Abstract
Purpose
This study aimed to explore the mechanism of Zuogui Jiangtang Shuxin formula (ZGJTSXF) in the treatment of diabetic cardiomyopathy (DCM) by an integrative strategy combining serum pharmacochemistry, network pharmacology analysis, and experimental validation.
Methods
An Ultra high performance liquid chromatography-high resolution mass spectrometry (UPLC-Q-Exactive-Orbitrap-MS) method was constructed to identify compounds in rat serum after oral administration of ZGJTSXF. A component-target network between the targets of ZGJTSXF ingredients and DCM was established using Cytoscape. Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway enrichment analyses were performed to deduce ZGJTSXF-associated targets and pathways. The DCM model mice were treated with ZGJTSXF, and the predicted important signaling pathways were verified using quantitative PCR and Western blot.
Results
We identified 78 compounds in serum of medicated rats, which mainly included flavonoids, small peptides, nucleosides, organic acids, phenylpropanoids, alkaloids, phenanthrenequinones, iridoids, phenols, and saponins. Network pharmacology analysis revealed that ZGJTSXF may regulate targets including ALB, TNF, AKT1, GAPDH, VEGFA, EGFR, SRC, CASP3, MAPK3, JUN, and PI3K/AKT signaling pathway in the treatment of DCM. ZGJTSXF administration improved blood sugar levels, heart function, and cardiac morphological changes in DCM mice. Notably, ZGJTSXF inhibited cardiomyocytes apoptosis, which was associated with restored PI3K/Akt signaling and upregulated Bcl-2 and Bcl-xL proteins expression.
Conclusion
Our preliminary results proposed the material basis and possible mechanisms of ZGJTSXF in treating DCM, which is related to the activation of the PI3K/AKT signaling pathway and apoptosis inhibition. These findings shed new light in developing ZGJTSXF-based therapeutics in treating DCM.
Keywords: Zuogui Jiangtang Shuxin formula, diabetic cardiomyopathy, pharmacochemistry, network pharmacology, PI3K/Akt pathway, apoptosis
Introduction
Diabetic cardiomyopathy (DCM) is a unique clinical disease that appears as a structural abnormality of the heart caused by diabetes mellitus (DM) in the absence of other cardiac risk factors.1 The 10th edition of the International Diabetes Federation (IDF) Diabetes Atlas shows that there are currently 537 million adults with diabetes, and the number of patients is expected to grow to 700 million worldwide by 2045.2 As the prevalence of DM increases globally, the incidence of DCM is also increasing. Studies have shown that DCM is the leading cause of death in DM, with around 50–80% of DM patients dying from DCM, which is a very grim fact.3 The pathogenesis of DCM is closely linked to many factors, such as systemic metabolic disorders, mitochondrial damage, oxidative stress, impaired microcirculation, inflammation, and cardiac remodeling.4 However, the exact pathogenesis of DCM remains unclear as it involves multifaceted mechanisms. Currently, effective treatments for diabetic cardiomyopathy are limited.5 Although more drugs have been shown to be effective in treating DCM, the morbidity and mortality rates of DCM remain high, partially due to drug-related adverse effects and complications.5 For example, metformin, a drug used for treating diabetes mellitus, can adversely affect renal functions, induce fatal lactic acidosis when accumulated, and act on diseases with opposite characteristics.6,7 Thus, it is recommended that metformin should be used carefully in the elderly and in patients who have trauma, fever, surgery, heart failure, impaired kidney or liver functions.6,7 Therefore, there is an urgent need to develop novel effective and safe treatment strategies for DCM.
Traditional Chinese Medicine (TCM) has the unique advantages of multi-component, multi-target and multi-pathway regulation, which can be used to effectively prevent and treat DM and its complications including DCM.8 In addition, TCM is receiving more and more attention largely due to its precise efficacy, low price and low side effects.8 Zuogui Jiangtang Shuxin formula (ZGJTSXF) is an herbal compound created by Professor Chen Da Shun of Hunan University of Traditional Chinese Medicine. ZGJTSXF is composed of nine herbs, including Panax ginseng C. A. Meyer, Astragalus membranaceus (Fisch.), Bunge, Rehmannia glutinosa (Gaetn.) Libosch. ex Fisch. et Mey., Pueraria lobata (Willd.), Ohwi, Cornus officinalis Sieb. et Zucc., Salvia miltiorrhiza Bunge, Coptis chinensis Franch., Ophiopogon japonicus (Linn. f.) Ker-Gawl. and Crataegus pinnatifida Bge. ZGJTSXF is effective in nourishing Yin and benefiting Qi, invigorating blood, and removing toxins. Previous results in our research group showed that ZGJTSXF could improve glucolipid metabolism, reduce serum inflammatory factor production and protect the myocardium through anti-lipid peroxidation in MKR mice with type 2 diabetes.9,10 Although studies have shown the efficacy of ZGJTSXF in metabolism diseases, the material basis and mechanism of action of ZGJTSXF in the treatment of DCM are not fully understood, which limits the further development and clinical application of ZGJTSXF.
The composition of TCM is complex, but it is generally believed that only those components that can be absorbed into the bloodstream are the active ingredients for TCM to exert its medicinal effects.11 New research approaches to reveal the interaction between components of TCM and biological system networks are urgently required. Featured with the advantages of high sensitivity, high resolution and high quality accuracy, ultra high performance liquid chromatography - high resolution mass spectrometry (UPLC-Q-Exactive-Orbitrap-MS) is a powerful tool for the separation and identification of the chemical components of TCMs.12 Network pharmacology integrates the techniques and theories of systems biology, multidirectional pharmacology, computer biology and network analysis to analyse the interactions between drugs, targets and diseases in the network with the objective of biological networks, providing a new direction for deciphering the complex pharmacological mechanisms of action of TCM.13 The combinational use of UPLC-Q-Exactive-Orbitrap-MS, network pharmacology, and other bioinformatics approaches could definitely facilitate the deciphering of mechanisms of action for TCMs.
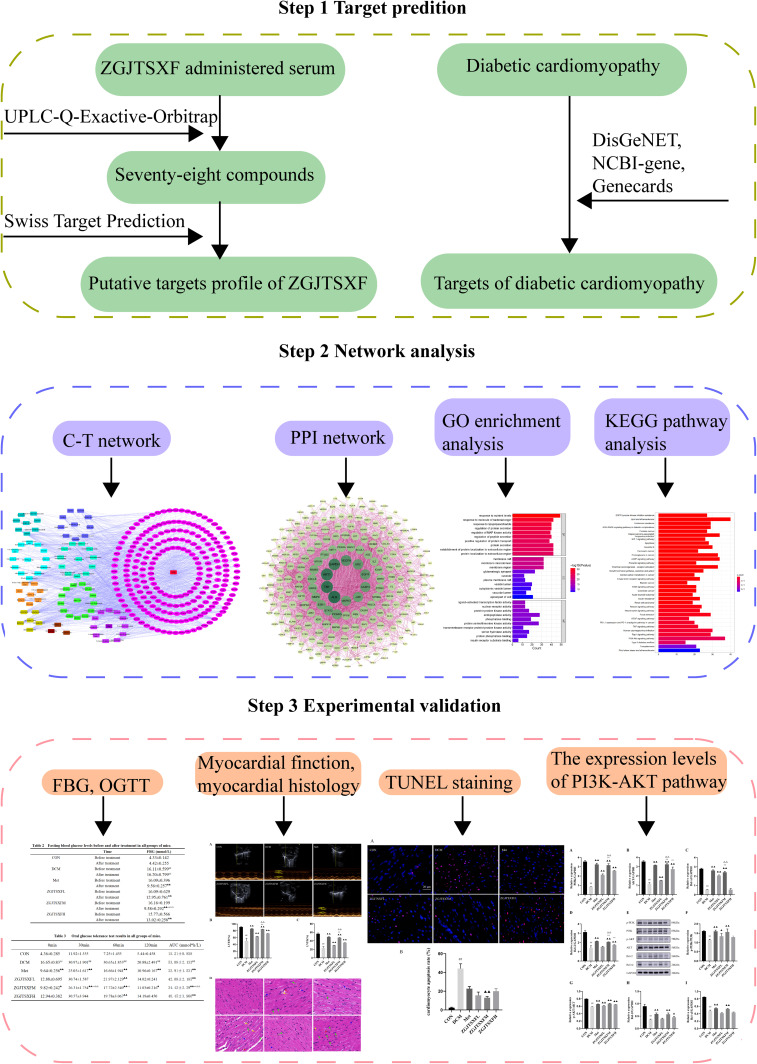
In this study, we employed the UPLC-Q-Exactive-Orbitrap technique to analyze the active components in rat serum containing the drug ZGJTSXF, and then combined the output results with the network pharmacology analysis to predict the targets of the blood components of ZGJTSXF. In addition, gene and pathway enrichment analyses were performed to further reveal the targets and pathways related to the therapeutic effects of ZGJTSXF. Moreover, a DCM mouse model was used to verify the predicted potential biological mechanisms in vivo. Our study with both in silicon network pharmacology analysis and experimental validation (Figure 1) helps elucidate the material basis and mechanism of action of ZGJTSXF for the treatment of DCM, and provides valuable insight in guiding the clinical application of ZGJTSXF and the development of new drugs for DCM patients.
Figure 1.
A schematic representation shows the outlines on studying the potential molecular mechanisms of action of Zuogui Jiangtang Shuxin formula (ZGJTSXF). The main active compounds of ZGJTSXF were identified by UPLC-Q-Exactive-Orbitrap-MS analysis of medicated rat serum. The potential targets for ZGJTSXF in treating DCM were also identified through analysis of multiple databases. The common targets were subjected to in silicon network pharmacology (C-T network and PPI network) analysis and pathway enrichment analysis. The mouse DCM model was established to experimentally validate the predicted targets and pathways through multiple cellular and molecular approaches.
Materials and Methods
Animals
Male Sprague-Dawley rats (weighing 240±10 g) were purchased from Hunan SJA Laboratory Animal Co., Ltd (Hunan, China). The MKR mice, which were first established by Fernandez et al and bear a dominant-negative IGF-1R in skeletal muscle,14 were obtained from Dr. Derek LeRoith in National Institutes of Health Diabetes Research Center (Bethesda, MD, USA). The animals were kept in an environmentally controlled room (temperature 22±2 ℃, humidity 50±10%) with access to food and water ad libitum in the specific pathogen-free (SPF) facility at the Animal Center of Hunan University of Traditional Chinese Medicine (Hunan, China). Homozygous MKR mice were used for breeding and their offspring were used for experiments. This study was carried out in accordance with the principles of the Basel Declaration and the guidelines of the National Institutes of Health (Maryland, USA). All animal experiments were approved by the Ethics Committee of Hunan University of Chinese Medicine (Hunan, China) with Animal Experiment Ethics Approval No. ZYFY20201229.
UPLC-Q-Exactive-Orbitrap-MS Analysis
ZGJTSXF was purchased from the herbal pharmacy of the First Affiliated Hospital of Hunan University of Traditional Chinese Medicine (Hunan, China). Twenty rats were randomly divided into 2 groups: the control group (n=10) received 0.9% saline IG, and the ZGJTSXF group (n=10) received 57.81 g/kg ZGJTSXF IG. The dose was determined according to the calculation method of equivalent dose coefficient of experimental animals in Methodology of Pharmacological Experiment,15 and was equivalent to 5 times of the clinical effective dose. Rats were orally dosed every day for 7 consecutive days. All rats fasted for 12 h before the experiment and then the serum samples were collected at 120 min after oral administration, via the abdominal aorta. The serum samples were centrifuged at 12,000 rpm for 10 min at 4℃. Then, 300μL of serum was mixed with 900μL methanol, which was subjected to vertexing for 10 min, and subsequent centrifugation (12,000 rpm, 15 min). The supernatant was collected for UPLC-Q-Exactive-Orbitrap-MS analysis.
