Abstract
Paris polyphylla is often used in Chinese medicine to treat conditions such as carbuncles, trauma, snake bites, and mosquito bites. In the present study, we investigated the effect and mechanism of the morphological transition and extracellular phospholipase activity of Candida albicans treated with polyphyllin I (PPI). First, the minimum inhibitory concentration and antifungal activity of PPI were evaluated using the multiple microdilution method and time-killing assays. Then, the effect of PPI on the morphological transition of Candida albicans in Spider liquid medium and Sabouraud-dextrose liquid medium containing 10% fetal bovine serum was observed under an inverted microscope and by scanning electron microscopy. Finally, egg yolk agar plates were used to evaluate extracellular phospholipase activity. Gene expression was detected by real-time quantitative polymerase chain reaction analysis. Our results suggest that PPI inhibited the transition from the yeast to the hyphal stage and decreased secreted aspartyl proteinase activity. We further confirmed that PPI significantly downregulated the expression of extracellular phospholipase genes and cAMP-PKA signaling pathway-related genes. Taken together, our results suggest that PPI exerts anti-Candida albicans activity by inhibiting virulence characteristics, including the yeast-to-hyphal transition and the secretion of aspartyl proteases and phospholipases. The study results also indicated that PPI could be a promising therapeutic strategy for Candida albicans.
1. Introduction
Candida albicans (C. albicans) is one of the most common opportunistic human fungal pathogens in healthy individuals and causes a wide spectrum of diseases [1, 2], from superficial mucosal infections to life-threatening systemic disorders in immunocompromised human hosts due to virulence factors [3–5], such as protease production and hyphae [6, 7]. As a concurrent infection with other diseases, C. albicans is associated with high morbidity, prolonged hospital stays, high relapse rates, and substantial healthcare costs. The recent emergence of COVID-19 in patients with C. albicans coinfection has been increasingly described in the literature. Among patients with COVID-19 admitted to the intensive care unit (ICU) in one United States (US) hospital, 8.9% developed candidemia, which resulted in longer ICU stays than in patients with COVID-19 without candidemia, with C. albicans being the most common Candida infections [8]. C. albicans can assume at least three distinct morphologies according to the environmental conditions: yeast-like, pseudohyphal, and true hyphal. It adapts well to different environmental niches. Regardless of the specific niche or site of infection, the ability of C. albicans to cause disease has been closely linked to its ability to undergo morphogenic transitions to either pseudohyphal or true hyphal stages [9]. The current therapeutic options for C. albicans are highly limited owing to problems of insufficient drug efficacy, severe toxicity, adverse side effects, high cost, and the emergence of drug-resistant strains to clinically available antifungal drugs [10, 11]. Therefore, the discovery of novel antifungal agents and strategies is necessary.
Natural products have gained increasing attention for their potential use against fungi [12]. The rhizome of Paris polyphylla is called Chong-Lou, and it is a traditional antipyretic detoxicate Chinese medicinal herb, which has been used for a broad range of applications in clinical practice for thousands of years [13]. Paris polyphylla has been applied to manage parotitis, mastitis, sore throat, snakebites, convulsion, fractures, abscesses, and various human malignancies [14]. Polyphyllin I (PPI), as illustrated in Figure 1, also called Chong Lou saponin I, a steroidal saponin, is one of the main active ingredients extracted from Paris polyphylla. In recent years, researchers have investigated the active ingredients and pharmacological mechanisms of PPI. PPI has many pharmacological effects, such as antitumor [15, 16], anti-inflammatory [17], antibacterial [18, 19], anti-Alzheimer's disease, and immune regulation [13]. PPI is also effective against C. albicans, but the antifungal mechanism remains unclear.
Figure 1.

Chemical structure of polyphyllin I (PPI).
2. Materials and Methods
2.1. Strains, Media, and Reagents
CA23, CA187, CA5408, CA3511, CA800, CA602, CA799, CA558, and CA3816 were generous gifts from Prof. Yu-ye Li, The First Affiliated Hospital of Kunming Medical University (Yunnan, China). The fluconazole (FLC)-sensitive C. albicans strain SC5314 was purchased from the American Type Culture Collection (ATCC). ATCC10231 and SC5314 were treated successively with FLC to obtain the FLC-resistantC. albicans strains (ATCC10231FR and SC5314FR, respectively). These strains were refreshed from storage at −80°C and subcultured on Sabouraud-dextrose-agar (SDA) at least twice at 37°C, and a single colony was inoculated into Sabouraud-dextrose-broth (SDB) at 37°C for 16–18 h before each experiment to ensure viability. For all experiments, only cultures in the logarithmic growth phase were used. SDB was used for C. albicans culture, and Spider liquid medium (1% mannitol, 1% nutrient broth, 0.2% K2HPO4) and SD + 10% fetal bovine serum (FBS) liquid medium (10 g of peptone and 40 g of dextrose in 1000 mL of ddH2O supplemented with 10% FBS) were used for hyphal induction. Polyphyllin I (Chonglou Saponin I, purity ≥ 98%) was purchased from Plant Origin Biological, and FLC was purchased from HelioEast Company (Nanchang, China). All drug solutions were dissolved in dimethyl sulfoxide (DMSO) (50 mg/mL). Stock solutions were stored at −20°C until use.
2.2. Antifungal Susceptibility Testing
The MIC50 of PPI and FLC against C. albicans was determined according to Clinical and Laboratory Standards Institute (CLSI) guidelines [20]. Briefly, PPI and FLC were serially diluted 5-fold to final concentrations of 200-0.064 μg/mL. Then, a volume of 100 microliters of 2 × 105 CFU/mL C. albicans (CA23, CA187, CA5408, CA3511, CA800, CA602, CA799, CA558, CA3816, ATCC10231FR, and SC5314FR) were added to 96-well plates. Cell culture wells without drugs were used as experiment controls, and medium-only wells were used as blank controls. The final volume in each well was 200 μL. The plate was then placed at 37°C for 24 hours. Absorbance at 630 nm was measured using a microplate reader. The inhibition ratio of the drugs was calculated as follows: inhibition ratio = [1 − (ODTreated − ODBlank)/(ODControl − ODBlank)] × 100%. MIC50 was calculated using GraphPad Prism 8.0 software. Three biological replicates were performed for each strain, and the experiments were repeated three times.
2.3. Time-Killing Curves
CA23 yeast cells were grown until the log phase, diluted to a final concentration of 1 × 105 CFU/mL, and coincubated with PPI (at final concentrations of 1 μg/mL, 2 μg/mL, and 4 μg/mL) or FLC (at a final concentration of 4 μg/mL) in a 37°C shaking incubator. C. albicans without drug treatment was used as a control. After 0, 2, 4, 8, 12, 24, 36, 48, and 72 h of incubation, the solution in each group at each time period was placed on a mixing shaker to assure complete mixing. Then, 200 μL of samples were placed in a sterile 96-well plate, and OD630 values were determined with a microplate reader. Triplicate wells of each group were assayed, and the experiment was repeated three times. The growth curve was plotted according to the OD630 values at each time point.
