Abstract
While the lifespan advantage of small body size and mixed breed status has been documented repeatedly, evidence for an effect of genetic diversity across dog breeds is equivocal. We hypothesized that this might be due to a strong right-censoring bias in available breed-specific lifespan estimates where early-dying dogs from birth cohorts that have not died off completely at the time of data collection are sampled disproportionately, especially in breeds with rapidly growing populations. We took advantage of data on owner reported lifespan and cause of death from a large public database to quantify the effect of size and genetic diversity (heterozygosity) on mortality patterns across 118 breeds based on more than 40,000 dogs. After documenting and removing the right-censoring bias from the breed-specific lifespan estimates by including only completed birth cohorts in our analyses, we show that small size and genetic diversity are both linked to a significant increase in mean lifespan across breeds. To better understand the proximate mechanisms underlying these patterns, we then investigated two major mortality causes in dogs — the cumulative pathophysiologies of old age and cancer. Old age lifespan, as well as the percentage of old age mortality, decreased with increasing body size and increased with increasing genetic diversity. The lifespan of dogs dying of cancer followed the same patterns, but while large size significantly increased proportional cancer mortality, we could not detect a significant signal for lowered cancer mortality with increasing diversity. Our findings suggest that outcross programs will be beneficial for breed health and longevity. They also emphasize the need for high-quality mortality data for veterinary epidemiology as well as for developing the dog as a translational model for human geroscience.
Supplementary Information
The online version contains supplementary material available at 10.1007/s11357-022-00653-w.
Keywords: Lifespan, Old age, Cancer, Size, Genetic diversity, Dog breeds
Introduction
Artificial selection has made the domestic dog the most phenotypically diverse mammal in the world, as illustrated by the tremendous variation in morphology and behavior among breeds. This also holds true for life history traits such as adult lifespan, which varies about two-fold across dog breeds [1–3]. Given that dogs suffer from many of the same diseases as humans, share our environment, and have a sophisticated level of medical care, what we learn about lifespan in dogs is likely to have a high degree of relevance to humans as well [4–6].
The strongest determinant of lifespan in dogs is body size, with small dogs outliving their large counterparts (by about 5 years on average and up to 8 years) [7–10]. Another factor that has the potential to affect canine lifespan is genetic diversity or the level of inbreeding [7, 11–13]. The genetic diversity found across dogs as a species is comparable to humans, but within dog breeds it is much smaller [14, 15]. Reproductive isolation (i.e., closed stud books), founder effects, bottlenecks, and genetic drift as well as breeding practices such as selection for extremes, inbreeding, and the use of popular sires have led to strong genetic differentiation among dog breeds and high levels of homozygosity in purebred dogs [15–20]. The detrimental effects of inbreeding on fitness-related traits have been documented in a wide variety of wild, captive, and domestic populations [21–27]. Populations with small effective population sizes and high levels of homzygosity such as modern dog breeds are at a particularly high risk of inbreeding depression though the benign living environment of these dogs and purging might mitigate it to some extent [21, 25, 27–29]. In line with this, the lifespan advantage of mixed-breed dogs which typically have a much higher genetic diversity compared to purebred dogs has been robustly documented based on various data sources, including owner surveys [1, 2], cemetery data [30], teaching hospital data [7, 11] and primary veterinary practices ([31] but see [12]). Mixed breed dogs live on average about 1.2 years longer than similarly-sized purebred dogs [11, 31]. There is also some evidence that individual inbreeding level negatively affects juvenile survival and adult lifespan within breeds [11, 13].
Despite the effects of genetic diversity on lifespan on those scales, we could not confirm the expected negative correlation between inbreeding levels and lifespan across dog breeds when accounting for body size in an earlier study [11], even though more inbred breeds have recently been shown to have a higher burden of overall morbidity [32]. There are several possible reasons for this counterintuitive finding. One possible explanation discussed in the study [11] is that the effect of inbreeding on lifespan is primarily due to loss of allelic diversity and a concomitant accumulation of deleterious mutations caused by bottlenecks during breed formation, with breeds exhibiting a relatively similar degree of historic inbreeding depression. This idea is consistent with the fact that bottlenecks during domestication and breed formation, rather than further inbreeding of established breeds, explain the increased genetic load in dogs compared with wolves [33]. Nevertheless, data on genetic diversity across breeds suggests substantial variation from highly inbred breeds to breeds with levels of genetic diversity quite close to that of mixed breed dogs ([32], see table S1).
