Abstract
Depending on sequence, bacterial and synthetic DNAs can activate the host immune system and influence the host response to infection. The purpose of this study was to determine the abilities of various phosphorothioate oligonucleotides with cytosine-guanosine-containing motifs (CpG DNA) to activate macrophages to produce nitric oxide (NO) and prostaglandin E2 (PGE2) and to induce expression of NO synthase 2 (NOS2) and cyclooxygenase 2 (COX2). As little as 0.3 μg of CpG DNA/ml increased NO and PGE2 production in a dose- and time-dependent fashion in cells of the mouse macrophage cell line J774. NO and PGE2 production was noted by 4 to 8 h after initiation of cultures with the CpG DNA, with the kinetics of NO production induced by CpG DNA being comparable to that induced by a combination of lipopolysaccharide and gamma interferon. CpG DNA-treated J774 cells showed enhanced expression of NOS2 and COX2 proteins as determined by immunoblotting, with the relative potencies of the CpG DNAs generally corresponding to those noted for the induction of NO and PGE2 production as well as to those noted for the induction of interleukin-6 (IL-6), IL-12, and tumor necrosis factor. Extracts from CpG DNA-treated cells converted l-arginine to l-citrulline, but the NOS inhibitor NG-monomethyl-l-arginine (NMMA) inhibited this reaction. The COX2-specific inhibitor NS398 inhibited CpG DNA-induced PGE2 production and inhibited NO production to various degrees. The NOS inhibitors NMMA, 1400W, and N-iminoethyl-l-lysine effectively blocked NO production and increased the production of PGE2 in a dose-dependent fashion. Thus, analogues of microbial DNA (i.e., CpG DNA) activate mouse macrophage lineage cells for the expression of NOS2 and COX2, with the production of NO and that of PGE2 occurring in an interdependent manner.
DNA is a complex macromolecule whose immunological properties vary with base sequence and backbone structure. While mammalian DNA is inactive, DNA from bacteria can induce potent immunological effects (12, 18, 32). Bacterial DNA can activate several different cell types (e.g., B lymphocytes, macrophages, and dendritic cells). As has been shown through the use of synthetic oligodeoxynucleotides (ODNs), the activity of bacterial DNA results from the presence of short, 6-base sequence motifs characterized by an unmethylated cytosine-guanosine dinucleotide within the context of flanking bases of two 5′ purines and two 3′ pyrimidines. These motifs are called immunostimulatory stimulatory sequences or CpG motifs, with DNA containing these sequences being termed CpG DNA. Because of its immune stimulatory properties, CpG DNA can activate innate immunity. In addition to being important in the context of infection and inflammation, these properties are relevant to the activities of DNA vaccines and to the use of synthetic DNA as an adjuvant and immunomodulator (13).
While CpG DNA has beneficial effects in stimulating host defense, it can also induce inflammation and may promote tissue injury during infection or local administration. For example, CpG DNA from bacteria (or synthetic CpG DNA) causes severe arthritis when it is injected into the joints of mice. This arthritis is characterized by synovial tumor necrosis factor (TNF) mRNA expression and monocyte influx (3, 4). Similarly, the instillation of CpG DNA into the lungs of mice elicits a marked exudate and inflammatory reaction (23). The proinflammatory effects of CpG DNA may result from the production of cytokines (e.g., TNF-α), chemokines, and other mediators such as nitric oxide (NO). The contribution of these various mediators in the response to CpG is not well understood and may reflect both local and systemic effects.
To further characterize immune activities of CpG DNA in the present study, we investigated its effects on macrophages for the production of two important mediators (NO and prostaglandins [PGs]). NO is a gaseous molecule with widespread biological activities, including inflammation, while the PGs have a number of important immunomodulatory activities. As a model for CpG DNA, we have used synthetic phosphorothioate ODNs (Ps ODNs) containing CpG motifs and tested their abilities to induce the production of NO and PG E2 (PGE2) by J774 mouse macrophage cell line cells. These ODNs contain the substitution of a sulfur for one of the nonbridging oxygens in the phosphodiester backbone. This substitution leads to nucleic resistance to degradation and increased immunological activities. Ps ODNs are now being investigated as adjuvants as well as immunomodulators to promote host defense and alter the Th1/Th2 balance.
We report here that certain Ps ODNs induce NO and PGE2 production as well as NO synthase 2 (NOS2) and cyclooxygenase 2 (COX2) expression by these cells in a time- and dose-dependent fashion. Furthermore, we show that inhibition of NO production leads to an increase in ODN-stimulated PGE2 production. These results indicate that pathways for the production of NO and PGE2 may be interdependent. Since NO and PGE2 can modulate immune responses, the in vivo and in vitro activities of CpG DNA may reflect the actions of these mediators, as well as those of induced cytokines and chemokines.
MATERIALS AND METHODS
Cells and culture methods.
Cells of the mouse macrophage cell lines J774 and RAW264 (American Type Culture Collection, Manassas, Va.) were used as models of macrophages. They were cultured in Dulbecco's modified Eagle medium with 10% heat-inactivated fetal bovine serum. For these experiments, cells were generally seeded into 11-mm-diameter wells of 24-well plates at a concentration of 106 cells in 1 ml of medium with the various additives. For selected experiments as noted in Results, some inhibitors were cultured with the cells before addition of CpG DNA.