2.4. C. albicans Hyphal Morphology Assay
For yeast-to-hyphae transition, yeast cells were induced in Spider liquid medium and SD + 10% FBS liquid medium [21]. A 1 mL suspension of CA23 cells (1 × 105 CFU/mL) treated with FLC (4 μg/mL) or PPI (4 μg/mL, 2 μg/mL, or 1 μg/mL) alone was incubated in 24-well plates at 37°C. At 4 h and 8 h, all wells were observed and photographed using an inverted microscope.
2.5. Scanning Electron Microscopy
The morphology of CA23 cells was observed by scanning electron microscopy (SEM) [22]. Briefly, C. albicans cells (1 × 105 CFU/mL) were coincubated with PPI or FLC in a 6-well flat-bottomed microplate at 37°C for 8 h, and the cells were collected by centrifugation. The cells were washed three times with sterile phosphate-buffered saline (PBS). The resulting suspension (10 μL) was dropped on sterile slides and allowed to dry. Then, the cells were fixed with 5% glutaraldehyde overnight, and the slides were washed gently three times with PBS. The cells were dehydrated in a series of ethanol solutions (30%, 10 min; 70%, 10 min; 90%, 10 min; and 100%, 10 min) and allowed to air dry. The slide was adhered to a metal plate with carbon tape and placed in a high-vacuum sputter coater for gold plating. After the sample was prepared, it was placed in a scanning electron microscope in the high-vacuum mode at 15 kV for observation and image acquisition.
2.6. Effect of Antifungal Agents on Phospholipase Activity
The phospholipase activity of CA23 cells was determined according to a previous method [23]. CA23 cells were harvested in the logarithmic growth phase and diluted to 1 × 105 CFU/mL. A 10 μL sample of the cells (treated and untreated with antifungal agents) was spotted onto the center of egg yolk agar plates containing 1% peptone, 3% glucose, 5.73% NaCl, 0.055% CaCl2, and 10% of egg yolk emulsion and incubated for 72 h at 37°C. Three replicate samples were designed for each group. Then, precipitation zones of different sizes were observed around the colonies, and the diameter of the colonies and precipitation zones was measured with digital Vernier calipers. To assess the effect of antifungal agents on phospholipase activity, Pz values were calculated according to the formula: Pz = colony diameter/(colony diameter + precipitation zone). Pz = 1.00 indicated no activity, Pz = 0.90–0.99 indicated weak enzymatic activity, Pz = 0.70–0.89 indicated moderate activity, and Pz ≤ 0.69 indicated strong enzymatic activity [24]. Therefore, higher Pz values indicated lower C. albicans phospholipase activity. Experiments were performed in triplicate with at least three independent repetitions, and the data obtained were averaged.
2.7. cAMP Rescue Assay
To verify the effect of cAMP on the cAMP-PKA-Efg1 pathway after PPI treatment, C. albicans was prepared in cell culture medium (1 × 105 CFU/mL) and coincubated with PPI (final concentration, 2 μg/mL) or FLC (final concentration, 4 μg/mL) in a 24-well plate at 37°C for 4 h and 8 h with or without dibutyryl-cAMP (db-cAMP). The db-cAMP-free group served as the control. Images were acquired using an inverted microscope.
2.8. Quantitative Real-Time Polymerase Chain Reaction Analysis
To evaluate the molecular mechanism of PPI, quantitative real-time polymerase chain reaction (qRT-PCR) analysis was performed [25, 26]. CA23 cells (1 × 105 CFU/mL) were coincubated with FLC (final concentration, 4 μg/mL) or PPI (final concentration, 2 μg/mL) with shaking at 37°C for 16 h. A drug-free sample served as the growth control. Fungal cells were harvested by centrifugation at 3500 rpm for 5 min and washed with PBS three times. The fungal cells were ground in liquid nitrogen, and total RNA was extracted using Trizol reagent. RNA concentration and purity were determined using a NanoDrop Lite spectrophotometer and by electrophoresis. Total RNA (1 μg) was reverse-transcribed into cDNA with random primers in a 20 μL reaction volume using a GoScript Reverse Transcription Kit following the manufacturer's instructions. After cDNA was synthesized, the expression levels of secreted aspartyl proteinase-related genes (SAP1, SAP2, SAP3, and SAP4) and phospholipase B1, phospholipase B2, and cAMP-PKA signaling pathway-related genes (GPR1, GPA2, CYR1, TPK1, EFG1, ECE1, and HWP1) were assessed by qRT-PCR. qRT-PCR mixtures (20 μL) containing cDNA, GoTaqR qPCR Master Mix, sterile nuclease-free water, and gene primers were freshly prepared. The primer sequences used for the amplification of specific genes are shown in Table 1. Quantitative PCR reactions were performed using a Lightcycler® 96 fluorescence quantitative PCR system (Roche) with the following cycles: 95°C for 60 s for predenaturation, then 95°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s for a total of 40 cycles. The signal from each sample was normalized to ACT1 gene expression. Relative quantitation analysis of the gene expression data was conducted according to the 2−(ΔΔCt) method. Independent experiments were repeated three times with similar results. The data are from one representative experiment.
Table 1.
Primer sequences for qRT-PCR reactions.
| Oligo Name | Sequence (5′to 3′) | Length (bp) |
|---|---|---|
| GPR1-forward | TTCATCTCGCCAGCAACAGT | 178 |
| GPR1-reverse | ATTACATTTCGGTGGGGGCT | |
|
| ||
| GPA2-forward | CCACCACCAAAACAACGCAA | 148 |
| GPA2-reverse | CTTTCACTTCAGGGGTCTCGT | |
|
| ||
| CYR1-forward | ACTTGGTGACTGCAGACTGG | 110 |
| CYR1-reverse | ACCCATACGAACCGACAACC | |
|
| ||
| TPK1-forward | GCTGCCGAAGTATTTTTGGCT | 194 |
| TPK1-reverse | GCCACCACTTCAGGAGCAAT | |
|
| ||
| EFG1-forward | AATGTGGCCCAAATGACACG | 131 |
| EFG1-reverse | GCCATGGCCAATGCTCTTTC | |
|
| ||
| ECE1-forward | GCCACTGGTGTTCAACAATCC | 123 |
| ECE1-reverse | AGTTTCCAGGACGCCATCAA | |
|
| ||
| HWP1-forward | CCGGAATCTAGTGCTGTCGT | 185 |
| HWP1-reverse | GCAGCACCGAAAGTCAATCTC | |
|
| ||
| SAP1-forward | GCTACGCTAACGGTCAACCT | 170 |
| SAP1-reverse | AGCAGCAATGTTTGAAGCAGA | |
|
| ||
| SAP2-forward | CAATGAAGCCGGTGGTAG | 108 |
| SAP2-reverse | GTGGCAGCATCTGGAGAA | |
|
| ||
| SAP3-forward | TCAAGCTGGTCAAGGACAAGA | 196 |
| SAP3-reverse | ATCGGCAAATTGTTGCTTTGTG | |
|
| ||
| SAP4-forward | TGCCGATGGTTCTGTTGC | 154 |
| SAP4-reverse | CCTGGTGGCTTCGTTGCT | |
|
| ||
| PLB1-forward | CATTCAGTGGCGGAGGGTAT | 155 |
| PLB1-reverse | TCCAACTAACCACGATCCACC | |
|
| ||
| PLB2-forward | TGGGAGAGCTTTGAGTCACC | 154 |
| PLB2-reverse | GAGCACAGTGTTTGGTTCCC | |
|
| ||
| ACT1-forward | ACGGTGAAGAAGTTGCTGCT | 180 |
| ACT1-reverse | TGGATTGGGCTTCATCACCA | |
2.9. Cytotoxicity
Drug cytotoxicity tests were evaluated using the MTS (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium) [27, 28]. Human bronchial epithelial cells (16HBE) were obtained from the American Type Culture Collection. They were cultured in Roswell Park Memorial Institute (RPMI) 1640 medium (Gibco) supplemented with 10% FBS and 1% penicillin-streptomycin. 16HBE cells were seeded into 96-well culture plates at a density of 1 × 104 cells per well and incubated for 24 h at 37°C and 5% CO2. The cell culture supernatant was aspirated, and cells were washed and then treated with FLC (4 μg/mL), PPI (2 μg/mL), or DMSO for 24 h. Cell culture wells without drug supplementation were used as the experiment controls, and medium-only served as the blank controls. The cells were further incubated for 24 h before determining cell viability by the MTS assay according to the manufacturer's instructions. Then, the OD values of each well were measured with a microplate reader at 492 nm. The percentage of growth inhibition was calculated as follows: growth inhibition (%) = [1 − (ODexperiment − ODBlank)/(ODcontrol − ODBlank)] × 100% [29].