We might also fail to detect an expected relationship between inbreeding and lifespan due to (a) the fact that we do not have estimates of lifespan and inbreeding from the same individual dogs; and (b) the relatively low quality of available lifespan estimates in veterinary studies, most of which suffer from bias due to incomplete birth cohorts (i.e., right-censored data [34]). Studies that include only those dogs that die before a specified date can lead to underestimating lifespan, if only dogs dying relatively young are captured in the estimate for birth cohorts, such that a large proportion of dogs might still be alive at the time of sampling. This bias can be exacerbated when population numbers, and hence age structure, change over time, as is often the case with dog breeds gaining or losing popularity (see, e.g., [35] for a low median lifespan in a rapidly increasing population of Chihuahuas). If the bias across breeds is relatively similar and/or the effect of the variable of interest is very strong, as in the case of body size, its relationship with lifespan will still be detectable. For traits with smaller effects on lifespan, especially if cohort bias is related to the trait of interest, it might mask the effect of the trait.
To better understand how size and genetic diversity shape lifespan across breeds, we need to analyze their relationship with the underlying causes of death. Large size seems to incur lifespan costs mainly via an increased rate of aging [9], whereas the mixed breed lifespan advantage seems to be caused by both a decreased age-independent mortality hazard as well as a decreased rate of aging [11], suggesting that these two factors might differ in their effect on the major causes of death. The impact of a given cause of death on mean lifespan depends on two parameters—the age at which dogs succumb to this cause of death, and its prevalence in the population. Here we investigate the effect of body size and genetic diversity on two major causes of death in dogs, one usually deemed desirable — “dying of old age,” the other one often dreaded — cancer.
Strictly speaking, old age is not a cause of death as aging is not a disease (but see [36]), though for practical reasons it has been termed as such in surveys on dog mortality before, and we will do so here as well. While old age strongly increases the risk of dying from many diseases, it is not a terminal process itself. Given that dog owners typically choose to euthanize their pets once they consider the pet’s quality of life to be too poor for continued survival, their owner-determined lifespan has been suggested as a measure of the end of their healthspan [4, 5]. Hence, the lifespan of dogs that reportedly died of old age can be viewed as a measure of the breed-specific healthspan potential. With a reported prevalence of around 25% based on owner reports including purebred and mixed breed dogs [1, 2], old age mortality in dogs seems of a similar magnitude as cancer mortality, but so far, no analysis of factors affecting its prevalence has been undertaken. Cancer is usually an age-related disease and the single most common cause of death in dogs, accounting for about 15–30% of all deaths but with prevalence varying widely across breeds (reviewed in [37]). Because of the similar prevalence and biology of canine cancers as in humans, dogs have become a valuable model in comparative oncology [38–44]. Several studies have documented an increased cancer risk in larger breeds [1, 38] and Fleming et al. [45] showed that larger size is linked to an increased risk of dying of neoplasia, but not of other pathophysiological processes based on a large sample of dogs dying at veterinary teaching hospitals. Evidence for an effect of genetic diversity on cancer mortality is more ambiguous. While some evidence points to increased cancer incidence in purebred compared to mixed breed dogs [46, 47], Bellumori et al. [48] did not find a significant mixed breed advantage in cancer prevalence. None of these studies, however, accounted for size effects on cancer mortality. We are not aware of any analyses that have looked at the effect of size or genetic diversity on cause-of-death-specific lifespan.
Taking advantage of a large public database for purebred dogs (koiranet of the Finnish Kennel Club), we address the problem of bias in lifespan and prevalence estimates by only including completed birth cohorts (i.e., all dogs of these cohorts have died off) in our mortality parameter estimates. Additionally, we quantify the bias in breed-specific lifespan estimates caused by incomplete birth cohorts and changing registration numbers and show that this indeed can explain our earlier failure to detect the relationship between genetic diversity and lifespan across dog breeds. We then quantify the effect of body size and genetic diversity on breed-specific mean lifespan, as well as lifespan and the proportion of old age and cancer mortality. We also show that our findings hold true when accounting for the relatedness among breeds.
Methods
Mortality data
We used average breed-specific lifespan and cause of death data from the public database koiranet of the Finnish kennel club (https://jalostus.kennelliitto.fi/frmEtusivu.aspx?Lang=en&R=57). To avoid the problem of underestimating lifespan due to the inclusion of incomplete birth cohorts (i.e., individuals born and registered within a specific calendar year), and additional bias due to changing registration numbers, we restricted the data to the birth cohorts of 1988 to 2002 (data extraction January 2020). This means that only the few dogs of the later birth cohorts that completed their 18th year of life (for 2002), 19th year of life (for 2001) and so on, are not included in the data set, which might lead to minor underestimates of lifespan, especially in small breeds where such long lifespans are more frequent. Restricting the included birth cohorts even further would have decreased the number of breeds with a large enough sample size substantially.
Table 1 lists the mortality parameters we extracted from the data. To exclude extrinsic mortality from our corrected mean lifespan estimate, we recalculated the average lifespan given for each breed without the causes of death categories “accident,” “damage done by large carnivores,” “euthanasia due to behavioural problems,” “lost,” “wash-outs” (dogs euthanized because of unsuitability for the intended working purpose), and “cause of death not specified” for all breeds. We did not exclude the categories “dead without diagnosis of illness,” “euthanasia, non-diagnosed,” because the percentage of dogs in these categories that might have died due to extrinsic causes of deaths is likely very small (results hold qualitatively if these are excluded, and also if “cause of death unspecified” is included).
