Abstract
The pathogenicity and prevalence of Mycoplasma penetrans, a Mycoplasma species recently isolated from humans, are still debated. A major P35 antigen, which is used as target epitope in serological assays, was shown to be a phase-variable lipid-associated membrane protein (LAMP). In this study, we performed a comparative analysis of the LAMP patterns from five M. penetrans clinical isolates and from the type strain. Sodium dodecyl sulfate-polyacrylamide gel electrophoresis profiles and immunoblots with sera serially collected from an M. penetrans-infected patient indicated that these strains expressed different LAMP repertoires. Furthermore, the intraclonal variation in the expression of LAMPs (P34A, P34B, P35, and P38) was monitored by immunoblot analysis with three specific monoclonal antibodies (MAbs) developed in this study and MAb 7 to P35. The phase variation of these LAMPs occurs in an independent manner, with frequencies of variation ranging from 10−2 to 10−4 per cell per generation. Consistent with their amphipathic nature, the P34B and P38 antigens were found exposed at the cell surface. The DNA sequence encoding the P38 antigen was defined and found to be related to those of the P35 gene and other putative LAMP-encoding genes, suggesting that these variable antigens are encoded by a family of related genes. Finally, the serum samples from an M. penetrans-infected patient contained antibodies that reacted with a P36 antigen expressed in different M. penetrans strains but not in the isolate recovered from this patient. This result suggested that in vivo phase variation of P36 occurred, which would support a role for these LAMP variations in avoiding the host's immune vigilance.
Members of the genus Mycoplasma have several characteristic features, including small genome size in comparison to most other prokaryotes (10, 16, 17). The drastic reduction in their genome size during evolution was only possible by the adoption of a parasitic lifestyle, allowing these bacteria to acquire many nutrients from their hosts. As obligatory parasites, they were also forced to develop efficient strategies to avoid their host's immune surveillance. A number of studies have demonstrated that mycoplasmas which colonize animals and humans are able to change their surface antigen profile at high frequency (for reviews, see references 5, 8, and 35). Such antigenic variation is proposed to play a key role in the evasion from the host immune response, a notion that has received some support from in vitro experiments in which phase and size variation of Mycoplasma hyorhinis surface antigens provided protection from growth inhibition by polyclonal serum (7). A second characteristic feature of mycoplasmas is the absence of a cell wall or outer membrane. Consequently, the mycoplasma surface lacks lipopolysaccharide and peptidoglycan. The major variable antigens at the mycoplasma cell surface are lipoproteins, which are also the dominant immunogens during mycoplasmal infections. The mechanisms underlying the variation of these lipoproteins are diverse and have been deciphered only for a limited number of mycoplasma species (for a review, see references 8 and 28).
Mycoplasma penetrans was initially isolated from a urine sample collected from a human immunodeficiency virus type 1 (HIV-1)-infected patient (22). This mycoplasma grows fastidiously, and so far only a few isolates have been obtained from clinical samples. M. penetrans has an elongated flask shape morphology with a tip-like structure (21) that is predicted to allow the mycoplasma to adhere and invade host cells (12, 23). Epidemiological studies indicated an association between M. penetrans and HIV infection (13, 33, 34), suggesting that M. penetrans could act as a cofactor in the progression of the HIV-associated disease (2). Indeed, Grau et al. (14) described an association between active M. penetrans infections and an accelerated progression of disease among HIV-infected patients. However, it is not proven whether M. penetrans was the true cause of the accelerated disease or only persisted in patients who were particularly immunocompromised (14). There is a single report describing the isolation of this mycoplasma from a patient who was not coinfected with HIV. In this clinical case, the M. penetrans infection was associated with a primary antiphospholipid syndrome (36), but no other study has since substantiated this finding.
There is clearly a need to better understand the potential role of M. penetrans infection in human disease. The epidemiological studies performed thus far are based on an M. penetrans-specific serological assay (13, 33). In this assay, the diagnostic target molecules were lipid-associated membrane proteins (LAMPs) which were extracted by using the neutral detergent Triton X-114 (TX-114). The LAMPs from the type strain M. penetrans GTU showed in immunoblots two major antigenic proteins, P35 and P38, and less-abundant 61- and 103-kDa polypeptides (33). The P35 protein was shown to be a surface-exposed variable lipoprotein undergoing high-frequency phase variation (9, 26, 27). These studies were performed by using the M. penetrans type strain and the heterogeneity of LAMPs between strains was not evaluated.
In the present study, we investigated the heterogeneity of LAMPs between different M. penetrans clinical isolates and among their isogenic progenies. For this purpose, serial sera from an HIV-M. penetrans-coinfected patient were used together with four newly developed monoclonal antibodies (MAbs) with unique LAMP specificities. These immunological tools allowed us to demonstrate that, in addition to P35, other putative lipoproteins, including P34B and the sequenced P38, vary among isogenic variants in an independent manner.
MATERIALS AND METHODS
Mycoplasma culture condition.
M. penetrans isolate ARA was kindly provided by Christiane Bebear (University Victor Segalen, Bordeaux, France), and isolates HF-1, HF-2, and HF-3 were as described elsewhere (36). M. penetrans 75 was isolated from the urine sediment of an HIV-infected patient (Freiburg, Germany) as previously described (18). The type strain GTU was kindly provided by S. Lo (Armed Forces Institute of Pathology, Bethesda, Md.). Mycoplasmas were grown at 37°C in liquid PPLO broth or PPLO agar (Difco, Detroit, Mich.) supplemented with 20% heat-inactivated horse serum and 10% yeast extract (15).
MAbs.
In order to establish specific MAbs to M. penetrans LAMP, BALB/c mice were immunized by the method of Cianfriglia et al. (6) with 108 cells of M. penetrans 75 per immunization. The splenocytes were fused with myeloma cells as previously described (20). Hybridomas were grown in 24-well microtiter plates in RPMI 1640 medium (Biochrom KG, Berlin, Germany) supplemented with 15% fetal calf serum, 5 μl of 2-mercaptoethanol (Gibco-BRL, Life Technologies, Karlsruhe, Germany)/liter, 20,000 IE of penicillin/liter, and 20 mg of streptomycin (Gibco-BRL)/liter. After 10 days the hybridoma supernatants were tested for M. penetrans-specific MAbs in Western immunoblots. Antibody-producing cells reacting with M. penetrans LAMPs were cloned by dilution with hybridoma medium supplemented with 2% hypoxanthine-aminopterin-thymidine medium supplement (Sigma Chemical Co., St. Louis, Mo.). This procedure was repeated twice with in-between monitoring of antibody production by Western blot analysis as described below. MAbs were also tested for their reactivity in colony immunoblots of M. penetrans 75. By using this approach, three MAbs (MAb C5 of immunoglobulin G2b [IgG2b] isotype, MAb D10 of IgG1 isotype, and MAb G9 of IgG3 isotype) were selected. MAb 7 of IgG1 isotype, which was previously shown to react with the P35 protein of M. penetrans GTU (26), was also used in this study. MAb 14, which reacts with the IMP14 peptide, is described below.
