Abstract
Germline mutations in ETV6 are associated with a syndrome of thrombocytopenia and leukemia predisposition, and ETV6 is among the most commonly mutated genes in leukemias, especially childhood B cell acute lymphoblastic leukemia. However, the mechanisms underlying disease due to ETV6 dysfunction are poorly understood. In order to address these gaps in knowledge, using CRISPR/Cas9, we developed a mouse model of the most common recurrent, disease-causing germline mutation in ETV6. We found defects in hematopoiesis, primarily related to abnormalities of the multipotent progenitor population 4 (MPP4) subset of hematopoietic progenitor cells and evidence of sterile inflammation. Expression of ETV6 in Ba/F3 cells altered the expression of several cytokines, some of which were also detected at higher levels in the bone marrow of the mice with Etv6 mutation. Among these, IL-18 and IL-13 abrogated B cell development of sorted MPP4 cells, but not common lymphoid progenitors, suggesting that inflammation contributes to abnormal hematopoiesis by impairing lymphoid development. These data, along with that from humans, support a model in which ETV6 dysfunction promotes inflammation, which adversely affects thrombopoiesis and promotes leukemogenesis.
Introduction
While the genomic landscape of acute lymphoblastic leukemia is being more clearly defined with large-scale, high-throughput sequencing studies,1,2 the molecular and cellular consequences of many of the identified aberrations are not fully understood. For example, ETV6 is among the most commonly altered genes in childhood B precursor acute lymphoblastic leukemia (B ALL), but our knowledge about the mechanisms by which ETV6 dysfunction promotes leukemogenesis is limited.3,4 This is of particular importance in light of the recent discovery of a rare autosomal dominant syndrome of thrombocytopenia and leukemia predisposition due to germline mutation in ETV6.5-9 This syndrome is somewhat unique in the high prevalence of B cell malignancies, whereas most other leukemia predispositions are primarily associated with myeloid disease.10,11
ETV6 is a member of the ETS family of transcription factors and functions primarily as a transcriptional repressor in complex with SIN3A, NCOR and HDAC3.12-14 The ETV6 protein has highly conserved Pointed (PNT) and ETS DNA-binding domains. PNT is essential for homo- and hetero- oligomerization, while the ETS domain is essential for DNA binding at the core GGA(A/T) sequence recognized by other ETS family members.15 The two domains are joined by a central linker domain.12 The c.641C>T mutation, the most common germline mutation, is in the central domain and results in a protein variant, p.P214L, with dominant-negative activity.5 Other variants cluster in the DNA binding domain.4 While the molecular function of the central domain is not fully defined, it is known to be involved in nuclear export16,17 and is required for the binding of NCOR and HDAC3.14 ETV6 p.P214L interferes with the transcriptional repression function of WT ETV6 and impairs megakaryocyte development.5 Furthermore, data suggest that the dominant negative function is in part due to sequestration of WT ETV6 and the HDAC3/NCOR repressor complex out of the nucleus.3-5 We recently demonstrated that ETV6 p.P214L leads to overexpression of HDAC3 regulated interferon response genes in human hematopoietic cells,3 raising the possibility that the altered state of inflammation contributes to the syndrome of thrombocytopenia and leukemia predisposition.18,19
Leukemia is a malignancy of disordered hematopoiesis, initiated by epigenetic changes, oncogenic mutations and structural alterations, often in genes involved in normal hematopoiesis.1,20 While models of hematopoiesis continue to evolve, experimental data continue to support hierarchical organization with long-term hematopoietic stem cells (LT-HSC) at the apex and fully differentiated platelets, erythroid, myeloid and lymphoid subsets at the base.21 A number of progenitor populations with varying lineage priming, potential and commitment lie in between, including short-term hematopoietic stem cells (ST-HSC), multipotent progenitors (MPP) subsets 1-6 common myeloid progenitors (CMP) and common lymphoid progenitors (CLP).22 Etv6 is highly expressed in early hematopoietic progenitors and its deletion in LT-HSC demonstrates that it is essential for adult hematopoiesis.23 But whether point mutations in Etv6 have as profound effect on hematopoiesis remains to be demonstrated.
We sought to examine the effects of Etv6 dysfunction in hematopoiesis by modeling the human ETV6 p.P214L (h.P214L-Etv6) in mice and subsequently analyzed hematopoietic progenitor cell function with complementary assays, including flow cytometry analyses, colony formation assays and competitive transplantation. We found that the mice with h.P214L-Etv6 have a mild hematopoietic defect, including deficiencies in lymphopoiesis. The functional deficit is most pronounced in the MPP4 population, from which B lymphocyte development is impaired, and gene expression analyses suggest an altered state of inflammation. In addition, Ba/F3 cells expressing wild type or P214L ETV6 had significant differences in the levels of secreted cytokines and chemokines, some of which were also detected at higher levels in the bone marrow (BM) of mice with h.P214L-Etv6. Both IL-18 and IL-13 impaired B lymphocyte development from sorted MPP4 cells, indicating that these cytokines influence lymphoid development from this early progenitor population, prior to lymphoid commitment at the CLP level. Together, these data indicate that the ETV6 p.P214L variant alters the function of early hematopoietic cells, leading to impaired lymphopoiesis, perhaps in part due to altered inflammation.
Methods
Mice
Mice were generated using CRISPR/Cas9 at the University of Utah, to create a p.P>L ETV6 in mice at the conserved site in the central domain (Supplementary Figure 1). The conserved site in the mouse ortholog is at position 216. The founder mice were bred to the C57BL/6J background. Rag1−/− mice, B6 (Ly5.2) mice and Ly5.1-congenic B6 mice were originally purchased from the Jackson Laboratory and bred in the Emory Animal Care Facility. The experiments were performed in compliance with laws and guidelines, and with approval of the Institutional Animal Care and Use Committees at the University of Utah and Emory University.
