Abstract
Objective:
To evaluate the association of genetic risk scores (GRS) of LDLR, APOB and proprotein convertase subtilisin-kexin type 9 (PCSK9) SNP and plasma LDL concentrations and to identify lifestyle interactions with the GRS in Korean middle-aged adults.
Design:
Korean genome and epidemiology study (KoGES) was conducted to determine genetic variants and lifestyle factors, including nutrient intakes, in a retrospective hospital-based city cohort conducted by the Korean Center for Disease and Control during 2004–2013.
Settings:
Hospitals in Korea.
Participants:
Adults aged 40–77 years (n 28 445) without serious diseases.
Results:
Subjects with the major alleles (risk allele) of LDLR rs1433099 and rs11557092, APOB rs13306194 and PCSK9 rs11583723 had higher plasma LDL concentration by 1·20-folds than those with the minor alleles. Subjects with High-GRS (major alleles) of the four SNP had higher adjusted OR for plasma total and LDL-cholesterol and TAG concentrations by 1·24-, 1·203- and 1·167-folds, respectively, but not HDL-cholesterol, than those with Low-GRS. Western-style flour-rich dietary patterns, but not balanced Korean-style and rice-based dietary patterns, had interactions with GRS to increase plasma LDL concentrations. Daily energy intake also interacted with GRS. In the high intake of Western-style flour-rich dietary patterns, carriers with High-GRS had much higher plasma LDL concentrations than the Low-GRS. With high energy intake, carriers with High-GRS had much higher plasma LDL concentrations than those with Low-GRS.
Conclusions:
Adults with major alleles of four SNP are recommended to have low-energy intakes with a balanced Korean diet need to avoid high-energy intakes especially with Western-style flour-rich diet patterns.
Keywords: LDLR, APOB, PCSK9, Dietary patterns, LDL-cholesterol, Genetic risk score
Cardiovascular diseases (CVD) remain a leading cause of death among adults in developed countries, although their morbidity and mortality rates have decreased in recent decades(1). However, CVD prevalence has continuously increased over the last decade in Korea(2). Dyslipidaemia and hyperglycaemia are well-known risk factors for CVD(3). In the body cholesterol and TAG are circulated bound to lipoproteins in the bloodstream, since they cannot dissolve in the blood. Cholesterol contents in the lipoproteins, mainly LDL and HDL, can be changed by lecithin cholesterol acyltransferase and cholesterol ester transfer protein. The LDL receptor (LDLR), a cell surface-glycoprotein, plays an important role in LDL metabolism(4). The LDLR recognizes and binds to apolipoprotein B100 (APO B) on the surface of the LDL particle and facilitates the uptake of LDL into the cells by endocytosis. Intracellular LDLR is degraded by proprotein convertase subtilisin-kexin type 9 (PCSK9) in the cell to regulate the LDLR levels on the cell membrane(5). LDLR deletion induces a moderate increase in plasma LDL concentrations of animals fed a normal chow diet, but it leads to a severe elevation of plasma LDL concentrations and subsequent atherosclerosis in animals fed high-fat diets(6). The mutation of APOB (R3500Q, R3500W) decreases the binding affinity of APOB-100 in mice and potentially increases serum LDL-cholesterol concentrations(7). In humans, mutations in LDLR cause elevated plasma LDL-cholesterol concentrations and carriers who are homozygous for risk alleles have a 4–5-fold increase of plasma LDL-cholesterol concentrations(8). They also have greatly increased cholesterol deposition in the arteries which causes atherosclerosis.
Dyslipidaemia is modifiable by environmental factors such as lifestyles and nutrient intake(3). A high carbohydrate (CHO) diet induces dyslipidaemia, particularly high TAG. High CHO diets with the low glycaemic index, resulting in a significant weight loss and a reduction of serum LDL concentrations but they raise serum TAG levels and reduce HDL levels(9,10). A very-low-fat diet (high CHO diet) results in metabolic syndrome risk by decreasing plasma HDL concentrations and increasing plasma TAG concentrations in Koreans(11). However, low-fat diets decrease serum LDL- and HDL-cholesterol concentrations but did not lower TAG in women in a mainly Caucasian meta-analysis(12). The inconsistent effects of high-fat diets on dyslipidaemia are associated with types of CHO and fat and race. A balanced high-fat diet reduced serum LDL concentrations and LDL particle size in European American women, not in African Americans(13). These results indicated that dietary fat and CHO contents interact with genetic variants.
Here, we hypothesised that the genetic variants related to LDL metabolism would have a strong association with plasma LDL concentrations in middle-aged adults and that they would interact with lifestyles and dietary patterns. This hypothesis was assessed in 28 342 middle-aged Korean adults in hospital-based city cohorts.
Methods
Subjects
Middle-aged Korean adults volunteered to participate in this hospital-based city cohort study. During 2004–2013, a total of 28 342 adults aged 40–77 years participated in the hospital-based city cohorts of Korean genome and epidemiology study that were organised by the Korean Center for Disease and Control. The genetic variants of 28 342 subjects were identified by Korean Chip and made available to scientists. Korean Chip was implemented for studying Korean genetic variants and includes important SNP related to diseases(14). This study was approved by the institutional review board of the Korean National Institute of Health for the Korean genome and epidemiology study and Hoseo University. The written informed consents were also collected from each subject.
Data collection
General characteristics, including social factors, were obtained by taking a survey of the subjects(15). Anthropometric and biochemical parameters and nutrient intake were measured. Height, weight and waist and hip circumference were measured, and BMI was calculated as previously described(16). Plasma was separated from the collected blood after an overnight fast(14). Plasma total and HDL-cholesterol, TAG and glucose concentrations in a fasting state were measured using a Hitachi 7600 Automatic Analyzer (Hitachi). Plasma LDL concentrations were calculated by the Friedewald Equation (plasma LDL concentrations = plasma total cholesterol concentrations – plasma HDL concentrations – plasma TAG concentrations/5). HbA1c contents in the blood were determined using an automatic analyser (ZEUS 9.9; Takeda). Each participant answered a question about whether they regularly take lipid-lowering medicine.
The education level was divided into less than high school, high school and college or more. Smoking status was categorised as current smoker, past smoker and never-smoker. Alcohol consumption was assessed by alcohol beverage types, its intake amounts and frequencies during the past month in the questionnaires, and daily alcohol intake amount was calculated by multiplying the frequencies of alcohol drinking by the amount of alcohol consumed at once. Alcohol consumption was divided into light drinkers (<0·3 g), moderate drinkers (0·3–10 g) and heavy drinkers (>10 g). Coffee intake was assessed in the same way as was alcohol intake and it was categorised into three groups by the tertiles of daily coffee intake.
Physical activity was estimated by the summation of multiplying each activity level by physical activity periods, while daily physical activity level was categorised into light, moderate and heavy activity by the tertiles.
Dietary assessment and dietary patterns
Usual food intake was assessed by semi-quantitative FFQ (SQFFQ). The SQFFQ was composed of 106 food items, and the frequencies and the average serving size were checked for each food item. Options for a serving size of each food item were 0·5-, 1- and 2-folds of one serving size displayed as a reference. The average intake of each food item was calculated by multiplying the intake of frequency and average selected serving size per day. The daily intake of each food item was calculated based on the midpoint of its frequency categories. The average intake of each food item was presented per day. The validity and reproducibility of the SQFFQ were confirmed by a 3-d food record(17). Using SQFFQ results in dietary patterns to explain the subjects’ eating pattern were generated using principal component analysis. Daily intake of energy and nutrients was calculated using the Can-Pro 2.0 nutrient intake assessment software generated by the Korean Nutrition Society(18).