2.10. Statistical Analysis
All statistical analyses were performed using Prism 8 (GraphPad) software. All experiments were independently repeated at least three times. The data are expressed as the means ± SD of triplicate experiments and differences between groups were evaluated by ANOVA. P values of <0.05 was considered statistically significant.
3. Results
3.1. Antifungal Susceptibility Testing
The results are shown in Table 2. PPI exhibited potent antifungal activity against CA23, CA187, CA5408, CA3511, CA800, CA602, 10231FR, and SC5314FR, with MIC50 values of 0.90–47.30 μg/mL, whereas the MIC50 for FLC of all the tested strains was >200 μg/mL, indicating no antifungal activity. No antifungal effects on the C. albicans clinical strains (CA799, CA558, and CA3816) were observed by treatment with PPI. All data are the average of triplicate experiments. The MIC50 values of the 11 strains were similar, as was the CA23 strain compared to the other 10 strains. CA23 is more stable and easy to culture, so CA23 was selected as the research strain.
Table 2.
MIC50s of PPI and FLC against C. albicans.
| C. albicans | MIC50 (μg/mL) | |
|---|---|---|
| FLC | PPI | |
| CA23 | >200 | 2.00 ± 0.31 |
| CA187 | >200 | 0.95 ± 0.06 |
| CA5408 | >200 | 6.36 ± 0.94 |
| CA3511 | >200 | 4.37 ± 0.01 |
| CA800 | >200 | 3.26 ± 0.17 |
| CA602 | >200 | 46.00 ± 1.30 |
| CA799 | >200 | >200 |
| CA558 | >200 | >200 |
| CA3816 | >200 | >200 |
| ATCC10231-FR | >200 | 0.90 ± 0.01 |
| SC5314-FR | >200 | 0.98 ± 0.01 |
FLC, fluconazole; PPI, polyphyllin I; CA, C. albicans. MIC50 was defined as the 50% inhibition of fungal growth compared to control group growth; the data are shown as mean ± SD.
3.2. Time-Killing Curves
The OD630nm values of C. albicans cultures containing FLC (4 μg/mL) and PPI (1, 2, and 4 μg/mL) were determined to assess the dynamic antifungal effect of PPI on the growth of C. albicans. The results are shown in Figure 2. After 4 h of PPI treatment, there was no significant difference compared to the control group. After 8 h and 12 h of PPI treatment (2 and 4 μg/mL), C. albicans growth was significantly inhibited (P < 0.0001). Twelve hours later, C. albicans treated with PPI (2 μg/mL) grew rapidly and then slowed before reaching a plateau at 24 h. The PPI (4 μg/mL) group exhibited fungicidal activity. Our results showed that PPI dose-dependently inhibited the growth of C. albicans. The OD values were not significantly different between the FLC group and the control group. OD values were measured at different time points of growth and shown on the Y-axis and time on the X-axis in Figure 2. The curves represent the trends in C. albicans growth. The results demonstrated that PPI could actively inhibit C. albicans, which is consistent with the microdilution assay results.
Figure 2.

Growth curve of C. albicans strain CA23 treated with PPI. C. albicans cells were incubated with different doses of PPI at 37°C with shaking. The OD630 of each group was detected at specific time points using a microplate reader. The results are shown as means ± standard deviation (SD). The PPI groups were compared to the growth control group. ∗∗∗∗P < 0.0001.
3.3. C. albicans Hyphal Morphology Assay
The effect of PPI on the hyphal formation of C. albicans in Spider liquid medium (Figure 3(a)) and SD + 10% FBS medium (Figure 3(b)) was observed using an inverted microscope. The control group formed hyphae at 4 h. The control group formed dense and long hyphae in Spider medium at 8 h. The FLC treatment did not significantly inhibit the hyphal formation of C. albicans compared to the control group at any time. The hyphae growth of C. albicans was effectively inhibited after PPI treatment; only yeast-like cells appeared but no obvious hyphae. This result suggests that PPI could effectively inhibit the transition of yeast to hyphae in Spider liquid medium. Interestingly, we found that PPI and FLC did not inhibit serum-induced C. albicans hyphae growth at any time.
Figure 3.

Effect of PPI and FLC on the yeast-to-hyphal transition of C. albicans in two different hyphae-inducing media. (a) Effect of PPI or FLC on CA23 hyphal growth induced by Spider medium at 37°C without shaking for 4 h and 8 h. PPI was diluted in Spider medium to final concentrations of 1, 2, and 4 μg/mL. FLC was diluted in Spider medium to a final concentrations of 4 μg/mL. The untreated strain in medium-only was set as the control. Images were photographed at ×40 magnification. (b) Effect of PPI or FLC on CA23 hyphal growth induced by SD + 10% FBS medium at 37°C without shaking for 4 h and 8 h. Treatments: control group (no treatment, medium-only), FLC group (4 μg/mL), and PPI group (1, 2, and 4 μg/mL). Images were photographed at ×40 magnification. Three independent experiments were performed.
3.4. Scanning Electron Microscopy
To further confirm the antifungal activity of PPI, the morphological appearance of C. albicans was examined by scanning electron microscopy. SEM images (Figure 4) of the untreated control group showed mixtures of pseudohyphae, crisscrossing hyphae, and few yeast cells. Compared to the untreated control, FLC reduced the length of C. albicans hyphae and increased the number of yeast-like cells. C. albicans exposed to 1, 2, and 4 μg/mL PPI only showed yeast cells. Our results show that PPI inhibits the transition from yeast to hyphae.