UPLC-Q-TOF/MS grade acetonitrile and HPLC grade acetonitrile, methanol, formic acid, were provided by Merck KGaA (Darmstadt, Germany). UPLC-Q-Exactive-Orbitrap-MS analysis was analyzed on a Waters Corporation Xbridge BEH C18 (2.1 mm×100 mm, 2.6 m) system, which was maintained at 40 ℃. The flow rate was set at 0.3mL / min, and the injection volume was 10 µL. The mobile phase was consisted of 0.1% formic acid water (A) - 0.1% formic acid acetonitrile (B) (0~1.5 min, 2%~2% B; 1.5~20 min, 2%~45% B; 20~27 min, 45%~95% B; 27~32 min, 95%~95% B; 32~32.1 min, 95%~2% B; 32.1~35 min, 2%~2% B). The eluent was detected by a quadrupole orbitrap high resolution mass spectrometer in the ESI positive and negative ion mode. The raw data were processed using Xcalibur 4.3 and Compound Discoverer 3.2 software (Thermo Fisher Scientific, USA).
Construction of the Compound-Target and Disease-Target Networks
To obtain the potential targets of the blood-entry compounds of ZGJTSXF, the SMILES strings obtained from the PubChem database, were imported into the Swiss Target Prediction Database (http://www.swisstargetprediction.ch/). The DisGeNET (https://www.disgenet.org/), NCBI-gene (https://www.ncbi.nlm.nih.gov/), and GeneCards database (https://www.genecards.org/) were used to search for DCM targets. The common potential target of ZGJTSXF in the treatment of DCM was obtained by comparing the component targets with the disease targets lists. Then, the component-target network between the targets of blood-entry compounds components in ZGJTSXF and DCM was established using Cytoscape (Version 3.7.2).
Protein-Protein Interaction (PPI) Analysis
The common potential target of ZGJTSXF in the treatment of DCM was uploaded to the STRING database (https://string-db.org/cgi/input.pl) with the species limited as “Homo sapiens” and Combined score>0.4. The results were imported into Cytoscape (Version 3.7.2) software to establish a protein-protein interaction network. Afterward, a network topology analysis was performed by using the Cytoscape plugin Network Analyzer to count the degrees. According to the degree sorting procedure, genes with scores greater than the two times of the average score were selected as key targets.
Gene Ontology (GO) Enrichment and Kyoto Encyclopedia of Genes and Genomes (KEGG) Pathway Analysis
The common targets were introduced into R packages (Clusterprofiler, DOSE, and Pathview) following the instructions of developers. To obtain the representative biological processes and pathways (p≤0.05) of ZGJTSXF in treating DCM, Kyoto Encyclopedia of Genes and Genomes signaling pathway enrichment analysis and Gene ontology enrichment analysis were performed according to the standard procedures from the developers.
Ethical Considerations
All datasets in this study were downloaded from public databases including PubChem database, Swiss Target Prediction Database, DisGeNET, NCBI-gene, GeneCards database, STRING database. These public databases allowed researchers to download and analyze public datasets for scientific purposes and thus ethics approval was not required. Ethics Review Committee of the First Affiliated Hospital of Hunan University of Traditional Chinese Medicine exempted the study from ethical review.
Animal Model and Drug Treatments
Thirty MKR mice (male, 8 weeks old) were randomly divided into the following groups: 1) diabetic cardiomyopathy model group (DCM, n = 6); 2) metformin treatment group (Met, n = 6); 3) ZGJTSXF low dose treatment group (ZGJTSXFL, n = 6); 4) ZGJTSXF medium dose treatment group (ZGJTSXFM, n = 6); 5) ZGJTSXF high dose treatment group (ZGJTSXFH, n = 6). To induce DCM in this model, mice were injected with 1% streptozotocin (STZ; Sigma Aldrich Co., USA) dissolved in citrate buffer (pH=4.5) at a dose of 40 mg/kg/day for 5 days after 4 weeks on a high fat diet (Jiangsu Xietong, XTHF60, 60% kcal from fat). Fasting blood glucose (FBG) values were tested regularly to determine the development of diabetes in the desired groups. Six additional C57BL/6 mice of the similar sex and age were used as control group (CON, n = 6).
The human adult dose of ZGJTSXF was converted to the mouse dose in terms of body surface area based on the pharmacological test methodology.15 Met group were treated with metformin (0.25g/kg/d; CSPC OUYI Pharmaceutical Co., Ltd., China); ZGJTSXFL group were treated with ZGJTSXF (16.84 g/kg/d); ZGJTSXM group were treated with ZGJTSXF (33.67 g/kg/d); and ZGJTSXFH group were treated with ZGJTSXF (67.34 g/kg/d). The CON and DCM group were treated with equal volume of distilled water. All mice were dosed by gavage once daily for 4 weeks. After the last gavage, the mice were fasted for 12 h. Then, blood was collected from the tail vein, and the fasting blood glucose (FBG) levels were measured using a blood glucose monitor (GT-1980. Aikelai Medical Electronics (Pinghu) Co., Ltd., China). All the protocols in these experiments involving the use of animals were approved by the Animal Ethical Committee of the Hunan University of Chinese Medicine.
Glucose Tolerance Test
A glucose tolerance test was conducted one week before the end of the experiment. The mice were fasted overnight and were administered 2 g/kg glucose solution by gavage. The blood samples were collected from the tail vein at 0, 30, 60 and 120 min after glucose loading. The blood glucose levels were measured using the blood glucose monitor (GT-1980. Aikelai Medical Electronics (Pinghu) Co., Ltd., China) and blood glucose test strip (Sinocare Inc, Changsha, China). The areas under the curve (AUC) values for glucose usage and metabolism were calculated using the trapezoidal method.
Echocardiography Analysis
Echocardiographic assessment was performed by a high resolution ultrasound imaging system (VINNO 6, Vinno Corporation, Suzhou, China) with a 23 MHz probe. The animals were anesthetized with 1.5% isoflurane in 95% oxygen and 5% carbon dioxide, and the hair was removed with depilatory cream 1 day before the examination. M-mode recordings were obtained from the parasternal short axis views. The left ventricular fractional shortening (FS) and ejection fraction (EF) were measured and recorded.
Hematoxylin and Eosin (H&E) Staining
After echocardiographic analysis, all mice were sacrificed, and heart tissues were rapidly extracted from each mouse, stored at −80℃ until analysis. One part of the heart tissues was placed into a flask containing 4% paraformaldehyde. The heart samples were immersed in 10% neutral buffered formaldehyde at room temperature for 48 h, and the fixed samples were then embedded in liquid paraffin and sectioned into 5μm thickness. The sections were stained with hematoxylin and eosin, and the cardiac morphological changes were observed under a light microscope (Motic China Group Co., Ltd.).
Terminal Deoxynucleotidyl Transferase dUTP Nick End Labeling (TUNEL) Staining
TUNEL assay kit (KGA704) was purchased from KGI Biotechnology Co., Ltd. (Nanjing, China). TUNEL staining was performed according to the manufacturer’s instructions in order to determine myocardial apoptosis. The apoptotic cells showed red fluorescence and the nucleus showed blue fluorescence, six high-power fields were selected from each sample to estimate apoptosis. All cells and positive stained cells were counted, and the percentage of positively stained cells was considered as apoptotic index (AI) (AI=number of apoptotic cells/total number of nucleated cells).
Real-Time Quantitative PCR Analysis
The total RNA of cells and tissues was extracted using the Total RNA Extracting Kit (Foregene Co. Ltd., China). Total RNA was reverse-transcribed to synthesize single strand complementary DNA (cDNA) using RT EasyTM II (with gDNase) (RT-01032) kit (Foregene Co. Ltd., China). Real Time PCR EasyTM-SYBR Green I (QP-01014; Foregene Co. Ltd., China) and LightCycler 96 Instrument (Roche, Mannheim, Germany) were used for real-time quantitative PCR (qPCR). GAPDH was used as the internal reference gene for qPCR, and gene expression levels were calculated with the 2−ΔΔCT method. The primers of each gene are as follows: Forward-PI3K, 5’-CCGGAGGATGAAGCCACGCA-3’; Reverse-PI3K, 5’- CAGGGCCGTTTCCGGTGTCT-3’; Forward-AKT, 5’-TAACGGACTCGGGCTGT-3’; Reverse-AKT, 5’-TTCTCGTGGTCCTGGTTGT-3’; Forward-Bcl-xl, 5’-TCTTCTCCTTTGGCGGGGCA-3’; Reverse-Bcl-xl, 5’-CAAGGGGCGGACACACAAGG-3’; Forward-BCL-2, 5’-GCCTCTACGGCCCCTTGTCG-3’; Reverse-BCL-2, 5’-CTCGCGGTGAAGGGCGTCAG-3’; Forward-GAPDH, 5’-ACTCTTCCACCTTCGATGCC-3’; Reverse-GAPDH, 5’-TGGGATAGGGCCTCTCTTGC-3’.
Western Blot Analysis
Hearts tissues were mechanically homogenized in lysis buffer (Biyuntian Biotech Co. Ltd., China), and centrifuged at 12,000 rpm for 10 min at 4℃. The proteins-containing supernatant was collected. BCA (Bicinchoninic Acid) protein assay with the kit from Biyuntian Company was used to determine protein concentrations. Equal amounts of protein were resolved on 10% sodium dodecyl-sulfate polyacrylamide gel electrophoresis (SDS-PAGE) gels, and were transferred onto polyvinylidene difluoride (PVDF) membranes. These membranes were soaked in 1× Tris-Buffered Saline, 0.1% Tween® 20 Detergent (TBST buffer) supplemented with 5% bovine serum albumin (BSA) for 1 h, and then these PVDF membranes were incubated with the primary antibodies overnight at 4℃. Subsequently, the membrane was washed for 3 times with TBST, followed by incubation with horseradish peroxidase (HRP)-conjugated secondary antibodies for 1 h at room temperature. After rewashing with TBST, the membranes were scanned on X-ray film through chemiluminescence reaction. The super-sensitive ECL chemiluminescent substrate kit was purchased from Biosharp Life Science Co., Ltd. (China). The band intensity was analyzed using ImageJ software (National Institutes of Health, Maryland, USA). The primary antibodies used in this study were as follows: PI3 Kinase p85 alpha monoclonal antibody (60225-1-Ig), AKT polyclonal antibody (10176-2-AP), human BCL2 polyclonal antibody (12789-1-AP), Bcl-XL polyclonal antibody (26967-1-AP), and phospho-AKT (Ser473) polyclonal antibody (28731-1-AP) were provided by Proteintech Group, Inc. (Wuhan, China); anti-PI3 Kinase p85 alpha (phospho Y607) antibody (ab182651) and anti-GAPDH antibody (EPR16891) were supplied by Abcam Biotech (Shanghai, China).
Statistical Analysis
All data are expressed as the mean ± standard deviation. When the measurement data conformed to normal distribution and the homogeneity of variance test was homogeneous, the comparison between groups was performed by one-way ANOVA, while paired samples t-test was used for comparisons of data corresponding to before and after treatments. Otherwise, Kruskal Wallis test and Wilcoxon rank test are used. P<0.05 was considered to indicate a statistically significant difference. All analyses were performed using SPSS 22.0 software (IBM, USA).
