Abstract
Differential display-PCR (DDPCR) was used to identify a Streptococcus pneumoniae gene with enhanced transcription during growth in the murine peritoneal cavity. Northern dot blot analysis and comparative densitometry confirmed a 1.8-fold increase in expression of the encoded sequence following murine peritoneal culture (MPC) versus laboratory culture or control culture (CC). Sequencing and basic local alignment search tool analysis identified the DDPCR fragment as pstS, the phosphate-binding protein of a high-affinity phosphate uptake system. PCR amplification of the complete pstS gene followed by restriction analysis and sequencing suggests a high level of conservation between strains and serotypes. Quantitative immunodot blotting using antiserum to recombinant PstS (rPstS) demonstrated an approximately twofold increase in PstS production during MPC from that during CCs, a finding consistent with the low levels of phosphate observed in the peritoneum. Moreover, immunodot blot and Northern analysis demonstrated phosphate-dependent production of PstS in six of seven strains examined. These results identify pstS expression as responsive to the MPC environment and extracellular phosphate concentrations. Presently, it remains unclear if phosphate concentrations in vivo contribute to the regulation of pstS. Finally, polyclonal antiserum to rPstS did not inhibit growth of the pneumococcus in vitro, suggesting that antibodies do not block phosphate uptake; moreover, vaccination of mice with rPstS did not protect against intraperitoneal challenge as assessed by the 50% lethal dose.
Acquisition of inorganic phosphate (Pi) in Esherichia coli is principally carried out by Pst and Pit (34), two independent transport systems that are coregulated as members of the phosphate regulon (34). When Pi is in excess, the expression of the phosphate regulon is inhibited and phosphate uptake is primarily the result of the low-affinity transporter Pit (30). Conversely, under phosphate limitation, most phosphate regulon genes are upregulated (34), including the high-affinity Pi-specific transporter Pst (3, 34). The Pst transporter complex is composed of five proteins whose genes, pstSCAB and phoU (7), are collectively transcribed. pstS, the first gene transcribed from the pst operon, encodes a phosphate-binding protein belonging to the family of ATP-binding cassette (ABC) transporters. pstC and pstA encode transmembrane proteins, while pstB encodes an ATP-binding protein. Mutations in any of these genes abolishes Pi uptake (7, 27, 33). Finally, phoU, while not required for phosphate transport, is responsible for repression of the phosphate regulon (12, 28).
Recently, in Streptococcus pneumoniae, a locus with homology to the Pst system was identified (18). Examination of the locus revealed five genes with characteristics similar to those previously described for E. coli. The pneumococcal Pst system includes a phosphate-binding protein (pstS), two transmembrane proteins (pstC and pstA), and an ATP-binding protein (pstB), all of which are putatively cotranscribed. Immediately downstream and under the control of a presumed independent promoter is a fifth gene homologous to the phoU gene of E. coli. Mutational analysis of the pneumococcal pst locus revealed that, as in E. coli (12, 28), mutagenesis of the ABC gene pstB resulted in decreased rates of phosphate uptake, decreased growth rates (18), and reduced pathogenicity in a septicemia model of infection (20). Moreover, mutagenesis of pstB resulted in decreased levels of transformation and resistance to penicillin-induced lysis (18).
In this report, we examine the gene expression of pst and the protective efficacy of vaccination with purified recombinant PstS (rPstS). Having identified a peritoneally enhanced differential display-PCR (DDPCR) product, we confirm enhanced pstS transcription and PstS production during murine peritoneal culture (MPC) (19). Further, we demonstrate that pst transcription and Pst production are increased in response to decreasing levels of Pi. Finally, we determine that, while pstS is conserved among multiple pneumococcal isolates, vaccination of mice with rPstS is not protective in a septicemia model and that polyclonal antiserum does not inhibit pneumococcal growth in vitro.
MATERIALS AND METHODS
Bacterial strains and in vitro growth conditions.
Bacterial strains and plasmids used are listed in Table 1. Pneumococci were grown on tryptic soy agar plates supplemented with sheep blood to a final concentration of 5% (vol/vol) (Becton Dickinson Microbiology Systems, Cockeysville, Md.). For growth in liquid media, bacteria were grown in Todd-Hewitt broth supplemented with 5% yeast extract (wt/vol) (THY). For experiments using assorted concentrations of phosphate, the bacteria were grown in casein hydrosylate media (C+Y medium) supplemented by 1.0, 3.0, 10, and 30 mM Pi (pH 8.0). C+Y media were prepared as indicated by Lacks et al. (13) with the exception of sodium phosphate. Phosphate concentration in the basal media was determined using a Vitros 950 System (Johnson & Johnson Clinical Diagnostics, Inc., Rochester, N.Y.) and a Vitros Phos Slide (Johnson & Johnson) available through the Department of Clinical Chemistry at the John Sealy Hospital, University of Texas Medical Branch at Galveston. Once determined, phosphate levels were then brought to the appropriate concentration (1.0, 3.0, 10, and 30 mM Pi) with the addition of sodium phosphate, the buffer capacity of which was maintained by substituting Tris-HCl.
TABLE 1.
Listing of S. pneumoniae isolates and plasmids
| Strain or plasmid | Description | Reference or source |
|---|---|---|
| Strains | ||
| D39 | Serotype 2; NTC 7466 | 15 |
| R6 | Serotype 2; D39 derivative | 16 |
| R6x | Serotype 2; R6 derivative | 18 |
| WU2 | Serotype 3 | 6 |
| ATCC 6303 | Serotype 3; ATCC reference strain | 17 |
| Seattle | Serotype 3; clinical isolate | This study |
| DW4.1 | Serotype 4; clinical isolate | This study |
| DW11 | Serotype 11A; clinical isolate | This study |
| DW14.1 | Serotype 14; clinical isolate | This study |
| DW19 | Serotype 19F; clinical isolate | This study |
| Plasmids | ||
| pMOSBlue | Vector (AmpR) | Amersham |
| pBad/Thio-Topo | Vector (AmpR) | Invitrogen |
| pWU2PSTS | pBad/Thio-Topo derivative carrying pstS from WU2 | This study |
MPC model.