Flow cytometry analysis
BM cells were harvested from femurs and tibias of mice. Cells were stained with fluorochrome-conjugated antibodies (eBiosciences, BD Biosciences, Biolegend) in 100 ul of PBS plus 2% FBS and 1 mM EDTA for 20 min on ice. The cells were washed twice and then analyzed on a LSRII flow cytometry. For cell sorting, BM cells were first lineage-depleted with lineage depletion kit (StemCell Technologies) or with lineage cocktail (biotin-Mac-1, biotin-Gr-1, biotin-B220, Biotin-CD19 and Biotin-CD5). Lineage depleted cells were sorted on a FacsAria cell sorter at the Emory Pediatric Flow Cytometry Core. The following anti mouse antibodies were used in this study: PE-Cy5 linked Mac-1 (M1/70), Gr-1 (RB6-8C5), Ter-119 (Ter-119), CD3e (145-2C11), CD8a (53-6.7), B220 (RA3-6B2), AF700-CD34 (RAM34), APC-Cy7-CD48 (HM48-1), FITC-Sca-1 (D7), BV605-CD117 (2B8), PE-CD150 (9D1), BV421-CD135 (A2F10.1), PE-Cy7-CD127 (A7R34), BV510-B220 (RA3-6B2), PE-CD43 (S7), APC-CD93 (AA4.1), BV711-Ly-6C (HK1.4), FITC-BP-1 (6C3), APC-eF780-CD24 (M1/69), eF450-CD19 (eBio1D3) and FITC-FcγR (93).
Hematopoietic stem cell transplantation
Freshly harvested whole BM cells (CD45.2+) from Etv6wt/wt, Etv6wt/mt, and Etv6mt/mt mice were transplanted into lethally irradiated (500 rad x 2, 4 hours apart) Rag1−/− mice with or without equal numbers of whole BM cells harvested from BoyJ mice (CD45.1+). Donor cell reconstitution was monitored up to 4 months after transplantation by analysis of periphery blood with flow cytometry. At 4 months after transplantation, the recipient’s BM cells were isolated and injected into lethally irradiated secondary recipients, which were similarly monitored.
CFU assay of HPSCs
The colony-forming unit assay was performed according to the manufacture’s protocol in MethoCult-3434 or 3630 media (StemCell Technologies) using whole BM cells or sorted hematopoietic progenitor and stem cells.
In vitro B cell differentiation assay
Sorted hematopoietic stem and progenitor cells (1000 long-term HSC and 2000 MPP4 or CLP) were cultured on irradiated OP9 cells in Opti-MEM supplemented with 5% FBS, SCF (10 ng/ml), FLT3L (10 ng/ml) and IL-7 (5 ng/ml). After sequential withdrawal of FLT3L and SCF at two-day intervals, the culture was maintained with IL-7. The medium was changed twice weekly and the cells were re-plated on fresh OP9 cells every 6 days. Differentiation was monitored every six days by flow cytometry.24,25
Short-term, in vivo hematopoietic reconstitution assay
The assay was performed as previously described with some modifications.26,27 Eight thousand isolated MPP4 cells were injected intravenously to lethally irradiated (400 Rad x 2, 4 hours apart) B6 mice. Ten days after transplantation, cells were harvested from spleen and BM, stained with anti B220, CD19 and Mac-1/Gr-1 antibodies and analyzed by flow cytometry. Control mice were injected with PBS without cells.
Luminex assay
The lymphoblastoid cell line, Ba/F3, (ATCC) was grown in RPMI1640 medium supplemented with 10% FBS ad WEHI3 conditioned media, and transduced with wild type ETV6, ETV6-P214L or empty vector (MSCV-iresGFP). Cell culture supernatant from the BaF/3 cells or the supernatant of BM aspirates were assayed with the Immune Monitoring 48-Plex ProcartaPlex panel (Thermo Fisher Scientific) according to the manufacture’s protocol.
RNA-seq
For each cell type, 1000 cells were sorted directly into RLT buffer (Qiagen) containing 1% BME and total RNA was purified with Quick-RNA Microprep kit (Zymo Research). All subsequent RNA was used as input for the SMART-seq v4 cDNA synthesis kit (Takara) with 12 cycles of PCR amplification. 400 pg of cDNA was used as input for the NexteraXT kit (Illumina) using 12 cycles of PCR amplification. Final libraries were qPCR quantitated, pooled at equimolar ratio, and sequenced on a NextSeq500 using 75bp PE chemistry. Raw fastq files were mapped to the mm10 version of the mouse genome using STAR28 with the default parameters and the UCSC KnownGene reference transcriptome.29 Duplicate reads were flagged with PICARD v1.127 (http://broadinstitute.github.io/picard/) and removed from downstream analyses. The coverage at each exon for all unique ENTREZ genes was summarized using the GenomicRanges v1.22.4 package30 and normalized to reads per kilobase per million reads (RPKM) using custom R/Bioconductor scripts. Genes were filtered for detection based on being expressed at greater than 3 reads per million (rpm) in all samples for at least one group. Differential gene expression for detected genes was determined using edgeR v3.18.131 and for each comparison genes with a greater than 2-fold change and FDR < 0.05 were considered significantly differentially expressed. For each heatmap the RPKM values for the indicated genes and GO pathways were z-score normalized and hierarchical clustering performed using the ward method. All code and data processing scripts are available upon request and at https://github.com/cdschar.
Statistics
All data are presented as the mean ± SD of at least three separate experiments, unless otherwise specified. All statistical analyses were performed with GraphPad Prism, version 8. Unless otherwise noted, one-way ANOVA was used for comparisons between more than two groups.
Data Sharing Statement
RNA-seq data has been deposited to the Gene Expression Omnibus (GEO) database (GSE183064).
Results
Mice with mutation in central domain of Etv6 do not have an overt hematopoietic defect
The ETV6 gene is highly conserved around the c.641C>T mutation that has been observed in multiple pedigrees with thrombocytopenia and leukemia predisposition.5-8 To address the impact of ETV6 mutation on hematopoietic function, we generated a mouse model using CRISPR-Cas9 to introduce a single nucleotide variant in Etv6 (p.P216L) corresponding to the c.641C>T (p.P214L) mutation in humans (Supplemental Figure 1A, B, C). Genotyping of progeny generated from mating mice heterozygous for the Etv6 mutation (Etv6mt/wt) demonstrated mendelian inheritance, indicating that this mutation did not confer embryonic lethality in the heterozygous or homozygous state (Supplemental Figure 1D), in contrast to homozygous deletion of Etv6.32 Mice with heterozygous and homozygous mutation in Etv6 appeared to develop normally and had no differences in weight at 12 weeks old (Supplemental Figure 1E). Analyses of complete blood counts (CBC) from these mice as compared to littermate controls (Etv6wt/wt) showed only inconsistent decreases in white blood cell counts in Etv6wt/mt and homozygotes (Etv6mt/mt) in Etv6mt/mt compared to Etv6wt/wt (Supplemental Figure 1F, G). No leukemias or premature deaths were observed in a small cohort of Etv6wt/wt (n=7), Etv6wt/mt (n=11), and Etv6mt/mt (n=6) mice monitored over the course of 30 weeks. Thus, the orthologous mutation in the central domain of Etv6 (h.P214L) does not confer an overt hematopoietic phenotype in mice.
h.P214L-Etv6 causes deficiencies in stress hematopoiesis.