Figure 4.

Effect of PPI or FLC on the morphological changes of C. albicans in Spider medium. Morphology was observed by SEM (3000x) after incubation at 37°C for 8 h treatments: control group (no treatment, medium-only), FLC group (4 μg/mL), and PPI group (1, 2, and 4 μg/mL).
3.5. Effect of Antifungal Agents on Phospholipase Activity
Lower Pz values indicate higher phospholipid activity. Therefore, the more enzyme that is produced, the lower the Pz value. FLC showed inhibitory effects on CA23 phospholipase activity (Table 3). Phospholipase activity was inhibited by PPI, as the Pz value was 0.85, compared to the very strong activity of the control sample with a Pz value of 0.67. These results indicated that PPI decreased the virulence of C. albicans by repressing phospholipase activity, thereby exerting antifungal effects.
Table 3.
Effect of PPI on the phospholipase activity of CA23.
| Groups | Pz value | Phospholipase activity |
|---|---|---|
| Control | 0.67 ± 0.02 | Strongly |
| FLC | 0.76 ± 0.03 | Moderate |
| PPI | 0.85 ± 0.04 | Moderate |
Each value is the average of three independent experiments ± SD.
3.6. cAMP Rescue Assay
We confirmed that the addition of exogenous db-cAMP rescued the inhibitory effect of PPI on hyphal formation in Spider liquid medium and restored the ability of C. albicans to form hyphae (Figure 5(a)). Thus, PPI could reduce cAMP levels in fungal cells in Spider medium. However, it is interesting to note that PPI did not affect C. albicans hyphal formation in SD + 10% FBS medium (Figure 5(b)) before or after the addition of exogenous cAMP.
Figure 5.
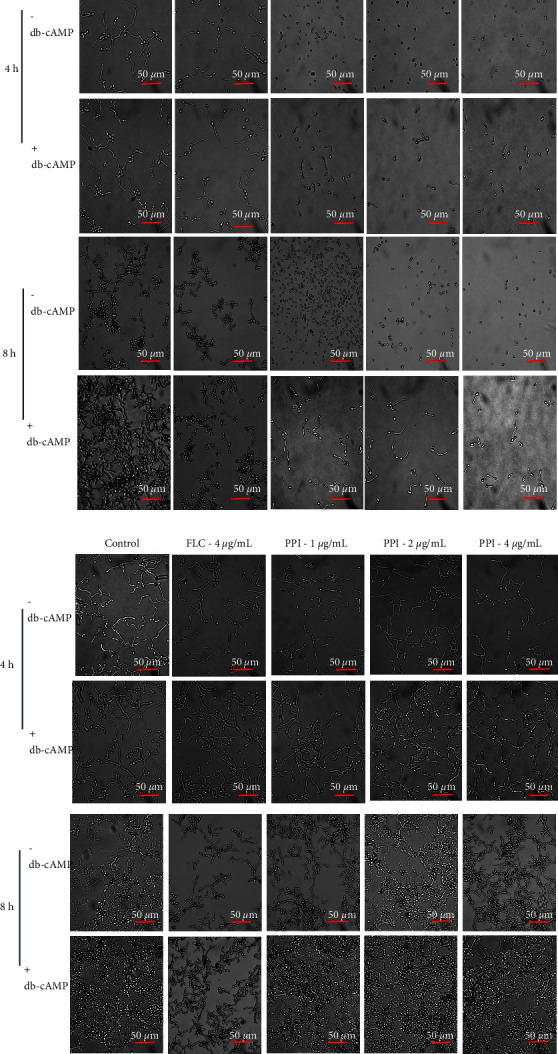
Exogenous cAMP restores the hyphal formation of C . albicans, which is inhibited by PPI. Images were photographed (40x magnification) after incubation in two different hyphae-inducing media at 37°C for 4 h and 8 h. (a) Exogenous cAMP restored PPI-inhibited hyphal formation in Spider medium with or without db-cAMP for 4 h and 8 h. (b) Exogenous cAMP restored PPI-inhibited hyphal formation in SD + 10% FBS medium with or without db-cAMP for 4 h and 8 h.
3.7. Quantitative Real-Time qPCR
The mechanism of filamentation inhibition by PPI was further investigated, and qRT-PCR was conducted to explore the effect of PPI on the cyclic adenosine monophosphate-protein kinase A (cAMP-PKA) pathway-related genes: GPR1, GPA2, CYR1, TPK1, enhanced filamentation growth factor-1 (EFG1), cell elongation protein 1 (ECE1), hyphal wall protein-1 (HWP1), and aspartyl proteases-encoding-related and phospholipase-encoding-related genes. Regarding gene expression changes in C. albicans (Figure 6), cAMP-PKApathway-related genes (GPR1, GPA2, CYR1, TPK1, EFG1, ECE1, and HWP1) were significantly downregulated after PPI treatment compared to the control group. The expression of the protease-encoding secreted aspartyl protease SAP1, SAP2, SAP3, and SAP4 genes, as well as the phospholipase-encoding gene phospholipase B1 (PLB1) and PLB2, were strongly inhibited by PPI.
Figure 6.
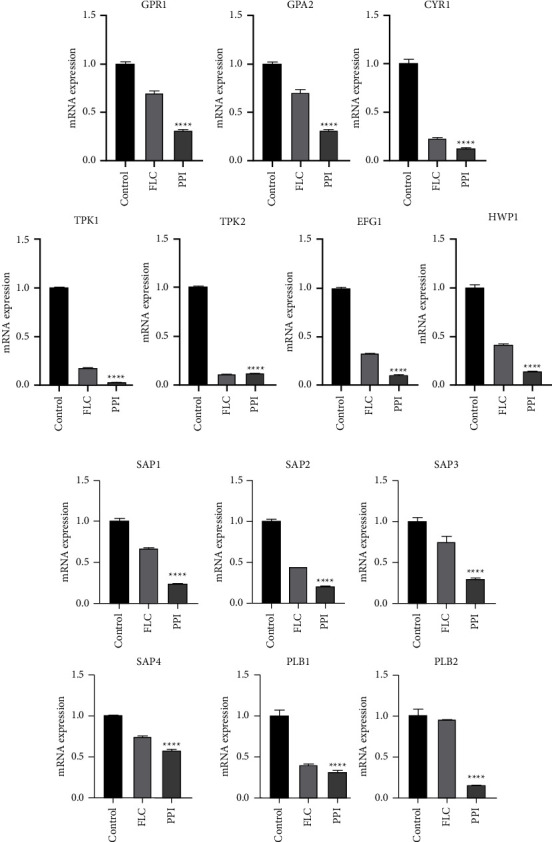
Changes in the expression of cAMP-PKApathway-related genes and hydrolase-related genes in the CA23 strain by PPI treatment. (a) The expression levels of Ras1-cAMP-Efg1 pathway-related genes GPR1, GPA2, CYR1, TPK1, TPK2, EFG1, and HWP1. (b) The expression levels of protease-encoding genes SAP1, SAP2, SAP3, and SAP4, and phospholipase-encoding genes PLB1 and PLB2. Gene expression was detected by qRT-PCR. The untreated strain in medium-only was used as the control. ACT1 was used as the internal reference gene and quantified using the 2−ΔΔCT method. ∗∗∗∗P < 0.0001 compared to the control group.