Electron microscopy.
Localization of the MAb target epitope at the surface of M. penetrans was examined by using the immunogold labeling technique described by Neyrolles et al. (26). Briefly, M. penetrans cells were washed once in 25 mM HEPES (Sigma) and resuspended in phosphate-buffered saline (PBS). Nickel grids were laid onto a droplet of a M. penetrans cell suspension for 20 min and fixed with 1% paraformaldehyde at room temperature for 3 min. The grids were incubated in PBS for 10 min and then with a PBS solution containing 50 μg of MAbs G9 or D10/ml for 1 h at room temperature. After five washings in a solution containing PBS–0.5% bovine serum albumin–0.1% gelatin, the grids were incubated for 1 h at room temperature with an anti-mouse IgG and IgM antibody labeled with 10-nm gold particles (Amersham) diluted 1:25 in PBS. The grids were washed once in PBS and five times in water and then examined with a JEOL EX 1200 electron microscope at an accelerating voltage of 80 kV.
SDS-PAGE and Western immunoblotting.
Mycoplasma cells from a broth culture were collected by centrifugation for 20 min at 5,400 × g and washed twice in PBS. M. penetrans membrane-associated proteins were fractionated by using TX-114 (3) (Serva, Heidelberg,Germany) as previously described (31) with slight modifications. Briefly, mycoplasmas from 50-ml cultures were resuspended in 1 ml of PBS containing 2% (vol/vol) TX-114 and incubated for 2 h at 4°C with shaking. After centrifugation at 4°C for 10 min at 13,000 × g, the supernatant containing the solubilized proteins was subjected to three cycles of phase fractionation, including incubation for micelle formation at 37°C for 5 min, followed by a centrifugation step (13,000 × g, 3 min, room temperature) for phase separation, resulting in an upper aqueous and a lower detergent TX-114 phase. After collection of the detergent phase, the concentration of TX-114 was readjusted with PBS to 2% and incubated on ice for 10 min. This procedure was repeated twice. Prior to sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) analysis, proteins that partitioned into the TX-114 detergent phase were concentrated by ethanol precipitation (80% [vol/vol]). SDS-PAGE was performed according to the method of Laemmli (19).
After electrophoresis, the proteins were stained with Coomassie blue or transferred to nitrocellulose membrane (Protran BA83; Schleicher & Schuell, Dassel, Germany) by semidry electroblotting in a Multiphor II electrophoresis unit (LKB Bromma). Membranes were blocked with PBS–0.1% (vol/vol) Tween 20 (Serva) containing 10% fetal calf serum. The immunostaining was performed by using the hybridoma supernatants of MAb 7, MAb C5, MAb D10, MAb G9, or human sera as the primary antibody and peroxidase-conjugated anti-mouse immunoglobulin (Dako A/S, Glostrup, Denmark) or anti-human immunoglobulin (Sigma) as secondary antibody.
Colony immunoblotting and isolation of clonal variants derived from M. penetrans
Colony immunoblotting was performed as previously described (31), except that 4-chloro-1-naphthol was used as the peroxidase substrate. Colonies that did not bind the MAbs were stained with Ponceau S. To establish a clonal lineage, MAb-positive or -negative colonies were selected after colony immunoblotting, propagated in 1 ml of culture and, after further Western blot or colony blot analyses, were used to generate isogenic progeny that exhibited alternative surface phenotypes. The frequency of a particular epitope to undergo phase variation was assessed by colony immunoblotting by calculating the number of colonies derived from a single variant that had switched ON or OFF the expression of the epitope.
Expression of the putative lipoprotein pepIMP14 in Escherichia coli.
The IMP14 gene sequence (GenBank-EMBL accession no. AJ006697) described by Neyrolles et al. (26) contained an open reading frame (ORF) encoding the putative lipoprotein pepIMP14. The sequence between nucleotides 49 and 1069 of the IMP14 ORF was amplified by using a forward primer, 5′-TACGGGATCCTCTTCTTGTTCTTCAAC-3′, located in the conserved 5′-terminal coding region and a reverse primer, 5′-AATTGTCCCGGGCATCAAATGAAACATCTTTGCTTCTCTAG-3′. The restriction sites for BamHI and XmaI (underlined nucleotides) were engineered for further directional cloning of the PCR fragment. First, the PCR product was directly inserted into the pCR2.1-TOPO vector (Invitrogen, Groningen, The Netherlands), and the four TGA triplets that code for tryptophan in mycoplasmas were mutated to TGG codons for expression in E. coli as described in the QuickChange Site-Directed Mutagenesis Kit (Stratagene, Heidelberg, Germany). The mutated insert was then digested with BamHI and XmaI and ligated into the pQE30 vector (Qiagen, Hilden, Germany) cut with the same enzymes, and the resulting ligation mixture was transformed into E. coli M15 (Qiagen).
A recombinant plasmid containing the pepIMP14 ORF in the correct orientation was selected, and its sequence was verified by DNA sequencing. The clone harboring the selected recombinant plasmid was used to produce the IMP14 polypeptide fused at the N terminus with a His6 tag to facilitate its purification by using nickel-nitrilotriacetic acid (NTA) chromatography (Qiagen). The purified IMP14 recombinant product was then used to produce the MAb designated MAb 14 (IgM) according to the procedure described above. Each immunization was performed with 100 μg of the purified recombinant product.
Peptide sequencing.
Proteins of M. penetrans 75 that partitioned into the TX-114 detergent phase were separated by electrophoresis and transferred to an Immobilon-P membrane (Schleicher & Schuell). Proteins were detected by staining with Coomassie blue R-250 (Ferak, Berlin, Germany), and the bands of interest were excised. In collaboration with P. Rücknagel (Max Planck Institute, Halle, Germany), proteins were digested with endoprotease Lys C (0.15 μg/20 μl of 25 mM Tris [pH 8.5]; Boehringer). The supernatants were separated, and the membranes were extracted twice with 20 μl of 50% formic acid in acetonitrile. The supernatant and extracts were concentrated to 20 μl under nitrogen. The peptides were separated by reversed-phase high-pressure liquid chromatography on a Shimadzu LC10 unit with an ET 125-by-2-mm Nucleosil 500-5 C3PPN column (Macherey-Nagel) and linear gradient elution (eluent A, 0.09% trifluoroacetic acid in water; eluent B, 0.08% trifluoroacetic acid in acetonitrile; gradient, 1 to 40% B in 60 min; flow rate, 200 μl min−1; column temperature, 40°C). Peptide-containing fractions were collected manually. Aliquots of the fractions were used for MALDI-mass spectrometry and Edman sequencing with an Applied Biosystems 476A Sequencer according to the manufacturer's instructions.