As Etv6 function is essential for normal hematopoietic stem cell (HSC) function,23 we sought to determine if HSC function is impaired in Etv6wt/mt, and Etv6mt/mt mice by challenging hematopoiesis. We first treated mice with a single injection of 5-fluorouracil (5FU) and found delayed hematopoietic recovery in Etv6wt/mt and Etv6mt/mt mice as compared to Etv6wt/wt mice, as measured by CBC and flow cytometry 10 and 14 days later (Figure 1A and Supplemental Figure 1H). We next serially transplanted whole BM from Etv6wt/wt, Etv6wt/mt, and Etv6mt/mt mice into lethally irradiated primary and secondary recipients. Rag1−/− mice, which lack mature B and T cells, were used in an attempt to amplify any differences in lymphoid development, but no differences were observed in the representation of B- or T- lymphoid or myeloid populations in primary or secondary recipients (Figure 1B, C, D). The efficiency of hematopoietic reconstitution in secondary recipients indicates that long-term HSC repopulating function is intact in Etv6wt/mt and Etv6mt/mt mice.
Figure 1. Mice with h.P214L-ETV6 have impaired hematopoiesis.
A. Etv6wt/wt (WT), Etv6wt/mt (HT), and Etv6mt/mt (HO) mice were treated with 5FU (150 mg/kg IP). Peripheral blood was periodically analyzed with complete blood counts and flow cytometry. Numbers of B220+ and Gr1+ cells in the peripheral blood are demonstrated. (n=4 per genotype from a single experiment; *P<0.05, **P<0.01; Mixed-effects model of repeated measures with Dunnett correction for multiple comparisons.). B, C, D. Whole BM from WT, HT, and HO mice was serially transplanted into lethally irradiated Rag1−/− recipients. Peripheral blood was analyzed by flow cytometry every 4 weeks. The percentage of live B220+ (B), CD3+ (C), and Gr1/Mac1+ (D) cells are depicted over time. E, F, G, H. Whole BM from WT, HT, and HO mice was mixed 1:1 with whole BM from B6.SJL (BoyJ) mice and serially transplanted into lethally irradiated Rag1−/− recipients. Peripheral blood was analyzed by flow cytometry every 4 weeks. Graphs present the compiled data from 13 primary recipients and 7 secondary recipients per group from 3 independent experiments. (*P<0.05, **P<0.01, ***P<0.001 and P>0.05; Mixed-effects model of repeated measures with Dunnett correction for multiple comparisons.) The percentage of live CD45.2+ cells among all (E), B220+ (F), CD3+ (G), and Gr1/Mac1+ (H) cells are depicted.
To identify more subtle hematopoietic progenitor cell defects, we next performed competitive transplantation. Whole BM cells harvested from Etv6wt/wt, Etv6wt/mt, and Etv6mt/mt mice (all CD45.2+) were mixed with equal numbers of whole BM cells from B6.SJL mice (CD45.2neg) and transplanted into lethally irradiated Rag1−/− recipients (CD45.2+). In this context, Etv6wt/mt and Etv6mt/mt cells initially engrafted equally as well as Etv6wt/wt cells, as measured by the percentage of CD45.2+ cells in the peripheral blood. However, beginning at 8 weeks after transplantation and persisting through secondary transplantation, a smaller percentage of CD45.2+ cells was detected in the peripheral blood of recipients of Etv6wt/mt and Etv6mt/mt cells mixed with CD45.2neg cells (Figure 1E). Examination of B- and T- lymphoid and myeloid compartments, demonstrated that the difference is primarily in the lymphoid compartment, as CD45.2+ contributions to both B and T populations were reduced in the peripheral blood of recipients of Etv6wt/mt and Etv6mt/mt BM (Figure 1F, G, H). Combining CBC and flow cytometry data demonstrated similar differences in circulating cell numbers (Supplemental Figure 2). Taken together, these data suggest that h.P214L Etv6 does not impair the repopulating function of long-term HSC but does impair hematopoietic stem and/or progenitor cell function, resulting in reduced lymphoid potential.
Mice with h.P214L-Etv6 have fewer MPP4 cells in the BM
To determine if h.P214L-Etv6 impacts the number or percentage of specific hematopoietic cell compartments, we analyzed the BM at steady state by counting nucleated cells and multiparametric flow cytometry (Figure 2A).33,34 There were no differences in number of nucleated cells or the percentages of LT-HSC (cKit+Sca-1+CD48−CD150+CD34−), MPP1 (cKit+Sca-1+CD48−CD150+CD34+), ST-HSC (cKit+Sca-1+CD48−CD150−), MPP2 (cKit+Sca-1+CD48+CD150+) and MPP3 (cKit+Sca-1+CD48+CD150−) among Etv6wt/wt, Etv6wt/mt, and Etv6mt/mt mice (Figure 2B and Supplemental Figure 3A, B). However, we did observe a consistent and significant decrease in the MPP4 population in Etv6mt/mt BM compared to wild type littermates. Further gating CD48+ cells within MPP4 showed a decrease in both Etv6wt/mt, and Etv6mt/mt mice. Notably, the decrease in MPP4 and CD48+ MPP4 populations was greater in degree in younger mice (4-6 weeks) as compared to older mice (Supplemental Figure 3C). There were no differences in lineage-committed populations, including CMP, CLP, megakaryocyte erythroid progenitor (MEP), and granulocyte monocyte progenitor (GMP) (Supplemental Figure 3D), regardless of age. In addition, there were no differences in B cell progenitor fractions A, B and C35 among Etv6wt/wt, Etv6wt/mt, and Etv6mt/mt mice (Supplemental Figure 3E). Thus, h.P214L-Etv6 confers a specific defect in the MPP4 population in mouse BM.
Figure 2. Mice with h.P214L-Etv6 have reduced proportion of MPP4 cells in the bone marrow.