Table 1.
Mortality parameters computed for each breed and their meaning. Further explanations can be found in the text
| Mortality parameter | Explanation |
|---|---|
| Uncorrected mean lifespan | Average age at death with extrinsic causes of death excluded, all available data included until January 2021 |
| Corrected mean lifespan | Average age at death with extrinsic causes of death excluded, data restricted to birth cohorts from 1988 to 2002 |
| Bias in mean lifespan | Difference between corrected and uncorrected mean lifespan |
| Old age lifespan | Average age at death of dogs with the reported cause of death ‘old age (natural or euthanasia)’ |
| % Old age mortality | Percentage of dogs dying of the reported cause of death ‘old age (natural or euthanasia)’ |
| Cancer lifespan | Average age at death of dogs with the reported cause of death ‘tumor, cancer’ |
| % Cancer mortality | Percentage of dogs dying of the reported cause of death ‘tumor, cancer’ |
We additionally extracted average breed lifespan without the birth cohort restriction (complete causes of death data available for a given breed up to the end of January 2021). As with the restricted data set, we recalculated this parameter excluding clearly extrinsic causes of mortality and deaths with unspecified cause (uncorrected mean lifespan). To estimate the bias in breed lifespan estimates introduced by incomplete birth cohorts and changes in population numbers, we computed the difference between the breed-specific mean lifespan estimate based on the complete and the restricted data set (bias in mean lifespan). For all breeds, we calculated the percentage change in annual registration numbers (i.e., the number of new individuals in a breed that are registered by the Finnish Kennel Club) over the years only included in the estimate of uncorrected mean lifespan, based on the difference between mean annual registration numbers for the years 2002–2006 and the years 2016–2020.
As a rough measure or proxy of breed-specific healthspan potential, we used the average age at death of dogs with the cause of death reported as “old age (natural or euthanasia),” which we refer to as old age lifespan. Additionally, we computed the proportion of dogs that reportedly died of old age as the % old age mortality. To characterize breed-specific cancer mortality, we extracted the average age at death for dogs with the reported cause of death “tumor, cancer” (i.e., cancer lifespan). Furthermore, we calculated the % cancer mortality as the proportion of dogs that died of cancer. In this case, we only included deaths with a specific diagnosis in the denominator, i.e., we excluded dogs in the cause of death categories “dead without diagnosis of illness,” “euthanasia, non-diagnosed.” It seems plausible that a non-negligible percentage of deaths in these categories were due to cancer, and therefore including them would have underestimated cancer rates (results hold if these are not excluded). The next most common cause of death in the database was heart disease, but case numbers for this and the other categories of death were too limited for many breeds to obtain meaningful cause-specific mortality estimates.
We included data from 119 breeds for which at least 80 deaths were documented. These breed-specific estimates of corrected mean lifespan are based on a total of 40,841 dogs.
Body size, genetic diversity, and genetic relatedness data
When available, we used the average of the female and male size (i.e., body mass) described in the Fédération Cynologique Internationale (FCI) breed standard. For breeds without an expected body mass in the breed standard, we used the expected body mass found in other breed descriptions from the internet (mainly American Kennel Club [AKC] standards). As an estimate of breed-specific genetic diversity, we used median heterozygosity measured by MyDogDNA® (Genoscoper Laboratories Oy, or Optimal Selection ®, Wisdom Health, Vancouver, WA, USA) and published by [32]. Since Genoscoper was originally located in Finland, the majority of the dogs tested so far come from Europe and many from Finland. Hence, the dogs from which the genetic diversity data come are not identical to those used for the mortality estimates, but most come from the same, albeit temporally derived European breed populations. Estimates were provided for breeds with at least 30 genotyped individuals. In contrast to the study by Yordy et al. (2019), levels of genetic diversity were not significantly correlated with body size (Pearson correlation: r =—0.062, p = 0.504).
We used genetic data from a recent large-scale dog genotyping analysis [49] that was generated using the Illumina Canine HD SNP chip (Illumina, San Diego, CA), which covers 172,000 SNPs spread out across the dog genome. We estimated genetic relatedness among individuals by calculating identity-by-state (IBS) for all pairs of individuals using PLINK v.1.9 [default settings: plink—file datafile—cluster—matrix;,50]. Using this matrix, we then generated breed-average IBS matrices by calculating the mean IBS within breeds and between pairs of breeds (Table S2).