DNA extraction, Southern blot analysis and cloning.
Genomic DNA of M. penetrans 75 was extracted by conventional procedures (24). For Southern blot analysis, 10 μg of DNA was digested with the appropriate restriction enzyme and subjected to agarose gel electrophoresis, and the DNA fragments were transferred to Hybond-N+ membranes (Amersham International, plc.). Southern blot hybridization was performed as previously described (26). Briefly, the membranes were prehybridized with hybridization solution containing 6× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate), 5× Denhardt solution, 0.1% SDS, and 100 μg of salmon sperm DNA ml−1 for 2 h and incubated overnight in hybridization solution with 32P-labeled oligonucleotide probes. The probes were labeled by end labeling with T4 polynucleotide kinase. The membranes were then washed twice with 6× SSC–0.1% SDS for 20 min at room temperature. The oligonucleotide probes used in this study were based on the peptide sequences derived from the fragments of the 38-kDa protein and were as follows : P38-1, 5′-TTATAWATWCCWAAWGCWGC-3′; P38-2, 5′-TTWATATCTTCWSWWATWAATTCATT-3′; and P38-3, 5′-CATAWGGWGCWGCATCWACWACWACWGTATAWAC-3′. Since tryptophan is preferentially coded in mycoplasmas by the codon TGA rather than TGG, this triplet was used to design the oligonucleotides even though it corresponds to a stop condon in the universal genetic code.
A PstI-DNA fragment of 3.5 kb hybridized with more than one probe (P38-1 and P38-3 of the probes described above). This DNA fragment was excised from the agarose gel and purified by electroelution (32). The eluted fragments were then ligated with PstI-pUC18 vector, and the ligation mixture was used to transform E. coli XL1-Blue competent cells. Identification the recombinant plasmid of interest was performed by Southern blot hybridization and was designated MP24.
For the expression of a recombinant P38 protein, a strategy similar to that adopted for pepIMP14 expression was used, except that P38-specific primer sequences allowed the amplification of the P38-encoding gene and the mutagenesis of TGA triplets into TGG triplets. The forward primer 5′-TACGGGATCCTCTTCTTGTTCTTCTTACTGATAATGG-3′ and the reverse primer 5′-ATTG TCCCGGGAT TCACAG T TAAAC TAACATCAAAAGAAAAC TC-3′ used for amplification included the restriction sites for BamHI and XmaI (underlined nucleotides) for cloning of the PCR fragment into the expression vector pQE30 (Qiagen).
RAPD analysis.
DNA of M. penetrans strains were isolated with a QIAamp DNA Mini kit (Qiagen) according to the manufacturer's instructions. The DNA concentration was determined spectrometrically. Randomly amplified polymorphic DNA (RAPD) analysis was performed as described by Geary et al. (11) and Marois et al. (25), with slight modifications. Briefly, the PCR mixture with a total reaction volume of 50 μl consists of Taq polymerase buffer (Perkin-Elmer); a final concentration of 3 mM MgCl2; 250 μM concentrations each of dATP, dTTP, dCTP, and dGTP (Hybaid AGS, Heidelberg, Germany); 2.5 U of AmpliTaq Gold DNA polymerase (Perkin-Elmer); 300 ng of primer (primer 1281 [5′-AACGCGCAAC-3′] and/or primer 1254 [5′-CCGCAGCCAA-3′]); and 40 ng of DNA. The PCR was performed in a Perkin-Elmer GenAmp PCR System 9600. The following reaction conditions were used: 1 cycle of 97°C for 5 min; 4 cycles at 94°C for 5 min, 36°C for 5 min, and 72°C for 5 min; followed by 31 cycles of 94°C for 1 min, 36°C for 1 min, and 72°C for 1 min; and terminated by 1 cycle of 72°C for 10 min. The amplified products (15 μl) were electrophoresed in 1,5% agarose gels in TBE (10.8 g of Tris-HCl liter−1, 5.5 g of borate liter−1, 0.74 g of Na2EDTA liter−1) for 1 h at 0.15 A. Then, 5 μl of Smart Ladder (Eurogentec) was run in each gel as the molecular size standard. The gels were stained with ethidium bromide and visualized by UV transillumination.
Nucleotide sequencing and sequence analysis.
The DNA insert of the recombinant plasmid MP24 was sequenced by using BigDye Terminator RR Mix (PE Applied Biosystems) and an automated sequencer (ABI Prism 377 DNA Sequencer, Perkin-Elmer). Sequence data were analyzed by using the software programs provided by the sequencer and by using the GenBank and EMBL databases. Published sequences P35 and P30 (L38250) and IMP14 (AJ006699) were obtained from GenBank-EMBL. The GenBank-EMBL accession number of the DNA sequence encoding P38 is AF260642.
RESULTS
Analysis of putative LAMPs of different M. penetrans isolates.
In the present study, LAMP profiles of uncloned M. penetrans isolates that had originally been cultured from (i) the urine sediment of an AIDS patient in Germany (isolate 75), (ii) the urine sediment of an HIV-infected patient in France (isolate ARA), and (iii) the blood (HF-1), the tracheal aspirate (HF-2), and the throat (HF-3) of an HIV-negative patient with primary antiphospholipid syndrome (36) were analyzed by SDS-PAGE and Western blotting (Fig. 1 and 2).
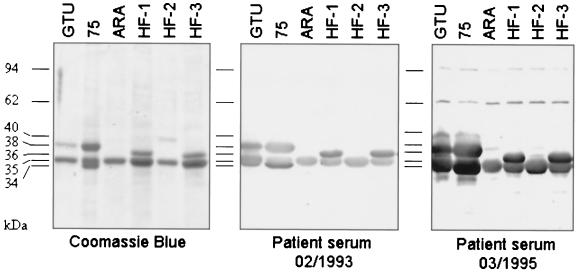
FIG. 1.
LAMP profile heterogeneity between M. penetrans isolates. TX-114 extraction of different M. penetrans patient isolates showing the heterogeneity of LAMP patterns. The results of SDS-PAGE of TX-114-fractionated LAMPs of M. penetrans type strain GTU and patient isolates 75, ARA, HF-1, HF-2, and HF-3 stained with Coomassie blue and corresponding immunoblots incubated with serum of patient 75 collected in either February 1993 or March 1995 are shown.
