Abstract
A Shiga toxin (Stx)-encoding temperate bacteriophage of Shigella sonnei strain CB7888 was investigated for its morphology, DNA similarity, host range, and lysogenization in Shigella and Escherichia coli strains. Phage 7888 formed plaques on a broad spectrum of Shigella strains belonging to different species and serotypes, including Stx-producing Shigella dysenteriae type 1. With E. coli, only strains with rough lipopolysaccharide were sensitive to this phage. The phage integrated into the genome of nontoxigenic S. sonnei and laboratory E. coli K-12 strains, which became Stx positive upon lysogenization. Moreover, phage 7888 is capable of transducing chromosomal genes in E. coli K-12. The relationships of phage 7888 with the E. coli Stx1-producing phage H-19B and the E. coli Stx2-producing phage 933W were investigated by DNA cross-hybridization of phage genomes and by nucleotide sequencing of an 8,053-bp DNA region of the phage 7888 genome flanking the stx genes. By these methods, a high similarity was found between phages 7888 and 933W. Much less similarity was found between phages H-19B and 7888. As in the other Stx phages, a regulatory region involved in Q-dependent expression is found upstream of stxA and stxB (stx gene) in phage 7888. The morphology of phage 7888 was similar to that of phage 933W, which shows a hexagonal head and a short tail. Our findings demonstrate that stx genes are naturally transferable and are expressed in strains of S. sonnei, which points to the continuous evolution of human-pathogenic Shigella by horizontal gene transfer.
The production of Shiga toxins, also called verocytotoxins, is associated with strains of Shigella dysenteriae type 1 and with certain serotypes of Escherichia coli, such as enterohemorrhagic E. coli O157:H7 and others (22, 29). S. dysenteriae type 1 produces the prototypical Shiga toxin (Stx), whereas different variants of Stx, designated Stx1 and Stx2, are produced by Shiga toxin-producing E. coli (STEC) strains. In E. coli, the genes encoding the different Stx types are frequently found associated with temperate lambdoid bacteriophages, which are integrated in the chromosome of the host bacterium. Shiga toxin-converting bacteriophages can multiply and lyse their bacterial host upon induction and are able to infect Stx-negative E. coli strains, which can thus be converted into STEC. At present, more than 200 different serotypes of STEC have been described (22).
Bacteriophage-specific DNA sequences are also found in the neighborhood of the stxA and stxB genes (stx gene) in the chromosomes of S. dysenteriae type 1 strains. However, intact Stx-converting phages are not produced by S. dysenteriae type 1, which is probably due to the loss of essential phage genes caused by transposition and recombination events (17, 37).
Other enterobacteria, such as Citrobacter freundii and Enterobacter cloacae, were occasionally found to be Stx producers (25, 32, 36). However, the stx genes are not stably inherited in these bacteria, which is also the case with certain strains of E. coli (13, 25, 32).
A human isolate of Shigella sonnei (strain CB7888) which carries a bacteriophage bearing the gene for production of Shiga toxin was described recently (9). Nucleotide sequence analysis revealed a high similarity between the stx gene of the S. sonnei phage (EMBL accession no. AJ132761) and a published stx sequence of S. dysenteriae type 1 (EMBL accession no. M19437), suggesting that the S. sonnei phage might have acquired its stx gene from an S. dysenteriae type 1 strain. In order to gather more information about the origin of Shiga toxin-producing phage 7888 of S. sonnei, we examined its host range and integration in Shigella and E. coli and studied its relationship to the Stx1- and Stx2-encoding phages of E. coli.
MATERIALS AND METHODS
Bacteria.
The bacterial strains which were used as sources of phages and for transduction experiments are listed in Table 1. The reference strains for the different subgroups and serotypes of Shigella and E. coli are described elsewhere (4, 24).
TABLE 1.
Bacterial strains
| Strain | Species | Relevant characteristic(s) | Reference or source |
|---|---|---|---|
| CB7888 | S. sonnei | Lysogenic for Stx phage 7888 | 9 |
| C600 | E. coli K-12 | Stx negative, Q gene negative, O rough; Lac− Trp+ His+ | 30 |
| HB101 | E. coli K-12 | O rough, Stx negative, Q gene negative | 30 |
| P400 | E. coli K-12 | O rough, Stx negative, Q gene negative | P. Manninga |
| JC3272 | E. coli K-12 | O rough, Stx negative, Q gene negative; Lac− Lys− Trp− His− | 6 |
| C600(H19) | E. coli K-12 | C600 lysogenic for Stx1 phage H-19B | 8 |
| C600(933W) | E. coli K-12 | C600 lysogenic for Stx2 phage 933W | 8 |
| 737/85 | S. sonnei | Stx negative, Q gene negative | Human isolateb |
| 508/75 | S. sonnei | Stx negative, Q gene negative | Human isolateb |
University of Adelaide, Adelaide, Australia.
Collection of the Robert Koch-Institut, Berlin, Germany.
Detection of Shiga toxins and phages bearing Q and Rz genes.
Production of Stx was investigated with the Vero cell test and the VTEC-RPLA test (10). stx1- and stx2-specific DNA sequences were detected by PCR as described previously (9). PCRs for the Q gene and the Rz gene were developed on the basis of published nucleotide sequences (GenBank accession no. AF125520, AP000363, AF034975, LO4539, and Y10775) using McVector software (Oxford Molecular Group). The oligonucleotides Q-up (5′ GTC CTG TGA CGA TGA TGC GAT C 3′) and Q-down (5′ ATG CCT TCA ACA ATC CCC TCC G 3′) were selected as common primers for the amplification of Q genes present on different Shiga toxin-encoding bacteriophages. The PCR was run for 30 cycles (1 min at 94°C, 1 min at 57°C, and 1 min at 72°C for each cycle) and yielded a 195-bp amplification product. Primers RZ-up (5′ GCT GAA AAT GAA ACT CTT CGC 3′) and RZ-down (5′ GCT CTC TGA GGG TGA AAT AAT CC 3′) were used for amplification of a 172-bp internal segment of the phage Rz gene. The PCR conditions were the same as for the Q-gene-specific PCR.
