Abstract
Agriculture is particularly vulnerable to climate change. To cope with the risks posed by climate-related stressors to agricultural production, global population growth, and changes in food preferences, it is imperative to develop new climate-smart crop varieties with increased yield and environmental resilience. Molecular genetics and genomic analyses have revealed that allelic variations in genes involved in phytohormone-mediated growth regulation have greatly improved productivity in major crops. Plant science has remarkably advanced our understanding of the molecular basis of various phytohormone-mediated events in plant life. These findings provide essential information for improving the productivity of crops growing in changing climates. In this review, we highlight the recent advances in plant hormonomics (multiple phytohormone profiling) and discuss its application to crop improvement. We present plant hormonomics as a key tool for deep physiological phenotyping, focusing on representative plant growth regulators associated with the improvement of crop productivity. Specifically, we review advanced methodologies in plant hormonomics, highlighting mass spectrometry- and nanosensor-based plant hormone profiling techniques. We also discuss the applications of plant hormonomics in crop improvement through breeding and agricultural management practices.
Keywords: Biosensor, Biostimulant, Breeding, Mass spectrometry, Phytohormone
Introduction
Agriculture is particularly vulnerable to climate change. According to a recent Intergovernmental Panel on Climate Change report, climate change has already negatively affected agricultural food production through climate-related factors such as rising temperatures, changing precipitation patterns and an increase in the frequency and severity of extreme weather events (Van Meijl et al. 2018, Zandalinas et al. 2021). Moreover, non-climate stressors such as global population growth, rapid urbanization and the associated lifestyle changes have led to changes in food consumption patterns, placing further pressure on sustainable food security (Seto and Ramankutty 2016, Prosekov and Ivanova 2018, FAO 2019, FAO, IFAD, UNICEF, WFP and WHO 2020). To cope with the combined risks of climate- and non-climate-related stressors to agricultural production, there is a need to develop new climate-smart crop varieties with increased environmental resilience and improved yield.
Plants endogenously produce small organic molecules—known as phytohormones or plant hormones—that function as signaling molecules and regulate almost all known biological phenomena in plants, including growth and developmental processes and responses to various biotic and abiotic environmental stresses. To the best of our knowledge, there are nine groups of plant hormones: auxins (AUXs), cytokinins (CKs), gibberellins (GAs), brassinosteroids (BRs), abscisic acid (ABA), jasmonates (JAs), salicylic acid (SA), ethylene (ET) and strigolactones (SLs) (Santner et al. 2009, Waters et al. 2017). The physiological functions of these plant hormones have been extensively studied. In short, AUX is primarily implicated in cell differentiation and elongation (Woodward and Bartel 2005), CK regulates cell differentiation and the progression of the cell division cycle (Kieber and Schaller 2018), GA is involved in cell expansion and differentiation (Hedden and Sponsel 2015), BR is required for cell and tissue growth (Clouse and Sasse 1998), SL modulates developmental processes (Waters et al. 2017), ABA is implicated in bud and seed dormancy and abiotic stress responses (Cutler et al. 2010), JA plays a pivotal role in injury response and abiotic stress responses (Schilmiller and Howe 2005, Dar et al. 2015), ET is required for low-oxygen stress response (Bakshi et al. 2015) and SA is involved in plant defense responses to pathogens (Vlot et al. 2009). Although these plant hormones share certain physiological functions, they generally have distinct roles (Nemhauser et al. 2006). Moreover, these hormones constitute a complex response network (see below) as they exhibit both positive and negative interactions. Plants make use of and are dependent on these hormones to orchestrate their biological activities in order to adapt to environmental fluctuations.
Following remarkable advances in the molecular genetics and genomics of crop plants, studies have highlighted the role of plant hormones in various agronomically important traits during crop breeding. A prominent example is the emergence of semi-dwarf varieties of major crops due to defective GA biosynthesis, which contributed to the Green Revolution in the 1960s. Over the 30 years following the Green Revolution, the major genes responsible for reduced plant height were identified in wheat and rice (Spielmeyer et al. 2002). In ‘usu’ varieties of barley, a mutation in the gene encoding the BR receptor can produce a semi-dwarf mutant. This occurs independently of GA-related mutations and results in a BR-insensitive phenotype (Chono et al. 2003), which is often observed in crop landraces in East Asian countries (Saisho et al. 2004). In rice, the genes responsible for flooding tolerance—the Snorkel genes and Submergence-1A—encode ET-responsive factor-type transcription factors that regulate the escape strategy for deep-water tolerance and the quiescence strategy for flash flooding tolerance by promoting and inhibiting the GA-mediated internode elongation, respectively (Singh et al. 2017). These examples demonstrate that allelic variations in genes that participate in phytohormone-mediated growth and regulation can contribute to improved productivity in major crops.
Molecular genetics has advanced our understanding of the molecular basis of various phytohormone-mediated life events in plants. Numerous studies have illustrated the cross talk networks among multiple phytohormones in various circumstances, including growth, development and biotic and abiotic stress responses in plants (Santner and Estelle 2009, Jaillais and Chory 2010, Peleg and Blumwald 2011, Ross et al. 2011, Vanstraelen and Benková 2012). Recent studies have also demonstrated that many plant hormones participate in the systemic regulation of responses to ambient conditions, which maximizes plant fitness through long-distance transport and signaling networks across plant organs (Ruffel 2018). In addition to classical phytohormones, various small peptides that function as signaling molecules (peptide hormones) have received increased attention, as they are involved in the systemic regulation of stress adaptation and development through long-distance signaling (Andrews and Rothnagel 2014, Oh et al. 2018, J. S. Kim et al. 2021, M.-J. Kim et al., 2021). Studies on plant–microbe interactions have also revealed that several phytohormones produced by plants and/or plant-associated microbes are vital components for plant–microbe communication; these findings may help develop new strategies for regulating plant growth by manipulating plant microbiota (Gao et al. 2019, Fitzpatrick et al. 2020, Eichmann et al. 2021). Therefore, phytohormones could function as a useful set of biomarkers indicating the physiological state of crops, which would facilitate crop breeding and management and help to improve the resilience of agricultural production.
In this mini review, we highlight the recent advances in plant hormonomics and their applications for crop improvement. Following our brief review of recent advances in plant hormone research, we showcase advanced methodologies and emerging approaches in plant hormonomics, including mass spectrometry (MS)-based hormone analysis and bio-nanosensor-based plant hormone profiling. Moreover, we discuss plant hormonomics and its application in crop improvement through selective breeding and agricultural management practices.
Plant Hormone Levels Are Associated with the Developmental and Physiological Status of Plants
Plants systematically regulate the levels of plant hormones through de novo synthesis and degradation or inactivation, thus ensuring the optimal functioning of phytohormones according to the internal or external environment (see below). Although plant hormones seem to exist ubiquitously in plant bodies, their concentrations are spatiotemporally regulated in various tissues and differentiated cell types. For example, plant hormones involved in developmental processes—such as indole acetic acid (IAA), CK, GA and BR—are localized in specific tissues (Novák et al. 2017). Moreover, all plant hormones have shown to be implicated in either the positive or the negative regulation of photosynthesis, which is crucial for plant growth (Gururani et al. 2015). Therefore, the quantification of plant hormone levels across tissues and developmental stages can offer insights into the functions of tissues during the various developmental stages.
The biosynthesis and degradation pathways of plant hormones are largely understood, and the genes responsible for the critical steps of these metabolic and catabolic pathways have been described (Vlot et al. 2009, Wasternack and Song 2017, Mostofa et al. 2018, Wei and Li 2020, Casanova-Sáez et al. 2021, Pattyn et al. 2021, Vukašinović et al. 2021). For example, CK levels are strictly regulated during development. CK refers to a group of chemicals, including isopentenyl adenine (iP), trans-zeatin (tZ), cis-zeatin and dihydrozeatin. In plants, these CKs are produced from adenosine monophosphate or transfer RNA and differ only in the side-chain structure at the N6 position of the adenine ring (Kamada-Nobusada and Sakakibara 2009, Kieber and Schaller 2018). A cytochrome P450 monooxygenase, CYP735A, which converts iP to tZ, is required for shoot growth in Arabidopsis, indicating a strong link between structural conversion and developmental regulation (Kiba et al. 2013). The genes for LONELYGUY (LOG)—a phosphoribohydrolase that catalyzes the final step of CK synthesis (Kurakawa et al. 2007)—are strictly regulated during development in Arabidopsis (Kuroha et al. 2009). CKs are reversibly inactivated through glucosylation (Bajguz and Piotrowska 2009) or irreversibly inactivated by cytokinin oxidase (CKX) (Werner et al. 2006). In Arabidopsis, seven CKX genes are controlled by developmental cues and environmental stimuli, supporting the notion that hormone levels can be useful biomarkers for the physiological state of plants (Schmülling et al. 2003, Werner et al. 2003, Nishiyama et al. 2011).
As aforementioned, ABA, JA, ET and SA are all involved in environmental stress responses in plants (Bailey-Serres et al. 2009, Vlot et al. 2009, Cutler et al. 2010, Dar et al. 2015, Kazan 2015). For example, ABA (an isoprenoid) plays a vital role in developmental processes, such as seed development and dormancy, as well as adaptive responses to abiotic environmental stresses, such as dehydration, cold and salinity. The bottleneck of the ABA synthetic pathway is the cleavage of violaxanthin and neoxanthin to xanthoxin by nine-cis-epoxycartenoid-dioxygenese (NCED) (Nambara and Marion-Poll 2005). Among the five NCED genes implicated in ABA synthesis in Arabidopsis, NCED3 responds to abiotic stresses and plays a pivotal role in ABA biosynthesis under conditions of abiotic stress (Nambara and Marion-Poll 2005). The CYP707A-catalyzed hydroxylation of ABA is a critical step in the irreversible inactivation of ABA. The balance between NCED and CYP707A activities is important; for example, ABA levels under drought stress were maintained through the upregulation of both NCED3 and CYP707A (Kushiro et al. 2004). Upon watering, NCED3 was downregulated, whereas the CYP707A genes were further activated to reduce ABA levels to basal levels.
In addition to the regulation of biosynthesis and degradation pathways, plants have developed sophisticated systems to control the functions of plant hormones as signaling molecules. The actions of plant hormones are regulated through their translocation by specific transporters (Park et al. 2017). Transporters for IAA, a major AUX, have been studied extensively, and are known to contribute to the polar accumulation of IAA (Adamowski and Friml 2015, Geisler et al. 2017, Park et al. 2017). Recent studies have shown that other plant hormones are spatially controlled by specific transporters. Plants have several different types of CK transporters, whereby some regulate CK traffic at the intracellular levels while others are implicated in intercellular or long-distant translocation (Gillissen et al. 2000, Bernard et al. 2011, Ko et al. 2014, K. Zhang et al. 2014, Kang et al. 2017, Liu et al. 2019). ABA is transported across cells or tissues by several transporters to regulate physiological responses such as stomata movement, stress responses of distal organs and seed germination (Kang et al. 2010, Kuromori et al. 2010, Kanno et al. 2012, H. Zhang et al. 2014, Kang et al. 2015, Kuromori et al. 2018). The ET precursor, 1-aminocyclopropane-1-carboxylic acid (ACC), is transported by an amino acid transporter (Shin et al. 2015, Choi et al. 2019). GAs are transported across the cell membrane by a nitrate transporter 1/peptide transporter family (NPF) and by sugar transporter-like transporters to regulate the spatial distribution of these hormones (Saito et al. 2015, Kanno et al. 2016, Tal et al. 2016). The tissue or intracellular distribution of JA is also regulated by transporters of the JA precursors, 12-oxophytodienoic acid and JA/JA-Ile (Footitt et al. 2007, Li et al. 2017). An ATP Binding Casette (ABC) transporter has been shown to regulate the tissue specific distribution and the exudation of this hormone to rhizophores (Kretzschmar et al. 2012, Sasse et al. 2015). Collectively, plant hormones are actively, spatiotemporally regulated to orchestrate the developmental processes and physiological states of the whole plant.