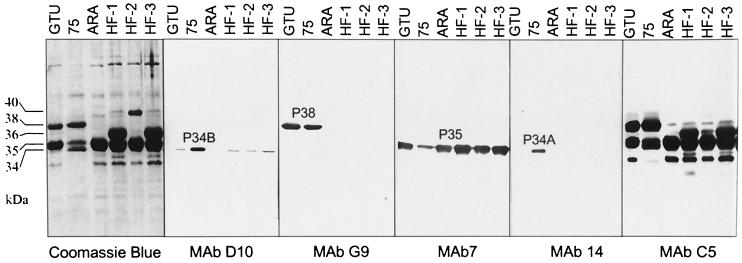
FIG. 2.
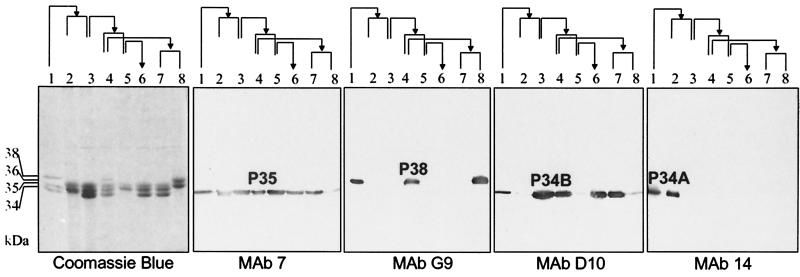
Different expressions of LAMP profiles in several M. penetrans patient isolates (75, ARA, HF-1, HF-2, and HF-3) and the type strain GTU determined by using Coomassie blue staining and immunostaining with MAbs D10, G9, 7, 14, or C5. MAb D10 reacted with a 34-kDa molecule (P34B). MAb G9 recognized a 38-kDa molecule (P38) in M. penetrans GTU and 75. MAb 7 stained P35. MAb 14 recognized a 34-kDa molecule in M. penetrans 75, named P34A. MAb C5 reacted with a common epitope of LAMPs with molecular masses ranging between 30 and 38 kDa.
The LAMP profiles were markedly different between the isolates, although in each case Coomassie blue staining revealed that the most abundant products migrated between 34 and 40 kDa, with products of 30, 62, 94, and 96 kDa only detectable when the sample load was increased. All M. penetrans strain and isolates tested in this study expressed a 35-kDa products which reacted with the MAb 7, an antibody which recognizes the P35 antigen previously identified in the type strain GTU (26). Other than this common P35 antigen, no LAMP profiles were identical except for two (HF-1 and HF-3) isolates obtained from different body sites of the same patient.
By using two sera collected at two different times from patient 75, the humoral response to the M. penetrans LAMPs was monitored by Western blot analyses. The first serum sample (collected in February 1993) contained antibodies that recognized LAMPs of 34, 35, and 38 kDa expressed by the M. penetrans isolate recovered from this patient (isolate 75). Beside the P35 shared antigen, immunoprofiles observed with this serum differed among the other strain and isolates. More specifically, this patient serum recognized a product of 38 kDa in strain GTU and of 36 kDa in isolates HF-1 and HF-3. This later 36-kDa reactivity was not detected in isolates ARA and HF-2.
The second serum sample from patient 75 was collected 2 years later at the time of isolation of M. penetrans 75 in March 1995. This serum showed a stronger reactivity with molecules of 34 to 38 kDa but also recognized additional LAMPs of 62 and 94 kDa in M. penetrans isolate from patient 75. This serum sample also recognized additional proteins in other isolates. The GTU profile resembled that of isolate 75. In the LAMPs from isolates HF-2 and HF-3, additional faintly labeled bands were observed.
To further characterize the different LAMPs of isolate 75, MAbs to M. penetrans 75 surface molecules were generated by using mycoplasma cells as antigen. Three MAbs were established based on their specific reactivity in Western blot analysis and recognized the major LAMPs. Western blot analysis revealed that the MAbs D10 and G9 specifically recognized a single, distinct product of 34 (P34B) and 38 (P38) kDa, respectively (Fig. 2). Only the type strain GTU and isolate 75 expressed the MAb G9-specific product. The third MAb, C5, strongly reacted with LAMPs of 38, 36, 35, 34, and 30 kDa, suggesting that the MAb C5-specific epitope is shared by multiple lipoproteins. MAb 14 (generated from a recombinant pepIMP14 antigen [26]) recognized a 34-kDa molecule similar to that stained by MAb D10. However, MAb 14 and MAb D10 recognized distinct molecules, since reactivity of MAb 14 was found only with isolate 75, while MAb D10 reacted with the type strain and all of the isolates except ARA. As a control, the MAb 7 (26) was also included in this analysis, and the results demonstrated that P35 is expressed in all of the isolates analyzed, confirming that the common 35-kDa band was indeed P35 (Fig. 2).
Typing of M. penetrans isolates with RAPD.
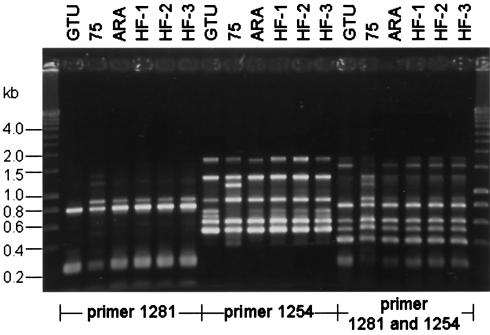
RAPD patterns generated with primer 1281 alone showed a limited banding, whereas the amplification with primer 1254 or the combination of both primers according to the method of Marois et al. (25) resulted in a more complex RAPD banding pattern. The RAPD patterns of M. penetrans strain GTU and isolates ARA, HF-1, HF-2, and HF-3 generated with primer 1254 were identical with slight differences in the intensity (Fig. 3), but isolate 75 showed an additional band of 1.3 kb and two lacking bands of 0.75 and 0.8 kb. Comparison of the RAPD patterns generated with the combination of primers 1254 and 1281 M. penetrans GTU showed no 1.4- and 0.9-kb fragments. M. penetrans 75 showed an additional amplification product of 1.3 kb. The overall banding patterns of isolates HF-1, HF-2, and HF-3 and ARA showed identical patterns with the amplification protocols but were distinguishable from those of strain GTU and isolate 75.
FIG. 3.
RAPD patterns of M. penetrans strain GTU and isolates 75, ARA, HF-1, HF-2, and HF-3 after amplification with primer 1281, 1254 or a combination of both primers. DNA basepair size standards are shown on the left and on the right.
P34B and P38 undergo antigenic variation in vitro
Immunostaining of M. penetrans 75 colonies with MAbs D10 and G9 revealed a pattern composed of positive, negative and sectored colonies (Fig. 4B), indicating that the corresponding target epitopes are surface exposed and may undergo variation in expression.