Isolation of Stx-encoding phages and preparation of phage lysates.
A single colony of S. sonnei strain CB7888 was grown in L broth to the exponential growth phase. For induction of lysogenic phages, mitomycin C was added to a final concentration of 0.5 μg/ml and the culture was further incubated for 3 h. The bacteria were then harvested by centrifugation, and the culture fluid was filtered through 0.22-μm-pore-size membranes (Schleicher and Schüll, Dassel, Germany). Dilutions of the supernatant were titrated on E. coli K-12 strain C600 and on the Stx-negative S. sonnei strain 737/85. Single plaques grown on either strain were used for preparation of high-titer phage stocks. High-titer lysates (1010 PFU/ml) of phage 7888 were grown on E. coli K-12 strain C600 on agarose plates as described previously (30). Stx1 (H-19B)- and Stx2 (933W)-encoding E. coli phages were isolated by mitomycin C induction from the lysogenic host strains C600(H19) and C600(933W), respectively (7). Phage H-19B and 933W lysates were propagated on E. coli C600 as described above.
Electron microscopy.
Phages were isolated in CsCl step gradients according to standard protocols (30). Phage suspensions were prepared by negative staining with 1% uranyl acetate on carbon films according to the method of Steven et al. (34) and examined using a Philips CM100 transmission electron microscope. Phage DNA was prepared by the microdiffusion technique for size determination as previously described (15).
Isolation of phage DNA.
Phage lysates were incubated with 1 μg of DNase (Qiagen, Hilden, Germany) per ml and 1 μl of RNase C (Qiagen) per ml for 30 min at 37°C. Phage DNA was prepared with a Qiagen lambda kit according to the instructions of the manufacturer.
Phage 7888 sensitivity test.
Cultures of E. coli and Shigella strains were grown overnight in L broth at 37°C. The next day, a 100-μl overnight culture of a bacterial test strain was added to 3 ml of molten LC-top agar (6) and the mixture was poured on Luria-Bertani agar plates. Sensitivity to phage 7888 was tested by spotting 10 μl of phage 7888 lysate (108 PFU/ml) onto the inoculated top agar overlay. The plates were then incubated for 22 h at 37°C. Lysis of bacteria was recorded after 5 and 22 h of incubation time.
Lysogenization of E. coli and S. sonnei with phage 7888.
Ten-microliter portions of diluted phage 7888 lysates were spotted on Luria-Bertani plates covered with 3 ml of LC-top agar containing 100 μl of an overnight culture of nonverotoxigenic Shigella or E. coli strains, which were negative for stx- and Q-gene-specific DNA sequences. The plates were incubated for 22 h at 37°C, and bacteria which had grown within the lysis zones were further subcultured to single colonies. The bacterial isolates were examined for verocytotoxicity and for stx-specific DNA sequences as described previously (9). stx-positive colonies were confirmed for the specific phenotype of the recipient host strain and for the presence of temperate phage 7888 by Q gene PCR and by induction of infectious phages as described above.
Generalized transduction experiments.
Bacteriophage P1vir was from the collection of the Robert Koch-Institut. Transduction of chromosomal DNA using phages P1vir and 7888 was performed as described previously (19). Phage 7888 lysates grown on E. coli strain C600 (Trp+ His+) were used to transduce E. coli K-12 strain JC3272, which carries mutations in the chromosomal genes involved in the synthesis of tryptophan (Trp) and histidine (His) (6). JC3272 requires exogenous lysine, histidine, and tryptophan for growth, and Trp+ and His+ transductants were selected on M9 medium containing glucose, vitamin B1, and lysine but lacking tryptophan or histidine, respectively. His+ and Trp+ transductant colonies were confirmed as JC3272 derivatives by their phenotypical properties and were examined for cotransduction of the phage 7888-associated stx and Q genes by PCR as described above.
Hybridization of XbaI-digested total bacterial DNA separated by PFGE.
Preparation of total DNA, XbaI digestion, and pulsed-field gel electrophoresis (PFGE) were performed as described before (8). Southern blots of XbaI-digested total bacterial DNA were hybridized with HincII-digested digoxigenin-11–dUTP-labeled phage 7888 DNA or with an stx- and stx1-specific gene probe which was prepared from PCR-amplified DNA from strain C600(H19) as described previously (8).
Cross-hybridization of phage genomes.
Restriction endonuclease-digested phage DNA was separated in 0.8% agarose gels and transferred to Hybond N+ membranes (Amersham Pharmacia Biotech, Freiburg, Germany) by capillary blotting. Hybridization was performed at 65°C with a mixture of 5× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 0.1% sodium dodecyl sulfate (SDS), 0.5% blocking reagent, and 5% dextran sulfate. Washing steps were done at 65°C with 2× SSC–0.1% SDS and then with 0.2× SSC–0.1% SDS.
Nucleotide sequencing.
Small PCR products of several hundred base pairs were purified with a QIAquick PCR purification kit (Qiagen) and used directly for sequencing. Larger PCR products were cloned using the pGEM-T Easy vector system (Promega, Madison, Wis.) and transformed into E. coli JM109. Plasmids were isolated using a NucleoSpin Plus plasmid miniprep kit (Clontech Laboratories, Inc., Palo Alto, Calif.). Nucleotide sequencing was carried out with universal primers for pBluescript SK+ and commercially synthesized primers. Sequencing reactions were carried out by the dideoxynucleotide termination cycle sequencing method using a Prism BigDye terminator cycle sequencing ready reaction kit (Perkin-Elmer/Applied Biosystems, Weiterstadt, Germany). Sequencing products were run on an automated DNA sequencer (ABI 377). The sequences were analyzed with Auto Assembler software (Perkin-Elmer/Applied Biosystems).