Statistical analyses
To test for the difference between uncorrected and corrected mean lifespan (i.e., the bias), we used a paired t test. We used linear regression to analyze the relationship between bias as well as our mortality parameters and our predictor variables body size, genetic diversity (heterozygosity), and percentage change in registration numbers. For the three percentage variables, we first used logistic regression, but model assumptions were violated (significant overdispersion and the response was not linear on a logit scale). In contrast, linear model assumptions were met relatively well, and therefore we used linear regression for these, too. To identify statistical outliers, we used studentized residuals and Bonferroni corrected p-values [51]. We checked for influential points visually using Cook’s distances and changes in the regression coefficients [51]. One breed, the Basenji, proved to be a highly influential point in several of the analyses, leading to a change of around 20% in the β coefficient for heterozygosity, and was subsequently removed from all analyses except the bias and the percentage change analyses. This resulted in a sample size of 118 breeds for analyses of mortality variables. None of the statistical outliers were influential points, so we did not remove them. We also did not remove influential points identified in only one of our analyses, because we wanted to keep the data set constant for our mortality analyses to ensure comparability of R2 values and β coefficients. Table S3 gives an overview of outliers and their direction as well as influential points and their effects on the model statistics. None of these decisions affected our basic results. Because sample sizes varied widely among breeds, we also checked for the need to use weighted regression using the DuMouchel-Duncan test of model change with weights. None of the tests indicated a significant change for the weighted model.
Due to a shared phylogenetic history, dog breeds are not strictly independent data points. To ensure that our results are robust with respect to breed relatedness, we repeated our analyses, employing phylogenetic comparative methods that controlled for breed-level identity by descent (studies that used this approach for research on dogs include [52–54]). First, we used a mixed model approach (package EMMREML [55]). Second, we used a Bayesian regression model framework (package brms [56, 57]). Although models did converge with this latter approach, results need to be interpreted with caution since for most models a number of divergent transitions and low effective sample sizes were reported. Nevertheless, estimated model parameters were very similar to the EMMREML approach. Because genetic information was not available for all breeds for which we had data on mortality, size, and genetic diversity, only 96 breeds could be included in these analyses. After accounting for breed relatedness, the results were qualitatively the same and quantitatively very similar to those based on the analyses including all breeds (Tables 2, S4, and S5).
Table 2.
Statistical effects of body size, heterozygosity, and the percentage change in registration numbers (% change) on our parameters of interest.
| Statistic | β | se(β) | t | P | Partial R2 (%) |
|---|---|---|---|---|---|
| Bias in mean lifespan (n = 119, Radj2 = 49.8%) | |||||
| Intercept | 0.178 | 0.273 | 0.650 | 0.517 | |
| Size | − 0.005 | 0.002 | − 2.075 | 0.040 | 3.6 |
| Heterozygosity | 0.031 | 0.008 | 3.819 | 0.0002 | 11.3 |
| % change | 0.006 | 0.001 | 7.733 | 4.37e-12 | 34.2 |
| Percentage change in registration numbers (n = 119, Radj2 = 12.3%) | |||||
| Intercept | − 112.312 | 33.958 | − 3.307 | 0.001 | |
| Size | − 0.386 | 0.298 | − 1.293 | 0.199 | (1.4) |
| Heterozygosity | 4.017 | 1.002 | 4.010 | 0.0001 | 12.2 |
| Uncorrected mean lifespan (n = 118, Radj2 = 51.3%) | |||||
| Intercept | 10.463 | 0.683 | 15.318 | < 2e-16 | |
| Size | − 0.063 | 0.006 | − 10.932 | < 2e-16 | 51.0 |
| Heterozygosity | 0.030 | 0.021 | 1.514 | 0.133 | (2.0) |
| Corrected mean lifespan (n = 118, Radj2 = 61.0%) | |||||
| Intercept | 10.000 | 0.663 | 15.08 | < 2e-16 | |
| Size | − 0.070 | 0.006 | − 12.49 | < 2e-16 | 57.6 |
| Heterozygosity | 0.084 | 0.019 | 4.34 | 3.07e-05 | 14.1 |
| Old age lifespan (n = 118, R2 = 56.5%) | |||||
| Intercept | 11.357 | 0.577 | 19.676 | < 2e-16 | |
| Size | − 0.053 | 0.005 | − 10.915 | < 2e-16 | 50.9 |
| Heterozygosity | 0.084 | 0.017 | 4.958 | 2.48e-06 | 17.6 |
| % Old age mortality (n = 118, Radj2 = 39.5%) | |||||
| Intercept | 26.433 | 5.318 | 4.970 | 2.35e-06 | |
| Size | − 0.323 | 0.045 | − 7.212 | 6.29e-11 | 31.1 |
| Heterozygosity | 0.707 | 0.156 | 4.529 | 1.45e-05 | 15.1 |
| Cancer lifespan (n = 118, Radj2 = 56.9%) | |||||
| Intercept | 9.675 | 0.633 | 15.290 | < 2e-16 | |
| Size | − 0.062 | 0.005 | − 11.607 | < 2e-16 | 53.9 |
| Heterozygosity | 0.069 | 0.019 | 3.716 | 0.0003 | 10.7 |
| % Cancer mortality (n = 118, Radj2 = 13.5%) | |||||
| Intercept | 25.929 | 5.807 | 4.465 | 1.88e-05 | |
| Size | 0.195 | 0.049 | 3.996 | 0.0001 | 12.2 |
| Heterozygosity | − 0.295 | 0.170 | − 1.730 | 0.086 | (2.5) |
Provided are sample size n and adjusted R2 for the model, the β estimate, its standard error, the test statistic t, the P-value, and partial R2 values for all predictors
All statistical analyses were performed in R version 4.0.3 [58]. Effect plots showing partial residuals were created using package jtools [59].