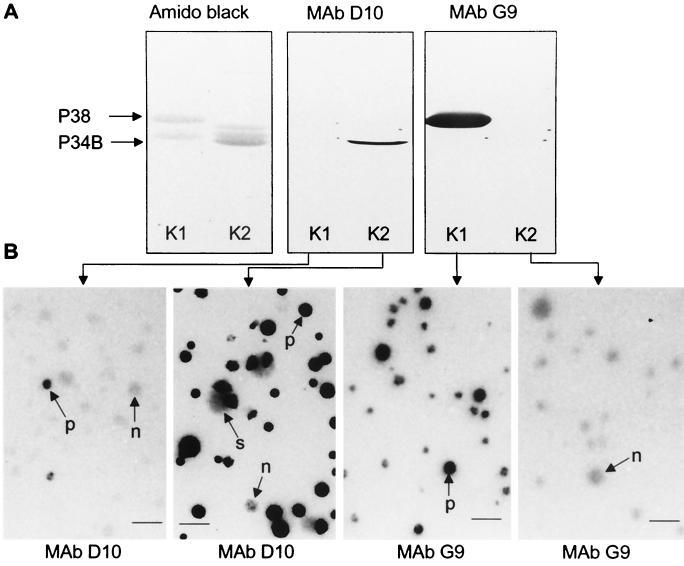
FIG. 4.
Western and colony immunoblots of clonal variants derived from M. penetrans isolate 75 showing phase-variable expression of P34B and P38. (A) SDS-PAGE analysis of LAMPs from two selected clones of M. penetrans 75. Clone K1 was positive with P38 (MAb G9) and negative with MAb D10, and clone 2 reacted positively with MAb D10 (P34B) and negatively with MAb G9. (B) Clones K1 and K2 were grown on agar plates, and colony immunoblots were labeled with MAb D10 or MAb G9. Staining of clone K1 colonies with MAb D10 revealed the presence of colonies among the P34B negative population that have reverted to the P34B-positive phenotype (arrow p). Colonies generated with clone K2 are predominantly positive for P34B, though a few present a negative (arrow n) and sectoring (arrow s) staining. Staining of colonies generated with clone K1 with MAb G9 showed predominantly the P38-positive phenotype. Colonies of clone K2 were negative. Bar, 100 μm.
To assess phase variation of D10 and of G9 target epitopes, cloned D10- and G9-positive or -negative variants from M. penetrans 75 were selected and tested by colony and Western immunoblotting (Fig. 4). Results obtained with clones K1 and K2 revealed that expression of P38 is only detected in clone K1 (G9 positive and D10 negative in colony immunoblotting), while P34B was expressed in its sibling, K2 (D10 positive and G9 negative in colony immunoblotting). Variations in expression of P34B and P38 occurred with high frequency in propagating populations, resulting in the appearance of minor populations that presented a different phenotype (Fig. 4B, for clone K1 and K2). The variation rate of P34B and P38 differed. It was estimated by colony immunoblotting to be 10−2 per cell per generation for P34B and 10−4 per cell per generation for P38.
To investigate independent ON-OFF switches of P34A, P34B, P35, and P38, clonal lineages of M. penetrans 75 were established. When these progeny were analyzed, the results showed that isogenic variants of the M. penetrans 75 clonal lineage differed in their expression profiles (Fig. 5). M. penetrans 75 expressed P34A, P34B, P35, and P38 (Fig. 5, lane 1), while subclones showed a change in the expression of P34B and P38 from OFF to ON (P34B, lane 5→6; P38, lane 3→4) and vice versa (P34B, lane 3→5; P38, lane 4→7). Only a switch from ON to OFF was observed for P34A, and it is not yet known whether reversion from OFF to ON can occur. Colony immunoblotting could not be used as a method to screen for P34A phase variants since the only available P34A-specific MAb does not recognize a surface-accessible epitope. P35 was constantly detected, with one clonal variant (lane 8) showing a weak immunoreaction with P35 and no 35-kDa band by Coomassie blue staining.
FIG. 5.
Immunoblots of TX-114-extracted LAMPs of a clonal lineage of M. penetrans 75 incubated with MAb 7, G9, D10, or 14. Lane 1 represents M. penetrans 75, and descendants are shown in lanes 2 to 8. The pedigree of the descendants is indicated by the arrows. The clonal lineage shows variability in protein expression of P34B from ON to OFF (lane 3 to lane 5) and vice versa (lane 5 to lane 6) and revealed ON and OFF switching of P38 (lane 3 [OFF] to lane 4 [ON] → lane 7 [OFF]).
Surface exposure of M. penetrans P34B and P38.
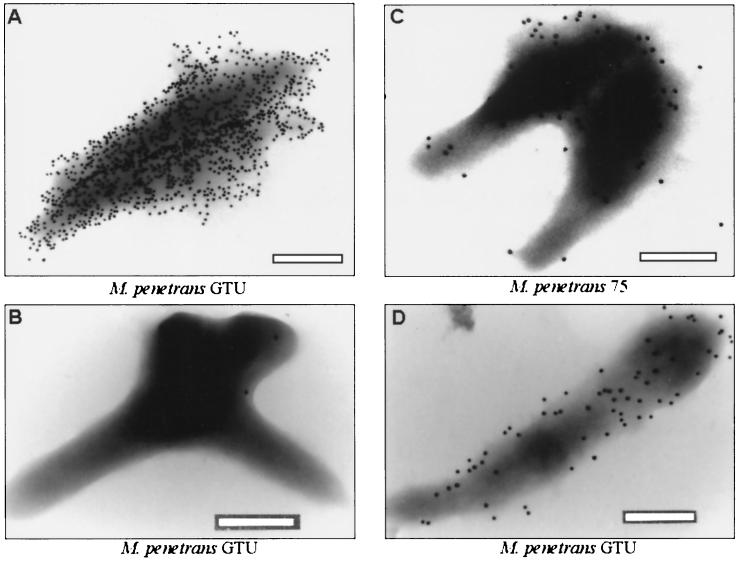
The cell surface exposure of P34B and P38 was further assessed by immunoelectron microscopy of M. penetrans GTU and 75 with specific MAbs D10 and G9 and gold-labeled secondary antibodies (Fig. 6). The MAbs D10 (Fig. 6A) and G9 (Fig. 6C and D) target epitopes were evenly distributed over the mycoplasma cell surfaces. Additionally, M. penetrans GTU and 75 cultures showed cells with bound P34B- and P38-specific MAbs; however, both cultures also contained cells without labeled surfaces.
FIG. 6.
Variable surface expression of P34B and P38 of M. penetrans cells by immunogold labeling electron microscopy. P34B-positive (A) and P34B-negative (B) mycoplasmas were examined in one culture of M. penetrans GTU. Cells of isolate 75 (C) and M. penetrans GTU (D) were labeled with MAb G9. Differences in the quantity of MAb binding sites at the cell surface are shown for MAbs D10 (A) and G9 (C and D). Bar, 200 nm.
Sequence analysis of the P38 coding sequence.