Genotyping and quality control
Genomic DNA was extracted from peripheral blood monocytes by standard procedures. SNP were calculated using Korean Chip, which contained 830 000 SNP customised for Koreans and designed by the Center for Genome Science of Korea National Institute of Health, Korea (http://www.cdc.go.kr/CDC/eng/mobile/CdcKrContentView.jsp?cid=74266&menuIds=HOME001-MNU1130-MNU1890).
The genotyping and quality-control processes were described in detail previously(11). DNA was isolated from their peripheral blood of the participants, and genotyping was conducted using the Affymetrix Genome-Wide Human SNP array for Koreans (Affymetrix). The genotyping accuracy was assessed using the Bayesian Robust Linear Modeling with Mahalanobis Distance genotyping algorithm(11,19). The exclusion criteria for genotyping DNA were as follows: low genotyping accuracies (<98 %), high missing genotype call rates (≥ 4 %), high heterozygosity (>30 %) and gender bias. Some SNP were not used for the following reasons: posterior probability score <0·90, Hardy–Weinberg equilibrium (P < 0·05), low genotype information content (info <0·5), minor allele frequency <1 % and SNP missing rate >0·1.
Genetic variants that influence plasma LDL concentrations by genome-wide association study in Korean adults
SNP that influence LDL concentrations were explored by the genome-wide association study. The clinical data were used to classify the risk group (Case) and the normal-group (Control) based on the plasma LDL concentrations (4·1376 mmol/l) in middle-aged subjects. The participants who answered ‘yes’ for the question about regularly taking lipid-lowering medicine were included in the case, even if their plasma LDL-cholesterol concentrations were lower than 4·1376 mmol/l. Age, gender, residence area and BMI of the participants were used as covariates to select SNP that are linked to higher plasma LDL concentrations by genome-wide association study using PLINK. The genetic variants were selected with P < 5·0 × 10−8, and linkage disequilibrium was calculated to confirm the correlation between LDLR two SNP (r 2 value <0·3). If they were found not to be correlated, they were included in the genetic risk score (GRS) of SNP to explain the association with plasma LDL concentrations.
GRS has the advantage to combine all relevant gene variants to assess their contribution to plasma LDL concentrations. GRS was calculated from the selected genetic variants by adding the number of risk alleles of each genetic variant. The non-risk (minor allele), heterozygous and risk (major allele) alleles of each of the selected four SNP (two SNP of LDLR, one SNP of APOB and one SNP of PCSK9) were assigned the numbers of 0, 1 and 2, and GRS was calculated by unweighted GRS since the OR of each SNP was similar (OR 0·8423, 0·8927). Accordingly, the allele numbers were added to calculate the GRS in each subject without multiplying the OR value of each SNP. The GRS was divided into three groups by the calculated values as Low-GRS, Medium-GRS and High-GRS.
Interaction between genetic risk scores and lifestyles on plasma LDL concentrations
The interaction between the genetic variants and lifestyles, including nutrient intakes, that influence plasma LDL levels was analysed. Daily energy, protein, fat, CHO, Ca, Na, coffee and alcohol intake, smoking status and physical activity were included in the analysis. These dietary intakes, smoking status and physical activity were divided into high and low groups by the median of the parameters. The interaction was adjusted by area, age, gender, BMI, smoking and drinking status, coffee intake, energy, fat, CHO and cholesterol intake, physical activity and plasma total cholesterol concentration.
Statistical analysis
Statistical analysis was performed using plink version 3.3 (http://pngu.mgh.harvard.edu/~purcell/plink) and SAS (version 9.4; SAS Institute). Allele frequencies were calculated by allele counting, and SNP were selected to influence plasma LDL concentrations by genome-wide association study using PLINK.
The 106 food items in SQFFQ were categorised into 30 predefined food groups (Supplemental Table 1). The predefined food groups were used as variables for the factor analysis using the FACTOR procedure. We determined the number of factors to retain based on eigenvalues >1·5, and the orthogonal rotation procedure (varimax) was used with principal components analysis(20). Post-rotated factor loadings showed that three factors described the distinct dietary patterns in the subjects. Foods with factor-loading values ≥0·40 were considered to have major contributions to the distinctive dietary pattern(21). Factor scores for each pattern and each individual were determined by summing the intake of all food groups weighted by the factor loading. Factor scores of each dietary pattern were categorised into tertiles for comparison of lifestyle factors and nutrient intake.
Frequency distributions by classification variables were analysed using the χ 2 test. Two-sampled t test was used to analyse the mean and sd of the continuous variables, such as age, BMI and plasma TAG concentrations. Significant differences among the groups were determined by one-way ANOVA with and without the adjustments for covariates. The multiple comparisons among the groups were determined by the Tukey test.
The association of GRS to influence dyslipidaemia was examined using logistic regression analysis, after adjustment for covariates. The adjusted OR and 95 % CI were estimated based on the non-risk allele of the genetic variants and their GRS calculated by summing the number of risk alleles was analysed to modulate plasma LDL concentrations in the logistic regression analysis after adjusting covariates. Model 1 included the covariates of residence area, gender, age, BMI and energy intake. Model 2 included adjustments for residence area, gender, age, BMI, smoking status, coffee intake, drinking status, total physical activity and plasma total cholesterol concentrations.
To determine the interaction between the GRS and lifestyles, subjects were categorised into the low intake and high intake groups with the classification criterion described above. The daily physical activity was divided into low physical activity and high physical activity. A multivariate interaction model including the main effects and their interaction term was used to evaluate the interactions between the GRS genotype and lifestyles for the risk of hyper-LDL cholesterolemia after adjustment for covariates. P-value ≤ 0·05 was considered statistically significant.
Results
General characteristics of the subjects according to plasma LDL concentrations
Middle-aged subjects were divided into two groups by 160 mg/dl plasma LDL concentrations and whether or not taking lipid-lowering medicine. Subjects in the group with plasma LDL concentrations ≥160 mg/dl (4·138 mM) and/or taking lipid-lowering medicine (High-LDL groups) were significantly older and had higher BMIs and waist and hip circumferences than those with <160 mg/dl plasma LDL concentrations (Low-LDL) (Table 1). Interestingly, women accounted for a higher percentage of the High-LDL group than did men. Plasma glucose concentrations were also higher in the High-LDL group than the Low-LDL group (Table 1). Plasma total cholesterol, LDL and TAG concentrations were higher in the High-LDL than the Low-LDL, but plasma HDL concentrations were also higher in the High-LDL than the Low-LDL (Table 1). Cardiovascular incidences were also higher in the high-LDL than the Low-LDL (Table 1).
Table 1.