Bone marrow from Etv6wt/wt (WT), Etv6wt/mt (HT), and Etv6mt/mt (HO) was analyzed by multiparameter flow cytometry. A. Gating strategy for analyses of hematopoietic stem and progenitor cell populations, after gating on singlets and live cells. B. The percentage of hematopoietic stem and progenitor cell populations is depicted (*P<0.05; **P<0.01; ANOVA with Dunnett’s multi-comparison test; n ≥ 13/genotype compiled from 3 independent experiments).
h.P214L-Etv6 disturbs the balance of myeloid/lymphoid differentiation potential of MPP4
As we observed decreased lymphoid reconstitution activity by whole BM cells from Etv6wt/mt and Etv6mt/mt in the competitive transplantation assay (Figure 1G, H), we next examined the lineage potential of HSPC cultured ex vivo. With whole BM, we did not observe any differences in the formation of erythroid, myeloid or B lymphoid colony formation in methylcellulose supplemented with cytokines (Supplemental Figure 4A). However, when we sorted for specific hematopoietic stem and progenitor cell populations, we observed significantly greater numbers of myeloid colonies, CFU-GM/G/GM, from Etv6wt/mt and Etv6mt/mt MPP3 and MPP4 populations as compared to Etv6wt/wt (Figure 3A). In liquid co-culture experiments highly favorable to B cell development, sorted MPP4 cells from Etv6wt/mt and Etv6mt/mt mice trended toward slower B cell commitment with less lineage fidelity while differences from sorted ST-HSC and common lymphoid progenitors (CLP) were more subtle (Figure 3B and Supplemental Figure 4B, C). Next, MPP4 cells from Etv6wt/wt, Etv6wt/mt and Etv6mt/mt were sorted and transplanted into lethally irradiated wild type recipients and after 10 days, BM and spleens were analyzed by flow cytometry. In the spleens of recipients of Etv6wt/mt and Etv6mt/mt MPP4 cells, there were significantly fewer B220+ cells, as compared to recipients of Etv6wt/wt MPP4 cells (Figure 3C and Supplemental Figure 4D). Together, these data indicate that mutation in Etv6 impairs the lymphoid potential of the MPP4 population.
Figure 3. Impaired lymphoid potential of early hematopoietic progenitor cells from mice with h.P214L-Etv6.
Hematopoietic stem and progenitor cell populations were sorted from Etv6wt/wt (WT), Etv6wt/mt (HT), and Etv6mt/mt (HO) mice. A. Sorted populations were used for colony forming assays in methylcellulose. The number of colonies, as a percentage of WT is depicted. (*P<0.05; **P<0.01; ***P<0.001; ANOVA with Dunnett’s multiple comparison test.) B. Sorted MPP4 cells were cultured in conditions to promote B cell development. Dot plots of flow cytometry data from days 6, 12 and 18. Data are representative of 3 independent experiments with very similar results. C. Sorted MPP4 cells were transplanted into lethally irradiated recipients. Controls (Ctl) were injected with PBS without cells. The relative proportions of B220+ (Left) and Mac1+ and/or Gr1+ (Right) cells, compared to WT, in the spleens of mice 10 days after transplantation are depicted (**P<0.01; Kruskall-Wallis ANOVA with Dunn’s multiple comparison test: n ≥ 7/genotype compiled from 3 independent experiments).
h.P214L-Etv6 alters gene expression in murine MPP4 cells in the BM
To begin to understand the mechanisms underlying functional differences in early hematopoietic progenitor cells due to Etv6 mutation, we performed RNA sequencing from sorted long term hematopoietic stem cells, MPP4 and common lymphoid progenitors from Etv6wt/wt and Etv6mt/mt mice. While there were thousands of genes differentially expressed when comparing different hematopoietic progenitor cell populations, regardless of genotype, there were far fewer differences across genotypes. Specifically, there were only 8, 18 and 1 genes significantly differently expressed in LT-HSC, MPP4 and CLP populations, respectively, when comparing populations from Etv6wt/wt and Etv6mt/mt mice (Supplemental Table 1). Notably, most of the significantly differently expressed genes are related to inflammation. Indeed, analyses of the entire transcriptome from each of these populations using GSEA identifies several inflammation related pathways, including TNF and STAT5 gene signatures, among others (Supplemental Table 2; Figure 4A). Unsupervised hierarchical clustering demonstrates that the gene expression clustered most closely by genotype in the MPP4 population for a subset of these gene sets (Figure 4B). Thus, mutation in Etv6 affects gene expression most consistently and significantly in the MPP4 population, as compared to LT-HSC and CLP, particularly among inflammation-related genes.
Figure 4. Inflammation-related gene expression changes due to h.P214L-Etv6 in MPP4 cells.
LT-HSC, MPP4 and CLP cells from Etv6wt/wt and Etv6mt/mt mice were sorted prior to RNA-seq. A. Representative enrichment plots of gene sets from MPP4 with nominal P<0.05. B. Hierarchical clustering and heatmaps of genes in the Inflammatory Response gene set from LT-HSC, MPP4 and CLP populations.
Perturbed ETV6 alters cytokine release from hematopoietic cells and cytokine levels in the BM
As many of the genes in the inflammation related gene sets are cytokines and cytokine receptors, we sought to determine whether mutated ETV6 alters cytokine release from hematopoietic cells. The lymphoblastoid cell line, Ba/F3, was transduced with vectors expressing human, wild type or P214L ETV6, or empty vector. After sorting for vector expressing cells, cell culture supernatants were analyzed for soluble factor levels using Luminex. We found that expression of wild type ETV6 led to decreased levels of several cytokines and chemokines relative to empty vector, which were also higher with expression of P214L ETV6 (Figure 5A). To determine if mutated Etv6 affected inflammation profiles in vivo, we used the same assay to measure cytokines and chemokines in BM aspirates from Etv6wt/wt, Etv6wt/mt, and Etv6mt/mt mice. While there was much more variability and no statistically significant difference, there was a trend toward higher levels of several cytokines in the BM of Etv6wt/mt and Etv6mt/mt mice as compared to Etv6wt/wt (Supplemental Figure 5A, B). Thus, mutation in Etv6 disrupts ETV6 dependent regulation of cytokines and chemokines.