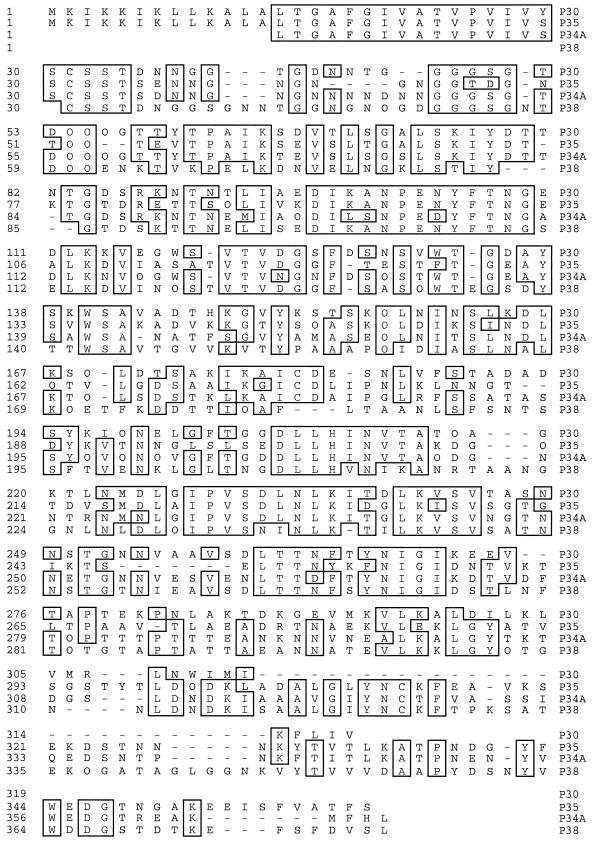
Amino acid sequencing of peptides derived from enzymatic digestion of the isolated 38-kDa protein led to the generation of three peptide sequences: p381 (ISAALGIYN), p382 (TTNELISEDIK), and p383 (VYTVVVDAAPYDSNYVWDDGSTDTK). Southern blots of digested genomic DNA of M. penetrans 75 were hybridized with three specific oligonucleotide probes corresponding to each sequenced peptide. Southern blot analysis revealed that a 3.5-kb PstI DNA fragment bound the two oligonucleotide probes corresponding to p381 and p383, while no hybridization signal was obtained with the third probe (data not shown). Cloning and sequencing of this 3.5-kb PstI DNA fragment revealed the presence of a 500-bp region containing a partial ORF that included the peptide sequences p381 and p383. A stop codon was found 11 amino acids downstream of the region encoding p383, indicating that the 500 bp localized on the 3.5-kb DNA fragment encodes the P38 C-terminal region. The deduced P38 sequence present DNA homologies with the translation products of several recently described genes (designated mpl for M. penetrans lipoprotein [26]): P35, P30, P34A (pepIMP14), pep1IMP12, and pepIMP13. One feature of these genes is that they possess identical sequences encoding for the signal peptide at the N terminus. Therefore, a forward primer (5′-CAGTTCCAGTAATTGTTTCTTC-3′) corresponding to the conserved N-terminal region and a reverse primer (5′-TTATATCTATGAAAAGGAAATGTA-3′) located at the 3′ end of the identified ORF were used for the amplification of the entire P38 gene. An amplification product of 1 kb was obtained and sequenced. The translated amino acid sequence encoded by this 1-kb DNA fragment contained, in addition to p381 and p383, the third p382 peptide sequence. The molecular mass for the mature lipoprotein deduced from the DNA sequence was 37 kDa. Further analysis (Fig. 7) revealed that P38 exhibits a high degree of sequence similarity with other members of the mpl family of LAMPs (26). Among this family, P34A had the greatest identity (36.4% identity) to P38. P38 had no homology to any other protein recorded in the databases. The genes encoding P30, P35, and P34A are known to be clustered in the M. penetrans genome (26). The relative location of the P38 gene to this cluster has not been ascertained, although a putative lipoprotein gene is also present downstream of the P38 gene, within the cloned DNA fragment. Although analysis of this lipoprotein gene is incomplete, the preliminary data suggest that it is new member of the mpl family.
FIG. 7.
Alignment of P38 deduced amino acid sequence with that of the previously identified P30, P35, and P34A (pepIMP14) of M. penetrans by using the DNAstar software. Boxed letters represent amino acids that are common to three of the four sequences.
Recombinant fusion proteins representing P38 and P34A (pepIMP14) were expressed in E. coli and, as expected, MAb G9 recognized the recombinant protein P38. The recombinant forms of both P38 and P34A were recognized by MAb C5. Since both of the recombinant proteins lack a signal peptide required for their lipid modification in E. coli, this result indicates that the epitope recognized by MAb C5 is encoded by both genes and is proteinaceous in character.
DISCUSSION
Comparative analysis of the LAMP profiles of the few available clinical isolates of M. penetrans indicated that this species possesses a complex family of related but distinct variable surface lipoproteins and that these lipoproteins showed a high degree of heterogeneity in their expression. Interestingly, our analysis also revealed that M. penetrans HF isolated from two distinct body sites of the same patient, expressed different combinations of LAMPs. A major 36-kDa band could be detected in the isolates HF-1 and HF-3, respectively, but was lacking in the Coomassie blue staining of isolate HF-2. In Western blot analysis, a faint 36-kDa band could be detected also in HF-2 (Fig. 1, patient serum 03/1995) most likely indicating that a minor population of HF-2 expressed the 36-kDa protein. The three M. penetrans HF isolates presented an identical banding pattern in RAPD analysis, supporting the hypothesis that these isolates had the same genetic origin. This result suggested either that mycoplasma populations presenting different LAMP phenotypes coexisted at the same site within a single host, with a given variant being selected during the isolation procedure, or that some of the phenotypic variations correspond to an adaptation to specific colonization sites. The ability of an M. penetrans population to express different LAMP repertoires in a single colonized host could also provide an explanation regarding the nature of the humoral response observed in the patient 75. In patient 75, LAMPs related to 34-, 35-, and 38-kDa molecules are highly immunogenic. This patient developed also a strong humoral response against P36 even though it was not detectable with Coomassie staining in the clinical isolate 75. Here we were able to demonstrate by in vitro subcloning that this isolate retained the genetic ability to revert in vitro to a P36-positive phenotype as a major part of the LAMP pattern (Fig. 5). Based on this result, it is more likely that P36 was expressed in vivo at some point during the infection and represented a potent antigen eliciting an immune response in patient 75. Altogether, these data strongly support that phase variation of M. penetrans LAMPs occurs in vivo.