Results
UPLC-Q-Exactive-Orbitrap-MS Analysis of Medicated Rat Serum Identified 78 Main Active Compounds of ZGJTSXF
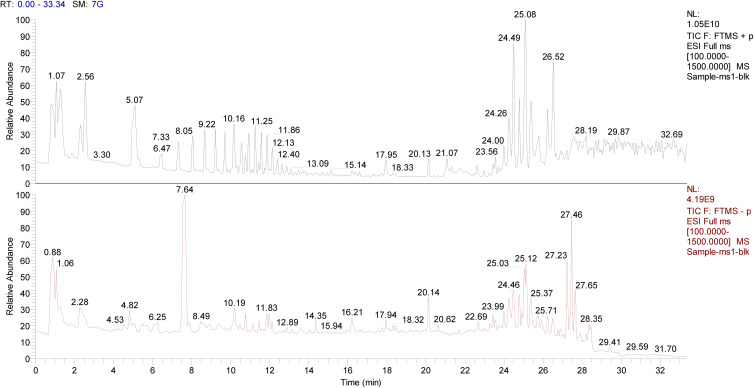
To investigate the molecular mechanisms underlying the therapeutic potential of ZGJTSXF, we dosed rats with the drug and performed UPLC-Q-Exactive-Orbitrap-MS analysis using serum samples from the medicated rats and control rats. As shown in Figure 2, the total ion chromatography in the positive and negative ion modes by UHPLC-Q-Orbitrap-MS did demonstrate different components, which were considered as the potential blood-entry compounds of ZGJTSXF. Based on the established UPLC-Q-Exactive-Orbitrap-MS method, 78 blood-entry compounds were identified (Table 1). These components can be divided into ten categories: flavonoids, small peptides, nucleosides, organic acids, phenylpropanoids, alkaloids, phenanthrenequinones, iridoids, phenols, and saponins. These 78 compounds of ZGJTSXF detected in the serum were determined to be the main active components and were further selected to predict the targets and pathways using network analysis.
Figure 2.
Total ion chromatography of the blood-entry compounds of ZGJTSXF in the positive and negative ion mode by UHPLC-Q-Orbitrap-MS.
Table 1.
Identification of Blood-Entry Components from ZGJTSXF in Serum of Rats
| No | RT [Min] | Precursor Ions | Formula | Identification | Fragment Ions | Compound Class | Origin Plant |
|---|---|---|---|---|---|---|---|
| 1 | 0.931 | 175.11911 | C6 H14 N4 O2 | DL-Arginine | 70.06522, 175.11899, 60.05593, 116.07062, 130.09756 | Small peptides | Astragalus membranaceus |
| 2 | 0.959 | 244.09302 | C9 H13 N3 O5 | Cytidine | 112.05056, 95.02399, 69.04485, 94.04008, 68.34165 | Nucleosides | Rehmannia glutinosa |
| 3 | 1.074 | 330.05869 | C10 H12 N5 O6 P | Adenosine 3’5’-cyclic monophosphate | 136.06184, 330.05981, 98.98419, 69.03362, 119.03532 | Nucleosides | Rehmannia glutinosa |
| 4 | 1.096 | 344.04007 | C10 H12 N5 O7 P | Guanosine cyclic monophosphate | 150.04204, 133.01555, 344.04007, 108.02032, 107.03622 | Nucleosides | Rehmannia glutinosa |
| 5 | 1.101 | 191.01984 | C6 H8 O7 | Citric acid | 111.00878, 87.00878, 85.02955, 191.01991, 129.01939 | Organic acids | Crataegus pinnatifida Bge/Panax ginseng C. A. Meyer |
| 6 | 1.12 | 113.03468 | C4 H4 N2 O2 | Uracil | 113.03469, 70.02885, 96.00801, 113.02348, 113.0598 | Nucleosides | Rehmannia glutinosa |
| 7 | 1.126 | 173.00916 | C6 H6 O6 | Trans-Aconitic acid | 85.0295, 129.01935, 111.00871, 173.00954, 86.03286 | Organic acids | Cornus officinalis Sieb |
| 8 | 1.139 | 190.07121 | C7 H11 N O5 | N-Acetyl-L-glutamic acid | 130.04996, 84.04443, 102.05501, 148.06041, 61.03994 | Small peptides | / |
| 9 | 1.232 | 247.12917 | C10 H18 N2 O5 | L-Isoleucyl-L-aspartic acid | 86.09642, 69.06999, 247.12881, 72.08086, 56.04981 | Small peptides | / |
| 10 | 1.243 | 117.01933 | C4 H6 O4 | Succinic acid | 73.02949, 117.0193, 99.00874, 74.03288, 118.0227 | Organic acids | Panax ginseng C. A. Meyer/Rehmannia glutinosa /Crataegus pinnatifida Bge |
| 11 | 1.443 | 169.01428 | C7 H6 O5 | Gallic acid | 125.02444, 169.01434, 69.03465, 97.02953, 81.03461 | Organic acids | Cornus officinalis Sieb |
| 12 | 1.592 | 298.11487 | C11 H15 N5 O5 | 7-Methylguanosine | 166.07243, 149.04585, 167.05634, 153.04074, 298.1373 | Nucleosides | Rehmannia glutinosa |
| 13 | 1.75 | 282.11987 | C11 H15 N5 O4 | 2’-O-Methyladenosine | 136.06183, 282.11948, 119.03525, 69.03362, 137.04579 | Nucleosides | Rehmannia glutinosa |
| 14 | 1.758 | 361.11414 | C15 H22 O10 | Catalpol | 169.01424, 125.02438, 271.0462, 361.07776, 59.01383 | Iridoids | Rehmannia glutinosa |
| 15 | 1.974 | 298.11487 | C11 H15 N5 O5 | 2’-O-Methylguanosine | 152.05681, 153.04082, 135.03024, 101.05983, 110.035 | Nucleosides | Rehmannia glutinosa |
| 16 | 2.277 | 731.22583 | C27 H42 O20 | Rehmannioside D | 179.05615, 263.07733, 71.01385, 59.01382, 89.02444 | Iridoids | Rehmannia glutinosa |
| 17 | 2.356 | 166.08641 | C9 H11 N O2 | L-Phenylalanine | 120.0808, 103.05421, 166.08623, 93.0699, 131.04916 | Small peptides | / |
| 18 | 4.095 | 312.13034 | C12 H17 N5 O5 | N2-Dimethylguanosine | 180.08795, 110.03492, 153.04073, 84.04442, 69.03363 | Nucleosides | Rehmannia glutinosa |
| 19 | 4.209 | 113.05981 | C6 H8 O2 | Sorbic acid | 113.05974, 67.0544, 95.04918, 71.04924, 85.06485 | Organic acids | Crataegus pinnatifida Bge |
| 20 | 5.077 | 158.08224 | C7 H13 N O3 | N-Acetyl-DL-norvaline | 116.0717, 158.08226, 114.09244, 159.02986, 73.02948 | Small peptides | / |
| 21 | 5.334 | 298.09698 | C11 H15 N5 O3 S | 5’-S-Methyl-5’-thioadenosine | 136.06181, 298.09689, 61.01092, 75.02634, 119.03523 | Nucleosides | Rehmannia glutinosa |
| 22 | 5.581 | 299.11374 | C14 H20 O7 | Salidroside | 59.01384, 71.01385, 89.02445, 119.05005, 119.03504 | Phenols | Astragalus membranaceus |
| 23 | 5.633 | 229.15492 | C11 H20 N2 O3 | Leucylproline | 116.07057, 86.09637, 70.06519, 229.15466, 68.88325 | Small peptides | / |
| 24 | 5.715 | 261.14462 | C11 H20 N2 O5 | Glu-Ile | 84.04442, 86.09641, 132.10196, 261.14468, 198.11235 | Small peptides | / |
| 25 | 6.015 | 337.09302 | C16 H18 O8 | 3-O-p-Coumaroylquinic acid | 163.0401, 119.0502, 191.05614, 337.09348, 173.04591 | Phenylpropanoids | Crataegus pinnatifida Bge |
| 26 | 6.47 | 405.14008 | C17 H26 O11 | Morroniside | 101.02437, 155.03502, 141.05577, 243.08743, 179.0562 | Iridoids | Cornus officinalis Sieb |
| 27 | 6.689 | 433.11285 | C21 H20 O10 | Pueraria glycoside | 433.11295, 313.07065, 283.06009, 415.10242, 255.06516 | Flavonoids | Pueraria lobata |
| 28 | 7.49 | 263.13931 | C14 H18 N2 O3 | Phe-Pro | 120.08077, 116.07059, 70.06522, 263.13895, 103.05418 | Small peptides | / |
| 29 | 7.637 | 417.11777 | C21 H20 O9 | Puerarin | 417.11801, 297.07574, 267.06512, 239.07022, 399.10742 | Flavonoids | Pueraria lobata |
| 30 | 7.741 | 291.08630 | C15 H14 O6 | Catechin | 139.039, 123.04406, 147.04404, 165.05464, 291.08691 | Flavonoids | Astragalus membranaceus |
| 31 | 7.839 | 172.09808 | C8 H15 N O3 | N-Acetyl-D-alloisoleucine | 130.08727, 172.09793, 93.03456, 128.10809, 131.09084 | Small peptides | / |
| 32 | 7.933 | 337.09302 | C16 H18 O8 | 4-O-p-Coumaroylquinic acid | 173.0455, 93.03454, 163.04004, 119.05011, 337.09344 | Phenylpropanoids | Crataegus pinnatifida Bge |
| 33 | 7.956 | 449.10788 | C21 H20 O11 | Orientin | 449.1077, 299.05484, 329.06549, 431.09714, 283.06006 | Flavonoids | Salvia miltiorrhiza Bunge |
| 34 | 8.041 | 359.13345 | C16 H22 O9 | Sweroside | 197.08084, 127.03897, 179.07016, 111.08046, 95.04912 | Flavonoids | Cornus officinalis Sieb |
| 35 | 8.075 | 447.12839 | C22 H22 O10 | 3’-Methoxypuerarin | 447.12863, 327.0863, 297.07581, 429.11816, 134.03622 | Flavonoids | Pueraria lobata |
| 36 | 8.13 | 549.15985 | C26 H28 O13 | Puerarin apioside | 297.07578, 417.11804, 267.06516, 239.07022, 549.16022 | Flavonoids | Pueraria lobata |
| 37 | 8.144 | 342.16986 | C20 H23 N O4 | (+)-Magnoflorine | 342.17007, 58.06546, 297.11212, 265.08594, 282.08862 | Alkaloids | Coptis chinensis Franch |
| 38 | 8.236 | 211.14403 | C11 H18 N2 O2 | Cyclo (pro-leu) | 211.14412, 86.09641, 70.06522, 136.07574, 98.06007 | Small peptides | / |
| 39 | 8.32 | 174.11270 | C8 H15 N O3 | N-Acetyl-D-leucine | 86.09642, 132.10196, 128.10703, 174.07648, 128.07059 | Small peptides | / |
| 40 | 8.434 | 369.11832 | C17 H20 O9 | 3-O-feruloyl-D-quinic acid | 177.05463, 145.02843, 117.03352, 89.03857, 149.05978 | Phenylpropanoids | Salvia miltiorrhiza Bunge |
| 41 | 8.561 | 367.10330 | C17 H20 O9 | 4-O-feruloyl-D-quinic acid | 173.0455, 93.03452, 134.03734, 193.05067, 137.02432 | Phenylpropanoids | Salvia miltiorrhiza Bunge |
| 42 | 8.745 | 211.14403 | C11 H18 N2 O2 | Cyclo (leucylprolyl) | 211.1441, 70.06524, 86.09643, 98.06008, 114.09142 | Small peptides | / |
