Abstract
Streptococcus suis is an economically important pathogen of pigs responsible for a variety of diseases including meningitis, septicemia, arthritis, and pneumonia, although little is known about the mechanisms of pathogenesis or virulence factors associated with this organism. Here, we report on the distribution and genetic diversity of the putative virulence factor suilysin, a member of the thiol-activated toxin family of gram-positive bacteria. On the basis of PCR analysis of over 300 isolates of S. suis, the suilysin-encoding gene, sly, was detected in 69.4% of isolates. However, sly was present in a considerably higher proportion of isolates obtained from cases of meningitis, septicemia, and arthritis (>80%) and isolates obtained from asymptomatic tonsillar carriage (>90%) than lung isolates associated with pneumonia (44%). With the exception of serotypes 1, 14, and 1/14, there was no strong correlation between the presence of suilysin and serotype. Analysis of the genetic diversity of suilysin by restriction fragment length polymorphism and sequence analysis found that the suilysin gene, where present, is highly conserved with a maximum of 1.79% diversity at the nucleotide level seen between sly alleles. Assays of hemolytic activity and hybridization analysis provided no evidence for a second member of the thiol-activated toxin family in S. suis. Inverse PCR was used to characterize regions flanking sly, which in turn allowed the first characterization of the equivalent region in a strain lacking sly. Sequence comparison of these regions from sly-positive (P1/7) and sly-negative (DH5) strains indicated that two alternative arrangements are both flanked by genes with highest similarity to haloacid dehalogenase-like hydrolases (5′ end) and putative N-acetylmannosamine-6-phosphate epimerases (3′ end). However, sly appears to be completely absent from the alternative arrangement, and a gene of unknown function is located in the equivalent position. Finally, PCR analysis of multiple sly-positive and -negative strains indicated that these two alternative genetic arrangements are conserved among many S. suis isolates.
Streptococcus suis is an important pathogen associated with a range of diseases in pigs including meningitis, pneumonia, septicemia, and arthritis, although the organism is also frequently carried asymptomatically. The organism has substantial implications for the swine industry in terms of both animal welfare concerns and economic impact. Occasionally, S. suis causes serious zoonotic infections in humans, where it has been associated with septicemia, meningitis, and endocarditis (4, 42). There are currently 35 serotypes of S. suis recognized based on capsular antigens (11–13, 19, 34). Although most clinical disease is associated with only a few of these (notably serotypes 1, 2, and 14), the importance of particular serotypes can vary both geographically and over time (17).
Currently, S. suis is controlled largely by the use of prophylactic and therapeutic antibiotics. However, an increase in isolation of antibiotic-resistant isolates over recent years and growing public concern over the use of prophylactic antimicrobials in farming indicates that alternative strategies are required to prevent a rise in incidence (1, 36, 46). The development of successful vaccines has been hampered by lack of knowledge of specific virulence factors contributing to the pathogenesis of S. suis disease, the large number of serotypes, and by apparent differences in virulence both within and between serotypes (42). Little is known about S. suis pathogenesis and potential virulence factors although the capsular polysaccharide has been demonstrated by insertional mutation to be a virulence determinant (6, 40). Despite this, most avirulent strains are encapsulated, indicating that other determinants are essential for virulence. Potential virulence determinants identified to date include a S. suis hemolysin (suilysin) and two proteins of unknown function, muramidase-released protein and extracellular protein factor (47), although recent studies have indicated that mutational deletion of these determinants does not abrogate virulence (39). It should be noted that while there is general agreement in the field that virulent and avirulent isolates of S. suis do exist, there is no clear consensus about how to define virulence in this organism, especially as animal infection models for S. suis can be misleading. Various studies have defined virulence based on the clinical condition of the animal from which the strain was derived, on the presence of certain virulence-associated proteins (although no definite role for these in virulence has been proven), or on various different animal infection models which often give contradictory results (see references 16 and 17 for discussion).
Suilysin is a secreted protein (21) and is a member of the thiol-activated, membrane-damaging toxin family, members of which are found in many gram-positive bacteria (14). Thiol-activated toxins have been implicated in the disease processes of many of these bacteria (5), although any role in S. suis pathogenesis remains largely undefined, and possess various activities in addition to their ability to lyse virtually all eukaryotic cells, which may be important in pathogenesis (see references 5 and 28 for reviews). There is evidence to indicate that suilysin may be a virulence determinant as it provides some protection to both mice and pigs against lethal challenge with a serotype 2 strain (21, 22), although a recent report has suggested that a defined mutant lacking suilysin in a porcine model of systemic infection was only marginally attenuated (2). Evidence to date suggests that the gene encoding suilysin, sly, is not present in all isolates of S. suis (33, 38), and a number of studies have correlated either the presence of the suilysin gene (44) or in vitro hemolytic activity expression (3, 20, 43, 45) with virulent isolates. Most previous studies of suilysin gene distribution have been limited to small numbers of isolates with substantial variation in the proportions of isolates reported to harbor suilysin (33, 38).
Virtually nothing is known about the genetic diversity of the suilysin-encoding gene. The sequences of sly from two strains, P1/7 (38) and 1933 (33), are currently available and reveal that the coding sequence is highly conserved with only four nucleotide differences, only one of which results in an amino acid alteration. Thus, while suilysin may contribute to virulence and be a potentially useful vaccine component, there is a clear need to understand more about the relationship of this protein to virulence, its genetic stability, and the extent of diversity of the gene. To address these issues, the aims of this study were as follows: first, to examine the presence of the suilysin-encoding gene, sly, in a large sample of field isolates and relate the presence of sly with the clinical background of the isolates and with other phenotypic characteristics such as serotype; second, to examine the nature and extent of genetic diversity of sly and any association of particular allelic variants with clinical background; and third, to attempt to determine the genetic basis of the apparent absence of sly from many isolates of S. suis.
MATERIALS AND METHODS
Isolates.
A total of 310 S. suis isolates were used in this study. Reference strains of 30 serotypes (serotypes 2 through 7, 9, 10, 12 through 19, and 21 through 34) were supplied by L. A. Devriese (Faculty of Veterinary Medicine, University of Ghent, Ghent, Belgium) and M. Gottschalk (Faculté de Médicine Vétérinaire, Université de Montréal, Montréal, Canada) (7). Twenty-two well-characterized serotype 2 isolates, including many isolates characterized in previous virulence studies, and 12 serotype 14 meningitis isolates previously screened for the presence of suilysin were supplied by P. Norton (Institute for Animal Health, Newbury, United Kingdom). A total of 138 field isolates obtained in the United Kingdom by the Veterinary Laboratories Agency, Bury St. Edmunds, United Kingdom, from diverse geographical sources were included in this study. These were selected to represent isolates from diverse serotypes, sites of isolation, and clinical background, including invasive disease isolates (meningitis, septicemia, and arthritis) and isolates from cases of pulmonary pneumonia. A similar sample of 87 field isolates obtained in Spain by C. Tarradas and I. Luque was included in this study. The sample consisted of 41 carriage isolates from the tonsils of healthy pigs and 46 clinical isolates from a variety of clinical backgrounds (25). In addition, eight Canadian serotype 2 isolates provided by M. Gottschalk were included, as was one pig isolate, previously described as atypical (24), provided by C. Lammler, Institut für Tierarztliche, Giessen, Germany.
Twelve isolates obtained from humans with S. suis disease were included—these isolates were obtained from Augustine Cheng, Department of Microbiology, Faculty of Medicine, The Chinese University of Hong Kong, Hong Kong (6 isolates); M. Gottschalk (3 isolates); C. Lammler (2 isolates); and B. François, Hospital Universitaire Dupuytren, Limoges, France (1 isolate [9]). Space limitations prevent the inclusion of a full isolate table herein; however, a full list of isolates, their clinical backgrounds, and the characteristics described in this paper is available on request.
Preparation of chromosomal DNA.
Chromosomal DNA was prepared from all isolates as described previously (50).
PCR analysis.
