. (Ref. 63) | 2021 | A549, H1437 | post | 0–12 | ~185(±35) | – | N/A | N/A | 14–86 ms | 145.74 MeV/u | Helium ions | no | yes |
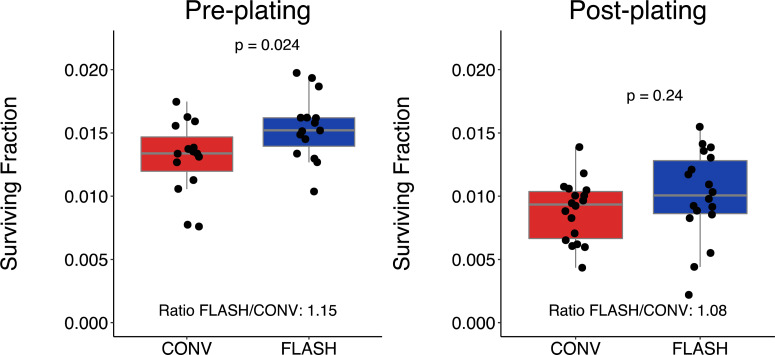
Challenges also arise from performing the clonogenic assay with irradiation at higher total doses. Depending on the cell line's plating efficiency and radiation sensitivity it may not be possible to achieve more than a few surviving colonies. Thereby, statistical uncertainties arise. In addition, the high inoculation density may cause difficulties to evaluate the samples. Together, clonogenic assays performed at higher dose ranges may be less reliable. The number of plated cells per flask or dish, the cell density, also affects the radiobiological response (Refs 38–40). In addition, the clonogenic assay can be performed in two principally different ways; pre-plating or post-plating (Ref. 64). In the pre-plating method, cells are plated in appropriate densities in individual flasks, dishes or wells, and each sample is irradiated. The cells are then incubated, without re-trypsinisation, to grow and form colonies. In the post-plating methods, a densely seeded flask is irradiated, trypsinised, cells are counted, plated in appropriate cell densities in dishes and allowed to grow and form colonies. Depending on the method used, different radiation response has been reported (Ref. 65). To investigate if plating methods could influence some of the reported inconsistent FLASH findings, we have performed pre- and post-plating experiments for the melanoma cell line MM576. Our current results suggest that both plating methods may detect a FLASH sparing, although the magnitude of the sparing and the power to detect statistical differences could differ (Fig. 2). Our lab has investigated clonogenic survival in normoxia after FLASH and CONV for nine human cell lines (including the MM576 in this publication) (Refs 19, 51). Five of the nine cell lines were found to have significantly increased survival fractions after FLASH irradiation. Although it is possible that statistical uncertainty caused the observed discrepancy between cell lines, it is also possible that a difference in biological factors affected the FLASH response. It has been found that the response to FLASH irradiation in vivo is cell line specific (Ref. 66), which is likely to also be the case in vitro. Further studies comparing different experimental set-ups could reveal additional phenomenological insights of the radiation response. For instance, by varying the time before re-plating after irradiation (delayed plating), the effect of ‘potentially lethal damage’ could be assessed (Ref 67).
Fig. 2.
Clonogenic survival for the melanoma cell line MM576 after irradiation with 9 Gy of CONV (red, measured delivered doses 9.2–9.4 Gy) or FLASH (blue, measured delivered doses 9.4–9.6 Gy) using the pre-plating (left panel) or post-plating method (right panel). The box and whisker plots illustrate median (grey line), interquartile range (box), the lowest/highest observation within ± 1.5 × interquartile range from the box (whiskers), and individual flasks as black dots. Irradiation was performed using a modified clinical accelerator (Ref. 28) with beam characteristics and the experimental pre-plating protocol as previously described (Ref. 14). For the post-plating, 500 000 cells were plated in T12.5 flasks the day before irradiation, and 1 h after irradiation the cells were trypsinised, counted, re-plated in appropriate densities and then incubated and evaluated as the pre-plating flasks. Statistical comparisons were made in RStudio (version 1.2.5042) using the unpaired Student's T-test after testing for normality using the Shapiro-Wilk's test. ‘Ratio FLASH/CONV’ was calculated as the mean (Survival FractionFLASH) divided by the mean (Survival FractionCONV). Data from three independent experiments with triplicate-sextuplicate flasks per condition.
Despite potential pitfalls with the clonogenic assay, it is still considered as the ‘gold standard’ and will undoubtedly help verify important findings in future FLASH studies. If care is taken to ascertain identical experimental conditions (e.g. time outside the incubator, cell medium volume, cell density and well-matched CONV and FLASH beam characteristics and irradiation doses), the assay will provide robust and reproducible data. For instance, the importance of beam line characteristics for the FLASH effect, such as pulse repetition frequency, dose per pulse, total delivery time, and average dose rate can be investigated in large-scale experiments. Detailed studies of the FLASH-response in physoxic and/or hypoxic/anoxic conditions (physoxia: oxygen levels of 3–7%, hypoxia: oxygen level ⩽2%, anoxia: 0% oxygen (Ref. 68)) are possible using the clonogenic assay, if care is taken to control the oxygen tension at the time of irradiation. Confirmatory studies in labs with different beam lines, using the same cell line, will be important. Besides varying oxygen concentration, the addition of scavengers such as superoxide dismutase might provide indirect mechanistic evidence for the underlying mechanism of the FLASH effect.
The current data suggest that the clonogenic assay can resolve at least part of the FLASH effect. Due to the clonogenic assay's central role in radiobiological research and the great advantage of large-scale experiments without involving living mammals, the continued use of clonogenic assays to investigate the FLASH effect will be important in our search for the underlying mechanisms.
Antibody-based assays
Radiation changes the physical and chemical environment of the cell, which leads to changes in protein phosphorylation, enzymatic activity, localisation, and the formation of protein-protein complexes that participate in cell cycle arrest, apoptosis, and DNA repair (Refs 69–72). Ionising radiation can induce DNA damage by direct interaction with the DNA or indirectly through the production of free radicals, which can react with the DNA. This causes a variety of DNA lesion types including base damage, single-strand breaks (SSBs) and double-strand breaks (DSBs) (Refs 73, 74). The generation of these DNA lesions, particularly DSBs, triggers sophisticated and highly regulated DNA damage response and repair (DDR) pathways (Ref. 75) that can easily be assessed in vitro by antibody-based techniques such as western blotting (WB), enzyme-linked immunosorbent assay (ELISA), flow cytometry, immunocytochemistry (ICC), or immunohistochemistry (IHC).
Both IHC and ICC provide spatial information such as distribution or localisation of specific cellular or subcellular components. IHC is generally applied on biological tissues from in vivo samples either embedded by paraffin or frozen to maintain the morphology, while ICC is generally applied on cells. They are especially useful in studying the DNA damage foci, which are subnuclear foci formed by DDR proteins near the DNA lesion site. IHC has provided some of the recent evidence identifying an in vivo FLASH effect. IHC staining for immune, apoptotic, and DDR markers showed compared to CONV irradiation, FLASH irradiation causes less apoptosis, DNA damage, and immune response in guts (Ref. 12), lung (Refs 1, 7), and brain (Refs 8, 9, 15). WB refers to the transfer of biological samples (mixture of proteins from cells or tissue samples) from a gel to a membrane and their subsequent detection on the surface of the membrane. It allows for the semi-quantification of a protein of interest amid a complex protein mixture, which can be achieved by ELISA, involving the use of standard curves with known protein concentrations. While WB detects the protein of interest from a mixture of cells, flow cytometry enables the study of intracellular and cell surface proteins at single-cell resolution.