| 43 | 8.767 | 417.11777 | C21 H20 O9 | Daidzin | 255.06526, 199.07542, 137.02344, 181.06476, 91.05426 | Flavonoids | Pueraria lobata |
| 44 | 9.262 | 447.12839 | C22 H22 O10 | Glycitin | 285.07578, 270.05237, 253.04944, 213.05446, 137.02332 | Flavonoids | Cornus officinalis Sieb |
| 45 | 9.275 | 431.09836 | C21 H20 O10 | Vitexin | 311.0564, 431.09839, 283.06122, 133.02931, 135.04515 | Flavonoids | Crataegus pinnatifida Bge |
| 46 | 9.294 | 208.09691 | C11 H13 N O3 | N-Acetyl-L-phenylalanine | 120.08078, 166.0862, 162.09125, 208.09569, 103.05431 | Small peptides | / |
| 47 | 9.566 | 300.99893 | C14 H6 O8 | Ellagic acid | 300.99905, 283.99643, 229.01419, 299.99142, 185.02452 | Phenols | Salvia miltiorrhiza Bunge |
| 48 | 9.587 | 417.11777 | C21 H20 O9 | Daidzein-6-C-glucoside | 417.1178, 297.07562, 267.06506, 399.10727, 239.07013 | Flavonoids | Pueraria lobata |
| 49 | 9.729 | 417.11777 | C21 H20 O9 | Daidzein 4’-O-glucoside | 255.06503, 417.11786, 199.07524, 85.02837, 137.02332 | Flavonoids | Pueraria lobata |
| 50 | 9.864 | 447.12839 | C22 H22 O10 | Calycosin-7-O-β-D-glucoside | 285.07578, 270.05219, 137.02328, 213.05438, 225.05461 | Flavonoids | Astragalus membranaceus |
| 51 | 10.052 | 417.11777 | C21 H20 O9 | Daidzein-5-C-glucoside | 417.11823, 297.07587, 267.06522, 239.07028, 399.10757 | Flavonoids | Pueraria lobata |
| 52 | 10.064 | 257.08078 | C15 H12 O4 | Liquiritigenin | 137.02333, 257.08078, 147.04399, 81.03351, 119.04916 | Flavonoids | Pueraria lobata |
| 53 | 11.175 | 283.06122 | C16 H12 O5 | Biochanin A | 283.06134, 268.03787, 211.04012, 239.03543, 240.04276 | Flavonoids | Crataegus pinnatifida Bge |
| 54 | 11.429 | 336.12277 | C20 H17 N O4 | Epiberberine | 336.12283, 320.09164, 292.09665, 262.08618, 290.08102 | Alkaloids | Coptis chinensis Franch |
| 55 | 11.558 | 338.13852 | C20H19 N O4 | Dihydroberberine | 338.13855, 322.1073, 279.08902, 294.11218, 265.07349 | Alkaloids | Coptis chinensis Franch |
| 56 | 11.803 | 338.13852 | C20 H19 N O4 | Jatrorrhizine | 338.13855, 322.10736, 279.08896, 294.11224, 265.07336 | Alkaloids | Coptis chinensis Franch |
| 57 | 11.866 | 359.07739 | C18 H16 O8 | Rosmarinic acid | 161.02443, 197.04559, 135.04517, 72.9931, 133.02951 | Phenylpropanoids | Salvia miltiorrhiza Bunge |
| 58 | 12.16 | 537.10394 | C27 H22 O12 | Lithospermic acid | 295.06131, 109.0294, 185.02446, 135.0451, 159.0452 | Phenylpropanoids | Salvia miltiorrhiza Bunge |
| 59 | 12.709 | 717.14648 | C36 H30 O16 | Salvianolic acid B | 321.04053, 519.09344, 109.02946, 339.05112, 295.06131 | Phenylpropanoids | Salvia miltiorrhiza Bunge |
| 60 | 12.88 | 255.06524 | C15 H10 O4 | Daidzein | 255.06509, 199.0753, 137.02332, 91.0542, 227.07022 | Flavonoids | Pueraria lobata |
| 61 | 12.981 | 159.10268 | C8 H16 O3 | 3-Hydroxyoctanoic acid | 59.01382, 159.10268, 159.0298, 73.02939, 129.01939 | Organic acids | Cornus officinalis Sieb |
| 62 | 13.005 | 285.04062 | C15 H10 O6 | Luteolin | 285.04053, 217.05058, 175.04002, 133.02946, 105.01924 | Flavonoids | Astragalus membranaceus |
| 63 | 13.083 | 257.08090 | C15 H12 O4 | Isoliquiritigenin | 257.08099, 137.02338, 147.04405, 81.03354, 91.05424 | Flavonoids | Pueraria lobata |
| 64 | 13.339 | 285.07596 | C16 H12 O5 | Glycitein | 285.07565, 270.05225, 137.0233, 213.05452, 253.04939 | Flavonoids | Pueraria lobata |
| 65 | 13.569 | 336.12277 | C20 H17 N O4 | Berberine | 336.12311, 320.09192, 292.09692, 278.08133, 318.07626 | Alkaloids | Coptis chinensis Franch |
| 66 | 13.58 | 493.11417 | C26 H22 O10 | Salvianolic acid A | 295.0611, 109.0294, 185.02438, 135.0451, 159.04507 | Phenylpropanoids | Salvia miltiorrhiza Bunge |
| 67 | 13.678 | 463.16034 | C23 H26 O10 | Methylnissolin-3-O-glucoside | 167.07025, 301.10709, 134.03622, 152.0468, 106.04133 | Phenols | Astragalus membranaceus |
| 68 | 13.688 | 201.11340 | C10 H18 O4 | Sebacic acid | 201.11331, 139.1129, 183.10269, 111.02, 202.11624 | Organic acids | Cornus officinalis Sieb |
| 69 | 13.759 | 285.07596 | C16 H12 O5 | Calycosin | 285.07568, 270.05222, 213.05452, 137.0233, 253.04942 | Flavonoids | Astragalus membranaceus |
| 70 | 15.102 | 269.04559 | C15 H10 O5 | Genistein | 269.04559, 133.02943, 63.02398, 224.04774, 181.06627 | Flavonoids | Pueraria lobata |
| 71 | 17.279 | 269.08072 | C16 H12 O4 | Formononetin | 269.08075, 197.05965, 253.04947, 237.05455, 254.05742 | Flavonoids | Pueraria lobata |
| 72 | 18.803 | 991.54892 | C48 H82 O18 | Ginsenoside Re | 71.01385, 101.02438, 783.48871, 621.44098, 459.38547 | Saponins | Panax ginseng C. A. Meyer |
| 73 | 20.255 | 283.06122 | C16 H12 O5 | Texasin | 283.06128, 268.03787, 267.03006, 239.03503, 211.04021 | Flavonoids | Cornus officinalis Sieb |
| 74 | 22.545 | 279.10162 | C18 H14 O3 | Dihydrotanshinone I | 279.10175, 233.09631, 205.10136, 190.07774, 261.09125 | Phenanthrenequinones | Salvia miltiorrhiza Bunge |
| 75 | 24.25 | 297.14853 | C19 H20 O3 | Cryptotanshinone | 297.14862, 251.14304, 254.09386, 282.12518, 279.13794 | Phenanthrenequinones | Salvia miltiorrhiza Bunge |
| 76 | 24.874 | 279.10178 | C18 H14 O3 | Dihydroisotanshinone II | 261.09116, 190.07771, 233.09622, 205.10127, 189.06987 | Phenanthrenequinones | Salvia miltiorrhiza Bunge |
| 77 | 25.112 | 295.22690 | C18 H30 O3 | 9-Oxo-10 (E), 12 (E)-octadecadienoic acid | 277.21631, 81.03353, 67.0544, 81.06989, 79.05424 | Organic acids | Crataegus pinnatifida Bge |
| 78 | 25.648 | 295.13300 | C19 H18 O3 | Tanshinone IIA | 295.13315, 249.1275, 277.12247, 191.08549, 206.10921 | Phenanthrenequinones | Salvia miltiorrhiza Bunge |
Component-Target Network Construction and PPI Network Plotting Identified Core Target Protein of ZGJTSXF
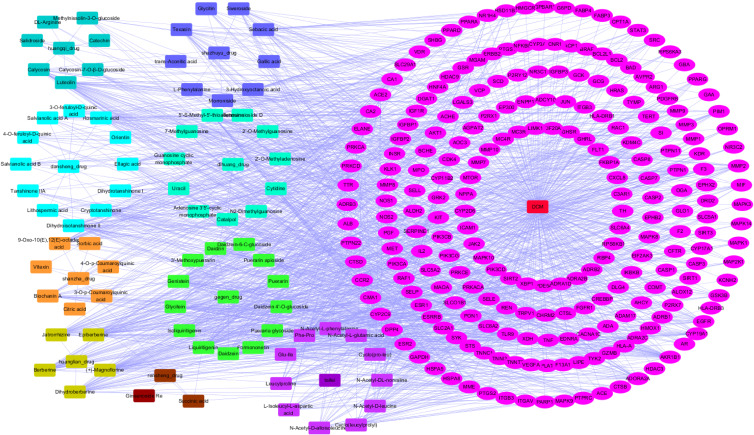
We next searched the potential targets of these selected 78 compounds of ZGJTSXF by exploring different datasets. By using Pubchem database and Swiss Target Prediction method, 827 targets for the 78 components (the SMILES strings cannot be found for one component and the therapeutic targets cannot be found for five components) were predicted, as shown in Supplementary Table S1. In addition, through searching the DisGeNET, NCBI-gene, and GeneCards databases, 1357 candidate targets of DCM were obtained, as shown in Supplementary Table S2. Taking the intersection of blood-entry compounds and candidate targets associated with DCM, 228 common targets were generated as potential targets for ZGJTSXF in treating DCM, which were used to construct a component-target network. As shown in Figure 3, the network comprised 78 components and 228 targets, and included 319 nodes and 1491 edges.
Figure 3.
The component-target network of ZGJTSXF in the treatment of DCM.
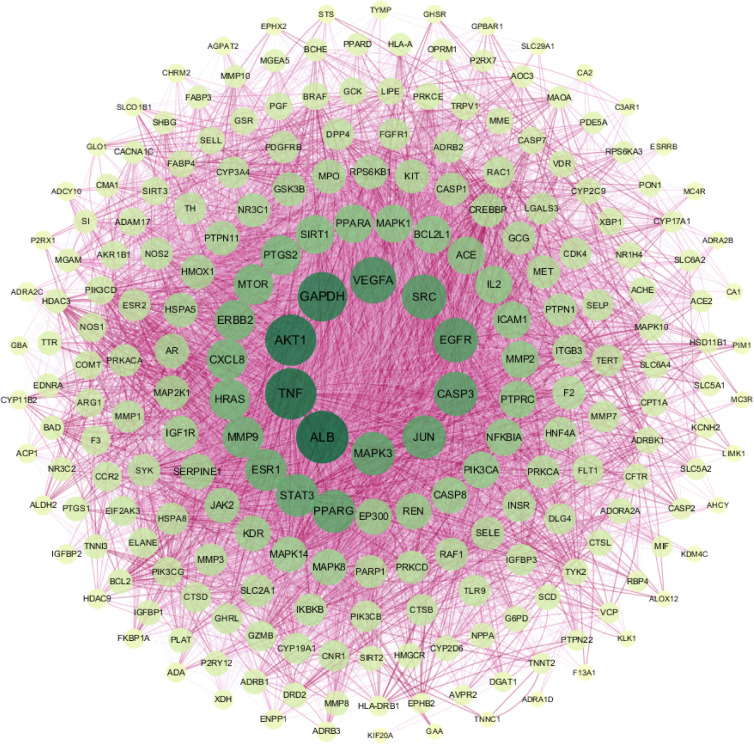
We then input the common targets into the STRING database, and the targets with no interactive relationship with others were hidden. As shown in Figure 4, the protein-protein interaction network was obtained after importing the output data from the STRING database into Cytoscape. While target genes were represented by individual nodes, their interactions were represented by edges. The color and thickness of the configuration edge follow the combine_score to continuously change, and finally the target genes were laid out into six concentric circles according to the node degree. According to the target protein interaction network diagram, the average of the degree of the relevant node was 32, and the target point with the node degree greater than twice of the average was selected as core target proteins. We identified a total of 29 key targets, including ALB, TNF, AKT1, GAPDH, VEGFA, EGFR, SRC, CASP3, MAPK3, JUN, PPARG, STAT3, ESR1, MMP9, HRAS, ERBB2, CXCL8, MTOR, PTGS2, SIRT1, PPARA, MAPK1, ACE, BCL2L1, IL2, MMP2, ICAM1, PTPRC, and NFKBIA. These targets were considered to be more relevant to ZGJTSXF in the treatment of DCM, and were predicted as the core target proteins (Supplementary Table S3).