Demographic characteristics and nutrient intake of the study population according to serum LDL concentrations
| LDL < 160 mg/dl (n 17 584) | LDL ≥ 160 mg/dl* (n 3787) | P value | |||||
|---|---|---|---|---|---|---|---|
| Mean | sd | Mean | sd | ||||
| Age (years) | 53·4 | 8·1 | 56·0 | 7·1 | <0·0001 | ||
| Gender | <0·0001 | ||||||
| n | 9235 | 1026 | |||||
| Male % | 37·5 | 27·1 | |||||
| BMI (kg/m2) | 23·8 | 2·9 | 24·5 | 2·8 | <0·0001 | ||
| Waist circumference (cm) | 80·8 | 8·7 | 81·0 | 8·2 | 0·0432 | ||
| Hip circumference (cm) | 94·5 | 5·7 | 94·0 | 5·8 | <0·0001 | ||
| Serum total cholesterol (mg/dl) | 190 | 29 | 238 | 30 | <0·0001 | ||
| Serum HDL (mg/dl) | 52·7 | 13 | 53·8 | 12 | <0·0001 | ||
| Serum LDL (mg/dl) | 112 | 26 | 158 | 40 | <0·0001 | ||
| Serum TAG (mg/dl) | 125 | 88 | 130 | 75 | 0·0023 | ||
| Plasma glucose (mg/dl) | 94·1 | 19·2 | 95·9 | 21·5 | <0·0001 | ||
| CVD | <0·0001 | ||||||
| n | 992 | 108 | |||||
| % | 3·65 | 8·62 | |||||
| Total activity | |||||||
| Little (<10 min/d) | 0·5927 | ||||||
| n | 10 900 | 1643 | |||||
| % | 46·1 | 45·6 | |||||
| Moderate (10–90 min/d) | 942 | 26·1 | |||||
| n | 5983 | ||||||
| % | 25·3 | ||||||
| Heavy (>90 min/d) | |||||||
| n | 6740 | 1022 | |||||
| % | 28·5 | 28·3 | |||||
| Alcohol intake | |||||||
| Little (<0·3 g/d) | <0·0001 | ||||||
| n | 14 304 | 2503 | |||||
| % | 58·5 | 65·5 | |||||
| Moderate (0·1–10 g/d) | |||||||
| n | 3895 | 583 | |||||
| % | 15·9 | 15·5 | |||||
| Heavy (>10 g/d) | |||||||
| n | 6260 | 677 | |||||
| % | 25·6 | 18·0 | |||||
| Smoking status | |||||||
| Non-smoking | <0·0001 | ||||||
| n | 17 616 | 2933 | |||||
| % | 71·8 | 77·9 | |||||
| Past-smoking | |||||||
| n | 4090 | 499 | |||||
| % | 16·7 | 13·3 | |||||
| Current smoking | |||||||
| n | 2846 | 331 | |||||
| % | 11·6 | 8·8 | |||||
| Energy intake (EER%) | 88·6 | 27·6 | 88·7 | 27·9 | 0·2597 | ||
| Carbohydrate intake (En%) | 72·0 | 6·8 | 71·5 | 7·0 | <0·0001 | ||
| Protein intake (En%) | 13·4 | 2·6 | 13·5 | 2·7 | 0·0040 | ||
| Fat intake (En%) | 13·6 | 5·2 | 14·1 | 5·3 | <0·0001 | ||
| Cholesterol (mg/d) | 169 | 124 | 168 | 123 | 0·3933 | ||
EER, estimated energy requirement; En%, energy percentage.
CVD was adding the incidence of myocardial infarction and stroke. Conversion factors of serum TAG, cholesterol and glucose concentrations into the SI unit are 0·01129, 0·02586 and 0·0556, respectively.
Participants with ≥plasma LDL-cholesterol plus those with lipid-lowering medicine.
There was no significant difference in daily physical activity between the High-LDL and Low-LDL groups. Significantly more subjects consuming no alcohol were in the Low-LDL group than the High-LDL group, but those with moderate intake were more prevalent in the High-LDL group (P < 0·0001). Unexpectedly, a higher percentage of subjects consuming high alcohol intake were in the Low-LDL group than in the High-LDL group (Table 1). A higher percentage of non-smokers were in the High-LDL than in the Low-LDL group, and a higher percentage of smokers were in the Low-LDL group. The frequencies of alcohol intake and smoking status were not adjusted for covariates including age and gender since they were categorical variables. Subjects in both High-LDL and Low-LDL groups had similar energy intake based on dietary reference intake, and it was about 88 % of the estimated energy requirement according to age and gender (Table 1). CHO intake (En%) was significantly higher in the Low-LDL group than the High-LDL group and protein and fat intake (En%) was opposite to the CHO intake. Daily cholesterol intake was not significantly different between the High- and Low-LDL groups (Table 1).
LDLR, PCSK9 and APOB genetic variants that influence plasma LDL concentrations
LDLR rs1433099 and rs11557092 were negatively associated with plasma LDL concentrations (P = 6·37 E-12 and 1·39 E-09, respectively) (Table 2). APOB rs13306194 was also negatively associated with plasma LDL concentration (P = 1·46 E-09). The rs11583723 of PCSK9 is involved with the regulation of LDLR protein contents in the cell membrane by its degradation and has a negative association with plasma LDL concentrations. The two genetic variants of LDLR are located in chromosome 19, and APOB and PCSK9 are located in chromosomes 2 and 1, respectively (Table 2). Carriers with the minor allele of the four SNP (rs1433099, rs11557092, rs13306194 and rs11583723) had lower OR for lower plasma LDL concentrations by 0·868-, 0·877-, 0·891- and 0·8927-folds, respectively, in comparison to those with major allele (Table 2). The minor alleles of each genetic variant had a protective effect against high plasma LDL concentration compared with the major allele. They were located in the utr-3 for LDLR rs1433099 and rs11557092, in the near 3’ for rs13306194 of APOB and the intron variant for PCSK9 rs11583723. All SNP met the criteria of minor allele frequency (≥0·05 %) and Hardy–Weinberg equilibrium (P > 0·05) criteria (Table 2). The linkage disequilibrium of the selected genetic variants was not satisfied (r 2 < 0·3) to include them in the GRS.
Table 2.
The characteristics of three genetic variants that have a strong association with hyper-LDL cholesterolemia*
| CHR | SNP | Position | Mi | Ma | OR | P adjusted | MAF | P-value HWE | Gene | Functional consequence |
|---|---|---|---|---|---|---|---|---|---|---|
| 19 | rs1433099 | 11 242 658 | T | C | 0·868 | 6·37 E-12 | 0·2702 | 0·1703 | LDLR | utr-3 |
| 19 | rs11557092 | 11 257 018 | T | C | 0·877 | 1·39 E-09 | 0·2317 | 0·0551 | LDLR | Near-gene-3 |
| 2 | rs13306194 | 21 252 534 | A | G | 0·8905 | 1·46 E-09 | 0·1193 | 0·955 | APOB | Reference |
| 1 | rs11583723 | 55 505 926 | T | C | 0·8927 | 2·22 E-05 | 0·1339 | 1·0000 | PCSK9 | Intron variant |
CHR, chromosome; Mi, minor allele; Ma, Major allele, P adjusted, P-value for OR adjusted for BMI, residence area, gender, age; MAF, minor allele frequency; HWE, Hardy–Weinberg equilibrium; LDLR, LDL receptor; APOB, apo B; PCSK9, proprotein convertase subtilisin-kexin type 9.
For serum LDL concentration in the reference of the major allele. P value for OR with adjustment.