To obtain pneumococci grown intraperitoneally, the MPC model was used (19). Briefly, overnight bacterial cultures were diluted to 105 to 106 CFU/ml in RPMI 1640 (Mediatech, Inc., Herndon, Va.) supplemented with glucose to a concentration of 0.4% (RPMI+). Dialysis tubing (Spectrum, Houston, Tex.) with a 25-kDa molecular-mass cutoff was tied at one end, sterilized overnight with 0.1% sodium azide, and prior to use rinsed extensively with RPMI+. Aliquots (∼1.0 ml) of the bacterial suspension were added to the dialysis tubing, the dialysis tubing was sealed, and the surface of the bags was rinsed with sterile RPMI+. Swiss outbred mice weighing 25 to 30 g were anesthetized, and the dialysis bags were surgically implanted into the peritoneal cavity through a 1-cm abdominal incision. After insertions, incisions were closed using surgical staples. Unless stated otherwise, bags containing pneumococci were incubated for 8 h at which time the mice were sacrificed, pneumococci were collected, and the bacteria were processed in the appropriate experiments. Parallel control cultures (CCs) were grown in RPMI+ at 37°C in a candle extinction jar.
RNA isolation.
Pneumococcus from individual culture conditions was pelleted and subsequently lysed in 100 μl of a 1% sodium deoxycholate Tris-EDTA buffer (10 mM Tris and 1 mM EDTA, pH 8.0) solution. Total RNA was collected using a Qiagen RNeasy MiniKit (Qiagen, Valencia, Calif.) and frozen at −80°C. Only mRNA samples whose absorbance ratio (260 nm/280 nm) was greater than 1.8 were used. All samples were used within 1 week of collection. The quality of the RNA was assessed for each sample by examination of the RNA on a formaldehyde gel (data not shown).
DDPCR.
DDPCR was performed using a modified method of Welsh and McClelland (32) previously described by Robb et al. (23). Briefly, 1.0 μg of RNA from both MPC and CCs was DNase I treated and heat inactivated using protocols described by the manufacturer (Gibco-BRL, Gaithersburg, Md.). Aliquots of RNA both at 100 and 200 ng were reverse transcribed for each set of culture condition at 42°C using Mcloney murine leukemia virus reverse transcriptase (RT) in the presence of 1× Promega RT Buffer, 0.25 mM concentrations of deoxynucleoside triphosphates, 2.25 mM first-strand synthesis primer, and 10 U of RT (Promega, Madison, Wis.). Reactions were performed in duplicate and with different concentrations of RNA to control for variations between reactions. After use of RT, 5.0 mCi (800 Ci/mM) of [32P]dCTP was added, and the reaction conditions were adjusted to 4 mM MgCl2, 2 mM Tris-HCl, and 5 mM KCl. Taq polymerase (Perkin-Elmer, Foster City, Calif.) was added after an initial 1-min 92°C denaturing cycle. PCR was performed as follows: one low-stringency cycle (40°C, 5 min) followed by 30 high-stringency cycles (60°C, 0.5 min; and 72°C, 1 min). Primers were designed using a 17-mer design approach consistent with that reported by Zhao et al. (35). DDPCR products were separated on a 12% polyacrylamide gel containing 8.0 M urea for 2.5 h by using a Bio-Rad PowerPac 3000 power source at 60 W of maximum output. Gels were dried under vacuum and were exposed overnight to low-background X-ray film (β-max; Amersham, Arlington Heights, Ill.).
Cloning of DDPCR product.
DDPCR products representing putative MPC-enhanced genes were identified and localized by densitometric analysis of autoradiographs from dried DDPCR gels (Applied Imaging System Densitometer; Applied Imaging Systems, Santa Clara, Calif.). DNA fragments from bands with enhanced expression under both MPC RNA concentrations (100 and 200 ng) were localized by overlaying the autoradiograph and were excised using a clean scalpel blade. Products were eluted in water and reamplified by PCR using the primer specific to each DDPCR. Amplified products were cloned using a TA cloning vector (pMOSBlue; Amersham, Little Chalfont, Buckinghamshire, England), according to the manufacturer's protocol.
Confirmation by Northern dot blot.
RNA dot blotting was performed using RNA derived from MPC- and CC-grown pneumococci. RNA from each set of conditions was serially diluted and blotted onto nylon membranes using a dot blot apparatus and vacuum manifold. RNA was fixed to the nylon membranes by soaking in 0.05 M NaOH for 20 min followed by rinsing with 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) (4). The membranes were prehybridized with Rapid-hyb buffer (Amersham Pharmacia Biotech Inc., Piscataway, N.J.) for a minimum of 1 h and were hybridized under stringent conditions using the standard protocol (24). Probes were obtained by PCR amplification of pstS from a recombinant plasmid containing the full-length gene and were labeled using Ready-To-Go DNA Labeling Beads (-dCTP) (Amersham). To confirm equal loading of RNA, parallel duplicate membranes were hybridized against a probe to the S. pneumoniae DNA gyrase A subunit. Previously, it has been shown that DNA gyrase A is constitutively expressed (10, 11). Analysis of DNA gyrase A transcription is an appropriate control for determination of the equalized RNA load. Northern dot blot analysis of pstS expression was performed in triplicate, with signal intensity determined by comparative densitometry of autoradiographs using a Gel-Doc 2000 (Gel-Doc 2000; Bio-Rad, Hercules, Calif.). Signal intensity was determined for spots in the linear detection range. Statistical significance was determined using a t test to see if the mean ratio exceeds 1, 1 being the no-effect hypothesis.