Plant hormones can be stored in inactive forms after conjugation with various metabolites, such as sugars and amino acids. When required, the hormones are detached from the conjugated form and activated. For example, ABA–glucose conjugates are stored in vacuoles (Dietz et al. 2000, Lee et al. 2006, Xu et al. 2012). It should be noted that in the case of JA signaling, JA-Ile is an endogenous signaling molecule that is recognized by the JA receptor complex (Fonseca et al. 2009). Collectively, hormonomics—an approach in which multiple spatiotemporally regulated plant hormones are monitored simultaneously—offers a great opportunity to obtain crucial data on the physiological and developmental conditions of plant cells, tissues and organs. Moreover, this approach can help identify useful biomarkers that can monitor the developmental and physiological states of plants under fluctuating conditions.
Importance of Simultaneous Profiling of Multiple Plant Hormones
As aforementioned, each phytohormone has specific physiological roles, established from numerous physiological and molecular studies. However, it is now known that individual hormones cannot manage most suggested biological processes. There is intense and intricate cross talk among plant hormones (Santner and Estelle 2009, Jaillais and Chory 2010, Peleg and Blumwald 2011, Ross et al. 2011), which regulates biological phenomena in plants. Plant hormones can act synergistically and antagonistically in a unidirectional or bidirectional manner. Moreover, they affect biosynthesis, signaling and output processes at multiple levels, including that of gene expression (Altmann et al. 2020, Aerts et al. 2021). A prominent and extensively investigated cross talk comprises the relationship between plant hormones involved in defense responses. Plants typically use SA and JA/ET to combat biotrophic and necrotrophic pathogens, respectively (Glazebrook 2005, Pieterse et al. 2009, Vlot et al. 2009). Due to their mutually antagonistic interactions, SA and JA/ET signaling pathways exhibit a negative regulatory relationship (Pieterse et al. 2012, Thaler et al. 2012, Caarls et al. 2015). In Arabidopsis, SA treatment reduces the JA-dependent expression of the plant defensin encoding gene, PDF1.2, upon infection with necrotrophic pathogens (Koornneef et al. 2008). In contrast, the phytotoxin coronatine—a JA analog produced by Pseudomonas syringae—inhibits SA synthesis through the JA signaling pathway and JA-dependent gene expression (Zheng et al. 2012). Moreover, these hormones interact with other plant hormones such as IAA, CK and ABA, suggesting that SA and JA also affect growth and abiotic stress response (Bari and Jones 2008, Robert-Seilaniantz et al. 2011, Shigenaga and Argueso 2016, Verma et al. 2016, Cortleven et al. 2019, Yang et al. 2019).
Presumably, the evolution of this broad spectrum of hormonal cross talk has allowed plants to increase their environmental fitness by maintaining a balanced trade-off between energetically expensive processes, such as growth and defense responses (Pieterse et al. 2012, Denance et al. 2013, Lozano-Durán and Zipfel 2015, Vos et al. 2015, Berens et al. 2017, Shigenaga et al. 2017, van Butselaar and Van den Ackerveken 2020). Consistent with this notion, there is evidence for hormonal cross talk among various biological activities in plants, including growth (Depuydt and Hardtke 2011, Su et al. 2011, El-Showk et al. 2013) and abiotic stress responses (Fahad et al. 2015, Bücker-Neto et al. 2017, Yu et al. 2020). Moreover, hormonal cross talk may help optimize competing biological functions mediated by antagonistic hormones in plants. As such, the simultaneous profiling of plant hormones—that is, hormonome analysis—is important for elucidating hormonal cross talk in plants.
Plant Hormones in Plant–Microbe Communications
The rhizosphere microbiota, including endophytes, strongly affect the growth and physiological state of the host plant, as exemplified by the relationship between legumes and N-fixing Rhizobium (Hayat et al. 2010, Berendsen et al. 2012, Santoyo et al. 2016, Trivedi et al. 2020, Vries et al. 2020). Beneficial rhizosphere microorganisms enhance plant growth by supplying nutrition and increasing tolerance to abiotic stress. Moreover, these microorganisms strengthen the immune response of plants by activating or priming the defense response of the host plant or by inhibiting the growth of pathogens in the rhizosphere. Metagenomic analyses have revealed that a flavobacterium found in the rhizosphere of a disease-resistant tomato accession conferred enhanced resistance to susceptible accessions (Kwak et al. 2018). The interaction between plants and the rhizosphere microbiota largely depends on the organic compounds produced by and exchanged between them. Microorganisms in the rhizosphere utilize organic chemicals found in plant exudates, such as sugars, amino acids and siderophores. By controlling root metabolites, plants may actively regulate the microbial composition of the rhizosphere so as to retain beneficial microorganisms, such as plant growth-promoting rhizobacteria (PGPR; Gao et al. 2019, Fitzpatrick et al. 2020, Lyu et al. 2021). Plant hormones play a pivotal role in this process (Tsukanova et al. 2017, Rosier et al. 2018, Eichmann et al. 2021). For instance, SLs released from roots induce hyphal branching and plant colonization in arbuscular mycorrhizal fungi, which enhance nutrient uptake by host plants (Akiyama et al. 2010, Lanfranco et al. 2018). Certain PGPR produce plant hormones and/or modulate hormone production in plants (Yang et al. 2009, Dodd et al. 2010, Chanclud and Morel 2016). It is presumed that the upregulation of endogenous phytohormone levels enhances plant growth ability or improves the physiological state of plants under conditions of stress. IAA produced by fungi requires a plant host (Chanclud and Morel 2016). Several rhizobacteria can synthesize and release ABA into the rhizosphere (Tsukanova et al. 2017, Eichmann et al. 2021). When the rhizobacterium, Azospirillum brasilense, was inoculated in the roots of Arabidopsis, increases in endogenous ABA levels and drought tolerance of the host plant were exhibited (Cohen et al. 2015). PGPR produce ACC deaminase, which catabolizes the ET precursor ACC and improves the abiotic stress tolerance of plants (Gamalero and Glick 2015). Therefore, the examination and/or manipulation of plant hormones in the rhizosphere could be an effective approach for improving plant growth (Zhang et al. 2015, Dessaux et al. 2016, Lu et al. 2021).
Hormone-Like Peptides
Over the preceding decade, small peptides secreted into extracellular spaces have been recognized as crucial signaling molecules among developmental regulation and stress responses in plants. Many hormone-like peptides participate in developmental processes, such as the regulation of cell differentiation in the shoot or root meristem (Katsir et al. 2011, Oh et al. 2018, Jeon et al. 2021) and stomata (Torii 2012). Remarkably, the reproductive process involves several signaling peptides that establish cell-to-cell communications (Kim et al. 2021b). A most famous example of which can be seen in Arabidopsis through CLAVATA3 (CLV3), which participates in the maintenance of the shoot apical meristem (Clark et al. 1995). CLV3 is a 96-aa peptide comprising a potential signal peptide used for secretions into the extracellular space (Fletcher et al. 1999). CLV3 is perceived by the receptor complex composed of CLAVATA1 and CLAVATA2, repressing WUSHEL encoding, a mobile homeodomain transcription factor that positively regulates CLV3, constituting a feedback loop that maintains the shoot apical meristem (Brand et al. 2000, Schoof et al. 2000). Moreover, hormone-like peptides have also been implicated in the systemic responses of plants to environmental conditions, which ensue nutrient deficiencies (Ohkubo et al. 2017, Gautrat et al. 2021), to drought stress (Takahashi et al. 2018, Takahashi and Shinozaki 2019), to salinity stress (Nakaminami et al. 2018) and to pathogen attacks (Huffaker 2015). The CLV3/EMBRYO-SURROUNDING REGION-RELATED25 peptide produced in Arabidopsis roots under drought stress conditions transmits the dehydration signal to shoots and activates the ABA biosynthetic genes via the cellular signaling pathway, which utilizes a minimum number of meristem receptors (Takahashi et al. 2018). Comprehensive peptide and genome analyses of putative signaling peptides or their genes have revealed several undescribed secretory hormone-like peptides in plants (Hanada et al. 2013, Hsu and Benfey 2018, S. Wang et al. 2020). In addition to these putative secretory peptide hormones, many novel short ORF genes have also been identified. Among these, several genes have been shown to have vital physiological functions (Narita et al. 2004, Hirayama et al. 2018, Grillet and Schmidt 2019). Further studies on these putative signaling peptides should improve our understanding of plant behavior at the molecular level.
Field Hormonomics for Monitoring Physiological Dynamics in Plants under Fluctuating Conditions
Plant hormones are known to play pivotal roles in the regulation of growth and environmental stress responses. This suggests that investigating the modulation of plant hormone levels or functions would be an efficient approach for improving the growth and productivity of crops (Cramer et al. 2011, Peleg and Blumwald 2011, Atkinson and Urwin 2012, Wilkinson et al. 2012, Wani et al. 2016, Asami and Nakagawa 2018, Vaidya et al. 2019, Chesterfield et al. 2020). The plant hormone-based intervention could potentially improve crop performance and resilience even under unfavorable field conditions. However, its effectiveness under field conditions has not yet been investigated. Indeed, plant hormones interact in a complex manner; therefore, changes in the functions of a plant hormone may positively or negatively affect the functions of other plant hormones. Long-term observations are needed to determine how plant hormone levels fluctuate and how these changes may affect other hormones in crops under field conditions, which may help to maximize the impact of these interventions (Mochida et al. 2015). A life-course hormonomic analysis of field-grown barley accessions revealed significant changes in endogenous hormone levels (Hirayama et al. 2020). Interestingly, the levels of plant hormones that have been implicated in developmental regulation—such as IAA and CKs—fluctuate vigorously, suggesting that environmental factors have a significant impact on crop growth via modulating phytohormone levels. Long-term phenomic analyses are a powerful tool for obtaining data that can be utilized to elucidate the interactions between genetic and environmental factors, which is necessary for crop improvement and design. Moreover, such analyses can be used to investigate the effects of intervention techniques based on the knowledge accumulated through interdisciplinary efforts in plant science.
Advances in Plant Hormonomic Methodology
Several methods have been developed to accurately measure plant hormones, which exhibit diverse structures and endogenous levels. During the initial period of plant hormone research, plant hormones were concentrated using various chromatography techniques and were detected or measured by various physical, chemical or biological detection methods such as ultraviolet light, fluorescence or immune assays (Weiler 1984, Hedden 1993, Dobrev et al. 2005, Zhao et al. 2006). However, these techniques are limited in their accuracy and specificity. MS has high detection performance in the analysis of any type of chemical compound (Glassbrook et al. 2000). The utilization of stable isotope-labeled compounds as internal standards for experimental steps including hormone extraction and ionization in the MS analysis can provide a highly accurate quantification of endogenous plant hormones in MS-based methods. Therefore, MS-based measurements have been implemented in recent decades to detect various plant hormones, as reviewed by Novák et al. (2017).