Figure 4.
The PPI network was constructed using the common targets to predict core target proteins of ZGJTSXF.The color and thickness of the configuration edge follow the combine_score to continuously change, and finally the target genes were laid out into six concentric circles according to the node degree. The degree of the target gene node is between 21–32; the degree of the node in the fourth layer is between 33 and 40; the degree of the node in the third layer is between 41 and 60; the degree of the node in the second layer is between 60 and 100; and the degree of the node in the innermost layer is between 33 and 40. The larger the value of the parameter Degree in the figure, the larger the shape of the corresponding node and the darker the color.
GO Enrichment and KEGG Pathway Analysis Identified Critical Pathways Associated with ZGJTSXF
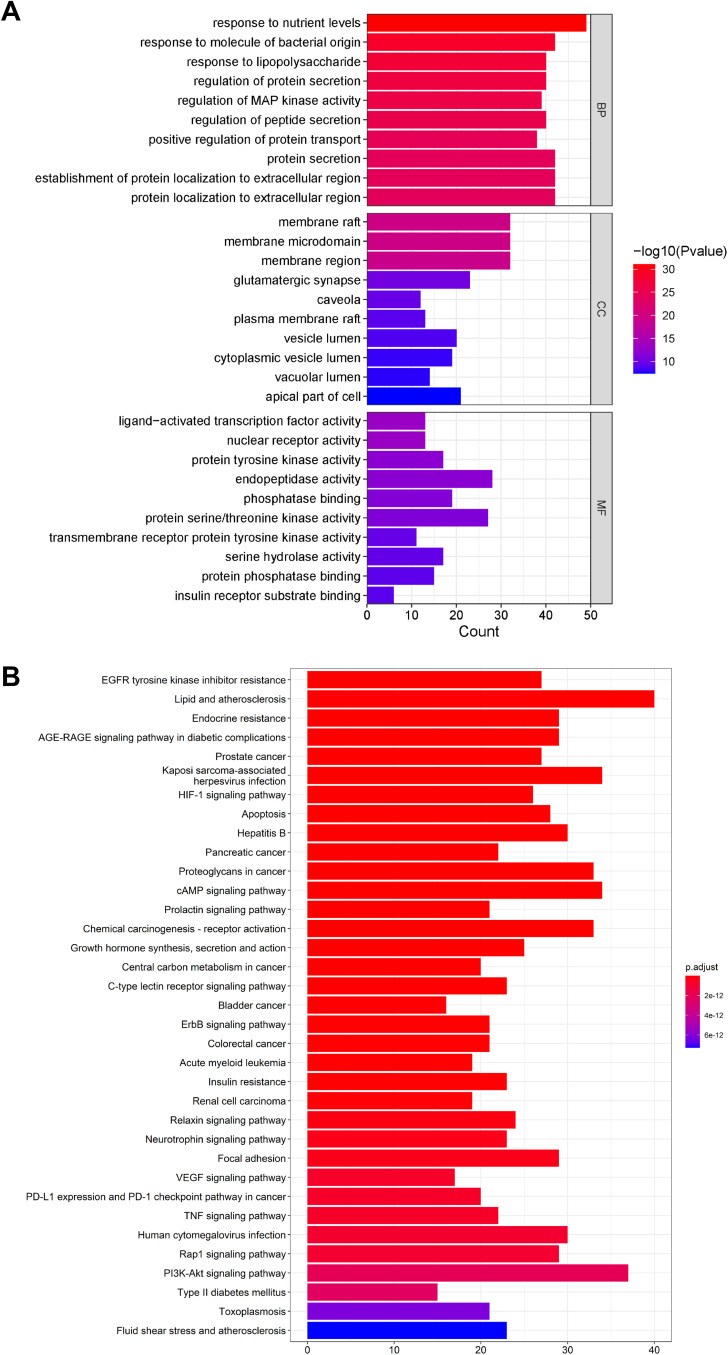
As a widely used gene annotation bioinformatics method, GO is mainly used to describe the biological process (BP) and cellular component (CC) and molecular function (MF) of gene products.16 GO analysis showed 2845 enriched processes (Supplementary Table S4), including 2550 in BP, 181 in MF, and 114 in CC. GO enrichment suggested that BP mainly involved response to nutrient levels, protein secretion, protein localization to extracellular region; CC mainly involved membrane raft, membrane microdomain, membrane region and glutamatergic synapse; MF mainly involved endopeptidase activity, protein serine/threonine kinase activity, and protein tyrosine kinase activity (Figure 5A).
Figure 5.
GO enrichment and KEGG pathway analysis identified critical pathways associated with ZGJTSXF. (A) The top 10 enrichment items of Biological Process, Molecular Function and Cellular Components from GO enrichment analysis. (B) The top 35 pathways of KEGG pathway analysis.
KEGG pathway enrichment analysis is often applied to the related functions and pathways of differentially expressed genes.17 We identified 185 signaling pathways in KEGG pathway enrichment analysis, as shown in Supplementary Table S5. The main enrichment pathways included lipid and atherosclerosis, PI3K-AKT signaling pathway, cAMP signaling pathway, Kaposi sarcoma-associated herpesvirus infection, hepatitis B, and apoptosis. The top 35 pathways were filtered out based on the P value, and the results were visualized to show the critical path diagram (Figure 5B), which indicated the pivotal involvement of the PI3K/AKT pathway and apoptosis pathway.
ZGJTSXF Administration Lowered Down Fasting Plasma Glucose and Enhanced Oral Glucose Tolerance in DCM Mice
To further verify the findings from our in silicon network pharmacology analysis, we sought to evaluate the therapeutic effects of ZGJTSXF dosing, as well as its impact on the predicted targets, in a mouse model of DCM. First, we evaluated the impacts of ZGJTSXF administration on FBG in mice with DCM. The results showed that the FBG levels of the model group were significantly higher than those of the CON group (P<0.01). After 4 weeks of intervention, compared with the DCM group, the Met group and the groups with different dosing levels of ZGJTSXF showed significantly reduced serum FBG levels in DCM mice (P<0.01). Compared with the ZGJTSXFL group and the ZGJTSXFH group, the ZGJTSXFM group seemed to be more effective, and the difference was statistically significant (P<0.01) (Table 2). Therefore, we confirmed that ZGJTSXF administration did exhibit significant benefits in terms of lowering FBG.
Table 2.
Fasting Blood Glucose Levels Before and After Treatment in All Groups of Mice
| Group | Time | FBG (mmol/L) |
|---|---|---|
| CON | Before treatment | 4.33±0.142 |
| After treatment | 4.42±0.255 | |
| DCM | Before treatment | 16.11±0.599## |
| After treatment | 16.20±0.799## | |
| Met | Before treatment | 16.09±0.396 |
| After treatment | 9.56±0.257▲▲ | |
| ZGJTSXFL | Before treatment | 16.09±0.629 |
| After treatment | 12.95±0.761▲▲ | |
| ZGJTSXFM | Before treatment | 16.16±0.199 |
| After treatment | 9.58±0.292▲▲**ΔΔ | |
| ZGJTSXFH | Before treatment | 15.77±0.566 |
| After treatment | 13.02±0.258▲▲ |
Notes: ##P<0.01, compared with the CON group; ▲▲P<0.01, compared with the DCM group; **P<0.01, compared with the ZGJTSXFL group; ΔΔP<0.01, compared with the ZGJTSXFH group.
Next, we evaluated the impacts of ZGJTSXF administration on oral glucose tolerance in mice with DCM. As shown in Table 3, after the intake of glucose, compared with the CON group, the DCM group had significantly increased blood glucose level at each time point (P<0.01), while the Met group and ZGJTSXFM group after treatment significantly alleviated the blood glucose increase in DCM mice (P<0.01). Moreover, the ZGJTSXFL and ZGJTSXFH treatments significantly alleviated the blood glucose level increase in DCM mice at the 60min point (P<0.01). Compared with the DCM group, each administration group showed a significant difference in the AUC (P < 0.01). Compared with the ZGJTSXFL group and the ZGJTSXFH group, the ZGJTSXFM group was more effective in enhancing oral glucose tolerance, and the difference was statistically significant (P<0.01) (Table 3). Collectively, these results confirmed that ZGJTSXF administration exhibited beneficial effects on oral glucose tolerance in mice with DCM.
Table 3.
Oral Glucose Tolerance Test Results in All Groups of Mice
| Group | 0Min | 30Min | 60Min | 120Min | AUC (mmol*h/L) |
|---|---|---|---|---|---|
| CON | 4.36±0.285 | 11.92±1.555 | 7.25±1.455 | 5.44±0.458 | 15.21±0.858 |
| DCM | 16.65±0.83## | 30.97±1.901## | 30.65±1.853## | 20.88±2.491## | 53.08±2.133## |
| Met | 9.64±0.256▲▲ | 25.05±1.617▲▲ | 16.66±1.941▲▲ | 10.96±0.167▲▲ | 32.91±1.531▲▲ |
| ZGJTSXFL | 12.88±0.695 | 30.74±1.587 | 21.97±2.129▲▲ | 14.02±0.241 | 42.08±2.185▲▲ |
| ZGJTSXFM | 9.82±0.242▲ | 26.31±1.754▲▲**ΔΔ | 17.72±2.549▲▲** | 11.03±0.216▲ | 34.42±2.29▲▲**ΔΔ |
| ZGJTSXFH | 12.94±0.382 | 30.57±3.944 | 19.78±3.067▲▲ | 14.19±0.450 | 40.45±3.966▲▲ |
Notes: ##P<0.01, compared with the CON group;▲▲P<0.01,▲P<0.05, compared with the DCM group; **P<0.01, compared with the ZGJTSXFL group; ΔΔP<0.01, compared with the ZGJTSXFH group.