Association between genetic risk scores and plasma lipid profiles after adjusting covariates
The summation of the minor (non-risk), heterozygote, and major (risk) alleles of four SNP was divided into three groups: Low-GRS (0; n 11 688), Medium-GRS (1–2; n 11 997) and High-GRS (3–8; n 4603); since all SNP included in the GRS had similar magnitudes of effect and similar OR, the GRS was unweighted (Table 3). Plasma total and LDL-cholesterol concentrations had significant positive associations with GRS after adjusting for age, gender, residence area, BMI and energy intake (model 1) and covariates plus intake of coffee, alcohol, cholesterol, fat, and CHO, smoking status, physical activity, and menopause (model 2). Adjusted OR for plasma total cholesterol and LDL concentrations were higher in the carriers with the High-GRS group by 1·242 (P < 0·0001) and 1·203 (P < 0·01), respectively, than those with the Low-GRS (Table 3). There was no significant interaction of gender and GRS for hyper-LDL cholesterolemia risk (P = 0·639). However, there was no significant association with plasma HDL concentrations according to the alleles of GRS (Table 3). Plasma TAG concentrations were higher by 1·183- and 1·167-folds in the High-GRS group than the Low-GRS in models 1 and 2, respectively (Table 3). The ratio of plasma LDL:HDL was significantly lower by 1·229- and 1·236-folds in subjects carrying the High-GRS allele in comparison to subjects carrying the Low-GRS alleles in models 1 and 2, respectively (P < 0.0001).
Table 3.
Adjusted odds ratio (OR) and 95 % confidence intervals (CI) for the risk of different lipid profiles according to the genetic risk score (GRS) of LDLR, PCSK9 and APOB SNP after covariate adjustments†
| Model 1‡ | Model 2§ | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Medium-GRS (n 13 644)¶ | High-GRS (n 10 076)¶ | Medium-GRS (n 13 644)¶ | High-GRS (n 10 076)¶ | ||||||
| Low-GRS‖ (n 4666) | OR | 95 % CI | OR | 95 % CI | OR | 95 % CI | OR | 95 % CI | |
| Total-C†† (mg/dl) | 1 | 1·153 | 1·046, 1·271 | 1·260* | 1·099, 1·444 | 1·184 | 1·069, 1·311 | 1·242*** | 1·079, 1·430 |
| LDL-cholesterol†† (mg/dl) | 1 | 1·067 | 0·989, 1·152 | 1·210** | 1·088, 1·345 | 1·075 | 0·993, 1·163 | 1·203** | 1·079, 1·343 |
| HDL-cholesterol†† (mg/dl) | 1 | 0·955 | 0·884, 1·032 | 0·988 | 0·890, 1·096 | 0·943 | 0·870, 1·022 | 0·986 | 0·884, 1·099 |
| Triglyceride†† (mg/dl) | 1 | 1·070 | 0·974, 1·126 | 1·183** | 1·070, 1·307 | 1·050 | 0·974, 1·131 | 1·167** | 1·053, 1·294 |
| Ratio of LDL-cholesterol: HDL-cholesterol†† | 1 | 1·062 | 1·000, 1·128 | 1·229*** | 1·131, 1·335 | 1·060 | 0·996, 1·129 | 1·236*** | 1·134, 1·348 |
Total-C, total cholesterol.
Non-risk allele (minor allele), heterozygote allele, risk allele (major allele) in each SNP were counted as 0, 1 and 2 and the scores of four selected SNP were added and GRS from 2 SNP of LDLR, 1 SNP of APOB and 1 SNP of PCSK9 was divided into three categories (0–1, 2–3 and 4–8) as the Low-GRS, Medium-GRS and High-GRS, respectively.
Model 1: adjusted for BMI, residence age, area, gender, BMI, and energy.
Model 2: adjusted for BMI, residence age, gender, area, BMI, intake of energy, coffee, and alcohol, fat percentage, CHO percentage, smoking status, physical activity, menopause and serum total cholesterol concentration.
Low-GRS was the reference for both model 1 and model 2.
Values represent OR and 95 % CI.
The cut-off points of serum total cholesterol (total-C), LDL-cholesterol, HDL-cholesterol and TAG were 200, 160, 40 for men (50 for women), and 150 mg/dl, respectively. The cut-off point of the ratio of LDL-cholesterol:HDL-cholesterol was 2·85.
Significantly different from the major allele in logistic regression analysis at *P < 0·05, **P < 0·01, ***P < 0·001.
Characteristics of subjects in the three dietary patterns and macronutrient intake
Three different dietary patterns were established by the factor analysis with principal component analysis and >1·5 eigenvector values. Factor loadings of food groups in dietary patterns are given in Supplemental Table 1. Factor loadings with ≥0·4 were used (Supplemental Table 2). The dietary patterns included the ‘balanced Korean dietary pattern’ with high consumption of fish, vegetables, mushrooms, potatoes, fruits, kimchi and pickles; the ‘Western-style flour-rich dietary pattern’ with high consumption of bread, cake, cookie and fast foods and the ‘rice-based dietary pattern’ with diets composed of mainly rice. The balanced Korean dietary pattern, Western-style flour-rich dietary pattern and rice-based dietary pattern explained 44·4, 23·2 and 18·0 % of the total variance in food intake, respectively (Supplemental Table 2). These three patterns explained 85·6 % of the total variance in food intake.
The subjects in each dietary pattern were sub-divided into three groups (tertiles) according to how much of each pattern was consumed, and the general characteristics of the subjects were given in Supplemental Table 3. The participants in the high tertile group of the balanced Korean dietary pattern were older, more frequently male, and had higher BMI than those in the low tertile. The subjects in the high tertile group of the balanced Korean dietary pattern group had higher energy intake, lower CHO and fat intake, higher protein and Na intakes and drank more coffee than those in the low tertile group (Supplemental Table 3). In the Western-style flour-rich diet pattern, the subjects in the high tertile group were younger and included males than those in the low tertile group. There was no significant difference in BMI and the percentage of energy intake based on the dietary reference intake (Supplemental Table 3). Similar to the balanced Korean dietary pattern, the subjects in the high tertile of the Western-style flour-rich diet pattern consumed less CHO and fat and had higher protein intake based on daily energy intake and lower in Na and alcohol intake based on energy intake (Supplemental Table 3). However, coffee intake was higher in the high tertile of the Western-style flour-rich pattern. In the rice diet pattern, fewer men were in the high tertile group than the low tertile group (Supplemental Table 3). Subjects in the high rice tertile group were younger with lower BMI, but daily energy intakes were not significantly different. Subjects in the high rice tertile group had lower alcohol and coffee intakes than those in the low rice tertile group (Supplemental Table 3).
Lipid profiles of subjects in different dietary patterns
The associations of plasma total cholesterol, LDL, HDL and TAG concentrations with tertiles of a balanced Korean diet, Western-style flour-rich diet and rice-based diet patterns were determined (Table 4). Plasma total cholesterol, LDL, HDL and TAG concentrations and the ratio of LDL:HDL were not significantly associated with tertiles of the balanced Korean dietary pattern. Intake of foods made of flours and fast foods had higher OR for plasma total (1·140) and LDL-cholesterol (1·172) concentrations and the ratio of LDL:HDL (1·181) in high tertile group than low tertile group (Table 4). This indicated that a high intake of Western-style flour-rich patterns increased plasma total and LDL-cholesterol concentrations and the ratio of LDL:HDL in comparison with the low intake. However, interestingly, a rice-based dietary pattern had no significant association with plasma total, LDL- and HDL-cholesterol and TAG concentrations with the tertiles of the intake (Table 4). Thus, the Western-style flour-rich diet pattern was associated with harmful effects on dyslipidaemia.
