Abstract
Pathogenic Yersinia spp. counteract host defense mechanisms by modulating the cellular signal relay in response to infection. Subversion of the antiapoptotic NF-κB signaling pathway by the Yersinia enterocolitica virulence protein YopP crucially determines the induction of apoptosis in Yersinia-infected macrophages. Here, we analyzed a panel of pathogenic, phylogenetically distinct Y. enterocolitica serotypes for their abilities to trigger macrophage apoptosis. Y. enterocolitica from the highly pathogenic serogroup O8 was substantially more effective in apoptosis induction than Yersinia from the serogroups O3 and O9. Complementation of yopP-knockout mutants revealed that this effect was specifically conferred by the serogroup O8 YopP. The amino acid sequences of YopPO8 and YopPO9 share 94% identity, and both YopP isotypes were found to interact with the NF-κB-activating kinase IKKβ in macrophages. However, selectively, YopPO8 mediated efficient inhibition of IKKβ activities, which led to substantial suppression of NF-κB activation. To localize the YopPO8-related effector domain, we interchanged stretches of amino acids and single amino acid residues between YopPO8 and YopPO9. Functional characterization of the resulting mutants revealed a major role of the arginine-143 residue in determining the inhibitory impact of YopP on IKKβ activity and survival of macrophages.
Pathogenic Yersinia spp. have evolved a series of strategies for evasion and neutralization of host defense mechanisms. The ability to escape the host immune response largely depends on the presence of the 70-kb virulence plasmid pYV, which is common to the three Yersinia species that are pathogenic for rodents and humans. Y. pestis is the causative agent of plague, and Y. enterocolitica and Y. pseudotuberculosis cause gastrointestinal syndromes, lymphadenitis, and septicemia (6, 9). The Yersinia virulence plasmid pYV encodes a sophisticated bacterial virulence system for subverting eukaryotic cells (9). It encompasses the genes for a type III protein secretion machinery and for a set of at least six Yersinia effector proteins (Yersinia outer proteins [Yops] YopE, YopH, YopM, YopT, YopO/YpkA, and YopP/YopJ). The type III protein secretion system is activated upon host-cell contact and specifically mediates the delivery of the Yersinia effector proteins inside eukaryotic cell, there perturbing key cellular functions. By interference with the actin cytoskeleton dynamics, Yersinia blocks its phagocytosis by macrophages and polymorphonuclear neutrophils (4, 9). Furthermore, the action of Yops prevents killing of Yersinia by the phagocytic oxidative burst (4, 9). Besides these immediate effects on the phagocyte, Yersinia inhibits the production of the proinflammatory cytokine tumor necrosis factor alpha (TNF-α) and triggers macrophage apoptosis (5, 23, 25, 29, 33, 35, 36). Both effects are conducted by YopP (Y. enterocolitica) or by its homologue, YopJ (Y. pseudotuberculosis and Y. pestis) (5, 23, 25, 29, 36). Yersinia suppresses the macrophage TNF-α production by down-regulating the signaling cascades of transcription factor NF-κB and of the mitogen-activated protein kinases (MAPK) (5, 29, 32, 33, 36). These signaling cascades synergistically control the TNF-α production in response to bacterial infection and lipopolysaccharide (LPS) treatment. YopP/YopJ disrupts the MAPK pathways by binding and inhibiting MAPK kinases (MKK) 1 to 5 of the MKK superfamily, which function as upstream MAPK activators (27). Subversion of the NF-κB cascade is accomblished by targeting YopP/YopJ to the NF-κB-activating IκB kinase-β (IKKβ) (27).
NF-κB acts as a key regulator of the inflammatory response. It rapidly upregulates the synthesis of cytokines, acute-phase proteins, and adhesion molecules and mediates cellular survival by the prevention of apoptosis (3, 15). Disruption of the antiapoptotic functions of NF-κB plays a crucial role in the mechanism of apoptosis induction by Yersinia (32, 34). A number of extracellular stimuli, such as TNF-α and ionizing radiation, activate pro- and antiapoptotic signaling pathways in eukaryotic cells. NF-κB functions to up-regulate the synthesis of proteins that counteract the proapoptotic signals, such as inhibitor of apoptosis proteins and Bcl-2 family members. Hence, NF-κB activation provides protection against apoptotic killing, otherwise induced by these stimuli (2, 30). Analogously, activation of NF-κB is essential for self-defense and survival of macrophages when encountered with bacteria or LPS (1, 21, 32). The suppression of NF-κB activation by YopP/YopJ and the simultaneous activation of LPS-induced signaling processes trigger severe apoptosis in macrophages (34). Thus, the impact of YopP/YopJ on NF-κB and the activation of proapoptotic signals by LPS or bacterial infection crucially determine the fate of the Yersinia-infected macrophage. It was recently reported that the Yersinia-mediated proapoptotic process involves cleavage of Bid, a Bcl-2 family member that relays the apoptotic response (10).
NF-κB activation depends on liberation of NF-κB from its inhibitory proteins IκBα, IκBβ, and IκBɛ, which sequester preformed NF-κB in the cytoplasm (3, 15). Phosphorylation and degradation of IκBs by the ubiquitin-proteasome pathway release NF-κB, which translocates to the nucleus and activates transcription. The critical step in NF-κB activation is phosphorylation of the IκBs, which is conferred by the IKK complex (3, 15). This complex is composed of at least three proteins: IKKα, IKKβ, and IKKγ. IKKα and IKKβ encode the catalytic kinase subunits, mediating IκB phosphorylation. After an initial NF-κB activation, Y. enterocolitica downregulates NF-κB activities in J774A.1 macrophages as little as 60 to 90 min after onset of infection, a lag time necessary for YopP to reach its targets and to exert its effects (32, 34). YopP selectively interacts with macrophage IKKβ, but not with IKKα, and simultaneously suppresses IKKβ activities (34). This points out a strategy evolved by Yersinia that specifically targets IKKβ, which is the major LPS-responsive NF-κB-activating kinase in monocytes/macrophages (26).
In this study, we analyzed the impact of the different pathogenic Y. enterocolitica serotypes on apoptosis in macrophages. We report that Y. enterocolitica serotype O8 exhibits an outstanding efficiency in apoptosis induction and NF-κB suppression in comparison to other Y. enterocolitica serogroups. These features are specifically conferred by the serogroup O8 YopP. To localize the serotype-related effector domain of YopPO8, we conducted site-directed mutagenesis, by using an approach in which multiple or single amino acids between YopPO8 and YopPO9 from the well-characterized strains WA (serogroup O8) and E40 (serogroup O9) are interchanged. Our data show that an individual amino acid, the arginine-143 residue, plays a predominant role in determining YopP effector functions by impairing IKKβ activities.
MATERIALS AND METHODS
Bacterial strains, cell culture, and stimulation conditions.
The Y. enterocolitica strains used in this study are listed in Table 1. The Y. enterocolitica serotype O9 wild-type strain E40 and the respective yopP-negative mutant E40-ΔyopP were kindly provided by G. R. Cornelis (Microbial Pathogenesis Unit, Université Catholique de Louvain, Brussels, Belgium). Overnight cultures grown at 26°C were diluted 1:20 in fresh Luria-Bertani broth and grown for 2 h at 37°C as described previously (32). The bacteria were then washed once and resuspended in phosphate-buffered saline (PBS) at the desired concentration, which was adjusted by measuring the optical density at 600 nm and checked by plating serial dilutions of the samples on agar (32). The murine macrophage cell line J774A.1 was cultured in RPMI 1640 cell growth medium supplemented with 10% heat-inactivated fetal calf serum and 5 mM l-glutamine (32). Human blood monocytes were isolated from heparinized blood with magnetic anti-CD14 antibody beads and differentiated to macrophages as previously reported (22). Infections were performed at a ratio of 50 bacteria per cell. For incubation times longer than 90 min, bacteria were killed by addition of gentamicin (100 μg/ml) after 90 min of infection.
TABLE 1.