Nucleotide sequence accession number.
The 8,053-bp nucleotide sequence of phage 7888 has been submitted to the EMBL data library under accession no. AJ279086 and includes the nucleotide sequences of the stxA and stxB genes, which were deposited previously under EMBL accession no. AJ132761.
RESULTS
Morphology of the phage 7888 virion and genome size.
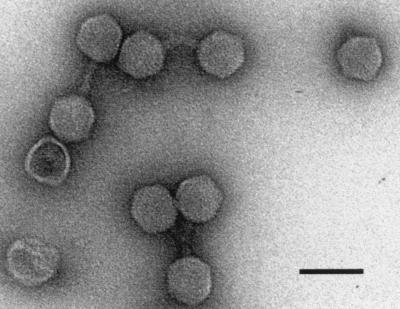
Phage 7888 was isolated from the Stx-producing S. sonnei strain CB7888, which was from feces from a human patient with diarrhea (9). Phages released from S. sonnei CB7888 after mitomycin C induction were purified in a CsCl step gradient and examined by transmission electron microscopy. Electron micrographs (Fig. 1) depicted phage particles with regular hexagonal heads about 65 nm wide. Phage 7888 particles showed short tails, approximately 26 nm long and 13 nm wide, which seemed to possess small base plates. As described for the stx2-carrying phage 933W, we also found aggregation of the phages due to interactions of the tail tips (26).
FIG. 1.
Electron micrograph of CsCl-purified phage 7888 particles. Bar, 100 nm.
The size of the phage 7888 genome was calculated to be 63.4 ± 2.4 kb from the sum of the restriction fragments obtained by endonuclease digestion of the complete phage DNA. Additionally, a size determination deduced from 47 genomes based on electron micrographs suggested a length of 64.2 ± 2.0 kb for the virion DNA.
Analysis of the stxA- and stxB (stx gene)-flanking region of phage 7888.
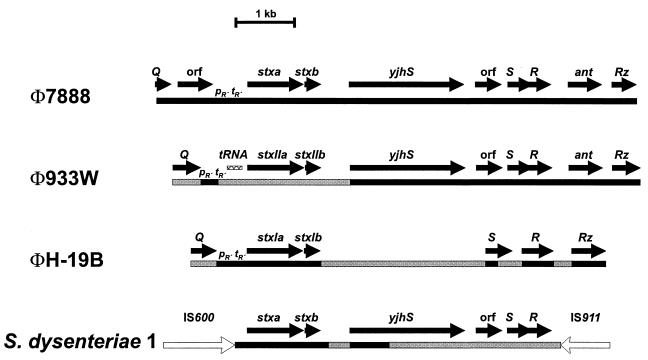
The stx gene of phage 7888 was described previously (EMBL accession number AJ132761) (9). In the present work, the region of phage 7888 flanking the stx coding region was analyzed by sequencing PCR products obtained by partial amplification of phage 7888 DNA as well as of genomic DNA of strain CB7888. PCR products containing the 5′ upstream region of the stx gene were obtained by using primer Q-up and an internal primer located in the stxA gene. The region downstream of the stx gene was amplified using a primer binding in the stxB gene and the RZ-down primer. The physical map of the sequenced part of the phage genome with a length of 8,053 bp is shown in Fig. 2. Downstream of the Q gene of phage 7888, an open reading frame (ORF) which may encode a putative protein with a deduced size of 197 amino acids was identified. However, analysis of protein databases did not identify proteins with significant homology to the deduced gene product. The stx region is located downstream of this ORF and contains regulatory elements consisting of a promoter, pR′, and a terminator, tR′, involved in antitermination by the Q transcription activator, followed by the stxA and stxB genes. The region downstream of the stx gene contains a number of genes known from other Shiga toxin-producing phages in the order yjhS, S, R, ant, and Rz.
FIG. 2.
Structures of Shiga toxin genes and flanking regions of phages 7888, 933W, and H-19B and of S. dysenteriae type 1. Black-shaded bars in the diagrams of 933W, H-19B, and S. dysenteriae indicate a DNA similarity of more than 95% to the corresponding region of phage 7888, while grey-shaded regions indicate a similarity of less than 95%. White arrows in the diagram of S. dysenteriae represent insertion elements IS600 and IS911. pR′ tR′, regulatory region upstream of Shiga toxin genes.
Relationship of phage 7888 to Stx-converting coliphages H-19B and 933W.
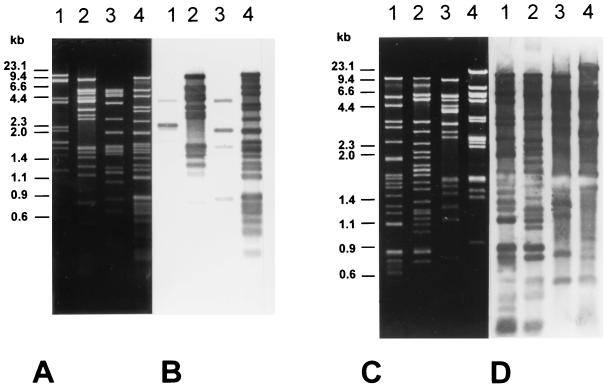
To assess the relationship of S. sonnei phage 7888 to other characterized Stx-converting phages, Stx2-converting phage 933W and Stx1-converting phage H-19B were chosen for comparison. All three phage genomes were subjected to restriction endonuclease digestions and to cross-hybridization experiments. The HincII and AccI restriction patterns for all virion DNAs differed considerably (Fig. 3A and C).
FIG. 3.