Sequence analysis of DDPCR products.
Plasmids containing putative enhanced gene products were subjected to automated sequencing available at the World Health Organization Centers for Tropical Disease Core Laboratory, University of Texas Medical Branch at Galveston, Galveston. DNA sequences were analyzed for homology in the The Institute for Genomic Research (TIGR) S. pneumoniae database (http://www.tigr.org) using the Basic Local Alignment Search Tool (BLASTN) (1). Once the corresponding open reading frame was identified, BLASTN and BLASTX (2) analysis available at GenBank, the National Center for Biotechnology server (http://www.ncbi.nlm.nih.gov/), was used to identify the fragment or determine the closest homologue of the gene.
PCR amplification of pstS, restriction analysis, cloning, and sequencing of pstS.
Whole-length pstS with exception of the terminal stop codon was PCR amplified from the 10 S. pneumoniae strains listed in Table 1. The primers used were designed from sequence data available through GenBank (accession number AF118229) and are as follows: upstream, 5′-ATGAAATTCAAAAAAATGCTTACTCTTGC; and downstream, 5′-TCTTGTCCCAGGTGGTTAATTTTGCC. Restriction endonuclease analysis was done using EcoRI, HincII, HindIII, and PvuII (Promega). Once confirmed, pstS from WU2 was cloned into the TA cloning vector pBad/Thio-Topo (Invitrogen, Carlsbad, Calif.). Transformants were selected in which the recombinant plasmid had an insert of the appropriate characteristics (i.e., size and restriction sites) and orientation. Recombinant plasmid (pWU2PSTS) was purified using a S.N.A.P. MiniPrep kit (Invitrogen) in preparation for double-stranded, automated sequencing. Sequencing was completed using automated sequencing available at the Centers for Tropical Disease Core Laboratory (GenBank accession number AY039745).
Expression, purification of PstS, and development of PstS antiserum.
Ligation of pstS into pBad/Thio-Topo created a thioredoxin::PstS::histidine tag fusion construct (rPstS) regulated by an l-arabinose-inducible promoter. Briefly, 100 ml of Luria broth was inoculated with E. coli containing pWU2PSTS and was incubated for 4 h at 37°C. l-Arabinose (Sigma, St. Louis, Mo.) was added to a final concentration of 0.2%, and the bacteria were incubated for an additional 4 h. After incubation bacteria were pelleted and the rPstS was collected using the Express Purification Kit (Invitrogen). Purification of the recombinant protein was performed as indicated by the manufacturer and was confirmed by Coomassie blue staining of a sodium dodecyl sulfate–10% polyacrylamide gel electrophoresis (SDS-PAGE) gel loaded with the recombinant protein (data not shown). Antiserum to rPstS was developed by intraperitoneal injection of mice with 1 μg of rPstS in 100 μl of Freund's complete adjuvant, followed by two subsequent injections of at 3 and 6 weeks in Freund's incomplete adjuvant. Serum was collected from the mice by retro-orbital bleeding prior to immunization and 3 weeks after the third immunization. Following collection, the serum was pooled and tested by Western blot analysis to insure specificity of the antiserum to rPstS and native PstS.
Western blot analysis.
Western blotting was performed as indicated earlier (4), with antiserum to rPstS used at a dilution of 1:1,000, and secondary antibody was used at a dilution of 1:3,500.
Quantitative immunodot blot analysis.
WU2 isolated from MPCs and CCs was pelleted and suspended in 500 μl of phosphate-buffered saline (PBS) (Sigma). Bacteria were disrupted by bead beating using 0.1-mm-diameter zirconia/glass beads in a Mini-Beadbeater (Bio-spec, Bartlesville, Okla.) and centrifuged to remove cell wall debris. The protein concentration of the supernatant was determined by bicinchoninic acid (26) (BCA-200 Protein Assay Kit; Pierce, Rockford, Ill.) with samples of whole-cell lysate (WCL) subsequently diluted to 50 μg/ml in transfer buffer (25 mM Tris, 192 mM glycine, and 20% [vol/vol] methanol, pH 8.3). Samples were blotted onto nitrocellulose in duplicate twofold serial dilutions using a 96-well dot blot apparatus. Additionally, purified rPstS was also serially blotted with an initial concentration of 100 ng/ml. Membranes were blocked in PBS supplemented with 0.1% Tween 20 and 5% nonfat dry milk for 1 h, with development of the membranes following standard immunodot blot protocol (4): namely, rPstS murine antiserum (1:1,000) in PBS and horseradish peroxidase-conjugated antibody (1:7,500). Blots were developed using the enhanced chemoluminescence system (Amersham). PstS production under different concentrations of phosphate was then quantitated in terms of nanograms per microgram of WCL with the purified rPstS serving as the known standard. Protein concentration was determined using bicinchoninic acid (BCA-200 Protein Assay Kit, Pierce). To further ensure equivalent loading of protein for immunodot blot analysis, protein were visualized on a Coomassie-stained SDS-PAGE gel for each sample (data not shown). Statistical analysis was performed using Student's t test (two samples, assuming equal variances).
Determination of phosphate concentration within peritoneal cavity.
To determine phosphate levels within the MPCs and CCs, dialysis bags loaded with RPMI+ were surgically implanted into the peritoneal cavity of mice or alternatively submerged in RPMI+. After 8 h, bags were collected and supernatant was analyzed. Phosphate levels were determined using a Vitros 950 System and the Vitros Phos Slide (Johnson & Johnson) in the Clinical Chemistry laboratory of John Sealy Hospital. Six samples were collected for each set of culture conditions, and statistical analysis was performed using Student's t test (paired two samples for means).