Gas chromatography equipped with tandem MS (GC–MS/MS) shows significantly improved performance in the identification and quantification of volatile plant hormones such as JA, SA and ET, among others (Engelberth et al. 2003, Schmelz et al. 2003). However, the GC–MS/MS analysis requires chemical derivatization. High-performance liquid chromatography (LC) equipped with tandem MS (LC–MS/MS) also provides highly sensitive, specific and accurate quantitative detection of plant hormones. At present, all plant hormones—except gaseous ET—can be measured by LC–MS/MS (Kojima et al. 2009, Pan et al. 2010). In addition, more than 100 phytohormone-related compounds, including phytohormone derivatives, such as the ET precursor, ACC, can also be detected using this technique (Šimura et al. 2018). The advanced methodologies of plant hormonomics with LC–MS have been summarized in recent reviews (Novák et al. 2017, L. Wang et al. 2020). As a result of continuous improvements in the experimental methodology and the technology utilized for these techniques, plant samples weighing less than 10 mg are sufficient for the analysis of most plant hormones (Cao et al. 2020). Such enhanced techniques allow for the life-course hormonomic analysis of various crop accessions under field conditions. Tiny tissue samples obtained by the laser micro-dissection of cryosections can also be analyzed by LC–MS, which increases the spatial resolution of hormonomics (Yamada et al. 2021). Moreover, the levels of ABA and JA in a single cell of Vicia faba leaves can be measured using these techniques (Shimizu et al. 2015). Furthermore, nanoparticle-assisted laser desorption/ionization MS now enables the determination of the distribution and the concentration of multiple hormones simultaneously from sections. Shiono and Taira (2020) recently reported on the successful detection of IAA, BR, ABA, tZ, iP, JA, SA and ACC in rice root sections using the latter technique.
Real-Time Monitoring of Plant Hormones with Biosensors
Various hormone sensors developed using state-of-the-art technologies can be used to monitor endogenous plant hormones in a non-invasive manner (Okumoto et al. 2012, Novák et al. 2017, Isoda et al. 2021). Although MS-based hormonomics enables highly sensitive and the accurate quantification of plant hormones, this approach is destructive, labor intensive and time consuming. Therefore, hormone sensors could provide new opportunities for the real-time monitoring of the physiological states of plants under fluctuating conditions. For example, real-time monitoring sensors for ABA would allow for observing physiological conditions related to water in plants more precisely and for more effective water management. Real-time monitoring of JA, SA or ET would provide a great opportunity to protect plants from infectious diseases and herbivory.
One approach for implementing biosensors is to construct genetically engineered biosensors based on native hormone receptors using Förster resonance energy transfer (FRET) technology, which allows for real-time quantitative analysis by measuring fluorescence. All the genes encoding plant hormone receptors have been identified, and their 3D structures at the atomic level are currently available. FRET-based biosensors using native hormone receptors for ABA and GA have been used to detect hormone levels in vivo (Jones et al. 2014, Waadt et al. 2014, 2020, Rizza et al. 2017). It is expected that direct biosensors can be developed using similar sensor design strategies for other plant hormones. Biosensors for IAA have been developed recently using bacterial proteins. Based on the tertiary structure, Herud-Sikimić et al. (2021) modified the tryptophan sensor of E. coli to recognize IAA. Using this IAA sensor, the latter authors successfully visualized the dynamics of IAA at the cellular level in Arabidopsis roots. Additionally, there are several indirect biosensing systems in which hormone levels in the cell or tissue are converted to other detectable biomarkers, such as protein or gene expression levels. Such indirect biosensors have been reported for most plant hormones (Novák et al. 2017, Isoda et al. 2021). Thus, capturing the dynamics of plant hormones in real time and continuously using these direct or indirect biosensors should improve our understanding of the complex functions of plant hormones. However, using these genetically engineered biosensors requires the development of transgenic plants containing the genes for these biosensors or biosensor systems. In addition, the stable introduction of foreign genetic materials is difficult in some plant species, and growing transgenic plants in the field is either restricted or regulated in most countries.
Sensors using nanomaterials, such as carbon nanotubes (CNTs), for monitoring biomolecules have recently received increased attention (Kwak et al. 2017, Giraldo et al. 2019, Anzar et al. 2020, Lew et al. 2020b). Nanomaterials emit fluorescence in the near-infrared region, where the autofluorescence from chlorophyll does not interfere. The fluorescence-emitting property of CNTs is affected by physical and chemical conditions (Barone et al. 2005). Thus, covalent or non-covalent modification of the CNT surface confers sensor properties by changing the sensitivity and selectivity of the ligand (Kruss et al. 2013, Yang et al. 2015). CNTs occur at a size of less than 100 nm and can be easily introduced into plant tissues irrespective of species. CNT nano-biosensors, non-covalently modified with DNA, have been shown to detect H2O2 in injured leaves (Wu et al. 2020, Lew et al. 2020a). Using a similar approach, endogenous glucose levels have been detected using a nanoparticle-based sensor (Li et al. 2018). Recently, an artificial metalloenzyme (ArM) biosensor for ET has been developed (Vong et al., 2019). In the presence of ET, the ArM biosensor catalyzes a compound intrinsically captured in ArM and produces fluorescence. Using this ET biosensor, the authors successfully detected ET production during the ripening of various fruits, such as apples, kiwifruit, Asian pears and grapes, and upon infection of pathogenic bacteria (Vong et al., 2019). This study offers a sound example for the usage of biosensors for plant hormones.
Although these nano- or ArM-biosensor technologies have opened new avenues for plant science, the detection of fluorescence by these sensors is challenging under field conditions. Thus, the development of other optical devices is required for the practical application of these sensors in agriculture. As an alternative, wearable devices have been developed using these nanomaterial sensors. Nanosensors equipped with fluorescence-detecting devices on flexible sheets can detect volatile organic compounds or moisture levels when mounted on plant organs (Lee et al. 2014, Lu et al. 2020, Li et al. 2021). Therefore, the combination of various advanced technologies in nanomaterial science and engineering should empower the development of nano-biosensors for real-time monitoring of the dynamics of key biochemical substances, including plant hormones.
Hormonomics for Crop Improvement
Twenty years after the first genome sequencing of a higher order plant (Arabidopsis), the genome data of several other crops have become available through revolutionary advances in DNA sequencing technologies in recent decades (Purugganan and Jackson 2021). Datasets of genome-scale polymorphism of diverse varieties, combined with datasets on phenotypes, have allowed researchers to explore a range of factors associated with agronomically important traits in crops. Key genetic factors associated with the agronomically important traits of crops under field conditions have been addressed using the quantitative trait locus analysis and genome-wide association studies. These findings may help to develop new crop varieties with higher yields and enhanced stress tolerance through improved breeding methods (such as genomic selection and speed breeding) and advanced genome editing-based strategies (including base editing and prime editing) (Mickelbart et al. 2015, Bevan et al. 2017, Varshney et al. 2020). Thus, genomic analyses can help to accelerate crop improvement.
Plant hormonomics is expected to facilitate the genetic manipulation of targeted genes and alleles in crop breeding from two major aspects. Plant hormonomics may be a fine addition to multi-omics approaches that facilitate gene discovery and allele mining by mapping intermediate phenotypes (Varshney et al. 2020) and thus allow us to dissect the complex terminal phenotypes of agronomic traits. In this regard, hormonome-based phenotypes may provide useful information to illustrate the diversity of developmental and physiological states across germplasm collections. However, precise genetic manipulation requires an in-depth understanding of traits. To design crop varieties that can cope with changing climates, it is essential to decode the genotype–environment interaction (G × E) in order to reverse engineer the agronomic traits of crops grown under fluctuating conditions. In this regard, hormonomics would play a key role in directly illustrating the profiles of signaling molecules that mediate physiolog-ical responses to environmental stimuli in plants. Combined with hormonome-based phenotype mapping, the causal loci of environmental fitness can provide the genetic factors in genotype-to-phenotype modeling in crops. Thus, these analyses can facilitate the prioritization of target genes that can be engineered to improve the environmental fitness of plants (Fig. 1).
Fig. 1.
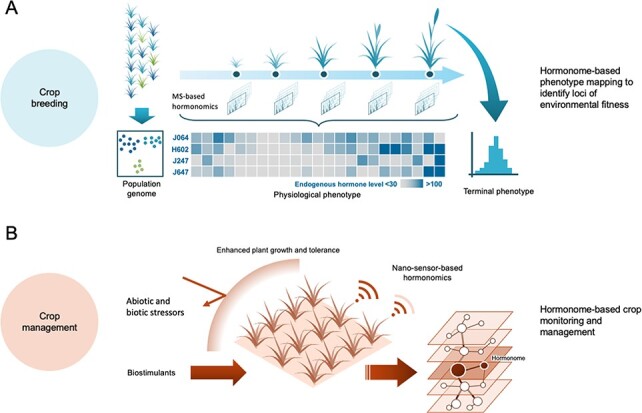
Crop breeding and management through hormonomics-based physiological phenotyping. In crop breeding, high-throughput analytical systems for plant hormonomics offer the simultaneous quantification of molecular species of plant hormones, which facilitate the monitoring of spatial–temporal dynamics of hormone profiles throughout the crop lifecycle as well as their diversity among crop varieties (A). Since plant hormones regulate and coordinate various biological phenomena that influence agronomic traits in crops, highly sensitive and high-throughput methods for plant hormone profiling may enable us to monitor the physiological state of crops through the simultaneous quantification of endogenous plant hormone levels that fluctuate in response to internal and environmental conditions. Profiles of endogenous ABA levels measured in four barley accessions (J064, H602, J247, J647, Hirayama et al. 2020) are represented throughout their life course as a heatmap (blue high, gray low). The hormone profiles from mapping populations (e.g. diverse panel, nested association mapping population, recombinant inbreed lines and chromosome segment substitution lines) could represent a kind of chemical phenotype that can be used to explore their genotype–phenotype association. The exploration of phenotypic association between hormone levels and agronomic traits (even across different developmental stages) may enable us to identify useful hormone-based biomarkers that prevision terminal phenotypes. For crop management, plant hormone profiling provides convenient avenues to monitor the physiological state of crops in response to various stressors (B). In this context, plant hormone profiling may facilitate the monitoring of the efficiency of agricultural management, including the application of biostimulants with the aim to enhance crop growth and tolerance. Moreover, various studies that aim to develop non-invasive plant hormone monitoring such as nanomaterial-based hormone sensors may provide new avenues for real-time plant hormone profiling. Such nanosensor-based crop physiological state monitoring may serve to alleviate costs for crop management.
Hormonomics for Crop Management
The exogenous application of chemical and biological substances often enhances the resource use efficiency, stress tolerance and quality of plants, thus potentially improving crop productivity. Plant biostimulants are usually defined as any substance or microorganism that enhances plant growth and tolerance to various stresses (Rouphael and Colla 2020). Biostimulants are roughly categorized as humic and fulvic acids, protein hydrolysates, seaweed extracts, chitosan and other biopolymers, inorganic compounds and microorganisms such as fungi or bacteria (du Jardin 2015). Some biostimulants can activate or prime the defense response of plants to pathogenic agents (Martinez-Medina et al. 2016, Vargas-Hernandez et al. 2017, Shukla et al. 2019, Nephali et al. 2020, Ali et al. 2021). For example, the exogenous application of l-histidine inhibits wilt disease in tomato and Arabidopsis and upregulates genes related to ET biosynthesis and signaling (Seo et al. 2016). Biostimulants are also effective in enhancing tolerance to abiotic stresses such as high temperatures and drought (Van Oosten et al., 2017, Drobek et al., 2019, Kerchev et al., 2020, Nephali et al. 2020, Ali et al. 2021). For example, the exogenous application of acetic acid enhances drought tolerance in Arabidopsis and a wide range of crop species, also promoting JA signaling and histone acetylation (Kim et al. 2017). These examples indicate that some biostimulants enhance stress tolerance through the activation of specific hormone-signaling networks; therefore, hormonomics can allow researchers to examine the physiological function of biostimulants, which would help to improve their function and efficiency in crop management. However, apart from the accumulated evidence for the remarkable effects of a few stimulants in improving crop productivity, the molecular mechanisms of biostimulants that enhance growth and stress tolerance remain largely elusive. Biostimulants derived from seaweed or microorganisms often contain various plant hormones or hormone-like substances (du Jardin 2015); however, their physiological activities have not been fully elucidated. Therefore, exploring these bioactive components and elucidating their molecular functions are important prerequisites for formulating biostimulant-based agricultural materials and for optimizing their efficiency of utilization in various crops.