Y. enterocolitica strains used in this study
| Strain | Relevant characteristics | Source or reference |
|---|---|---|
| WA | Wild-type strain WA-314; serogroup O8; clinical isolate harboring the virulence plasmid pYVO8 | 8, 19 |
| WA-ΔpYV | Plasmidless derivative of strain WA; identical to strain WA-C | 19, 33 |
| WA-ΔyopP | yopPO8-negative mutant of WA; insertional inactivation of yopP | 34 |
| WA-ΔyopP/+PO8 | WA-ΔyopP complemented with yopPO8 | 34 |
| WA-ΔyopP/+PO9 | WA-ΔyopP complemented with yopPO9 | This study |
| WA-ΔΣyop | Strain harboring plasmid pLCR encoding the secretion/translocation apparatus of WA, but no Yop effector gene; identical to strain WA-C-pLCR | 34 |
| WA-ΔΣyop/+PO8 | WA-ΔΣyop complemented with yopPO8 | 34 |
| WA-ΔΣyop/+PO9 | WA-ΔΣyop complemented with yopPO9 | This study |
| WA-ΔΣyop/+PO8(R143S) | WA-ΔΣyop complemented with yopPO8 in which arginine-143 was replaced by serine | This study |
| WA-ΔΣyop/+PO9(S143R) | WA-ΔΣyop complemented with yopPO9 in which serine-143 was replaced by arginine | This study |
| E40 | Wild-type strain; serogroup O9; harboring the serogroup O9 virulence plasmid pYV40 | 23 |
| E40-ΔyopP | yopPO9-negative mutant of E40; identical to strain E40(pMSK41); inactivation of yopP by deletion | 23 |
| E40-ΔyopP/+PO8 | E40-ΔyopP complemented with yopPO8 | This study |
| E40-ΔyopP/+PO9 | E40-ΔyopP complemented with yopPO9 | This study |
| E40-ΔyopP/+PO8(44-214PO9) | E40-ΔyopP complemented with yopPO8 in which the internal BamHI region was replaced by the BamHI region from yopPO9 | This study |
| E40-ΔyopP/+PO9(44-214PO8) | E40-ΔyopP complemented with yopPO9 in which the internal BamHI region was replaced by the BamHI region from yopPO8 | This study |
| E40-ΔyopP/+PO8(R143S) | E40-ΔyopP complemented with yopPO8 in which arginine-143 was replaced by serine | This study |
| E40-ΔyopP/+PO9(S143R) | E40-ΔyopP complemented with yopPO9 in which serine-143 was replaced by arginine | This study |
| E40-ΔyopP/+PO8(A144T) | E40-ΔyopP complemented with yopPO8 in which alanine-144 was replaced by threonine | This study |
| E40-ΔyopP/+PO9(T144A) | E40-ΔyopP complemented with yopPO9 in which threonine-144 was replaced by alanine | This study |
| E40-ΔyopP/+PO8(RA143/144ST) | E40-ΔyopP complemented with yopPO8 in which RA143/144 were replaced by ST143/144 | This study |
| E40-ΔyopP/+PO9(ST143/144RA) | E40-ΔyopP complemented with yopPO9 in which ST143/144 were replaced by RA143/144 | This study |
| E40-ΔyopP/+PO9(44-129PO8) | E40-ΔyopP complemented with yopPO9 in which the region for amino acids 44–129 was replaced by the respective region from yopPO8 | This study |
| E40-ΔyopP/+PO9(130-214PO8) | E40-ΔyopP complemented with yopPO9 in which the region for amino acids 130–214 was replaced by the respective region from yopPO8 | This study |
| E40-ΔyopP/+PO9(141-214PO8) | E40-ΔyopP complemented with yopPO9 in which the region for amino acids 141–214 was replaced by the respective region from yopPO8 | This study |
| E40-ΔyopP/+pYVyopPO8 | E40-ΔyopP complemented by a 5.5-kb pYVO8 EcoRI-KpnI fragment encoding the wild-type yopO/yopPO8 operon | This study |
| E40-ΔyopP/+pYVyopPO8(R143S) | E40-ΔyopP complemented by a 5.5-kb pYVO8 EcoRI-KpnI fragment encoding a yopO/yopPO8 operon in which yopPO8 arginine-143 was replaced by serine | This study |
| Y-8081 | Wild-type strain, serogroup O8; clinical isolate | 31 |
| Y-96-P | Wild-type strain, serogroup O9; clinical isolate | 18 |
| Y.e.-93 | Wild-type strain, serogroup O9; clinical isolate from the institute | This study |
| Y-108-P | Wild-type strain, serogroup O3; clinical isolate | 18 |
| Y.e.-88 | Wild-type strain, serogroup O3; clinical isolate from the institute | This study |
Construction of plasmids and complementation of Y. enterocolitica strains and mutants.
The yopP-negative mutants WA-ΔyopP and E40-ΔyopP were complemented with plasmids encoding diverse yopP constructs. The yopP constructs were engineered by PCR. To obtain full-length yopP from serogroup O8 or O9, DNA fragments encompassing the entire yopP open reading frames from strain WA or E40 were amplified and subcloned in plasmid pCJYE138-G3 (20), replacing the gfp gene of pCJYE138-G3 in frame by yopP, as described previously (34). In the resulting plasmid, the yopP gene is located downstream of the yopE promoter and fused to regions encoding the first 138 amino acids of YopE. Induction of gene expression mediates production of a YopE138-YopP fusion protein that is intrabacterially stabilized by the YopE-specific chaperone SycE. Translocated YopE138 hybrid proteins do not exert a YopE effect (20). Complementation of WA-ΔyopP and E-40ΔyopP with these contructs generated Yersinia strains that selectively produce YopP from serogroup O8 (WA-ΔyopP/+PO8 and E40-ΔyopP/+PO8) or from serogroup O9 (WA-ΔyopP/+PO9 and E40-ΔyopP/+PO9). To enable complementation of the chloramphenicol-resistant strain WA-ΔyopP, we disrupted the endogenous chloramphenicol resistance marker of the complementator plasmid by introducing a spectinomycin resistance fragment. For modulation and comparison of YopP effects by the different serogroups, we exchanged a central 514-bp BamHI fragment of the yopP gene between yopP from serogroups O8 and O9 [resulting in strains E40-ΔyopP/+PO8(44–214PO9) and E40-ΔyopP/+PO9(44–214PO8)]. The 514-bp DNA fragment encodes amino acid residues 44 to 214 of YopP. Mapping of the YopPO8-specific effector domain and site-directed mutagenesis of single amino acids were accomplished by generating mutated yopP BamHI fragments by the splicing and overlap extension method. Accordingly, two PCR fragments with homologous overlapping sequences at one terminus were engineered with appropriate complementary primers. For instance, for fusing a sequence from yopPO8 to the respective C-terminal coding region from yopPO9, one PCR was run on yopPO9 with a forward primer annealing outside the BamHI fragment and the reverse primer determining the fusion region within the BamHI fragment. The second PCR was run on yopPO8, with the forward primer being complementary to the reverse primer used for the PCR on yopPO9 and the reverse primer annealing outside the BamHI fragment. Both PCR products were fused by subsequent PCR. The resulting DNA fragment encompasses the yopPO9/yopPO8 chimeric DNA. Site-directed mutagenesis of amino acids was conducted by the same approach, except that both primary PCRs were performed with either yopPO9 or yopPO8, by using overlapping complementary mutation primers. The resulting PCR products, encompassing either mutagenized yopP or yopPO9/yopPO8 hybrid BamHI fragments, were introduced into the internal BamHI restriction sites of yopP, thereby replacing the endogenous BamHI fragment. The primers used for fusion of amino acids 44 to 129 from YopPO9 to amino acids 130 to 214 from YopO8 and vice versa were 5′-AAATGGGAAAACATCTCTGATATTGTTTGAACCAGC-3′ (forward) and 5′-GCTGGTTCAAACAATATCAGAGATGTTTTCCCATTT-3′ (reverse). The mutation primers for exchange of arginine to serine at position 143 in YopPO8 were 5′-GGCAATAAGTGCAAAAACGGCCATTGAACGTT-3′ (forward) and 5′-AACGTTCAATGGCCGTTTTTGCACTTATTGCC-3′ (reverse), for exchange of serine 143 to arginine in YopPO9 we used 5′-GGCAATAAGGACAAAAACGGCCATTGAACGTT-3′ (forward) and 5′-AACGTTCAATGGCCGTTTTTGTCCTTATTGCC-3′ (reverse); the mutations are underlined. The resulting strains, E40-ΔyopP/+PO8(R143S) and E40-ΔyopP/+PO9(S143R), produce the respective mutagenized YopP proteins. The same strategy was used to generate YopPO9(141–214PO8) hybrid protein and mutagenized YopPO8(A144T), YopPO8(RA143/144ST), YopPO9(T144A), and YopPO9(ST143/144RA). In an additional approach, E40-ΔyopP was complemented by a 5.5-kb EcoRI-KpnI DNA fragment of the Yersinia pYV plasmid that encompasses the yopO/yopP operon encoding either wild-type or mutagenized yopPO8. Accordingly, a 4.6-kb EcoRI-KpnI DNA fragment of pYVO8, which encodes the yopO/yopP promoter region and the yopO gene, was subcloned. The KpnI restriction site of this fragment is located 60 bp upstream of the yopP start codon in the pYV plasmid (37). The 4.6-kb EcoRI-KpnI DNA fragment was extended in frame at the KpnI site by introduction of a 930-bp KpnI fragment that encodes the yopP gene and the preceding DNA sequence, including the endogenous KpnI restriction site. The 930-bp KpnI-yopP DNA fragments were engineered by PCR and by the splicing and overlap extension method with wild-type yopPO8 and mutagenized yopPO8(R143S) as described above. The resulting Yersinia strains, E40-ΔyopP/+pYVyopPO8 and E40-ΔyopP/+pYVyopPO8(R143S), produce YopPO8 or YopPO8(R143S) under control of the yopO/yopPO8 endogenous promoter. All PCR products were checked for sequence accuracy by sequencing. To generate Yersinia strains that secret and translocate the YopP constructs as unique effector Yops, the respective plasmids were introduced in strain WA-ΔΣyop. WA-ΔΣyop solely harbors a plasmid encoding the Y. enterocolitica secretion and translocation machinery and the gene for the adhesin YadA. It does not bear any effector Yop gene and thus does not produce any effector Yop (34).