Table 4.
The adjusted odds ratio (OR) and 95 % confidence intervals (CI) for the risk of lipid profiles according to dietary patterns after covariate adjustments
| Balanced Korean diet | Western flour-rich diet | Rice-based diet | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| T2 (n 9213) | T3 (n 9421) | T2 (n 9480) | T3 (n 9659) | T2 (n 9278) | T3 (n 9681) | ||||||||||
| T1† (n 9811) | Adjusted OR | 95 % CI | Adjusted OR | 95 % CI | T1‡ (n 9306) | Adjusted OR | 95 % CI | Adjusted OR | 95 % CI | T1§ (n 9486) | Adjusted OR | 95 % CI | Adjusted OR | 95 % CI | |
| Total cholesterol | 1 | 0·977 | 0·914, 1·044 | 0·962 | 0·888, 1·042 | 1 | 1·115** | 1·048, 1·186 | 1·140*** | 1·062, 1·224 | 1 | 0·977 | 0·914, 1·044 | 0·962 | 0·888, 1·042 |
| LDL | 1 | 0·9701 | 0·904, 1·0402 | 0·9446 | 0·868, 1·026 | 1 | 1·117** | 1·048, 1·191 | 1·172*** | 1·088, 1·261 | 1 | 1·032 | 0·964, 1·105 | 1·043 | 0·961, 1·105 |
| HDL | 1 | 1·003 | 0·909, 1·107 | 0·961 | 0·853, 1·083 | 1 | 0·993 | 0·908, 1·087 | 1·034 | 0·932, 1·148 | 1 | 0·943 | 0·880, 1·011 | 0·935 | 0·859, 1·017 |
| Triglyceride | 1 | 0·970 | 0·886, 1·062 | 0·995 | 0·922, 1·073 | 1 | 1·030 | 0·962, 1·104 | 1·056 | 0·975, 1·143 | 1 | 1·008 | 0·925, 1·100 | 0·931 | 0·838, 1·034 |
| Ratio of LDL:HDL | 1 | 0·959 | 0·887, 1·037 | 0·878 | 0·799, 0·964 | 1 | 1·074* | 1·000, 1·154 | 1·181*** | 1·088, 1·281 | 1 | 0·987 | 0·916, 1·065 | 0·953 | 0·870, 1·045 |
Adjusting for BMI, residence age, gender, area, BMI, intake of energy, cholesterol, coffee, and alcohol, fat and CHO, smoking status, physical activity, menopause and serum total cholesterol concentration.
T1, T2 and T3 indicated the lowest, middle and highest tertiles of the intake of each diet pattern.
Reference was the T1.
Significantly different from major allele in logistic regression analysis at *P < 0·05, **P < 0·01, ***P < 0·001.
The cutpoint of dietary pattern scores for T1 and T2 was −0·470 and 0·146 in the balanced Korean dietary pattern†, −0·405 and 0·09 for Western flour-rich diet pattern‡ and −0·451 and 0·07 in rice-based diet pattern§, respectively.
Interactions of genetic risk score with dietary patterns, nutrient intake and lifestyles
There were interactions of GRS of LDLR, APOB and PCSK9 with a Western-style flour-rich diet pattern (P = 0·0413). However, the balanced Korean and rice-based dietary patterns had no significant interaction with GRS (Table 5). High intake of Western-style flour-rich diet pattern increased adjusted OR by 1·212-fold in the High-GRS alleles compared with the Low-GRS allele. In subjects with low intakes of Western-style flour-rich dietary patterns, carriers with High-GRS had less increase of plasma LDL concentrations than in those with its high intake (Fig. 1A). The results indicated that a high intake of Western-style flour-rich diet patterns exacerbated High-GRS on increasing plasma LDL concentrations.
Table 5.
Interaction of genetic risk score (GRS) and lifestyles and adjusted odds ratio (OR) and 95 % confidence intervals (CI) for the risk of serum LDL concentrations of GRS after covariate adjustments according to the patterns of lifestyles†
| Medium-GRS (n 13 644) |
High-GRS (n 10 076) |
|||||
|---|---|---|---|---|---|---|
| Low-GRS | OR | 95 % CI | OR | 95 % CI | GRS-nutrient interaction P value |
|
| Low balanced Korean diet‡ | 1 | 1·094 | 0·951, 1·258 | 1·157 | 0·957, 1·398 | 0·2895 |
| High balanced Korean diet | 1·064 | 0·967, 1·171 | 1·231** | 1·076, 1·408 | ||
| Low Western flour-rich diet§ | 1 | 1·114 | 0·986, 1·285 | 1·180 | 0·972, 1·434 | 0·0413 |
| High Western flour-rich diet | 1·057 | 0·961, 1·162 | 1·212** | 1·061, 1·384 | ||
| Low rice-based diet‖ | 1 | 1·102 | 1·001, 1·231 | 1·278** | 1·117, 1·462 | 0·3474 |
| High rice-based diet | 1·108 | 1·008, 1·219 | 1·229*** | 1·117, 1·352 | ||
| Low energy¶ | 1 | 1·017 | 0·926, 1·116 | 1·179* | 1·035, 1·342 | 0·0492 |
| High energy | 1·151 | 1·035, 1·279 | 1·104 | 0·957, 1·274 | ||
| Low CHO percentage†† | 1 | 0·995 | 0·871, 1·138 | 1·164 | 0·967, 1·400 | 0·5824 |
| High CHO percentage | 1·082 | 0·972, 1·204 | 1·169* | 1·009, 1·355 | ||
| Low protein percentage‡‡ | 1 | 1·084 | 0·968, 1·213 | 1·192* | 1·020, 1·393 | 0·8751 |
| High protein percentage | 1·057 | 0·947, 1·180 | 1·213* | 1·041, 1·414 | ||
| Low fat percentage§§ | 1 | 1·079 | 0·978, 1·191 | 1·219* | 1·063, 1·397 | 0·2671 |
| High Fat percentage | 1·046 | 0·917, 1·193 | 1·166 | 0·972, 1·399 | ||
| Low alcohol drinking‖‖ | 1 | 1·110 | 1·107, 1·121 | 1·261** | 1·116, 1·425 | 0·7667 |
| High alcohol drinking | 1·097 | 0·914, 1·316 | 1·024 | 0·801, 1·309 | ||
| Low coffee intake¶¶ | 1 | 1·068 | 0·983, 1·161 | 1·187* | 1·058, 1·331 | 0·6037 |
| High coffee intake | 1·097 | 0·849, 1·417 | 1·374 | 0·958, 1·969 | ||
| Non- and past-smoking††† | 1 | 1·075 | 0·993, 1·163 | 1·203** | 1·079, 1·343 | 0·0644 |
| Smoking | 1·092 | 1·005, 1·187 | 1·193* | 1·064, 1·338 | ||
| Low physical activity‡‡‡ | 1 | 1·038 | 0·927, 1·163 | 1·073 | 0·918, 1·253 | 0·4593 |
| High physical activity | 1·105 | 0·990, 1·232 | 1·342*** | 1·151, 1·565 | ||
Non-risk allele (minor allele), heterozygote allele, risk allele (major allele) in each SNP were counted as 0, 1 and 2 and the scores of 4 selected SNP were added and GRS from 2 SNP of LDLR, 1 SNP of APOB and 1 SNP of PCSK9 was divided into three categories (0–1, 2–3 and 4–8) as the Low-GRS, Medium-GRS and High-GRS, respectively. Reference was the Low-GRS (n4666). Multivariate regression models include the corresponding main effects, interaction terms of GRS and lifestyles including nutrient intake and potential confounders such as BMI, residence area, gender, age, smoking, physical activity, energy, carbohydrate (CHO), fat, protein, cholesterol, coffee and alcohol intake and serum total cholesterol concentrations.
