Abstract
Typhoid is a major public health concern. Even though antibiotics are usually used to treat typhoid fever, the spread of multi drug resistant Salmonella typhi is making antibiotics much less effective. This study was conducted to assess the prevalence of multidrug-resistant Salmonella typhi from the clinical samples. During this study, 154 blood samples of suspected typhoid patients were collected from the hospital and diagnostic center located in Chattogram City, Bangladesh. Isolation and identification of Salmonella typhi was done by both biochemical tests. PCR analysis was also done for the confirmation of biochemical result. Antimicrobial susceptibility test was performed according to the Kirby-Bauer disk diffusion method against ampicillin, chloramphenicol, cefepime, cotrimoxazole, ceptriaxone, ciprofloxacin, nalidixic acid, and azithtomycin. Out of 154, 21 (13.64%) isolates were identified as Salmonella typhi and the prevalence of typhoid in Chattogram, Bangladesh was 13.64% (n = 21). It was also found that children under the age of 5 are the more vulnerable target of Salmonella typhi infection. Antibiotic resistance profiling revealed 85% isolates were Multi-Drug Resistant (MDR) and highest resistance was found in case of Nalidixic acid. Although, most of the isolated Salmonella typhi were MDR, first generation antibiotics Co-trimoxazile, Chloramphenicol, and Ampicillin were found effective against Salmonella typhi.
Keywords: Salmonella typhi, antibiotics, resistance, PCR assay, typhoid
Introduction
Salmonella typhi is a gram-negative, rod-shaped, facultative anaerobic bacteria that belongs to the family of Enterobacteriae and only humans are the reservoir for it. Typhoid fever, also known as enteric fever, is a potentially fatal systemic infection caused mainly by Salmonella entericaserovartyphi (Salmonella typhi).1-3 It is one of the most prevalent bacterial infections caused worldwide.4 The people of low and middle-income countries are at a high risk of typhoid infection.5,6 In South Asia, every year more than 7 million people are infected,2 with a death rate of 10%. Antimicrobial resistance is a natural phenomenon that occurs due to the adaptation of infectious agents to the exposure of antimicrobial agents. Most antimicrobial resistance is due to a genetic change in the organism, either the acquisition of foreign genes through a plasmid or a chromosomal mutation.3 Over-prescription and indiscriminate use of antibiotics are referred to as the main reasons behind the initial development of antimicrobial resistance in Salmonella typhi.7 The use of antibiotics unnecessarily may result in the emergence of multi-drug resistant bacteria.8,9 In a developing country like Bangladesh, typhoid fever is considered one of the major public health concerns due to the lack of proper treatments.10 This problem has been further complicated due to the emergence of multidrug-resistant Salmonella typhi, and the treatment of typhoid fever has become more and more difficult. Epidemiological studies suggest that MDR Salmonella serotypes are more virulent than susceptible strains, as they increase the severity of infection.11 MDR Salmonella typhi has emerged all over the world, especially in the Indian sub-continent, Africa, and Southeast Asia. Extensive use of second-generation fluoroquinolones such as ciprofloxacin and levofloxacin has given rise to the attenuated susceptibility of Salmonella typhi.2 Antimicrobial resistance to antibiotics from different classes, like β-lactams, fluoroquinolones, and aminoglycosides, is usually caused by many genes.12 The Widal test and blood culture tests are commonly used by clinicians in South Asia to diagnose typhoid. But it is common to find false-positive results in these methods, which leads to over-diagnosis of typhoid fever. On the other hand, the introduction of molecular methods, particularly PCR, has proved to be rapid, sensitive, and specific for the diagnosis and analysis of typhoid. The flagellin gene, fliC, of Salmonella typhi is used as an identifiable targeted gene to confirm the presence of Salmonella typhi.13 As antibiotic use as well as overuse vary from region to region and time to time, accurate diagnosis and understanding of the antibiotic resistance pattern according to regions and time intervals is a crying need for providing proper treatment against typhoid. So, this study is designed to reveal the limitation of existing diagnosis process in the detection of Salmonella typhi among typhoid patients and also to analyze the consequences of antibiotic resistance development in Salmonella typhi in clinical samples.