Immunodot blot analysis of PstS production versus phosphate concentration.
PstS production in various concentrations of phosphate was determined using quantitative immunodot blotting. Exponential-phase S. pneumoniae strains D39, R6, R6x, WU2, ATCC 6303, DW4.1, and DW14.1 grown in Tedd-Hewitt broth supplemented with 5% yeast extract were washed and diluted 1:10 in C+Y media supplemented by 1.0, 3.0, 10, and 30 imM Pi. After incubation for 100 min at 37°C, the bacteria were pelleted and the supernatant was removed. PstS production was then determined as previously described, with equivalent loading of protein also assessed as previously described. Samples were collected in triplicate for each set of culture conditions, with statistical analysis performed using Student's t test (paired two samples for means.)
Northern blot analysis of pst transcription versus phosphate concentration.
Northern dot blot analysis of the pst locus in R6x and WU2 was performed using radiolabeled pstS. RNA was collected from the bacteria grown in C+Y media supplemented by 3.0, 10, and 30 mM Pi for 100 min. Hybridization was performed according to standard protocol (24). The equalization of RNA load was determined as previously described, with exception of the probe. A probe to 23S rRNA was used. Previously we have shown that levels of 23S rRNA are expressed in equivalent amounts during MPC and CC (data not shown). Experiments were performed in duplicate.
Pneumococcal growth in presence of rPstS antibodies.
WU2 was grown in C+Y media supplemented by 3.0, 10, and 30 mM Pi in the presence of rPstS antiserum at dilutions of 1:100, 1:500, and 1:1,000. Growth rates were determined by serial dilution and subsequent CFU counting at 2-h intervals for 8 h.
LD50 assays.
The 50% lethal dose (LD50) was calculated by the statistical approach of Reed and Muench (22). Briefly, two groups of 40 mice representing four cohorts of 10 were challenged intraperitoneally with pneumococci suspended in RPMI medium at concentrations of 101 to 104 CFU/ml. Mice were observed for 7 days. The experiment was done in parallel with one group vaccinated with rPstS, while the second was vaccinated with rOma90. Briefly, rOma90 is a Shigella flexneri outer membrane protein expressed from the same vector as rPstS (23). rOma90 served as a control for antibodies that may develop to the thioredoxin and histidine motifs present on rPstS. Western blot analysis of murine preimmune-phase serum and postvaccination antiserum verified development of antibodies (data not shown).
RESULTS
In an attempt to identify S. pneumoniae genes expressed during intraperitoneal growth, RNA was collected from WU2 after MPC and CC and was analyzed by DDPCR. Examination of the DDPCR autoradiographs identified several DDPCR products present at higher levels during MPC than during CC. Conversely, products were also identified at higher levels during CC than during MPC. Products present at higher levels during MPC were amplified using PCR, cloned into pMOSBlue, and subjected to automated sequencing. Nucleotide sequences were searched against the TIGR S. pneumoniae database using BLASTN analysis that located the fragment within a defined contiguous sequence and identified the complete gene sequence. Subsequently, the full-length gene was then searched against GenBank using BLASTN and BLASTX analyses. Of the multiple products identified, DDPCR product P6-1 was identified as pstS, the S. pneumoniae phosphate-binding protein and first gene of the pst operon. Moreover, because previous reports have indicated that pst plays an important role in vivo (12, 18, 20, 28), we decided to examine DDPCR product P6-1 further.
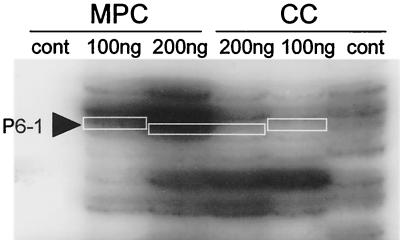
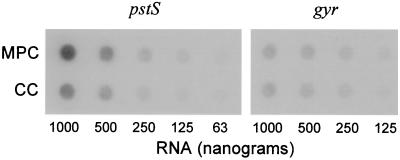
Comparative densitometry of the P6-1 autoradiograph revealed a 3.2-fold increase in signal strength during MPC when compared to CC (Fig. 1). To verify that pstS was in fact MPC enhanced, total RNA from MPC and CC was compared by RNA dot blot analysis using radiolabeled full-length pstS as a probe. Densitometric analysis of three Northern dot blots revealed 1.9-, 1.6-, and 2.0-fold increases in pstS transcription using equivalents amounts of RNA for analysis (Fig. 2). This difference (∼1.8-fold) was determined to be statistically significant (P = 1.0 × 10−4) using a t test to see if the mean ratio of MPC enhanced transcription exceeded 1, 1 being the no-effect hypothesis. PCR amplification of full-length pstS from 10 pneumococcal isolates representing seven serotypes amplified a single band at approximately 875 bp for each isolate (data not shown). Diagnostic restriction analysis of these fragments demonstrated a conserved restriction map regardless of isolate or serotype and was consistent with the predicted pstS restriction map of P394, the serotype 4 clinical isolate in the TIGR database and GenBank (data not shown). Sequencing of the cloned WU2 PCR fragment revealed that 870 out of 873 (99.6%) nucleotides were conserved between WU2 and P394 (data not shown). The 3-nucleotide differences in the sequence do not result in alterations in the predicted amino acid sequence. Southern blot analysis of pstS using radiolabeled full-length pstS as a probe identified only a single copy of the gene within the WU2 chromosome (data not shown). Presently, the WU2 pstS sequence is available through GenBank accession number AY039745.
FIG. 1.
Sectional autoradiograph of S. pneumoniae DDPCR P6 indicating differentially amplified (3.2-fold) DDPCR product P6-1. Reactions using total RNA from both MPC and CC were done in duplicate with 100 and 200 ng of total RNA. As a negative control, reactions without RT are shown in the designated lanes (cont).