ZGJTSXF Administration Promoted Myocardial Function and Improved Myocardial Histology in DCM Mice
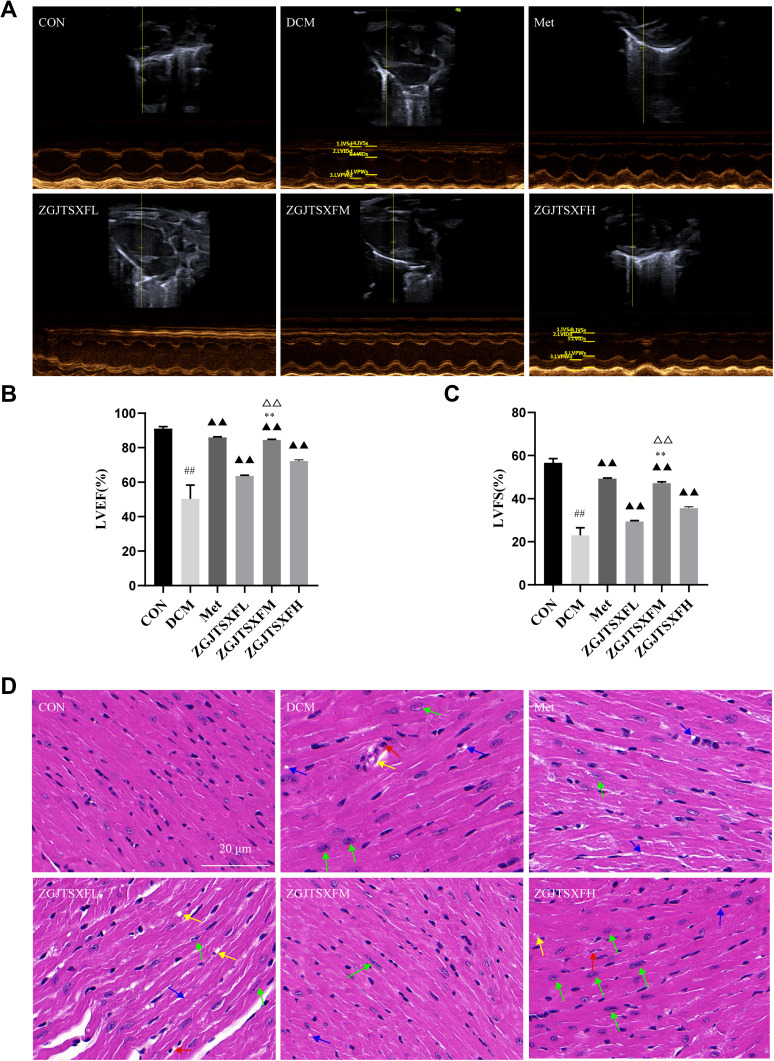
We also evaluated the effects of ZGJTSXF on echocardiography in DCM mice. As shown in Figure 6A–C, echocardiography analysis found that mice in DCM group had significantly reduced left ventricular ejection fraction (LVEF) and left ventricular fractional shortening (LVFS), compared with mice in CON group. Notably, mice in the Met, ZGJTSXFL, ZGJTSXFM, and ZGJTSXFH groups showed significantly increased LVEF (Figure 6B) and LVFS (Figure 6C) (P<0.01). In addition, the LVEF and LVFS of mice in the ZGJTSXFM group significantly increased compared to those of mice in the ZGJTSXFL and ZGJTSXFH groups (P<0.01).
Figure 6.
ZGJTSXF administration changed myocardial function and myocardial histology in DCM mice. (A–C) Echocardiographic observation of changes in cardiac function in various groups of mice. (A) Representative M-mode echocardiograms in mice of the indicated groups. (B) Summary on the left ventricular fractional shortening (FS). (C) Summary on the left ventricular ejection fraction (EF). ##P<0.01, compared with the CON group; ▲▲P<0.01, compared with the DCM group; **P<0.01, compared with the ZGJTSXFL group; ΔΔP<0.01, compared with the ZGJTSXFH group. (D) Histopathological changes in myocardial tissues of mice in the indicated groups were observed by HE staining (Scale bar, 20 μm). (a) CON group; (b) DCM group; (c) Met group, (d) ZGJTSXFL group; (e) ZGJTSXFM group; (f) ZGJTSXFH group. Green arrow, cardiomyocyte hypertrophy; red arrow, small focal inflammatory cell infiltrates in the interstitial myocardium; blue arrow, vacuolar degeneration of cardiomyocytes; yellow arrow, thickening of the capillary basement membrane.
We further checked how ZGJTSXF administration could influence the histological changes in myocardial tissues. As shown in Figure 6D, in the CON group of mice, the ventricular wall and papillary muscle cells were arranged neatly and regularly; the nuclei were of uniform size; the cell gaps were not widened or narrowed; the morphology and structure were good. No pathological changes were detected in the CON group. In the DCM group, there was a widening of the interstitial space, hypertrophy of cardiomyocytes (green arrow), inflammatory cell infiltration (red arrow) with vacuole-like degeneration (blue arrow), thickening of the capillary basement membrane (yellow arrow) and coagulative necrosis of cardiomyocytes. However, cardiomyocyte hypertrophy, inflammatory infiltration and vacuolation, and capillary basement membrane thickening were significantly improved in the Met and ZGJTSXF groups, compared to the DCM group. Among the ZGJTSXF treatment groups, the most significant pathological improvement was seen in the ZGJTSXFM group (Figure 6D). Taken together, these results indicated that ZGJTSXF administration promoted myocardial function and improved myocardial histology in DCM mice.
ZGJTSXF Administration Inhibited Apoptosis and Activated the PI3K-AKT Pathway in Myocardial Tissues of DCM Mice
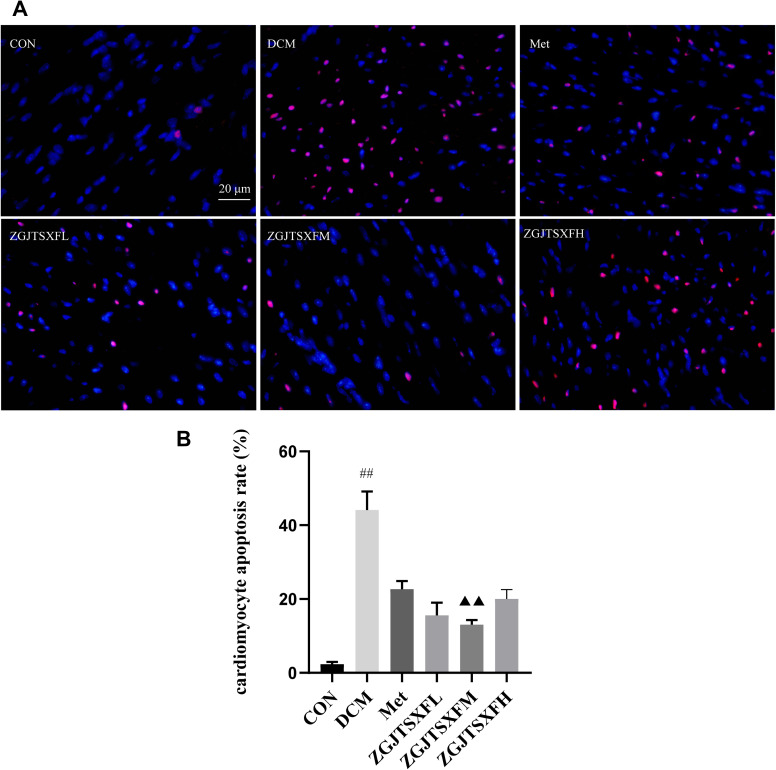
We further examined whether the predicted target pathways of ZGJTSXF can be experimentally validated in the DCM mice model, with a focus on the apoptosis and PI3K-AKT pathways in myocardial tissues. We first quantitated the apoptosis of mouse cardiomyocytes through TUNEL assay. As shown in Figure 7A and B, the apoptosis rate of cardiomyocytes in the DCM group was significantly higher than that in the CON group (P<0.01). Compared with the DCM group, the Met, ZGJTSXFL, ZGJTSXFM and ZGJTSXFH groups demonstrated significantly reduced cardiomyocyte apoptosis (P<0.01). Among the three ZGJTSXF treatment groups, the ZGJTSXFM group had the lowest cardiomyocyte apoptosis rate (P<0.01 and P<005). However, all these three groups did not show a higher cardiomyocyte apoptosis rate than the Met group (Figure 7B).
Figure 7.
Apoptosis of mouse myocardial tissues was evaluated by TUNEL staining. (A) Representative images of apoptosis of mice myocardial tissues after TUNEL staining (Scale bar, 20 μm). (B) Apoptosis rate of cardiomyocytes in mice of the indicated group was summarized. n=6 for each group; ##P<0.01, compared with the CON group; ▲▲P<0.01, compared with the DCM group; **P<0.01, *P<0.05, compared with the ZGJTSXFM group.
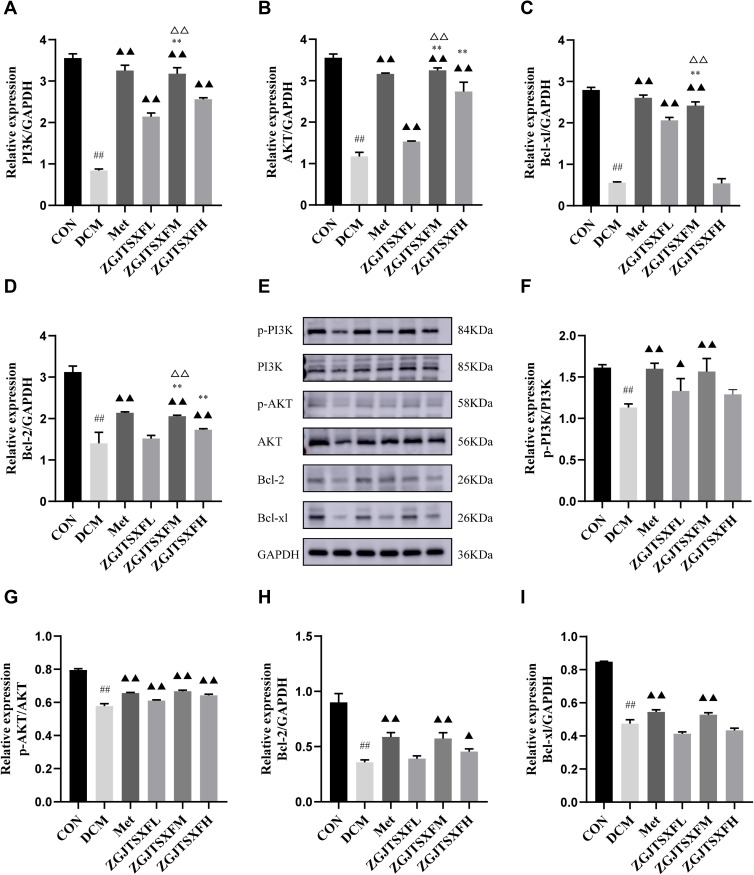
The transcription and protein levels of PI3K-AKT pathway molecules and apoptosis-associated molecules (Bcl-2 and Bcl-xL) were quantitated by qPCR and Western blot assays (Figure 8), respectively, to verify the previous findings on network pharmacology analysis of ZGJTSXF. Compared with the CON group, the DCM group had significantly decreased mRNA levels of PI3K (Figure 8A), AKT (Figure 8B), Bcl-2 (Figure 8C), and Bcl-xL (Figure 8D) in the myocardial tissues (P<0.01). After the 4-week treatment period, compared with the DCM group, both the Met group and the ZGJTSXFM group showed significantly elevated mRNA expression of PI3K, AKT, Bcl-2, and Bcl-xL in mice with DCM (P < 0.01) (Figure 8A–D). Compared with the DCM group, significant upregulated expression levels of PI3K, Akt and Bcl-xL in myocardial tissues were observed in the ZGJTSXFH group (P<0.01), while there were no significant differences in the expression of Bcl-xL. The Western blot assay (Figure 8E) showed largely consistent expression patterns of these molecules among the groups. Both Met treatment and ZGJTSXF treatment could significantly restore the ratios of p-PI3K/PI3K (Figure 8F) and p-AKT/AKT (Figure 8G) that were downregulated in the DCM group (P<0.05). Similarly, the Met group and the ZGJTSXFM group had significantly higher protein expressions of Bcl-2 (Figure 8H) and Bcl-xL (Figure 8I) than the DCM group (P<0.05). Compared with the ZGJTSXFL group and the ZGJTSXFH group, the ZGJTSXFM group was more effective in upregulating the expression of Bcl-2 and Bcl-xL, and the differences were statistically significant (P<0.01). Taken together, these results demonstrated that ZGJTSXF administration inhibited apoptosis and activated the PI3K-AKT pathway in myocardial tissues of DCM mice.