Materials.
Endotoxin-free media were from GIBCO-BRL (Gaithersburg, Md.). The Ps ODNs (Table 1) were purchased from Midland Certified Reagent Company (Midland, Tex.). All ODNs had undetectable endotoxin (lipopolysaccharide [LPS]) contents as determined by the Limulus amebocyte lysate assay (<0.1 endotoxin unit [EU]/ml). Escherichia coli LPS was from Sigma-Aldrich (St. Louis, Mo.), and gamma interferon (IFN-γ) was from R&D Systems (Minneapolis, Minn.). The NOS-nonspecific inhibitor NG-monomethyl-l-arginine (NMMA) and the NOS2-specific inhibitors N-iminoethyl-l-lysine (l-NIL) and 1400W were from Alexis Biochemicals (San Diego, Calif.), and the COX2-specific inhibitor NS398 was from Cayman Chemicals (Ann Arbor, Mich.). All other chemicals were from Sigma-Aldrich.
TABLE 1.
CpG ODNsa
| CpG ODN | Nucleotide sequence |
|---|---|
| 74 | A12AACGTTA12 |
| 75 | G12AACGTTG12 |
| 115 | T12AACGTTT12 |
| 139 | C12AACGTTC12 |
| SAK2 | TCCATGACGTTCCTGACGTT |
| SAK1 | TCCATGAGCTTCCTGAGTCT |
Nucleotide sequences are listed, and CpG is denoted by a boldface CG. All nucleotides have a Ps backbone.
NO, PG, and cytokine assays.
The NO oxidation products nitrate and nitrite (NOx) were measured using nitrate reductase and the Griess method as described before (33). PGE2, interleukin-6 (IL-6), IL-12 p40/p70, and TNF were measured using enzyme-linked immunoassays (R&D Systems).
NOS enzyme assay and immunoblots.
Cells were harvested by scraping, washed, and suspended in a buffer containing 1 mM phenylmethylsulfonyl fluoride, 5 μg of aprotinin/ml, 1 μg of chymostatin/ml, and 5 μg of pepstatin A/ml. Cells were then lysed by 3 cycles of freezing and thawing. The lysate was centrifuged at 14,000 × g, and the supernatant was assayed (24). Protein content was determined by the Bradford assay (Bio-Rad, Hercules, Calif.). NOS activity was determined by an assay converting l-[14C]arginine to l-[14C]citrulline as noted previously (34). In brief, the assay buffer contained 50 mM HEPES (pH 7.5), 200 μM NADPH, 1 mM dithiothreitol, 10 μM flavin adenine dinucleotide, 100 μM tetrahydrobiopterin, and 10 μM l-arginine with l-[14C]arginine labeled in the guanido position (NEN, Wilmington, Del.). The specificity of the reaction was determined by inhibition with NMMA.
For immunoblots, cells were lysed in 50 μl of 40 mM EPPS (N-hydroxyethyl]piperazine-N′-[3-propanesulfonic acid) buffer containing 10% glycerol, 150 mM NaCl, 50% Beeper II detergent (Pierce Chemicals, Rockford, Ill.), 1 mM phenylmethylsulfonyl fluoride, and leupeptin and aprotinin (5 μg/ml each) by incubating on ice with occasional shaking for 30 min. The lysate was centrifuged at 14,000 × g, and the supernatant was analyzed by immunoblotting as noted above by using the ECL technique (Amersham, Piscataway, N.J.). Anti-mouse NOS2 and COX2 antibodies were from Transduction Laboratories (Lexington, Ky.).
RESULTS
NO and PGE2 production and NOS2 and COX2 expression.
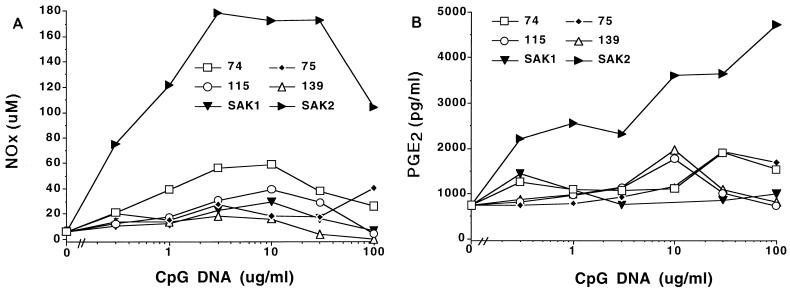
To assess the effects of CpG DNA on the production of NO and PGE2, we treated J774 cells with a panel of Ps ODNs and assessed mediator production. As shown in Fig. 1, certain of the ODNs tested increased the production of both NO and PGE2. Increased production was noted with as little as 0.3 μg of CpG DNA/ml and occurred without preactivation of the cells with either LPS or IFN-γ. Activation of the cells for NO and PGE2 production was sequence specific, with the 74, 75, 115, and SAK2 ODNs showing the highest activity. SAK2 was the most potent inducer of NO and PGE2 production. SAK1, an ODN that does not contain a CpG motif, was generally the least effective of the agents. CpG DNA enhanced NO and PGE2 production by cells of the mouse macrophage line RAW 264 (data not shown) as well as by J774 cells.