Restriction patterns and Southern hybridization of DNA of phages H-19B, 933W, and 7888. (A) Agarose gel showing restriction enzyme-digested DNA of phages 7888 and H-19B. Lane 1, AccI digest of H-19B; lane 2, AccI digest of phage 7888; lane 3, HincII digest of phage H-19B; lane 4, HincII digest of phage 7888. (B) Southern blot of the gel shown in panel A after hybridization with labeled DNA of phage 7888. (C) Agarose gel showing restriction enzyme-digested DNAs of phages 7888 and 933W. Lane 1, HincII digest of phage 7888; lane 2, HincII digest of phage 933W; lane 3, AccI digest of phage 7888; lane 4, AccI digest of phage 933W. (D) Southern blot of the gel shown in panel C after hybridization with labeled DNA of phage 7888.
The hybridization experiments were carried out under stringent conditions using the complete genome of phage 7888 as the probe. The hybridization patterns obtained from Southern blots of HincII and AccI digests of the complete genomes of the Shiga toxin-converting phages are shown in Fig. 3B and D. Whereas hybridization signals between the DNAs of phages 7888 and H-19B were restricted to a few bands (Fig. 3B, lanes 1 and 3), the hybridization between the genomes of 933W and 7888 revealed a large pattern of hybridizing signals (Fig. 3D, lanes 2 and 4). The observed hybridization patterns clearly indicate a very close relationship between phages 7888 and 933W, while the relationship of phage 7888 to phage H-19B is far more distant.
Host range of phage 7888.
Reference strains of S. dysenteriae (n = 12), Shigella boydii (n = 19), Shigella flexneri (n = 31), and S. sonnei (n = 7) belonging to different serotypes were tested for their sensitivities to phage 7888 (Table 2). The phage formed plaques on a wide range of Shigella strains belonging to different groups and serotypes. With E. coli, only O-rough strains (E. coli K-12 and O-rough wild-type strains) were sensitive to phage 7888. Wild-type strains of E. coli belonging to different O serogroups, including strains belonging to O groups O1, O53, O79, O87, O129, which are serologically related to certain O groups of Shigella (4), did not form plaques with the phage.
TABLE 2.
Host range of phage 7888
| Species | Serotype(s) of strains
|
|
|---|---|---|
| Sensitive to phage 7888 | Resistant to phage 7888 | |
| S. dysenteriae | 1, 4, 6, 7, 9 | 2, 3, 5, 8, 10, 11, 12 |
| S. boydii | 2, 3, 4, 5, 6, 7, 9, 17 | 1, 8, 10, 11, 12, 13, 14, 15, 16, 18 |
| S. flexneri | 1a, 1b, 2a, 2b, 3a, 3b, 4a, 4b, 5, 6 | 1a, 1b, 4a, 5a |
| S. sonnei | O rough and smooth | Strain CB7888b |
| E. coli | O roughc | O1, O53, O55, O79, O87, O129, O157 |
Some S. flexneri 1a, 1b, 4a, and 5 strains were resistant to phage 7888.
Source of phage 7888.
E. coli K-12 strains C600 and P400, E. coli HB101, and E. coli O-rough wild-type strains.
Lysogenization of Shigella with phage 7888.
Lysogenization of Shigella with phage 7888 was performed as described in Materials and Methods. Stable lysogens were obtained with S. sonnei strains 737/85 and 508/75. Phage 7888 lysogeny was demonstrated by the production of Stx and by a positive stx- and Q-gene PCR result. Infectious phages could be induced by mitomycin C treatment of lysogenized Shigella strains.
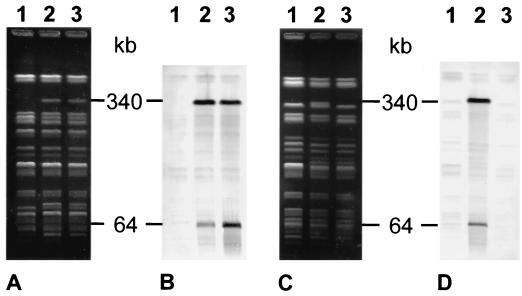
The integration of phage 7888 in the genomes of Stx-negative S. sonnei strains 737/85 and 508/75 resulted in changes in their PFGE patterns, which led in both strains to the generation of an approximately 340-kb-size XbaI fragment which hybridized with digoxigenin-labeled HincII-digested phage 7888 DNA (Fig. 4A, lanes 2 and 3, and C, lane 2). This band was not present in the original S. sonnei host strains 737/85 and 508/75, which were also negative for phage 7888-specific DNA sequences (Fig. 4A and C, lanes 1). In addition, a second hybridizing XbaI fragment corresponding to the size of the phage 7888 genome (64 kb) was detected in each of the two S. sonnei strains which were lysogenized with phage 7888 (Fig. 4B, lanes 2 and 3, and D, lane 2).
FIG. 4.
Detection of prophage 7888 in the genomes of lysogenized strains of S. sonnei. (A) XbaI-digested total DNA of S. sonnei 737/85 and derivative strains. Lane 1, S. sonnei 737/85 (Stx negative); lanes 2 and 3, two derivatives of S. sonnei 737/85 lysogenized with phage 7888 (Stx positive). (B) Southern blot of the PFGE gel shown in panel A after hybridization with labeled DNA of phage 7888 as the DNA probe. The positions and sizes of the hybridizing fragments are indicated in kilobases. (C) XbaI-digested total DNA of S. sonnei 508/75 and derivative strains. Lane 1, S. sonnei 508/75 (Stx negative); lane 2, S. sonnei 508/75 carrying phage 7888 (Stx positive); lane 3, a derivative of an S. sonnei 508/75 transductant which has spontaneously lost phage 7888 (Stx negative). (D) Southern blot of the PFGE gel shown in panel C after hybridization with labeled DNA of phage 7888 as the DNA probe. The positions and sizes of the hybridizing fragments are indicated in kilobases.
Lysogenization and transduction of E. coli chromosomal genes by phage 7888.