Deep Phenotyping of Plants and Crops
To build accurate crop models that can be used to design crops with high yield and stress tolerance and to understand the molecular basis of biostimulant action, it is necessary to collect data regarding the information relay from genes to phenotypes. Comprehensive genetic data—including reference genome data and data regarding genetic variation among accessions—can be used to deduce gene performance and are available for mathematical or computational usage. What about phenotypic data?
In biology, the term ‘phenotype’ refers to the observable traits of organisms (including morphology and physiology) at various levels of biological organization, including the cell, organ and whole body. In plant science, phenotyping has generally been used to describe mutants, environmental cues and disease symptoms that have considerable effects on plant growth or morphology. However, these phenotypic descriptions are occasionally inaccurate and lack direct links to biological phenomena. Owing to these limitations, these data are less valuable for the elucidation of connections between genes and phenotypes. In addition, the phenotypic data of organisms usually contain large amounts of noise (Raser and O’Shea 2005). Several researchers have reported that genetically identical individuals can have different phenotypes. Obtaining quantitative data on the phenotypes or traits of crops is important for understanding gene-to-phenotype linkages. However, to overcome the various problems associated with obtaining these data, it is important to establish a feasible and accurate methodology for assessing phenotypic traits.
Recent studies have made outstanding progress in analytical technologies, which allowed us to obtain various omics data for the elucidation of the biological system of any given organism, ranging from bacteria to humans. Transcriptome analyses using RNA sequencing or microarray techniques provide comprehensive data on the expression (usage) of each gene (Morozova et al. 2009, Tang et al. 2009). Further, metabolomic analysis, which involves LC–MS, GC, NMR, or capillary electrophoresis, provides comprehensive information regarding the metabolite composition and level of each metabolite (Zhang et al. 2012, Johnson et al. 2016). These data can reflect cellular biochemical activities such as respiration, photosynthesis and various other biological processes. All organisms require various ions or minerals for nutrition. Accordingly, ionomic analysis using inductively coupled plasma–MS can measure the levels of over 50 ions in biological samples and offer crucial information on the correlation of physiological status with ion usage and distribution (Salt et al. 2008). Proteomics allows for the comprehensive assessment of proteins produced in cells and of the levels of post-translational modifications and also provides valuable information on the cellular functions of proteins (Cox and Mann 2011, Larance and Lamond 2015). All these omics data provide valuable information on the developmental and physiological status of the cells, tissues and organs of plants. Moreover, omics data at various levels of biological organization offer important clues regarding the connections between genes and phenotypes. A combined analysis of these multi-dimensional biological data using advanced data science tools (aided by artificial intelligence and statistical models) can allow us to visualize a network comprising genes, metabolites, proteins and visible symptoms or morphological phenotypes. Such multi-dimensional phenotyping or ‘phenomics’ enables us to capture the phenotype more precisely and to a greater degree (in other words, ‘deep phenotyping’).
Deep phenotyping has been used to describe health conditions in humans, including disease and pre-disease conditions (Tracy 2008, Robinson 2012, Delude 2015). The concept of deep phenotyping has been applied widely in basic life sciences, including plant science (Houle et al. 2010). In addition to the biochemical or biophysical multi-omics analyses described above, the non-invasive phenotyping of crops under field conditions using computer vision technology has accelerated the practical application of deep phenotyping in crops and plants (Taghavi Namin et al. 2018, Harfouche et al. 2019, Mochida et al. 2019, Lürig et al. 2021). Computer vision technology fueled by deep learning can extract crucial information from RGB and hyperspectral images, including not only morphological data but also information on physiological marks, such as nutritional conditions or disease symptoms. The combination of the multi-disciplinary data obtained by various analytical technologies and by computer vision technology and analysis using artificial intelligence could offer the key discoveries on the linkages between the biological processes at different hierarchal levels that enable a comprehensive understanding of plant biological phenomena. Under the implementation of such monitoring strategies, data and outcomes, plant phenomics is expected to have a profound impact on plant sciences and agriculture. The deep phenotyping of crops in the field will also improve our mechanistic understanding of gene-to-phenotype connections in plants. Among the various biochemical phenotyping methods (such as metabolomics and ionomics), plant hormone profiling (or hormonomics) has received more attention in recent decades. This approach offers the most critical information on the developmental and physiological states of plants, which is indispensable for describing their growth processes.
Conclusion and Future Perspectives
Plant science is expected to provide techniques for enhancing crop production to meet the demands of an increasing population and should also present solutions to counteract decreasing crop yields due to global climate change. Moreover, research in this field is expected to increase crop yields with lower input and to improve plant tolerance to abiotic and biotic stresses. Researchers have conducted extensive studies to understand the mechanisms underlying these traits and have accumulated considerable knowledge on the functional characteristics of genes. However, the application of this knowledge to crops in the field faces some difficulties, indicating the limits of the aforementioned reductionistic approaches. Over the preceding decades, rapid advances in high-throughput and multi-omics methodologies, along with the development of data handling techniques and mathematical modeling, have opened new avenues for life science research. We now have access to high-quality genome data of crop species and the variations among accessions. Thus, we can describe the physiological status of plants more precisely using high-dimensional phenomics. Among the various molecular or biochemical phenotyping methodologies, hormonomics provides additional information regarding the developmental and physiological states of plants or crops. However, measuring endogenous plant hormones requires destructive, labor-intensive and time-consuming sample preparations and specialized analytical equipment. Therefore, hormonomics might not be an easily accessible methodology at present. In addition, the physiological function of plant hormones is not yet fully elucidated. To overcome these issues, various sensors have been developed to monitor plant hormone dynamics in real time. The long-term or life-course monitoring of plant hormones or other key biomarkers in field crops should provide data that can advance our understanding of the complex gene–phenotype–environment interactions and the roles of plant hormones in this domain. Data collected and deduced from these monitoring endeavors are crucial for crop design and for improving agricultural technology, including the development of biostimulants.
Acknowledgements
This work was supported by Grants-in-Aid for Scientific Research (nos. 21K19117 and 21H02509 to T.H.) from the Ministry of Education, Culture, Sports, Science and Technology Japan and was partly supported by grants from the Core Research for Evolutionary Science and Technology of the Japan Science and Technology Agency (no. JPMJCR16O4) provided to T.H. and K.M. This work was partially supported by Cabinet Office, Government of Japan, Moonshot R&D Program for Agriculture, Forestry and Fisheries (funding agency: Bio-oriented Technology Research Advancement Institution, No. JPJ009237).
Contributor Information
Takashi Hirayama, Institute of Plant Science and Resources, Okayama University, 2-20-1 Chuo, Kurashiki, Okayama, 710-0046 Japan.
Keiichi Mochida, RIKEN Center for Sustainable Resource Science, 1-7-22 Suehirocho, Tsurumiku, Yokohama, Kanagawa, 230-0045 Japan; Kihara Institute for Biological Research, Yokohama City University, 641-12 Maiokacho, Totsukaku, Yokohama, Kanagawa, 244-0813 Japan; School of Information and Data Sciences, Nagasaki University, 1-14 Bunkyo-machi, Nagasaki, 852-8521 Japan; RIKEN Baton Zone Program, RIKEN Cluster for Science, Technology and Innovation Hub, 1-7-22 Suehirocho, Tsurumiku, Yokohama, Kanagawa 230-0045 Japan.
Disclosures
The authors have no conflicts of interest to declare.
References
- Adamowski M. and Friml J. (2015) PIN-dependent auxin transport: action, regulation, and evolution. Plant Cell 27: 20–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aerts N., Pereira Mendes M. and Van Wees S.C.M. (2021) Multiple levels of crosstalk in hormone networks regulating plant defense. Plant J. 105: 489–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akiyama K., Ogasawara S., Ito S. and Hayashi H. (2010) Structural requirements of strigolactones for hyphal branching in AM fungi. Plant Cell Physiol. 51: 1104–1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali O., Ramsubhag A. and Jayaraman J. (2021) Biostimulant properties of seaweed extracts in plants: implications towards sustainable crop production. Plants 10: 531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altmann M., Altmann S., Rodriguez P.A., Weller B., Elorduy Vergara L., Palme J., et al. (2020) Extensive signal integration by the phytohormone protein network. Nature 583: 271–276. [DOI] [PubMed] [Google Scholar]
- Andrews S.J. and Rothnagel J.A. (2014) Emerging evidence for functional peptides encoded by short open reading frames. Nat. Rev. Genet. 15: 193–204. [DOI] [PubMed] [Google Scholar]
- Anzar N., Hasan R., Tyagi M., Yadav N. and Narang J. (2020) Carbon nanotube—a review on synthesis, properties and plethora of applications in the field of biomedical science. Sens Int. 1: 100003. [Google Scholar]
- Asami T. and Nakagawa Y. (2018) Preface to the special issue: brief review of plant hormones and their utilization in agriculture. J. Pestic. Sci. 43: 154–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atkinson N.J. and Urwin P.E. (2012) The interaction of plant biotic and abiotic stresses: from genes to the field. J. Exp. Bot. 63: 3523–3543. [DOI] [PubMed] [Google Scholar]
- Bailey-Serres J., Sorenson R. and Juntawong P. (2009) Getting the message across: cytoplasmic ribonucleoprotein complexes. Trends Plant Sci. 14: 443–453. [DOI] [PubMed] [Google Scholar]