FIG. 1.
NOx (A) and PGE2 (B) production by J774 cells after stimulation with CpG DNAs. Each symbol represents the mean of results for triplicate samples. Cells were cultured with the indicated additives, and supernatants were analyzed after 48 h of culture. These results are from one experiment, which is representative of four that were done. Variation was small, and error bars fall within the areas of the symbols.
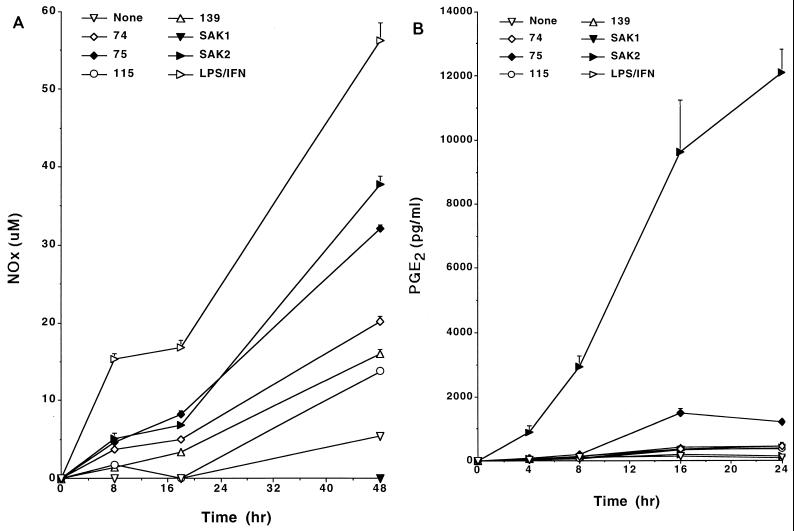
We tested the kinetics of the response to analyze further the production of these mediators and the relationship of this response to responses induced by other stimulants (Fig. 2A and B). As these data indicate, production of NO and PGE2 was noted by 4 to 8 h after initiation of cultures with SAK2, with the kinetics of NO production induced by CpG DNA being comparable to that induced by LPS–IFN-γ. Under these conditions, however, LPS, IFN-γ, and LPS–IFN-γ generally induced very little PGE2 production or none at all.
FIG. 2.
Time course for production of NOx (A) and PGE2 (B) by J774 cells after stimulation with CpG DNAs. Each symbol represents the mean of results from triplicate samples. Cells were cultured with 10 μg of the indicated ODN/ml and with 100 ng of LPS/ml plus 100 U of IFN-γ/ml. Supernatant media were assayed at the indicated times. These results are from one experiment, which is representative of three that were done. Error bars show the SEMs. Some SEMs fall within the areas of the symbols and are not seen.
Production of NO was paralleled by induction of NOS enzyme activity. Lysates of SAK2- or LPS–IFN-γ-simulated J774 cells had an increased ability to convert l-arginine to l-citrulline, while SAK1-stimulated cells showed little or no increase in their ability. For example, NOS activity in lysates of control cells was 659 ± 29 pmol of l-citrulline/mg (mean ± standard error of the mean [SEM]); in cells treated with LPS–IFN-γ, it was 1,438 ± 58; in cells treated with SAK2, it was 1,609 ± 90; and in cells treated with SAK1, it was 714 ± 38. Increases in NO production induced by the ODNs 74, 75, 115, 139, and SAK2 were inhibited by more than 90% after inclusion of the NOS2-specific inhibitor 1400W (0.5 mM) during the culture period. 1400W was not toxic for the cells (data not shown). Similarly, inclusion of 0.5 mM 1400W or 2 mM NMMA in the NOS activity assay using cell extracts inhibited activity by more than 95%.
NOS2 and COX2 protein expression.