Although the serum collected in 1993 from the HIV-positive patient 75 contained circulating antibodies that strongly bound to different LAMPs, M. penetrans was isolated 2 years later, indicating that the strong humoral response was not sufficient to eradicate mycoplasmas from the urogenital tract. It is possible that the HIV-associated immunodeficiency of the patient was a factor predisposing for the mycoplasma persistence. However, this observation is also in accordance with data obtained by Cartner et al. (4) showing that mice infected with the respiratory pathogen Mycoplasma pulmonis develop an humoral response against the mycoplasma that does not block the colonization of the target tissue but rather plays a role in preventing the systemic dissemination of the pathogen.
Phase variation of surface antigens was previously demonstrated for M. penetrans type strain GTU, with a focus on the P35 antigen (26). By using newly established MAbs, we documented that at least three further prominent lipoproteins (P34A, P34B, and P38) can undergo phase variation in vitro. It could be demonstrated with the different clonal variants of M. penetrans 75 that MAb D10, MAb G9, MAb 7, and MAb 14 reacted with distinct LAMPs. The ON and OFF switching of P34B and P38 was demonstrated with the clonal lineage. The switching frequency of P34B of 10−2 per cell per generation is among the highest rates of variation measured thus far for lipoproteins of animal pathogenic mycoplasmas (29). The different combinations of expressed LAMPs in the clonal variants revealed that variation of P34A, P34B, P35, and P38 appeared to be independent of each other.
P38 is a new member of a family of related surface proteins in M. penetrans 75 that undergo intraclonal variation and that share a common immunogenic region, as demonstrated by using the newly established MAb C5.
The development of the MAb C5 is of particular interest because it allows the detection of a spectrum of related surface antigens. The MAb C5 epitope is linked to the protein moiety because the MAb C5 cross-reacts with the recombinant proteins P38 and P34A expressed in E. coli in absence of their signal peptide required for the lipid modification. Unlike a large number of other variable proteins identified in other mycoplasmas (1, 30), mpl genes do not contain repeated sequence which might be involved in size variation. Therefore, the ability of MAb C5 to detect the TX-114-extracted proteins in the molecular mass range of 30 to 38 kDa might help in future studies to monitor a distinct ON or OFF switched status of each member of this family without establishing specific MAbs for each individual LAMP. Furthermore, this effective reagent together with the other MAb developed in this study might be of relevance to define which set of expressed molecules should be necessary to support an in vivo colonization of the lower or even upper urogenital tract and to define a changing surface pattern of M. penetrans in later stages of a developing immune response.
Based on the finding that major components of M. penetrans cells undergo phase variations, it is noteworthy to speculate that LAMP diversity generated by the mycoplasma, despite a limited amount of genetic information, may contribute to host adaptation as a pathogenic mechanism and participate in establishing successful chronic infection of the human host.
ACKNOWLEDGMENTS
We thank Tsuguo Sasaki (National Institute of Infectious Diseases, Tokyo, Japan) for the gift of MAb 7. We thank P. Rücknagel for the sequencing of peptides. We also thank S. Ferris and S. Gäbler for excellent technical assistance.
This work was supported by the Deutsche Forschungsgemeinschaft (Ja 399/8-1).
REFERENCES
- 1.Behrens A, Heller M, Kirchhoff H, Yogev D, Rosengarten R. A family of phase- and size-variant membrane surface lipoprotein antigens (Vsps) of Mycoplasma bovis. Infect Immun. 1994;62:5075–5084. doi: 10.1128/iai.62.11.5075-5084.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blanchard A, Montagnier L, Gougeon M L. Influence of microbial infections on the progression of HIV disease. Trends Microbiol. 1997;5:326–331. doi: 10.1016/S0966-842X(97)01089-5. [DOI] [PubMed] [Google Scholar]
- 3.Bordier C. Phase separation of integral membrane proteins in Triton X-114 solution. J Biol Chem. 1981;256:1604–1607. [PubMed] [Google Scholar]
- 4.Cartner S C, Lindsey J R, Gibbs-Erwin J, Cassell G H, Simecka J W. Roles of innate and adaptive immunity in respiratory mycoplasmosis. Infect Immun. 1998;66:3485–3491. doi: 10.1128/iai.66.8.3485-3491.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambaud I, Wroblewski H, Blanchard A. Interactions between mycoplasma lipoproteins and the host immune system. Trends Microbiol. 1999;7:493–499. doi: 10.1016/s0966-842x(99)01641-8. [DOI] [PubMed] [Google Scholar]
- 6.Cianfriglia M, Armellini D, Massone A, Mariani M. Simple immunization for high-frequency production of soluble antigen-specific hybridomas. Hybridoma. 1983;2:451–457. doi: 10.1089/hyb.1983.2.451. [DOI] [PubMed] [Google Scholar]
- 7.Citti C, Kim M F, Wise K S. Elongated versions of Vlp surface lipoproteins protect Mycoplasma hyorhinis escape variants from growth-inhibiting host antibodies. Infect Immun. 1997;65:1773–1785. doi: 10.1128/iai.65.5.1773-1785.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Citti C, Rosengarten R. Mycoplasma genetic variation and its implication for pathogenesis. Wien Klin Wochenschr. 1997;109:562–568. [PubMed] [Google Scholar]
- 9.Ferris S, Watson H L, Neyrolles O, Montagnier L, Blanchard A. Characterization of a major Mycoplasma penetrans lipoprotein and of its gene. FEMS Microbiol Lett. 1995;130:313–319. doi: 10.1111/j.1574-6968.1995.tb07737.x. [DOI] [PubMed] [Google Scholar]
- 10.Fraser C M, Gocayne J D, White O, Adams M D, Clayton R A, Fleischmann R D, Bult C J, Kerlavage A R, Sutton G, Kelley J M, Fritchman J L, Weidman J F, Small K V, Sandusky M, Fuhrman J, Nguyen D, Utterback T R, Saudek D M, Phillips C A, Merrick J M, Tomb J-F, Dougherty B A, Bott K F, Hu P-C, Lucier T S, Petterson S N, Smith H O, Hutchison C A, I. I I, Venter J C. The minimal gene complement of Mycoplasma genitalium. Science. 1995;270:397–403. doi: 10.1126/science.270.5235.397. [DOI] [PubMed] [Google Scholar]