- Bajguz A. and Piotrowska A. (2009) Conjugates of auxin and cytokinin. Phytochemistry 70: 957–969. [DOI] [PubMed] [Google Scholar]
- Bakshi A., Shemansky J.M., Chang C. and Binder B.M. (2015) History of research on the plant hormone ethylene. J. Plant Growth Regul. 34: 809–827. [Google Scholar]
- Bari R. and Jones J. (2008) Role of plant hormones in plant defence responses. Plant Mol. Biol. 69: 473–488. [DOI] [PubMed] [Google Scholar]
- Barone P.W., Baik S., Heller D.A. and Strano M.S. (2005) Near-infrared optical sensors based on single-walled carbon nanotubes. Nat. Mater 4: 86–92. [DOI] [PubMed] [Google Scholar]
- Berendsen R.L., Pieterse C.M.J. and Bakker P.A.H.M. (2012) The rhizosphere microbiome and plant health. Trends Plant Sci. 17: 478–486. [DOI] [PubMed] [Google Scholar]
- Berens M.L., Berry H.M., Mine A., Argueso C.T. and Tsuda K. (2017) Evolution of hormone signaling networks in plant defense. Annu Rev Phytopathol 55: 401–425. [DOI] [PubMed] [Google Scholar]
- Bernard C., Traub M., Kunz H.-H., Hach S., Trentmann O. and Möhlmann T. (2011) Equilibrative nucleoside transporter 1 (ENT1) is critical for pollen germination and vegetative growth in Arabidopsis. J. Exp. Bot. 62: 4627–4637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bevan M.W., Uauy C., Wulff B.B.H., Zhou J., Krasileva K. and Clark M.D. (2017) Genomic innovation for crop improvement. Nature 543: 346–354. [DOI] [PubMed] [Google Scholar]
- Brand U., Fletcher J.C., Hobe M., Meyerowitz E.M. and Simon R. (2000) Dependence of stem cell fate in Arabidopsis on a feedback loop regulated by CLV3 activity. Science 289: 617–619. [DOI] [PubMed] [Google Scholar]
- Bücker-Neto L., Paiva A.L.S., Machado R.D., Arenhart R.A. and Margis-Pinheiro M. (2017) Interactions between plant hormones and heavy metals responses. Genet. Mol. Biol. 40: 373–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caarls L., Pieterse C.M.J. and Van Wees S.C.M. (2015) How salicylic acid takes transcriptional control over jasmonic acid signaling. Front. Plant Sci. 6: 170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao D., Barbier F., Yoneyama K. and Beveridge C.A. (2020) A rapid method for quantifying RNA and phytohormones from a small amount of plant tissue. Front. Plant Sci. 11: 1832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova-Sáez R., Mateo-Bonmatí E. and Ljung K. (2021) Auxin metabolism in plants. Cold Spring Harb. Perspect. Biol. 13: a039867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chanclud E. and Morel J.-B. (2016) Plant hormones: a fungal point of view. Mol. Plant Pathol. 17: 1289–1297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chesterfield R.J., Vickers C.E. and Beveridge C.A. (2020) Translation of strigolactones from plant hormone to agriculture: achievements, future perspectives, and challenges. Trends Plant Sci. 25: 1087–1106. [DOI] [PubMed] [Google Scholar]
- Choi J., Eom S., Shin K., Lee R.-A., Choi S., Lee J.-H., et al. (2019) Identification of lysine histidine transporter 2 as an 1-aminocyclopropane carboxylic acid transporter in Arabidopsis thaliana by transgenic complementation approach. Front. Plant Sci. 10: 1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chono M., Honda I., Zeniya H., Yoneyama K., Saisho D., Takeda K., et al. (2003) A semidwarf phenotype of barley uzu results from a nucleotide substitution in the gene encoding a putative brassinosteroid receptor. Plant Physiol. 133: 1209–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S.E., Running M.P. and Meyerowitz E.M. (1995) CLAVATA3 is a specific regulator of shoot and floral meristem development affecting the same processes as CLAVATA1. Development 121: 2057–2067. [Google Scholar]
- Clouse S.D. and Sasse J.M. (1998) Brassinosteroides: essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49: 427–451. [DOI] [PubMed] [Google Scholar]
- Cohen A.C., Bottini R., Pontin M., Berli F.J., Moreno D., Boccanlandro H., et al. (2015) Azospirillum brasilense ameliorates the response of Arabidopsis thaliana to drought mainly via enhancement of ABA levels. Physiol. Plant. 153: 79–90. [DOI] [PubMed] [Google Scholar]
- Cortleven A., Leuendorf J.E., Frank M., Pezzetta D., Bolt S. and Schmülling T. (2019) Cytokinin action in response to abiotic and biotic stresses in plants. Plant Cell Environ. 42: 998–1018. [DOI] [PubMed] [Google Scholar]
- Cox J. and Mann M. (2011) Quantitative, high-resolution proteomics for data-driven systems biology. Annu. Rev. Biochem. 80: 273–299. [DOI] [PubMed] [Google Scholar]
- Cramer G.R., Urano K., Delrot S., Pezzotti M. and Shinozaki K. (2011) Effects of abiotic stress on plants: a systems biology perspective. BMC Plant Biol. 11: 163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cutler S.R., Rodriguez P.L., Finkelstein R.R. and Abrams S.R. (2010) Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61: 651–679. [DOI] [PubMed] [Google Scholar]
- Dar T.A., Uddin M., Khan M.M.A., Hakeem K.R. and Jaleel H. (2015) Jasmonates counter plant stress: a review. Environ. Exp. Bot. 115: 49–57. [Google Scholar]
- Delude C.M. (2015) Deep phenotyping: the details of disease. Nature 527: S14–15. [DOI] [PubMed] [Google Scholar]
- Denance N., Sanchez-Vallet A., Goffner D. and Molina A. (2013) Disease resistance or growth: the role of plant hormones in balancing immune responses and fitness costs. Front. Plant Sci. 4: 155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Depuydt S. and Hardtke C.S. (2011) Hormone signalling crosstalk in plant growth regulation. Curr. Biol. 21: R365–73. [DOI] [PubMed] [Google Scholar]
- Dessaux Y., Grandclément C. and Faure D. (2016) Engineering the rhizosphere. Trends Plant Sci. 21: 266–278. [DOI] [PubMed] [Google Scholar]
- Dietz K., Sauter A., Wichert K., Messdaghi D. and Hartung W. (2000) Extracellular β‐glucosidase activity in barley involved in the hydrolysis of ABA glucose conjugate in leaves. J. Exp. Bot. 51: 937–944. [PubMed] [Google Scholar]
- Dobrev P.I., Havlíček L., Vágner M., Malbeck J. and Kamínek M. (2005) Purification and determination of plant hormones auxin and abscisic acid using solid phase extraction and two-dimensional high performance liquid chromatography. J. Chromatogr. A 1075: 159–166. [DOI] [PubMed] [Google Scholar]
- Dodd I.C., Zinovkina N.Y., Safronova V.I. and Belimov A.A. (2010) Rhizobacterial mediation of plant hormone status. Ann. Appl. Biol. 157: 361–379. [Google Scholar]
- Drobek M., Frąc M. and Cybulska J. (2019) Plant biostimulants: importance of the quality and yield of horticultural crops and the improvement of plant tolerance to abiotic stress—a review. Agronomy. 9: 335. [Google Scholar]
- du Jardin P. (2015) Plant biostimulants: definition, concept, main categories and regulation. Sci. Hortic. 196: 3–14. [Google Scholar]
- Eichmann R., Richards L. and Schäfer P. (2021) Hormones as go-betweens in plant microbiome assembly. Plant J. 105: 518–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El-Showk S., Ruonala R. and Helariutta Y. (2013) Crossing paths: cytokinin signalling and crosstalk. Development 140: 1373–1383. [DOI] [PubMed] [Google Scholar]
- Engelberth J., Schmelz E.A., Alborn H.T., Cardoza Y.J., Huang J. and Tumlinson J.H. (2003) Simultaneous quantification of jasmonic acid and salicylic acid in plants by vapor-phase extraction and gas chromatography-chemical ionization-mass spectrometry. Anal. Biochem. 312: 242–250. [DOI] [PubMed] [Google Scholar]
- Fahad S., Hussain S., Matloob A., Khan F.A., Khaliq A., Saud S., et al. (2015) Phytohormones and plant responses to salinity stress: a review. Plant Growth Regul. 75: 391–404. [Google Scholar]
- FAO . (2019) FAO framework for the urban food agenda.doi: 10.4060/ca3151en. [DOI]
- FAO, IFAD, UNICEF, WFP and WHO . (2020) The state of food security and nutrition in the world 2020. Transforming food systems for affordable healthy diets. Rome.doi: 10.4060/ca9692en. [DOI] [Google Scholar]
- Fitzpatrick C.R., Salas-González I., Conway J.M., Finkel O.M., Gilbert S., Russ D., et al. (2020) The plant microbiome: from ecology to reductionism and beyond. Annu. Rev. Microbiol. 74: 81–100. [DOI] [PubMed] [Google Scholar]
- Fletcher J.C., Brand U., Running M.P., Simon R. and Meyerowitz E.M. (1999) Signaling of cell fate decisions by CLAVATA3 in Arabidopsis shoot meristems. Science 283: 1911–1914. [DOI] [PubMed] [Google Scholar]
- Fonseca S., Chini A., Hamberg M., Adie B., Porzel A., Kramell R., et al. (2009) (+)-7-iso-Jasmonoyl-L-isoleucine is the endogenous bioactive jasmonate. Nat. Chem. Biol. 5: 344–350. [DOI] [PubMed] [Google Scholar]
- Footitt S., Dietrich D., Fait A., Fernie A.R., Holdsworth M.J., Baker A., et al. (2007) The COMATOSE ATP-binding cassette transporter is required for full fertility in Arabidopsis. Plant Physiol. 144: 1467–1480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gamalero E. and Glick B.R. (2015) Bacterial modulation of plant ethylene levels. Plant Physiol. 169: 13–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Z., Karlsson I., Geisen S., Kowalchuk G. and Jousset A. (2019) Protists: puppet masters of the rhizosphere microbiome. Trends Plant Sci. 24: 165–176. [DOI] [PubMed] [Google Scholar]
- Gautrat P., Laffont C., Frugier F. and Ruffel S. (2021) Nitrogen systemic signaling: from symbiotic nodulation to root acquisition. Trends Plant Sci. 26: 392–406. [DOI] [PubMed] [Google Scholar]
- Geisler M., Aryal B., di Donato M. and Hao P. (2017) A critical view on ABC transporters and their interacting partners in auxin transport. Plant Cell Physiol. 58: 1601–1614. [DOI] [PubMed] [Google Scholar]
- Gillissen B., Bürkle L., André B., Kühn C., Rentsch D., Brandl B., et al. (2000) A new family of high-affinity transporters for adenine, cytosine, and purine derivatives in Arabidopsis. Plant Cell 12: 291–300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraldo J.P., Wu H., Newkirk G.M. and Kruss S. (2019) Nanobiotechnology approaches for engineering smart plant sensors. Nat. Nanotechnol 14: 541–553. [DOI] [PubMed] [Google Scholar]
- Glassbrook N., Beecher C. and Ryals J. (2000) Metabolic profiling on the right path. Nat. Biotechnol. 18: 1142–1143. [DOI] [PubMed] [Google Scholar]
- Glazebrook J. (2005) Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol. 43: 205–227. [DOI] [PubMed] [Google Scholar]
- Grillet L. and Schmidt W. (2019) Iron acquisition strategies in land plants: not so different after all. New Phytol. 224: 11–18. [DOI] [PubMed] [Google Scholar]
- Gururani M.A., Mohanta T.K. and Bae H. (2015) Current understanding of the interplay between phytohormones and photosynthesis under environmental stress. Int. J. Mol. Sci. 16: 19055–19085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K., Higuchi-Takeuchi M., Okamoto M., Yoshizumi T., Shimizu M., Nakaminami K., et al. (2013) Small open reading frames associated with morphogenesis are hidden in plant genomes. Proc. Natl. Acad. Sci. USA 110: 2395–2400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harfouche A.L., Jacobson D.A., Kainer D., Romero J.C., Harfouche A.H., Scarascia Mugnozza G., et al. (2019) Accelerating climate resilient plant breeding by applying next-generation artificial intelligence. Trends Biotechnol. 37: 1217–1235. [DOI] [PubMed] [Google Scholar]