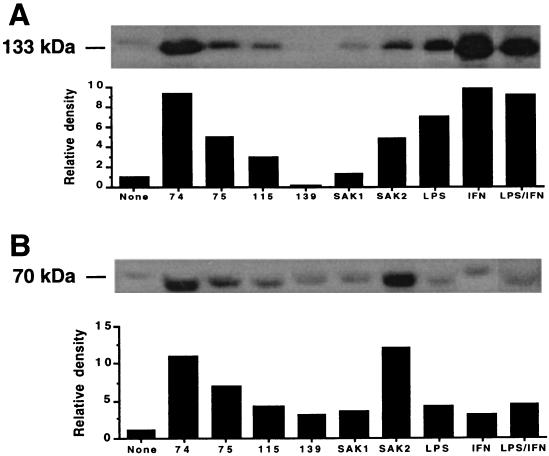
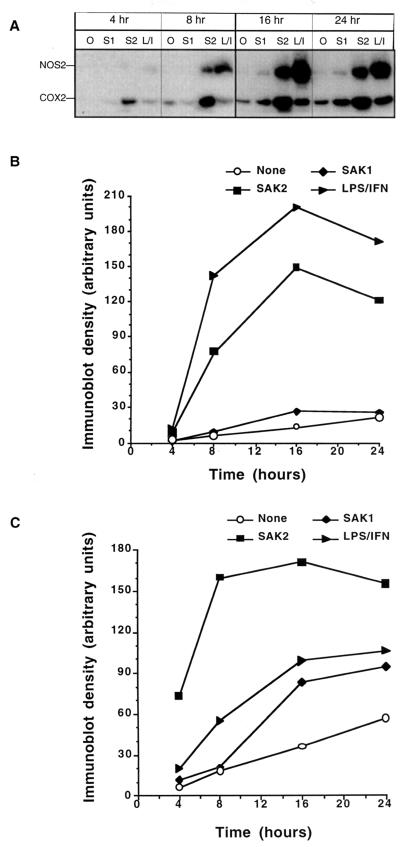
We studied the effects of CpG DNA on enzyme protein expression to investigate the mechanisms for the increase in NO and PGE2 production. J774 cells treated with CpG DNA showed enhanced expression of NOS2 and COX2 protein as determined by immunoblotting (Fig. 3A and B). In general, the relative potencies of the ODNs corresponded to those noted for the induction of NO and PGE2 production. Levels of NOS2 and COX2 protein expression did not correspond directly with the levels of NO and PGE2 produced, respectively. Such discrepancies have been noted by others and may relate to numerous factors, including mRNA and protein stability and cofactor abundance (17, 30). They may also relate to differences in the times of the various measurements. The intensity of NOS2 expression was greatest in IFN-γ- and LPS–IFN-γ-treated cells. The ODNs 74 and SAK2 were the most potent inducers of COX2 protein. As noted in Fig. 4, COX2 and NOS2 proteins induced by SAK2 or LPS–IFN-γ were noted as early as after 4 to 8 h of culture, with expression peaking at about 16 h. Cultures with no addition showed no increase in NOS2 protein over time (Fig. 4A and B) but showed a small increase in COX2 protein over time. SAK1 induced no increase in NOS2, but it did cause an increase in COX2. Overall, LPS–IFN-γ and SAK2 were the most potent inducers of NOS2 and COX2 protein.
FIG. 3.
NOS2 and COX2 expression in J774 cells after treatment with CpG DNAs or LPS–IFN-γ. Immunoblots for NOS2 (A) and COX2 (B) proteins are shown for extracts of cells cultured for 48 h with 10 μg of the indicated ODN/ml and with 100 ng of LPS/ml plus 100 U of IFN-γ/ml. The blot and a bar graph showing the relative density of each band are shown.
FIG. 4.
Immunoblot analysis of the time course for NOS2 and COX2 induction after treatment of J774 cells with CpG DNAs or LPS–IFN-γ. (A) Shown is the time course for the appearance of NOS2 (133 kDa) and COX2 (70 kDa) in cells after treatment with 10 μg of SAK1 (S1) or SAK2 (S2)/ml or 100 ng of LPS/ml plus 100 U of IFN-γ/ml (L/I). Lanes O, no addition. (B and C) Relative densities of the bands for NOS2 (B) and COX2 (C).
IL-6, IL-12, and TNF expression.
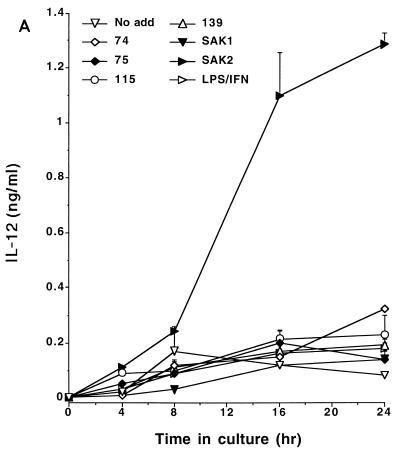
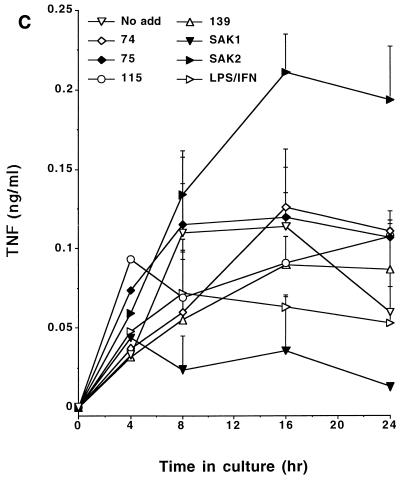
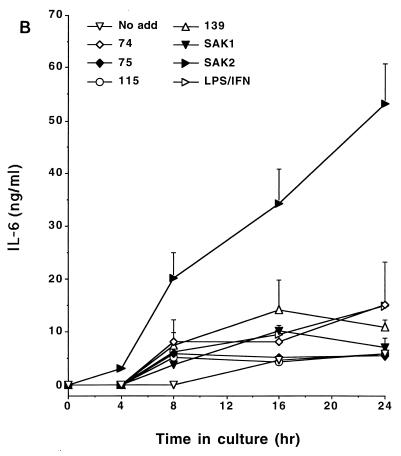
CpG DNA has the capacity to induce cytokine expression by macrophages (14, 19). To determine the relationship between the inductions of NO, PGE2, and cytokines, we tested the CpG DNAs for their abilities to induce the production of IL-12, IL-6, and TNF by J774 cells. As these data indicate, SAK2 was a potent inducer of these cytokines while SAK1 had low activity; the other ODNs showed intermediate levels of activity (Fig. 5). The relative abilities of ODNs to induce the cytokines were thus comparable to those to induce NO and PGE2 production, while SAK2 was much more potent than LPS–IFN-γ in inducing cytokine production.