EMSAs and IKK assays.
To analyze nuclear translocation of NF-κB, nuclear proteins were extracted and electrophoretic mobility shift assays (EMSAs) were performed as described previously (32, 34). In brief, the cells were washed with ice-cold PBS and lysed with hypotonic buffer on ice (5 mM HEPES [pH 7.9], 1.5 mM MgCl2, 10 mM KCl, 0.5 mM dithiothreitol [DTT] and protease inhibitors). Proteins were extracted from the nuclei by resuspension of the nuclear pellets in ice-cold extraction buffer (20 mM HEPES [pH 7.9], 25% glycerol, 1 M NaCl, 1.5 mM MgCl2, 0.5 mM EDTA, 0.5 mM DTT and protease inhibitors). Seven micrograms of the extracted nuclear proteins was incubated with 2 to 5 ng of a radiolabeled oligonucleotide probe that encompasses the consensus binding site for NF-κB dimeric complexes (Santa Cruz Biotechnology). The DNA-binding reactions were performed in the presence of a combination of 25 mM HEPES (pH 7.9), 0.5 mM EDTA, 0.5 mM DTT, and 5% glycerol for 30 min on ice. The DNA-protein complexes were subsequently separated by 5% polyacrylamide gel electrophoresis (PAGE) and analyzed by autoradiography (32, 34).
Immunoprecipitations with anti-IKKβ antibody and kinase assays were carried out with 1.25 × 107 or 1 × 108 cells per sample according to the method described previously (11, 34). Following treatment with LPS or yersiniae, the cells were lysed with lysis buffer (10 mM HEPES [pH 7.8], 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1% NP-40, 1 mM DTT, and phosphatase and protease inhibitors) and diluted 1/1 with precipitation buffer (20 mM Tris [pH 7.5], 200 mM NaCl, 1 mM DTT, and phosphatase and protease inhibitors). The lysates were precleared with an irrelevant polyclonal rabbit antibody and protein A-agarose (Santa Cruz Biotechnology) and subsequently were incubated with a polyclonal rabbit anti-IKKβ antibody (Santa Cruz Biotechnology) for 4 h at 4°C. Immune complexes were collected with protein A-agarose, washed three times with precipitation buffer and then kinase buffer, and subsequently subjected to kinase assay. The kinase reactions were performed with 1 μg of glutathione S-transferase (GST)-ΙκBα for 30 min at 30°C in the presence of 20 μM ATP (4 μCi of [γ-32P]ATP per sample) and kinase buffer (20 mM HEPES [pH 8], 10 mM MgCl2, 50 mM NaCl, 2 mM DTT, and phosphatase and protease inhibitors). The plasmid encoding GST-ΙκBα was kindly provided by U. Siebenlist (National Institute of Allergy and Infectious Diseases, Bethesda, Md.). Proteins were separated by sodium dodecyl sulfate (SDS)-PAGE and electrotransferred to polyvinylidene difluoride (PVDF) membrane. The upper part of the membrane was immunoblotted with anti-IKKβ antibody to determine the amount of precipitated IKKβ; the lower part, including GST-ΙκBα, was analyzed by autoradiography.
Coimmunoprecipitations and Western immunoblotting.
Coimmunoprecipitations were performed as described previously (34). Briefly, 108 J774A.1 cells were infected with bacteria for 60 min, washed, scraped, and lysed on ice in lysis buffer containing 10 mM HEPES (pH 7.8), 10 mM KCl, 2 mM MgCl2, 0.1 mM EDTA, 1% NP-40, and 1 mM DTT, and phosphatase and protease inhibitors. These conditions selectively lyse the cells, but not the bacteria. In addition, these conditions do not extract critical amounts of Yops from the bacteria. The cellular lysates were diluted 1/1 with precipitation buffer (20 mM Tris [pH 7.5], 200 mM NaCl, 1 mM DTT, and phosphatase and protease inhibitors) and precleared with an irrelevant polyclonal rabbit antibody and protein A-agarose (Santa Cruz Biotechnology). For immunoprecipitation, cell lysates were incubated with a polyclonal rabbit anti-YopE antibody (20) for 4 h at 4°C. Immune complexes were collected with protein A-agarose, washed with precipitation buffer, separated by SDS-PAGE, electrotransferred to PVDF membrane, and probed with either rabbit anti-YopE or polyclonal rabbit anti-IKKβ antibody (which exhibits partial cross-reactivity with IKKα; Santa Cruz Biotechnology). Immunoreactive bands were visualized by incubation with goat anti-rabbit antibodies conjugated to horseradish peroxidase (Amersham Pharmacia) by using enhanced chemiluminescence reagents (Amersham Pharmacia). For quantification of translocated YopP constructs, cytoplasmic lysates of J774A.1 cells were produced 60 min after onset of infection. The lysates were separated by SDS-PAGE, electrotransferred to PVDF membrane, and probed with either rabbit anti-YopE, or polyclonal rabbit anti-MEK1 antibody (Santa Cruz Biotechnology). Nonsaturated immunoreactive bands were visualized as described above and processed for quantification by using Image-Master 1D software (Amersham Pharmacia).
Assessment of apoptosis by fluorescence microscopy.
To quantify apoptosis of J774A.1 cells in response to bacterial infection, apoptotic cells were specifically labeled with fluorescein-conjugated annexin V (Boehringer, Mannheim, Germany) as described previously (35). Annexin V binds with high affinity to phosphatidylserine exposed on the outer leaflet of apoptotic cells and confers green fluorescence to cells undergoing apoptosis. The simultaneous application of the DNA stain propidium iodide (Sigma, St. Louis) allows the discrimination of apoptotic from necrotic cells. The rate of apoptosis was determined by counting a minimum of 200 cells per sample in a fluorescence microscope. Results are expressed as mean percentages of fluorescing apoptotic cells ± standard deviation from three independent experiments.
RESULTS
YopP from Y. enterocolitica serotype O8 is substantially more efficient in suppression of the NF-κB pathway and mediation of apoptosis than serotype O9 YopP.
In order to analyze the impact of Y. enterocolitica on macrophage apoptosis, we investigated a panel of wild-type strains from diverse human pathogenic Y. enterocolitica serotypes (listed in Table 1). Surprisingly, Y. enterocolitica strains from the serogroup O8 (strains WA and Y-8081) were by far more efficient at triggering apoptosis in murine J774A.1 macrophages than Yersinia strains from the serotypes O3 (strains Y-108-P and Y.e.-88) and O9 (strains E40, Y-96-P, and Y.e.-93). Accordingly, serogroup O8 Y. enterocolitica killed more than 90% of infected macrophages 6 h after onset of infection, whereas only 20 to 40% of the cells treated with serogroup O3 or O9 Y. enterocolitica were apoptotic within the same time. To reveal whether these discrepant apoptotic responses were due to more active YopPO8 versus YopPO9, we amplified and subcloned the yopP genes from the well-characterized strains WA (serotype O8) and E40 (serotype O9) in order to complement the yopP-negative knockout mutants. Accordingly, yopPO8 and yopPO9 were introduced in trans into the yopPO8-negative mutant WA-ΔyopP. The resulting strains, WA-ΔyopP/+PO8 and WA-ΔyopP/+PO9, synthesize YopPO8 or YopPO9 as a fusion protein, with the first 138 amino acids of YopE controlled by the yopE promoter. This procedure allows efficient production and secretion of Yop hybrid proteins by Yersinia in comparable amounts. This is a useful method to complement yopP-negative mutants (34). yopP is located on an operon behind yopO; therefore, a complementation procedure using the endogenous yopP promoter is complicated (13). In all further approaches, we used this tool to introduce yopP constructs into Yersinia and to analyze their impact on the host cell.