The phosphorylation of the histone H2AX (γH2AX) is one of the most well-characterised DNA DSB markers. It generally occurs within minutes at the DSB sites in the nucleus and shows a maximum number of foci around 30 minutes post-irradiation (Ref. 76). Therefore, γH2AX can be used as a marker for exploring the spatial and temporal dynamics of DNA repair in cells following irradiation (Refs 77–80). In addition, γH2AX has been suggested as a prognostic biomarker to predict the radiotherapy response of patients (Ref. 81). Another classic DDR marker is 53BP1, which becomes hyperphosphorylated and colocalises with γH2AX near the site of DNA DSBs (Ref. 82).
WB, ICC, and IHC have previously been used to assess DNA damage level after FLASH or CONV irradiation. Using IHC, acute apoptosis was quantified with caspase-3 cleavage and TUNEL labelling in histological sections of irradiated lungs (Ref. 1), brain (Ref. 83) and intestines (Ref. 12). Using flow cytometry, Ehlert et al. showed increased γH2AX signal intensity in Jurkat and Ramos cells with increased radiation dose, using laser-accelerated protons at a dose rate of 108 Gy/s (Ref. 84). Fouillade et al. (Ref. 7) investigated γH2AX and recruitment of 53BP1 at sites of DNA damage by immunofluorescence microscopy in two normal lung fibroblast cell lines (MRC5 and IMR90) and A549 lung cancer cells. Though no difference in γH2AX foci per nucleus was found among the cell lines between CONV and FLASH irradiation, less 53BP1 was observed in the two normal lung fibroblasts after FLASH irradiation but not in the cancer cell line. However, Adrian et al. (Ref. 51) found no significant difference in 53BP1-foci number after FLASH and CONV irradiation for three studied cell lines.
Comet assay
Another method that has been implemented in many radiobiology studies to measure DNA damage and repair is the comet assay (Refs 85, 86). It was first introduced by Ostling and Johanson using neutral lysis (Ref. 87) in 1984 and then modified by Singh et al. to an alkaline version to increase the sensitivity (Ref. 88). The alkaline comet assay is an inexpensive, time efficient, highly sensitive method for assessing DNA damage formation and repair at the level of single cells (Refs 89, 90). In its simplest form, it enables the detection of SSBs, DSBs, alkali-labile sites (ALS), as well as SSB sites associated with incomplete DNA excision repair (Ref. 91). The comet assay can be further extended to detect DNA-DNA/DNA-protein cross-links, adding to the versatility in its applications (Refs 92–96).
In the alkaline comet assay, cells embedded in low melting point agarose on microscope slides are lysed in a solution containing high salt and detergents to remove cell membranes, cytoplasm, nucleoplasm and histones leaving nucleoids of supercoiled DNA linked to the nuclear matrix (Ref. 97). Relaxed in alkaline buffer, broken negatively charged DNA is free to migrate towards the anode upon electrophoresis. These stretched nucleoids resemble comets upon staining with an intercalating DNA dye which may be visualised via fluorescence microscopy (Ref. 89). The intensity of the comet tail relative to the amount of DNA residing in the ‘head’ indicates the level of induced strand breaks; with a reduction in DNA migration indicative of DNA crosslinks (Ref. 91). Furthermore, the addition of lesion-specific bacterial repair enzymes post-lysis enables the detection of a variety of DNA lesions other than the standard SSBs, DSBs, and ALSs (Refs 98–100). Therefore, the assay is useful in the profiling of DNA damage following different delivery modalities of ionising radiation, for instance different dose rates or beam particles.
The comet assay may also be used as a diagnostic, prognostic, and predictive biomarker in oncology (Refs 101, 102). It has been used as a tool for predicting an individual's tumour sensitivity to radiation with and without chemotherapeutic regimes (Refs 85, 103–105). A key feature of the assay is that only a small cell sample is required, making it possible to analyse cells from biopsy prior to and post treatment (Refs 101, 104), or from lymphocyte cells obtained via finger prick (Refs 106, 107). Historically, the assay has been used to identify the hypoxic fraction of tumours in mice and humans obtained from fine-needle aspiration (Refs 108, 109). This method also allows for monitoring changes to the hypoxic fraction of cells during a course of fractionated RT (Ref. 108).
The comet assay has recently been used by us to assess the difference in DNA damage formation of human peripheral blood lymphocytes (PBL) irradiated with 6 MeV electrons at FLASH (2 kGy/s) or CONV (0.1 Gy/s) dose rates, under a variety of oxygen concentrations. This was achieved by incubating cells embedded in low melting point agarose gels mounted on glass slides for 2 h in a humidified hypoxia chamber, prior to irradiation. PBL were used as a representative body-wide systemic normal tissue susceptible to irradiation, to assess DNA damage formation following FLASH or CONV irradiations (Refs 110–112). We found that the difference in DNA damage was modulated by the oxygen concentration, with a maximum difference of 30–40% seen at 0.25–0.5% oxygen tension. We also utilised the method to show how dose rate and total dose modulated the ex vivo DNA damage sparing effect observed between FLASH and CONV irradiation at a low (0.5%) oxygen tension, with significant sparing observed at average dose rates ⩾ 30 Gy/s and total doses ⩾ 20 Gy (Ref. 113).
Genomic approach
We and other groups have used RNA sequencing to identify potential novel biomarkers relevant to the differential RT response between FLASH and CONV irradiation (Refs 7, 114, 115). In addition, functional genomic screening using techniques like CRISPR (Ref. 116) or RNA interference (Ref. 117), which provides a large-scale genetic loss-of-function experimental approach, can be used to systematically identify the genes responsible for the FLASH effect. However, such methods are generally very time-consuming, resource demanding, and sometimes challenging to perform as they require high standards in quality control to give robust and reliable results, i.e. it requires identical irradiations (FLASH and CONV) of numerous identically prepared samples. In addition, the results will also need to be carefully validated in subsequent experiments using assays such as the ones described above.
Assessing oxygen content, free radicals and oxidative stress
It has been observed by us and several other groups that the FLASH effect is oxygen dependent (Refs 8, 19, 29, 63, 118). Therefore, being able to measure and control the oxygen concentration is of importance when conducting FLASH experiments. Techniques to measure oxygen tension in tissues in vivo have been previously reviewed (Ref. 119), some of which may be applied in vitro. Due to the nature of oxygen depletion during FLASH irradiation, the oxygen measurement technique of choice should ideally have a temporal resolution in the order of milliseconds. Quenching of fluorescent or phosphorescent dyes by oxygen is a common technique that has been applied to measure the oxygen consumption during FLASH irradiation. We and other groups have used fibre optic probes coated with oxygen-sensitive fluorescent compounds to measure the local oxygen tension in vitro during FLASH irradiation (Refs 31, 120). Using a water-soluble molecular probe Oxyphor 2P, Cao et al. measured and compared the oxygen consumption during FLASH and CONV irradiation, both in vitro and in vivo (Ref. 121). Other oxygen measurement techniques could be used to study the in vitro oxygen dynamic in FLASH irradiation, e.g. cellular tracking of oxygen concentration can be achieved using soluble oxygen probes and fluorescence/phosphorescence lifetime imaging microscopy (Refs 122, 123).