The cut-off points were as follows: ‡0·146 factor for balanced Korean diet, §0·09 factor for Western flour-rich diet, ‖0·07 factor for rich-based diet, ¶estimated energy intake, ††70 energy % CHO, ‡‡13 energy % of protein, §§15 energy % fat, ‖‖10 g alcohol per day, ¶¶1 cup of coffee per day; †††smoking status and ‡‡‡90 min physical activity per day.
Significantly different from the major allele in logistic regression analysis at * P < 0·05, ** P < 0·01, *** P < 0·001.
Fig. 1.
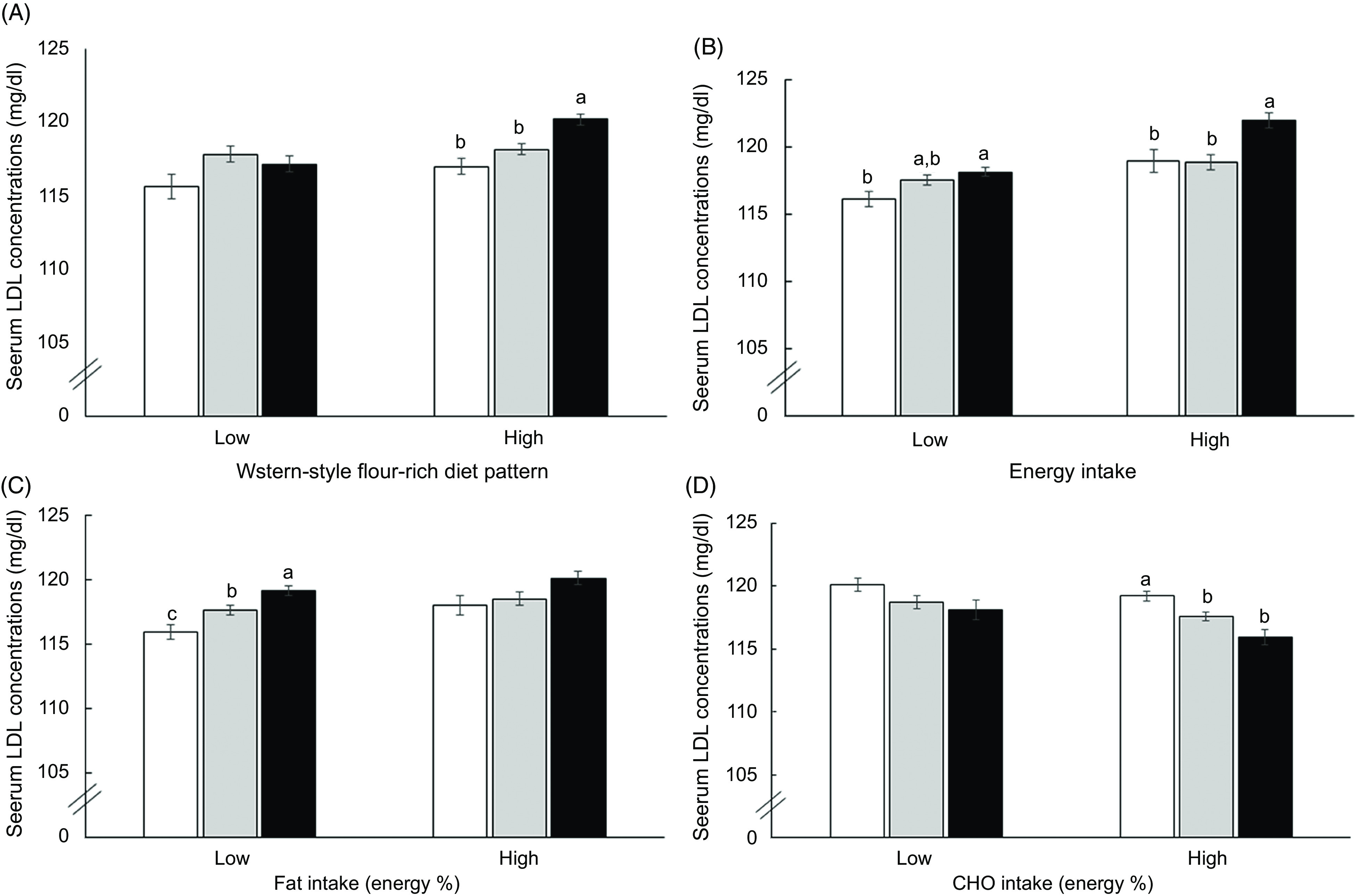
Serum LDL concentrations of subjects with Low-, Medium- and High-genetic risk score (GRS) generated with LDLR rs1433099 and rs11557092, PCSK9 rs11583723 and APOB rs13306194 according to high and low nutrient intake and smoking status. (A) Western-style flour-rich diet pattern. (B) Energy intake (dietary reference intake; DRI). (C) Fat intake (reference: 15 %). (D) Carbohydrate intake (reference: 70 %). P-value indicated the significance of the interaction of GRS and nutrient intake to affect serum LDL concentration. Each bar and error bar represents means ± se. a,b,c Unlike superscript letters on the bars indicate significant differences at P < 0·05. (A) to (D) ( ), Low-GRS; (
), Low-GRS; ( ), Medium-GRS; (
), Medium-GRS; ( ), High-GRS
), High-GRS
Energy intake also had a significant interaction with GRS to influence plasma LDL concentrations (P = 0·0492; Table 5). With high energy intake, adjusted OR was higher by 1·254-fold for plasma LDL concentrations in the High-GRS than the Low-GRS and plasma LDL concentrations in the High-GRS were much higher in a high energy intake than those in the Low-GRS (P < 0·0001; Fig. 1C). In nutrient intake analysis, fat and CHO intakes had no interaction with GRS to modulate plasma LDL concentrations. However, there were different effects of fat and CHO intake on GRS in a low and high intake of CHO and fat. In low fat intake (<15 fat energy percentage), but not high fat intake (≥15 %), subjects with the High-GRS had a positive association with plasma LDL concentration compared with the Low-GRS (Table 5). Plasma LDL concentrations were lower in subjects with Low-GRS than those with Medium- and High-GRS only in the low fat intake, but there were no significant differences among the GRS groups in low fat intake (Fig. 1C). CHO intake exhibited an opposite pattern to fat intake to modulate plasma LDL concentrations. Plasma LDL concentrations had a positive association with the High-GRS compared with the Low-GRS only in subjects with high CHO intake (<70 energy %; Fig. 1D). However, there was no significant association in smoking status, alcohol and coffee intake and physical activity (Table 5).