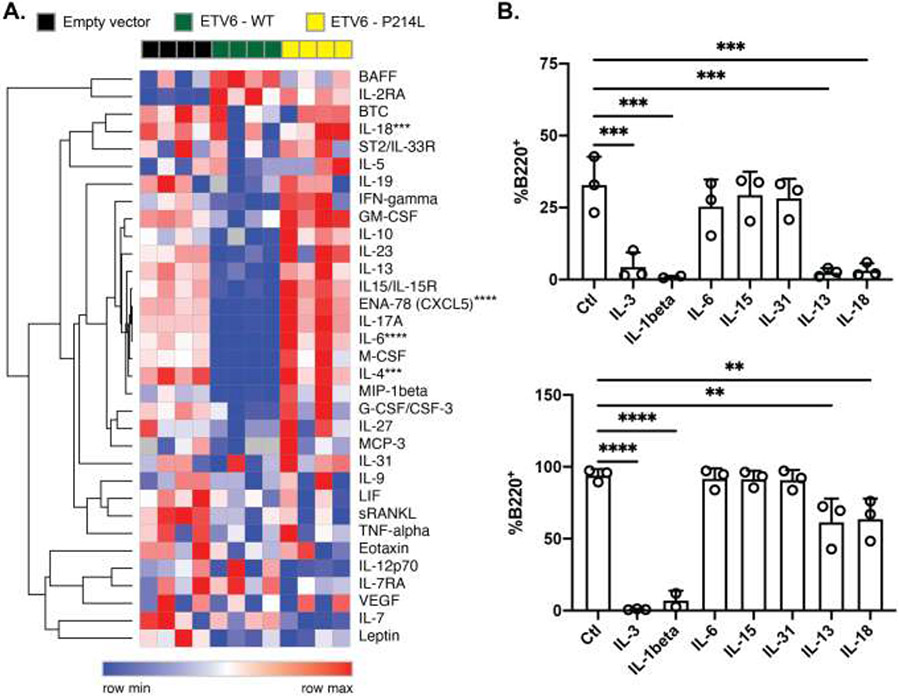
Figure 5. h.P214L-Etv6 alters the secretion of cytokines which impair MPP4 differentiation to B cells.
A. The supernatant of Ba/F3 cells transduced with empty vector, wild type ETV6 or P214L ETV6 expressing plasmids was analyzed by Luminex for cytokine levels. Relative cytokine levels are depicted as a heatmap (***P<0.001; ****P<0.0001, ANOVA with Dunnett’s multiple comparison test). B. MPP4 cells from WT mice were sorted and cultured in conditions to promote B cell development. The percentage of B220+ cells after 6 (Top) and 12 (Bottom) days is depicted (**P<0.01; ***P<0.001; ****P<0.0001; ANOVA with Dunnett’s multiple comparison test; data are compiled from 3 independent experiments performed in triplicate).
IL-13 and IL-18 impair MPP4 lymphoid differentiation
To determine the influence of these cytokines on hematopoietic progenitor cell function, sorted MPP4 and CLP from wild type Bl6 mice were cultured in liquid culture in conditions highly favorable for B cell differentiation in the presence or absence of additional cytokines, including IL-6, IL-15, IL-31, IL-13 or IL-18. IL-1β and IL-3 were included as controls that are known to promote myeloid development and showed abrogation of B cell development, as expected (Figure 5C).36,37 In contrast, IL-6, IL-15 and IL-31 had no effect on B cell differentiation from MPP4 cells. IL-13 and IL-18, however, had profound inhibitory effect on B cell development from MPP4, and to a lesser extent CLP, through 6 days in culture, which was less pronounced by 12 days (Figure 5C and Supplemental Figure 4E). Taken together, these data indicate that ETV6-regulated cytokines IL-13 and IL-18 negatively influence B lymphoid differentiation from the MPP4 population.
Discussion
Using CRISPR-Cas9, we have developed a novel model of ETV6 dysfunction demonstrating hematopoietic deficits and altered inflammation. Notably, we observed differences in mice both heterozygous and homozygous h.P214L-Etv6, perhaps supporting previous findings of dominant negative effects of this mutation.5 Interestingly, the h.P214L-Etv6 appears to cause a functional defect in a very specific hematopoietic progenitor cell population, MPP4, to a greater extent than others. This is evidenced by fewer MPP4 cells in mice with h.P214L-Etv6 and impaired B lymphocyte development from sorted MPP4 in culture and transplant assays. MPP4 also exhibit more extensive alterations in gene expression as compared to LT-HSC and CLP. h.P214L-Etv6 MPP4 cells are enriched in inflammation related genes, and cells with P214L ETV6 have different patterns of cytokine secretion, particularly elevated levels of IL-18 and IL-13. As IL-18 and IL-13 impair B cell development from sorted MPP4 cells, the altered inflammation may contribute to the observed phenotype of the mice.
ETV6 is known to have a critical function in long-term hematopoietic stem cells, as its deletion leads to failure of hematopoiesis in the bone marrow, whereas deletion in committed lineages had much less effect.23,38 An important distinction between prior studies using genetic knockouts and our current work is that our system expresses a full-length ETV6 protein variant that may have multiple functions. Indeed, while the function of ETV6 as a DNA binding transcription factor is well documented,39-42 this function is dependent upon self-association and association with co-repressors SIN3A, NCOR and HDAC3,14,42 and we have demonstrated that the P214L variant sequesters co-repressors in the cytoplasm and alters histone acetylation and gene expression.3 Thus, ETV6 has direct and indirect effects on the regulation of gene expression. While serial transplantation has some limitations in measuring LT-HSC function,43 the lack of effect on LT-HSC in this model of ETV6 dysfunction, at least in their capacity to maintain hematopoiesis in secondary recipients, suggests that the presence of a mutated protein is sufficient to maintain LT-HSC. On the other hand, the P214L variant fails to properly regulate lymphoid lineage output in our assays.