Phage 7888 lysates grown on E. coli C600 (His+ Trp+) were used for generalized transduction with E. coli strain JC3272 (His− and Trp−) as the recipient. His+ and Trp+ recombinants of JC3272 were obtained according to the selection. The numbers of JC3272 His+ and Trp+ recombinants obtained were proportional to the amounts of phage lysates used for the transduction assays. We calculated that approximately 0.002% of the phage particles carried chromosomal DNA. Cotransduction of His+ and Trp+ was not observed with any of the His+ (n = 174) or Trp+ (n = 231) recombinants. However, 50 to 80% of the His+ and Trp+ transductional recombinants of JC3272 also became positive for production of Stx and were shown to carry stx- and Q-gene-specific DNA sequences. Intact phage 7888 could be isolated from these strains but not from Trp+ or His+ recombinants of strain JC3272, which were negative for Stx production and for the Q gene. In order to investigate whether the stx gene was genetically linked to the chromosomal genes encoding histidine and tryptophan, P1 lysates were made on Trp+ Stx+ and on His+ Stx+ JC3272 derivatives. The P1 lysates were used for transduction of JC3272 (His− Trp− to His+ and to Trp+ prototrophy, respectively). However, none of the resulting His+ or Trp+ recombinants were positive for stx- and Q-gene-specific DNA sequences, which indicates that the stx gene was not linked genetically to the chromosomal genes. This finding indicates that the Stx-positive JC3272 transductants were the result of double infections with wild-type and E. coli chromosomal markers transducing phage 7888 particles.
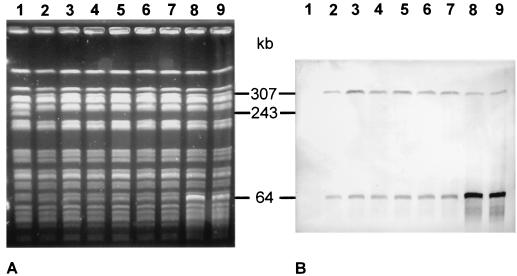
The integration of phage 7888 into the genome of E. coli JC3272 was investigated by PFGE and by DNA hybridization with an stx- and stx1-specific gene probe. Nine independently isolated phage 7888 lysogens from transductional experiments with JC3272 were analyzed for their XbaI patterns by PFGE. The lysogenic JC3272 isolates were identical in their XbaI patterns but showed differences in two bands from the original host strain, JC32372 (Fig. 5A). A 243-kb XbaI fragment present in JC3272 was replaced by a 307-kb fragment, which was present in all nine JC3272 lysogens but absent in the parental strain JC3272. The 307-kb XbaI fragment hybridized with the stx- and stx1-specific gene probe, which indicates that it was generated by integration of the phage 7888 genome (Fig. 5B). A second band 64 kb in size which hybridized with the stx- and stx1-specific gene probe was found in all nine lysogenic JC3272 derivatives (Fig. 5B). The 64-kb fragment might represent free phage DNA, because it was overexpressed in two JC3272 lysogenic strains which were were growing poorly and which spontaneously liberated free plaque-forming phages during growth (Fig. 5, lanes 8 and 9).
FIG. 5.
Integration of phage 7888 in the genome of E. coli K-12 strain JC3272. (A) XbaI-digested total DNA of JC3272 and lysogenized derivatives separated by PFGE. Lane 1, JC3272; lanes 2 to 9, independent isolates of JC3272 carrying prophage 7888. (B) Southern blot of the PFGE gel shown in panel A after hybridization with an stx- and stx1-specific DNA probe. The positions and sizes of the hybridizing fragments are indicated in kilobases.
Presence of stx genes in Shigella strains from human patients.
Patient isolates of S. sonnei (n = 149), S. flexneri (n = 118), S. boydii (n = 7), and S. dysenteriae (n = 8), including two S. dysenteriae type 1 strains, were investigated for carriage of stx- and stx1-specific DNA sequences by DNA hybridization. PCR-amplified DNA from strain C600(H19) was used as an stx- and stx1-specific gene probe (8). Of the 282 strains, only the two S. dysenteriae type 1 strains and S. sonnei CB7888 were found to be positive for stx.
DISCUSSION
Bacteriophages are increasingly recognized as vectors of the horizontal transfer of virulence determinants in many bacterial species (for reviews, see references 11 and 18). In Shigella, some virulence functions are associated with bacteriophages, such as the genes for O-antigen modification of S. flexneri (3) and the aerobactin genes in S. flexneri and S. boydii strains (3, 28). Recent studies have shown that the stx gene of S. dysenteriae type 1 is associated with bacteriophage-related genes, which indicates that the stx gene was initially introduced by integration of an Stx-encoding phage (17). Stx1- and Stx2-encoding lambdoid phages are frequently associated with STEC strains, and in contrast to S. dysenteriae type 1, viable Stx phages can be isolated from serologically different STEC strains (21, 33, 35). These phages can spread their stx genes to Stx-negative E. coli strains by transduction and lysogenization (2, 20, 31). These findings therefore suggest that bacteriophages have contributed to the evolution and genetic diversity of the STEC group by the horizontal spread of stx determinants in E. coli (2, 31).
The Stx-encoding S. sonnei phage 7888 showed a broad host range for many Shigella strains and types, which indicates that it is well adapted to Shigella as a natural host and may have acquired its stx gene from S. dysenteriae type 1. On the other hand, the comparison of S. sonnei phage 7888 to the well-characterized stx-carrying E. coli phages H-19B and 933W suggested a close relationship of phage 7888 and 933W. Electron micrographs of 7888 virions revealed a morphology similar to that of the Stx2-converting phage 933W. Phage 933W shows a head 70 nm in diameter and a tail 27 nm long and 13 nm wide (26). In contrast, the stx1-carrying H-19B virion has a considerably longer head and a flexible tail resembling those of phage lambda (39). The genome size of phage 7888 was found to be very similar to that of the 933W genome, i.e., 61,670 bp as determined by nucleotide sequence analysis (26).