FIG. 2.
RNA dot blot analysis of DDPCR product P6-1. Total RNA was isolated from S. pneumoniae cultured in MPCs and CCs and spotted in parallel onto duplicate nylon membranes in the indicated amounts. Membranes were hybridized with either radiolabeled pstS or a probe to pneumococcal DNA gyrase subunit A. Hybridization of DNA gyrase subunit A served as a control to ensure equal loading of RNA. These experiments were performed in triplicate. pstS in MPC and in CC demonstrated a 1.8-fold difference.
To express and purify the pstS gene product, the full-length pstS gene sequence was amplified from WU2 and cloned into pBad/Thio-Topo (Invitrogen). Ligation into pBad/Thio-Topo created a thioredoxin::PstS::histidine tag fusion construct under the regulation of an l-arabinose-inducible promoter. Pilot studies found optimal protein production at a 0.2% concentration of arabinose and confirmed the size of the recombinant PstS (rPStS) protein at 49 kDa (data not shown). Purification of rPstS was performed on an NiCl2 column following the manufacturer's directions and resulted in the isolation of 49- and 37-kDa bands on an SDS-PAGE gel (data not shown). The 37-kDa band was later ascertained by Western blotting to most likely be a degradation product of rPstS (see below).
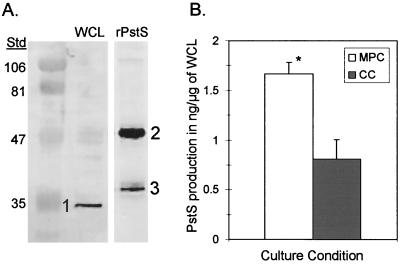
Polyclonal antiserum to rPstS was generated in mice using 49-kDa rPstS purified from E. coli harboring pWU2PSTS. Sera collected from these mice contained antibody to rPstS, as was demonstrated by the 1:1,000 dilution of antiserum utilized for subsequent Western blot analyses. Specificity of the antiserum to native PstS was determined by Western blot analysis using polyclonal serum from the immunized mice and WCL from S. pneumoniae. Antiserum to rPstS identified a single 33-kDa band present in the WCL lane at the native size of PstS (Fig. 3A), which demonstrated the specificity of the antiserum. Antiserum to rPstS also identified the 49- and 37-kDa proteins present in the purified rPstS lane (Fig. 3A). This indicates that the 37-kDa band is most likely a degradation product. Preimmune-phase sera did not identify any bands in the MPC and CC and the purified rPstS lanes (data not shown).
FIG. 3.
(A) A Western blot analysis of pneumococcal WCL demonstrating specificity of the rPstS antiserum to native PstS (33 kDa) (1) and rPstS (49 kDa) (2). The 37-kDa protein (3) is believed to be a product of degradation. Kilodaltons are indicated on left. (B) PstS production during MPCs and CCs as determined by quantitative immunodot blot analyses. Note the 2.2-fold difference between PstS production during MPCs (n = 4) and that during CCs (n = 6). This difference was determined to be statistically significant using a Student's t test (two samples, assuming equal variances) (P = 3.0 × 10−3).
To quantitate PstS production during MPC and CC, quantitative immunodot blot analysis was performed using WCL from WU2 cultured under both MPC and CC conditions. Analysis of PstS production demonstrated that PstS production during MPCs was approximately 1.67 ng per μg of WCL. PstS production during CCs was determined to be 0.81 ng per μg of WCL (Fig. 3B). PstS production overall was 2.1-fold greater during MPC (n = 4) than during CC (n = 6); this difference was statistically significant using Student's t test (two samples; assuming equal variances) (P = 3.0 × 10−3). Quantitative analysis of Pi determined phosphate levels in the murine peritoneum to be half of those present in CC. Specifically, phosphate levels within MPC (n = 6) were determined to be 2 mM Pi, whereas those of CCs (n = 6) were found to be 4 mM Pi. These results indicate that pstS expression and protein production are phosphate responsive. This difference in phosphate levels was found to be statistically significant using Student's t test (paired two samples for means) (P = 8.0 × 10−4).
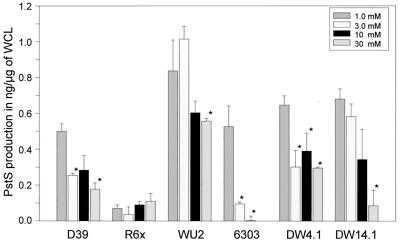
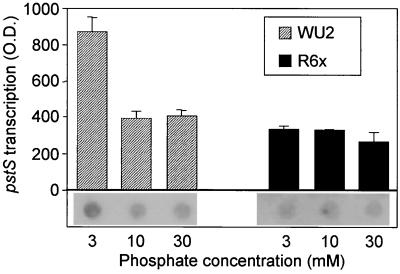
To determine if the Pi concentration affected production of PstS, quantitative immunodot blot analysis was performed using WCL from six pneumococcal isolates: D39, R6x, WU2, ATCC 6303, DW4.1, and DW14.1, grown in media containing 1.0, 3.0, 10, or 30 mM Pi (Fig. 4). Of the six isolates, five demonstrated statistically significant increases in PstS production during growth in 1.0 mM Pi versus that in 30 mM Pi. Additionally D39, ATCC 6303, and DW4.1 showed statistically significant increases in PstS production at 3.0 and 10 mM Pi from that at 1.0 mM Pi. Of the six isolates, R6x was the only strain that did not demonstrate a significant increase in PstS production in response to changes in phosphate concentration. To ascertain if this lack of response was strain specific, the R6x parental strain R6 was tested and was found to have statistically significant, phosphate-dependent production of PstS in response to growth at 3.0 mM versus 30 mM Pi, with 1.0 mM Pi not tested (data not shown). Likewise, transcriptional analyses of R6x using Northern dot blot analyses determined that pstS transcription in R6x did not respond to Pi concentrations, whereas strain WU2, the control, did respond with a 2.1-fold increase in pstS RNA levels at 3.0 mM Pi from those at 30 mM Pi (Fig. 5), consistent with the 1.8-fold increase in PstS production observed by immunodot blot analysis.