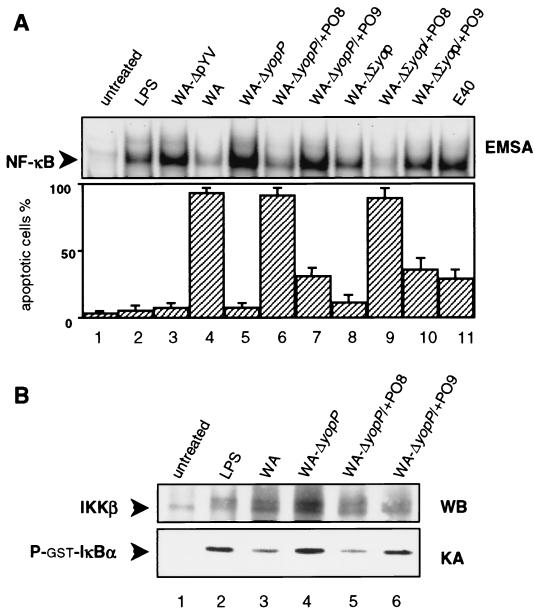
Figure 1A shows that only Yersinia strains producing YopP derived from the serogroup O8 (the wild-type strain WA and the yopPO8-complemented mutant WA-ΔyopP/+PO8; lanes 4 and 6) efficiently triggered macrophage apoptosis. Apoptosis mediated by YopPO9-producing Yersinia was markedly reduced (the yopPO9-complemented mutant WA-ΔyopP/+PO9 and the wild-type strain E40; lanes 7 and 11). The virulence plasmid-cured strain WA-ΔpYV and the yopP-negative mutant WA-ΔyopP did not induce cell death (lanes 3 and 5). We furthermore compared two Yersinia strains that secret either YopPO8 (WA-ΔΣyop/+PO8; lane 9) or YopPO9 (WA-ΔΣyop/+PO9; lane 10) as a single effector Yop. Again, the YopPO8-producing strain was more efficient in apoptosis induction than the strain producing YopPO9. The original strain WA-ΔΣyop, which solely bears the Yersinia type III protein secretion apparatus, but does not produce any effector Yop, only marginally mediated apoptosis (lane 8). In a previous study, we demonstrated that Yersinia-induced apoptosis mainly results from inhibition of the NF-κB survival pathway by YopP (34). To substantiate the relationship between apoptosis induction and NF-κB inhibition, we analyzed the capabilities of the different Yersinia strains to interfere with NF-κB activation. Therefore, we investigated nuclear translocation of NF-κB in Yersinia-infected macrophages by EMSA. The results, depicted in the upper panel of Fig. 1A, revealed a striking correlation between apoptosis induction by YopPO8-producing Yersinia and substantial suppression of nuclear translocation of NF-κB after 90 min of infection (lanes 4, 6, and 9). NF-κB inhibition mediated by YopPO9-producing Yersinia was considerably less pronounced (lanes 7, 10, and 11). The NF-κB response induced by WA-ΔΣyop was reduced compared with NF-κB stimulation mediated by WA-ΔΣyop/+PO9 (compare lanes 8 and 10). This probably is the result of lytic affection of a portion of the infected cells, which is conferred by pore-forming proteins of the Yersinia translocation apparatus under conditions in which no effector Yops are translocated (14).
FIG. 1.
YopPO8 efficiently triggers macrophage apoptosis and interferes with NF-κB activation. (A) NF-κB suppression and apoptosis induction. J774A.1 cells were left untreated (lane 1) or were stimulated with LPS (lane 2), virulence plasmid-cured WA-ΔpYV (lane 3), wild-type WA from serogroup O8 (lane 4), yopPO8-negative WA-ΔyopP (lane 5), YopPO8-producing WA-ΔyopP/+PO8 (lane 6), YopPO9-producing WA-ΔyopP/+PO9 (lane 7), WA-ΔΣyop producing no effector Yops (lane 8), WA-ΔΣyop/+PO8 producing only YopPO8 (lane 9), WA-ΔΣyop/+PO9 producing only YopPO9 (lane 10), or wild-type E40 from serogroup O9 (lane 11). The NF-κB activities were determined by EMSA 90 min after infection (upper panel). Only sections of the autoradiogram containing the NF-κB–DNA complexes are shown. The EMSA shows data from one experiment representative of five performed. Apoptosis was assayed 6 h after onset of infection by staining cells with annexin V and counting apoptotic cells by fluorescence microscopy (lower panel). Results are expressed as mean percentages ± standard deviations from three independent experiments. (B) IKKβ activity assay. Cell extracts from 1.25 × 107 untreated cells (lane 1) or cells treated with LPS (lane 2), wild-type WA from serogroup O8 (lane 3), yopPO8-negative WA-ΔyopP (lane 4), YopPO8-producing WA-ΔyopP/+PO8 (lane 5), or YopPO9-producing WA-ΔyopP/+PO9 (lane 6) for 30 min were incubated with anti-IKKβ antibodies to precipitate IKKβ. IKKβ activities were assayed by measuring the abilities of the immunocomplexes to radioactively phosphorylate recombinant GST-IκBα. Kinase reaction samples were subjected to SDS-PAGE and transferred to PVDF membrane. The upper part of the membrane was immunoblotted with anti-IKK antibodies (upper panel). The double band appearing in lanes 2 to 6 may reflect phosphorylated and nonphosphorylated forms of IKKβ. The lower part of the membrane including GST-ΙκBα was analyzed by autoradiography (lower panel). The results shown are from one representative experiment out of three performed. WB, Western blot; KA, kinase assay.
To find out whether these effects on the NF-κB pathway directly result from distinct actions of YopPO8 and YopPO9 on IKK activities, we performed in vitro kinase assays with immunoprecipitated IKKβ (Fig. 1B). LPS and yopP-negative Yersinia (WA-ΔyopP) induced a substantial increase in phosphotransferase activities toward recombinant GST-ΙκBα by immunoprecipitated IKKβ after 30 min of stimulation (lanes 2 and 4), indicating IKKβ activation. In contrast, precipitated IKKβ from macrophages infected with YopPO8-producing Yersinia (WA and WA-ΔyopP/+PO8; lanes 3 and 5) was impaired in phosphorylating GST-ΙκBα. In relation to YopPO8-producing Yersinia, suppression of IKKβ activities conferred by YopPO9-producing Yersinia (WA-ΔyopP/+PO9; lane 6) was considerably less established. These data indicate stronger efficiency of YopPO8 in subversion of the NF-κB pathway and mediation of apoptosis compared to YopPO9.
To investigate whether these effects specifically occur in mouse macrophages or whether they are a more general phenomenon, we analyzed apoptosis induction in macrophages derived from human monocytes (35). Whereas the YopPO8-producing strain WA-ΔyopP/+PO8 triggered 60 to 80% apoptosis, cell death mediated by WA-ΔyopP/+PO9 was markedly reduced (30 to 50% apoptosis). The yopP-negative mutant WA-ΔyopP did not confer cell death (<10% apoptosis). Thus, enhanced apoptosis through serogroup O8 Y. enterocolitica is not a unique characteristic of engagement of mouse macrophages, but also occurs in human macrophages, although to a slighter extent.
Arginine-143 is an essential residue of the YopPO8-specific effector domain.
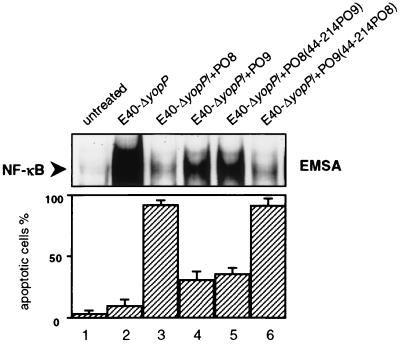
We next attempted to analyze the relationship between structural domains and the serotype-related functions of YopP isotypes. For that reason, we compared the deduced amino acid sequences of YopPO8 (GenBank accession no. AF336309) and YopPO9 (GenBank accession no. AF102990). YopPO8 from strain WA, which was investigated in our studies, displays a sequence corresponding to the published sequence of YopPO8 from strain Y-8081 (37). Figure 2 shows that the sequences of YopPO8 and YopPO9 are highly homologous (94% identity). Single amino acids that differ between these two serogroups are marked in boldface. In the first approach to localize the YopPO8 effector domain, we excised an internal 514-bp region located between two BamHI restriction sites of yopPO8 and yopPO9 and exchanged the fragment between the two yopP genes (Fig. 2; the corresponding amino acids of the BamHI restriction sites are underlined). The 514-bp DNA fragment encodes amino acids 44 to 214 of YopP. The respective yopPO8/yopPO9 chimeric DNA constructs were transferred into the yopPO9-negative mutant E40-ΔyopP. We utilized strain E40-ΔyopP instead of WA-ΔyopP in this set of experiments, because the lack of any antibiotic resistance marker in E40-ΔyopP facilitated the complementation procedure. Both E40-ΔyopP and WA-ΔyopP exhibited identical yopP-negative phenotypes. With the exchange of the 514-bp yopP BamHI region, the YopP effector functions were completely reversed in the resulting strains (Fig. 3). In contrast to the original strain E40-ΔyopP/+PO8, strain E40-ΔyopP/+PO8(44–214PO9) harboring the yopPO9 BamHI fragment, provoked a strong NF-κB signal that was associated with substantial cellular survival (compare lanes 3 and 5). On the other hand, insertion of the yopPO8 BamHI fragment into E40-ΔyopP/+PO9 strongly suppressed the NF-κB response to the resulting strain E40-ΔyopP/+PO9(44–214PO8), which led to fulminant apoptosis (compare lanes 4 and 6). Thus, the serogroup O8-specific effector domain of YopP and the corresponding phenotype are interchangeable between the two Y. enterocolitica serotypes.