FIG. 5.
Time course for production of IL-12 (A), IL-6 (B), and TNF-α (C) by J774 cells after treatment with CpG DNAs or LPS–IFN-γ. Cells were cultured with 10 μg of the indicated ODN/ml and with 100 ng of LPS/ml plus 100 U of IFN-γ/ml. Supernatant media were assayed at the indicated times. These results are from one experiment, which is representative of two that were done. Results are displayed as means + 1 SEM. No add, no addition.
Influence of NOS and COX inhibitors on NO and PGE2 production.
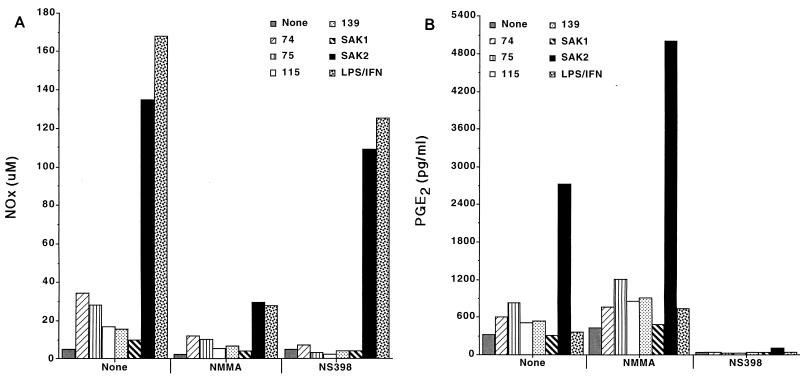
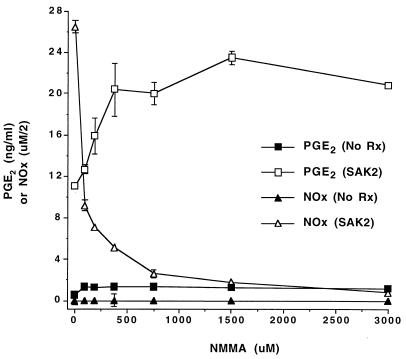
We next assessed the effects of inhibitors of NO and PGE2 production. Inclusion of the nonspecific NOS inhibitor NMMA or of the NOS2-specific inhibitor 1400W or l-NIL (data not shown) in cultures inhibited CpG DNA-induced NO production. The COX2-specific inhibitor NS398 also inhibited CpG DNA- and LPS–IFN-γ-induced NO production to various degrees (Fig. 6A). NS398 completely blocked PGE2 production induced by any of the stimuli (Fig. 6B). CpG DNA-induced PGE2 production was enhanced in cells incubated with either NMMA (Fig. 6B) or 1400W (data not shown). Figure 7 displays the effect of a wide range of concentrations of NMMA on CpG DNA-induced PGE2 production. As the amount of NMMA included in the cultures increased, there was a reduction in SAK2-induced NO production and a reciprocal increase in PGE2 production, with doses as low 92 μM having an enhancing effect.
FIG. 6.
Influence of treatment with a NOS inhibitor (NMMA) or a COX2 inhibitor (NS398) on production of NO (A) or PGE2 (B) by J774 cells after treatment with CpG DNAs or LPS–IFN-γ. Cells were cultured with 10 μg of the indicated ODN/ml and with 100 ng of LPS/ml plus 100 U of IFN-γ/ml for 48 h. The NOS inhibitor NMMA (2 mM) or the COX2 inhibitor NS398 (100 μM) was present in the cultures as indicated. Cells were cultured with the indicated additives, and supernatants were analyzed after 48 h of culture. These results are from one experiment, which is representative of three that were done. Results are displayed as the means of results from triplicate samples.
FIG. 7.
Effect of various doses of NMMA on PGE2 and NO production by J774 cells after treatment with SAK2 CpG DNA. Cells were cultured with no treatment or with 10 μg of SAK2/ml. “No Rx” signifies cultures to which no SAK2 was added. Various concentrations of NMMA were present throughout the culture period as indicated. Supernatant media were assayed after 48 h of culture. These results are from one experiment, which is representative of two that were done. NOx values are expressed as NOx divided by 2 to allow easy comparison to PGE2 values. Results are displayed as the means ± SEMs of results from triplicate samples. For some data points, the error bars are obscured by the symbol.
DISCUSSION
Our results provide new insights into the immunological properties of CpG DNA and show that this stimulator can induce both NO and PGE2 production by mouse macrophage lineage cells. Bacterial DNA as well as synthetic ODNs with certain sequences can potently stimulate B and T lymphocytes, NK cells, mononuclear phagocytes, and dendritic cells (see references 11 and 18 for reviews). These activities may be beneficial in inducing innate immunity and providing adjuvant activity. They may also promote inflammation, however, and may induce or exacerbate conditions such as arthritis or pneumonitis. Although cytokines such as TNF have been implicated as mediators of CpG DNA-induced inflammation, our findings suggest that PGE2 and NO may also contribute to this action. Furthermore, PGE2 and NO, by virtue of their effects on various lymphoid and myeloid populations, may influence the overall host response to CpG DNA.