The close genetic relationship between phages 933W and 7888 was demonstrated by DNA cross-hybridization analysis using the complete genome of phage 7888 as a gene probe (Fig. 3). In order to gain more information on the possible origin of Stx-encoding phage 7888 and on the genetic similarity between phages 7888 and 933W, we sequenced an 8,053-bp DNA region located between the Q and Rz genes of phage 7888. This part of the phage genome encompasses the stx gene and carries regulatory sites involved in the regulation of Shiga toxin expression (23, 26). By comparing this region to the corresponding regions of phages H-19B (AF034975) and 933W (AF125520), we found a regulatory site, pR′ tR′, located upstream of the stx gene which is involved in expression of stx genes by Q antitermination and is highly conserved (more than 95% identity) in all three phage genomes (Fig. 2). The presence of the PR′ tR′ site in the genome of phage 7888 suggests that the expression of the stx gene is also regulated by the action of the Q protein as a transcriptional activator, as was reported for phages H-19B and 933W (23, 38).
The largest stretch of DNA with sequence identity of >98% between phages 7888 and H-19B is a 1,895-bp region encompassing the stx and stx1 genes and adjacent sequences. In both phages, the Q-dependent regulatory region PR′ tR′ is located directly upstream of the stx or stx1 gene, respectively. In phage 933W, the PR′ tR′ region is separated by an approximately 300-bp stretch of tRNA genes from the adjacent stx2 gene. The tRNA genes are not present in the corresponding regions of phages H-19B and 7888.
More than 95% similarity is found between the genomes of phages 7888 and 933W in an approximately 4,800-bp DNA stretch downstream of the stx gene. The region of high DNA similarity starts shortly upstream of a 1,938-bp ORF encoding an analogue of the E. coli K-12 YjhS protein, followed by an unknown ORF and the genes S, R, and Rz. In contrast, in the corresponding region on the genome of phage H-19B, small stretches of high sequence similarity to phage 933W and 7888 are found only in the coding regions of the S, R, and Rz genes. The complete genomic sequence of another Stx1-encoding prophage (VT1-Sakai) from an enterohemorrhagic E. coli O157 outbreak in Japan has been reported (GenBank accession no. AP000400) (40), though no Stx1 phage particles could be isolated from the strain. The Q-to-Rz region, which includes the stx1 gene of this prophage, is very similar to that of phage 7888. As other parts of the genomic sequence of this prophage do not possess any DNA similarity to phage 933W, however, we suppose that the relationship to phage 7888 is minimal.
A specific feature of the Q to Rz region of phage 7888 is the presence of a 594-bp ORF of unknown function located downstream of the Q gene, which is absent in strains H-19B and 933W and in the VT1-Sakai prophage. Another difference concerns the 3′ coding region of the partially sequenced Q gene of phage 7888, which encodes a Q protein that is slightly different from the Q proteins of the H-19B and 933W phages; i.e., it has a carboxy-terminal region that is truncated by 18 amino acids.
A sequence comparison of the stx region of phage 7888 with the chromosomally located stx region of S. dysenteriae type 1 (AF153317), which are enclosed by IS600 and IS911 elements, revealed that the coding regions of the stxA and stxB genes as well as 187 bp 5′ upstream and 136 bp 3′ downstream of the stx gene are nearly identical (four nucleotide differences in a stretch of 1,550 bp). The stx gene of S. dysenteriae is likely to have been derived from an integrated prophage which became defective due to the integration of insertion sequence elements and rearrangements and thus resulted in a stable integration of the stx gene in the chromosome (17). The regulatory region for Q-dependent activation, however, is missing in S. dysenteriae. Downstream of the stx region, high sequence similarities (>98% identity) between phage 7888 and the S. dysenteriae stx region are found only in the 5′ coding region of yhjS genes.
The alignment of 22 published stx and stx1 sequences (http://www.ncbi.nlm.nih.gov/BLASTN) originating from different S. dysenteriae type 1 strains, from E. coli, and from S. sonnei phage 7888 revealed only a few base exchanges in the stxA and stxA1 genes which cannot be related to the source of the toxin genes, Shigella, or E. coli. It is, therefore, not possible to determine whether the stx gene present on the genome of phage 7888 was originally acquired from S. dysenteriae type 1 or from an E. coli phage. However, our findings indicate that phage 7888 belongs to the family of lambdoid 933W-like phages, which are known to have a mosaic structure due to frequent gene exchange by recombination (12, 14, 37). It is possible that these lambdoid phages also differ in their host ranges and that similar phages might be more adapted to Shigella, which is highly related to E. coli.
Our finding that the lambdoid phage 7888 can mediate generalized transduction of chromosomal genes in E. coli points to its general role in the evolution of bacteria. It is known that bacteriophage lambda can behave as a generalized transducer in E. coli (16). The uniform hybridization patterns found with independent isolates of JC3272 phage 7888 lysogens indicate that phage 7888, like lambda, has a specific integration site in the bacterial chromosome. Further work is necessary to analyze the attachment site of phage 7888 in E. coli and S. sonnei.
The detection of an stx gene on a phage of S. sonnei, which can transfer the Stx-encoding characteristic to nontoxigenic S. sonnei strains, points to the continuous evolution of Shigella by horizontal gene transfer. In our study, we could not detect stx or stx1 sequences in 280 Shigella strains other than S. dysenteriae type 1. However, older reports indicate that Stx production might occasionally occur in non-S. dysenteriae type 1 Shigella strains (5, 27). As S. sonnei isolates are not routinely examined for production of Shiga toxins in clinical laboratories, it is possible that Stx production in patient isolates of S. sonnei remains undetected in diagnosis. Among the different Shigella species, S. sonnei is the major isolate in the developed countries (1). In contrast to S. dysenteriae type 1, S. sonnei is distributed worldwide and the emergence of Stx-producing S. sonnei strains may contribute to an increase in the virulence of these widely occurring human pathogens.
ACKNOWLEDGMENTS
We thank Dorothea Knabner for technical assistance.
This work was supported by the German Federal Ministry of Health.