Materials and Methods
Sample collection
Ethical approval was obtained from the ethical review committee of Chittagong Medical College, Bangladesh (Ref. no.: CMC/PG/2020/89). Blood samples were collected from suspected typhoid fever patients at Chittagong Medical College Hospital, Popular Diagnostic Ltd, Lancet Diagnostic and Research Center, and Chevron Clinical Laboratory Pte. Ltd. located in Chittagong, Bangladesh with written informed consent. Ethylenediaminetetra-acetic acid (EDTA from Sigma-Aldrich, Germany) tubes were used to collect blood samples from patients with the help of medical technologists. To maintain aseptic conditions, collected samples were placed in an icebox and stored at 4°C.
Sample processing and standardization of bacteria
The samples were prepared on Tryptone Soya Broth (TSB from Hi Media, India). One milliliter of blood sample was added to the test tube containing 10 ml of TSB solution to maintain the ratio of 1:10. Then the culture was shaken at 250 revolutions per minute (RPM) in a shaking incubator (at 37°C for 16 to 20 hours.14,15
Selective plating
For the isolation of Salmonella from clinical samples, Xylose Lysine Deoxycholate Agar (XLD, Hi Media, India) and MacConkey agar (Hi Media, India) were used.16,17 The transparent colonies on the MacConkey agar and the red colonies with a black center on the XLD agar were selected for the pure culture of the bacteria. These colonies were sub-cultured to the XLD agar plate through the streak plate method.
Biochemical test (TSI test)
Triple sugar iron (TSI from Hi Media, India) agar was used to identify organisms based on properties related to carbohydrate fermentation and the ability to reduce sulfur.17
Genomic DNA extraction and PCR analysis
Genomic DNA of isolated Salmonella typhi was extracted by using the boiling method of DNA extraction.18 During this study, the flagellin gene, fliC of Salmonella typhi, was targeted to confirm the presence of Salmonella typhi.18,19 Salmonella typhi possesses unique nucleotide sequences in the hypervariable region of the flagellin gene, which are distinct from those sequences in other strains of Salmonella.20 Through a review of the literature, the forward primer: 5′-ACT GCT AAA ACC ACT ACT-3′ and the reverse primer5′-TGG AGA CTT CGG TCGCGTAG-3′ were chosen for fliC gene PCR amplification.13 The specificity of the primer was determined by the Basic Local Alignment Search Tool (BLAST). The PCR conditions were: 1 minutes at 94°C; 36 cycles of 94°C for 1 minutes, 55°C for 1 minutes, and 72°C for 2 minutes; 10 minutes for72°C. The PCR mixture was a total volume of 20 µl: 10 µl master mix, 4 µl template, 2 µl forward primer, 2 µl reverse primer, 3 µl water. In each experiment, positive and negative controls were used. A PCR thermal cycler (NYX Technik) was used for the amplification of extracted DNA. The PCR products were run through a 1.5% agarose gel using12 150 mA current for 45 minutes and the desirable band of 367 bp was visualized on a UV illuminator (Thermo Scientific, USA).
Sequencing
The PCR product fliC gene of 1 sample was sequenced at “First Base Research Laboratory,” Malaysia. After sequencing, the quality of the sequences was assessed manually based on the corresponding electrochromatogram (Chromas software, version 2.6.6). After that, sequences were submitted to Genbank using Bankit.
Antimicrobial susceptibility test
The antimicrobial susceptibility test was performed according to the Kirby-Bauer disk diffusion method with the McFarland Standard (Kirby-Bauer Disk Diffusion Susceptibility Test Protocol), American Society for Microbiology) using 8 types of selected antibiotic disks of Hi-media : ampicillin (30 ug), chloramphenicol (30 ug), cefepime(5 ug), cotrimoxazole (25 ug), ceptriaxone (30 ug), ciprofloxacin (5 ug), nalidixic acid (30 ug), and azithtomycin (15 ug). Ampicillin, cefepime, and ceptriaxone are β-lactams antibiotics; ciprofloxacin and nalidixic acid are quinolone antibiotics; chloramphenicol has a nitrobenzene ring; co-trimoxazole is a sulfonamides class antibiotic; and azithtomycin is a macrolide. Interpretation of antibiotic resistance was done according to the BSAC Methods for Antimicrobial Susceptibility Testing, 2013).