Discussion
Hyper-LDL cholesterolemia is atherosclerotic and increases the risk of CVD. Mutations in LDLR, APOB, PCSK9 and LDL adapter protein 1 (LDLRAP1) are major genetic risk factors for familial hypercholesterolemia, indicating that these genes play an important role in cholesterol metabolism, especially LDL(22). Plasma LDL concentrations were influenced by plasma TAG levels since plasma VLDL and LDL-cholesterols were not directly measured. However, fewer than 5 % of the subjects had high plasma TAG concentrations, so they might not affect the study of their genetic variants. To our knowledge, this is the first study of the interaction of GRS with dietary patterns to modulate plasma LDL concentrations. We investigated whether a GRS derived from LDLR, APOB and PCSK9 SNP has a strong association with plasma LDL concentrations and identified lifestyle interactions with the GRS in 28 445 middle-aged Korean adults. In this study, subjects with the major alleles had a higher risk of high plasma LDL concentrations, and the minor alleles were protective against elevated LDL-cholesterol. Therefore, subjects with the major alleles were assigned a high GRS for elevated LDL-cholesterol. Subjects with the High-GRS from summing LDLR rs1433099 and rs11557092, APOB rs13306194 and PCSK9 rs11583723 had higher plasma LDL and TAG concentrations by 1·137- and 1·153-folds, respectively, compared to those with the Low-GRS. The balanced Korean diet was not associated with dyslipidaemia. With a high intake of Western-style flour-rich diet and rice-based diet patterns, carriers with Low-GRS had much lower plasma LDL concentrations than those with High-GRS. Energy and fat intake also had an interaction with GRS. In low energy and low fat intake, carriers with Low-GRS had much lower plasma LDL concentrations compared to those with High-GRS. The results suggested that adults with a Low-GRS of LDLR rs1433099 and rs11557092, APOB rs13306194 and PCSK9 rs11583723 should be advised to have low energy intakes with the low fat intake (<15 % energy from fat) to protect against the increase of plasma LDL concentrations. However, subjects with High-GRS should avoid a Western-style flour-rich-based diet.
Comparison with previous studies in genetic impact on plasma LDL concentrations
Hypercholesterolemia is a strong determinant of CVD. LDL particles transport the cholesteryl esters in the bloodstream and are mainly taken up by the liver and some by peripheral tissues(22). The LDL uptake is dependent on the binding of LDL particles to LDL receptors of the hepatocyte surface receptors. The LDLR specifically recognises ApoB in the LDL particle and binds to it. The complex of LDL and LDLR is internalised by endocytosis into the liver and peripheral tissues. LDLR and APOB mutations are known to cause familial hypercholesterolemia and it is inherited(22). However, hypercholesterolemia is a polygenic disease that influences the susceptible genotypes and their genetic susceptibility to hypercholesterolemia, mainly LDL, and consists of cumulative effects of each genetic variant with each having a small effect on elevating LDL in the bloodstream. The cumulative effects of the genetic variants are estimated by GRS. The genetic variants have interactions with environmental factors including nutrient intake and lifestyles. PCSK9 is involved in LDLR degradation after endocytosis of the LDL-LDLR complex. Previous European and African ancestry studies have shown that some SNP of LDLR, rs6511720, rs6511721, rs2738447, rs72658867 and rs5742911, APOB rs533617, rs533617 and rs1367117 and PCSK9 rs11591147 and rs639750 have associations with plasma LDL concentrations(23). In the present study, the GRS for influencing plasma LDL concentrations was calculated by pooling LDLR rs1433099 and rs11557092, APOB rs13306194 and PCSK9 rs11583723, and pooled GRS had a negative association with plasma LDL concentrations in Koreans (5·0 × 10E-8). Plasma LDL concentration had lower OR in the Low-GRS than in the High-GRS. As a result, carriers with the minor alleles of the four SNP had lower plasma LDL concentrations than those with the major allele-GRS.
Comparison with previous studies in dietary patterns on plasma LDL concentrations
Dietary patterns were categorised into a balanced Korean diet, a Western-style flour-rich diet and a rice-based diet. The balanced Korean dietary pattern explained about 52 % of all middle-aged adult subjects. From previous studies, a balanced Korean dietary pattern is similar to the other prudent or Korean traditional patterns using KNHANES(24,25) although the studies explore the food patterns in adults aged >20 years. A balanced Korean dietary pattern is composed of grains, beans, nuts, vegetables and fruits, and it explained more 50 % of the variation of consuming foods, which is similar to the present study(24). It contains the high total antioxidant capacity, and it has an inverse association with hypertriglyceridaemia from KNHANES(24). The present study showed that the balanced Korean dietary pattern did not have any association with lipid profiles among the tertiles. Consistent with a balanced Korean dietary pattern, the subjects with rice-rich dietary patterns, there was no significant association with lipid profiles. A previous study has reported that a kimchi-rich diet was associated with a low prevalence of hypercholesterolemia and a high prevalence of hypertriglyceridaemia and hypoHDL cholesterolemia in KNHANES(24). In the Western-style flour-rich diet pattern, plasma total and LDL-cholesterol concentrations and the ratio of LDL:HDL increased in high tertile by 1·14-, 1·17- and 1·18-folds, respectively (P < 0·001), compared with the low tertile. Plasma TAG concentrations have been shown to increase with a high CHO intake and a low fat intake in Koreans(11,26). However, plasma LDL concentrations increased with high fat and low CHO diets in the present study. Thus, not all subjects should be recommended, and susceptible genetic variants need to be considered to suggest the diet patterns.
Strengths and limitations
To the best of our knowledge, this is the first study to demonstrate that there was the interaction of GRS of LDLR, APOB and PCSK9 SNP with a Western-style flour-rich dietary pattern and energy intake. In a high intake of Western-style flour-rich diet and energy intake subjects with the High-GRS increased the risk of hyper-LDL cholesterolemia compared to those with the Low-GRS in Koreans. However, this study had some limitations. First, the results cannot show a cause-and-effect relationship since the subjects were recruited from a cohort study containing several city areas. This study did not apply a stratification method and survey design in the statistical analysis although the sample size was big (n 28 445). Second, plasma LDL concentrations were not directly measured, but they were estimated using the Friedewald equation and can be underestimated in subjects with hypertriglyceridaemia. Third, the nutrient intake was computed from the food intakes measured by SQFFQ. It is known that with SQFFQ some nutrient intake may be under- or over-estimated. The SQFFQ included 103 foods that Koreans frequently consume, and it had good validity and reproducibility as compared with the 3-d food records in the four seasons study(27).
Conclusions
The results of the present study demonstrated that adults with major alleles (High-GRS) of LDLR rs1433099 and rs11557092, APOB rs13306194 and PCSK9 rs11583723 are recommended to restrict energy intake (less than dietary reference intake) with a balanced Korean diet containing <15 % fat and avoid the Western-style flour-rich diet pattern. Since 84 % of the participants had Medium or High-GRS, most Korean people have a high risk of hyper-LDL cholesterolemia. Most Koreans may benefit from the traditional Korean diet for preventing hyper-LDL cholesterolemia. The results can be applied to prevent hyper-LDL cholesterolemia in middle-aged adults with high genetic risk as personalised nutrition.