It has been hypothesised that the differential tissue response between FLASH and CONV irradiation is due to difference in damage from radiation-induced free radicals, caused by the higher temporal concentration of radicals produced for FLASH leading to an increase in radical-radical interactions and consequently less indirect damage of DNA (Fig. 1) (Refs 32, 124). Therefore, detection of free radicals in cells and tissues will be important to decipher if this is the mechanism responsible for the FLASH effect. A comprehensive review on free radical detection has previously been published by Halliwell and Whiteman (Ref. 125). Direct measurement of reactive oxygen species (ROS, a subset of free radicals containing oxygen) in cells is generally achieved by fluorogenic probes (Ref. 126). In addition, cells genetically encoded with fluorescent protein-based redox sensors can be used to capture the ROS dynamics within cells, in real-time (Ref. 126). Electron paramagnetic resonance (EPR) is the only technique that can detect specific free radicals directly, and it can also be used to measure oxygen concentration (Ref. 127). However, many of the free radicals generated in vitro or in vivo have very short half-life times and will require the use of ‘spin-traps’, chemicals that can form stable radicals, to enable detection by EPR. Incorporation of antibody-based technology with EPR, the ‘immuno-spin trapping,’ can detect DNA radicals with high sensitivity and subcellular information (Ref. 128). These techniques allow for spatiotemporal detection of free radicals following in vitro FLASH and CONV irradiation.
Compared to detecting free radicals, the oxidative stress, the damage of cells and tissues caused by ROS, is less technically challenging to detect since the markers are generally more stable. Oxidative stress can be assessed by DNA damage, lipid peroxidation, and protein damage using antibody-based or chromatographic assays (Ref. 129). In addition, fluorogenic lipophilic probes have been developed to study the spatiotemporal information of lipid metabolism (Ref. 130).
Novel in vitro assays
The radiobiological assays used for studying the FLASH effect have so far mostly been conducted with 2-dimensional (2D) cell cultures. Recent advancement in 3-dimensional (3D) cultures has offered more ways to model/mimic physiological conditions. Therefore, these technologies may be useful to create second-order events such as oxygen gradient and immune response, which could enhance the FLASH effect. The characteristics of each technology and the potential second-order events each can model to enhance the FLASH effect are summarised in Table 2.
Table 2.
Novel cell culture technology and the potential secondary events to enhance the FLASH effect
| Type | Characteristics | Secondary event | ||
|---|---|---|---|---|
| Oxygen gradient | Immune effect | Blood flow | ||
| Spheroids | Single cell population | Yes | No | No |
| Organoids | Multi-cell populations Have stem cells Self-renewal (biobanking possible) |
Yes | No | No |
| Tissue slices | Multi-cell populations Recapitulate spatial cellular information |
No | Yes | No |
| Organ-on-chip | Single or multi-cell population Recapitulate mechanical microenvironment High flexibility |
Yes | Yes | Yes |
| Engineered tissues | Single or multi-cell population High flexibility |
Yes | Yes | Yes |
Spheroids
Tumour spheroids are aggregates of cancer cells with 3D cell-cell contact. They can be either derived by permanent cancer cell lines or patient-derived tumour cells. Generation of spheroids by mixing cancer cells with other cell types (e.g. stromal cells) is also possible and widely used in drug screening. The 3D geometry of the tumour spheroids allows for the generation of oxygen and nutrient gradients from the outer layers to the centre, similar to in vivo tumours though being avascular. The outer cells of the tumour spheroid are generally proliferating, and the inner quiescent or sometimes necrotic. The application of tumour spheroids in radiobiology can be found from an earlier review by Santini et al. (Ref. 131). However, the potential of this technology was not fully realised until the recent development of high throughput imaging and high content screening (Ref. 132). Compared to in vivo models, tumour spheroids present a simple but still physiologically relevant model for radiobiology as their radiation response is independent of vasculatures, the normal tissues, and the immune cells. Therefore, the model can be used to understand the direct radiation response of cancer cells under simple environmental cues of oxygen and nutrients. With its spherical geometry, the environmental gradient and growth of tumour spheroids can be modelled in silico (Refs 133, 134), making it an easy model to be utilised with radiochemical or radiobiological modelling.
Use of tumour spheroids as a model for oxygen depletion during FLASH irradiation has been reported by Khan et al. (Ref. 22). Tumour spheroids were irradiated before they formed necrotic cores. After irradiation, tumour spheroids were subsequently dissociated for clonogenic assay. Using this method, they showed that irradiating tumour spheroids can result in a large difference in cell survival between FLASH and CONV irradiation, with a maximum dose-modifying factor of 1.3, while they observed no difference in survival in 2D cell culture. The enhanced FLASH effect in tumour spheroids is likely caused by the relative increase in the hypoxic cell population during the FLASH irradiation, due to radiochemical oxygen depletion, and the differential radiosensitivity of normoxic and hypoxic cells within the spheroids. Interestingly, their study also showed that the growth kinetics of the intact spheroids is not a good indicator of radiation damage due to the confounding effect of senescent cells. Currently, the growth kinetics of tumour spheroids is the most studied end-point in drug and therapy screening studies. After irradiation, a large portion of cells in the spheroids will enter the senescent stage, but still contribute to the growth in spheroid size, making this method less relevant for radiobiological studies. As senescent cells are generally less dense and as FLASH irradiation was shown to produce less senescent cells (Ref. 20), it is possible that the difference between FLASH and CONV irradiation can be evaluated by measuring the weight or density of the tumour spheroids (Ref. 135) or analysing the senescent biomarkers using IHC (Ref. 136). In addition, high content imaging with important biomarkers for different stages of cell cycles and DDR signalling may also highlight the biological difference between FLASH and CONV irradiation (Refs 137, 138). Overall, spheroids provide a simple 3D tumour model to study the FLASH effect under a hypoxic tumour microenvironment.
Organoids
Organoids are 3D miniature and self-organised cultures showing physiological micro-anatomy. They can be derived from adult stem cells, embryonic or pluripotent stem cells. Unlike spheroids, organoids consist of heterogeneous cell types including stem cells or progenitor cells that are crucial for normal tissue response to radiation damage. The organoid culture also requires extracellular matrix and growth factors. The advantage of organoid is that they can be passaged in vitro and cryopreserved, facilitating biobanking for further preclinical studies. The application of organoids in FLASH-RT has not yet been reported, but they have been used to study the radiosensitivity and radiation damage of normal tissues. Martin et al. showed that small and large intestine organoids were able to recapitulate the radiosensitivity profile of the intact organ (Ref. 139). Das et al. used human induced pluripotent stem cell-derived cerebral organoids to study the radiation-induced DNA repair (Ref. 140). Martinez et al. used parotid salivary gland organoids to study the radiation response (Ref. 141). Mammary organoids have also been used to study the immune cell recruitment (Ref. 142). Patient-derived tumour specimens can also be a source for tumour organoids (tumouroids). Tumouroids can reflect the genetic and phenotypic heterogeneity of their original sources, making them a valuable tool for personalised medicine (Refs 143–145). In addition to cellular phenotype, organoids can mimic the hypoxic microenvironment of the original tissues. Hubert et al. showed that patient-derived glioblastoma organoids demonstrated hypoxia gradient and cancer stem cell heterogeneity similar to in vivo conditions (Ref. 146). Multi-parametric imaging of oxygen tension and cell cycle stage has also been developed in intestinal organoids (Ref. 147). Therefore, organoids can be a useful tool to study the dynamic cellular response in tumours and normal tissues after radiation. Both tumouroids and normal tissue organoids can be derived from the same patients, which could allow for the development of safe and efficient personalised treatments using this protocol.