PCR was performed under standard conditions with 30 cycles of 95°C for 1 min, x°C for 1 min, and 72°C for 1 min per kilobase of predicted product size, where x°C represents an annealing temperature appropriate for the particular primer set used. Products were visualized by agarose gel electrophoresis on 1.0% agarose in the presence of ethidium bromide (1 μg ml−1). Details of all oligonucleotides used in this study are shown in Table 1.
TABLE 1.
PCR primers utilized in this study
| Primer | Sequence (5′ to 3′) | Binding site (bp) |
|---|---|---|
| 1 | CAGCTCGTTGCCTTGTACTA | 11–30a |
| 2 | ACTCTATCACCTCATCCGC | 1,811–1,829a |
| 3 | ATGAGAAAAAGTTCGCACTTG | 338–358a |
| 4 | ATGTCTATTCATTTCATTAG | 294–313a |
| 5 | AATTTGAATGGTTTCACTCCAGT | 1,641–1,663a |
| 6 | CAAGTGGTTCATCATGCTGCC | |
| 7 | GCGGATGAGGTGATAGAGTA | 3,603–3,622b |
| 8 | CTTCCTCAAAGGTTGAAATA | 3,060–3,079c |
| 9 | TAGAAGAAGAGCTGGAGG | 1–18c |
| 10 | GAATTATTGAACATGGGAAG | 691–717c |
| 11 | CCTGCTTCTTGGGCAGCCTT | 2,770–2,789c |
| 12 | TCCATACTCTAGAGACTGTT | 1,713–1,732c |
| 13 | ACACCACATCCAAGGCATTT | 2,140–2,159c |
| 14 | GGGATGTCAGGCGAAGATAT | 1,337–1,356c |
| 15 | TTCGCCTGGCAAAGCCTGAC | 2,708–2,727c |
Analysis of suilysin genetic diversity.
Approximately 5 μl of PCR product was digested by restriction enzymes in a total volume of 25 μl according to the manufacturer's instructions. Digests were then separated on 4 and 8% polyacrylamide gels and visualized under UV illumination, after being stained for 15 min with ethidium bromide (0.3 μg ml−1). Alleles were designated by visual comparison of restriction fragment length polymorphism (RFLP) profiles. A fraction of PCR products used in RFLP analysis were purified by passage through QiaQuick PCR product purification columns and directly sequenced using primers 1 to 3 and a series of internal primers. Sequencing was performed using the Beckman CEQ2000 system according to the manufacturer's instructions.
Analysis of the regions flanking the suilysin gene.
To extend the known sequence flanking suilysin, inverse PCR (IV-PCR) was performed using primers 4 and 5. Chromosomal DNA from strain P1/7 was restricted with XbaI and religated prior to IV-PCR. The resulting IV-PCR product was cloned into pGEM-T (Promega) and miniprep DNA (Qiagen) was sequenced to obtain the flanking sequence. Additional sequence downstream of suilysin was obtained by sequencing a PCR product, obtained using primers 6 and 7, which overlapped the IV-PCR product. Strain P1/7, originally isolated in the United Kingdom, is considered a virulent strain that has been widely used for the characterization of suilysin and for pathogenesis studies (21, 31, 38). Once the flanking region was obtained, this was used to design primers 8 and 9, allowing amplification and characterization of the corresponding region of a sly-negative isolate. Strain DH5, a well-studied isolate, was chosen for this purpose although it should be noted that there is controversy in the literature over the pathogenic potential of this strain, which highlights some of the problems in defining virulence in this species, as outlined in the introduction (16). Strain DH5 was originally isolated from the brain of a pig during an outbreak of meningitis (30), and while considered highly virulent by some authors (43) other authors (31, 32) have described it as avirulent on the basis of studies by Galina et al. (10).
Bioinformatics.
Primary sequence analysis was performed using the package DNASTAR. The similarity of sequences identified in this study to those already present in the databases was examined by BLAST analysis (http://www.ncbi.nlm.nih.gov/BLAST). Analysis of variable sites was performed using the MEGA software package, version 1.0 (23).
Dot blot analysis.
Dot blot analysis was performed using the Boehringer digoxigenin labeling system according to the manufacturer's instructions and using approximately 0.5 μg of target DNA. In brief, two sly probes were constructed by labeling fragments obtained by restricting a PCR product amplified from strain P1/7 with primers 2 and 3 (the entire coding region) with EcoRV. This restriction resulted in a 614-bp probe to the 5′ end of sly and an 879-bp probe to the 3′ end of the gene. The orfC probe was obtained from strain DH5 using primers 12 and 13. All probes were obtained by using DIG-High Prime (Boehringer) to label amplified fragments, and hybridization was allowed to proceed overnight at 42°C in EasyHyb solution (Boehringer). Posthybridization washes were performed at room temperature using 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate)–0.1% (wt/vol) sodium dodecyl sulfate, followed by 0.1× SSC–0.1% (wt/vol) sodium dodecyl sulfate.
Hemolytic assays.
Hemolytic assays were performed essentially as described previously (51). Assays were performed using serial doubling dilutions of the supernatant of cultures grown in brain heart infusion broth to an optical density at 600 nm of approximately 0.3. A positive result was recorded when the erythrocyte cell pellet could not be visualized in one or more supernatant dilutions. Negative results were recorded when the erythrocyte pellet in all wells was indistinguishable from that seen in the control wells (sterile brain heart infusion broth plus dithiothreitol buffer). Some isolates consistently gave a reduced erythrocyte pellet size, though not complete lysis, indicative of weak hemolytic activity. Representative isolates that gave a negative result were screened for activity throughout the growth cycle as described in Results.
Nucleotide sequence accession numbers.
Sequences of the 21 suilysin alleles described here have been submitted to GenBank and assigned the accession numbers AJ416287 through AJ416307. Sequences of the genomic regions flanking suilysin were assigned accession numbers AJ416309 and AJ416310 (sly positive) and AJ416308 (sly negative).
RESULTS
Investigation of the distribution of suilysin by PCR.
Chromosomal DNA was purified from 310 isolates of S. suis and screened for sly using primers 1 and 2. A PCR product of approximately the predicted 1,818 bp (on the basis of a published sequence) was amplified from 210 of 310 isolates screened. Primer 1 binds 307 to 327 bp upstream of the suilysin open reading frame. To detect any strains possessing the gene but divergent in this region, primer 3 was designed to correspond to the start of the open reading frame. Amplification with primers 2 and 3 resulted in a band of approximately 1,492 bp from all 210 previously identified sly-positive isolates and 5 additional isolates. These 5 strains included isolates from multiple countries, serotypes, and disease states. In total, therefore, 69.4% (215 of 310) of isolates screened gave a PCR product with at least one sly primer pair. Of the serotype type strains screened by PCR, a band of the predicted size was visualized for serotypes 2, 4, 5, 14, 15, 17, 18, 19, 23, and 28.
Assessment of allelic diversity of sly and upstream region by RFLP.
Allelic diversity of 208 of the 210 sly PCR products obtained by amplification with primers 1 and 2 was assessed by restricting each independently with six frequently cutting restriction enzymes. Restriction with HaeIII/DdeI, HinfI, MseI, MwoI, RsaI, and Tsp509I resulted in the identification of 19 distinct RFLP profiles, although the vast majority of isolates possessed profile 1. The frequency of these 19 profiles and their distribution with respect to serotype and country of origin are shown in Table 2. RFLP was also used to assess the diversity within the five PCR products obtained using primers 2 and 3; as a result, two additional profiles (20 and 21) were identified.
TABLE 2.