Results
Identification of Salmonella typhi among patients
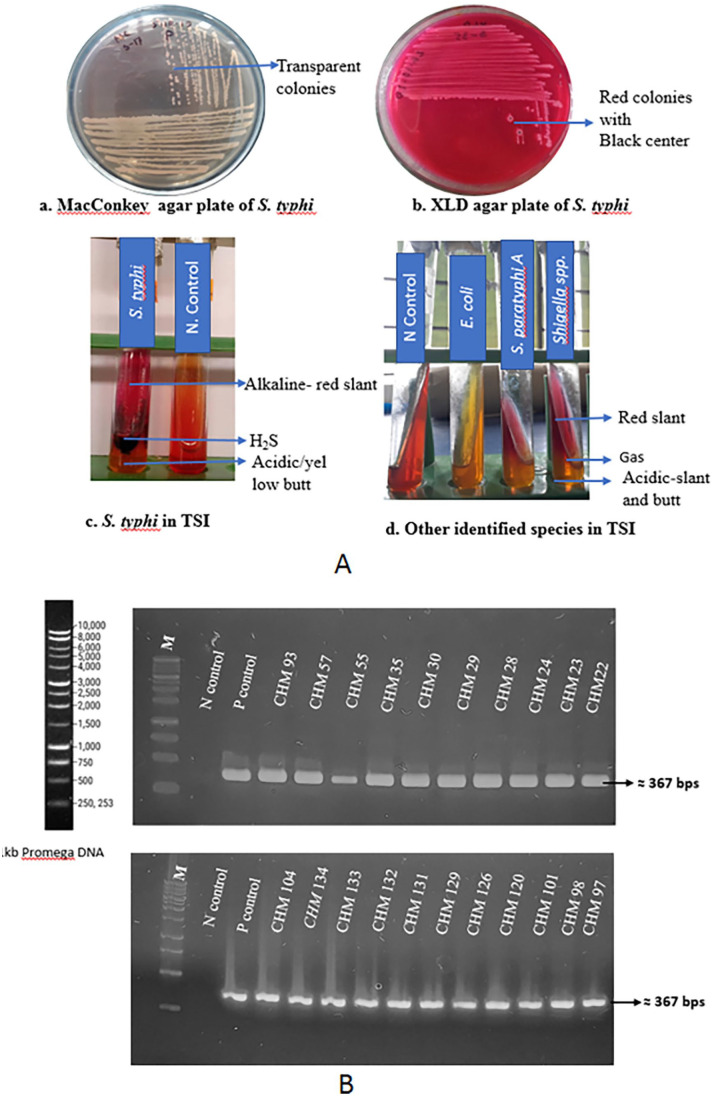
Blood samples were collected from 154 typhoid suspected patients from the different Diagnostic Centers mentioned in the methodology section. Among the 154 collected samples, 43 were found positive on MacConkey and XLD agar media Figure 1A (a + b). Finally, out of 43 expected positive colonies, 23 isolates of Salmonella typhi were identified through the TSI test Figure 1A(c). Besides, 4 E. coli, 3 Salmonella paratyphi A, 11 Shigella spp., and 2 Pseudomonas aeruginosa samples were suspected based on selective plating and biochemical test Figure 1A(d); Table 1.
Figure 1.
Identification of Salmonella typhi: A. Selective plating of Salmonella typhi on MacConky agar (a), XLD agar (b) and biochemical test on TSI (c); Other species on TSI (d). B. Molecular identification of Salmonella typhi through electrophoretic (1.5%) separation of fliC gene. Here, Salmonella typhi CHM 55 was sequenced and used as positive control (GenBank accession number: MW819865). P- Positive; N-Negative; M-1 kb Promega DNA marker.
Table 1.
Bacteria identified by biochemical (TSI) test.
| Slant | Butt | H2S | Gas | Expected organism | No. |
|---|---|---|---|---|---|
| Alkaline (red) | Acidic (yellow) | Yes | No | Salmonella typhi | 23 |
| Alkaline (red) | Acidic (yellow) | No | Yes | Salmonella paratyphi A | 3 |
| Acidic (yellow) | Acidic (yellow) | No | Yes | E. coli | 4 |
| Acidic (yellow) | Alkaline (red) | No | No | Shigella spp. | 11 |
| Alkaline (red) | Alkaline (red) | No | No | Pseudomonas aeruginosa | 2 |
To identify and confirm whether biochemically identified isolates were Salmonella typhi, a PCR amplification of the identifiable gene fliC was performed to identify the expected band of 367 bp Figure 1B.13,19 The fliC gene was detected in 21 isolates out of 23 biochemically identified bacterial genomic DNA. The remaining 2 false positive samples were discarded. One sample, CHM-55, was sequenced and confirmed as Salmonella typhi through BLAST analysis that showed 99% similarity with the reference sequence (CP053702.1). This sequence was submitted and accepted in the GenBank database (GenBank accession number: MW819865).