Figure 8.
The expression levels of PI3K-AKT pathway molecules and apoptosis-related molecules in mouse myocardial tissues were quantitated. (A–D) The mRNA levels of PI3K (A), AKT (B), Bcl-2 (C), and Bcl-xL (D) in mouse myocardial tissues of the indicated groups were quantitated by qPCR. n=6 for each group; ##P<0.01, Compared with the CON group; ▲▲P<0.01, compared with the DCM group; **P<0.01, compared with the ZGJTSXFL group; ΔΔP<0.01, compared with the ZGJTSXFH group. (E–I) The protein levels of phosphorylated PI3K (p-PI3K; (F), phosphorylated AKT (p-AKT; (G), Bcl-2 (H), and Bcl-xL (I) in mouse myocardial tissues of the indicated groups were quantitated by Western blot assay. Representative images of Western blot bands are shown (E), and the relative expressions were summarized. n=6 for each group; ##P<0.01, compared with the CON group; n=6 for each group; ▲▲P<0.01, ▲P<0.05, compared with the DCM group.
Discussion
Diabetic cardiomyopathy is the leading cause of death among diabetics and a major complication of diabetes mellitus.1,4 Because of the complex mechanisms, multiple protein targets, and pathways involved during the development and progression of DCM, a single targeted drug cannot achieve the desired therapeutic effect.1,4 ZGJTSXF, characterized by multiple components and targets, has been shown to have a relatively satisfactory therapeutic effect in treating DCM.9,10 Here, we found that ZGJTSXF improved blood glucose and reduced diabetic cardiomyocyte damage by suppressing myocardial apoptosis, which coincided with the holistic view and synergistic action theory of TCM. However, it is difficult to accurately identify the active compounds and mechanisms of ZGJTSXF in treating DCM solely by using conventional pharmacological methods. Thus, an integrated strategy was established by combining serum pharmacochemistry and network pharmacology to investigate the components and mechanisms of ZGJTSXF in treating DCM in a comprehensive and systematical manner, which was followed by experimental validation. We found that ZGJTSXF could protect the myocardium in diabetic cardiomyopathy mice by regulating the PI3K/Akt pathway and suppressing apoptosis of cardiomyocytes.
We identified 78 compounds in rat serum after oral administration of ZGJTSXF, and these main active components of ZGJTSXF were demonstrated to exert beneficial roles in various diseases. Among them, flavonoids, such as puerarin, daidzin, luteolin, calycosin-7-O-β-D-glucoside, calycosin and formononetin, are the most abundant. They can alleviate diabetic cardiomyopathy or other cardiovascular complications by their anti-hyperglycemia,18 anti-hyperlipidemia,19 anti-oxidant,20 anti-inflammatory21 and anti-apoptotic22 functions. Small peptides, such as N-Acetyl-D-leucine, Cyclo (leucylprolyl) and N-Acetyl-L-phenylalanine are considered to be a new generation of modulators of biological activity, which can prevent cellular oxidation and inflammation, and can be used to prevent the development of diabetes.23 Organic acids, such as gallic acid and sorbic acid, have been shown to have various biological activities including antioxidant, antimicrobial, and anticancer capacities. Gallic acid probably attenuate inflammation, oxidative stress and hypotension resulted from diabetes via increasing plasma miR-24 and miR-126 levels.24 Furthermore, phenylpropanoids, such as rosmarinic acid, salvianolic acid B and Salvianolic acid A, can also exert beneficial functions in multiple diabetic or obese animal models.25–28 Therefore, our UPLC-Q-Exactive-Orbitrap-MS analysis revealed the multi-component nature of ZGJTSXF, and the reported functions of major active components of ZGJTSXF support the advantage of ZGJTSXF in multi-target and multi-pathway regulation.
Our integrated network analysis identified multiple targets and signaling pathways that potentially contribute to the beneficial roles of ZGJTSXF, while we focused on apoptosis and the PI3K-AKT signaling pathway. An important step in the pathogenesis of diabetic cardiomyopathy is cardiomyocyte apoptosis, which involves a variety of complex molecular biological mechanisms.1,5 Through controlling the pathways involved in apoptotic signaling, cardiomyocyte apoptosis can be minimized. PI3K/Akt is a key intracellular signaling pathway involved in cellular survival, apoptosis, proliferation, etc. When activated, this pathway significantly reduces cardiomyocyte apoptosis and myocardial damage, and when inhibited, the opposite happens.29–31 When upstream protein kinases such as PDK1 are phosphorylated, Akt translocation takes place, revealing binding sites and resulting in catalytic activity.32 The activation of the Akt pathway provides cells with a survival signal, which allows them to withstand apoptotic stimuli. This is regulated through the balance between proapoptotic and antiapoptotic proteins.32
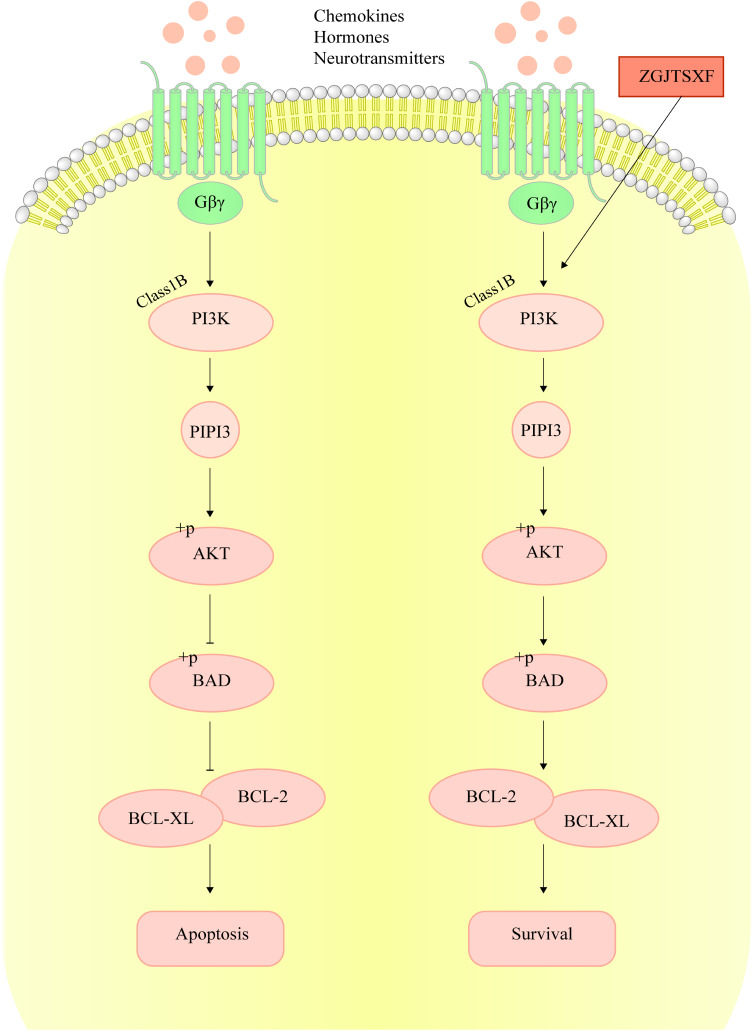
Bcl-2 family proteins play an important role in cell apoptosis, which include pro-apoptotic (like BAK, BAX, and BAD) and antiapoptotic (BCL-2 and BCL-XL) proteins.33 The increase in apoptosis was associated with decreased expression of anti-apoptotic genes BCL-2 and BCL-XL.34 Therefore, the increased expression of Bcl-xL and Bcl-2 in myocardial tissues upon ZGJTSXF treatment suggested the suppression of apoptosis. In our study, we found that ZGJTSXF administration positively regulated the PI3K/Akt signaling pathways, which was associated with increased expressions of anti-apoptotic proteins Bcl-2 and Bcl-xL (Figure 9). These findings are consistent with previous studies,29–31 and indicating a positive correlation between the activation of PI3K-AKT pathway and apoptosis inhibition in cardiomyocytes.
Figure 9.
ZGJTSXF inhibits cardiomyocyte apoptosis by regulating PI3K/Akt signaling pathway.
We used spontaneous nonobese T2DM MKR mice fed with a high-fat diet to establish the DCM model, as MKR mice is one of the ideal animal models for studying T2DM and diabetes complications.35 The results of this study showed that the FPG in the model group was significantly higher than that in the control group. Further histology and echocardiographic observation verified that the DCM model was successfully established in this study. The positive control drug, metformin, a derivative of biguanide, has an excellent hypoglycemic effect and is used a first-line treatment for type 2 diabetes mellitus.36 A previous study revealed that metformin can improve many clinical parameters and reduced cardiovascular disease events in Chinese type 2 diabetes patients as compared with lifestyle changes alone.37 Zhang et al found that metformin exerts cardioprotective effect by alleviating myocardial structural lesions, suppressing myocardial apoptosis, and inhibiting inflammatory response in vivo and in vitro.38 Metformin can mediate the expression of multiple biomarkers and pathways, such as AMPK, endothelial nitric oxide synthase (eNOS), NF-κB, and the PI3K/AKT signaling pathways, etc., thereby exerting anti-inflammatory, anti-apoptotic, and anti-oxidant stress effects, etc.39
In this study, we discovered that the Met and ZGJTSXF groups can reduce hypertrophy, vacuolar degeneration of cardiomyocytes, myocardial interstitial infiltration of inflammatory cells, capillary basement membrane thickened, and myocardial fibrosis caused by diabetic cardiomyopathy, thus improving cardiac function. As predicted by network pharmacology analysis, we focused on the role of PI3K/AKT signaling pathway in the mechanisms of ZGJTSXF against myocardial apoptosis in diabetic cardiomyopathy. Our results showed that ZGJTSXF exerted similar effects as metformin in activating the PI3K/Akt pathway and regulating Bcl-2 and Bcl-xL proteins expression. It is worth noting that the ZGJTSXF medium-dose group exhibited superior therapeutic effects in multiple aspects to the ZGJTSXF high-dose group, implying possible side effect elicited by a high amount of ZGJTSXF dosing.
This study has some limitations. First, we conducted research on the role of ZGJTSXF in suppressing apoptosis using only mouse tissues. Due to the difficulty of obtaining human myocardial tissue, we have not started clinical trials for the time being, which means that the clinical transformation capabilities of these findings need to be strengthened. Therefore, in the future, we will further carry out randomized controlled clinical studies. Second, one limitation of the current study is the lack of evaluation on the dynamic changes of the PI3K/AKT pathway and apoptosis pathway in cardiomyocytes of DCM mice following ZGJTSXF administration. In addition, since the tissue samples were obtained after 4-week treatment, the short-term and long-term effects of ZGJTSXF treatments are also needed to be elucidated in more in-depth studies.
Conclusion
In this study, the active components and molecular mechanisms of ZGTSXF against DCM were illustrated by adopting an integrated strategy, which employed UPLC-Q-Exactive-Orbitrap technique, network pharmacological analysis, and experimental validation. A total of 78 components of ZGJTSXF was identified by UPLC-Q-Exactive-Orbitrap study and adopted for further network pharmacology analysis. Through experimental validating the hub targets and hub signaling pathways by network pharmacological analysis, we demonstrated that ZGJTSXF could exert its antidiabetic cardiomyopathy effect through suppressing cardiomyocytes apoptosis and restoring PI3K/AKT signaling in mouse myocardial tissues. Our work sheds new light in further developing ZGJTSXF-based therapeutics and guiding clinical application of these therapeutics in treating DCM. Additionally, our study also suggests that the combination of network pharmacology prediction and experimental validation can effectively elucidate the multi-target and multilink pharmacological mechanisms of TCM formula.