Tissue slice cultures
Tissue slice cultures, or organotypic culture, are generated by cutting non-fixed normal tissues or tumours into thin slices. These slices can preserve the morphology and microenvironment of the original tissues for several days. They have been commonly used in drug screening and in a few radiobiological studies. Suckert et al. used tumour slices of head and neck squamous cell carcinoma and slice culture of adult mice brain to study the radiation response of proton beam RT (Ref. 148). They found that tumour slice culture demonstrated DNA damage and morphology results similar to the in vivo condition, while brain slice culture failed to reveal relevant radiation response, potentially due to change in cell morphology and phenotype after long term culture. On the other hand, the use of neonatal brain slice culture has proven more successful in maintaining the morphology and phenotype after long term culture (Ref. 149). With the preservation of immune cells in the tumour microenvironment, tumour slice cultures have been commonly used to study immunotherapy. Innate immunity can be induced by PD-1 inhibitors on patient-derived lung tumour slices (Ref. 150). In addition, the use of patient-derived lung tumour slices can be predictive of the immunotherapy outcome (Ref. 151). In summary, tissue slice cultures can be used to study short term radiation effect on the bona fide cellular microenvironment and in physiological conditions.
Organ-on-Chip models
One of the major limitations in 2D and 3D cell culture models is the ability to simulate flow and relevant mechanical properties (e.g. shear stress and tensile strain). Organ-on-chip models provide the solution to incorporate fluid dynamics and biomechanics into the culture systems. Organ-on-chip models are created by culturing cells of 2D or 3D formats in a microfluidic device that can control the physical or chemical microenvironment of the cells. Though it has not been widely applied in studying FLASH-RT, several projects have been established to use organ-on-chip systems as a countermeasure of radiotoxicity (Ref. 152). Patient-derived head and neck squamous cell carcinoma has been used in an organ-on-chip model to assess the radiation-induced cell death and could predict the clinical outcome (Refs 153, 154). Radiation-induced cell death and protection by radioprotectors were also demonstrated on a Gut-on-a-Chip model (Ref. 155). An oxygen gradient can be easily created in the organ-on-chip model by using chemical oxygen quenchers in the medium or pumping nitrogen/carbon dioxide through the device (Refs 156, 157). It is also possible to link multiple organ-on-chip models together to create a body-on-chip system. This may also open the door to studies of systemic radiation effects such as long-range bystander (Ref. 158) and abscopal effects, which is not possible using simpler in vitro models. The highly tailorable nature of organ-on-chip models make them a versatile tool to study the effect of both cellular and environmental factors following irradiation. Biochemical sensors can also be integrated into the organ-on-chip systems to provide in situ monitoring of cellular and microenvironmental parameters.
Engineered tissue models by 3D bioprinting
3D bioprinting allows for the construction of complex multi-cell models layer by layer, with high flexibility and precision. The cells are printed into bio-inks, which are generally biocompatible materials. The bio-inks determine the physical and chemical microenvironment of the printed tissues. The choice of bio-ink is important when using the engineered tissue for modelling radiation damage response as it can affect the cellular response (Ref. 159). Vascularised engineered tissues can be created by 3D bioprinting to simulate oxygen and nutrient gradients (Ref. 160). Oxygen released bio-ink can also be used to modulate the local oxygen tension inside the engineered construct (Ref. 161). Today 3D bioprinting focuses mostly on regenerative medicine but with the development of high throughput printing technology, it is expected that the engineered tissues can also be used for tumour/normal tissue modelling to study the FLASH effect. The combination of patient-derived cells and stimuli-responsive bio-inks, which can change the physical or biochemical environment, offer a platform to study the dynamic response of physiologically relevant tissues following FLASH irradiation.
Concluding remarks/translational implications
The FLASH effect is an intriguing phenomenon that is currently being studied by many research labs around the world. The magnitude of the effect is such that it could be a ‘game changer’ for the treatment of many tumours. However, as we do not yet understand the effect or know the mechanisms behind the effect, we cannot translate it into the clinic in an optimal way. Consequently, more preclinical studies are needed. Specifically, in vitro studies to complement the more challenging and expensive in vivo studies. Here, we have described several in vitro assays that have been used or could be used to finally elucidate the mechanisms behind the FLASH effect. The doses often required to show a significant FLASH effect (⩾10 Gy) make some of the mentioned assays more suitable than others, e.g. the comet assay compared to γH2AX.
FLASH-RT is a very promising new radiotherapy technique that we would like our cancer patients to benefit from as soon as possible. However, a successful clinical translation of the technique hinges on a better understanding of the FLASH sparing effect of normal tissues. If we understand this phenomenon, we can optimise our FLASH treatments to maximise the effect, e.g. by using hypofractionated approaches (Ref. 17), non-homogeneous irradiation (Ref. 162), and combination with drugs that enhances the effect (Ref. 163). Preclinical assays, such as the ones described above, will be essential tools in identifying the radiobiological mechanisms behind the effect and finally being able to fully exploit the benefits associated with FLASH-RT.
Conflict of interest
None.
Financial support
CRC's PhD is funded by the Leicester Cancer Research Centre, University of Leicester. JLR and SP positions are funded through Cancer Research UK (RadNet Grant [C6078/A28736]) and KP position is funded through the Medical Research Council (MRC [MC_UU_00001/9]). GA is supported by Mrs Berta Kamprad Foundation (grant no 2020-19-301), Swedish Cancer Society (grant no 20 1298), John and Augusta Persson's Foundation and governmental research funding (ST-ALF).
References
- 1.Favaudon V et al. (2014) Ultrahigh dose-rate FLASH irradiation increases the differential response between normal and tumor tissue in mice. Science Translational Medicine 6, 245ra93. [DOI] [PubMed] [Google Scholar]
- 2.Loo BW et al. (2017) (P003) delivery of ultra-rapid flash radiation therapy and demonstration of normal tissue sparing after abdominal irradiation of mice. International Journal of Radiation Oncology • Biology • Physics 98, E16. [Google Scholar]
- 3.Montay-Gruel P et al. (2017) Irradiation in a flash: unique sparing of memory in mice after whole brain irradiation with dose rates above 100Gy/s. Radiotherapy & Oncology 124, 365–369. [DOI] [PubMed] [Google Scholar]
- 4.Montay-Gruel P et al. (2018) X-rays can trigger the FLASH effect: ultra-high dose-rate synchrotron light source prevents normal brain injury after whole brain irradiation in mice. Radiotherapy & Oncology 129, 582–588. [DOI] [PubMed] [Google Scholar]
- 5.Bourhis J et al. (2019) Clinical translation of FLASH radiotherapy: why and how? Radiotherapy & Oncology 139, 11–17. [DOI] [PubMed] [Google Scholar]
- 6.Bourhis J et al. (2019) Treatment of a first patient with FLASH-radiotherapy. Radiotherapy & Oncology 139, 18–22. [DOI] [PubMed] [Google Scholar]
- 7.Fouillade C et al. (2019) FLASH Irradiation spares lung progenitor cells and limits the incidence of radio-induced senescence. Clinical Cancer Research 26, 1497–1506. [DOI] [PubMed] [Google Scholar]