3.8. Cytotoxicity
To assess cell viability following the treatments, cytotoxicity was analyzed by the MTS assay. PPI exerted inhibitory effects on the proliferation of 16HBE cells at high concentrations. At concentrations ≤10 μg/mL, the toxicity of PPI in 16HBE cells was low, at about 79.26% viability compared to the control groups (Table 4, Figure 7).
Table 4.
Cell survival rates.
| Group | Concentration (μg/mL) | OD values | Survival rate (%) |
|---|---|---|---|
| Blank | — | 0.1134 ± 0.0053 | — |
|
| |||
| Drug control | 40 | 0.1340 ± 0.009 | — |
| 20 | 0.1431 ± 0.0081 | — | |
| 10 | 0.1357 ± 0.0143 | — | |
| 5 | 0.1378 ± 0.0162 | — | |
| 2.5 | 0.1453 ± 0.0197 | — | |
| 1.25 | 0.1403 ± 0.0257 | — | |
|
| |||
| Control | 0 | 1.1460 ± 0.0110 | 100.00 |
|
| |||
| Drug-treated | 40 | 0.5739 ± 0.0754 | 54.59∗∗ |
| 20 | 0.7818 ± 0.0628 | 79.26∗ | |
| 10 | 0.9046 ± 0.0554 | 95.42 | |
| 5 | 0.9658 ± 0.0789 | 102.76 | |
| 2.5 | 0.9706 ± 0.0573 | 102.41 | |
| 1.25 | 0.9852 ± 0.0299 | 104.85 | |
Figure 7.

Effect of PPI on the cell survival rate of 16HBE cells. 16HBE cells were incubated with different concentrations of PPI (1.25, 2.5, 5, 10, 20, and 40 μg/mL) for 24 h, and the cell survival rate was measured by the MTS assay kit. All data are presented as mean ± SD. ∗indicates statistical significance between control and PPI-treated groups.
The data in Table 4 represent the mean ± SD. ∗indicates statistical significance between the control and PPI-treated groups. The blank group contained cell-free medium. The viability of control cells cultured in pure medium was 100%. The drug control group was a drug-containing medium without cells. In the drug-treated group, cells were treated with different concentrations of PPI for 24 h, and the cell survival rate was determined by the MTS assay.
4. Discussion and Conclusion
C. albicans is a dimorphic commensal fungus commonly present in the healthy microbiota of the oral, gut, and vaginal mucosae. It can become pathogenic when the balance between the fungus, mucosa, and host defense mechanisms is interrupted, leading to the emergence of candidiasis [30, 31]. Treating C. albicans is challenging due to its close evolutionary relationship with the human host, the limited efficacy and side effects of antifungal drugs, and the emergence of drug-resistant strains. It poses a threat not only to global health but also to the economy. New antifungal agents that are effective against fungal pathogens are urgently needed [32–34].
The generation of filamentous hyphae is a defining feature of C. albicans pathogenesis [1, 35]. The transition, which depends on its environment, is important for C. albicans infection, colonization, and the evasion of the host immune system. Yeast-form cells facilitate dissemination through the bloodstream, while hyphal cells cause tissue damage and invasion, as well as aid in the escape of phagocytic cells [36]. The cAMP-PKA pathway in C. albicans plays a role in yeast-to-hyphal transition [37]. In the cAMP-PKA pathway, GPR1 (encoding the G protein-coupled receptor Gpr1) acts through the G-protein Gpa2 on the cAMP pathway in response to environmental stimuli. The deletion of CaGPR1 causes strong defects in the yeast-to-hyphal transition on various solid hypha-inducing media [38]. C. albicans expresses a single adenylyl cyclase, which is encoded by CYR1 (also known as CDC35) and can be activated by the G protein Gpa2 to form cAMP [37]. The formation of hyphae is defective in cyr1Δ mutants [39]. cAMP acts as a key second messenger, and the binding of Bcy1 by cAMP activates PKA isoforms thiamin pyrophosphokinase 1 (Tpk1) and Tpk2 to activate Efg1 and other PKA targets. TPK1 and TPK2 encode both isoforms of PKA catalytic subunits, and different cAMP-dependent pathways determine the cellular activity of the catalytic subunits depending on the nature of the inducing medium [40]. Tpk1p is required for the derepression of certain amino acid biosynthetic genes and plays a positive role in the morphogenetic process. Studies showed that the TPK1Δ strain exhibited a delayed morphogenetic shift in several liquid-inducing media compared to the CAI4Δ and TPK2Δ strains [41]. The EFG1 gene, which encodes for enhanced filamentous growth protein 1 (Efg1), is a transcription factor that regulates filamentation, metabolism, biofilm formation, and virulence [42, 43]. Hyphal wall protein 1 (HWP1) and candidalysin (ECE1) are the core filamentation genes and are highly expressed in C. albicans hyphae [44]. Hyphal elongation or yeast-to-hyphal transition has been shown to contribute to fungal invasion, which is mediated by the cell elongation 1 gene (ECE1), or by commensalism, which is mediated by Efg1p during Candida mucosal infections [31]. Efg1 is responsible for the positive regulation of the expression of several hyphal-specific genes, including SAP3-6, ALS3, and HWP1. Mutants lacking Efg1 significantly decreased the expression of SAP3 and SAP5, and the virulence of the strain decreased [45–47]. HWP1, ECE1, and SAP3 were regulated by Efg1. To sum up, hyphal growth is very important for C. albicans host invasion, and the molecular mechanism is relatively clear. Thus, the prevention of hyphal formation is considered an effective treatment option.