This is consistent with our findings specific to the MPP4 compartment of hematopoietic progenitor cells. While subtle, this was the only compartment in which we observed altered population abundance in the bone marrow, changes in gene expression patterns and altered lineage potential. The MPP4 population is the largest progenitor cell population and is primed for lymphopoiesis, but maintain the capacity for megakaryocytic, erythroid and myeloid development.33,44 As hierarchical models of hematopoiesis have evolved to include a more plastic continuum with functional heterogeneity within populations of stem and progenitor cells,21,45,46 transcription factors have been demonstrated to play a key role in determining lineage potential of early progenitors.47-49 Our results suggest that ETV6 contributes to the fine tuning of lineage commitment in the MPP4 population, either specifically in that population or by reducing lymphoid bias in HSC that is manifest in assays of MPP4 function. This fine tuning may allow for adjustments in the production of specific immune components needed in response to immune challenges.19
Our findings of ETV6-dependent altered expression of inflammation related genes and cytokine secretion is consistent with data from our studies of human blood cells,3 and raise the possibility that this fine tuning is not entirely cell autonomous. Inflammation has been shown to influence hematopoietic stem and progenitor commitment in a number of ways, often with skewing toward the myeloid lineages. Interferons, tumor necrosis factor and inflammasome signaling have all been demonstrated to have direct effects in hematopoietic stem and progenitor cell function.18,19 For example, short- and long- term exposure to IL-1 have distinct effects on HSC function, with chronic exposure promoting myeloid skewing and reduced self-renewal.50 IL-18 is a member of the IL-1 family of cytokines, activated by the inflammasome along with IL-1. Its role is most commonly associated with a pro-inflammatory response, stimulating interferon gamma expression and Type I immunity, typically in cooperation with STAT-inducing cytokines.51 IL-18 was demonstrated to mobilize hematopoietic stem and progenitor cells to the peripheral blood in mice52 reminiscent of our observation of higher levels of circulating progenitor cells in patients with ETV6 mutations.3 However, when mice are challenged with 5FU or transplantation, IL-18 constrains progenitor cell proliferation.53 In addition, higher levels of IL-18 are associated with delayed hematopoietic recovery, particularly platelets, after allogeneic stem cell transplantation.54 IL-13 promotes interferon expression in some contexts and promotes myelopoiesis from hematopoietic progenitor cells.55,56 Although the differences in concentrations in the bone marrow were not statistically significant, the published literature led us to focus on whether these cytokines have direct effects on the MPP4 population, and both IL-18 and IL-13 significantly diminished B cell development in our model. To our knowledge, the impact of IL-18 and IL-13 on the lineage potential of MPP4 has not been previously reported and adds additional complexity to the role of inflammation in orchestrating the response to dangerous stimuli at the level of hematopoietic progenitors. It also raises questions about the impact of even small differences in concentrations of potent cytokines over long periods of time in individuals with constitutional dysregulation, such as with ETV6 mutation,3 particularly in light of the demonstration that altered cytokine levels, including IL-1 family members, can promote the expansion of leukemia cells.57-59
Accumulating evidence implicates inflammation in promoting the development of myelodysplastic syndrome and leukemia.60,61 In contrast to most leukemia predisposition syndromes, B cell malignancies seem to be more prevalent in individuals with ETV6 mutations.10 The suppression of B lymphopoiesis in the context of germline ETV6 mutation may provide a unique context that promotes the selection of B cell progenitors harboring mutations that confer a selective advantage and drive leukemogenesis, as we have observed in the context of aging.62,63 In this setting, mutation(s) that confer resistance to the suppressive effects of inflammatory cytokines on B lymphopoiesis may play a key role in initiating oncogenic selection. While we have not observed spontaneous leukemia development in mice with Etv6 h.P214L, whether additional genetic perturbations, such as RAS activating mutations or PAX5 loss,4 will be more leukemogenic in these mice as compared to wild type Etv6 remains to be demonstrated.
In summary, we have developed a mouse model of ETV6 mutation that may be useful to study the ETV6 thrombocytopenia and leukemia predisposition syndrome, although the hematopoietic deficit is only apparent in specific contexts. Our observations indicate that disruption of Etv6 in these mice leads to a pro-inflammatory state, with elevated levels of cytokines in the bone marrow. The MPP4 population of the bone marrow appears to be the most affected in these mice, demonstrating impaired lymphoid potential, but defects in lymphoid bias in HSC were not excluded. Notably, we found that the MPP4 population is sensitive to IL-18 and IL-13, which abrogate B cell development, although their role in impaired hematopoiesis in the context of ETV6 dysfunction remains to be formally demonstrated. These data complement those from humans with this disease, who also have evidence of unprovoked inflammation.3 This model should prove useful for further studies of thrombocytopenia and leukemogenesis due to ETV6 dysfunction.
Highlights.
Germline mutation in ETV6 impairs hematopoiesis in a specific progenitor population (MPP4)
ETV6 mutation alters expression of inflammation related genes including cyotkines and receptors
IL-18 & IL-13 abrogate B cell development from MPP4 cells
Key points:
Germline mutation in ETV6 impairs hematopoiesis in a specific progenitor population (MPP4)
ETV6 mutation alters the secretion of several cytokines including IL-18 & IL-13, which abrogate B cell development from MPP4 cells
Acknowledgements
This work was supported in part by infrastructure and training grants from the National Institutes of Health (CA138292, CA046934, TR001081, CA082086, HL139443) and the American Cancer Society (to CLJ), research grants from the National Heart Lung & Blood Institute (HL120728, HL141794 to JDP) and the National Institute for Allergy & Infectious Disease (AI148471, AI125180 to JMB and AI148471 to CDS), Children’s Healthcare of Atlanta, and the Cancer League of Colorado (to CCP). We would like to acknowledge the University of Utah Transgenic and Gene Targeting Core for providing transgenic and/or gene targeted mice that were used in this research.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of Interest Disclosures
The authors have no conflicts of interest to disclose.