Results
Table 2 provides statistics for all regressions performed and Table S1 shows the breed-specific values for all variables used in the analyses.
Bias in lifespan estimates
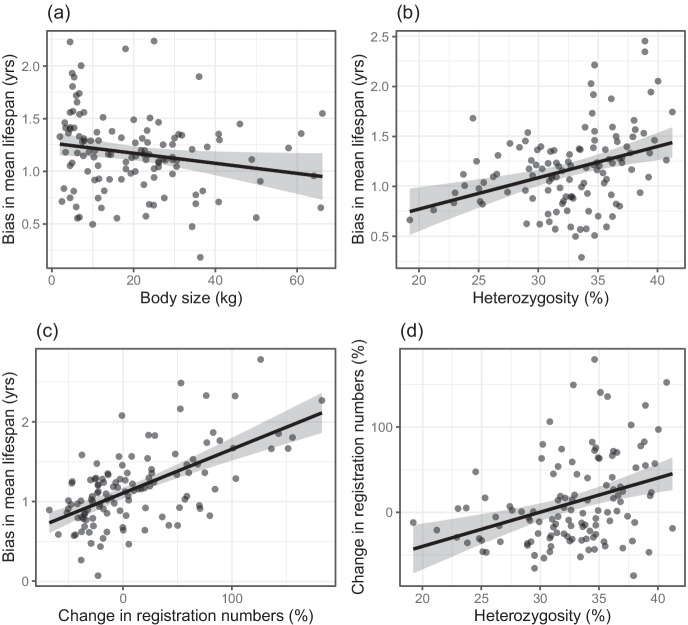
Lifespan estimates for the dataset including only completed birth cohorts were higher than those based on the unrestricted data set for all breeds with the difference in means being statistically significant (paired t test: mean bias = 1.17 years, t118 = 24.227, P < 2.2e-16). The estimated bias ranged from 0.02 to 3.0 years (Table S1) and was statistically linked to our variables of interest. It increased significantly with decreasing size and increasing heterozygosity (Fig. 1a,b). As expected, it also increased significantly with the percentage change in registration numbers, which explained 34.2% of its variance (Table 2, Fig. 1c). This shows that the mean lifespan of small and more heterozygous breeds as well as those rising in popularity is underestimated. The percentage change in registration numbers was significantly larger for breeds with higher heterozygosity (Table 2, Fig. 1d).
Fig. 1.
(a) The estimated bias decreases with body size (P = 0.040, partial R2 = 3.6%), (b) increases with heterozygosity (P < 0.001, partial R2 = 11.3%), and (c) with the percentage change in registration numbers (P < 0.001, partial R2 = 34.2%). (d) Breeds with higher heterozygosity showed a stronger percentage increase in registration numbers (P < 0.001, partial R2 = 51.2%). Depicted are predicted values with 95% confidence intervals and partial residuals accounting for the effect of the other variables in the regression models (n = 119)
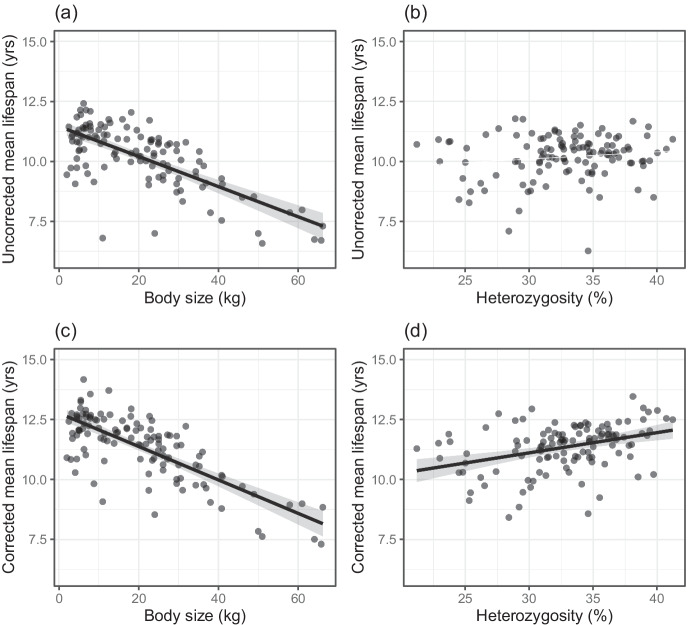
Mean lifespan
For uncorrected mean lifespan, which varied between 6.4 and 12.3 years (Table S1), the model predictors accounted for 51.3% of the variance (Table 2). While size was a statistically significant predictor, heterozygosity was not significantly associated with this parameter (Fig. 2a b, Table 2). Corrected mean lifespan varied between 7.0 and 13.9 years (Table S1). When the bias introduced by incomplete birth cohorts was removed, corrected mean lifespan decreased significantly with increasing body size, and increased significantly with increasing heterozygosity (Fig. 2c d, Table 2). The two factors together explained 61.0% of the variance in mean lifespan across breeds. Judged by partial R2 values, size was a relatively more important predictor than heterozygosity (Table 2). For each additional 1 kg of body mass of the averaged breed standard mass, mean breed lifespan is estimated to decrease by 25.6 days, and for each additional one percent of heterozygosity, a breed is predicted to gain 30.7 days of lifespan. Over the observed size range, that translates into a difference of 4.5 years in predicted mean lifespan from the smallest to the largest breeds, and a difference of 1.7 years from the lowest to the highest observed heterozygosity value.