FIG. 2.
Alignment of the deduced amino acid sequences from YopPO8 (GenBank accession no. AF336309) and YopPO9 (GenBank accession no. AF102990). Amino acid differences are marked in boldface. The corresponding amino acids of the internal yopP BamHI restriction sites are underlined. Dark shading denotes the proposed isotype-related YopP effector site (arginine-143, YopPO8; serine-143, YopPO9). Light shading indicates the amino acid residues of the proposed cysteine protease-related catalytic triad (histidine-109, glutamic acid-128, and cysteine-172).
FIG. 3.
Interchange of amino acids 44 to 214 reverses YopPO8 and YopPO9 isotype-dependent effector functions. J774A.1 cells were left untreated (lane 1) or were infected with yopPO9-negative E40-ΔyopP (lane 2), YopPO8-producing E40-ΔyopP/+PO8 (lane 3), YopPO9-producing E40-ΔyopP/+PO9 (lane 4), E40-ΔyopP/+PO8(44–214PO9) producing a YopPO8 construct in which the internal amino acids 44 to 214 were substituted for by the respective amino acids from YopPO9 (lane 5), and E40-ΔyopP/+PO9(44–214PO8) producing a YopPO9 construct in which internal amino acids 44 to 214 were substituted for by the respective amino acids from YopPO8 (lane 6). The NF-κB activities were determined 90 min after infection by EMSA (upper panel). Only sections of the autoradiogram containing the NF-κB–DNA complexes are shown. The EMSA shows data from one experiment representative of three performed. Apoptosis was assayed 6 h after onset of infection by staining cells with annexin V and counting apoptotic cells by fluorescence microscopy (lower panel). Results are expressed as mean percentages ± standard deviations from three independent experiments.
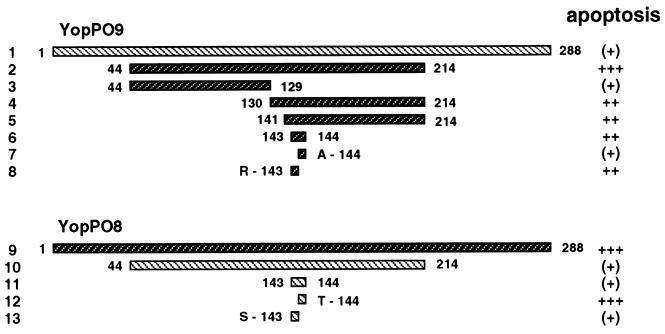
We took advantage of this finding for design of our further experimental strategy. In order to map the YopPO8-related effector domain, we generated yopPO9/yopPO8 chimeric DNA, in which regions from the BamHI fragment of yopPO9 were substituted for by the respective regions from yopPO8. The hybrid DNA was engineered by a PCR-based splicing and overlap extension method with complementary primers that overlap in a distinct region of the BamHI fragment (for details, see the Materials and Methods section). The synthesized PCR products were fused by subsequent PCR, digested with BamHI restriction endonuclease, and inserted into the internal BamHI restriction sites of yopPO9, thereby replacing the endogenous BamHI fragment. The plasmids were transferred into the yopPO9-negative mutant E40-ΔyopP. The resulting strains, which produce diverse YopPO9/YopPO8 chimeric proteins, were screened on apoptosis induction, as displayed in Fig. 4. A Yersinia strain that produces YopPO9 encompassing the amino acids 130 to 214 from YopPO8 triggered significant apoptosis (lane 4). Thus, the major apoptosis-inducing domain localizes to the C-terminal coding region of the yopPO8 BamHI fragment. Within this region, we found potentially critical arginine-143 and alanine-144 residues in YopPO8, which differed from serine-143 and threonine-144 in YopPO9. By mutation of either or both of the position 143 and 144 residues, we identified arginine-143 as an important component of the YopPO8 effector domain. Replacement of serine-143 in YopPO9 by YopPO8-derived arginine strongly enhanced the ability of YopPO9 to mediate apoptosis (from 20 to 40% to 60 to 80%; compare lanes 1 and 8). Conversely, the mutation of YopPO8 arginine-143 to serine completely abrogated YopPO8-specific augmentation of cell death (compare lanes 9 and 13). This indicates a predominant role of arginine-143 in triggering the apoptotic response. The mutations of the neighboring threonine-144 (YopPO9) to alanine-144 (YopPO8) and vice versa did not exert any effects on YopP activity (lanes 7 and 12).
FIG. 4.
Mapping of the YopPO8 isotype-related effector domain by interchange of amino acids between YopPO9 and YopPO8. Mutagenesis of YopPO9 (lane 1) was accomplished by replacement of an internal 514-bp yopPO9 DNA fragment by diverse yopPO9/yopPO8 hybrid sequences for this region (lanes 2 to 8). The DNA region encodes amino acids 44 to 214 of YopP. The fragments were generated by PCR and introduced into the internal yopPO9 BamHI restriction sites, which encompass the 514-bp region. In lanes 2 to 8, only the amino acid sequences introduced from YopPO8, but not the endogenous YopPO9 amino acid sequences, are displayed. Mutagenesis of YopPO8 (lane 9) was conducted by the same method, resulting in substitution of multiple or single amino acids of YopPO8 by residues from YopPO9 (lanes 10 to 13). Apoptosis was assayed 6 h after onset of infection by staining cells with annexin V and analyzing apoptotic cells by fluorescence microscopy [apoptosis: (+), 20 to 40%; +, 40 to 60%; ++, 60 to 80%; +++, >80%].
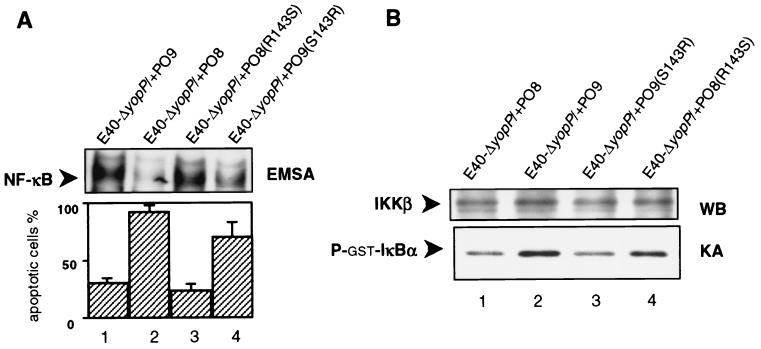
The data obtained by the NF-κB and IKK assays correlated with the results on apoptosis (Fig. 5). The inability of YopPO8(R143S) to mediate substantial apoptosis coincided with a strong NF-κB response in infected macrophages [strain E40-ΔyopP/+PO8(R143S)] (Fig. 5A, lane 3). Conversely, the mutation of serine-143 to arginine in YopPO9(S143R) induced considerable NF-κB suppression in comparison to wild-type YopO9 [strains E40-ΔyopP/+PO9 and E40-ΔyopP/+PO9(S143R)] (Fig. 5A, lanes 1 and 4). The IKK assays produced corresponding data (Fig. 5B). Precipitated IKKβ from macrophages infected with E40-ΔyopP/+PO9 and E40-ΔyopP/+PO8(R143S) exhibited stronger phosphotransferase activities toward GST-IκBα than IKKβ precipitated from E40-ΔyopP/+PO8 or E40-ΔyopP/+PO9(S143R)-treated cells (Fig. 5B, compare lanes 1 and 3 to 2 and 4). Thus, the mutations of arginine-143 to serine and vice versa critically influence the action of YopP on IKKβ activities and NF-κB activation. Since functional characterizations of the respective YopP mutations were accomplished by transcomplementation studies with YopE138-YopP fusion proteins, we additionally analyzed the effects of nonfused, wild-type and mutated YopPO8 produced under control of the yopO/yopPO8 endogenous promoter. Accordingly, strain E40-ΔyopP was complemented by a 5.5-kb EcoRI-KpnI DNA fragment of the Yersinia pYVO8 plasmid, which encompasses the yopO/yopP operon encoding either wild-type or mutagenized yopPO8. In these experiments, strain E40-ΔyopP/+pYVyopPO8, which produces wild-type YopPO8, induced 70 to 80% apoptosis in J774A.1 cells 6 h after onset of infection. The strain producing YopPO8 mutagenized from arginine-143 to serine [E40-ΔyopP/+pYVyopPO8(R143S)] mediated considerably less pronounced apoptosis (20 to 25% cell death). Comparable differences were observed in terms of the capabilities of these strains to suppress activation of NF-κB (data not shown). These data indicate that the YopE138-YopP fusion proteins behave functionally similar to the nonfused versions of YopP.