REFERENCES
- 1.Acheson D W, Keusch G T. Shigella and enteroinvasive Escherichia coli. In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press; 1995. pp. 763–784. [Google Scholar]
- 2.Acheson D W K, Reidl J, Zhang X, Keusch G T, Mekalanos J J, Waldor M K. In vivo transduction with shiga toxin 1-encoding phage. Infect Immun. 1998;66:4496–4498. doi: 10.1128/iai.66.9.4496-4498.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Allison G E, Verma N K. Serotype-converting bacteriophages and O-antigen modification in Shigella flexneri. Trends Microbiol. 2000;8:17–23. doi: 10.1016/s0966-842x(99)01646-7. [DOI] [PubMed] [Google Scholar]
- 4.Anonymous. The genus Shigella. In: Ewing W H, editor. Edwards and Ewing's identification of Enterobacteriaceae. 4th ed. New York, N.Y: Elsevier Science Publishing, Co., Inc.; 1986. pp. 135–172. [Google Scholar]
- 5.Bartlett A V, Prado D, Cleary T G, Pickering L K. Production of Shiga toxin and other cytotoxins by serogroups of Shigella. J Infect Dis. 1986;154:996–1002. doi: 10.1093/infdis/154.6.996. [DOI] [PubMed] [Google Scholar]
- 6.Beutin L, Achtman M. Two Escherichia coli chromosomal cistrons, sfrA and sfrB, which are needed for expression of F factor tra functions. J Bacteriol. 1979;139:730–737. doi: 10.1128/jb.139.3.730-737.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beutin L, Geier D, Steinrück H, Zimmermann S, Scheutz F. Prevalence and some properties of verotoxin (Shiga-like toxin)-producing Escherichia coli in seven different species of healthy domestic animals. J Clin Microbiol. 1993;31:2483–2488. doi: 10.1128/jcm.31.9.2483-2488.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Beutin L, Geier D, Zimmermann S, Aleksic S, Gillespie H A, Whittam T S. Epidemiological relatedness and clonal types of natural populations of Escherichia coli strains producing Shiga toxins in separate populations of cattle and sheep. Appl Environ Microbiol. 1997;63:2175–2180. doi: 10.1128/aem.63.6.2175-2180.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beutin L, Strauch E, Fischer I. Isolation of Shigella sonnei lysogenic for a bacteriophage encoding gene for production of Shiga toxin. Lancet. 1999;353:1498. doi: 10.1016/S0140-6736(99)00961-7. [DOI] [PubMed] [Google Scholar]
- 10.Beutin L, Zimmermann S, Gleier K. Rapid detection and isolation of shiga-like toxin (verocytotoxin)-producing Escherichia coli by direct testing of individual enterohemolytic colonies from washed sheep blood agar plates in the VTEC-RPLA assay. J Clin Microbiol. 1996;34:2812–2814. doi: 10.1128/jcm.34.11.2812-2814.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boyd E F, Davis B M, Hochhut B. Bacteriophage-bacteriophage interactions in the evolution of pathogenic bacteria. Trends Microbiol. 2001;9:137–144. doi: 10.1016/s0966-842x(01)01960-6. [DOI] [PubMed] [Google Scholar]
- 12.Johansen B K, Wasteson Y, Granum P E, Brynestad S. Mosaic structure of Shiga-toxin-2-encoding phages isolated from Escherichia coli O157:H7 indicates frequent gene exchange between lambdoid phage genomes. Microbiology. 2001;147:1929–1936. doi: 10.1099/00221287-147-7-1929. [DOI] [PubMed] [Google Scholar]
- 13.Karch H, Meyer T, Rüssmann H, Heesemann J. Frequent loss of Shiga-like toxin genes in clinical isolates of Escherichia coli upon subcultivation. Infect Immun. 1992;60:3464–3467. doi: 10.1128/iai.60.8.3464-3467.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karch H, Schmidt H, Janetzki-Mittmann C, Scheef J, Kroger M. Shiga toxins even when different are encoded at identical positions in the genomes of related temperate bacteriophages. Mol Gen Genet. 1999;262:600–607. doi: 10.1007/s004380051122. [DOI] [PubMed] [Google Scholar]
- 15.Lang D, Mitani M. Simplified quantitative electron microscopy of biopolymers. Biopolymers. 1970;9:373–379. doi: 10.1002/bip.1970.360090310. [DOI] [PubMed] [Google Scholar]
- 16.Masters M. Generalized transduction. In: Neidhardt F C, Curtiss III R, Ingraham J L, Lin E C C, Low K B, Magasanik B, Resnikoff W S, Riley M, Schaechter M, Umbarger H E, editors. Escherichia coli and Salmonella: cellular and molecular biology. 2nd ed. Washington, D.C.: ASM Press; 1996. pp. 2421–2448. [Google Scholar]
- 17.McDonough M A, Butterton J R. Spontaneous tandem amplification and deletion of the shiga toxin operon in Shigella dysenteriae 1. Mol Microbiol. 1999;34:1058–1069. doi: 10.1046/j.1365-2958.1999.01669.x. [DOI] [PubMed] [Google Scholar]
- 18.Miao E A, Miller S I. Bacteriophages in the evolution of pathogen-host interactions. Proc Natl Acad Sci USA. 1999;96:9452–9454. doi: 10.1073/pnas.96.17.9452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Miller J H. A short course in bacterial genetics. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1992. pp. 263–278. [Google Scholar]