- Hayat R., Ali S., Amara U., Khalid R. and Ahmed I. (2010) Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60: 579–598. [Google Scholar]
- Hedden P. (1993) Modern methods for the quantitative analysis of plant hormones. Ann. Rev. Plant Physiol. Plant Biol. 44: 107–1029. [Google Scholar]
- Hedden P. and Sponsel V. (2015) A century of gibberellin research. J. Plant Growth Regul. 34: 740–760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herud-Sikimić O., Stiel A.C., Kolb M., Shanmugaratnam S., Berendzen K.W., Feldhaus C., et al. (2021) A biosensor for the direct visualization of auxin. Nature 592: 768–772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirayama T., Lei G.J., Yamaji N., Nakagawa N. and Ma J.F. (2018) The putative peptide gene FEP1 regulates iron deficiency response in Arabidopsis. Plant Cell Physiol. 59: 1739–1752. [DOI] [PubMed] [Google Scholar]
- Hirayama T., Saisho D., Matsuura T., Okada S., Takahagi K., Kanatani A., et al. (2020) Life-course monitoring of endogenous phytohormone levels under field conditions reveals diversity of physiological states among barley accessions. Plant Cell Physiol. 61: 1438–1448. [DOI] [PubMed] [Google Scholar]
- Houle D., Govindaraju D.R. and Omholt S. (2010) Phenomics: the next challenge. Nat. Rev. Genet. 11: 855–866. [DOI] [PubMed] [Google Scholar]
- Hsu P.Y. and Benfey P.N. (2018) Small but mighty: functional peptides encoded by small ORFs in plants. PROTEOMICS 18: 1700038. [DOI] [PubMed] [Google Scholar]
- Huffaker A. (2015) Plant elicitor peptides in induced defense against insects. Curr. Opin. Insect. Sci. 9: 44–50. [DOI] [PubMed] [Google Scholar]
- Isoda R., Yoshinari A., Ishikawa Y., Sadoine M., Simon R., Frommer W.B., et al. (2021) Sensors for the quantification, localization and analysis of the dynamics of plant hormones. Plant J. 105: 542–557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaillais Y. and Chory J. (2010) Unraveling the paradoxes of plant hormone signaling integration. Nat. Struct. Mol. Biol. 17: 642–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon B.W., Kim M.-J., Pandey S.K., Oh E., Seo P.J. and Kim J. (2021) Recent advances in peptide signaling during Arabidopsis root development. J. Exp. Bot. 72: 2889–2902. [DOI] [PubMed] [Google Scholar]
- Johnson C.H., Ivanisevic J. and Siuzdak G. (2016) Metabolomics: beyond biomarkers and towards mechanisms. Nat. Rev. Mol. Cell Biol. 17: 451–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones A.M., Danielson J.Å., ManojKumar S.N., Lanquar V., Grossmann G. and Frommer W.B. (2014) Abscisic acid dynamics in roots detected with genetically encoded FRET sensors. eLife 3: e01741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kamada-Nobusada T. and Sakakibara H. (2009) Molecular basis for cytokinin biosynthesis. Phytochemistry 70: 444–449. [DOI] [PubMed] [Google Scholar]
- Kang J., Hwang J.-U., Lee M., Kim -Y.-Y., Assmann S.M., Martinoia E., et al. (2010) PDR-type ABC transporter mediates cellular uptake of the phytohormone abscisic acid. Proc. Natl. Acad. Sci. USA 107: 2355–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J., Lee Y., Sakakibara H. and Martinoia E. (2017) Cytokinin transporters: go and stop in signaling. Trends Plant Sci. 22: 455–461. [DOI] [PubMed] [Google Scholar]
- Kang J., Yim S., Choi H., Kim A., Lee K.P., Lopez-Molina L., et al. (2015) Abscisic acid transporters cooperate to control seed germination. Nat. Commun. 6: 8113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y., Hanada A., Chiba Y., Ichikawa T., Nakazawa M., Matsui M., et al. (2012) Identification of an abscisic acid transporter by functional screening using the receptor complex as a sensor. Proc. Natl. Acad. Sci. USA 109: 9653–9658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanno Y., Oikawa T., Chiba Y., Ishimaru Y., Shimizu T., Sano N., et al. (2016) AtSWEET13 and AtSWEET14 regulate gibberellin-mediated physiological processes. Nat Commun. 7: 13245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsir L., Davies K.A., Bergmann D.C. and Laux T. (2011) Peptide signaling in plant development. Curr. Biol. 21: R356–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kazan K. (2015) Diverse roles of jasmonates and ethylene in abiotic stress tolerance. Trends Plant Sci. 20: 219–229. [DOI] [PubMed] [Google Scholar]
- Kerchev P., van der Meer T., Sujeeth N., Verlee A., Stevens C.V., Van Breusegem F., et al. (2020) Molecular priming as an approach to induce tolerance against abiotic and oxidative stresses in crop plants. Biotechnol Adv. 40: 107503. [DOI] [PubMed] [Google Scholar]
- Kiba T., Takei K., Kojima M. and Sakakibara H. (2013) Side-chain modification of cytokinins controls shoot growth in Arabidopsis. Dev. Cell 27: 452–461. [DOI] [PubMed] [Google Scholar]
- Kieber J.J. and Schaller G.E. (2018) Cytokinin signaling in plant development. Development 145: dev149344. [DOI] [PubMed] [Google Scholar]
- Kim J.-M., To T.K., Matsui A., Tanoi K., Kobayashi N.I., Matsuda F., et al. (2017) Acetate-mediated novel survival strategy against drought in plants. Nat Plants 3: 1–7. [DOI] [PubMed] [Google Scholar]
- Kim J.S., Jeon B.W. and Kim J. (2021) Signaling peptides regulating abiotic stress responses in plants. Front. Plant Sci. 12: 704490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M.-J., Jeon B.W., Oh E., Seo P.J. and Kim J. (2021) peptide signaling during plant reproduction. Trends Plant Sci. 26: 822–835. [DOI] [PubMed] [Google Scholar]
- Ko D., Kang J., Kiba T., Park J., Kojima M., Do J., et al. (2014) Arabidopsis ABCG14 is essential for the root-to-shoot translocation of cytokinin. Proc. Natl. Acad. Sci. USA 111: 7150–7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kojima M., Kamada-Nobusada T., Komatsu H., Takei K., Kuroha T., Mizutani M., et al. (2009) Highly sensitive and high-throughput analysis of plant hormones using ms-probe modification and liquid chromatography–tandem mass spectrometry: an application for hormone profiling in Oryza sativa. Plant Cell Physiol. 50: 1201–1214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koornneef A., Leon-Reyes A., Ritsema T., Verhage A., Den Otter F.C., Van Loon L.C., et al. (2008) Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 147: 1358–1368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzschmar T., Kohlen W., Sasse J., Borghi L., Schlegel M., Bachelier J.B., et al. (2012) A petunia ABC protein controls strigolactone-dependent symbiotic signalling and branching. Nature 483: 341–344. [DOI] [PubMed] [Google Scholar]
- Kruss S., Hilmer A.J., Zhang J., Reuel N.F., Mu B. and Strano M.S. (2013) Carbon nanotubes as optical biomedical sensors. Adv. Drug Deliv. Rev. 65: 1933–1950. [DOI] [PubMed] [Google Scholar]
- Kurakawa T., Ueda N., Maekawa M., Kobayashi K., Kojima M., Nagato Y., et al. (2007) Direct control of shoot meristem activity by a cytokinin-activating enzyme. Nature 445: 652–655. [DOI] [PubMed] [Google Scholar]
- Kuroha T., Tokunaga H., Kojima M., Ueda N., Ishida T., Nagawa S., et al. (2009) Functional analyses of LONELY GUY cytokinin-activating enzymes reveal the importance of the direct activation pathway in Arabidopsis. Plant Cell 21: 3152–3169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T., Miyaji T., Yabuuchi H., Shimizu H., Sugimoto E., Kamiya A., et al. (2010) ABC transporter AtABCG25 is involved in abscisic acid transport and responses. Proc. Natl. Acad. Sci. USA 107: 2361–2366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuromori T., Seo M. and Shinozaki K. (2018) ABA transport and plant water stress responses. Trends Plant Sci. 23: 513–522. [DOI] [PubMed] [Google Scholar]
- Kushiro T., Okamoto M., Nakabayashi K., Yamagishi K., Kitamura S., Asami T., et al. (2004) The Arabidopsis cytochrome P450 CYP707A encodes ABA 8ʹ-hydroxylases: key enzymes in ABA catabolism. EMBO J. 23: 1647–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwak M.-J., Kong H.G., Choi K., Kwon S.-K., Song J.Y., Lee J., et al. (2018) Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36: 1100–1109. [DOI] [PubMed] [Google Scholar]
- Kwak S.-Y., Wong M.H., Lew T.T.S., Bisker G., Lee M.A., Kaplan A., et al. (2017) Nanosensor technology applied to living plant systems. Annu. Rev. Anal. Chem. 10: 113–140. [DOI] [PubMed] [Google Scholar]
- Lanfranco L., Fiorilli V., Venice F. and Bonfante P. (2018) Strigolactones cross the kingdoms: plants, fungi, and bacteria in the arbuscular mycorrhizal symbiosis. J. Exp. Bot. 69: 2175–2188. [DOI] [PubMed] [Google Scholar]
- Larance M. and Lamond A.I. (2015) Multidimensional proteomics for cell biology. Nat. Rev. Mol. Cell Biol. 16: 269–280. [DOI] [PubMed] [Google Scholar]
- Lee K., Park J., Lee M.-S., Kim J., Hyun B.G., Kang D.J., et al. (2014) In-situ synthesis of carbon nanotube–graphite electronic devices and their integrations onto surfaces of live plants and insects. Nano Lett. 14: 2647–2654. [DOI] [PubMed] [Google Scholar]
- Lee K.H., Piao H.L., Kim H.Y., Choi S.M., Jiang F., Hartung W., et al. (2006) Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126: 1109–1120. [DOI] [PubMed] [Google Scholar]
- Lew T.T.S., Koman V.B., Silmore K.S., Seo J.S., Gordiichuk P., Kwak S.-Y., et al. (2020a) Real-time detection of wound-induced H2O2 signalling waves in plants with optical nanosensors. Nat Plants 6: 404–415. [DOI] [PubMed] [Google Scholar]
- Lew T.T.S., Sarojam R., Jang I.-C., Park B.S., Naqvi N.I., Wong M.H., et al. (2020b) Species-independent analytical tools for next-generation agriculture. Nat Plants 6: 1408–1417. [DOI] [PubMed] [Google Scholar]
- Li J., Wu H., Santana I., Fahlgren M. and Giraldo J.P. (2018) Standoff optical glucose sensing in photosynthetic organisms by a quantum dot fluorescent probe. ACS Appl. Mater. Interfaces 10: 28279–28289. [DOI] [PubMed] [Google Scholar]
- Li Q., Zheng J., Li S., Huang G., Skilling S.J., Wang L., et al. (2017) Transporter-mediated nuclear entry of jasmonoyl-isoleucine is essential for jasmonate signaling. Mol Plant 10: 695–708. [DOI] [PubMed] [Google Scholar]
- Li Z., Liu Y., Hossain O., Paul R., Yao S., Wu S., et al. (2021) Real-time monitoring of plant stresses via chemiresistive profiling of leaf volatiles by a wearable sensor. Matter 4: 2553–2570. [Google Scholar]
- Liu C.-J., Zhao Y. and Zhang K. (2019) Cytokinin transporters: multisite players in cytokinin homeostasis and signal distribution. Front. Plant Sci. 10: 693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Durán R. and Zipfel C. (2015) Trade-off between growth and immunity: role of brassinosteroids. Trends Plant Sci. 20: 12–19. [DOI] [PubMed] [Google Scholar]
- Lu Y., Wang E., Tang Z., Rui J., Li Y., Tang Z., et al. (2021) Roots and microbiome jointly drive the distributions of 17 phytohormones in the plant soil continuum in a phytohormone‐specific manner. Plant Soil 470: 153–165. [Google Scholar]
- Lu Y., Xu K., Zhang L., Deguchi M., Shishido H., Arie T., et al. (2020) Multimodal plant healthcare flexible sensor system. ACS Nano 14: 10966–10975. [DOI] [PubMed] [Google Scholar]