- 8.Montay-Gruel P et al. (2019) Long-term neurocognitive benefits of FLASH radiotherapy driven by reduced reactive oxygen species. Proceedings of the National Academy of Sciences of the United States of America 116, 10943–10951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Simmons DA et al. (2019) Reduced cognitive deficits after FLASH irradiation of whole mouse brain are associated with less hippocampal dendritic spine loss and neuroinflammation. Radiotherapy & Oncology 139, 4–10. [DOI] [PubMed] [Google Scholar]
- 10.Vozenin MC et al. (2019) The advantage of FLASH radiotherapy confirmed in Mini-pig and Cat-cancer patients. Clinical Cancer Research 25, 35–42. [DOI] [PubMed] [Google Scholar]
- 11.Diffenderfer ES et al. (2020) Design, implementation, and in vivo validation of a novel proton FLASH radiation therapy system. International Journal of Radiation Oncology Biology Physics 106, 440–448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Levy K et al. (2020) Abdominal FLASH irradiation reduces radiation-induced gastrointestinal toxicity for the treatment of ovarian cancer in mice. Scientific Reports 10, 21600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Soto LA et al. (2020) FLASH Irradiation results in reduced severe skin toxicity compared to conventional-dose-rate irradiation. Radiation Research 194, 618–624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang Q et al. (2020) FLASH Investigations using protons: design of delivery system, preclinical setup and confirmation of FLASH effect with protons in animal systems. Radiation Research 194, 656–664. [DOI] [PubMed] [Google Scholar]
- 15.Alaghband Y et al. (2020) Neuroprotection of radiosensitive juvenile mice by ultra-high dose rate FLASH irradiation. Cancers (Basel) 12, 1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson JD et al. (2020) Ultra-high dose rate (FLASH) radiotherapy: silver bullet or fool's gold? Frontiers in Oncology 9, 1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Montay-Gruel P et al. (2021) Hypofractionated FLASH-RT as an effective treatment against glioblastoma that reduces neurocognitive Side effects in mice. Clinical Cancer Research 27, 775–784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zlobinskaya O et al. (2014) The effects of ultra-high dose rate proton irradiation on growth delay in the treatment of human tumor xenografts in nude mice. Radiation Research 181, 177–183. [DOI] [PubMed] [Google Scholar]
- 19.Adrian G et al. (2020) The FLASH effect depends on oxygen concentration. The British Journal of Radiology 93, 20190702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Buonanno M, Grilj V and Brenner DJ (2019) Biological effects in normal cells exposed to FLASH dose rate protons. Radiotherapy & Oncology 139, 51–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hopewell JW and van den Aardweg GJ (1988) Radiobiological studies with pig skin. International Journal of Radiation Oncology Biology Physics 14, 1047–1050. [DOI] [PubMed] [Google Scholar]
- 22.Khan S et al. (2021) Multicellular spheroids as In vitro models of oxygen depletion during FLASH irradiation. International Journal of Radiation Oncology Biology Physics 110, 833–844. [DOI] [PubMed] [Google Scholar]
- 23.Konradsson E et al. (2021) Establishment and initial experience of clinical FLASH radiotherapy in canine cancer patients. Frontiers in Oncology 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jaccard M et al. (2018) High dose-per-pulse electron beam dosimetry: commissioning of the oriatron eRT6 prototype linear accelerator for preclinical use. Medical Physics 45, 863–874. [DOI] [PubMed] [Google Scholar]
- 25.Lempart M et al. (2019) Modifying a clinical linear accelerator for delivery of ultra-high dose rate irradiation. Radiotherapy & Oncology 139, 40–45. [DOI] [PubMed] [Google Scholar]
- 26.Schuler E et al. (2017) Experimental platform for ultra-high dose rate FLASH irradiation of small animals using a clinical linear accelerator. International Journal of Radiation Oncology Biology Physics 97, 195–203. [DOI] [PubMed] [Google Scholar]
- 27.Maxim PG, Tantawi SG and Loo BW Jr. (2019) PHASER: a platform for clinical translation of FLASH cancer radiotherapy. Radiotherapy & Oncology 139, 28–33. [DOI] [PubMed] [Google Scholar]
- 28.Hall EJ and Brenner DJ (1991) The dose-rate effect revisited: radiobiological considerations of importance in radiotherapy. International Journal of Radiation Oncology Biology Physics 21, 1403–1414. [DOI] [PubMed] [Google Scholar]
- 29.Hendry JH et al. (1982) The constant low oxygen concentration in all the target cells for mouse tail radionecrosis. Radiation Research 92, 172–181. [PubMed] [Google Scholar]
- 30.Vozenin MC, Hendry JH and Limoli CL (2019) Biological benefits of ultra-high dose rate FLASH radiotherapy: sleeping beauty awoken. Clin Oncol (R Coll Radiol) 31, 407–415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moon EJ, Petersson K and Olcina MM (2021) The importance of hypoxia in radiotherapy for the immune response, metastatic potential and FLASH-RT. International Journal of Radiation Biology, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Labarbe R et al. (2020) A physicochemical model of reaction kinetics supports peroxyl radical recombination as the main determinant of the FLASH effect. Radiotherapy & Oncology 153, 303–310. [DOI] [PubMed] [Google Scholar]
- 33.Wardman P (2020) Radiotherapy using high-intensity pulsed radiation beams (FLASH): a radiation-chemical perspective. Radiation Research 194, 607–617. [DOI] [PubMed] [Google Scholar]
- 34.Durante M, Brauer-Krisch E and Hill M (2018) Faster and safer? FLASH ultra-high dose rate in radiotherapy. The British Journal of Radiology 91, 20170628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Jin JY et al. (2020) Ultra-high dose rate effect on circulating immune cells: a potential mechanism for FLASH effect? Radiotherapy & Oncology 149, 55–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Puck TT and Marcus PI (1956) Action of x-rays on mammalian cells. The Journal of experimental medicine 103, 653–666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sia J et al. (2020) Molecular mechanisms of radiation-induced cancer cell death: a primer. Frontiers in Cell and Developmental Biology 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Brix N et al. (2020) The clonogenic assay: robustness of plating efficiency-based analysis is strongly compromised by cellular cooperation. Radiation Oncology 15, 248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adrian G et al. (2018) Rescue effect inherited in colony formation assays affects radiation response. Radiation Research 189, 44–52. [DOI] [PubMed] [Google Scholar]
- 40.Brix N et al. (2021) Analysis of clonogenic growth in vitro. Nature Protocols 16, 4963–4991. [DOI] [PubMed] [Google Scholar]
- 41.Berry RJ et al. (1969) Survival of mammalian cells exposed to X rays at ultra-high dose-rates. The British Journal of Radiology 42, 102–107. [DOI] [PubMed] [Google Scholar]
- 42.Town CD (1967) Effect of high dose rates on survival of mammalian cells. Nature 215, 847–848. [DOI] [PubMed] [Google Scholar]
- 43.Berry RJ and Stedeford JBH (1972) Reproductive survival of mammalian cells after irradiation at ultra-high dose-rates: further observations and their importance for radiotherapy. The British Journal of Radiology 45, 171–177. [DOI] [PubMed] [Google Scholar]
- 44.Nias AHW et al. (1970) Survival of HeLa cells from 10 nanosecond pulses of electrons. International Journal of Radiation Biology and Related Studies in Physics, Chemistry and Medicine 17, 595–598. [DOI] [PubMed] [Google Scholar]
- 45.Epp ER et al. (1972) The radiosensitivity of cultured mammalian cells exposed to single high intensity pulses of electrons in various concentrations of oxygen. Radiation Research 52, 324–332. [PubMed] [Google Scholar]
- 46.Michaels HB et al. (1978) Oxygen sensitization of CHO cells at ultrahigh dose rates: prelude to oxygen diffusion studies. Radiation Research 76, 510–521. [PubMed] [Google Scholar]
- 47.Dewey DL and Boag JW (1959) Modification of the oxygen effect when bacteria are given large pulses of radiation. Nature 183, 1450–1451. [DOI] [PubMed] [Google Scholar]