We demonstrate here that CpG DNA can, in the absence of prior treatment with IFN-γ or LPS, directly activate mouse J774 macrophage cells for not only NOS2 expression and NO production but also COX2 expression and PGE2 production. These results contrast with those of studies by other investigators who showed that CpG DNA-induced NO production in murine macrophages requires priming with IFN-γ or LPS (6, 25, 27). The causes of the differences among these studies are not known, but they could relate to either the sequences of the Ps ODNs used, the cell population studied, or the conditions for cell culture.
Among Ps compounds tested, SAK2 (an ODN with two CpG sequences) was the most potent inducer of NO, PGE2, and cytokines. Other CpG compounds had less activity, and a control compound without a CpG was generally inactive. This result is consistent with those of other studies demonstrating that the stimulation by an ODN is determined by a CpG motif and its flanking sequences. Furthermore (although we did not study phosphodiester ODNs), Ps ODNs are in general more active than other ODNs. The activities of these compounds most likely relate to the stability conferred by the Ps backbone. Thus, both base sequence and backbone structure contribute to the overall activity of CpG DNA.
A variety of stimuli can activate cells for both NOS2 and COX2 expression, but in experiments reported here, we observed differences between the effects of CpG DNA and the effects of LPS and IFN-γ. CpG DNA induced high levels of COX2 and NOS2, but IFN-γ, LPS, and LPS–IFN-γ increased expression of COX2 in the J774 cells less well. These results suggest that, while the activation pathways induced by CpG DNA and LPS are similar, they are likely not identical. This conclusion is supported by previous studies of the kinetics of cytokine induction by LPS and CpG DNA and the differences in the responses of C3H/HeJ mice. These mice, with a mutation in TLR4, can respond to CpG DNA but not to LPS (9, 20).
Although NOS2 and COX2 differ in their levels of activation by CpG DNA, there is nevertheless significant cross talk between these systems. Eicosanoids can reduce NOS2 expression and NO production (5, 10, 15, 16, 29), and NO modulates PGE2 formation (22, 26, 28, 31). Stimuli that enhance NOS2 expression and NO formation may also induce COX2 expression, but the time courses for induction may differ. Furthermore, arginine analogues such as NMMA may lead to inhibition of both COX2 and NOS (21), while aspirin (in high doses) inhibits both COX and NOS2 (2). Studies to define these relationships further have produced somewhat divergent results that may in part be related to the use of different cells and experimental conditions.
An important finding in our study concerns the effects of NO inhibition on the production of PGE2 induced by CpG DNA. We found that while NMMA, 1400W, and l-NIL inhibited NO production, these agents increased PGE2 production in a dose-dependent fashion. These results suggest that blocking NO production with a NOS inhibitor reduces the inhibitory effect of NO on COX2 expression and activity. A variety of mechanisms could account for this response. The modulating effects of NO could result from altered transcription of COX genes, modulation of COX activity, or modification of PG-metabolizing enzymes. In this regard, peroxynitrite, derived from NO and O2−, activates the COX activities of COX1 and COX2 by serving as a substrate for the enzymes' peroxidase activities (7). Furthermore, NO may inhibit the enzymatic activity of COX2 by reacting with the iron in the COX2 heme group in a fashion comparable to that noted for NOS2 (8).
While the mechanism by which NOS inhibitors increase PGE2 production is unknown, this effect has implications for the therapy of inflammatory disease. To the extent that NO modulates PGE2 production, treatment of inflammation with a NOS2 inhibitor alone might result in increased PGE2 production and increased inflammation. An interrelationship between the NO and PG systems has also been observed in a study of cartilage (1). These findings suggest that the cross talk we have observed with CpG DNA stimulation may be a general phenomenon and not confined to this mode of stimulation.
In summary, we have found that CpG DNA coordinately induces high levels of expression of COX2 and NOS2 and high levels of production of PGE2 and NO. These mediators may contribute to the immune activity of CpG DNA and modulate the effects of the chemokines and cytokines induced by CpG DNA. Furthermore, we have shown that the inhibition of NO formation enhances PGE2 production. While the interplay of the PG and NO systems may influence the effects of CpG DNA, our findings suggest a broader relevance for the treatment of inflammatory disease with agents that modify the production or activities of these mediators.
ACKNOWLEDGMENTS
This work was supported in part by the Veterans Affairs Research Service, the James Swiger Hematology Research Fund, and NIH grant AR-39162.