Fig. 2.
(a) Uncorrected mean lifespan decreases with body size (P < 0.001, partial R2 = 51.0%), (b) but it lacks a statistically significant association with heterozygosity (P = 0.133, partial R2 = 2.0%). (c) Corrected mean lifespan decreases with size (P < 0.001, partial R2 = 57.6%) and (d) increases with heterozygosity (P < 0.001, partial R2 = 14.1%). Depicted are predicted values with 95% confidence intervals and partial residuals accounting for the effect of the other variables in the multiple regression model (n = 118)
Causes of death
Old age mortality
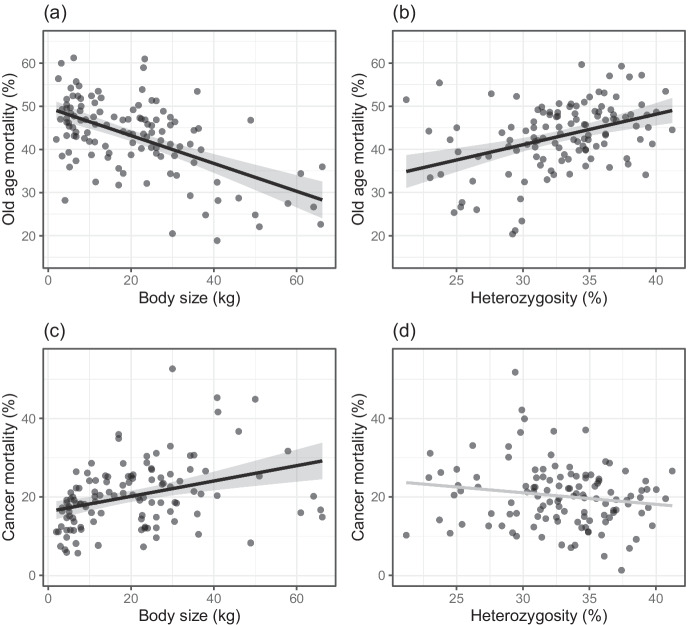
The average lifespan of dogs reportedly dying of old age varied between 9.3 and 14.9 years across breeds (Table S1). On average, old age lifespan was 1.7 years higher than mean lifespan across breeds, and dogs dying of disease died on average 2.9 years younger than those dying of old age. The findings observed for old age lifespan were similar to those for corrected mean lifespan. We found that old age lifespan was significantly negatively correlated with body size and positively correlated with heterozygosity (Table 2, Fig. S1a, b). The percentage of dogs dying of old age varied between 16.8 and 62.5% across breeds (Table S1) and was also negatively correlated with body size and positively correlated with heterozygosity (Fig. 3a b, Table 2). The full model explained 39.5% of the variance in the percentage old age mortality (Table 2), with heterozygosity explaining 15.1% of the variance, which increased to 22.6% when we removed two influential points, Rough Collie and Smooth Collie. We found that the predicted percentage old age mortality increased by 1% for every decrease of 3 kg in breed size or, respectively, each increase of 1.4% in heterozygosity. Over the observed size range, that translates into a difference of 20.7% in the predicted percentage old age mortality from the smallest to the largest breed, and a difference of 14.1% from the lowest to the highest observed heterozygosity value.
Fig. 3.