- 11.Geary S J, Forsyth M H, Aboul-Saoud S, Wang G, Berg D E, Berg C M. Mycoplasma gallisepticum strain differentiation by arbitrary primer PCR (RAPD) fingerprinting. Mol Cell Probes. 1994;8:311–316. doi: 10.1006/mcpr.1994.1042. [DOI] [PubMed] [Google Scholar]
- 12.Giron J A, Lange M, Baseman J B. Adherence, fibronectin binding, and induction of cytoskeleton reorganization in cultured human cells by Mycoplasma penetrans. Infect Immun. 1996;64:197–208. doi: 10.1128/iai.64.1.197-208.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grau O, Slizewicz B, Tuppin P, Launay V, Bourgeois E, Sagot N, Moynier M, Lafeuillade A, Bachelez H, Clauvel J P. Association of Mycoplasma penetrans with human immunodeficiency virus infection. J Infect Dis. 1995;172:672–681. doi: 10.1093/infdis/172.3.672. [DOI] [PubMed] [Google Scholar]
- 14.Grau O, Tuppin P, Slizewicz B, Launay V, Goujard C, Bahraoui E, Delfraissy J F, Montagnier L. A longitudinal study of seroreactivity against Mycoplasma penetrans in HIV-infected homosexual men: association with disease progression. AIDS Res Hum Retrov. 1998;14:661–667. doi: 10.1089/aid.1998.14.661. [DOI] [PubMed] [Google Scholar]
- 15.Hayflick L. Tissue cultures and mycoplasma. Tex Rep Biol Med. 1965;23:285–303. [PubMed] [Google Scholar]
- 16.Himmelreich R, Hilbert H, Plagens H, Pirkl E, Li B C, Herrmann R. Comparative analysis of the genomes of the bacteria Mycoplasma pneumoniae and Mycoplasma genitalium. Nucleic Acids Res. 1997;25:701–712. doi: 10.1093/nar/25.4.701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Himmelreich R, Plagens H, Hilbert H, Reiner B, Herrmann R. Complete sequence analysis of the genome of the bacterium Mycoplasma pneumoniae. Nucleic Acids Res. 1996;24:4420–4449. doi: 10.1093/nar/24.22.4420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Klär U, Jacobs E. Mycoplasma penetrans und Mycoplasma fermentans: Nachweise im Urogenitaltrakt immunsupprimierter Wirte. Klin Labor. 1996;6:540. [Google Scholar]
- 19.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 20.Lindl T, Bauer J. Zell und Gewebekultur. Stuttgart, Germany: Gustav Fischer Verlag; 1989. [Google Scholar]
- 21.Lo S C, Hayes M M, Wang R Y, Pierce P F, Kotani H, Shih J W. Newly discovered mycoplasma isolated from patients infected with HIV. Lancet. 1991;338:1415–1418. doi: 10.1016/0140-6736(91)92721-d. [DOI] [PubMed] [Google Scholar]
- 22.Lo S C, Hayes M M, Tully J G, Wang R Y, Kotani H, Pierce P F, Rose D L, Shih J W. Mycoplasma penetrans sp. nov., from the urogenital tract of patients with AIDS. Int J Syst Bacteriol. 1992;42:357–364. doi: 10.1099/00207713-42-3-357. [DOI] [PubMed] [Google Scholar]
- 23.Lo S C, Hayes M M, Kotani H, Pierce P F, Wear D J, Newton P B, Tully J G, Shih J W. Adhesion onto and invasion into mammalian cells by Mycoplasma penetrans: a newly isolated mycoplasma from patients with AIDS. Mod Pathol. 1993;6:276–280. [PubMed] [Google Scholar]
- 24.Marmur J. A procedure for the isolation of deoxyribonucleic acid from microorganisms. J Mol Biol. 1961;3:208–218. [Google Scholar]
- 25.Marois C, Dufour-Gesbert F, Kempf I. Comparison of puls-field gel electrophoresis with random amplified polymorphic DNA for typing of Mycoplasma synoviae. Vet Microbiol. 2001;79:1–9. doi: 10.1016/s0378-1135(00)00342-4. [DOI] [PubMed] [Google Scholar]
- 26.Neyrolles O, Chambaud I, Ferris S, Prevost M C, Sasaki T, Montagnier L, Blanchard A. Phase variations of the Mycoplasma penetrans main surface lipoprotein increase antigenic diversity. Infect Immun. 1999;67:1569–1578. doi: 10.1128/iai.67.4.1569-1578.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Neyrolles O, Eliane J P, Ferris S, Ayr Florio da Cunha R, Prevost M C, Bahraoui E, Blanchard A. Antigenic characterization and cytolocalization of P35, the major Mycoplasma penetrans antigen. Microbiology. 1999;145:343–355. doi: 10.1099/13500872-145-2-343. [DOI] [PubMed] [Google Scholar]
- 28.Razin S, Yogev D, Naot Y. Molecular biology and pathogenicity of mycoplasmas. Microbiol Mol Biol Rev. 1998;62:1094–1156. doi: 10.1128/mmbr.62.4.1094-1156.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rosengarten R, Wise K S. Phenotypic switching in mycoplasmas: phase variation of diverse surface lipoproteins. Science. 1990;247:315–318. doi: 10.1126/science.1688663. [DOI] [PubMed] [Google Scholar]
- 30.Rosengarten R, Wise K S. The Vlp system of Mycoplasma hyorhinis: combinatorial expression of distinct size variant lipoproteins generating high-frequency surface antigenic variation. J Bacteriol. 1991;173:4782–4793. doi: 10.1128/jb.173.15.4782-4793.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rosengarten R, Behrens A, Stetefeld A, Heller M, Ahrens M, Sachse K, Yogev D, Kirchhoff H. Antigen heterogeneity among isolates of Mycoplasma bovis is generated by high-frequency variation of diverse membrane surface proteins. Infect Immun. 1994;62:5066–5074. doi: 10.1128/iai.62.11.5066-5074.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. 2nd ed. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory; 1989. [Google Scholar]
- 33.Wang R Y, Shih J W, Grandinetti T, Pierce P F, Hayes M M, Wear D J, Alter H J, Lo S C. High frequency of antibodies to Mycoplasma penetrans in HIV-infected patients. Lancet. 1992;340:1312–1316. doi: 10.1016/0140-6736(92)92493-y. [DOI] [PubMed] [Google Scholar]
- 34.Wang R Y, Shih J W, Weiss S H, Grandinetti T, Pierce P F, Lange M, Alter H J, Wear D J, Davies C L, Mayur R K. Mycoplasma penetrans infection in male homosexuals with AIDS: high seroprevalence and association with Kaposi's sarcoma. Clin Infect Dis. 1993;17:724–729. doi: 10.1093/clinids/17.4.724. [DOI] [PubMed] [Google Scholar]
- 35.Wise K S, Yogev D, Rosengarten R. Antigenic variation. In: Maniloff J, McElhaney R N, Finch L R, Baseman J, editors. Mycoplasmas: molecular biology and pathogenesis. Washington, D.C.: American Society for Microbiology; 1992. pp. 473–490. [Google Scholar]
- 36.Yanez A, Cedillo L, Neyrolles O, Alonso E, Prevost M C, Rojas J, Watson H L, Blanchard A, Cassell G H. Mycoplasma penetrans bacteremia and primary antiphospholipid syndrome. Emerg Infect Dis. 1999;5:164–167. doi: 10.3201/eid0501.990122. [DOI] [PMC free article] [PubMed] [Google Scholar]