This study demonstrated that PPI markedly reduced the hyphal formation of C. albicans at a concentration of 2 μg/mL. The microscopic analysis demonstrated that PPI effectively inhibited hyphal development in a dose-dependent manner. Gene expression analysis revealed that PPI significantly downregulated the expression of hydrolase-related genes and cAMP-PKApathway-related genes, consistent with the phenotypic analysis. The expression of GPR1, GPA2, CYR1, EFG1, AlS3, ECE1, and HWP1 was significantly downregulated by PPI treatment. For further verification, C. albicans treated with PPI was also treated with db-cAMP, a functional analog of cAMP, to evaluate whether it could restore hyphal growth function in culture. db-cAMP partially restored the hyphae-forming ability of C. albicans, consistent with previous studies. Our results suggest that PPI exerts its anti-C. albicans effect mainly by inhibiting the cAMP-PKA pathway. Hyphae also express specific virulence factors, such as degradative enzymes (the Sap family of secreted aspartyl proteases), cell surface adhesins (adhesin agglutinin-like protein 3 (Als3) and Hwp1, and the pore-forming toxin candidalysin (Ece1). During the invasion, pathogenic fungi secrete various hydrolytic enzymes to facilitate host entry. Such secreted fungal enzymes are divided into three major groups: secreted aspartyl proteases, lipases, and phospholipases. Secreted hydrolytic enzymes disrupt host cell membranes, promote cell adhesion and biofilm formation, impair host barrier function, and damage host tissues [48]. Hydrolytic enzymes, aspartyl proteases, and phospholipases secreted by C. albicans are the best characterized [49, 50]. Secreted aspartyl proteinase (Sap) is an extracellular protease secreted by C. albicans. In human mucosal diseases, it is responsible for adhesion and invasion [51]. The transition from round budding cells to long hyphal forms and the production of Saps are considered the virulence-associated factors of C. albicans. Saps are the products of a family of 10 SAP genes divided into subfamilies based on amino acid sequence homology alignment (SAP1 to SAP3, SAP4 to SAP6, SAP9, and SAP10) [52, 53]. SAP1, SAP2, and SAP3 contribute to the overall virulence of C. albicans and presumably play an important role in the process of disseminated infection. SAP4, SAP5, and SAP6 form a group distinct from SAP1, SAP2, and SAP3. When guinea pigs and mice were injected intravenously with the delta saps 4, 5, and 6, triple-homozygous null mutant DSY459, their survival time was significantly longer than that of control animals infected with wild-type SC5314 [47, 54]. Saps 1–6 are required for invasive disease. In guinea pig and murine models of invasive disease, deletions in Sap1–6 attenuated virulence [47]. The SAP family genes encoding proteins Sap1, Sap2, Sap3, and Sap4 are required for hyphal formation and maintenance. Gene expression analysis revealed that PPI significantly downregulated the expression of Sap1, Sap2, Sap3, and Sap4, indicating that PPI could decrease the production of Saps and thus the virulence of the strain.
C. albicans phospholipase is an important virulence factor. In recent years, phospholipase has been increasingly reported [55]. Phospholipases are divided into four major classes, A, B, C, and D, according to the specific bond targeted in the phospholipid molecule. Phospholipase B mainly hydrolyzes lysophospholipid bonds [56]. The relative risk of death was 5.6-fold higher in mice infected with higher-phospholipase-secreting strains than with the low-phospholipase secretors [57]. Phospholipase B contributes to the pathogenicity of C. albicans by traversing host cell membranes, a process that may increase the rate of infection dissemination [58]. PLB1, which codes for phospholipase B/lysophospholipase in yeast, was required for virulence in an animal model of candidiasis; a gene-deleted strain produced less phospholipase in vitro and was less virulent than the wild-type [56, 59]. The study found that sodium houttuynate inhibited the relative expression of PLB1 and PLB2, thus exerting anti-C. albicans activity [60]. Our results also showed that PPI could decrease the relative expression of PLB1 and PLB2, inhibit the secretion of phospholipase by C. albicans, and reduce the virulence of the strain.
In summary, PPI converted the pathogenic hyphal form to the less-virulent yeast state of C. albicans by inhibiting the expression of hyphae-related virulence factors. Thus, PPI can be used not only for the prevention of C. albicans infections but also as an effective therapeutic agent.
Acknowledgments
The research was supported by the Yunnan Province Science and Technology Department (grant no. 202101AF070001) and (grant no. 202103AC100005).
Data Availability
The data used to support the findings of this study are included in the article.
Conflicts of Interest
The authors declare that they have no conflicts of interest.
Authors' Contributions
Rui-Rui Wang and Li Li participated in the research design. Ai-Mei Sun, Ying-Xian Wang, and Guo-Xian-Hu conducted experiments and performed data analysis. Ai-Mei Sun and Rui-Rui Wang were in charge of writing the manuscript. Ai-Mei Sun and Ying-Xian Wang contributed equally to this work.
References
- 1.Ho J., Yang X., Nikou S. A., et al. Candidalysin activates innate epithelial immune responses via epidermal growth factor receptor. Nature Communications . 2019;10(1):p. 2297. doi: 10.1038/s41467-019-09915-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chaillot J., Tebbji F., Mallick J., Sellam A. Integration of growth and cell size via the TOR pathway and the Dot6 transcription factor in Candida albicans. Genetics . 2019;211(2):637–650. doi: 10.1534/genetics.118.301872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fidel P. L., Sobel J. D., And J. D. Immunopathogenesis of recurrent vulvovaginal candidiasis. Clinical Microbiology Reviews . 1996;9(3):335–348. doi: 10.1128/cmr.9.3.335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pereira R., Santos F. R., Brito E., Morais S. Biofilm of Candida albicans: formation, regulation and resistance. Journal of Applied Microbiology . 2021;131(1):11–22. doi: 10.1111/jam.14949. [DOI] [PubMed] [Google Scholar]
- 5.Ponde N. O., Lortal L., Ramage G., Naglik J. R., Richardson J. P. Candida albicans biofilms and polymicrobial interactions. Critical Reviews in Microbiology . 2021;47(1):91–111. doi: 10.1080/1040841x.2020.1843400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sudbery P. E. Growth of Candida albicans hyphae. Nature Reviews Microbiology . 2011;9(10):737–748. doi: 10.1038/nrmicro2636. [DOI] [PubMed] [Google Scholar]
- 7.Calderone R. A., Fonzi W. A. Virulence factors of Candida albicans. Trends in Microbiology . 2001;9(7):327–335. doi: 10.1016/s0966-842x(01)02094-7. [DOI] [PubMed] [Google Scholar]
- 8.Seagle E. E., Jackson B. R., Lockhart S. R., et al. The landscape of candidemia during the coronavirus disease 2019 (COVID-19) pandemic. Clinical Infectious Diseases . 2022;74(5):802–811. doi: 10.1093/cid/ciab562. [DOI] [PubMed] [Google Scholar]