FIG. 5.
Arginine-143 critically determines YopP effector functions. (A) NF-κB suppression and apoptosis induction. J774A.1 cells were infected with YopPO9-producing E40-ΔyopP/+PO9 (lane 1), YopPO8-producing E40-ΔyopP/+PO8 (lane 2), E40-ΔyopP/+PO8(R143S) producing a YopPO8 construct in which arginine-143 was substituted for by serine (lane 3), or E40-ΔyopP/+PO9(S143R) producing a YopPO9 construct in which serine-143 was substituted for by arginine (lane 4). The NF-κB activities were determined 90 min after infection by EMSA (upper panel). Only sections of the autoradiogram containing the NF-κB–DNA complexes are shown. The EMSA shows data from one experiment representative of three performed. Apoptosis was assayed 6 h after onset of infection by staining cells with annexin V and counting apoptotic cells by fluorescence microscopy (lower panel). Results are expressed as mean percentages ± standard deviations from three independent experiments. (B) IKKβ activity assay. Cell extracts from 108 cells infected with YopPO8-producing E40-ΔyopP/+PO8 (lane 1), YopPO9-producing E40-ΔyopP/+PO9 (lane 2), YopPO9(S143R)-producing E40-ΔyopP/+PO9(S143R) (lane3), or YopPO8(R143S)-producing E40-ΔyopP/+PO8(R143S) (lane 4) for 30 min were incubated with anti-IKKβ antibodies to precipitate IKKβ. IKKβ activities were assayed by measuring the abilities of the immunocomplexes to radioactively phosphorylate recombinant GST-IκBα. Kinase reaction samples were subjected to SDS-PAGE and transferred to PVDF membrane. The upper part of the membrane was immunoblotted with anti-IKK antibodies (upper panel). The lower part of the membrane including GST-ΙκBα was analyzed by autoradiography (lower panel). The results shown are from one representative experiment out of three performed. WB, Western blot; KA, kinase assay.
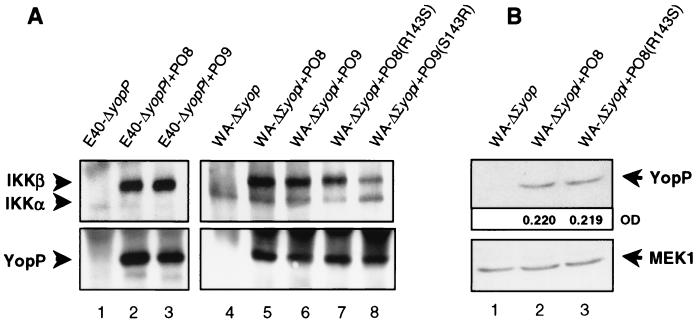
We wondered whether the distinct effects of the respective YopP constructs on NF-κB signaling may result from differential IKKβ targeting. Therefore, we immunoprecipitated translocated YopP from Yersinia-infected macrophages. Immunoprecipitations were conducted with polyclonal anti-YopE antibodies that recognize the N-terminal 138 amino acids of the YopE138-YopP fusion proteins. Precipitation with this antibody efficiently accumulated translocated YopP from macrophages infected with YopE138-YopP fusion protein-producing Yersinia strains [E40-ΔyopP/+PO8, E40-ΔyopP/+PO9, WA-ΔΣyop/+PO8, WA-ΔΣyop/+PO9, WA-ΔΣyop/+PO8(R143S), WA-ΔΣyop/+PO9(S143R)] (Fig. 6A, lanes 2 to 3 and 5 to 8), but not from macrophages infected with the control strains (E40-ΔyopP, WA-ΔΣyop) (Fig. 6A, lanes 1 and 4). Figure 6A shows that the diverse YopP constructs are translocated into the cells to comparable intensities. Immunostaining of the separated precipitates with antibodies directed against IKKβ revealed coprecipitation of IKKβ with the different YopP contructs. This demonstrates that both YopPO8 and YopPO9 interact with IKKβ in infected macrophages. This suggests that the stronger suppressive effect of YopPO8 on IKKβ activities directly results from an enhanced inhibitory action of YopPO8 on IKKβ, rather from solely better IKKβ targeting. This conclusion is further supported by the observation that YopPO9(S143R) substantially reduces IKKβ activities (Fig. 5B), although it apparently exhibits reduced IKKβ binding compared to YopPO8, YopPO9, and YopPO8(R143S) (Fig. 6A, lanes 5 to 8).
FIG. 6.
Targeting of macrophage IKKβ by the diverse YopP constructs. (A.) Interaction of YopP with macrophage IKKβ. J774A.1 cells were infected with yopPO9-negative E40-ΔyopP (lane 1), YopPO8-producing E40-ΔyopP/+PO8 (lane 2), YopPO9-producing E40-ΔyopP/+PO9 (lane 3), WA-ΔΣyop producing no effector Yops (lane 4), WA-ΔΣyop/+PO8 producing only YopPO8 (lane 5), WA-ΔΣyop/+PO9 producing only YopPO9 (lane 6), WA-ΔΣyop/+PO8(R143S) producing only the YopPO8 construct with the mutation of arginine-143 to serine (lane 7), or WA-ΔΣyop/+PO9(S143R) producing only the YopPO9 construct with the mutation of serine-143 to arginine (lane 8). After 60 min, cells were lysed, and YopP was immunoprecipitated with polyclonal anti-YopE antibody recognizing the YopE138-YopP fusion proteins. Immunocomplexes were subjected to SDS-PAGE and transferred to PVDF membrane. One part of the membrane was immunoblotted with anti-YopE antibodies, recognizing the YopP fusion proteins (lower panel), and the other part was immunoblotted with anti-ΙKK antibodies (upper panel). (B) Quantification of amounts of cytoplasmic YopP. J774A.1 cells were infected with WA-ΔΣyop producing no effector Yops (lane 1), WA-ΔΣyop/+PO8 producing only YopPO8 (lane 2), or WA-ΔΣyop/+PO8(R143S) producing only the YopPO8 construct with the mutation of arginine-143 to serine (lane 3). After 60 min, cells were lysed under conditions that selectively lyse the cells. The lysates were subjected to SDS-PAGE and transferred to PVDF membrane. One part of the membrane was immunoblotted with anti-YopE antibodies, recognizing the YopP fusion proteins (upper panel), and the other part was immunoblotted with control anti-MEK1 antibodies to confirm equal loading with cellular lysates (lower panel). Nonsaturated immunoreactive bands were visualized with enhanced chemiluminescence reagents. The optical densities (OD) of the YopP bands were quantified, and the values obtained are indicated. The results shown are from one representative experiment out of three performed.
Figure 6B additionally rules out that the discrepant activities of mutagenized versus nonmutagenized versions of YopP result from differential translocation of the respective proteins into the host cell. Both the YopE138-YopPO8 wild-type (lane 2) and YopE138-YopPO8(R143S) (lane 3) fusion proteins were detected to similar amounts in the host cell cytoplasm by immunoblotting with anti-YopE antibodies (upper panel). Quantification of the optical densities of the respective chemiluminescence signals produced nearly identical values (upper panel). Additional immunoblotting with anti-MEK1 antibodies confirmed equal loading of the gel by the cytoplasmic lysates (lower panel).
DISCUSSION
This study demonstrates that YopP isotypes from diverse pathogenic Y. enterocolitica serogroups differ in their abilities to affect macrophage NF-κB signaling and to trigger macrophage apoptosis. Yersiniae producing YopP from the Y. enterocolitica serotype O8 remarkably impaired IKKβ activities and nuclear translocation of NF-κB, whereas serotype O9 YopP-producing yersiniae evoked substantial IKKβ and NF-κB activation. The stronger inhibitory action of YopPO8 on IKKβ activities and NF-κB activation coincided with a substantially enhanced apoptotic response. In contrast, apoptosis due to YopPO9 was remarkably less pronounced. This indicates that subversion of the NF-κB pathway by YopP-producing yersiniae and the induction of macrophage apoptosis are tightly coupled. The close connection of these two effects supports the idea that macrophage apoptosis results from disruption of the NF-κB survival pathway by Yersinia (32, 34). Indeed, overexpression of the transcriptionally active NF-κB p65 subunit provides a protective effect against Yersinia-mediated apoptosis (34). These findings suggest that subversion of the NF-κB pathway by YopP critically determines apoptosis induction by Y. enterocolitica, although direct action of YopP on cell death signaling was recently proposed (10).