- Lürig M.D., Donoughe S., Svensson E.I., Porto A. and Tsuboi M. (2021) Computer vision, machine learning, and the promise of phenomics in ecology and evolutionary biology. Front. Ecol. Evol. 9: 642774. [Google Scholar]
- Lyu D., Zajonc J., Pagé A., Tanney C.A.S., Shah A., Monjezi N., et al. (2021) Plant holobiont theory: the phytomicrobiome plays a central role in evolution and success. Microorganisms 9: 675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Medina A., Flors V., Heil M., Mauch-Mani B., Pieterse C.M.J., Pozo M.J., et al. (2016) Recognizing plant defense priming. Trends Plant Sci. 21: 818–822. [DOI] [PubMed] [Google Scholar]
- Mickelbart M.V., Hasegawa P.M. and Bailey-Serres J. (2015) Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat. Rev. Genet. 16: 237–251. [DOI] [PubMed] [Google Scholar]
- Mochida K., Koda S., Inoue K., Hirayama T., Tanaka S., Nishii R., et al. (2019) Computer vision-based phenotyping for improvement of plant productivity: a machine learning perspective. GigaScience 8: giy153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochida K., Saisho D. and Hirayama T. (2015) Crop improvement using life cycle datasets acquired under field conditions. Front. Plant Sci. 6: 740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morozova O., Hirst M. and Marra M.A. (2009) Applications of new sequencing technologies for transcriptome analysis. Annu. Rev. Genomics Hum. Genet. 10: 135–151. [DOI] [PubMed] [Google Scholar]
- Mostofa M.G., Li W., Nguyen K.H., Fujita M. and Tran L.-S.P. (2018) Strigolactones in plant adaptation to abiotic stresses: an emerging avenue of plant research. Plant Cell Environ. 41: 2227–2243. [DOI] [PubMed] [Google Scholar]
- Nakaminami K., Okamoto M., Higuchi-Takeuchi M., Yoshizumi T., Yamaguchi Y., Fukao Y., et al. (2018) AtPep3 is a hormone-like peptide that plays a role in the salinity stress tolerance of plants. Proc. Natl. Acad. Sci. USA 115: 5810–5815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nambara E. and Marion-Poll A. (2005) Abscisic acid biosynthesis and catabolism. Annu Rev Plant Biol 56: 165–185. [DOI] [PubMed] [Google Scholar]
- Narita N.N., Moore S., Horiguchi G., Kubo M., Demura T., Fukuda H., et al. (2004) Overexpression of a novel small peptide ROTUNDIFOLIA4 decreases cell proliferation and alters leaf shape in Arabidopsis thaliana. Plant J. 38: 699–713. [DOI] [PubMed] [Google Scholar]
- Nemhauser J.L., Hong F. and Chory J. (2006) Different plant hormones regulate similar processes through largely nonoverlapping transcriptional responses. Cell 126: 467–475. [DOI] [PubMed] [Google Scholar]
- Nephali L., Piater L.A., Dubery I.A., Patterson V., Huyser J., Burgess K., et al. (2020) Biostimulants for plant growth and mitigation of abiotic stresses: a metabolomics perspective. Metabolites 10: 505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishiyama R., Watanabe Y., Fujita Y., Le D.T., Kojima M., Werner T., et al. (2011) Analysis of cytokinin mutants and regulation of cytokinin metabolic genes reveals important regulatory roles of cytokinins in drought, salt and abscisic acid responses, and abscisic acid biosynthesis. Plant Cell 23: 2169–2183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Novák O., Napier R. and Ljung K. (2017) Zooming in on plant hormone analysis: tissue- and cell-specific approaches. Annu Rev Plant Biol 68: 323–348. [DOI] [PubMed] [Google Scholar]
- Oh E., Seo P.J. and Kim J. (2018) Signaling peptides and receptors coordinating plant root development. Trends Plant Sci. 23: 337–351. [DOI] [PubMed] [Google Scholar]
- Ohkubo Y., Tanaka M., Tabata R., Ogawa-Ohnishi M. and Matsubayashi Y. (2017) Shoot-to-root mobile polypeptides involved in systemic regulation of nitrogen acquisition. Nat Plants 3: 17029. [DOI] [PubMed] [Google Scholar]
- Okumoto S., Jones A. and Frommer W.B. (2012) Quantitative imaging with fluorescent biosensors. Annu Rev Plant Biol 63: 663–706. [DOI] [PubMed] [Google Scholar]
- Pan X., Welti R. and Wang X. (2010) Quantitative analysis of major plant hormones in crude plant extracts by high-performance liquid chromatography–mass spectrometry. Nat Protoc 5: 986–992. [DOI] [PubMed] [Google Scholar]
- Park J., Lee Y., Martinoia E. and Geisler M. (2017) Plant hormone transporters: what we know and what we would like to know. BMC Biol. 15: 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pattyn J., Vaughan-Hirsch J. and Van de Poel B. (2021) The regulation of ethylene biosynthesis: a complex multilevel control circuitry. New Phytol. 229: 770–782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peleg Z. and Blumwald E. (2011) Hormone balance and abiotic stress tolerance in crop plants. Curr. Opin. Plant Biol. 14: 290–295. [DOI] [PubMed] [Google Scholar]
- Pieterse C.M.J., Leon-Reyes A., Van der Ent S. and Van Wees S.C.M. (2009) Networking by small-molecule hormones in plant immunity. Nat. Chem. Biol. 5: 308–316. [DOI] [PubMed] [Google Scholar]
- Pieterse C.M.J., Van der Does D., Zamioudis C., Leon-Reyes A. and Van Wees S.C.M. (2012) Hormonal modulation of plant immunity. Annu. Rev. Cell Dev. Biol. 28: 489–521. [DOI] [PubMed] [Google Scholar]
- Prosekov A.Y. and Ivanova S.A. (2018) Food security: the challenge of the present. Geoforum 91: 73–77. [Google Scholar]
- Purugganan M.D. and Jackson S.A. (2021) Advancing crop genomics from lab to field. Nat. Genet. 53: 595–601. [DOI] [PubMed] [Google Scholar]
- Raser J.M. and O’Shea E.K. (2005) Noise in gene expression: origins, consequences, and control. Science 309: 2010–2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizza A., Walia A., Lanquar V., Frommer W.B. and Jones A.M. (2017) In vivo gibberellin gradients visualized in rapidly elongating tissues. Nat Plants 3: 803–813. [DOI] [PubMed] [Google Scholar]
- Robert-Seilaniantz A., Grant M. and Jones J.D.G. (2011) Hormone crosstalk in plant disease and defense: more than just jasmonate-salicylate antagonism. Annu Rev Phytopathol 49: 317–343. [DOI] [PubMed] [Google Scholar]
- Robinson P.N. (2012) Deep phenotyping for precision medicine. Hum. Mutat. 33: 777–780. [DOI] [PubMed] [Google Scholar]
- Rosier A., Medeiros F.H.V. and Bais H.P. (2018) Defining plant growth promoting rhizobacteria molecular and biochemical networks in beneficial plant-microbe interactions. Plant Soil 428: 35–55. [Google Scholar]
- Ross J.J., Weston D.E., Davidson S.E. and Reid J.B. (2011) Plant hormone interactions: how complex are they? Physiol. Plant. 141: 299–309. [DOI] [PubMed] [Google Scholar]
- Rouphael Y. and Colla G. (2020) Toward a sustainable agriculture through plant biostimulants: from experimental data to practical applications. Agronomy 10: 1461. [Google Scholar]
- Ruffel S. (2018) Nutrient-related long-distance signals: common players and possible cross-talk. Plant Cell Physiol. 59: 1723–1732. [DOI] [PubMed] [Google Scholar]
- Saisho D., Tanno K., Chono M., Honda I., Kitano H. and Takeda K. (2004) Spontaneous brassinolide-insensitive barley mutants ‘uzu’ adapted to East Asia. Breed. Sci. 54: 409–416. [Google Scholar]
- Saito H., Oikawa T., Hamamoto S., Ishimaru Y., Kanamori-Sato M., Sasaki-Sekimoto Y., et al. (2015) The jasmonate-responsive GTR1 transporter is required for gibberellin-mediated stamen development in Arabidopsis. Nat. Commun. 6: 6095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salt D.E., Baxter I. and Lahner B. (2008) Ionomics and the study of the plant ionome. Annu Rev Plant Biol 59: 709–733. [DOI] [PubMed] [Google Scholar]
- Santner A., Calderon-Villalobos L.I.A. and Estelle M. (2009) Plant hormones are versatile chemical regulators of plant growth. Nat. Chem. Biol. 5: 301–307. [DOI] [PubMed] [Google Scholar]
- Santner A. and Estelle M. (2009) Recent advances and emerging trends in plant hormone signalling. Nature 459: 1071–1078. [DOI] [PubMed] [Google Scholar]
- Santoyo G., Moreno-Hagelsieb G., Del Carmen Orozco-mosqueda M. and Glick B.R. (2016) Plant growth-promoting bacterial endophytes. Microbiol. Res. 183: 92–99. [DOI] [PubMed] [Google Scholar]
- Sasse J., Simon S., Gübeli C., Liu G.-W., Cheng X., Friml J., et al. (2015) Asymmetric localizations of the ABC transporter PaPDR1 trace paths of directional strigolactone transport. Curr. Biol. 25: 647–655. [DOI] [PubMed] [Google Scholar]
- Schilmiller A.L. and Howe G.A. (2005) Systemic signaling in the wound response. Curr. Opin. Plant Biol. 8: 369–377. [DOI] [PubMed] [Google Scholar]
- Schmelz E.A., Engelberth J., Alborn H.T., O’Donnell P., Sammons M., Toshima H., et al. (2003) Simultaneous analysis of phytohormones, phytotoxins, and volatile organic compounds in plants. Proc. Natl. Acad. Sci. USA 100: 10552–10557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmülling T., Werner T., Riefler M., Krupková E. and Bartrina Y Manns I. (2003) Structure and function of cytokinin oxidase/dehydrogenase genes of maize, rice, Arabidopsis and other species. J. Plant Res. 116: 241–252. [DOI] [PubMed] [Google Scholar]
- Schoof H., Lenhard M., Haecker A., Mayer K.F.X., Jürgens G. and Laux T. (2000) The stem cell population of Arabidopsis shoot meristems is maintained by a regulatory loop between the CLAVATA and WUSCHEL genes. Cell 100: 635–644. [DOI] [PubMed] [Google Scholar]
- Seo S., Nakaho K., Hong S.W., Takahashi H., Shigemori H. and Mitsuhara I. (2016) l-histidine induces resistance in plants to the bacterial pathogen Ralstonia solanacearum partially through the activation of ethylene signaling. Plant Cell Physiol. 57: 1932–1942. [DOI] [PubMed] [Google Scholar]
- Seto K.C. and Ramankutty N. (2016) Hidden linkages between urbanization and food systems. Science 352: 943–945. [DOI] [PubMed] [Google Scholar]
- Shigenaga A.M. and Argueso C.T. (2016) No hormone to rule them all: interactions of plant hormones during the responses of plants to pathogens. Semin. Cell Dev. Biol. 56: 174–189. [DOI] [PubMed] [Google Scholar]
- Shigenaga A.M., Berens M.L., Tsuda K. and Argueso C.T. (2017) Towards engineering of hormonal crosstalk in plant immunity. Curr. Opin. Plant Biol. 38: 164–172. [DOI] [PubMed] [Google Scholar]
- Shimizu T., Miyakawa S., Esaki T., Mizuno H., Masujima T., Koshiba T., et al. (2015) Live single-cell plant hormone analysis by video-mass spectrometry. Plant Cell Physiol. 56: 1287–1296. [DOI] [PubMed] [Google Scholar]
- Shin K., Lee S., Song W.-Y., Lee R.-A., Lee I., Ha K., et al. (2015) Genetic Identification of ACC-RESISTANT2 Reveals Involvement of LYSINE HISTIDINE TRANSPORTER1 in the Uptake of 1-Aminocyclopropane-1-Carboxylic Acid in Arabidopsis thaliana. Plant Cell Physiol. 56: 572–582. [DOI] [PubMed] [Google Scholar]
- Shiono K. and Taira S. (2020) Imaging of multiple plant hormones in roots of rice (Oryza sativa) using nanoparticle-assisted laser desorption/ionization mass spectrometry. J. Agric. Food Chem. 68: 6770–6775. [DOI] [PubMed] [Google Scholar]
- Shukla P.S., Mantin E.G.Adil M., Bajpai S., Critchley A.T. and Prithiviraj B. (2019) Ascophyllum nodosum-based biostimulants: sustainable applications in agriculture for the stimulation of plant growth, stress tolerance, and disease management. Front. Plant Sci. 10: 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Šimura J., Antoniadi I., Široká J., Tarkowská D., Strnad M., Ljung K., et al. (2018) Plant hormonomics: multiple phytohormone profiling by targeted metabolomics. Plant Physiol. 177: 476–489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh A., Septiningsih E.M., Balyan H.S., Singh N.K. and Rai V. (2017) Genetics, physiological mechanisms and breeding of flood-tolerant rice (Oryza sativa L.). Plant Cell Physiol. 58: 185–197. [DOI] [PubMed] [Google Scholar]