Epidemiological study of typhoid in Chattogram
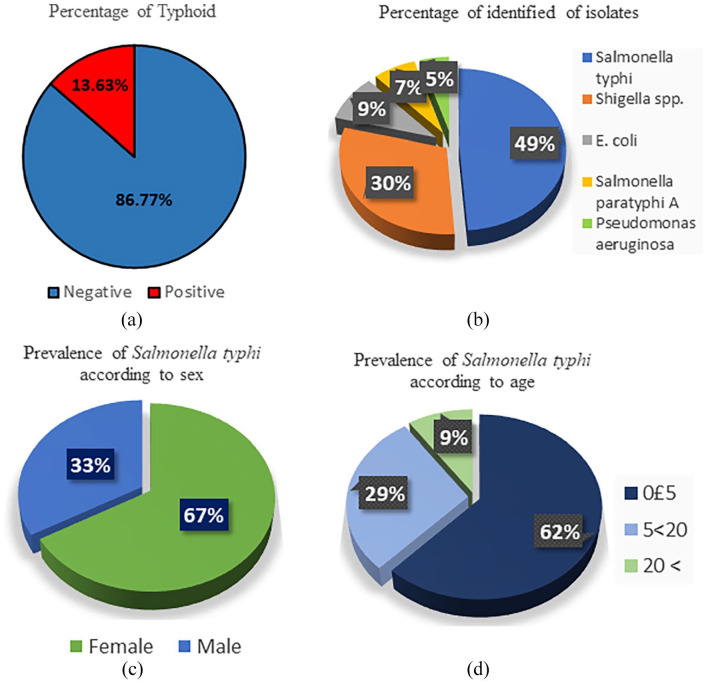
Among the 154 collected samples, 13.65% (n = 21) of Salmonella typhi isolates were identified through molecular identification Figure 2a. Overall, 28% (n = 43) of the patients carried bacteria in their blood. Besides, 49%of the isolates were Salmonella typhi among the 43 isolated organisms Figure 2b. Distribution of 21 identified Salmonella typhi according to sex and age of the clinical patients has been shown in Figure 2c and d, where prevalence rate was highest in female and 0 to 5 years old children according to sex and ages.
Figure 2.
Epidemiological study among the suspected 154 typhoid patients: (a) percentage of the typhoid patients among the suspected patients, (b) percentage of Salmonella typhi and other suspected organisms within the 43 isolated organisms, (c) percentage of male and female typhoid patients, and (d) Age intervals among the 21 identified typhoid patients.
Antimicrobial resistance profiling of Salmonella typhi
Antimicrobial resistance profiling was done according to the Kirby-Bauer disk diffusion method.21 Eight antibiotics of different generations were used to examine the level of antibiotic resistance against Salmonella typhi. These antibiotics were selected through the review of previously published articles22,23 and with the consultation of physicians.