- 48.Epp E, Weiss H and Santomasso A (1968) The oxygen effect in bacterial cells irradiated with high-intensity pulsed electrons. Radiation Research 34, 320–325. [PubMed] [Google Scholar]
- 49.Cygler JE et al. (1994) The survival of aerobic and anoxic human glioma and melanoma cells after irradiation at ultrahigh and clinical dose rates. Radiation Research 140, 79–84. [PubMed] [Google Scholar]
- 50.Zackrisson BU, Nyström UH and Ostbergh P (1991) Biological response In vitro to pulsed high dose rate electrons from a clinical accelerator. Acta Oncologica 30, 747–751. [DOI] [PubMed] [Google Scholar]
- 51.Adrian G et al. (2021) Cancer cells Can exhibit a sparing FLASH effect at Low doses under normoxic In vitro-conditions. Frontiers in Oncology 11, 686142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Venkatesulu BP et al. (2019) Ultra high dose rate (35 Gy/sec) radiation does not spare the normal tissue in cardiac and splenic models of lymphopenia and gastrointestinal syndrome. Scientific Reports 9, 17180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McMahon SJ (2018) The linear quadratic model: usage, interpretation and challenges. Physics in Medicine & Biology 64, 01T, R01. [DOI] [PubMed] [Google Scholar]
- 54.Potter MDE et al. (2011) Hidden stressors in the clonogenic assay used in radiobiology experiments. Australasian Physical & Engineering Sciences in Medicine 34, 345–350. [DOI] [PubMed] [Google Scholar]
- 55.Joshi GP et al. (1982) Discrimination of slow growth from non-survival among small colonies of diploid Syrian hamster cells after chromosome damage induced by a range of x-ray doses. International Journal of Radiation Biology and Related Studies in Physics, Chemistry, and Medicine 42, 283–296. [DOI] [PubMed] [Google Scholar]
- 56.Nias AH et al. (1965) Clone-size analysis in the study of cell growth following single or during continuous irradiation. International Journal of Radiation Biology and Related Studies in Physics, Chemistry, and Medicine 9, 275–290. [DOI] [PubMed] [Google Scholar]
- 57.Yohem KH, Bregman MD and Meyskens FL Jr. (1987) Effect of tumor colony definition on ionizing radiation survival curves of melanoma-colony forming cells. International Journal of Radiation Oncology Biology Physics 13, 1725–1733. [DOI] [PubMed] [Google Scholar]
- 58.Nias AH et al. (1969) Effects of pulses of radiation on the survival of mammalian cells. The British Journal of Radiology 42, 553. [DOI] [PubMed] [Google Scholar]
- 59.Auer S et al. (2011) Survival of tumor cells after proton irradiation with ultra-high dose rates. Radiation Oncology 6, 139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pommarel L et al. (2017) Spectral and spatial shaping of a laser-produced ion beam for radiation-biology experiments. Physical Review Accelerators and Beams 20. [Google Scholar]
- 61.Manti L et al. (2017) The radiobiology of laser-driven particle beams: focus on sub-lethal responses of normal human cells. Journal of Instrumentation 12. [Google Scholar]
- 62.Bayart E et al. (2019) Fast dose fractionation using ultra-short laser accelerated proton pulses can increase cancer cell mortality, which relies on functional PARP1 protein. Scientific Reports 9, 10132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Tessonnier T et al. (2021) FLASH dose rate helium ion beams: first In Vitro investigations. International Journal of Radiation Oncology Biology Physics 111, 1011–1022. [DOI] [PubMed] [Google Scholar]
- 64.Franken NAP et al. (2006) Clonogenic assay of cells in vitro. Nature Protocols 1, 2315–2319. [DOI] [PubMed] [Google Scholar]
- 65.Kriegs M et al. (2016) Radiosensitization of HNSCC cells by EGFR inhibition depends on the induction of cell cycle arrests. Oncotarget 7, 45122–45133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Chabi S et al. (2021) Ultra-high-dose-rate FLASH and conventional-dose-rate irradiation differentially affect human acute lymphoblastic leukemia and normal hematopoiesis. International Journal of Radiation Oncology Biology Physics 109, 819–829. [DOI] [PubMed] [Google Scholar]
- 67.Liu M et al. (2016) DNA Repair pathway choice at various conditions immediately post irradiation. International Journal of Radiation Biology 92, 819–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.McKeown SR (2014) Defining normoxia, physoxia and hypoxia in tumours-implications for treatment response. The British Journal of Radiology 87, 20130676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Levine AJ and Oren M (2009) The first 30 years of p53: growing ever more complex. Nature Reviews Cancer 9, 749–758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Meek DW (2009) Tumour suppression by p53: a role for the DNA damage response? Nature Reviews Cancer 9, 714–723. [DOI] [PubMed] [Google Scholar]
- 71.O'Driscoll M and Jeggo PA (2006) The role of double-strand break repair — insights from human genetics. Nature Reviews Genetics 7, 45–54. [DOI] [PubMed] [Google Scholar]
- 72.Prise KM et al. (2005) New insights on cell death from radiation exposure. The Lancet. Oncology 6, 520–528. [DOI] [PubMed] [Google Scholar]
- 73.Alloni D et al. (2011) Monte Carlo evaluation of DNA fragmentation spectra induced by different radiation qualities. Radiation Protection Dosimetry 143, 226–231. [DOI] [PubMed] [Google Scholar]
- 74.Alloni D et al. (2012) Track structure, radiation quality and initial radiobiological events: considerations based on the PARTRAC code experience. International Journal of Radiation Biology 88, 77–86. [DOI] [PubMed] [Google Scholar]
- 75.Santivasi WL and Xia F (2014) Ionizing radiation-induced DNA damage, response, and repair. Antioxidants & Redox Signaling 21, 251–259. [DOI] [PubMed] [Google Scholar]
- 76.Mariotti L et al. (2013) Use of the γ-H2AX assay to investigate DNA repair dynamics following multiple radiation exposures. PloS One 8, e79541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Costes SV et al. (2010) Spatiotemporal characterization of ionizing radiation induced DNA damage foci and their relation to chromatin organization. Mutation Research 704, 78–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Costes SV et al. (2007) Image-based modeling reveals dynamic redistribution of DNA damage into nuclear sub-domains. PLoS Computational Biology 3, e155–e155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Rothkamm K and Löbrich M (2003) Evidence for a lack of DNA double-strand break repair in human cells exposed to very low x-ray doses. Proceedings of the National Academy of Sciences of the United States of America 100, 5057–5062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Schmid TE et al. (2010) Differences in the kinetics of γ-H2AX fluorescence decay after exposure to low and high LET radiation. International Journal of Radiation Biology 86, 682–691. [DOI] [PubMed] [Google Scholar]
- 81.Ivashkevich A et al. (2012) Use of the γ-H2AX assay to monitor DNA damage and repair in translational cancer research. Cancer Letters 327, 123–133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Ward I et al. (2003) Accumulation of checkpoint protein 53BP1 at DNA breaks involves its binding to phosphorylated histone H2AX. The Journal of Biological Chemistry 278, 19579–19582. [DOI] [PubMed] [Google Scholar]
- 83.Allen BD et al. (2020) Maintenance of tight junction integrity in the absence of vascular dilation in the brain of mice exposed to ultra-high-dose-rate FLASH irradiation. Radiation Research 194, 625–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ehlert J et al. (2021) An experimental platform for studying the radiation effects of laser accelerated protons on mammalian cells. AIP Advances 11, 065208. [Google Scholar]
- 85.Olive PL (2009) Impact of the comet assay in radiobiology. Mutation Research 681, 13–23. [DOI] [PubMed] [Google Scholar]
- 86.Zhang M et al. (2018) A comet assay for DNA damage and repair after exposure to carbon-Ion beams or X-rays in saccharomyces Cerevisiae. Dose-response 16, 1559325818792467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Ostling O and Johanson KJ (1984) Microelectrophoretic study of radiation-induced DNA damages in individual mammalian cells. Biochemical and Biophysical Research Communications 123, 291–298. [DOI] [PubMed] [Google Scholar]