To molecularly characterize the various effects of YopPO8 and YopPO9, we complemented yopP-negative mutants in trans by using a vector construct that mediates synthesis and translocation of distinct YopP proteins under identical conditions. The mutants then complemented by the respective yopPO8 and yopPO9 constructs injected comparable amounts of YopP in infected macrophages and restored the wild-type-related phenotypes. Both YopPO8 and YopPO9 interacted with macrophage IKKβ. To relate the different effects of the YopP isotypes to certain structural domains, we compared the amino acid sequences of YopPO8 and YopPO9. These sequences are highly homologous (94% identity). We gradually mutagenized YopPO8 and YopPO9, thereby replacing endogenous amino acids of YopPO8 with residues from YopPO9 and vice versa. Functional characterization of the resulting YopPO8/YopPO9 chimeric proteins identified arginine-143 of YopPO8 as crucially involved in both the suppression of the NF-κB pathway and the mediation of apoptosis.
These data show that there exist functional differences between genetically highly homologous members of YopP from diverse Y. enterocolitica serotypes. Arginine-143 substantially enhances the inhibitory impact of YopP on NF-κB signaling, leading to severe apoptosis. Interestingly, YopP from the Y. enterocolitica serotype O3 strain Y-108-P, which induced a serogroup O9-like, modest apoptotic response, displays serine at position 143 identical to YopPO9 (data not shown). On the contrary, YopPO8 from the strains WA and Y-8081, which both triggered intense apoptosis, are both bearing arginine-143 (37). The published sequences of the YopP homologues YopJ from Y. pseudotuberculosis (GenBank accession no. L33833) and Y. pestis (GenBank accession no. AF074612) also display arginine at position 143, similar to Y. enterocolitica serogroup O8. In correlation with the proposed critical role of arginine-143 in apoptosis induction, the corresponding Y. pseudotuberculosis strain YPIII (kindly provided by H. Wolf-Watz; Department of Cell and Molecular Biology, Umea, Sweden) induced a strong apoptotic response (data not shown). This suggests that YopPO8 shares structural and functional similarities with YopJ that differ from the effects observed for YopPO9 or YopPO3.
In fact, Y. enterocolitica of serogroup O8 is, with respect to pathogenicity, more related to Y. pestis and Y. pseudotuberculosis than Y. enterocolitica of the serogroups O3 and O9 (6, 7, 9, 16). Y. enterocolitica serotype O8, Y. pseudotuberculosis, and Y. pestis are well adapted to rodents as animal hosts and display high pathogenicity in the mouse infection model even at low bacterial doses. The high virulence phenotype, leading to mouse lethality, is predominantly attributed to the chromosomal Yersinia high-pathogenicity island (HPI), which encodes the yersiniabactin siderophore system (7). The Y. enterocolitica serotypes O3 and O9 lack the HPI and are therefore of low mouse virulence. Although the Yersinia virulence plasmid pYV is also essentially required for mouse virulence, the exchange of pYV between the Y. enterocolitica serotypes O8 and O9 does not significantly alter mouse lethality of the respective serogroups (12, 17). In view of these observations, distinct phenotypic profiles due to diverse YopP isotypes can probably not be expected from the mouse model of infection. Such clear differences may be even more unlikely, since the virulence phenotypes of yopJ/yopP-negative mutants in comparison to those of the respective wild-type strains have been shown to be subtle (13, 24, 38). Two studies could not attribute significant virulence functions to YopJ from Y. pseudotuberculosis and Y. pestis (13, 38). Preliminary results from our laboratory suggest that YopPO8 does not display an immediate obvious virulence phenotype. On the contrary, Monack et al. demonstrated that YopJ helps yersiniae in colonizing deeper lymphatic tissues and in the establishment of a systemic infection (24). These studies suggest that YopP/YopJ may play a discreet role in the infectious process. YopP/YopJ may predominantly be involved in the maintenance of prolonged colonization and asymptomatic latent infection, rather than in mediating severe infectious disease. This hypothesis is supported by the in vitro functions of YopP/YopJ, which primarily are intended to dampen the host immune response, thereby preventing overwhelming immune reactions and inflammation. In this context, a more active YopP/YopJ, carrying arginine-143 in the case of Y. enterocolitica serogroup O8, Y. pseudotuberculosis, and Y. pestis, may have evolved to establish an advantage to yersiniae for better colonization and adaptation to rodents as primary hosts.
The molecular mechanism by which YopPO8 exerts a stronger inhibitory effect on IKKβ is still not clear. It was suggested that binding of YopP/YopJ blocks phosphorylation and activation of its target proteins (27). However, a recent report indicates that YopP/YopJ may act as a cysteine protease on cellular proteins modified by the ubiquitin-like molecule SUMO-1 (28). Mutations of the proposed catalytic residues histidine-109 and cysteine-172 abolish the ability of YopP/YopJ to inhibit the NF-κB pathway and to mediate apoptosis (10, 28). A link between reduced SUMO-1 conjugation of proteins and inhibition of IKKβ or MKKs is hitherto unclear. MKKs and IKKβ do not appear to be direct targets for SUMO-1 cleavage, and the proteins that are de-SUMOylated by YopP/YopJ still await identification (28). Orth and colleagues speculate that SUMO-1 conjugation may be an important posttranslational modification of signaling complexes, which directs their intracellular processing (28). This may involve the regulation of IKKβ- and MKK-dependent signaling pathways. The predicted secondary structure of YopP/YopJ perceives arginine-143 as a component of an α-helix, implying mainly a structural role of arginine-143 in YopP constitution (28). On the other hand, the sequences of YopPO8 and YopPO9 both include the cysteine protease-related catalytic residues histidine-109, glutamic acid-128, and cysteine-172 (Fig. 2). Arginine-143 is located centrally within this catalytic domain, which suggests that arginine-143 may directly affect the proposed enzymatic activity of YopP/YopJ on SUMO-1-conjugated proteins. Together, these data point out a crucial role of arginine-143 in the down-regulation of IKKβ function by YopP, which critically determines the fate of the Yersinia-infected macrophage.
ACKNOWLEDGMENTS
This work was supported by grants from the Bundesministerium für Forschung und Technologie and the Deutsche Forschungsgemeinschaft (grant DFG Ru788).
We thank G. Pfaffinger for expert technical assistance and M. Aepfelbacher, B. Rouot, C. Barz, and W. D. Hardt for constructive discussions. We thank A. Wiedemann for preparation and cultivation of human monocytes/macrophages, A. Schröder for quantification of immunoblot chemiluminescence signals, S. Linder for printing of figures, G. R. Cornelis for providing us with Yersinia strains, and U. Siebenlist for providing us with GST-IκBα cDNA.