References
- 1.Hunger SP, Mullighan CG. Acute Lymphoblastic Leukemia in Children. N Engl J Med. 2015;373(16):1541–1552. [DOI] [PubMed] [Google Scholar]
- 2.Tasian SK, Hunger SP. Genomic characterization of paediatric acute lymphoblastic leukaemia: an opportunity for precision medicine therapeutics. Br J Haematol. 2017;176(6):867–882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fisher MH, Kirkpatrick GD, Stevens B, et al. ETV6 germline mutations cause HDAC3/NCOR2 mislocalization and upregulation of interferon response genes. JCI Insight. 2020;5(18). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nishii R, Baskin-Doerfler R, Yang W, et al. Molecular Basis of ETV6-Mediated Predisposition to Childhood Acute Lymphoblastic Leukemia. Blood. 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Noetzli L, Lo RW, Lee-Sherick AB, et al. Germline mutations in ETV6 are associated with thrombocytopenia, red cell macrocytosis and predisposition to lymphoblastic leukemia. Nat Genet. 2015;47(5):535–538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhang MY, Churpek JE, Keel SB, et al. Germline ETV6 mutations in familial thrombocytopenia and hematologic malignancy. Nat Genet. 2015;47(2): 180–185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Melazzini F, Palombo F, Balduini A, et al. Clinical and pathogenic features of ETV6-related thrombocytopenia with predisposition to acute lymphoblastic leukemia. Haematologica. 2016;101(11):1333–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Poggi M, Canault M, Favier M, et al. Germline variants in ETV6 underlie reduced platelet formation, platelet dysfunction and increased levels of circulating CD34+ progenitors. Haematologica. 2017;102(2):282–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Topka S, Vijai J, Walsh MF, et al. Germline ETV6 Mutations Confer Susceptibility to Acute Lymphoblastic Leukemia and Thrombocytopenia. PLoS Genet. 2015;11(6):e1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Di Paola J, Porter CC. ETV6-related thrombocytopenia and leukemia predisposition. Blood. 2019;134(8):663–667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Godley LA, Shimamura A. Genetic predisposition to hematologic malignancies: management and surveillance. Blood. 2017;130(4):424–432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bohlander SK. ETV6: a versatile player in leukemogenesis. Semin Cancer Biol. 2005;15(3):162–174. [DOI] [PubMed] [Google Scholar]
- 13.Chakrabarti SR, Nucifora G. The leukemia-associated gene TEL encodes a transcription repressor which associates with SMRT and mSin3A. Biochem Biophys Res Commun. 1999;264(3):871–877. [DOI] [PubMed] [Google Scholar]
- 14.Wang L, Hiebert SW. TEL contacts multiple co-repressors and specifically associates with histone deacetylase-3. Oncogene. 2001;20(28):3716–3725. [DOI] [PubMed] [Google Scholar]
- 15.Poirel H, Oury C, Carron C, et al. The TEL gene products: nuclear phosphoproteins with DNA binding properties. Oncogene. 1997;14(3):349–357. [DOI] [PubMed] [Google Scholar]
- 16.Ca Hanson, Wood LD, Hiebert SW. Cellular stress triggers TEL nuclear export via two genetically separable pathways. J Cell Biochem. 2008;104(2):488–498. [DOI] [PubMed] [Google Scholar]
- 17.Arai H, Maki K, Waga K, et al. Functional regulation of TEL by p38-induced phosphorylation. Biochem Biophys Res Commun. 2002;299(1):116–125. [DOI] [PubMed] [Google Scholar]
- 18.Pietras EM. Inflammation: a key regulator of hematopoietic stem cell fate in health and disease. Blood. 2017;130(15):1693–1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.King KY, Goodell MA. Inflammatory modulation of HSCs: viewing the HSC as a foundation for the immune response. Nat Rev Immunol. 2011;11(10):685–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ntziachristos P, Mullenders J, Trimarchi T, Aifantis I. Mechanisms of epigenetic regulation of leukemia onset and progression. Adv Immunol. 2013;117:1–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jacobsen SEW, Nerlov C. Haematopoiesis in the era of advanced single-cell technologies. Nat Cell Biol. 2019;21(1):2–8. [DOI] [PubMed] [Google Scholar]
- 22.Sommerkamp P, Romero-Mulero MC, Narr A, et al. Mouse multipotent progenitor 5 cells are located at the interphase between hematopoietic stem and progenitor cells. Blood. 2021;137(23):3218–3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hock H, Meade E, Medeiros S, et al. Tel/Etv6 is an essential and selective regulator of adult hematopoietic stem cell survival. Genes Dev. 2004;18(19):2336–2341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kawano Y, Petkau G, Stehle C, et al. Stable lines and clones of long-term proliferating normal, genetically unmodified murine common lymphoid progenitors. Blood. 2018;131(18):2026–2035. [DOI] [PubMed] [Google Scholar]
- 25.Reynaud D, Demarco IA, Reddy KL, et al. Regulation of B cell fate commitment and immunoglobulin heavy-chain gene rearrangements by Ikaros. Nat Immunol. 2008;9(8):927–936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yang L, Bryder D, Adolfsson J, et al. Identification of Lin(−)Sca1(+)kit(+)CD34(+)Flt3-short-term hematopoietic stem cells capable of rapidly reconstituting and rescuing myeloablated transplant recipients. Blood. 2005;105(7):2717–2723. [DOI] [PubMed] [Google Scholar]
- 27.Adolfsson J, Borge OJ, Bryder D, et al. Upregulation of Flt3 expression within the bone marrow Lin(−)Sca1(+)c-kit(+) stem cell compartment is accompanied by loss of self-renewal capacity. Immunity. 2001;15(4):659–669. [DOI] [PubMed] [Google Scholar]
- 28.Dobin A, Gingeras TR. Mapping RNA-seq Reads with STAR. Curr Protoc Bioinformatics. 2015;51:11 14 11–11 14 19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC Known Genes. Bioinformatics. 2006;22(9):1036–1046. [DOI] [PubMed] [Google Scholar]
- 30.Lawrence M, Huber W, Pages H, et al. Software for computing and annotating genomic ranges. PLoS Comput Biol. 2013;9(8):e1003118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Robinson MD, McCarthy DJ, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wang LC, Kuo F, Fujiwara Y, Gilliland DG, Golub TR, Orkin SH. Yolk sac angiogenic defect and intra-embryonic apoptosis in mice lacking the Ets-related factor TEL. EMBO J. 1997; 16(14):4374–4383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pietras EM, Reynaud D, Kang YA, et al. Functionally Distinct Subsets of Lineage-Biased Multipotent Progenitors Control Blood Production in Normal and Regenerative Conditions. Cell Stem Cell. 2015;17(1):35–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wilson A, Laurenti E, Oser G, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135(6):1118–1129. [DOI] [PubMed] [Google Scholar]