Distribution of suilysin alleles identified by RFLP of sly PCR products by country of origin and serotype
| RFLP profile | No. of isolates (%) | Country of isolation | Serotype(s) |
|---|---|---|---|
| 1 | 170 (79.8) | United Kingdom, Germany, Canada, Hong Kong, Spain, The Netherlands, United States, Denmark, France | 1, 1/2, 1/14, 2–5, 7–9, 14, 16 |
| 2 | 1 (0.47) | Germany | Unknown |
| 3 | 8 (3.76) | The Netherlands, United Kingdom | 2, 4, 8, 28 |
| 4 | 1 (0.47) | The Netherlands | 2 |
| 5 | 2 (0.94) | Finland, Spain | 2, 15 |
| 6 | 1 (0.47) | United Kingdom | 14 |
| 7 | 1 (0.47) | United Kingdom | 7 |
| 8 | 1 (0.47) | United Kingdom | 1 |
| 9 | 2 (0.94) | United Kingdom | 11, 15 |
| 10 | 1 (0.47) | United Kingdom | 7 |
| 11 | 1 (0.47) | Canada | 14 |
| 12 | 2 (0.94) | Spain | 1/2, 2 |
| 13 | 1 (0.47) | The Netherlands | 14 |
| 14 | 1 (0.47) | Canada | 28 |
| 15 | 1 (0.47) | Spain | 5 |
| 16 | 8 (3.76) | Spain, Canada | 9, 10, 16, 17, 18, 19 |
| 17 | 1 (0.47) | Spain | 1/2 |
| 18 | 4 (1.88) | Spain | 3, 7 |
| 19 | 1 (0.47) | Spain | 15 |
| 20a | 2 (0.94) | Spain | 21, 28 |
| 21a | 3 (1.41) | United Kingdom | 15 |
Profiles from RFLP analysis of PCR products obtained from amplification with primers 2 and 3.
Sequencing of sly RFLP variants confirms limited genetic diversity.
To further understand the extent and nature of genetic diversity within sly and the upstream region, PCR products from a representative of each of the 21 variants identified by RFLP were directly sequenced. Nucleotide sequences obtained from these 21 representatives and the 2 published alleles (33, 38) resulted in the identification of 16 sly alleles corresponding to nine distinct predicted amino acid sequences (Fig. 1). The remaining RFLP profiles reflected alteration 5′ to the coding region, although whether these alterations may have any effect on the expression of suilysin is unknown. The diversity between the sequenced alleles varied from 0.07 to 1.03% at the nucleotide level with the exception of isolate 3, corresponding to RFLP profile 2, which had previously been characterized as an atypical S. suis strain (24) and showed a maximum divergence from the other alleles of 1.79%. The amino acid diversity ranged between 0.21 and 0.62% with the exception of the allele corresponding to RFLP profile 2, which showed a maximum diversity from the other alleles of 1.03%. The vast majority of nucleotide changes seen within alleles are synonymous (data not shown). It should be noted that the individual variants identified by RFLP could contain multiple sequence species, although the diversity within these is likely to be very low as illustrated in previous studies using this approach (49). This was indeed seen to be the case for RFLP profile 1, where the representative sequenced showed a single nucleotide alteration from the published sequence of P1/7, restriction of which also gave RFLP profile 1.
FIG. 1.
Alignments of the variable sites identified by sequencing sly PCR products from a representative strain of each distinct profile identified by RFLP (left) and the published sequences from isolates P1/7 and 1933 (33, 38). Panel A illustrates the nucleotide diversity of the 16 alleles identified, while panel B illustrates alterations within the predicted amino acid sequence. Identical sites are represented by periods (.) and numbering above the residues corresponds to their position within the sequence relative to P1/7.
Correlation between suilysin distribution, serotype, and disease state.
Table 3 shows the distribution of the sly gene among the cohorts of United Kingdom and Spanish field isolates from both asymptomatic carriage and different disease states. The data show that sly is present in a higher percentage of Spanish isolates (87.3%) than United Kingdom isolates (69.4%). Chi-square tests confirm that sly is present in a significantly higher number of isolates from pigs with meningitis, septicemia, and arthritis than isolates from pigs with pneumonia (P < 0.005). Of the 12 isolates from humans included in this study, sly was successfully amplified from nine isolates. The relationship between presence of sly by PCR and serotype was also investigated where adequate numbers of strains from individual serotypes had been examined. Within the study there were nine serotypes that had eight or more representatives, and the distribution of sly PCR positives varied between these groups. Amplification from only 25% (3 of 12) of serotype 3 isolates and 38.5% (5 of 13) of serotype 7 isolates resulted in a sly PCR product, whereas 97.4% (37 of 38) of serotype 14 isolates, 86.7% (13 of 15) of serotype 1 isolates, and 100% (11 of 11) of isolates cross-reactive to antisera against both serotypes 1 and 14 (17) gave a positive result. Serotype 2 isolates accounted for 141 of the 310 isolates, and 73.8% (104 of 141) were found by PCR to contain sly. Other serotypes contained similar numbers of isolates in which sly was shown to be present or absent by PCR; 50% (4 of 8) of serotype 9 isolates, 66.7% (6 of 9) of serotype 15 isolates, and 62.5% (5 of 8) of serotype 1/2 harbored sly. The suilysin gene was not detected in any representatives of serotypes 6, 12, 13, 22, 24 through 27, and 29 through 32 (for each serotype, one strain was screened, with the exception of serotypes 22 and 27, where two isolates each were screened).
TABLE 3.
Isolates from the United Kingdom and Spain obtained from carriage and different disease states which are positive for sly
| Country of origin | No. of
sly-positive isolates/no. studied (%) from:
|
||||
|---|---|---|---|---|---|
| Septicemia | Meningitis | Pneumonia | Arthritis | Carriage states | |
| United Kingdom | 23/29 (79.3) | 37/52 (71.2) | 7/19 (36.8) | 8/8 (100) | |
| Spain | 12/14 (85.7) | 20/22 (90.9) | 5/8 (62.5) | 2/2 (100) | 37/41 (90.2) |
| Total | 35/43 (81.4) | 57/74 (77) | 12/27 (44.4) | 10/10 (100) | 37/41 (90.2) |
Characterization of the sly-flanking sequence.
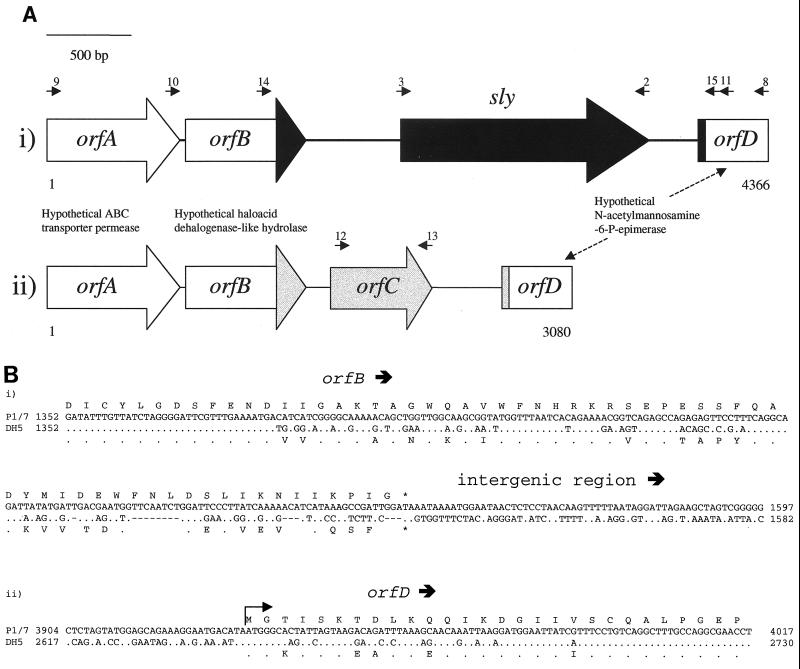
To investigate the region flanking sly and characterize the genetic basis of sly-negative strains, IV-PCR was performed. The restriction enzyme XbaI was selected, since Okwumabua et al. (32) reported that sly is located on a 4-kb XbaI fragment. Subsequent PCR amplification of strain P1/7 using primers 4 and 5 resulted in a fragment of approximately 2.9 kb. The product was cloned, sequenced, and found to correspond to published regions flanking sly (33, 38) and a novel additional sequence. An additional 143 bp of sequence was identified by sequencing an overlapping PCR product obtained from P1/7 using primers 6 and 7. This 900-bp PCR product was identical to the IV-PCR product with the exception of an additional 143 bp downstream of suilysin. A schematic of predicted genes within the sequence obtained can be seen in Fig. 2A. Sequence upstream of sly revealed two open reading frames. The first, orfA, is a 776-bp (corresponding to 257 amino acids) fragment of a gene, the 5′ region of which is presumed to lie outside the region characterized. The highest matching BLAST searches of both GenBank and available genome sequences revealed 64% amino acid identity and 78% similarity with a predicted ABC transporter (ATP binding protein) from Bacillus subtilis identified by sequence similarity to yheH (GenBank protein database accession no. H69828). The second open reading frame, orfB, with 711 bp (corresponding to 236 amino acids) shares high sequence similarity to OrfB of unknown function from Bacillus firmus (37% identity and 55% similarity at the amino acid level) (GenBank AAC45434). OrfB was found by conserved domain database search (52) to produce a significant alignment with a haloacid dehalogenase-like hydrolase consensus domain (Pfam 00702; Pfam [protein families] database; http://pfam.wustl.edu), suggesting that these three genes may be members of this family. Downstream of sly one partial open reading frame of 433 bp (corresponding to 144 amino acids), orfD, was identified, which has 76% identity and 85% similarity to a conserved hypothetical protein from Lactococcus lactis subsp. lactis (GenBank AAK05273). Other significant alignments with a putative N-acetylmannosamine-6-phosphate epimerase from Clostridium perfringens (48) indicate a possible function for orfD.