Funding Statement
This work was supported by grants from the National Natural Science Foundation of China (82074400), Hunan Provincial Technology Key Research and Development Program (2020SK2101), National Natural Science Foundation of China (82004185), National Natural Science Foundation of China (U21A20411), Postgraduate Research and Innovation Project of Hunan Province (CX20210692), and Hunan Provincial Key Laboratory of Translational Medicine for TCM Recipe and Syndrome Research (2018TP1021).
Author Contributions
All authors made a significant contribution to the work reported, whether that is in the conception, study design, execution, acquisition of data, analysis and interpretation, or in all these areas; took part in drafting, revising or critically reviewing the article; gave final approval of the version to be published; have agreed on the journal to which the article has been submitted; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare that they have no conflicts of interest in relation to this work.
References
- 1.Jia G, Hill MA, Sowers JR. Diabetic cardiomyopathy: an update of mechanisms contributing to this clinical entity. Circ Res. 2018;122(4):624–638. doi: 10.1161/CIRCRESAHA.117.311586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sun H, Saeedi P, Karuranga S, et al. IDF diabetes atlas: global, regional and country-level diabetes prevalence estimates for 2021 and projections for 2045. Diabetes Res Clin Pract. 2022;183:109119. doi: 10.1016/j.diabres.2021.109119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zheng H, Yang Z, Xin Z, et al. Glycogen synthase kinase-3β: a promising candidate in the fight against fibrosis. Theranostics. 2020;10(25):11737–11753. doi: 10.7150/thno.47717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Parim B, Sathibabu Uddandrao VV, Saravanan G. Diabetic cardiomyopathy: molecular mechanisms, detrimental effects of conventional treatment, and beneficial effects of natural therapy. Heart Fail Rev. 2019;24(2):279–299. doi: 10.1007/s10741-018-9749-1 [DOI] [PubMed] [Google Scholar]
- 5.Lorenzo-Almorós A, Cepeda-Rodrigo JM, Lorenzo Ó. Diabetic cardiomyopathy. Rev Clin Esp. 2022;222(2):100–111. doi: 10.1016/j.rceng.2019.10.012 [DOI] [PubMed] [Google Scholar]
- 6.DeFronzo R, Fleming GA, Chen K, Bicsak TA. Metformin-associated lactic acidosis: current perspectives on causes and risk. Metabolism. 2016;65(2):20–29. doi: 10.1016/j.metabol.2015.10.014 [DOI] [PubMed] [Google Scholar]
- 7.Thomas I, Gregg B. Metformin; a review of its history and future: from lilac to longevity. Pediatr Diabetes. 2017;18(1):10–16. doi: 10.1111/pedi.12473 [DOI] [PubMed] [Google Scholar]
- 8.Hao P, Jiang F, Cheng J, Ma L, Zhang Y, Zhao Y. Traditional Chinese Medicine for Cardiovascular Disease: evidence and Potential Mechanisms. J Am Coll Cardiol. 2017;69(24):2952–2966. doi: 10.1016/j.jacc.2017.04.041 [DOI] [PubMed] [Google Scholar]
- 9.Cheng X, Huang ZD. Effect of Zuogui Jiangtang Shuxin Recipe on glucose-lipid metabolism and inflammatory cytokines in high-fat diet MKR mice. Chin Tradit Herb Drugs. 2011;42(03):546–549. [Google Scholar]
- 10.Cheng X, Wu YJ. Protective effect of Zuogui Jiangtang Shuxin Recipe on myocardial impairment in MKR mice. Chin Tradit Herb Drugs. 2011;42(02):343–345. [Google Scholar]
- 11.Wang KX, Gao Y, Gong WX, et al. A novel strategy for decoding and validating the combination principles of huanglian jiedu decoction from multi-scale perspective. Front Pharmacol. 2020;11:567088. doi: 10.3389/fphar.2020.567088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wang K, Tian J, Li Y, et al. Identification of components in citri sarcodactylis fructus from different origins via UPLC-Q-exactive Orbitrap/MS. ACS Omega. 2021;6(26):17045–17057. doi: 10.1021/acsomega.1c02124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hopkins AL. Network pharmacology: the next paradigm in drug discovery. Nat Chem Biol. 2008;4(11):682–690. doi: 10.1038/nchembio.118 [DOI] [PubMed] [Google Scholar]
- 14.Fernández AM, Kim JK, Yakar S, et al. Functional inactivation of the IGF-I and insulin receptors in skeletal muscle causes type 2 diabetes. Genes Dev. 2001;15(15):1926–1934. doi: 10.1101/gad.908001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nair A, Morsy MA, Jacob S. Dose translation between laboratory animals and human in preclinical and clinical phases of drug development. Drug Dev Res. 2018;79(8):373–382. doi: 10.1002/ddr.21461 [DOI] [PubMed] [Google Scholar]
- 16.Carbon S, Douglass E, Good BM. The gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49(D1):D325–D334. doi: 10.1093/nar/gkaa1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Sato Y, Kawashima M, Furumichi M, Tanabe M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016;44(D1):D457–462. doi: 10.1093/nar/gkv1070 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ghorbani A. Mechanisms of antidiabetic effects of flavonoid rutin. Biomed Pharmacother. 2017;96:305–312. doi: 10.1016/j.biopha.2017.10.001 [DOI] [PubMed] [Google Scholar]
- 19.Musolino V, Gliozzi M, Scarano F, et al. Bergamot polyphenols improve dyslipidemia and pathophysiological features in a mouse model of non-alcoholic fatty liver disease. Sci Rep. 2020;10(1):2565. doi: 10.1038/s41598-020-59485-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Garcia JP, Santana A, Baruqui DL, Suraci N. The Cardiovascular effects of chocolate. Rev Cardiovasc Med. 2018;19(4):123–127. doi: 10.31083/j.rcm.2018.04.3187 [DOI] [PubMed] [Google Scholar]
- 21.Maleki SJ, Crespo JF, Cabanillas B. Anti-inflammatory effects of flavonoids. Food Chem. 2019;299:125124. doi: 10.1016/j.foodchem.2019.125124 [DOI] [PubMed] [Google Scholar]
- 22.Yu H, Chen B, Ren Q. Baicalin relieves hypoxia-aroused H9c2 cell apoptosis by activating Nrf2/HO-1-mediated HIF1α/BNIP3 pathway. Artif Cells Nanomed Biotechnol. 2019;47(1):3657–3663. doi: 10.1080/21691401.2019.1657879 [DOI] [PubMed] [Google Scholar]
- 23.Mesgari-Abbasi M, Valizadeh H, Mirzakhani N, Vahdatpour T. Protective effects of di- and tri-peptides containing proline, glycine, and leucine on liver enzymology and histopathology of diabetic mice. Arch Physiol Biochem. 2022;128(1):59–68. doi: 10.1080/13813455.2019.1662453 [DOI] [PubMed] [Google Scholar]
- 24.Ramezani Ali Akbari F, Badavi M, Dianat M, Mard SA, Ahangarpour A. Gallic acid improves oxidative stress and inflammation through regulating micrornas expressions in the blood of diabetic rats. Acta Endocrinol. 2019;15(2):187–194. doi: 10.4183/aeb.2019.187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ren Y, Tao S, Zheng S, et al. Salvianolic acid B improves vascular endothelial function in diabetic rats with blood glucose fluctuations via suppression of endothelial cell apoptosis. Eur J Pharmacol. 2016;791:308–315. doi: 10.1016/j.ejphar.2016.09.014 [DOI] [PubMed] [Google Scholar]
- 26.Runtuwene J, Cheng KC, Asakawa A, et al. Rosmarinic acid ameliorates hyperglycemia and insulin sensitivity in diabetic rats, potentially by modulating the expression of PEPCK and GLUT4. Drug Des Devel Ther. 2016;10:2193–2202. doi: 10.2147/DDDT.S108539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.An T, Zhang J, Lv B, et al. Salvianolic acid B plays an anti-obesity role in high fat diet-induced obese mice by regulating the expression of mRNA, circRNA, and lncRNA. PeerJ. 2019;7:e6506. doi: 10.7717/peerj.6506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li L, Li R, Zhu R, et al. Salvianolic acid B prevents body weight gain and regulates gut microbiota and LPS/TLR4 signaling pathway in high-fat diet-induced obese mice. Food Funct. 2020;11(10):8743–8756. doi: 10.1039/d0fo01116a [DOI] [PubMed] [Google Scholar]
- 29.Rajesh KG, Suzuki R, Maeda H, Yamamoto M, Yutong X, Sasaguri S. Hydrophilic bile salt ursodeoxycholic acid protects myocardium against reperfusion injury in a PI3K/Akt dependent pathway. J Mol Cell Cardiol. 2005;39(5):766–776. doi: 10.1016/j.yjmcc.2005.07.014 [DOI] [PubMed] [Google Scholar]
- 30.Dhanasekaran A, Gruenloh SK, Buonaccorsi JN, et al. Multiple antiapoptotic targets of the PI3K/Akt survival pathway are activated by epoxyeicosatrienoic acids to protect cardiomyocytes from hypoxia/anoxia. Am J Physiol Heart Circ Physiol. 2008;294(2):H724–735. doi: 10.1152/ajpheart.00979.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ebner B, Lange SA, Hollenbach D, et al. In situ postconditioning with neuregulin-1β is mediated by a PI3K/Akt-dependent pathway. Can J Cardiol. 2015;31(1):76–83. doi: 10.1016/j.cjca.2014.10.035 [DOI] [PubMed] [Google Scholar]
- 32.Kim D, Dan HC, Park S, et al. AKT/PKB signaling mechanisms in cancer and chemoresistance. Front Biosci. 2005;10:975–987. doi: 10.2741/1592 [DOI] [PubMed] [Google Scholar]
- 33.Niu F, Qian K, Qi H, Zhao Y, Jiang Y, Sun M. Antiapoptotic and anti-inflammatory effects of CPCGI in rats with traumatic brain injury. Neuropsychiatr Dis Treat. 2020;16:2975–2987. doi: 10.2147/NDT.S281530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Opferman JT, Kothari A. Anti-apoptotic BCL-2 family members in development. Cell Death Differ. 2018;25(1):37–45. doi: 10.1038/cdd.2017.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mallipattu SK, Gallagher EJ, LeRoith D, et al. Diabetic nephropathy in a nonobese mouse model of type 2 diabetes mellitus. Am J Physiol Renal Physiol. 2014;306(9):F1008–1017. doi: 10.1152/ajprenal.00597.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rena G, Lang CC. Repurposing metformin for cardiovascular disease. Circulation. 2018;137(5):422–424. doi: 10.1161/CIRCULATIONAHA.117.031735 [DOI] [PubMed] [Google Scholar]
- 37.Fung CS, Wan EY, Wong CK, Jiao F, Chan AK. Effect of metformin monotherapy on cardiovascular diseases and mortality: a retrospective cohort study on Chinese type 2 diabetes mellitus patients. Cardiovasc Diabetol. 2015;14:137. doi: 10.1186/s12933-015-0304-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang J, Huang L, Shi X, et al. Metformin protects against myocardial ischemia-reperfusion injury and cell pyroptosis via AMPK/NLRP3 inflammasome pathway. Aging. 2020;12(23):24270–24287. doi: 10.18632/aging.202143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Higgins L, Palee S, Chattipakorn SC, Chattipakorn N. Effects of metformin on the heart with ischaemia-reperfusion injury: evidence of its benefits from in vitro, in vivo and clinical reports. Eur J Pharmacol. 2019;858:172489. doi: 10.1016/j.ejphar.2019.172489 [DOI] [PubMed] [Google Scholar]