- 35.Hardy RR, Shinton SA. Characterization of B lymphopoiesis in mouse bone marrow and spleen. Methods Mol Biol. 2004;271:1–24. [DOI] [PubMed] [Google Scholar]
- 36.Kennedy DE, Knight KL. Inhibition of B Lymphopoiesis by Adipocytes and IL-1-Producing Myeloid-Derived Suppressor Cells. J Immunol. 2015;195(6):2666–2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hirayama F, Ogawa M. Cytokine regulation of early lymphohematopoietic development. Stem Cells. 1996;14(4):369–375. [DOI] [PubMed] [Google Scholar]
- 38.Ciau-Uitz A, Pinheiro P, Gupta R, Enver T, Patient R. Tel1/ETV6 specifies blood stem cells through the agency of VEGF signaling. Dev Cell. 2010;18(4):569–578. [DOI] [PubMed] [Google Scholar]
- 39.Fears S, Gavin M, Zhang DE, et al. Functional characterization of ETV6 and ETV6/CBFA2 in the regulation of the MCSFR proximal promoter. Proc Natl Acad Sci U S A. 1997;94(5):1949–1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Li L, Rispoli R, Patient R, Ciau-Uitz A, Porcher C. Etv6 activates vegfa expression through positive and negative transcriptional regulatory networks in Xenopus embryos. Nat Commun. 2019;10(1):1083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lopez RG, Carron C, Oury C, Gardellin P, Bernard O, Ghysdael J. TEL is a sequence-specific transcriptional repressor. J Biol Chem. 1999;274(42):30132–30138. [DOI] [PubMed] [Google Scholar]
- 42.Green SM, Coyne HJ 3rd, McIntosh LP, Graves BJ. DNA binding by the ETS protein TEL (ETV6) is regulated by autoinhibition and self-association. J Biol Chem. 2010;285(24):18496–18504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rundberg Nilsson A, Pronk CJ, Bryder D. Probing hematopoietic stem cell function using serial transplantation: Seeding characteristics and the impact of stem cell purification. Exp Hematol. 2015;43(9):812–817 e811. [DOI] [PubMed] [Google Scholar]
- 44.Naik SH, Perie L, Swart E, et al. Diverse and heritable lineage imprinting of early haematopoietic progenitors. Nature. 2013;496(7444):229–232. [DOI] [PubMed] [Google Scholar]
- 45.Carrelha J, Meng Y, Kettyle LM, et al. Hierarchically related lineage-restricted fates of multipotent haematopoietic stem cells. Nature. 2018;554(7690):106–111. [DOI] [PubMed] [Google Scholar]
- 46.Rodriguez-Fraticelli AE, Wolock SL, Weinreb CS, et al. Clonal analysis of lineage fate in native haematopoiesis. Nature. 2018;553(7687):212–216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dias S, Mansson R, Gurbuxani S, Sigvardsson M, Kee BL. E2A proteins promote development of lymphoid-primed multipotent progenitors. Immunity. 2008;29(2):217–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Arinobu Y, Mizuno S, Chong Y, et al. Reciprocal activation of GATA-1 and PU.1 marks initial specification of hematopoietic stem cells into myeloerythroid and myelolymphoid lineages. Cell Stem Cell. 2007;1(4):416–427. [DOI] [PubMed] [Google Scholar]
- 49.Ng SY, Yoshida T, Zhang J, Georgopoulos K. Genome-wide lineage-specific transcriptional networks underscore Ikaros-dependent lymphoid priming in hematopoietic stem cells. Immunity. 2009;30(4):493–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pietras EM, Mirantes-Barbeito C, Fong S, et al. Chronic interleukin-1 exposure drives haematopoietic stem cells towards precocious myeloid differentiation at the expense of self-renewal. Nat Cell Biol. 2016;18(6):607–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mantovani A, Dinarello CA, Molgora M, Garlanda C. Interleukin-1 and Related Cytokines in the Regulation of Inflammation and Immunity. Immunity. 2019;50(4):778–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lenkiewicz AM, Adamiak M, Thapa A, et al. The Nlrp3 Inflammasome Orchestrates Mobilization of Bone Marrow-Residing Stem Cells into Peripheral Blood. Stem Cell Rev Rep. 2019;15(3):391–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Silberstein L, Goncalves KA, Kharchenko PV, et al. Proximity-Based Differential Single-Cell Analysis of the Niche to Identify Stem/Progenitor Cell Regulators. Cell Stem Cell. 2016;19(4):530–543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Radujkovic A, Kordelas L, Bogdanov R, et al. Interleukin-18 and Hematopoietic Recovery after Allogeneic Stem Cell Transplantation. Cancers (Basel). 2020; 12(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Jacobsen SE, Okkenhaug C, Veiby OP, Caput D, Ferrara P, Minty A. Interleukin 13: novel role in direct regulation of proliferation and differentiation of primitive hematopoietic progenitor cells. J Exp Med. 1994;180(1):75–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Minty A, Chalon P, Derocq JM, et al. Interleukin-13 is a new human lymphokine regulating inflammatory and immune responses. Nature. 1993;362(6417):248–250. [DOI] [PubMed] [Google Scholar]
- 57.Carey A, Edwards DKt, Eide CA, et al. Identification of Interleukin-1 by Functional Screening as a Key Mediator of Cellular Expansion and Disease Progression in Acute Myeloid Leukemia. Cell Rep. 2017;18(13):3204–3218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fitch BA, Zhou M, Situ J, et al. Decreased IL-10 accelerates B-cell leukemia/lymphoma in a mouse model of pediatric lymphoid leukemia. Blood Adv. 2022;6(3):854–865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Beneforti L, Dander E, Bresolin S, et al. Pro-inflammatory cytokines favor the emergence of ETV6-RUNX1-positive pre-leukemic cells in a model of mesenchymal niche. Br J Haematol. 2020;190(2):262–273. [DOI] [PubMed] [Google Scholar]
- 60.Sallman DA, List A. The central role of inflammatory signaling in the pathogenesis of myelodysplastic syndromes. Blood. 2019;133(10):1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bellissimo DC, Speck NA. RUNX1 Mutations in Inherited and Sporadic Leukemia. Front Cell Dev Biol. 2017;5:111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Henry CJ, Casas-Selves M, Kim J, et al. Aging-associated inflammation promotes selection for adaptive oncogenic events in B cell progenitors. J Clin Invest. 2015;125(12):4666–4680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Henry CJ, Marusyk A, Zaberezhnyy V, Adane B, DeGregori J. Declining lymphoid progenitor fitness promotes aging-associated leukemogenesis. Proc Natl Acad Sci U S A. 2010;107(50):21713–21718. [DOI] [PMC free article] [PubMed] [Google Scholar]