FIG. 2.
Schematic representation of the sly genomic region and corresponding sequence in strains lacking the gene. The small arrows represent the approximate location of primers used in mapping this region. (A) Diagrammatic representation of the region containing sly and the corresponding region in strains lacking the gene. (i) The arrangement of the region in sly-positive isolates, as determined by the amalgamation of our sequencing and the published sequence (P1/7), is shown at the top. (ii) The sequence determined from a sly-negative S. suis isolate (DH5) is illustrated. The white blocks represent regions of essentially identical sequence while the grey and black areas indicate regions specific to sly-negative and sly-positive strains, respectively. (B) Sequence of the boundaries between regions of sequence conserved between the sly-positive strain P1/7 and sly-negative strain DH5 and those containing diverged sequences. The 3′ region of orfB (i) and the 5′ region of orfD (ii) are shown. In each case, the top line represents the predicted amino acid sequence of P1/7, while the lower amino acid sequence refers to DH5. The top line of the nucleotide sequence refers to P1/7, and the lower line refers to DH5. Identical residues are represented by periods (.) and gaps in the sequence are represented by dashes (-).
Identification of the genetic basis of sly-negative strains.
Primers 8 and 9 (see Fig. 2Ai) were designed to amplify a PCR product corresponding to the region between the open reading frames found to flank suilysin. Amplification using the sly-positive strain, P1/7, resulted in a product of approximately the predicted size (4,366 bp), while the sly-negative strain, DH5, yielded a product of approximately 3,080 bp. Sequencing of this product revealed that the entire suilysin gene is absent from strain DH5 and that an additional open reading frame, orfC, is present. A schematic of this region comparing the sly-containing strain P/1/7 with the sly-negative strain DH5 is shown in Fig. 2A. The hypothetical ABC transporter gene, orfA, is 98.6% identical at the nucleotide level and 99.2% identical at the amino acid level between the two isolates. The potential haloacid dehalogenase-like hydrolases (orfB) share 99.3% nucleotide identity and 99.5% amino acid identity over the first 555 bp (corresponding to 185 amino acids) of the open reading frame. Beyond this point, there is a high level of sequence divergence, as shown in Fig 2Bi. BLAST analysis of the amino acid sequence of DH5 from amino acid 186 identifies highest similarity to a hypothetical haloacid dehalogenase from Sulfolobus solfataricus (GenBank CAC23414), while that from the sly-positive strain (P1/7) has highest sequence similarity to another hypothetical haloacid dehalogenase from Pyrococcus horikoshii (GenBank G71405). Downstream of orfB all sequence similarity between P1/7, which possesses sly between orfB and orfD, and DH5, which harbors a distinct open reading frame, orfC, is lost (Fig. 2Aii and 2Bi). There are three alternative in-frame start codons for orfC located within 30 bp of each other, but on the basis of the most promising of these (the most 3′) orfC is 558 bp long, encoding a predicted protein of 186 amino acids. The putative protein encoded by orfC shares 50% identity and 66% similarity at the amino acid level with a hypothetical protein from Pseudomonas aeruginosa (GenBank D83354), which itself shares significant similarity with an unnamed putative protein of Legionella pneumophila (GenBank AAB31024). This protein was identified on the basis of a fourfold-increased level of expression within macrophage relative to liquid culture (29). To prevent confusion it should be noted that although this protein is referred to as a macrophage infectivity potentiator (designated Mip) in a database accession created by GenBank staff, it is distinct from another, far-better-characterized protein of L. pneumophila, also called Mip (8). Downstream of orfC, both the sly-positive sequence (in P1/7) and DH5 harbor a 433-bp 5′ fragment of a hypothetical N-acetylmannosamine-6-phosphate epimerase (orfD). Sequence divergence between P1/7 and DH5 gradually diminishes after the predicted start codon of orfD until 57 bp (19 amino acids) into the open reading frame, from which point the sequences are highly conserved (98.9% nucleotide identity and 98.4% amino acid identity). The G+C contents of the diverged sequences between P1/7 and DH5 were found to be 36.8 and 38.1%, respectively. These percentages are similar to the G+C contents seen in a number of other recently described S. suis genes (41). The G+C content of orfC alone is somewhat higher at 44.5%, although several S. suis open reading frames identified by Smith et al. (41) display a similar G+C content.
Confirmation of sly-negative and -positive genomic arrangements in other isolates.
To demonstrate that the genomic arrangements in DH5 and P1/7 are present in other S. suis isolates, a series of PCR-mapping experiments were performed The location of all primers used in these experiments is illustrated on Fig. 2A. All mapping experiments and all further work described in this paper were performed using a cohort of 32 isolates. The isolates selected included the representative strain of each of the 21 RFLP profiles from which sly had already been sequenced (see Fig. 1), P1/7, and 10 putative sly-negative isolates including DH5 (Table 4).
TABLE 4.
Hemolytic activity and confirmation of sly region genomic arrangements by PCRa
| Isolate or strain | Characteristic or test
|
||||||||
|---|---|---|---|---|---|---|---|---|---|
| sly | RFLP profile | HE activity | PCR with primer pairc
|
||||||
| 14 (orfB)-15 (orfD) | 3(sly)-11 (orfD) | 10(orfA)-2 (sly) | 12 (orfC)-11 (orfD) | 10 (orfA)-13 (orfC) | 12 (orfC)-13 (orfC) | ||||
| 47 | + | 1 | + | +d | + | + | − | − | − |
| 3 | + | 2 | + | − | + | − | − | − | − |
| 58 | + | 3 | + | +d | + | + | − | − | − |
| 35 | + | 4 | ±b | +d | + | + | − | − | − |
| 37 | + | 5 | + | +d | + | + | − | − | − |
| 73 | + | 6 | + | +d | + | + | − | − | − |
| 102 | + | 7 | − | +d | + | + | − | − | − |
| 122 | + | 8 | + | +d | + | + | − | − | − |
| 128 | + | 9 | + | − | + | + | − | − | − |
| 167 | + | 10 | − | +d | + | + | − | − | − |
| 230 | + | 11 | + | +d | + | + | − | − | − |
| 238 | + | 12 | + | − | + | + | − | − | − |
| 348 | + | 13 | + | +d | + | + | − | − | − |
| 352 | + | 14 | ±b | +d | + | + | − | − | − |
| 296 | + | 15 | − | +d | + | + | − | − | − |
| 242 | + | 16 | + | +d | + | + | − | − | − |
| 247 | + | 17 | + | − | + | + | − | − | − |
| 248 | + | 18 | + | +d | + | + | − | − | − |
| 275 | + | 19 | + | +d | + | + | − | − | − |
| 297 | + | 20 | + | +d | + | + | − | − | − |
| 99 | + | 21 | + | +d | + | + | − | − | − |
| P1/7 | + | + | +d | + | + | − | − | − | |
| DH5 | − | − | +e | − | − | + | + | + | |
| 36 | − | − | +e | − | − | + | + | + | |
| 44 | − | − | +e | − | − | + | + | + | |
| 91 | − | − | +e | − | − | + | + | + | |
| 109 | − | − | +e | − | − | + | + | + | |
| 119 | − | − | +e | − | − | + | + | + | |
| 141 | − | − | +e | − | − | + | + | + | |
| 151 | − | − | +e | − | − | + | + | + | |
| 160 | − | − | +e | − | − | + | + | + | |
| 346 | − | − | +e | − | − | + | + | + | |
Isolates shown represent 21 sly RFLP profiles, strain P1/7, and 10 sly-negative isolates (including DH5). HE, hemolytic; +, present; −, absent.