- 9.Wakade R. S., Ristow L. C., Stamnes M. A., Kumar A., Krysan D. J. The ndr/LATS kinase Cbk1 regulates a specific subset of Ace2 functions and suppresses the hypha-to-yeast transition in Candida albicans. mBio . 2020;11(4):p. 20. doi: 10.1128/mbio.01900-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lee Y., Puumala E., Robbins N., Cowen L. E. Antifungal drug resistance: molecular mechanisms in Candida albicans and beyond. Chemistry Review . 2021;121(6):3390–3411. doi: 10.1021/acs.chemrev.0c00199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mount H. O., Revie N. M., Todd R. T., et al. Global analysis of genetic circuitry and adaptive mechanisms enabling resistance to the azole antifungal drugs. PLoS Genetics . 2018;14(4) doi: 10.1371/journal.pgen.1007319.e1007319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Perfect J. R. The antifungal pipeline: a reality check. Nature Reviews Drug Discovery . 2017;16(9):603–616. doi: 10.1038/nrd.2017.46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tian Y., Gong G. Y., Ma L. L., Wang Z. Q., Song D., Fang M. Y. Anti-cancer effects of Polyphyllin I: an update in 5 years. Chemico-Biological Interactions . 2020;316 doi: 10.1016/j.cbi.2019.108936.108936 [DOI] [PubMed] [Google Scholar]
- 14.Negi J., Bisht V., Bhandari A., Bhatt V., Singh P., Singh N. Paris polyphylla: chemical and biological prospectives. Anti-Cancer Agents in Medicinal Chemistry . 2014;14(6):833–839. doi: 10.2174/1871520614666140611101040. [DOI] [PubMed] [Google Scholar]
- 15.Shen Z., Wang J., Ke K., et al. Polyphyllin I, a lethal partner of Palbociclib, suppresses non-small cell lung cancer through activation of p21/CDK2/Rb pathway in vitro and in vivo. Cell Cycle . 2021;20(23):2494–2506. doi: 10.1080/15384101.2021.1991121. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 16.Lai L., Shen Q., Wang Y., et al. Polyphyllin I reverses the resistance of osimertinib in non-small cell lung cancer cell through regulation of PI3K/Akt signaling. Toxicology and Applied Pharmacology . 2021;419 doi: 10.1016/j.taap.2021.115518.115518 [DOI] [PubMed] [Google Scholar]
- 17.Cai Y., Xie F., Wang H., Zhai Q., Lin F., Pan S. Polyphyllin I alleviates inflammatory injury in mice with gestational diabetes through AMPK pathway. General Physiology and Biophysics . 2022;41(2):159–164. doi: 10.4149/gpb_2022006. [DOI] [PubMed] [Google Scholar]
- 18.Meng Tingting L. J., Li L., He Y. Study on the Antibacterial Effect of Heavy Building . Vol. 11. Shijiazhuang, Technology wind; 2018. p. p. 166. [Google Scholar]
- 19.He Yinsheng Q. J., Yu Li, Yuan M., He M., Zhang M., Xu Y. Study on the volatile components and antibacterial activity of three medicinal plants of the genus Chonglou. Chinese Archives of Traditional Chinese Medicine . 2022;10 [Google Scholar]
- 20.Li X. N., Zhang L. M., Wang Y. Y., et al. SWL-1 reverses fluconazole resistance in Candida albicans by regulating the glycolytic pathway. Frontiers in Microbiology . 2020;11 doi: 10.3389/fmicb.2020.572608.572608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Su L. Y., Ni G. H., Liao Y. C., et al. Antifungal activity and potential mechanism of 6, 7, 4’-O-triacetylscutellarein combined with fluconazole against drug-resistant C. albicans. Frontiers in Microbiology . 2021;12 doi: 10.3389/fmicb.2021.692693.692693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wu J., Wu D., Zhao Y., et al. Sodium new houttuyfonate inhibits Candida albicans biofilm formation by inhibiting the ras1-cAMP-efg1 pathway revealed by RNA-seq. Frontiers in Microbiology . 2020;11:p. 2075. doi: 10.3389/fmicb.2020.02075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vaňková E., Kašparová P., Dulíčková N., Čeřovský V. Combined effect of lasioglossin LL-III derivative with azoles against Candida albicans virulence factors: biofilm formation, phospholipases, proteases and hemolytic activity. FEMS Yeast Research . 2020;20(3) doi: 10.1093/femsyr/foaa020.foaa020 [DOI] [PubMed] [Google Scholar]
- 24.Neji S., Hadrich I., Trabelsi H., et al. Virulence factors, antifungal susceptibility and molecular mechanisms of azole resistance among Candida parapsilosis complex isolates recovered from clinical specimens. Journal of Biomedical Science . 2017;24(1):p. 67. doi: 10.1186/s12929-017-0376-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Song Y. D., Hsu C. C., Lew S. Q., Lin C. H. Candida tropicalis RON1 is required for hyphal formation, biofilm development, and virulence but is dispensable for N-acetylglucosamine catabolism. Medical Mycology . 2021;59(4):379–391. doi: 10.1093/mmy/myaa063. [DOI] [PubMed] [Google Scholar]
- 26.Qadri H., Haseeb S. A., Ahmad M. M., Fazal Q. M., Prasad R. Quinidine drug resistance transporter knockout Candida cells modulate glucose transporter expression and accumulate metabolites leading to enhanced azole drug resistance. Fungal Genetics and Biology . 2022;161 doi: 10.1016/j.fgb.2022.103713.103713 [DOI] [PubMed] [Google Scholar]
- 27.Zhao Z., Li Y., Liu H., Jain A., Patel P. V., Cheng K. Co-delivery of IKBKE siRNA and cabazitaxel by hybrid nanocomplex inhibits invasiveness and growth of triple-negative breast cancer. Science Advances . 2020;6(29) doi: 10.1126/sciadv.abb0616.eabb0616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ni K., Lan G., Guo N., et al. Nanoscale metal-organic frameworks for x-ray activated in situ cancer vaccination. Science Advances . 2020;6(40) doi: 10.1126/sciadv.abb5223.eabb5223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park S., Ou J., Chamberlain L., et al. U2AF35(S34F) promotes transformation by directing aberrant ATG7 pre-mRNA 3’ end formation. Molecular Cell . 2016;62(4):479–490. doi: 10.1016/j.molcel.2016.04.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hermann P., Berek Z., Nagy G., Kamotsay K., Rozgonyi F. Molecular pathogenesis of oral candidiasis (candidosis) Orvosi Hetilap . 2001;142(47):2621–2625. [PubMed] [Google Scholar]
- 31.Iliev I. D., Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nature Reviews Immunology . 2017;17(10):635–646. doi: 10.1038/nri.2017.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Shapiro R. S., Chavez A., Porter C. B. M., et al. A CRISPR-Cas9-based gene drive platform for genetic interaction analysis in Candida albicans. Nature microbiology . 2017;3(1):73–82. doi: 10.1038/s41564-017-0043-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Iyer K. R., Camara K., Daniel-Ivad M., et al. An oxindole efflux inhibitor potentiates azoles and impairs virulence in the fungal pathogen Candida auris. Nature Communications . 2020;11(1):p. 6429. doi: 10.1038/s41467-020-20183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lehrnbecher T., Fisher B. T., Phillips B., et al. Clinical practice guideline for systemic antifungal prophylaxis in pediatric patients with cancer and hematopoietic stem-cell transplantation recipients. Journal of Clinical Oncology . 2020;38(27):3205–3216. doi: 10.1200/jco.20.00158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Witchley J. N., Penumetcha P., Abon N. V., Woolford C. A., Mitchell A. P., Noble S. M. Candida albicans morphogenesis programs control the balance between gut commensalism and invasive infection. Cell Host & Microbe . 