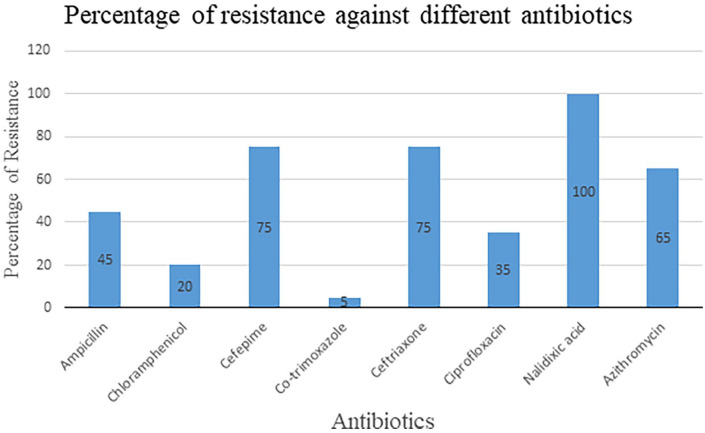
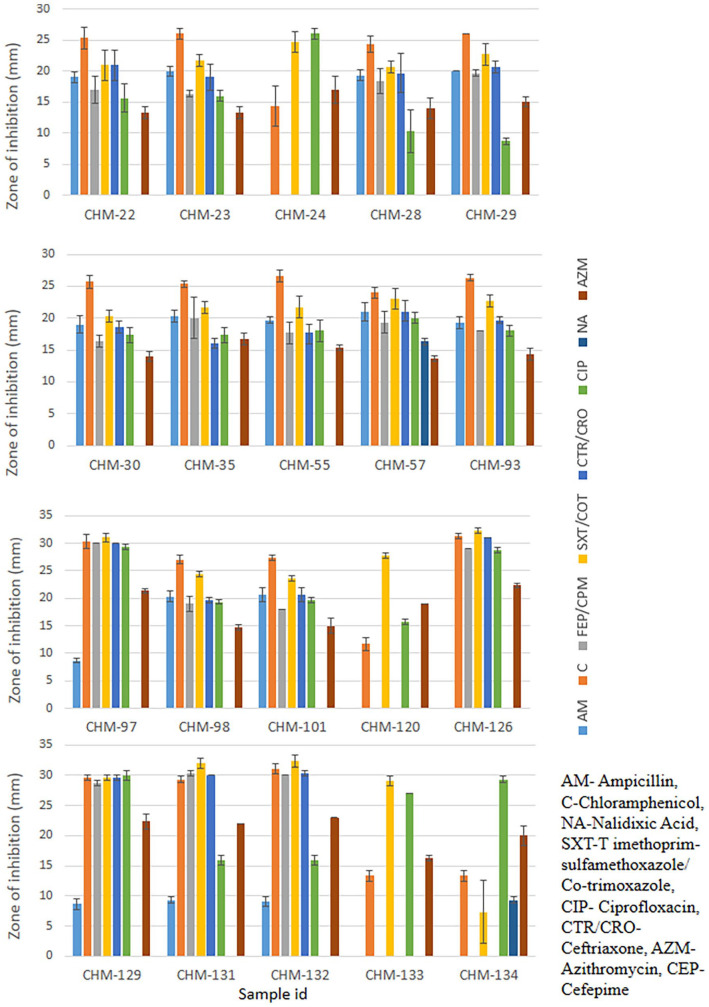
Among the 20 isolates, 85% (n = 17) were found to be multi-drug resistant (Figure 4). These multi-drug resistant (MDR) isolates displayed resistance to more than 2 antibiotics. Another 15% (n = 3) of isolates, including CHM-97, CHM-120, and CHM-129, were classified as double drug-resistant (DDR) because they were resistant to 2 different antibiotics (Figure 3). Among the 8 different antibiotics, Salmonella typhi isolates were shown to have maximum resistance to Nalidixic acid (Figure 3).
Figure 4.
Percentage of resistant Salmonella typhi against each selected antibiotic.
Figure 3.
Antibiotic resistant profiling of isolated Salmonella typhi. Antibiotic sensitivity test by Kirby-Bauer Disk diffusion method. Measurement of zone of inhibition by different Salmonella typhi isolates against selected antibiotics. The error bars represent data from 3 independent experiments (mean ± standard deviation).
Discussion
Salmonella typhi, a causative agent of typhoid fever, is considered the predominant infectious pathogen in South and Southeast Asia.24 Typhoid fever remains a significant public health concern, contributing to the severe economic burden through the costs associated with the management of the disease.25 The emergence of multidrug-resistant Salmonella typhi has had a great impact on the efficacy of antibiotic treatment, thus resulting in an increased number of deaths among typhoid patients. In Bangladesh, a majority of people take antibiotics without the prescription of a physician when they suffer from fever, diarrhea, cold, and other complications. Moreover, most physicians in Bangladesh prescribe antibiotics without an accurate diagnosis and antibiotic sensitivity test for fever, diarrhea, and other infectious diseases. As a result, a large portion of patients do not recover from the first, second, and even third dosage of antibiotics. Such types of misuse, overuse, and in-appropriate use of antimicrobial agents are the major risk factors for the development of antimicrobial resistance in Salmonella typhi. This scenario is very common in Chattogram City, Bangladesh.