- 88.Singh NP et al. (1988) A simple technique for quantitation of low levels of DNA damage in individual cells. Experimental Cell Research 175, 184–191. [DOI] [PubMed] [Google Scholar]
- 89.Collins AR (2004) The comet assay for DNA damage and repair: principles, applications, and limitations. Molecular Biotechnology 26, 249–261. [DOI] [PubMed] [Google Scholar]
- 90.Azqueta A et al. (2014) Comet assay to measure DNA repair: approach and applications. Frontiers in Genetics 5, 288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Tice RR et al. (2000) Single cell gel/comet assay: guidelines for in vitro and in vivo genetic toxicology testing. Environmental and Molecular Mutagenesis 35, 206–221. [DOI] [PubMed] [Google Scholar]
- 92.Pfuhler S and Wolf HU (1996) Detection of DNA-crosslinking agents with the alkaline comet assay. Environmental and Molecular Mutagenesis 27, 196–201. [DOI] [PubMed] [Google Scholar]
- 93.Hartley JM et al. (1999) Measurement of DNA cross-linking in patients on ifosfamide therapy using the single cell gel electrophoresis (comet) assay. Clinical Cancer Research 5, 507–512. [PubMed] [Google Scholar]
- 94.Almeida GM et al. (2006) Detection of oxaliplatin-induced DNA crosslinks in vitro and in cancer patients using the alkaline comet assay. DNA Repair (Amst) 5, 219–225. [DOI] [PubMed] [Google Scholar]
- 95.Spanswick VJ, Hartley JM and Hartley JA (2010) Measurement of DNA interstrand crosslinking in individual cells using the single cell Gel electrophoresis (Comet) assay. Methods in Molecular Biology 613, 267–282. [DOI] [PubMed] [Google Scholar]
- 96.Fikrova P et al. (2013) Detection of DNA crosslinks in peripheral lymphocytes isolated from patients treated with platinum derivates using modified comet assay. Neoplasma 60, 413–418. [DOI] [PubMed] [Google Scholar]
- 97.Azqueta A and Collins AR (2013) The essential comet assay: a comprehensive guide to measuring DNA damage and repair. Archives of Toxicology 87, 949–968. [DOI] [PubMed] [Google Scholar]
- 98.Ding W et al. (2016) In vivo alkaline comet assay and enzyme-modified alkaline comet assay for measuring DNA strand breaks and oxidative DNA damage in rat liver. Journal of Visualized Experiments: JoVE (111). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Azqueta A et al. (2019) Technical recommendations to perform the alkaline standard and enzyme-modified comet assay in human biomonitoring studies. Mutation Research 843, 24–32. [DOI] [PubMed] [Google Scholar]
- 100.Muruzabal D, Collins A and Azqueta A (2021) The enzyme-modified comet assay: past, present and future. Food and Chemical Toxicology 147, 111865. [DOI] [PubMed] [Google Scholar]
- 101.McKenna DJ, McKeown SR and McKelvey-Martin VJ (2008) Potential use of the comet assay in the clinical management of cancer. Mutagenesis 23, 183–190. [DOI] [PubMed] [Google Scholar]
- 102.Vodicka P et al. (2019) DNA Damage and repair measured by comet assay in cancer patients. Mutation Research. Genetic Toxicology and Environmental Mutagenesis 843, 95–110. [DOI] [PubMed] [Google Scholar]
- 103.Moneef MA et al. (2003) Measurements using the alkaline comet assay predict bladder cancer cell radiosensitivity. British Journal of Cancer 89, 2271–2276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Bowman KJ et al. (2014) Comet assay measures of DNA damage are predictive of bladder cancer cell treatment sensitivity in vitro and outcome in vivo. International Journal of Cancer 134, 1102–1111. [DOI] [PubMed] [Google Scholar]
- 105.Wood JP et al. (2015) Comet assay measures of DNA damage as biomarkers of irinotecan response in colorectal cancer in vitro and in vivo. Cancer Medicine 4, 1309–1321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Garcia O and Mandina T (2005) DNA Damage evaluated by the comet assay in lymphocytes of children with 137Cs internal contamination caused by the chernobyl accident. Mutation Research 565, 191–197. [DOI] [PubMed] [Google Scholar]
- 107.Al-Salmani K et al. (2012) Simplified method for the collection, storage, and comet assay analysis of DNA damage in whole blood. PubMed 51, 719–725. [DOI] [PubMed] [Google Scholar]
- 108.Olive PL et al. (1993) Gel electrophoresis of individual cells to quantify hypoxic fraction in human breast cancers. Cancer Research 53, 733–736. [PubMed] [Google Scholar]
- 109.Shibuya K et al. (1999) Detection of hypoxic cells in murine tumors using the comet assay: comparison with a conventional radiobiological assay. Japanese Journal of Cancer Research 90, 880–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Louagie H et al. (1999) Changes in peripheral blood lymphocyte subsets in patients undergoing radiotherapy. International Journal of Radiation Biology 75, 767–771. [DOI] [PubMed] [Google Scholar]
- 111.Petersson K et al. (2014) Haematological toxicity in adult patients receiving craniospinal irradiation--indication of a dose-bath effect. Radiotherapy & Oncology 111, 47–51. [DOI] [PubMed] [Google Scholar]
- 112.Heylmann D et al. (2021) Comparison of DNA repair and radiosensitivity of different blood cell populations. Scientific Reports 11, 2478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Cooper CR et al. (2022) FLASH irradiation induces lower levels of DNA damage ex vivo, an effect modulated by oxygen tension, dose, and dose rate. British Journal of Radiology published online. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Velalopoulou A et al. (2021) FLASH proton radiotherapy spares normal epithelial and mesenchymal tissues while preserving sarcoma response. Cancer Research 81, 4808–4821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Ruan J et al. (2021) Irradiation at ultra-high (FLASH) dose rates reduces acute normal tissue toxicity in the mouse gastrointestinal system. International Journal of Radiation Oncology, Biology, Physics 111, 1250–1261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.He C et al. (2021) CRISPR screen in cancer: status quo and future perspectives. American Journal of Cancer Research 11, 1031–1050. [PMC free article] [PubMed] [Google Scholar]
- 117.Mullenders J and Bernards R (2009) Loss-of-function genetic screens as a tool to improve the diagnosis and treatment of cancer. Oncogene 28, 4409–4420. [DOI] [PubMed] [Google Scholar]
- 118.Petersson K et al. (2020) A quantitative analysis of the role of oxygen tension in FLASH radiation therapy. International Journal of Radiation Oncology Biology Physics 107, 539–547. [DOI] [PubMed] [Google Scholar]
- 119.Springett R and Swartz H (2007) Measurements of oxygen in vivo: overview and perspectives on methods to measure oxygen within cells and tissues. Antioxidants & Redox Signaling 9, 1295–1301. [DOI] [PubMed] [Google Scholar]
- 120.Jansen J et al. (2021) Does FLASH deplete oxygen? Experimental evaluation for photons, protons, and carbon ions. Medical Physics 48, 3982–3990. [DOI] [PubMed] [Google Scholar]
- 121.Cao X et al. (2021) Quantification of oxygen depletion during FLASH irradiation In vitro and In vivo. International Journal of Radiation Oncology, Biology, Physics 111, 240–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Kurokawa H et al. (2015) High resolution imaging of intracellular oxygen concentration by phosphorescence lifetime. Scientific Reports 5, 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Penjweini R et al. (2020) Single cell-based fluorescence lifetime imaging of intracellular oxygenation and metabolism. Redox Biology 34, 101549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Favaudon V, Labarbe R and Limoli C (2021) Model studies of the role of oxygen in the FLASH effect. Medical Physics 00, 1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Halliwell B and Whiteman M (2004) Measuring reactive species and oxidative damage in vivo and in cell culture: how should you do it and what do the results mean? British Journal of Pharmacology 142, 231–255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Bai X et al. (2019) Small-molecule-based fluorescent sensors for selective detection of reactive oxygen species in biological systems. Annual Review of Biochemistry 88, 605–633. [DOI] [PubMed] [Google Scholar]