(a) The percentage of old age mortality decreases with body size (P < 0.001, partial R2 = 31.1%) and (b) increases with heterozygosity (P < 0.001, partial R2 = 15.1%). (c) The percentage of cancer mortality increases with body size (P < 0.001, partial R2 = 12.2%), (d) but there was only a weak trend for it decreasing with heterozygosity (P = 0.086, partial R2 = 2.5%). Depicted are predicted values with 95% confidence intervals and partial residuals accounting for the effect of the other variables in the multiple regression model (n = 118)
Cancer mortality
Cancer lifespan varied between 6.7 and 13.0 years (Table S1). It was on average 0.7 years lower than mean lifespan and 2.4 years lower than old age lifespan across breeds. With respect to size and heterozygosity, the pattern for cancer lifespan mirrored that for the other lifespan parameters — cancer lifespan decreased significantly with body size and increased significantly with heterozygosity (Table 2, Fig. S1c, d). The percentage cancer mortality varied between 5.8 and 53.7% (Table S1) and the full model explained 13.5% of the variance in the percentage of cancer mortality. It increased significantly with body size and tended to decrease with heterozygosity (Fig. 3c d, Table 2). However, the trend was not replicated in the reduced data set correcting for breed relatedness and removing an outlier (Flat-Coated Retriever) erased it. Each 1 kg of additional size increased the predicted percentage of cancer mortality by a bit more than 0.2%, which translates into a difference of 12.5% in the predicted percentage cancer mortality over the observed range of breed sizes.
Discussion
Our findings show that both body size and genetic diversity are important factors in shaping lifespan across dog breeds, with body size exerting a stronger effect than genetic diversity across all mortality measures. Genetic diversity played a larger role in old age mortality than in cancer mortality. However, both of these owner-reported cause of death categories are very heterogenous, so more detailed analyses are needed to improve the accuracy and interpretation of the quantitative estimates of these associations.
Mean lifespan
Our study shows that the bias in lifespan estimates due to right-censored age-at-death data as well as changes in population size and age structure can be substantial–up to 3 years in our set of breed populations. Both of our variables of interest affected the size of the estimated bias, with heterozygosity showing a tighter association than body size. The larger bias in small and more heterozygous breeds is consistent with the fact that right-censoring bias is expected to be higher in long-lived population [34]. As expected, we saw a strong correlation between population increase and the estimated bias, consistent with other studies that found low lifespan in rapidly growing breed populations [35, 60, 61] Interestingly, change in breed popularity was also linked to breed average heterozygosity, suggesting that heterozygosity affected right-censoring bias in our population via two mechanisms — increased longevity and popularity. Factors that might contribute to the increase in popularity of more genetically diverse breeds are that newly recognized breeds become fashionable over time and/or people might switch away from very inbred breeds due to health concerns. The causality of the relationship might also be reversed, with breeds losing genetic diversity as they become rarer. The fact that the bias in lifespan estimates was positively correlated with genetic diversity likely explains why we could not detect its expected effect on lifespan across dog breeds in an earlier study [11]. Once this bias is largely corrected for, not only size, but also genetic diversity emerges as an important factor in shaping lifespan across dog breeds.
The strong negative effect of body size on breed-specific mean lifespan we document here is consistent with earlier studies [8–11, 31, 62]. However, we found a larger percentage of variance in mean lifespan explained by size compared with earlier studies (this study 58%, 40% [10], 44% [9]). Comparing the partial R2 of size for uncorrected and bias corrected mean lifespan suggests that this difference is due in part to the fact that we corrected for right-censoring bias. Additionally, our high explanatory power might have been influenced by the fact that we took genetic diversity into account as an additional predictor, that we used a relatively larger sample size for many breeds, and that we considered a more representative sample population compared to, e.g., teaching hospital data. Despite size being the strongest determinant of lifespan in dogs, we still do not fully understand the proximate mechanisms underlying this relationship. The insulin-like growth factor (IGF-1) pathway, a known modulator of lifespan in many model organisms, likely contributes to the lifespan advantage of small dogs [63] which have been shown to have lower IGF-1 levels than their large counterparts [64–66]. More recent studies suggest that glycolytic metabolic rates [67], mitochondrial bioenergetics and thermoregulation [68] or tryptophan metabolism [69] might play a role in lifespan differences of small and large dogs. Jimenez [70] reviews the physiological mechanisms underlying canine aging.