±, weakly hemolytic activity.
Large product (ca. 2,700 bp).
Small product (ca. 1,400 bp).
First, reactions were performed to confirm the relative location of sly and the flanking genes. Primer pair 3 and 11 (sly-orfD) resulted in a PCR product of approximately the predicted 1,947 bp from all sly-positive isolates, with the exception of the isolate with the most divergent sly gene (isolate 3), while no product was seen in any sly-negative isolate. Similarly, amplification using primers 10 and 2 (orfA-sly) resulted in a product of approximately the predicted 2,931 bp for all sly-positive isolates; no product was seen from sly-negative isolates. To confirm the location of orfC in sly-negative isolates, primer pairs 10 and 13 (orfA-orfC) and 12 and 11 (orfC-orfD) were used. No PCR products were obtained from sly-containing strains, whereas sly-negative isolates gave PCR products of approximately the predicted sizes for both amplification from the upstream sequence to orfC (primers 10 and 13; 1,469 bp) and from orfC to the downstream sequence (primers 11 and 12; 1,077 bp).
Second, PCR across the entire divergent region from orfB to orfD (primers 14 and 15) was performed to confirm the predicted amplification of products of various sizes from sly-positive and sly-negative isolates. This resulted in a product corresponding to the predicted 1,391 bp from all 10 sly-negative isolates while 18 of 22 sly-positive isolates gave a product corresponding to the predicted 2,678 bp. As the four sly-positive strains that failed to produce an orfB-orfD product did give products when amplification was carried out between flanking genes and sly, as described above, the lack of a product may reflect minor changes in the sequence corresponding to the primer binding sites.
Third, all 32 isolates were screened for the presence of orfC using primers 12 and 13. Amplification from only the 10 putative sly-negative isolates resulted in a fragment of the predicted 447 bp (Table 4).
Confirmation of sly and orfC distribution by DNA dot blotting.
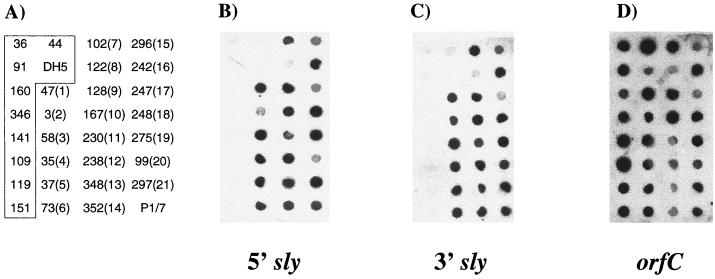
Despite the elucidation of the genetic basis for sly-negative S. suis, it remained possible that the suilysin gene is present elsewhere in the genome. To confirm the distribution of suilysin determined by PCR, DNA from the 32 isolates used to confirm the genomic arrangements of the sly region was used in a DNA dot blot. Two sly probes were constructed by labeling fragments obtained by restricting a PCR product amplified from strain P1/7 with primers 2 and 3 (the entire coding region) with EcoRV. This restriction resulted in a 614-bp probe to the 5′ of sly and a 879-bp probe to the 3′ of the gene. Both sly probes hybridized to chromosomal DNA from P1/7 used as a positive control. In addition, all strains that were positive for sly by PCR hybridized with both probes while there was little or no hybridization observed for either probe to DNA from sly-negative isolates (Fig. 3A and B). As confirmation that any weak signal seen represented background activity, Southern blot analyses were performed using chromosomal DNA from five of the PCR-negative isolates (DH5, 36, 44, 109, and 141) digested with BglII. In contrast to sly-positive isolates included on the same gel, digests originating from these five isolates showed absolutely no hybridization to an entire gene sly probe (data not shown). To investigate whether orfC or a closely related gene is present within the sly-positive isolates elsewhere in the genome, the same dot blot was probed with a PCR product obtained from DH5 with primers 12 and 13. Somewhat surprisingly, this orfC probe hybridized to chromosomal DNA from all 32 isolates, indicating that a related sequence is present within all S. suis isolates screened (Fig. 3C).
FIG. 3.
Dot blot analyses to determine to presence of sly and orfC in the 32 S. suis isolates described in Table 4. (A) Isolate numbers with allele designations (based on RFLP analysis) shown in parentheses. The boxed isolates represent those determined by PCR to be sly negative. (B) 5′ sly probe; (C) 3′ sly probe; (D) orfC probe.
Hemolytic assays.
Hemolytic assays were performed to determine if activity could be detected in all sly-positive isolates and to investigate if any activity could be identified in sly-negative isolates, indicating the possible presence of a novel hemolysin(s). A sample of culture supernatant collected at mid-exponential phase (optical density at 600 nm, 0.3) was used to assay for hemolytic activity in each of the cohort of 32 isolates from which sly had been sequenced where present and which were used in the PCR mapping described above. With this approach, no hemolytic activity was detected in any of the 10 sly-negative isolates screened, whereas activity was detected in 16 of 22 sly-positive strains, including P1/7. Further investigations were undertaken to ensure that hemolytic activity was not being missed in the remaining six sly-positive isolates in which no hemolytic activity was apparent as a result of variable expression during the in vitro growth cycle. Samples of culture supernatant were taken from the onset of exponential growth until 2 h into stationary phase. Supernatant from a positive control culture (isolate 348) was hemolytic at all time points, whereas no sample from two negative control isolates (36 and 141) displayed any hemolytic activity. Of the six test strains, one displayed clear hemolytic activity at all time points (strain 275) while supernatant from three others had no activity. The two remaining isolates, 35 and 352, were found to reduce the size of the red blood cell pellet at the lowest dilution tested, indicative of a very low level of hemolytic activity; hence, these isolates have been designated as possessing weak activity. Full results of hemolytic assays are included in Table 4.
DISCUSSION
The results presented in this study confirm previous reports suggesting that the suilysin-encoding gene is absent from a considerable proportion of clinical isolates. Only 69.4% of isolates examined in this study were found to be PCR positive for sly—these findings are in broad agreement with those of Segers et al. (38), who reported that 75% (72 of 96) of isolates from Europe and Asia possess sly, based on PCR and confirmatory Southern blot analysis. The type strains screened by PCR for sly in this study gave identical results to those previously reported on the basis of PCR and entire gene probes (33). A number of the serotype 2 and 14 reference isolates included in this study have also been examined previously by Norton et al. (31) who used Southern blots to determine the presence of sly. PCR amplification with primers 1 and 2 gave identical conclusions, supporting the conclusions based on the primer set used in this study.