- 127.Khan N et al. (2007) Repetitive tissue pO2 measurements by electron paramagnetic resonance oximetry: current status and future potential for experimental and clinical studies. Antioxidants & Redox Signaling 9, 1169–1182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Mason R (2016) Imaging free radicals in organelles, cells, tissue, and in vivo with immuno-spin trapping. Redox Biology 8, 422–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Katerji M, Filippova M and Duerksen-Hughes P (2019) Approaches and methods to measure oxidative stress in clinical samples: research applications in the cancer field. Oxidative Medicine and Cellular Longevity 2019, 1279250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Greene L, Lincoln R and Cosa G (2018) Spatio-temporal monitoring of lipid peroxyl radicals in live cell studies combining fluorogenic antioxidants and fluorescence microscopy methods. Free Radical Biology & Medicine 128, 124–136. [DOI] [PubMed] [Google Scholar]
- 131.Santini M, Rainaldi G and Indovina P (1999) Multicellular tumour spheroids in radiation biology. International Journal of Radiation Biology 75, 787–799. [DOI] [PubMed] [Google Scholar]
- 132.Li L et al. (2016) High-throughput imaging: focusing in on drug discovery in 3D. Methods (San Diego, Calif.) 96, 97–102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Gomes A et al. (2016) Oxygen partial pressure Is a rate-limiting parameter for cell proliferation in 3D spheroids grown in physioxic culture condition. PloS One 11, e0161239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Bull J et al. (2020) Mathematical modelling reveals cellular dynamics within tumour spheroids. PLoS Computational Biology 16, e1007961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Cristaldi D et al. (2020) A reliable flow-based method for the accurate measure of mass density, size and weight of live 3D tumor spheroids. Micromachines 11, 465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Al-Ramadan A et al. (2018) Analysis of radiation effects in two irradiated tumor spheroid models. Oncology Letters 15, 3008–3016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Onozato Y et al. (2017) Radiosensitivity of quiescent and proliferating cells grown as multicellular tumor spheroids. Cancer Science 108, 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Mondesert O et al. (2015) Monitoring the activation of the DNA damage response pathway in a 3D spheroid model. PloS One 10, e0134411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Martin ML et al. (2020) Organoids reveal that inherent radiosensitivity of small and large intestinal stem cells determines organ sensitivity. Cancer Research 80, 1219–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Das D et al. (2020) Human forebrain organoids from induced pluripotent stem cells: a novel approach to model repair of ionizing radiation-induced DNA damage in human neurons. Radiation Research 194, 191–198. [DOI] [PubMed] [Google Scholar]
- 141.Martinez S, et al. P (2021) Mouse parotid salivary gland organoids for the in vitro study of stem cell radiation response. Oral Diseases 27, 52–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Hacker B et al. (2019) Growth and characterization of irradiated organoids from mammary glands. Journal of Visualized Experiments : JoVE (147). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Sachs N et al. (2018) A living biobank of breast cancer organoids captures disease heterogeneity. Cell 172, 373–386. [DOI] [PubMed] [Google Scholar]
- 144.van de Wetering M et al. (2015) Prospective derivation of a living organoid biobank of colorectal cancer patients. Cell 161, 933–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Broutier L et al. (2017) Human primary liver cancer-derived organoid cultures for disease modeling and drug screening. Nature Medicine 23, 1424–1435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Hubert CG et al. (2016) A three-dimensional organoid culture system derived from human glioblastomas recapitulates the hypoxic gradients and cancer stem cell heterogeneity of tumors found In vivo. Cancer Research 76, 2465–2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Okkelman IA et al. (2017) Multi-parametric imaging of hypoxia and cell cycle in intestinal organoid culture. Advances in Experimental Medicine and Biology 1035, 85–103. [DOI] [PubMed] [Google Scholar]
- 148.Suckert T et al. (2020) Applying tissue slice culture in cancer research-insights from preclinical proton radiotherapy. Cancers 12, 1589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Mewes A et al. (2012) Organotypic brain slice cultures of adult transgenic P301S mice--a model for tauopathy studies. PloS One 7, e45017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Fan TWM et al. (2021) Innate immune activation by checkpoint inhibition in human patient-derived lung cancer tissues. eLife 10, e69578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151.Junk D et al. (2021) Human tissue cultures of lung cancer predict patient susceptibility to immune-checkpoint inhibition. Cell Death Discovery 7, 264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.@US_FDA (2019) Organs-On-Chips for Radiation Countermeasures FDA. @US_FDA, https://www.fda.gov/emergency-preparedness-and-response/mcm-regulatory-science/organs-chips-radiation-countermeasures. [Google Scholar]
- 153.Carr S et al. (2014) Analysis of radiation-induced cell death in head and neck squamous cell carcinoma and rat liver maintained in microfluidic devices. Otolaryngology – Head and Neck Surgery: Official Journal of American Academy of Otolaryngology-Head and Neck Surgery 150, 73–80. [DOI] [PubMed] [Google Scholar]
- 154.Cheah R et al. (2017) Measuring the response of human head and neck squamous cell carcinoma to irradiation in a microfluidic model allowing customized therapy. International Journal of Oncology 51, 1227–1238. [DOI] [PubMed] [Google Scholar]
- 155.Jalili-Firoozinezhad S et al. (2018) Modeling radiation injury-induced cell death and countermeasure drug responses in a human Gut-on-a-chip. Cell Death & Disease 9, 223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Tonon F et al. (2019) In vitro metabolic zonation through oxygen gradient on a chip. Scientific Reports 9, 13557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kang Y et al. (2020) Progressive hypoxia-on-a-chip: an in vitro oxygen gradient model for capturing the effects of hypoxia on primary hepatocytes in health and disease. Biotechnology and Bioengineering 117, 763–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Hu B et al. (2006) The time and spatial effects of bystander response in mammalian cells induced by low dose radiation. Carcinogenesis 27, 245–251. [DOI] [PubMed] [Google Scholar]
- 159.Lacombe J et al. (2020) Plant-Based scaffolds modify cellular response to drug and radiation exposure compared to standard cell culture models. Frontiers in Bioengineering and Biotechnology 8, 932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Sharma D et al. (2019) Upgrading prevascularization in tissue engineering: a review of strategies for promoting highly organized microvascular network formation. Acta Biomaterialia 95, 112–130. [DOI] [PubMed] [Google Scholar]
- 161.Erdem A et al. (2020) 3D Bioprinting of oxygenated cell-laden gelatin methacryloyl constructs. Advanced Healthcare Materials 9, e1901794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Griffin RJ et al. (2020) Understanding high-dose, ultra-high dose-rate and, spatially fractionated radiotherapy. International Journal of Radiation Oncology Biology Physics 107, 766–778. [DOI] [PubMed] [Google Scholar]
- 163.Zhang Y et al. (2021) Can rational combination of ultra-high dose rate FLASH radiotherapy with immunotherapy provide a novel approach to cancer treatment? Clin Oncol (R Coll Radiol) 33, 713–722. [DOI] [PubMed] [Google Scholar]