This study revealed that the PCR identification method is more effective for accurate identification of Salmonella typhi among suspected typhoid patients. For instance, in our study,23 isolates were found as Salmonella typhi by selective plating and biochemical analysis but PCR analysis revealed that out of 23 isolates 21 were Salmonella typhi as fliC gene was not available in 2 isolates. This result exhibits the limitations of culture-based diagnostic methods. Therefore, it is essential to develop some effective method that will help the rapid diagnosis of the bacteria. If it exists, a physician might be able to determine the exact condition of infection to suggest appropriate treatment. Another alarming finding of this study is the presence of other organisms such as E. coli, Salmonella paratyphi A, Shigella spp., and Pseudomonas aeruginosa in typhoid suspected patients, which might be a subject of severe concern in near future.
The previous study reported that the prevalence of Salmonella typhi infection in Bangladesh was estimated at 14% between 2014 and 2015.26 In this study, the prevalence of Salmonella typhi in Chattogram was identified as13.64% (n = 21) Figure 1. The high infection rate in Chattogram, Bangladesh could be due to the drinking of contaminated water, food, and the unhygienic environment. If this situation continues, typhoid will become worse in the near future. Therefore, it is necessary to take some effective measures to minimize the infection rate in Bangladesh.
Although, no evidence supports the sex-specific preference for typhoid from the previous study. In this study, we found that the females were more susceptible to typhoid. As 67% of patients were identified as female and the rest of 33% were male Figure 2. This result suggests that there is a significant level of sex-specific variation in illness with Salmonella typhi. More research can show whether there are any factors responsible for this sex-specific vulnerability of typhoid patients.
Patients age is always an important issue in any microbial infection. People of all ages are susceptible to typhoid. However, children under 5 years of age are considered more susceptible.27 From a previous study, we found that the infection rate among young children (age ⩽17 years) was 80.7% between 2014 and 2015.28 In another study, 45.8% of the patients were identified as children under age 5.29 In this study, it was found that 62% of the typhoid patients (n = 13) in Chattrogram are children under the age of 5. Moreover, 91% (n = 19) of patients infected by Salmonella typhi were observed under the age of 20 years Figure 2. These results suggest that the prevalence of multidrug-resistant Salmonella typhi has become more dangerous, especially for children. This occurrence is because young children usually suffer from malnutrition that may enhance susceptibility to typhoid infection by alterations in the intestinal flora or other host defenses. Moreover, other risk factors for children may be their unhygienic habits and dependence on food from adults, carrying multidrug-resistant typhoid strains.8
This study revealed that a significant number (85%) of Salmonella typhi are multi-drug resistant and the resistance pattern is changing day by day in Chattogram, Bangladesh. Usually, ampicillin, chloramphenicol, and co-trimoxazole are considered ineffective for curing typhoid in Bangladesh. However, this study revealed that those antibiotics which were almost ineffective against Salmonella typhi at previous time30 now showed significant effectiveness against Salmonella typhi (Figures 3 and 4). There have been some reports regarding this similar occurrence in other countries.7, 31-33 So, what is the reason behind these sudden changes in the resistance pattern? The removal of selective pressure against those antibiotics might be the cause of there-emergence of sensitivity.31 However, the exact mechanism behind this hypothesis is still unknown.
Conclusion
This study has revealed the limitation of typhoid diagnosis in Bangladesh and also the misuse of antibiotics in its treatment. The conventional culture method takes 4 to 5 days to detect typhoid. This constraint encourages physicians to prescribe broad-spectrum antibiotics prior to a specific diagnosis of typhoid. Furthermore, most typhoid patients, particularly low-and middle-income people, are accustomed to taking antibiotics before consultation with a physician. Therefore, it takes more than a week from the time of infection until the typhoid diagnosis is made. Such activities increase the suffering of the patients as well as make the treatment of typhoid more complicated. Using a quick detection method like PCR, these problems can be fixed. Early detection of typhoid will help to decrease the sufferings of the patients as well as reduce the misuse of antibiotics. We have also found that antibiotics from the first generation, like co-trimoxazile, chloramphenicol, and ampicillin, are now effective against Salmonella typhi. So, it may be possible to reuse these antibiotics for the treatment of typhoid in Bangladesh.
Acknowledgments
The authors are grateful to the authority of Chittagong Medical College Hospital, Popular diagnostic Ltd., Lancet Diagnostic and Research Center, and Chevron Clinical Laboratory Pte. Ltd. located in Chittagong, Bangladesh for giving permission to collect clinical samples. Authors also thank to patients for giving consent to collect samples. The authors also feel gratitude to the authority of Genetic Engineering and Biotechnology lab and HEQUP lab of Biological Science, University of Chittagong for providing lab facilities.