- 20.Muniesa M, Jofre J. Abundance in sewage of bacteriophages that infect Escherichia coli O157:H7 and that carry the Shiga toxin 2 gene. Appl Environ Microbiol. 1998;64:2443–2448. doi: 10.1128/aem.64.7.2443-2448.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Muniesa M, Recktenwald J, Bielaszewska M, Karch H, Schmidt H. Characterization of a Shiga toxin 2e-converting bacteriophage from an Escherichia coli strain of human origin. Infect Immun. 2000;68:4850–4855. doi: 10.1128/iai.68.9.4850-4855.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nataro J P, Kaper J B. Diarrheagenic Escherichia coli. Clin Microbiol Rev. 1998;11:142–201. doi: 10.1128/cmr.11.1.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Neely M N, Friedman D I. Functional and genetic analysis of regulatory regions of coliphage H-19B: location of shiga-like toxin and lysis genes suggest a role for phage functions in toxin release. Mol Microbiol. 1998;28:1255–1267. doi: 10.1046/j.1365-2958.1998.00890.x. [DOI] [PubMed] [Google Scholar]
- 24.Orskov F, Orskov I. Serotyping of Escherichia coli. Methods Microbiol. 1984;14:43–112. [Google Scholar]
- 25.Paton A W, Paton J C. Enterobacter cloacae producing a Shiga-like toxin II-related cytotoxin associated with a case of hemolytic-uremic syndrome. J Clin Microbiol. 1996;34:463–465. doi: 10.1128/jcm.34.2.463-465.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Plunkett G, III, Rose D J, Durfee T J, Blattner F R. Sequence of Shiga toxin 2 phage 933W from Escherichia coli O157:H7: Shiga toxin as a phage late-gene product. J Bacteriol. 1999;181:1767–1778. doi: 10.1128/jb.181.6.1767-1778.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Prado D, Cleary T G, Pickering L K, Ericsson C D, Bartlett A V, DuPont H L, Johnson P C. The relation between production of cytotoxin and clinical features in shigellosis. J Infect Dis. 1986;154:149–155. doi: 10.1093/infdis/154.1.149. [DOI] [PubMed] [Google Scholar]
- 28.Purdy G E, Payne S M. The SHI-3 iron transport island of Shigella boydii 0-1392 carries the genes for aerobactin synthesis and transport. J Bacteriol. 2001;183:4176–4182. doi: 10.1128/JB.183.14.4176-4182.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Riley L W. Acute inflammatory diarrhea (dysentery) In: Blaser M J, Smith P D, Ravdin J I, Greenberg H B, Guerrant R L, editors. Infections of the gastrointestinal tract. New York, N.Y: Raven Press, Ltd.; 1995. pp. 283–298. [Google Scholar]
- 30.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 31.Schmidt H, Bielaszewska M, Karch H. Transduction of enteric Escherichia coli isolates with a derivative of Shiga toxin 2-encoding bacteriophage ψ3538 isolated from Escherichia coli O157:H7. Appl Environ Microbiol. 1999;65:3855–3861. doi: 10.1128/aem.65.9.3855-3861.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schmidt H, Montag M, Bockemühl J, Heesemann J, Karch H. Shiga-like toxin II-related cytotoxins in Citrobacter freundii strains from humans and beef samples. Infect Immun. 1993;61:534–543. doi: 10.1128/iai.61.2.534-543.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Smith H W, Green P, Parsell Z. Vero cell toxins in Escherichia coli and related bacteria: transfer by phage and conjugation and toxic action in laboratory animals, chickens and pigs. J Gen Microbiol. 1983;129:3121–3137. doi: 10.1099/00221287-129-10-3121. [DOI] [PubMed] [Google Scholar]
- 34.Steven A C, Trus B L, Maizel J V, Unser M, Parry D A, Wall J S, Hainfeld J F, Studier F W. Molecular substructure of a viral receptor-recognition protein. The gp17 tail-fiber of bacteriophage T7. J Mol Biol. 1988;200:351–365. doi: 10.1016/0022-2836(88)90246-x. [DOI] [PubMed] [Google Scholar]
- 35.Strockbine N A, Marques L R M, Newland J W, Smith H W, Holmes R K, O'Brien A D. Two toxin-converting phages from Escherichia coli O157:H7 strain 933 encode antigenically distinct toxins with similar biologic activities. Infect Immun. 1986;53:135–140. doi: 10.1128/iai.53.1.135-140.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tschape H, Prager R, Streckel W, Fruth A, Tietze E, Bohme G. Verotoxinogenic Citrobacter freundii associated with severe gastroenteritis and cases of haemolytic uraemic syndrome in a nursery school: green butter as the infection source. Epidemiol Infect. 1995;114:441–450. doi: 10.1017/s0950268800052158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Unkmeir A, Schmidt H. Structural analysis of phage-borne stx genes and their flanking sequences in Shiga toxin-producing Escherichia coli and Shigella dysenteriae type 1 strains. Infect Immun. 2000;68:4856–4864. doi: 10.1128/iai.68.9.4856-4864.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wagner P L, Neely M N, Zhang X, Acheson D W K, Waldor M K, Friedman D I. Role for a phage promoter in Shiga toxin 2 expression from a pathogenic Escherichia coli strain. J Bacteriol. 2001;183:2081–2085. doi: 10.1128/JB.183.6.2081-2085.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Willshaw G A, Smith H R, Scotland S M, Field A M, Rowe B. Heterogeneity of Escherichia coli phages encoding Vero cytotoxins: comparison of cloned sequences determining VT1 and VT2 and development of specific gene probes. J Gen Microbiol. 1987;133:1309–1317. doi: 10.1099/00221287-133-5-1309. [DOI] [PubMed] [Google Scholar]
- 40.Yokoyama K, Makino K, Kubota Y, Watanabe M, Kimura S, Yutsudo C H, Kurokawa K, Ishii K, Hattori M, Tatsuno I, Abe H, Yoh M, Iida T, Ohnishi M, Hayashi T, Yasunaga T, Honda T, Sasakawa C, Shinagawa H. Complete nucleotide sequence of the prophage VT1-Sakai carrying the Shiga toxin 1 genes of the enterohemorrhagic Escherichia coli O157:H7 strain derived from the Sakai outbreak. Gene. 2000;258:127–139. doi: 10.1016/s0378-1119(00)00416-9. [DOI] [PubMed] [Google Scholar]