REFERENCES
- 1.Amin A R, Attur M, Patel R N, Thakker G D, Marshall P J, Rediske J, Stuchin S A, Patel I R, Abramson S B. Superinduction of cyclooxygenase-2 activity in human osteoarthritis-affected cartilage—influence of nitric oxide. J Clin Investig. 1997;99:1231–1237. doi: 10.1172/JCI119280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Amin A R, Vyas P, Attur M, Leszczynska-Piziak J, Patel I R, Weissmann G, Abramson S B. The mode of action of aspirin-like drugs: effect on inducible nitric oxide synthase. Proc Natl Acad Sci USA. 1995;92:7926–7930. doi: 10.1073/pnas.92.17.7926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Deng G M, Nilsson I M, Verdrengh M, Collins L V, Tarkowski A. Intra-articularly localized bacterial DNA containing CpG motifs induces arthritis. Nat Med. 1999;5:702–705. doi: 10.1038/9554. [DOI] [PubMed] [Google Scholar]
- 4.Deng G M, Tarkowski A. The features of arthritis induced by CpG motifs in bacterial DNA. Arthritis Rheum. 2000;43:356–364. doi: 10.1002/1529-0131(200002)43:2<356::AID-ANR15>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 5.Gaillard T, Mulsch A, Klein H, Decker K. Regulation by prostaglandin E2 of cytokine-elicited nitric oxide synthesis in rat liver macrophages. Biol Chem Hoppe-Seyler. 1992;373:897–902. doi: 10.1515/bchm3.1992.373.2.897. [DOI] [PubMed] [Google Scholar]
- 6.Gao J J, Zuvanich E G, Xue Q, Horn D L, Silverstein R, Morrison D C. Cutting edge: bacterial DNA and LPS act in synergy in inducing nitric oxide production in RAW 264.7 macrophages. J Immunol. 1999;163:4095–4099. [PubMed] [Google Scholar]
- 7.Goodwin D C, Landino L M, Marnett L J. Effects of nitric oxide and nitric oxide-derived species on prostaglandin endoperoxide synthase and prostaglandin biosynthesis. FASEB J. 1999;13:1121–1136. doi: 10.1096/fasebj.13.10.1121. . (Review.) [DOI] [PubMed] [Google Scholar]
- 8.Griscavage J M, Rogers N E, Sherman M P, Ignarro L J. Inducible nitric oxide synthase from a rat alveolar macrophage cell line is inhibited by nitric oxide. J Immunol. 1993;151:6329–6337. [PubMed] [Google Scholar]
- 9.Hacker H, Vabulas R M, Takeuchi O, Hoshino K, Akira S, Wagner H. Immune cell activation by bacterial CpG-DNA through myeloid differentiation marker 88 and tumor necrosis factor receptor-associated factor (TRAF)6. J Exp Med. 2000;192:595–600. doi: 10.1084/jem.192.4.595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Imai Y, Kolb H, Burkart V. Nitric oxide production from macrophages is regulated by arachidonic acid metabolites. Biochem Biophys Res Commun. 1993;197:105–109. doi: 10.1006/bbrc.1993.2447. [DOI] [PubMed] [Google Scholar]
- 11.Krieg A M. Mechanisms and applications of immune stimulatory CpG oligodeoxynucleotides. Biochim Biophys Acta. 1999;1489:107–116. doi: 10.1016/s0167-4781(99)00147-5. . (Review.) [DOI] [PubMed] [Google Scholar]
- 12.Krieg A M. The role of CpG motifs in innate immunity. Curr Opin Immunol. 2000;12:35–43. doi: 10.1016/s0952-7915(99)00048-5. . (Review.) [DOI] [PubMed] [Google Scholar]
- 13.Krieg A M, Yi A K, Hartmann G. Mechanisms and therapeutic applications of immune stimulatory CpG DNA. Pharmacol Ther. 1999;84:113–120. doi: 10.1016/s0163-7258(99)00023-6. . (Review.) [DOI] [PubMed] [Google Scholar]
- 14.Lipford G B, Sparwasser T, Bauer M, Zimmermann S, Koch E S, Heeg K, Wagner H. Immunostimulatory DNA: sequence-dependent production of potentially harmful or useful cytokines. Eur J Immunol. 1997;27:3420–3426. doi: 10.1002/eji.1830271242. [DOI] [PubMed] [Google Scholar]
- 15.Marotta P, Sautebin L, Di Rosa M. Modulation of the induction of nitric oxide synthase by eicosanoids in the murine macrophage cell line J774. Br J Pharmacol. 1992;107:640–641. doi: 10.1111/j.1476-5381.1992.tb14499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Milano S, Arcoleo F, Dieli M, D'Agostino R, D'Agostino P, De Nucci G, Cillari E. Prostaglandin E2 regulates inducible nitric oxide synthase in the murine macrophage cell line J774. Prostaglandins. 1995;49:105–115. doi: 10.1016/0090-6980(94)00004-g. [DOI] [PubMed] [Google Scholar]
- 17.Nathan C, Xie Q-W. Regulation of biosynthesis of nitric oxide. J Biol Chem. 1994;269:13725–13728. [PubMed] [Google Scholar]
- 18.Pisetsky D S. Immune activation by bacterial DNA: a new genetic code. Immunity. 1996;5:303–310. doi: 10.1016/s1074-7613(00)80256-3. [DOI] [PubMed] [Google Scholar]
- 19.Pisetsky D S. DNA and the immune system. Ann Intern Med. 1997;126:169–171. doi: 10.7326/0003-4819-126-2-199701150-00015. [DOI] [PubMed] [Google Scholar]