- Spielmeyer W., Ellis M.H. and Chandler P.M. (2002) Semidwarf (sd-1), “green revolution” rice, contains a defective gibberellin 20-oxidase gene. Proc. Natl. Acad. Sci. USA 99: 9043–9048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su Y.-H., Liu Y.-B. and Zhang X.-S. (2011) Auxin–cytokinin interaction regulates meristem development. Mol. Plant 4: 616–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taghavi Namin S., Esmaeilzadeh M., Najafi M., Brown T.B. and Borevitz J.O. (2018) Deep phenotyping: deep learning for temporal phenotype/genotype classification. Plant Methods 14: 66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takahashi F. and Shinozaki K. (2019) Long-distance signaling in plant stress response. Curr. Opin. Plant Biol. 47: 106–111. [DOI] [PubMed] [Google Scholar]
- Takahashi F., Suzuki T., Osakabe Y., Betsuyaku S., Kondo Y., Dohmae N., et al. (2018) A small peptide modulates stomatal control via abscisic acid in long-distance signalling. Nature 556: 235–238. [DOI] [PubMed] [Google Scholar]
- Tal I., Zhang Y., Jørgensen M.E., Pisanty O., Barbosa I.C.R., Zourelidou M., et al. (2016) The Arabidopsis NPF3 protein is a GA transporter. Nat. Commun. 7: 11486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang F., Barbacioru C., Wang Y., Nordman E., Lee C., Xu N., et al. (2009) mRNA-Seq whole-transcriptome analysis of a single cell. Nat. Methods 6: 377–382. [DOI] [PubMed] [Google Scholar]
- Thaler J.S., Humphrey P.T. and Whiteman N.K. (2012) Evolution of jasmonate and salicylate signal crosstalk. Trends Plant Sci. 17: 260–270. [DOI] [PubMed] [Google Scholar]
- Torii K.U. (2012) Mix-and-match: ligand–receptor pairs in stomatal development and beyond. Trends Plant Sci. 17: 711–719. [DOI] [PubMed] [Google Scholar]
- Tracy R.P. (2008) ‘Deep phenotyping’: characterizing populations in the era of genomics and systems biology. Curr. Opin. Lipidol. 19: 151–157. [DOI] [PubMed] [Google Scholar]
- Trivedi P., Leach J.E., Tringe S.G., Sa T. and Singh B.K. (2020) Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18: 607–621. [DOI] [PubMed] [Google Scholar]
- Tsukanova K.A., Сhеbоtаr V.К., Meyer J.J.M. and Bibikova T.N. (2017) Effect of plant growth-promoting Rhizobacteria on plant hormone homeostasis. South Afr. J. Bot. 113: 91–102. [Google Scholar]
- Vaidya A.S., Helander J.D.M., Peterson F.C., Elzinga D., Dejonghe W., Kaundal A., et al. (2019) Dynamic control of plant water use using designed ABA receptor agonists. Science 366: eaaw8848. [DOI] [PubMed] [Google Scholar]
- van Butselaar T. and Van den Ackerveken G. (2020) Salicylic acid steers the growth–immunity tradeoff. Trends Plant Sci. 25: 566–576. [DOI] [PubMed] [Google Scholar]
- Van Meijl H., Havlik P., Lotze-Campen H., Stehfest E., Witzke P., Domínguez I.P., et al. (2018) Comparing impacts of climate change and mitigation on global agriculture by 2050. Environ. Res. Lett. 13: 064021. [Google Scholar]
- Van Oosten M.J., Pepe O., De Pascale S., Silletti S. and Maggio A. (2017) The role of biostimulants and bioeffectors as alleviators of abiotic stress in crop plants. Chem. Biol. Technol. Agric. 4: 5. [Google Scholar]
- Vanstraelen M. and Benková E. (2012) Hormonal interactions in the regulation of plant development. Annu. Rev. Cell Dev. Biol. 28: 463–487. [DOI] [PubMed] [Google Scholar]
- Vargas-Hernandez M., Macias-Bobadilla I., Guevara-Gonzalez R.G., Romero-Gomez S.D.J., Rico-Garcia E., Ocampo-Velazquez R.V., et al. (2017) Plant hormesis management with biostimulants of biotic origin in agriculture. Front. Plant Sci. 8: 1762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varshney R.K., Sinha P., Singh V.K., Kumar A., Zhang Q. and Bennetzen J.L. (2020) 5Gs for crop genetic improvement. Curr. Opin. Plant Biol. 56: 190–196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Verma V., Ravindran P. and Kumar P.P. (2016) Plant hormone-mediated regulation of stress responses. BMC Plant Biol. 16: 86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vlot A.C., Dempsey D.A. and Klessig D.F. (2009) Salicylic acid, a multifaceted hormone to combat disease. Annu. Rev. Phytopathol. 47: 177–206. [DOI] [PubMed] [Google Scholar]
- Vong K., Eda S., Kadota Y., Nasibullin I., Wakatake T., Yokoshima S., et al. (2019) An artificial metalloenzyme biosensor can detect ethylene gas in fruits and Arabidopsis leaves. Nat. Commun. 10: 5746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos I.A., Moritz L., Pieterse C.M.J. and Van Wees S.C.M. (2015) Impact of hormonal crosstalk on plant resistance and fitness under multi-attacker conditions. Front. Plant Sci. 6: 639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vries F.T., de Griffiths R.I., Knight C.G., Nicolitch O. and Williams A. (2020) Harnessing rhizosphere microbiomes for drought-resilient crop production. Science 369: 270–274. [DOI] [PubMed] [Google Scholar]
- Vukašinović N., Wang Y., Vanhoutte I., Fendrych M., Guo B., Kvasnica M., et al. (2021) Local brassinosteroid biosynthesis enables optimal root growth. Nat. Plants 7: 619–632. [DOI] [PubMed] [Google Scholar]
- Waadt R., Hitomi K., Nishimura N., Hitomi C., Adams S.R., Getzoff E.D., et al. (2014) FRET-based reporters for the direct visualization of abscisic acid concentration changes and distribution in Arabidopsis. eLife 3: e01739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waadt R., Köster P., Andrés Z., Waadt C., Bradamante G., Lampou K., et al. (2020) Dual-reporting transcriptionally linked genetically encoded fluorescent indicators resolve the spatiotemporal coordination of cytosolic abscisic acid and second messenger dynamics in Arabidopsis. Plant Cell 32: 2582–2601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L., Zou Y., Kaw H.Y., Wang G., Sun H., Cai L., et al. (2020) Recent developments and emerging trends of mass spectrometric methods in plant hormone analysis: a review. Plant Methods 16: 54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Tian L., Liu H., Li X., Zhang J., Chen X., et al. (2020) Large-scale discovery of non-conventional peptides in maize and Arabidopsis through an integrated peptidogenomic pipeline. Mol. Plant 13: 1078–1093. [DOI] [PubMed] [Google Scholar]
- Wani S.H., Kumar V., Shriram V. and Sah S.K. (2016) Phytohormones and their metabolic engineering for abiotic stress tolerance in crop plants. Crop J. 4: 162–176. [Google Scholar]
- Wasternack C. and Song S. (2017) Jasmonates: biosynthesis, metabolism, and signaling by proteins activating and repressing transcription. J. Exp. Bot. 68: 1303–1321. [DOI] [PubMed] [Google Scholar]
- Waters M.T., Gutjahr C., Bennett T. and Nelson D.C. (2017) Strigolactone signaling and evolution. Annu. Rev. Plant Biol. 68: 291–322. [DOI] [PubMed] [Google Scholar]
- Wei Z. and Li J. (2020) Regulation of brassinosteroid homeostasis in higher plants. Front. Plant Sci. 11: 583622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiler E.W. (1984) Immunoassay of plant growth regulators. Ann. Rev. Plant Physiol. 35: 85–96. [Google Scholar]
- Werner T., Köllmer I., Bartrina I., Holst K. and Schmülling T. (2006) New insights into the biology of cytokinin degradation. Plant Biol. 8: 371–381. [DOI] [PubMed] [Google Scholar]
- Werner T., Motyka V., Laucou V., Smets R., Van Onckelen H. and Schmülling T. (2003) Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 15: 2532–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson S., Kudoyarova G.R., Veselov D.S., Arkhipova T.N. and Davies W.J. (2012) Plant hormone interactions: innovative targets for crop breeding and management. J. Exp. Bot. 63: 3499–3509. [DOI] [PubMed] [Google Scholar]
- Woodward A.W. and Bartel B. (2005) Auxin: regulation, action, and interaction. Ann. Bot. 95: 707–735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H., Nißler R., Morris V., Herrmann N., Hu P., Jeon S.-J., et al. (2020) Monitoring plant health with near-infrared fluorescent H2O2 nanosensors. Nano Lett. 20: 2432–2442. [DOI] [PubMed] [Google Scholar]
- Xu Z.-Y., Lee K.H., Dong T., Jeong J.C., Jin J.B., Kanno Y., et al. (2012) A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24: 2184–2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Nakanowatari M., Yumoto E., Satoh S. and Asahina M. (2021) Spatiotemporal plant hormone analysis from cryosections using laser microdissection-liquid chromatography-mass spectrometry. J. Plant Res. 135: 377–386. [DOI] [PubMed] [Google Scholar]
- Yang J., Duan G., Li C., Liu L., Han G., Zhang Y., et al. (2019) The crosstalks between jasmonic acid and other plant hormone signaling highlight the involvement of jasmonic acid as a core component in plant response to biotic and abiotic stresses. Front. Plant Sci. 10: 1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang J., Kloepper J.W. and Ryu C.-M. (2009) Rhizosphere bacteria help plants tolerate abiotic stress. Trends Plant Sci. 14: 1–4. [DOI] [PubMed] [Google Scholar]
- Yang N., Chen X., Ren T., Zhang P. and Yang D. (2015) Carbon nanotube based biosensors. Sens. Actuators B Chem. 207: 690–715. [Google Scholar]
- Yu Z., Duan X., Luo L., Dai S., Ding Z. and Xia G. (2020) How plant hormones mediate salt stress responses. Trends Plant Sci. 25: 1117–1130. [DOI] [PubMed] [Google Scholar]
- Zandalinas S.I., Fritschi F.B. and Mittler R. (2021) Global warming, climate change, and environmental pollution: recipe for a multifactorial stress combination disaster. Trends Plant Sci. 26: 588–599. [DOI] [PubMed] [Google Scholar]
- Zhang A., Sun H., Wang P., Han Y. and Wang X. (2012) Modern analytical techniques in metabolomics analysis. Analyst 137: 293–300. [DOI] [PubMed] [Google Scholar]
- Zhang H., Zhu H., Pan Y., Yu Y., Luan S. and Li L. (2014) A DTX/MATE-type transporter facilitates abscisic acid efflux and modulates aba sensitivity and drought tolerance in Arabidopsis. Mol. Plant 7: 1522–1532. [DOI] [PubMed] [Google Scholar]
- Zhang K., Novak O., Wei Z., Gou M., Zhang X., Yu Y., et al. (2014) Arabidopsis ABCG14 protein controls the acropetal translocation of root-synthesized cytokinins. Nat. Commun. 5: 3274. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Ruyter-Spira C. and Bouwmeester H.J. (2015) Engineering the plant rhizosphere. Curr. Opin. Biotechnol. 32: 136–142. [DOI] [PubMed] [Google Scholar]
- Zhao J., Li G., Yi G.-X., Wang B.-M., Deng A.-X., Nan T.-G., et al. (2006) Comparison between conventional indirect competitive enzyme-linked immunosorbent assay (icELISA) and simplified icELISA for small molecules. Anal. Chim. Acta 571: 79–85. [DOI] [PubMed] [Google Scholar]
- Zheng X., Spivey N.W., Zeng W., Liu -P.-P., Fu Z.Q., Klessig D.F., et al. (2012) Coronatine promotes Pseudomonas syringae virulence in plants by activating a signaling cascade that inhibits salicylic acid accumulation. Cell Host Microbe 11: 587–596. [DOI] [PMC free article] [PubMed] [Google Scholar]