2019;25(3):432–443 e6. doi: 10.1016/j.chom.2019.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Maciel E. I., Jiang C., Barghouth P. G., Nobile C. J., Oviedo N. J. The planarian Schmidtea mediterranea is a new model to study host-pathogen interactions during fungal infections. Developmental & Comparative Immunology . 2019;93:18–27. doi: 10.1016/j.dci.2018.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen H., Zhou X., Ren B., Cheng L. The regulation of hyphae growth in Candida albicans. Virulence . 2020;11(1):337–348. doi: 10.1080/21505594.2020.1748930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Maidan M. M., De Rop L., Serneels J., et al. The G protein-coupled receptor Gpr1 and the Galpha protein Gpa2 act through the cAMP-protein kinase A pathway to induce morphogenesis in Candida albicans. Molecular Biology of the Cell . 2005;16(4):1971–1986. doi: 10.1091/mbc.e04-09-0780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Parrino S. M., Si H., Naseem S., Groudan K., Gardin J., Konopka J. B. cAMP-independent signal pathways stimulate hyphal morphogenesis in Candida albicans. Molecular Microbiology . 2017;103(5):764–779. doi: 10.1111/mmi.13588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bockmuhl D. P., Krishnamurthy S., Gerads M., Sonneborn A., Ernst J. F. Distinct and redundant roles of the two protein kinase A isoforms Tpk1p and Tpk2p in morphogenesis and growth of Candida albicans. Molecular Microbiology . 2002;42(5):1243–1257. doi: 10.1046/j.1365-2958.2001.02688.x. [DOI] [PubMed] [Google Scholar]
- 41.Souto G., Giacometti R., Silberstein S., Giasson L., Cantore M. L., Passeron S. Expression of TPK1 and TPK2 genes encoding PKA catalytic subunits during growth and morphogenesis in Candida albicans. Yeast . 2006;23(8):591–603. doi: 10.1002/yea.1377. [DOI] [PubMed] [Google Scholar]
- 42.Ramage G., VandeWalle K., Lã³pez-Ribot J. L., Wickes B. L., Chen L. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development inCandida albicans. FEMS Microbiology Letters . 2002;214(1):95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- 43.Ramage G., Vandewalle K., Lã³pez-Ribot J. L., Wickes B. L. The filamentation pathway controlled by the Efg1 regulator protein is required for normal biofilm formation and development inCandida albicans. FEMS Microbiology Letters . 2002;214(1):95–100. doi: 10.1111/j.1574-6968.2002.tb11330.x. [DOI] [PubMed] [Google Scholar]
- 44.Muñoz J. F., Gade L., Chow N. A., et al. Genomic insights into multidrug-resistance, mating and virulence in Candida auris and related emerging species. Nature Communications . 2018;9(1):p. 5346. doi: 10.1038/s41467-018-07779-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Leng P., Lee P. R., Wu H., Brown A. J. P. Efg1, a morphogenetic regulator in Candida albicans, is a sequence-specific DNA binding protein. Journal of Bacteriology . 2001;183(13):4090–4093. doi: 10.1128/jb.183.13.4090-4093.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Moazeni M., Khoramizadeh M. R., Teimoori-Toolabi L., Noorbakhsh F., Rezaie S. The effect of EFG1 gene silencing on down-regulation of SAP5 gene, by use of RNAi technology. Acta Medica Iranica . 2014;52(1):9–14. [PubMed] [Google Scholar]
- 47.Hube B., Sanglard D., Odds F. C., et al. Disruption of each of the secreted aspartyl proteinase genes SAP1, SAP2, and SAP3 of Candida albicans attenuates virulence. Infection and Immunity . 1997;65(9):3529–3538. doi: 10.1128/iai.65.9.3529-3538.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tóth R. N., Nosek J., Mora-Montes H. M., et al. Candida parapsilosis: from genes to the bedside. Clinical Microbiology Reviews . 2019;32(2):18. doi: 10.1128/cmr.00111-18.e00111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Penha S. S., Birman E. G., Silveira F. R. X. d., Paula C. R. d. Frequency and enzymatic activity (proteinase and phospholipase) of Candida albicans from edentulous patients, with and without denture stomatitis. Pesquisa Odontológica Brasileira . 2000;14(2):119–122. doi: 10.1590/s1517-74912000000200005. [DOI] [Google Scholar]
- 50.Riceto É. B. d. M., Menezes R. d. P., Penatti M. P. A., Pedroso R. d. S. Enzymatic and hemolytic activity in different Candida species. Revista Iberoamericana De Micologia . 2015;32(2):79–82. doi: 10.1016/j.riam.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 51.Hoegl L., Ollert M., Korting H. C. The role of Candida albicans secreted aspartic proteinase in the development of candidoses. Journal of Molecular Medicine (Berlin) . 1996;74(3):135–142. doi: 10.1007/bf01575445. [DOI] [PubMed] [Google Scholar]
- 52.Staniszewska M., Bondaryk M., Siennicka K., et al. In vitro study of secreted aspartyl proteinases Sap1 to Sap3 and Sap4 to Sap6 expression in Candida albicans pleomorphic forms. Polish Journal of Microbiology . 2012;61(4):247–256. doi: 10.33073/pjm-2012-034. [DOI] [PubMed] [Google Scholar]
- 53.Tatsuki Sato T. W., Takeshi M. I. K. A. M. I., Matsumoto T. Farnesol, a morphogenetic autoregulatory substance in the dimorphic fungus Candida albicans, inhibits hyphae growth through suppression of a mitogen-activated protein kinase cascade. Biol. Pharm . 2004;10(1) doi: 10.1248/bpb.27.751. [DOI] [PubMed] [Google Scholar]
- 54.Sanglard D., Hube B., Monod M., Odds F. C., Gow N. A. A triple deletion of the secreted aspartyl proteinase genes SAP4, SAP5, and SAP6 of Candida albicans causes attenuated virulence. Infection and Immunity . 1997;65(9):3539–3546. doi: 10.1128/iai.65.9.3539-3546.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cotter G., Kavanagh K. Adherence mechanisms of Candida albicans. British Journal of Biomedical Science . 2000;57(3):241–249. [PubMed] [Google Scholar]
- 56.Ghannoum M. A. Potential role of phospholipases in virulence and fungal pathogenesis. Clinical Microbiology Reviews . 2000;13(1):122–143. doi: 10.1128/cmr.13.1.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ibrahim A. S., Mirbod F., Filler S. G., et al. Evidence implicating phospholipase as a virulence factor of Candida albicans. Infection and Immunity . 1995;63(5):1993–1998. doi: 10.1128/iai.63.5.1993-1998.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Leidich S. D., Ibrahim A. S., Fu Y., et al. Cloning and disruption of caPLB1, a phospholipase B gene involved in the pathogenicity of Candida albicans. Journal of Biological Chemistry . 1998;273(40):26078–26086. doi: 10.1074/jbc.273.40.26078. [DOI] [PubMed] [Google Scholar]
- 59.Merkel O., Fido M., Mayr J. A., et al. Characterization and function in vivo of two novel phospholipases B/lysophospholipases fromSaccharomyces cerevisiae. Journal of Biological Chemistry . 1999;274(40):28121–28127. doi: 10.1074/jbc.274.40.28121. [DOI] [PubMed] [Google Scholar]
- 60.Ying X. U., Wen-Jing M. O., Shuang F. U., Stomatology S. O., University J. Effect of sodium houttuyfonate on Candida albicans phospholipase in vitro. Journal of Oral Science Research . 2014;30(5):396–395. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data used to support the findings of this study are included in the article.