- 20.Pisetsky D S, Reich C F., III The influence of base sequence on the immunological properties of defined oligonucleotides. Immunopharmacology. 1998;40:199–208. doi: 10.1016/s0162-3109(98)00044-7. [DOI] [PubMed] [Google Scholar]
- 21.Salvemini D, Manning P T, Zweifel B S, Seibert K, Connor J, Currie M G, Needleman P, Masferrer J L. Dual inhibition of nitric oxide and prostaglandin production contributes to the anti-inflammatory properties of nitric oxide synthase inhibitors. J Clin Investig. 1995;96:301–308. doi: 10.1172/JCI118035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Salvemini D, Misko T P, Masferrer J L, Seibert K, Currie M G, Needleman P. Nitric oxide activates cyclooxygenase enzymes. Proc Natl Acad Sci USA. 1993;90:7240–7244. doi: 10.1073/pnas.90.15.7240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwartz D A, Wohlford-Lenane C L, Quinn T J, Krieg A M. Bacterial DNA or oligonucleotides containing unmethylated CpG motifs can minimize lipopolysaccharide-induced inflammation in the lower respiratory tract through an IL-12-dependent pathway. J Immunol. 1999;163:224–231. [PubMed] [Google Scholar]
- 24.Sharara A I, Perkins D J, Misukonis M A, Chan S U, Dominitz J A, Weinberg J B. Interferon (IFN)-alpha activation of human blood mononuclear cells in vitro and in vivo for nitric oxide synthase (NOS) type 2 mRNA and protein expression—possible relationship of induced NOS2 to the anti-hepatitis C effects of IFN-alpha in vivo. J Exp Med. 1997;186:1495–1502. doi: 10.1084/jem.186.9.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stacey K J, Sweet M J, Hume D A. Macrophages ingest and are activated by bacterial DNA. J Immunol. 1996;157:2116–2122. [PubMed] [Google Scholar]
- 26.Stadler J, Harbrecht B G, Di Silvio M, Curran R D, Jordan M L, Simmons R L, Billiar T R. Endogenous nitric oxide inhibits the synthesis of cyclooxygenase products and interleukin-6 by rat Kupffer cells. J Leukoc Biol. 1993;53:165–172. doi: 10.1002/jlb.53.2.165. [DOI] [PubMed] [Google Scholar]
- 27.Sweet M J, Stacey K J, Kakuda D K, Markovich D, Hume D A. IFN-γ primes macrophage responses to bacterial DNA. J Interferon Cytokine Res. 1998;18:263–271. doi: 10.1089/jir.1998.18.263. [DOI] [PubMed] [Google Scholar]
- 28.Swierkosz T A, Mitchell J A, Warner T D, Botting R M, Vane J R. Co-induction of nitric oxide synthase and cyclo-oxygenase: interactions between nitric oxide and prostanoids. Br J Pharmacol. 1995;114:1335–1342. doi: 10.1111/j.1476-5381.1995.tb13353.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tetsuka T, Daphna-Iken D, Srivastava S K, Baier L D, DuMaine J, Morrison A R. Cross-talk between cyclooxygenase and nitric oxide pathways: prostaglandin E2 negatively modulates induction of nitric oxide synthase by interleukin 1. Proc Natl Acad Sci USA. 1994;91:12168–12172. doi: 10.1073/pnas.91.25.12168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vane J, Bakhle Y, Botting R. Cyclooxygenase 1 and 2. Annu Rev Pharmacol Toxicol. 1998;38:97–120. doi: 10.1146/annurev.pharmtox.38.1.97. [DOI] [PubMed] [Google Scholar]
- 31.Vane J R, Mitchell J A, Appleton I, Tomlinson A, Bishop-Bailey D, Croxtall J, Willoughby D A. Inducible isoforms of cyclooxygenase and nitric-oxide synthase in inflammation. Proc Natl Acad Sci USA. 1994;91:2046–2050. doi: 10.1073/pnas.91.6.2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wagner H. Bacterial CpG DNA activates immune cells to signal infectious danger. Adv Immunol. 1999;73:329–368. doi: 10.1016/s0065-2776(08)60790-7. [DOI] [PubMed] [Google Scholar]
- 33.Weinberg J B, Granger D L, Pisetsky D S, Seldin M F, Misukonis M A, Mason S N, Pippen A M, Ruiz P, Wood E R, Gilkeson G S. The role of nitric oxide in the pathogenesis of spontaneous murine autoimmune disease: increased nitric oxide production and nitric oxide synthase expression in MRL-1pr/1pr mice, and reduction of spontaneous glomerulonephritis and arthritis by orally administered NG-monomethyl-l-arginine. J Exp Med. 1994;179:651–660. doi: 10.1084/jem.179.2.651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Weinberg J B, Misukonis M A, Shami P J, Mason S N, Sauls D L, Dittman W A, Wood E R, Smith G K, McDonald B, Bachus K E, Haney A F, Granger D L. Human mononuclear phagocyte inducible nitric oxide synthase (iNOS). Analysis of iNOS mRNA, iNOS protein, biopterin, and nitric oxide production by blood monocytes and peritoneal macrophages. Blood. 1995;86:1184–1195. [PubMed] [Google Scholar]