Acknowledgements
Acknowledgement: We sincerely appreciate the participants in the KoGES. Financial support: This research was supported by the National Research Foundation, Korea (NRF-2019R1A2C1007203). Conflict of interest: None. Authorship: S.P. formulated the research question, interpreted the data and wrote the first draft of the manuscript. S.K. designed the study and analysed the data. All authors read and approved the final draft of this manuscript. Ethics of human subject participation: This study was conducted according to the guidelines in the Declaration of Helsinki and approved by the institutional review board of the Korean National Institute of Health for the KoGES and Hoseo University. The written informed consents were also collected from each subject.
Supplementary material
For supplementary material accompanying this paper visit https://doi.org/10.1017/S1368980020001305.
click here to view supplementary material
References
- 1. Leong DP, Joseph PG, McKee M et al. (2017) Reducing the global burden of cardiovascular disease, part 2: prevention and treatment of cardiovascular disease. Circ Res 121, 695–710. [DOI] [PubMed] [Google Scholar]
- 2. Kim GS, Im E & Rhee JH (2017) Association of physical activity on body composition, cardiometabolic risk factors, and prevalence of cardiovascular disease in the Korean population (from the fifth Korea national health and nutrition examination survey, 2008–2011). BMC Public Health 17, 275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Stein R, Ferrari F & Scolari F (2019) Genetics, dyslipidemia, and cardiovascular disease: new insights. Curr Cardiol Rep 21, 68. [DOI] [PubMed] [Google Scholar]
- 4. Hori M, Ohta N, Takahashi A et al. (2019) Impact of LDLR and PCSK9 pathogenic variants in Japanese heterozygous familial hypercholesterolemia patients. Atherosclerosis 289, 101–108. [DOI] [PubMed] [Google Scholar]
- 5. Sánchez-Hernández RM, Di Taranto MD, Benito-Vicente A et al. (2019) The Arg499His gain-of-function mutation in the C-terminal domain of PCSK9. Atherosclerosis 289, 162–172. [DOI] [PubMed] [Google Scholar]
- 6. Ishibashi S, Goldstein JL, Brown MS et al. (1994) Massive xanthomatosis and atherosclerosis in cholesterol-fed low-density lipoprotein receptor-negative mice. J Clin Invest 93, 1885–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Vrablik M, Ceska R & Horinek A (2001) Major apolipoprotein B-100 mutations in lipoprotein metabolism and atherosclerosis. Physiol Res 50, 337–343. [PubMed] [Google Scholar]
- 8. Vallejo-Vaz AJ & Ray KK (2018) Epidemiology of familial hypercholesterolemia: community and clinical. Atherosclerosis 277, 289–297. [DOI] [PubMed] [Google Scholar]
- 9. Ma Y, Li Y, Chiriboga DE et al. (2006) Association between carbohydrate intake and serum lipids. J Am Coll Nutr 25, 155–163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Zafar MI, Mills KE, Zheng J et al. (2019) Low glycaemic index diets as an intervention for obesity: a systematic review and meta-analysis. Obes Rev 20, 290–315. [DOI] [PubMed] [Google Scholar]
- 11. Park S, Ahn J & Lee BK (2016) Very-low-fat diets may be associated with an increased risk of metabolic syndrome in the adult population. Clin Nutr 35, 1159–1167. [DOI] [PubMed] [Google Scholar]
- 12. Wu L, Ma D, Walton-Moss B et al. (2014) Effects of low-fat diet on serum lipids in premenopausal and postmenopausal women: a meta-analysis of randomized controlled trials. Menopause 21, 89–99. [DOI] [PubMed] [Google Scholar]
- 13. Niswender KD, Fazio S, Gower BA et al. (2018) Balanced high-fat diet reduces cardiovascular risk in obese women although changes in adipose tissue, lipoproteins, and insulin resistance differ by race. Metabolism 82, 125–134. [DOI] [PubMed] [Google Scholar]
- 14. Kim Y, Han B-G; KoGES Group (2016) Cohort profile: the Korean genome and epidemiology study (KoGES) consortium. Int J Epidemiol 46, e20–e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lim S, Jang H, Lee H et al. (2006) A rural–urban comparison of the characteristics of the metabolic syndrome by gender in Korea: the Korean Health and Genome Study (KHGS). J Endocrinol Invest 29, 313. [DOI] [PubMed] [Google Scholar]
- 16. Park S, Kim DS & Kang S (2019) Carrying minor allele of FADS1 and haplotype of FADS1 and FADS2 increased the risk of metabolic syndrome and moderate but not low-fat diets lowered the risk in two Korean cohorts. Eur J Nutr 58, 831–842. [DOI] [PubMed] [Google Scholar]
- 17. Park S, Daily JW, Zhang X et al. (2016) Interactions with the MC4R rs17782313 variant, mental stress and energy intake and the risk of obesity in Genome Epidemiology Study. Nutr Metab 13, 38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Park S, Ham JO & Lee BK (2015) Effects of total vitamin A, vitamin C, and fruit intake on risk for metabolic syndrome in Korean women and men. Nutrition 31, 111–118. [DOI] [PubMed] [Google Scholar]
- 19. Hong K-W, Kim SH, Zhang X et al. (2018) Interactions among the variants of insulin-related genes and nutrients increase the risk of type 2 diabetes. Nutr Res 51, 82–92. [DOI] [PubMed] [Google Scholar]
- 20. Steel RG, Torrie JH & Dickey DA (1997) Principles and Procedures of Statistics: A Biological Approach. New York, USA: McGraw-Hill. [Google Scholar]
- 21. Kim JO & Mueller CW (1978) Factor Analysis. Statistical Methods and Practical Issues. Thousand Oaks, CA: Sage Publications. [Google Scholar]
- 22. Paththinige CS, Sirisena ND & Dissanayake V (2017) Genetic determinants of inherited susceptibility to hypercholesterolemia – a comprehensive literature review. Lipids Health Dis 16, 103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Safarova MS, Satterfield BA, Fan X et al. (2019) A phenome-wide association study to discover pleiotropic effects of PCSK9, APOB, and LDLR. NPJ Genom Med 4, 3–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kim S-A, Joung H & Shin S (2019) Dietary pattern, dietary total antioxidant capacity, and dyslipidemia in Korean adults. Nutr J 18, 37–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Song Y & Joung H (2012) A traditional Korean dietary pattern and metabolic syndrome abnormalities. Nutr Metab Cardiovas Dis 22, 456–462. [DOI] [PubMed] [Google Scholar]
- 26. Park S, Ahn J, Kim NS et al. (2017) High carbohydrate diets are positively associated with the risk of metabolic syndrome irrespective to fatty acid composition in women: the KNHANES 2007–2014. Int J Food Sci Nutr 68, 479–487. [DOI] [PubMed] [Google Scholar]
- 27. Ahn Y, Kwon E, Shim JE et al. (2007) Validation and reproducibility of food frequency questionnaire for Korean genome epidemiologic study. Eur J Clin Nutr 61, 1435–1441. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
For supplementary material accompanying this paper visit https://doi.org/10.1017/S1368980020001305.
click here to view supplementary material