FIG. 4.
PstS production after 100 min of growth in 1, 3, 10, and 30 mM Pi as determined by quantitative immunodot blot analysis. Columns designated with asterisk indicate a statistically significant difference between the samples and PstS production during growth in 1.0 mM Pi (D39, 3 mM [P = 0.019] and 30 mM [P = 0.014]; WU2, 30 mM [P = 0.049]; ATCC 6303, 3 mM [P = 0.003] and 10 mM [P = 0.002]; DW4.1, 3 mM [P = 0.005], 10 mM [P = 0.017] and 30 mM [P = 0.0002]; and DW 14.1, 30 mM [P = 0.004]). Statistical analysis was performed using Student's t test (paired two samples for means).
FIG. 5.
Northern dot blot analysis examining pstS transcription of WU2 and R6x during growth in various phosphate concentrations. O.D., optical density.
Western blot analyses using pooled human convalescent-phase sera failed to identify rPstS (data not shown). In order to determine if antibodies to rPstS inhibit the ability of the pneumococcus to grow in vitro, WU2 was grown at various phosphate concentrations in the presence of rPstS antisera at dilutions of 1:100, 1:500, and 1:1,000. Growth was not inhibited by the presence of antiserum. In contrast, cultures exposed to high levels of antiserum (dilutions of 1:100 and 1:500) grew better than those at low antiserum concentrations (1:1,000), possibly indicating that components in the serum stimulated bacterial growth in a dose-dependent manner (data not shown). Phosphate levels did, however, affect growth rates, with WU2 growth rates peaking at 30 and 10 mM Pi versus that at 3.0 mM Pi (data not shown), consistent with the findings reported by Novak et al. (18). Along similar lines, LD50 determinations examining protection in mice vaccinated with rPstS did not demonstrate a significant difference either. The LD50 in mice challenged with pneumococci intraperitoneally was determined to be 2.0 × 103 for rPstS-vaccinated mice, whereas in controls it was found to be 2.5 × 103. This is despite control studies demonstrating that vaccinated mice developed antibodies to rPstS (data not shown).
DISCUSSION
Environmental signals present in vivo such as temperature, nutrient availability, pH, and osmolarity have all been shown to influence bacterial virulence gene expression (9). Previously, we have characterized pneumococcal gene expression and protein production during MPC (19). We have demonstrated changes in two-dimensional protein profiles, adhesive capacity, and virulence-associated gene expression. In this report, we focus on the characterization of DDPCR product P6-1, an MPC-enhanced product that encodes pstS, the phosphate-binding protein of the pneumococcal pst phosphate transport system. We demonstrate that pstS is highly conserved among S. pneumoniae isolates, that both transcription of pstS and production of PstS are enhanced in the murine peritoneum, and that PstS production is responsive to environmental levels of Pi.
PstS encodes a phosphate-binding protein/ABC transporter with a high level of homology to the same gene in Methanobacterium autotrophicum and E. coli. Previous studies with E. coli have shown that the pst locus is a member of the phosphate regulon and plays an important role in the acquisition of phosphate when levels of environmental Pi are low (31). Recently, studies in S. pneumoniae in vitro have demonstrated that mutagenesis of the pneumococcal pst locus resulted in decreased uptake of Pi, reduced rates of growth, resistance to penicillin-induced lysis, and an inability of the bacteria to undergo transformation (18). In a separate study, Polissi et al. identified what at the time was an unknown pneumococcal gene homologous to pstB in Methanococcus jannaschii. This gene was determined to be critical for the survival of S. pneumoniae in a septicemia model of infection (20). Our findings of both increased pstS transcription and increased PstS production in response to growth in the murine peritoneum support the notion that phosphate acquisition is necessary for survival in vivo. Because pstS is the first gene transcribed from the pst operon, increased pstS transcription and PstS production may be interpreted as indicators of transcription of the operon and production of the Pst transport complex. Overall, our findings suggest that the pneumococcus adjusts to the low concentration of phosphate (2 mM Pi) present in the peritoneum by increasing the production of this phosphate transporter.
Previously, Novak et al. (18) attempted to examine S. pneumoniae pst transcription in response to Pi levels. These studies using strain R6x determined that pst expression did not respond to decreasing levels of Pi, an unexpected result in light of other reports indicating that pst expression is phosphate regulated in other bacteria (21, 34). Our analysis of Pst production in seven isolates determined a statistically significant increase in PstS production in six strains in response to decreasing levels of Pi. Analysis of pstS transcription in R6x, the nonresponding isolate, established that this lack of response occurred at the transcriptional level. What is more, analyses of D39 and R6, the R6x parental strains, determined that this lack of response is strain specific and that extracellular levels of Pi regulate most isolates.
Restriction analyses and limited sequence analyses of pstS from 10 isolates of S. pneumoniae have shown that pstS is conserved between isolates at both the nucleotide and predicted amino acid level. Conservation of pstS may indicate the metabolic importance of the protein product. Similarly, the conserved nature of the protein and operon among different bacteria may point toward a conserved evolutionary lineage. Conservation of pst may indirectly point toward the existence of a Streptococcus spp. phosphate regulon, an idea which is supported by the failure of mutagenesis of the locus to completely ablate phosphate uptake (indicating the existence of a second transporter) (18) and by the existence of a phosphate regulon in other pst-conserved bacteria, including the gram-positive Bacillus subtilis (5, 14, 21, 25).