Footnotes
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was financially supported by planning and development office of Chittagong University, Grant Ref 166/P&D/7285(18)/2019.
Author Contributions: AMMAC and SAM designed this study. SAM manage the fund and administrated the project. MZH, SAM, and AKMZH performed the experiments. AB and MRM helped in sample collection and processing. AMMAC and SAM analyzed the data. SAM and MZH wrote the original draft. AMMAC and SAM reviewed the several versions of the manuscript. All authors read and approved the final manuscript
Ethical Approval: Ethical approval was obtained from the ethical review committee of Chittagong Medical College, Bangladesh. The reference number of the ethical approval letter was CMC/PG/2020/89.
References
- 1. Nabi AQ. Molecular study on some antibiotic resistant genes in Salmonella spp. isolates. AIP Conf Proc. 2017;1888:020037. [Google Scholar]
- 2. Parry CM, Ribeiro I, Walia K, et al. Multidrug resistant enteric fever in South Asia: unmet medical needs and opportunities. BMJ. 2019;22:364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ugboko H, De N. Mechanisms of antibiotic resistance in Salmonella typhi. Int J CurrMicrobiol App Sci. 2014;3(12):461-476. [Google Scholar]
- 4. Saha S, Tanmoy AM, Andrews JR, et al. Evaluating PCR-based detection of Salmonella Typhi and Paratyphi A in the environment as an enteric fever surveillance tool. Am J Trop Med Hyg 2019;100:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mogasale V, Maskery B, Ochiai RL, et al. Burden of typhoid fever in low-income and middle-income countries: a systematic, literature-based update with risk-factor adjustment. Lancet Glob Health 2014;2:e570-e580. [DOI] [PubMed] [Google Scholar]
- 6. Chau TT, Campbell JI, Galindo CM, et al. Antimicrobial drug resistance of SalmonellaentericaserovarTyphi in Asia and molecular mechanism of reduced susceptibility to the fluoroquinolones. Antimicro Agents Chemother. 2007;51:4315-4323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rashmi S, Chaman LS, Bhuvneshwar K. Antibacterial resistance: current problems and possible solutions. Indian J Med Sci. 2005;59:120-129. [PubMed] [Google Scholar]
- 8. Zaki SA, Karande S. Multidrug-resistant typhoid fever: a review. J Infect Dev Ctries. 2011;5:324-337. [DOI] [PubMed] [Google Scholar]
- 9. Tunger O, Karakaya Y, Cetin CB, et al. Rational antibiotic use. J Infect Dev Ctries. 2009;3: 88-93. [DOI] [PubMed] [Google Scholar]
- 10. Majowicz SE, Musto J, Scallan E, et al. International collaboration on enteric disease “burden of illness” studies. The global burden of non-typhoidal Salmonella gastroenteritis. Clin Infect Dis. 2010;50:882-889. [DOI] [PubMed] [Google Scholar]
- 11. Eng SK, Pusparajah P, Ab Mutalib NS, et al. Salmonella: a review on pathogenesis, epidemiology and antibiotic resistance. Front Life Sci. 2015;8:284-293. [Google Scholar]
- 12. Blair JM, Webber MA, Baylay AJ, Ogbolu DO, Piddock LJ. Molecular mechanisms of antibiotic resistance. Nat Rev Microbiol. 2015;13:42-51. [DOI] [PubMed] [Google Scholar]
- 13. Haque A, Haque A, Sarwar Y, et al. Identification of drug resistance genes in clinical isolates of salmonella typhi for development of diagnostic multiplex PCR. Pak J Med Sci. 2005;21:402-407. [Google Scholar]
- 14. Munir T, Lodhi M, Ali S, et al. Early diagnosis of typhoid by PCR for fliC-d gene of Salmonella Typhi in patients taking antibiotics. J Coll Physicians Surg Pak. 2015;25:662-666. [PubMed] [Google Scholar]
- 15. Zhou L, Pollard AJ. A fast and highly sensitive blood culture PCR method for clinical detection of Salmonella entericaserovartyphi. Ann Clin Microbiol. 2010;9:1-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Al-Blooshi SY, Latif MA, Sabaneh NK, et al. Development of a novel selective medium for culture of Gram-negative bacteria. BMC Res Notes. 2021;14:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Wanger A, Chavez V, Huang RS, et al. Media for the clinical microbiology laboratory. microbiology and molecular diagnosis in pathology. In: Microbiology and Molecular Diagnosis in Pathology. Elsevier; 2017:51-60. [Google Scholar]