REFERENCES
- 1.Aliprantis A O, Yang R B, Weiss D S, Godowski P, Zychlinsky A. The apoptotic signaling pathway activated by Toll-like receptor-2. EMBO J. 2000;19:3325–3336. doi: 10.1093/emboj/19.13.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Baichwal V R, Baeuerle P A. Apoptosis: activate NF-κB or die? Curr Biol. 1997;7:R94–R96. doi: 10.1016/s0960-9822(06)00046-7. [DOI] [PubMed] [Google Scholar]
- 3.Baldwin A S., Jr The transcription factor NF-κB and human disease. J Clin Investig. 2001;107:3–6. doi: 10.1172/JCI11891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bliska J B. Yop effectors of Yersinia spp. and actin rearrangements. Trends Microbiol. 2000;8:205–208. doi: 10.1016/s0966-842x(00)01738-8. [DOI] [PubMed] [Google Scholar]
- 5.Boland A, Cornelis G R. Role of YopP in suppression of tumor necrosis factor alpha release by macrophages during Yersinia infection. Infect Immun. 1998;66:1878–1884. doi: 10.1128/iai.66.5.1878-1884.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bottone E J. Yersinia enterocolitica: the charisma continues. Clin Microbiol Rev. 1997;10:257–276. doi: 10.1128/cmr.10.2.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Carniel E. The Yersinia high-pathogenicity island: an iron-uptake island. Microbes Infect. 2001;3:561–569. doi: 10.1016/s1286-4579(01)01412-5. [DOI] [PubMed] [Google Scholar]
- 8.Carter P B, Varga C F, Keet E E. New strain of Yersinia enterocolitica pathogenic for rodents. Appl Microbiol. 1973;26:1016–1018. doi: 10.1128/am.26.6.1016-1018.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M-P, Stainier I. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Denecker G, Declercq W, Geuijen C A, Benabdillah R, van Gurp M, Sory M P, Vandenabeele P, Cornelis G R. Yersinia enterocolitica YopP-induced apoptosis of macrophages involves the apoptotic signaling cascade upstream of Bid. J Biol Chem. 2001;276:19706–19714. doi: 10.1074/jbc.M101573200. [DOI] [PubMed] [Google Scholar]
- 11.Fischer C, Page S, Weber M, Eisele T, Neumeier D, Brand K. Differential effects of lipopolysaccharide and TNF on monocytic Ικβ kinase signalsome activation and Ικβ proteolysis. J Biol Chem. 1999;274:24625–24632. doi: 10.1074/jbc.274.35.24625. [DOI] [PubMed] [Google Scholar]
- 12.Gaede K I, Heesemann J. Arthritogenicity of genetically manipulated Yersinia enterocolitica serotype O8 for Lewis rats. Infect Immun. 1995;63:714–719. doi: 10.1128/iai.63.2.714-719.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Galyov E E, Hakansson S, Wolf-Watz H. Characterization of the operon encoding the YpkA Ser/Thr protein kinase and the YopJ protein of Yersinia pseudotuberculosis. J Bacteriol. 1994;176:4543–4548. doi: 10.1128/jb.176.15.4543-4548.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hakansson S, Schesser K, Persson C, Galyov E E, Rosqvist R, Homblé F, Wolf-Watz H. The YopB protein of Yersinia pseudotuberculosis is essential for the translocation of Yop effector proteins across the target cell membrane and displays a contact-dependent membrane disrupting activity. EMBO J. 1996;15:5812–5823. [PMC free article] [PubMed] [Google Scholar]
- 15.Hatada E N, Krappmann D, Scheidereit C. NF-κB and the innate immune response. Curr Opin Immunol. 2000;12:52–58. doi: 10.1016/s0952-7915(99)00050-3. [DOI] [PubMed] [Google Scholar]
- 16.Hayashidani H, Ohtomo Y, Toyokawa Y, Saito M, Kaneko K-I, Kosuge J, Kato M, Ogawa M, Kapperud G. Potential sources of sporadic human infection with Yersinia enterocolitica serovar O:8 in Aomori Prefecture, Japan. J Clin Microbiol. 1995;33:1253–1257. doi: 10.1128/jcm.33.5.1253-1257.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heesemann J, Algermissen B, Laufs R. Genetically manipulated virulence of Yersinia enterocolitica. Infect Immun. 1984;46:105–110. doi: 10.1128/iai.46.1.105-110.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Heesemann J, Gross U, Schmidt N, Laufs R. Immunochemical analysis of plasmid-encoded proteins released by enteropathogenic Yersinia sp. grown in calcium-deficient media. Infect Immun. 1986;54:561–567. doi: 10.1128/iai.54.2.561-567.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Heesemann J, Laufs R. Construction of a mobilizable Yersinia enterocolitica virulence plasmid. J Bacteriol. 1983;155:761–767. doi: 10.1128/jb.155.2.761-767.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jacobi C A, Roggenkamp A, Rakin A, Zumbihl R, Leitritz L, Heesemann J. In vitro and in vivo expression studies of yopE from Yersinia enterocolitica using the gfp reporter gene. Mol Microbiol. 1998;30:865–882. doi: 10.1046/j.1365-2958.1998.01128.x. [DOI] [PubMed] [Google Scholar]
- 21.Kitamura M. NF-κB-mediated self defense of macrophages faced with bacteria. Eur J Immunol. 1999;29:1647–1655. doi: 10.1002/(SICI)1521-4141(199905)29:05<1647::AID-IMMU1647>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 22.Linder S, Nelson D, Weiss M, Aepfelbacher M. Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages. Proc Natl Acad Sci USA. 1999;96:9648–9653. doi: 10.1073/pnas.96.17.9648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mills S D, Boland A, Sory M P, van-der-Smissen P, Kerbourch C, Finlay B B, Cornelis G R. Yersinia enterocolitica induces apoptosis in macrophages by a process requiring functional type III secretion and translocation mechanisms and involving YopP, presumably acting as an effector protein. Proc Natl Acad Sci USA. 1997;94:12638–12643. doi: 10.1073/pnas.94.23.12638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Monack D M, Mecsas J, Bouley D, Falkow S. Yersinia-induced apoptosis in vivo aids in the establishment of a systemic infection of mice. J Exp Med. 1998;188:2127–2137. doi: 10.1084/jem.188.11.2127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Monack D M, Mecsas J, Ghori N, Falkow S. Yersinia signals macrophages to undergo apoptosis and YopJ is necessary for this cell death. Proc Natl Acad Sci USA. 1997;94:10385–10390. doi: 10.1073/pnas.94.19.10385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.O'Connell M A, Bennett B L, Mercurio F, Manning A M, Mackman N. Role of IKK1 and IKK2 in lipopolysaccharide signaling in human monocytic cells. J Biol Chem. 1998;273:30410–30414. doi: 10.1074/jbc.273.46.30410. [DOI] [PubMed] [Google Scholar]
- 27.Orth K, Palmer L E, Bao Z Q, Stewart S, Rudolph A E, Bliska J B, Dixon J E. Inhibition of the mitogen-activated protein kinase kinase superfamily by a Yersinia effector. Science. 1999;285:1920–1923. doi: 10.1126/science.285.5435.1920. [DOI] [PubMed] [Google Scholar]
- 28.Orth K, Xu Z, Mudgett M B, Bao Z Q, Palmer L E, Bliska J B, Mangel W F, Staskawicz B, Dixon J E. Disruption of signaling by Yersinia effector YopJ, a ubiquitin-like protein protease. Science. 2000;290:1594–1597. doi: 10.1126/science.290.5496.1594. [DOI] [PubMed] [Google Scholar]
- 29.Palmer L E, Hobbie S, Galan J E, Bliska J B. YopJ of Yersinia pseudotuberculosis is required for the inhibition of macrophage TNFα production and downregulation of the MAP kinases p38 and JNK. Mol Microbiol. 1998;27:953–965. doi: 10.1046/j.1365-2958.1998.00740.x. [DOI] [PubMed] [Google Scholar]
- 30.Perkins N D. The Rel/NF-κB family: friends and foe. Trends Biochem Sci. 2000;25:434–440. doi: 10.1016/s0968-0004(00)01617-0. [DOI] [PubMed] [Google Scholar]
- 31.Portnoy D A, Moseley S L, Falkow S. Characterization of plasmids and plasmid-associated determinants of Yersinia enterocolitica pathogenesis. Infect Immun. 1981;31:775–782. doi: 10.1128/iai.31.2.775-782.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ruckdeschel K, Harb S, Roggenkamp A, Hornef M, Zumbihl R, Köhler S, Heesemann J, Rouot B. Yersinia enterocolitica impairs activation of transcription factor NF-κB: involvement in the induction of programmed cell death and in the suppression of the macrophage TNFα production. J Exp Med. 1998;187:1069–1079. doi: 10.1084/jem.187.7.1069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ruckdeschel K, Machold J, Roggenkamp A, Schubert S, Pierre J, Zumbihl R, Liautard J P, Heesemann J, Rouot B. Yersinia enterocolitica promotes deactivation of macrophage mitogen-activated protein kinases extracellular signal-regulated kinase-1/2, p38, and c-Jun NH2-terminal kinase: correlation with its inhibitory effect on TNFα production. J Biol Chem. 1997;272:15920–15927. doi: 10.1074/jbc.272.25.15920. [DOI] [PubMed] [Google Scholar]
- 34.Ruckdeschel K, Mannel O, Richter K, Jacobi C A, Trülzsch K, Rouot B, Heesemann J. Yersinia outer protein P of Yersinia enterocolitica simultaneously blocks the nuclear factor-kappaB pathway and exploits lipopolysaccharide signaling to trigger apoptosis in macrophages. J Immunol. 2001;166:1823–1831. doi: 10.4049/jimmunol.166.3.1823. [DOI] [PubMed] [Google Scholar]
- 35.Ruckdeschel K, Roggenkamp A, Lafont V, Mangeat P, Heesemann J, Rouot B. Interaction of Yersinia enterocolitica with macrophages leads to macrophage cell death through apoptosis. Infect Immun. 1997;65:4813–4821. doi: 10.1128/iai.65.11.4813-4821.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schesser K, Spiik A K, Dukuzumuremyi J M, Neurath M F, Pettersson S, Wolf-Watz H. The yopJ locus is required for Yersinia-mediated inhibition of NF-κB activation and cytokine expression: YopJ contains a eukaryotic SH2-like domain that is essential for its repressive activity. Mol Microbiol. 1998;28:1067–1079. doi: 10.1046/j.1365-2958.1998.00851.x. [DOI] [PubMed] [Google Scholar]
- 37.Snellings N J, Popek M, Lindler L E. Complete DNA sequence of Yersinia enterocolitica serotype 0:8 low-calcium-response plasmid reveals a new virulence plasmid-associated replicon. Infect Immun. 2001;69:4627–4638. doi: 10.1128/IAI.69.7.4627-4638.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Straley S C, Bowmer W S. Virulence genes regulated at the transcriptional level by Ca2+ in Yersinia pestis include structural genes for outer membrane proteins. Infect Immun. 1986;51:445–454. doi: 10.1128/iai.51.2.445-454.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]