While suilysin does not appear to be present in all isolates, where present it appears to display very limited genetic diversity, in contrast to many bacterial virulence factors, with almost 80% of isolates containing an identical allele as defined by RFLP. A maximum of only 1.79% nucleotide diversity was seen and this was in an apparently atypical isolate of S. suis whose phylogenetic relationship to the rest of the species is unclear (24). The paucity of nonsynonymous alterations in this gene is indicative of purifying selection preferentially eliminating amino acid changes. No genetic diversity was seen in any of the motifs previously identified as being important for the activity of members of the thiol-activated toxin family, including the undecapeptide sequence conserved between all thiol-activated toxins (35). Similarly, no sequence alterations were observed between the alleles in two further regions corresponding to those implicated in complement activation by pneumolysin (27). These are both present in suilysin, although in all alleles identified there is an alteration in the second sequence changing Asp385 in pneumolysin to an Asn residue. In pneumolysin, this alteration abolished complement activation (27), although the effect of this alteration in suilysin has not been investigated. In light of the lack of genetic diversity, suilysin would be a promising molecule for inclusion in a multitarget vaccine against S. suis.
In most cases the presence of sly does not correlate strongly with serotype. Of the serotypes from which a sufficient number of isolates were examined to allow comparisons to be drawn only serotypes 1, 14, and 1/14 displayed a convincing relationship with most, although not all, members of these serotypes possessing sly. In the case of all other serotypes there appears to be no strong correlation with either the presence or absence of sly. In light of the lack of genetic diversity and dominance of a single suilysin allele corresponding to RFLP profile 1 in the S. suis population, no link between particular alleles and clinical background could be established. However, one potentially interesting finding of this study was an apparent correlation between the absence of sly and pneumonia-associated isolates—this supports the very recent similar observation by Allgaier et al. (3) who examined the correlation between hemolytic activity and clinical background. There is ongoing debate about whether S. suis is a primary cause of pneumonia, as many isolations of S. suis are made in conjunction with organisms which are considered to be more significant respiratory pathogens, such as Actinobacillus pleuropneumoniae, Haemophilus parasuis, Pasteurella multocida, and swine influenza virus (18, 26, 37). However, reports that isolation of S. suis in pure culture from swine suffering acute respiratory distress or pneumonia is not infrequent do suggest a potential causative role for this organism in respiratory disease (3, 18). To further address this issue it would be of interest to screen a larger number of S. suis isolates obtained from diseased lungs in pure culture to determine the distribution of sly.
This study clearly supports suggestions that suilysin may play an important contributory role in the pathogenesis of the classic invasive diseases caused by S. suis. In our hands the vast majority of isolates associated with meningitis, septicemia, and arthritis harbor sly, supporting previous suggestions that sly is important for virulence of invasive isolates at least in Europe. The situation in North America may be somewhat different, as sly may be much less common in these isolates (15, 44) and as our study included only a small number of North American isolates. The presence of sly in a large number of carriage isolates suggests that there is no intrinsic difference between carriage isolates and those causing septicemia, meningitis, and arthritis and that the immune state of the host determines disease progression. However, it is pertinent to point out that over 25% of meningitis isolates characterized in this study, and apparently many North American isolates, do not possess sly. Although some of this discrepancy could reflect factors such as the presence of mixed populations of S. suis it is likely that the S. suis disease process involves multiple and potentially complementary processes yet to be elucidated. Three Canadian isolates from humans were shown by PCR not to contain sly, indicating that the presence of the gene is also not essential for infection of humans.
The results presented here represent the first characterization of the genetic basis of the absence of suilysin from some isolates of S. suis. A recombination event appears to have resulted in two alternative genetic structures located between the putative haloacid dehalogenase hydrolase (orfB) located upstream of sly and the hypothetical N-acetylmannosamine-6-phosphate epimerase (orfD) downstream of sly. In strains lacking sly its position is occupied by a novel gene (orfC) displaying similarity (28% identity and 46% similarity at the amino acid level) to an L. pneumophila protein of unknown function, characterized on the basis of its upregulation during growth within macrophages (29). The directionality of the orfC-sly alteration is unknown, as is the mechanism. There is no evidence of obvious repetitive structures flanking the variant sequence, and the G+C content of this region is not significantly different from that seen in other S. suis genes. In light of these observations it is impossible to be clear whether this arrangement is the result of an intragenomic recombination event or the result of horizontal transfer of DNA from another bacterial species. Similarly, it is not known whether the flanking genes (orfB and orfD) are functional in both sly-positive and sly-negative genetic arrangements. However, the fact that in spite of substantial diversity at the 3′ and 5′ ends of these genes, respectively, the genes are in frame in each case with appropriate initiation and termination codons and that the entire conserved domain of the hydrolase is present implies this is the case. Rather surprisingly, dot blot analysis showed that an orfC-related sequence is present elsewhere in the S. suis genome, although as orfC was not detected by PCR in sly-negative isolates, it appears that this represents hybridization to a distinct, if related, gene or gene fragment. Further hybridization studies involving fragments of orfC are necessary to clarify the basis of these findings. The PCR-mapping studies presented in Table 4 were carried out using a diverse range of isolates and suggest that all sly-positive and-negative isolates show similar genetic arrangements. They may therefore originate from a single recombination event, followed by horizontal movement between strains resulting in the current widespread distribution of both arrangements among multiple serotypes of S. suis. Further studies currently in progress examining the genetic arrangement of this region in a large number of isolates in conjunction with a large population genetic survey in progress using the same isolates will help address this issue.
Both assays of hemolytic activity, in which all sly-negative isolates tested lacked any evidence of hemolytic activity, and dot blot results provided no convincing evidence of the presence of a second hemolysin in S. suis belonging to the same protein family. It has previously been reported that there are regions with similarity to the 3′ region of sly present in PCR-negative isolates, including strain DH5, examined in this study (33, 44). For this reason, two sly probes were constructed to both the 5′ and 3′ regions of the gene. Chromosomal DNA from isolates that contain sly, on the basis of PCR results, bound both probes strongly; however, there was little or no hybridization observed for either probe to DNA from sly-negative isolates. As confirmation that any weak signal seen represented background activity, Southern blot analyses were performed using chromosomal DNA from five of the PCR-negative isolates and showed no hybridization to an entire gene sly probe. However, as shown in Table 4, a number of isolates that harbor sly showed either negligible or no hemolytic activity (5 of 22 sly-positive isolates examined). As these isolates were all from sly RFLP groups represented by a single isolate, their sly genes were sequenced, allowing determination of whether there are naturally occurring mutations in sly which impact on biological activity. Comparing the five sequences (RFLP profiles 4 and 14 from isolates with weak hemolytic activity and profiles 7, 10, and 15 from isolates with no hemolytic activity) (see Fig. 1), it can been seen that there are no changes in common between any of these isolates which might explain the observed low and/or absent hemolytic activity. Indeed, the predicted amino acid sequences of three of the alleles (4, 14, and 10) are identical to sequences of active alleles. These findings indicate that the low or absent hemolytic activity in these five isolates reflects changes outside sly, perhaps in upstream regulatory elements or alterations in other genes, including potential regulatory genes that may affect expression of sly rather than changes in the protein itself. While it is of interest to note that four of these five isolates were from lungs, with the fifth isolated from asymptomatic nasal carriage, it should be borne in mind that these isolates represented rare sly alleles, each seen only once despite examination of over 300 isolates. Thus, these isolates are not typical of those responsible for the vast majority of the S. suis disease burden.
In conclusion, we have demonstrated a strong association of the sly-encoding gene with S. suis isolates from invasive disease states. Where present, the gene is highly conserved with remarkably little allelic variation apparent. For the first time, we have defined the genetic basis of the absence of suilysin from many isolates of S. suis, particularly those associated with lung infections, and determined the presence of a novel gene, orfC, in these isolates. Clearly much further work is required to understand the relevance of these observations in the pathogenesis of the various clinical syndromes associated with S. suis.
ACKNOWLEDGMENTS
We gratefully acknowledge all of the colleagues listed in Materials and Methods who provided isolates for inclusion in this study and Jamie Leigh for helpful discussions.
This work was supported by a project grant from the BBSRC (grant no. 88/S11598). A.M.W. is supported by a Wellcome Trust Research Fellowship in Biodiversity (grant no. 053589).