- 18. Chowdhury AM, Akter S, Mina SA. Isolation, identification and functional characterization of Escherichia coli as probiotic against Shigella in Bangladesh. Indian J Microbiol Res. 2020;7:313-321. [Google Scholar]
- 19. Akter S, Chowdhury AM, Mina SA. Antibiotic resistance and plasmid profiling of Escherichia coli isolated from human sewage samples. Microbiol Insights. 2021;14:1-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Massi MN, Gotoh A, Bishnu A, et al. Rapid diagnosis of typhoid fever by PCR assay using one pair of primers from flagellin gene of Salmonella typhi.J. Infect. Chemother. 2003;9:233-237. [DOI] [PubMed] [Google Scholar]
- 21. Bauer AW. Antibiotic susceptibility testing by a standardized single disc method. Am J ClinPathol. 1966;45:149-158. [PubMed] [Google Scholar]
- 22. Akinyemi KO, Ajoseh SO, Iwalokun BA, et al. Antimicrobial resistance and plasmid profiles of Salmonella entericaserovars from different sources in Lagos, Nigeria. Health. 2018;10:758-772. [Google Scholar]
- 23. Djeghout B, Saha S, Sajib MS, et al. Ceftriaxone-resistant Salmonella Typhi carries an IncI1-ST31 plasmid encoding CTX-M-15. J Med Microbiol. 2018;67:620-627. [DOI] [PubMed] [Google Scholar]
- 24. Deen J, Von Seidlein L, Andersen F, et al. Community-acquired bacterial bloodstream infections in developing countries in south and southeast Asia: a systematic review. Lancet Infect Dis. 2012;12(6):480-487. [DOI] [PubMed] [Google Scholar]
- 25. Crump JA, Luby SP, Mintz ED. The global burden of typhoid fever. Bull. World Health Organ. 2004;82:346-353. [PMC free article] [PubMed] [Google Scholar]
- 26. Pouzol S, Tanmoy AM, Ahmed D, et al. Clinical evaluation of a multiplex PCR for the detection of Salmonella entericaserovarsTyphi and Paratyphi A from blood specimens in a high-endemic setting. Am J Trop Med Hyg. 2019;101:513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Naheed A, Ram PK, Brooks WA, et al. Burden of typhoid and paratyphoid fever in a densely populated urban community, Dhaka, Bangladesh. Int J Infect Dis. 2010;14:e93-e99. [DOI] [PubMed] [Google Scholar]
- 28. Antillon M, Saad NJ, Baker S, Pollard AJ, Pitzer VE. The relationship between blood sample volume and diagnostic sensitivity of blood culture for typhoid and paratyphoid fever: a systematic review and meta-analysis. J Infect Dis. 2018;218:S255-S267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Khanam F, Sayeed MA, Choudhury FK, et al. Typhoid fever in young children in Bangladesh: clinical findings, antibiotic susceptibility pattern and immune responses. PLoS Negl Trop Dis. 2015;9:e0003619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Parvin A, Hasan MM, Talukder KA, et al. Extended-spectrum β-lactamase-producing Salmonella species isolated from diarrhoeal patients in Bangladesh: characterization and their dissemination through conjugation. Br Microbiol Res J. 2015:6:41-53. [Google Scholar]
- 31. Gupta V, Kaur J, Kaistha N. Re-emerging chloramphenicol sensitivity and emerging low-level ciprofloxacin resistance among Salmonella enterica serotype typhi isolates in North India. Trop Doc. 2009;39:28-30. [DOI] [PubMed] [Google Scholar]
- 32. Prajapati B, Rai GK, Rai SK, et al. Prevalence of Salmonella typhi and paratyphi infection in children: a hospital-based study. Nepal Med Coll J. 2008;10:238-241. [PubMed] [Google Scholar]
- 33. Wasfy MO, Frenck R, Ismail TF, et al. Trends of multiple-drug resistance among Salmonella serotype Typhi isolates during a 14-year period in Egypt. Clin Infect Dis. 2002;35:1265-1268. [DOI] [PubMed] [Google Scholar]