Generally, it is believed that identification and characterization of in vivo-expressed genes will provide insight into the mechanisms that underlie bacterial pathogenesis. Furthermore, because in vivo-produced factors are those likely necessary for bacterial adaptation, survival, and disease progression, it is believed that in vivo-expressed genes are potential targets for antimicrobials, vaccines, and other pharmacological agents (9). Prior to this report, two studies have examined the importance of the Pst transporter for bacterial survival in an infection model. As previously discussed, Polissi et al. demonstrated that mutagenesis of pstB in S. pneumoniae resulted in an attenuated mutant incapable of surviving in a septicemia model of infection (20). Daigle et al. demonstrated that mutagenesis of pstC in E. coli resulted in a serum-sensitive mutant incapable of systemic survival after an intragastric challenge (8). Along this line, studies with Mycobacterium tuberculosis have determined that vaccination of mice with a DNA vaccine encoding the Mycobacterium pstS gene offered protection after intravenous challenge with M. tuberculosis (29).
Because of the requirement for Pst in vivo and the potential for a PstS vaccine, we elected to examine if antibodies to the phosphate-binding protein PstS inhibited bacterial growth in vitro and, more important, offered protection against pneumococcal challenge. Unfortunately, this was not the case, as rPstS antiserum failed to affect the growth rate of WU2. Additionally, mice vaccinated with rPst and controls did not demonstrate a difference in the LD50. These results suggest that either antibody to rPstS does not block Pi uptake or that PstS is inaccessible to rPstS antibodies; the latter hypothesis is supported in part by the conserved nature of the protein, indicating that PstS is not under antigenic pressure. Presently, it remains unknown if vaccination with PstS is efficacious in an intranasal challenge model.
In summary, DDPCR identified an S. pneumoniae gene with enhanced transcription during MPC. Analysis of the gene identified it as pstS, the first gene transcribed from the pst locus. Characterization of pstS by Northern and Western dot blot analysis confirmed that transcription of pstS and production of its gene product were enhanced during MPC and during growth in low concentrations of Pi. While the specific PstS role in virulence has not been specifically determined, previous studies examining mutagenesis of other components in pst indicate that the complete operon is required for phosphate uptake and that mutagenesis of pst is pleiotropic, having multiple effects at multiple levels (12, 18, 20, 28). Unfortunately, our studies indicate that antibodies to rPstS do not appear to be protective, nor do they appear to block phosphate uptake in vitro.
Finally, because pstS is the first gene transcribed from pst, enhanced expression of pstS may indicate that other phosphate-dependent genes may also be enhanced in vivo. It is, however, important that physiological factors other than phosphate may be present within the MPC model and may be capable of inducing pstS. Future studies will be necessary to determine this.
ACKNOWLEDGMENTS
Graduate Research Fellowship PGE 9818286 from the National Science Foundation, grant 98-HEDS02-291 from NASA, and a grant from the Texas Advanced Research Program (004952-0090) supported this work.
We thank Elbert Whorton for assistance with the statistical analysis.
REFERENCES
- 1.Altschul S F, Gish W, Miller W, Myers E W, Lipman D J. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 2.Altschul S F, Madden T L, Schaffer A A, Zhang J, Zhang Z, Miller W, Lipman D J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ames G F. Bacterial periplasmic transport systems: structure, mechanism, and evolution. Annu Rev Biochem. 1986;55:397–425. doi: 10.1146/annurev.bi.55.070186.002145. [DOI] [PubMed] [Google Scholar]
- 4.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1999. [Google Scholar]
- 5.Birkey S M, Liu W, Zhang X, Duggan M F, Hulett F M. Pho signal transduction network reveals direct transcriptional regulation of one two-component system by another two-component regulator: Bacillus subtilis PhoP directly regulates production of ResD. Mol Microbiol. 1998;30:943–953. doi: 10.1046/j.1365-2958.1998.01122.x. [DOI] [PubMed] [Google Scholar]
- 6.Briles D E, Nahm M H, Schroer K, Davie J, Baker P, Kearney J, Barletta R. Antiphosphorylcholine antibodies found in normal mouse serum are protective against intravenous infection with type 3 Streptococcus pneumoniae. J Exp Med. 1981;153:694–705. doi: 10.1084/jem.153.3.694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cox G B, Rosenberg H, Downie J A, Silver S. Genetic analysis of mutants affected in the Pst inorganic phosphate transport system. J Bacteriol. 1981;148:1–9. doi: 10.1128/jb.148.1.1-9.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Daigle F, Fairbrother J M, Harel J. Identification of a mutation in the pst-phoU operon that reduces pathogenecity of an Escherichia coli strain causing septicemia in pigs. Infect Immun. 1995;63:4924–4927. doi: 10.1128/iai.63.12.4924-4927.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finlay B B, Falkow S. Common themes in microbial pathogenicity revisited. Microbiol Mol Biol Rev. 1997;61:136–169. doi: 10.1128/mmbr.61.2.136-169.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Goerke C, Bayer M G, Wolz C. Quantification of bacterial transcripts during infection using competitive reverse transcription-PCR (RT-PCR) and LightCycler RT-PCR. Clin Diagn Lab Immunol. 2001;8:279–282. doi: 10.1128/CDLI.8.2.279-282.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goerke C, Campana S, Bayer M G, Doring G, Botzenhart K, Wolz C. Direct quantitative transcript analysis of the agr regulon of Staphylococcus aureus during human infection in comparison to the expression profile in vitro. Infect Immun. 2000;68:1304–1311. doi: 10.1128/iai.68.3.1304-1311.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haldimann A, Daniels L L, Wanner B L. Use of new methods for construction of tightly regulated arabinose and rhamnose promoter fusions in studies of the Escherichia coli phosphate regulon. J Bacteriol. 1998;180:1277–1286. doi: 10.1128/jb.180.5.1277-1286.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lacks S, Hotchkiss R D. A study of the genetic material determining an enzyme activity in pneumococcus. Biochim Biophys Acta. 1960;39:508–517. doi: 10.1016/0006-3002(60)90205-5. [DOI] [PubMed] [Google Scholar]