REFERENCES
- 1.Aarestrup F M, Rasmussen S R, Artursson K, Jensen N E. Trends in resistance to antimicrobial agents of Streptococcus suisisolates from Denmark and Sweden. Vet Microbiol. 1998;63:71–80. doi: 10.1016/s0378-1135(98)00228-4. [DOI] [PubMed] [Google Scholar]
- 2.Allen A G, Bolitho S, Lindsay H, Khan S, Bryant C, Norton P, Ward P, Leigh J, Morgan J, Riches H, Eastty S, Maskell D. Generation and characterization of a defined mutant of Streptococcus suislacking suilysin. Infect Immun. 2001;69:2732–2735. doi: 10.1128/IAI.69.4.2732-2735.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allgaier A, Goethe R, Wisselink H J, Smith H E, Valentin-Weigand P. Relatedness of Streptococcus suisisolates of various serotypes and clinical backgrounds as evaluated by macrorestriction analysis and expression of potential virulence traits. J Clin Microbiol. 2001;39:445–453. doi: 10.1128/JCM.39.2.445-453.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Arends J P, Zanen H C. Meningitis caused by Streptococcus suisin humans. Rev Infect Dis. 1988;10:131–137. doi: 10.1093/clinids/10.1.131. [DOI] [PubMed] [Google Scholar]
- 5.Billington S J, Jost B H, Songer J G. Thiol-activated cytolysins: structure, function and role in pathogenesis. FEMS Microbiol Lett. 2000;182:197–205. doi: 10.1016/s0378-1097(99)00536-4. [DOI] [PubMed] [Google Scholar]
- 6.Charland N, Harel J, Kobisch M, Lacasse S, Gottschalk M. Streptococcus suisserotype 2 mutants deficient in capsular expression. Microbiology. 1998;144:325–332. doi: 10.1099/00221287-144-2-325. [DOI] [PubMed] [Google Scholar]
- 7.Chatellier S, Harel J, Zhang Y, Gottschalk M, Higgins R, Devriese L A, Brousseau R. Phylogenetic diversity of Streptococcus suisstrains of various serotypes as revealed by 16S rRNA gene sequence comparison. Int J Syst Bacteriol. 1998;48:581–589. doi: 10.1099/00207713-48-2-581. [DOI] [PubMed] [Google Scholar]
- 8.Cianciotto N P, Eisenstein B I, Mody C H, Toews G B, Engelberg N C. A Legionella pneumophilagene encoding a species-specific surface protein potentiates initiation of intracellular infection. Infect Immun. 1989;57:1255–1262. doi: 10.1128/iai.57.4.1255-1262.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.François B, Gissot V, Ploy M C, Vignon P. Recurrent septic shock due to Streptococcus suis. J Clin Microbiol. 1998;36:2395. doi: 10.1128/jcm.36.8.2395-2395.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Galina L, Pijoan C, Sitjar M, Christianson W T, Rossow K, Collins J E. Interaction between Streptococus suisserotype 2 and porcine reproductive and respiratory syndrome virus in specific pathogen-free piglets. Vet Rec. 1994;134:60–64. doi: 10.1136/vr.134.3.60. [DOI] [PubMed] [Google Scholar]
- 11.Gottschalk M, Higgins R, Jacques M, Mittal K R, Henrichsen J. Description of 14 new capsular types of Streptococcus suis. J Clin Microbiol. 1989;27:2633–2636. doi: 10.1128/jcm.27.12.2633-2636.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gottschalk M, Higgins R, Jacques M, Beaudoin M, Henrichsen J. Isolation and characterization of Streptococcus suiscapsular types 9–22. J Vet Diagn Investig. 1991;3:60–65. doi: 10.1177/104063879100300113. [DOI] [PubMed] [Google Scholar]
- 13.Gottschalk M, Higgins R, Jacques M, Beaudoin M, Henrichsen J. Characterization of six new capsular types (23 through 28) of Streptococcus suis. J Clin Microbiol. 1991;29:2590–2594. doi: 10.1128/jcm.29.11.2590-2594.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gottschalk M, Lacouture S, Dubreuil J D. Characterization of Streptococcus suiscapsular type 2 haemolysin. Microbiology. 1995;141:189–195. doi: 10.1099/00221287-141-1-189. [DOI] [PubMed] [Google Scholar]
- 15.Gottschalk M, Lebrun A, Wisselink H, Dubreuil J D, Smith H, Vecht U. Production of virulence-related proteins by Canadian strains of Streptococcus suiscapsular type 2. Can J Vet Res. 1998;62:75–79. [PMC free article] [PubMed] [Google Scholar]
- 16.Gottschalk M, Higgins R, Quessy S. Dilemma of the virulence of Streptococcus suisstrains. J Clin Microbiol. 1999;37:4202–4203. doi: 10.1128/jcm.37.12.4202-4203.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gottschalk M, Segura M. The pathogenesis of the meningitis caused by Streptococcus suis: the unresolved questions. Vet Microbiol. 2000;76:259–272. doi: 10.1016/s0378-1135(00)00250-9. [DOI] [PubMed] [Google Scholar]
- 18.Heath P J, Hunt B W. Streptococcus suisserotypes 3 to 28 associated with disease in pigs. Vet Rec. 2001;148:207–208. doi: 10.1136/vr.148.7.207. [DOI] [PubMed] [Google Scholar]
- 19.Higgins R, Gottschalk M, Boudreau M, Lebrun A, Henrichsen J. Description of six new capsular types (29–34) of Streptococcus suis. J Vet Diagn Investig. 1995;7:405–406. doi: 10.1177/104063879500700322. [DOI] [PubMed] [Google Scholar]
- 20.Jacobs A A, van den Berg A J, Baars J C, Neilsen B, Johannsen L W. Production of suilysin, the thiol-activated haemolysin of Streptococcus suis,by field isolates from diseased pigs. Vet Rec. 1995;137:295–296. doi: 10.1136/vr.137.12.295. [DOI] [PubMed] [Google Scholar]
- 21.Jacobs A A C, Loeffen P L W, van den Berg A J G, Storm P K. Identification, purification, and characterization of a thiol-activated hemolysin (suilysin) of Streptococcus suis. Infect Immun. 1994;62:1742–1748. doi: 10.1093/benz/9780199773787.article.b00034458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobs A A C, van den Berg A J G, Loeffen P L W. Protection of experimentally infected pigs by suilysin, the thiol-activated haemolysin of Streptococcus suis. Vet Rec. 1996;139:225–228. doi: 10.1136/vr.139.10.225. [DOI] [PubMed] [Google Scholar]
- 23.Kumar S, Tamura K, Nei M. MEGA—molecular evolutionary genetics analysis, version 1.01. University Park, Pa: The Pennsylvania State University; 1993. [Google Scholar]
- 24.Lammler C, Weiss R. Characterisation of an unusual Streptococcus suisisolated from an aborted fetus of a pig. Med Sci Res. 1997;25:263–264. [Google Scholar]
- 25.Luque I, Tarradas C, Astorga R, Perea A, Wisselink H J, Vecht U. The presence of muramidase released protein and extracellular factor protein in various serotypes of Streptococcus suisisolated from diseased and healthy pigs in Spain. Res Vet Sci. 1999;66:69–72. doi: 10.1053/rvsc.1998.0242. [DOI] [PubMed] [Google Scholar]
- 26.Macinnes J I, Desrosiers R. Agents of the “suis-ide diseases” of swine: Actinobacillus suis, Haemophilus parasuis, and Streptococcus suis. Can J Vet Res. 1999;63:83–89. [PMC free article] [PubMed] [Google Scholar]
- 27.Mitchell T J, Andrew P W, Saunders F K, Smith A N, Boulnois G J. Complement activation and antibody binding by pneumolysin via a region of the toxin homologous to a human acute phase protein. Mol Microbiol. 1991;5:1883–1888. doi: 10.1111/j.1365-2958.1991.tb00812.x. [DOI] [PubMed] [Google Scholar]
- 28.Mitchell T J, Andrew P W. Biological properties of pneumolysin. Microb Drug Resist. 1997;3:19–26. doi: 10.1089/mdr.1997.3.19. [DOI] [PubMed] [Google Scholar]
- 29.Miyamoto H, Yoshida S, Taniguchi H, Hui Qin M, Fujio H, Mizuguchi Y. Protein profiles of Legionella pneumophila Philadelphia-1 grown in macrophages and characterization of a gene encoding a novel 24 Kda Legionellaprotein. Microb Pathog. 1993;15:469–484. doi: 10.1006/mpat.1993.1095. [DOI] [PubMed] [Google Scholar]
- 30.Mogollon J D, Pijoan C, Murtaugh M P, Collins J E, Cleary P P. Identification of epidemic strains of Streptococcus suisby genomic fingerprinting. J Clin Microbiol. 1991;29:782–787. doi: 10.1128/jcm.29.4.782-787.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Norton P M, Rolph C, Ward P N, Bentley R W, Leigh J A. Epithelial invasion and cell lysis by virulent strains of Streptococcus suisis enhanced by the presence of suilysin. FEMS Immunol Med Microbiol. 1999;26:25–35. doi: 10.1111/j.1574-695X.1999.tb01369.x. [DOI] [PubMed] [Google Scholar]
- 32.Okwumabua O, Staats J, Chengappa M M. Detection of genomic heterogeneity in Streptococcus suisisolates by DNA restriction fragment length polymorphisms of rRNA genes (ribotyping) J Clin Microbiol. 1995;33:968–972. doi: 10.1128/jcm.33.4.968-972.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Okwumabua O, Abdelmagid O, Chengappa M M. Hybridization analysis of the gene encoding a hemolysin (suilysin) of Streptococcus suistype 2: evidence for the absence of the gene in some isolates. FEMS Microbiol Lett. 1999;181:113–121. doi: 10.1111/j.1574-6968.1999.tb08833.x. [DOI] [PubMed] [Google Scholar]
- 34.Perch B, Pedersen K B, Henrichsen J. Serology of capsulated streptococci pathogenic for pigs: six new serotypes of Streptococcus suis. J Clin Microbiol. 1983;17:993–996. doi: 10.1128/jcm.17.6.993-996.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Pinkney M, Beachey E, Kehoe M. The thiol-activated toxin streptolysin O does not require a thiol group for cytolytic activity. Infect Immun. 1989;57:2553–2558. doi: 10.1128/iai.57.8.2553-2558.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prieto C, Garcia F J, Saurez P, Imaz M, Castro J M. Biochemical traits and antimicrobial susceptibility of Streptococcus suisisolated from slaughtered pigs. J Vet Med B. 1994;41:608–617. doi: 10.1111/j.1439-0450.1994.tb00271.x. [DOI] [PubMed] [Google Scholar]
- 37.Reams R Y, Harrington D D, Glickman L T, Thacker H L, Bowersock T L. Multiple serotypes and strains of Streptococcus suisin naturally infected swine herds. J Vet Diagn Investig. 1996;8:119–121. doi: 10.1177/104063879600800121. [DOI] [PubMed] [Google Scholar]
- 38.Segers R P A M, Kenter T, de Haan L A M, Jacobs A A C. Characterisation of the gene encoding suilysin from Streptococcus suisand expression in field strains. FEMS Microbiol Lett. 1998;167:255–261. doi: 10.1111/j.1574-6968.1998.tb13236.x. [DOI] [PubMed] [Google Scholar]
- 39.Smith H E, Vecht U, Wisselink H J, Stockhofe-Zurwieden N, Biermann Y, Smits M A. Mutants of Streptococcus suistypes 1 and 2 impaired in expression of muramidase-released protein and extracellular protein induce disease in newborn germfree pigs. Infect Immun. 1996;64:4409–4412. doi: 10.1128/iai.64.10.4409-4412.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Smith H E, Damman M, van der Velde J, Wagenaar F, Wisselink H J, Stockehofe-Zurwieden N, Smits M A. Identification and characterization of the cps locus of Streptococcus suisserotype 2: the capsule protects against phagocytosis and is an important virulence factor. Infect Immun. 1999;67:1750–1756. doi: 10.1128/iai.67.4.1750-1756.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Smith H E, Bujis H, de Vries R, Wisselink H J, Stockhofe-Zurwieden N, Smits M A. Environmentally regulated genes of Streptococcus suis: identification by the use of iron-restricted conditions in vitro and by experimental infection of piglets. Microbiology. 2001;147:271–280. doi: 10.1099/00221287-147-2-271. [DOI] [PubMed] [Google Scholar]
- 42.Staats J J, Feder I, Okwumabua O, Chengappa M M. Streptococcus suis: past and present. Vet Res Commun. 1997;21:381–407. doi: 10.1023/a:1005870317757. [DOI] [PubMed] [Google Scholar]
- 43.Staats J J, Plattner B L, Nietfeld J, Dritz S, Chengappa M M. Use of ribotyping and haemolysin activity to identify highly virulent Streptococcus suistype 2 isolates. J Clin Microbiol. 1998;36:15–19. doi: 10.1128/jcm.36.1.15-19.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Staats J J, Plattner B L, Stewart G C, Chengappa M M. Presence of the Streptococcus suis suilysin gene and expression of MRP and EF correlates with high virulence in Streptococcus suistype 2 isolates. Vet Microbiol. 1999;70:201–211. doi: 10.1016/s0378-1135(99)00147-9. [DOI] [PubMed] [Google Scholar]
- 45.Tarradas C, Borge C, Arenas A, Maldonado A, Astorga R, Miranda A, Luque I. Suilysin production by Streptococcus suisstrains isolated from diseased and healthy carrier pigs in Spain. Vet Rec. 2001;148:183–184. doi: 10.1136/vr.148.6.183. [DOI] [PubMed] [Google Scholar]
- 46.Turgeon P L, Higgins R, Gottschalk M, Beaudoin M. Antimicrobial susceptibility of Streptococcus suisisolates. Br Vet J. 1994;150:263–269. doi: 10.1016/S0007-1935(05)80006-5. [DOI] [PubMed] [Google Scholar]
- 47.Vecht U, Wisselink H J, Jellema M L, Smith H E. Identification of two proteins associated with virulence of Streptococcus suistype 2. Infect Immun. 1991;59:3156–3162. doi: 10.1128/iai.59.9.3156-3162.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Walters D M, Stirewalt V L, Melville S B. Cloning, sequence, and transcriptional regulation of the operon encoding a putative N-acetylmannosamine-6-phosphate epimerase (nanE) and sialic acid lyase (nanA) in Clostridium perfringens. J Bacteriol. 1999;181:4526–4532. doi: 10.1128/jb.181.15.4526-4532.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Whatmore A M, Dowson C G. The autolysin-encoding gene (lytA) of Streptococcus pneumoniaedisplays restricted allelic variation despite localized recombination events with genes of pneumococcal bacteriophage encoding cell wall lytic enzymes. Infect Immun. 1999;67:4551–4556. doi: 10.1128/iai.67.9.4551-4556.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Whatmore A M, Barcus V A, Dowson C G. Genetic diversity of the streptococcal competence (com) gene locus. J Bacteriol. 1999;181:3144–3154. doi: 10.1128/jb.181.10.3144-3154.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Whatmore A M, Efstratiou A, Pickerill A P, Broughton K, Woodard G, Sturgeon D, George R, Dowson C G. Genetic relationships between clinical isolates of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus mitis: characterization of “atypical” pneumococci and organisms allied to S. mitis harboring S. pneumoniaevirulence factor-encoding genes. Infect Immun. 2000;68:1374–1382. doi: 10.1128/iai.68.3.1374-1382.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wheeler D L, Church D M, Lash A E, Leipe D D, Madden T L, Pontius J U, Schuler G D, Schriml L M, Tatusova T A, Wagner L, Rapp B A. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2001;29:11–16. doi: 10.1093/nar/29.1.11. [DOI] [PMC free article] [PubMed] [Google Scholar]