- 14.Liu W, Hulett F M. Bacillus subtilis PhoP binds to the phoB tandem promoter exclusively within the phosphate starvation-inducible promoter. J Bacteriol. 1997;179:6302–6310. doi: 10.1128/jb.179.20.6302-6310.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Magee A D, Yother J. Requirement for capsule in colonization by Streptococcus pneumoniae. Infect Immun. 2001;69:3755–3761. doi: 10.1128/IAI.69.6.3755-3761.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Michel U, Zobotke R, Mader M, Nau R. Regulation of matrix metalloproteinase expression in endothelial cells by heat-inactivated Streptococcus pneumoniae. Infect Immun. 2001;69:1914–1916. doi: 10.1128/IAI.69.3.1914-1916.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Neeleman C, Geelen S P, Aerts P C, Daha M R, Mollnes T E, Roord J J, Posthuma G, van Dijk H, Fleer A. Resistance to both complement activation and phagocytosis in type 3 pneumococci is mediated by the binding of complement regulatory protein factor H. Infect Immun. 1999;67:4517–4524. doi: 10.1128/iai.67.9.4517-4524.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Novak R, Cauwels A, Charpentier E, Tuomanen E. Identification of a Streptococcus pneumoniae gene locus encoding proteins of an ABC phosphate transporter and a two-component regulatory system. J Bacteriol. 1999;181:1126–1133. doi: 10.1128/jb.181.4.1126-1133.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Orihuela C J, Janssen R, Robb C W, Watson D A, Niesel D W. Peritoneal culture alters Streptococcus pneumoniae protein profiles and virulence properties. Infect Immun. 2000;68:6082–6086. doi: 10.1128/iai.68.10.6082-6086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect Immun. 1998;66:5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Qi Y, Kobayashi Y, Hulett F M. The pst operon of Bacillus subtilis has a phosphate-regulated promoted and is involved in phosphate transport but not in regulation of the pho regulon. J Bacteriol. 1997;179:2534–2539. doi: 10.1128/jb.179.8.2534-2539.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reed L J, Muench H. A simple method for estimating fifty percent end points. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 23.Robb C W, Orihuela C J, Ekkelenkamp M B, Niesel D W. Identification and characterization of an in vivo regulated D15/Oma87 homologue in Shigella flexneri using differential display polymerase chain reaction. Gene. 2001;262:169–177. doi: 10.1016/s0378-1119(00)00537-0. [DOI] [PubMed] [Google Scholar]
- 24.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 25.Shi L, Liu W, Hulett F M. Decay of activated Bacillus subtilis pho response regulator, PhoP∼P, involves the PhoR∼P intermediate. Biochemistry. 1999;38:10119–10125. doi: 10.1021/bi990658t. [DOI] [PubMed] [Google Scholar]
- 26.Smith P K, Krohn R I, Hermanson G T, Mallia A K, Gartner F H, Provenzano M D, Fujimoto E K, Goeke N M, Olson B J, Klenk D C. Measurement of protein using bicinchoninic acid. Anal Biochem. 1985;150:76–85. doi: 10.1016/0003-2697(85)90442-7. [DOI] [PubMed] [Google Scholar]
- 27.Sprague G F, Jr, Bell R M, Cronan J E., Jr A mutant of Escherichia coli auxotrophic for organic phosphates: evidence for two defects in inorganic phosphate transport. Mol Gen Genet. 1975;143:71–77. doi: 10.1007/BF00269422. [DOI] [PubMed] [Google Scholar]
- 28.Steed P M, Wanner B L. Use of the rep technique for allele replacement to construct mutants with deletions of the pstSCAB-phoU operon: evidence of a new role for the PhoU protein in the phosphate regulon. J Bacteriol. 1993;175:6797–6809. doi: 10.1128/jb.175.21.6797-6809.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tanghe A, Lefevre P, Denis O, D'Souza S, Brailbant M, Lozes E, Singh M, Montgomery D, Content J, Huygen K. Immunogenicity and protective efficacy of tuberculosis DNA vaccines encoding putative phosphate transport receptors. J Immunol. 1999;162:1113–1119. [PubMed] [Google Scholar]
- 30.van Veen H W, Abee T, Kortstee G J, Konings W N, Zehnder A J. Translocation of metal phosphate via the phosphate inorganic transport system of Escherichia coli. Biochemistry. 1994;33:1766–1770. doi: 10.1021/bi00173a020. [DOI] [PubMed] [Google Scholar]
- 31.Wanner B L. Gene regulation by phosphate in enteric bacteria. J Cell Biochem. 1993;51:47–54. doi: 10.1002/jcb.240510110. [DOI] [PubMed] [Google Scholar]
- 32.Welsh J, McClelland M. Fingerprinting genomes using PCR with arbitrary primers. Nucleic Acids Res. 1990;18:7213–7218. doi: 10.1093/nar/18.24.7213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Willsky G R, Bennett R L, Malamy M H. Inorganic phosphate transport in Escherichia coli: involvement of two genes which play a role in alkaline phosphatase regulation. J Bacteriol. 1973;113:529–539. doi: 10.1128/jb.113.2.529-539.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Willsky G R, Malamy M H. Characterization of two genetically separable inorganic phosphate transport systems in Escherichia coli. J Bacteriol. 1980;144:356–365. doi: 10.1128/jb.144.1.356-365.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao S, Ooi S L, Pardee A B. New primer strategy improves precision of differential display. BioTechniques. 1995;18:842–846. , 848, 850. [PubMed] [Google Scholar]