More genetically diverse breeds outlived their more inbred counterparts, with each percent in median heterozygosity adding one month in predicted mean lifespan when accounting for body size. This finding is well in accordance with expectations based on population genetic theory and in line with previous studies reporting a lifespan advantage of mixed breed dogs [7, 11, 31] as well as a negative effect of individual pedigree-based inbreeding coefficient on lifespan documented in at least one breed (Golden Retrievers [11],but see [71] for Irish Wolfounds). Similarly, Urfer et al. [12] found that breeds with an inbreeding coefficient below the median enjoyed a lifespan that was 3 to 6 months longer than those above the median inbreeding coefficient, but, surprisingly, they did not detect a lifespan advantage of mixed breed dogs. Across the range of observed genetic diversity in our breed sample, the effect of genetic diversity translated into an average lifespan gap of about 1.7 years between the least and the most genetically diverse breed, which is at the higher end of the reported estimates of the lifespan gap between mixed breed and purebred dogs (1.8 years [7], 1.2 years [11, 31]). The relative importance of genetic diversity was smaller than that of body size, but it is possible that we are underestimating its explanatory power as measured by partial R2. First, the effect of inbreeding depression on lifespan also has a random component. Because of founder effects and subsequent breeding events (e.g., a specific disease-linked mutation in a popular sire), different deleterious alleles linked to mortality become enriched in the various breed populations and the ages at death for those causes will vary. For example, dilated cardiomyopathy in Dobermans kills dogs at a younger age than degenerative myelopathy in Corgis, (see also [72]). Second, the population on which our mortality parameter estimates are based is not the same as, but rather ancestral to, the one that has been genotyped for the heterozygosity estimates. This might introduce a variable amount of error in the median heterozygosity estimates across breeds.
“Dying of old age”
Old age was the most reported cause of death in our study population. Therefore, it is not surprising that lifespan patterns in this category largely match those for mean lifespan. Interestingly, the relative importance of size was somewhat smaller and the slope less steep than for mean lifespan. The expected lifespan gap across the range of sizes decreased to 3.4 years compared to 4.5 years for mean lifespan. Rapid growth has been conceptually linked to a higher probability of developmental errors and consequently “jerry-built” bodies [73, 74]. Kraus et al. [9] hypothesized that this might cause a higher variance in quality in large dogs compared to small ones. Hence, while on average the bodies of large dogs break down earlier, there will be some individuals that escape a high burden of errors. The size-related pattern we observed in our study (i.e., a lower percentage of large dogs dying of old age) combined with a somewhat attenuated effect of size on the lifespan of those dogs fits this idea well. However, so far there is no clear empirical evidence of a higher burden of replication error in large dogs. There is some indication for lowered production of reactive oxygen species (ROS) in cells of long-lived breeds [67, 68], but the link to size is less clear [70]. Contrary to expectations, a higher rate of damage in circulating lipids was found in small compared to large dogs [52].
Other factors might also contribute to the observed pattern. Large dogs with a similar morbidity burden to small dogs might be more difficult to manage and therefore the decision for euthanasia might be reached earlier, which could also contribute to the body size effect on old age lifespan. Moreover, while people are generally aware of the fact that large dogs have lower life expectancies, they might still be less likely to consider “old age” as the appropriate cause of death category, because ages of death below about 10 years might still seem young for dogs in general. This might decrease the old age mortality percentage for larger dog breeds.
Increased genetic diversity was positively correlated with old age lifespan as well as the percentage of dogs dying of old age. The latter makes intuitive sense. Founder effects during breed creation and subsequent breeding practices have inadvertently led to an enrichment of deleterious mutations in the closed populations of purebred dogs, resulting in a multitude of breed-specific disease predispositions [75, 76]. Inbreeding depression is thought to be mainly caused by deleterious recessive alleles becoming unmasked with increased homozygosity [26]. Consistently, while mixed breed dogs were found to more likely carry a common recessive disease-linked mutation, purebred dogs were more likely to be genetically at risk, i.e., homozygous for such mutations [77]. Hence, we would expect that we are more likely to find homozygosity of recessive disease-associated alleles in breeds with a low average heterozygosity, so more individuals are expected to succumb to disease vs. dying of age-related morbidities. The higher level of morbidity in more inbred dog breeds is in line with this expectation [32].
The shorter lifespan of the aged population in less genetically diverse breeds emphasizes that inbreeding not only affects canine lifespan via an increased likelihood of dying of inherited diseases, but also more generally in a cumulative way. Indeed, Marsden et al. [33] found that weakly deleterious mutations contributed most of the additive genetic load in dogs. There is some evidence that mixed breed dogs age slower than purebred dogs when size is accounted for [11] but to confirm that the rate of aging is negatively associated with genetic diversity across breeds we need to construct age-specific mortality and morbidity trajectories, which was outside the scope of this study. Consistent with an effect of genetic diversity on age-related morbidity, a study of the Hungarian dog population based on owner questionnaires found that purebred dogs started to suffer from health problems at an earlier age than mixed breed dogs, and suggests that mixed breed dogs might have a longer healthspan than their purebred counterparts [78]. Intriguingly, the explanatory power of genetic diversity on old age lifespan was larger than for mean lifespan or cancer death lifespan, perhaps because the variable age at onset of diseases associated with mortality adds variance to the relationship compared to the effect of genetic diversity on aging per se.
The group of dogs dying or being euthanized because of age-related morbidities rather than an unequivocal disease process can provide an especially valuable comparative model for human research into morbidity and mortality patterns in old age [4, 5]. Across breeds, dogs that reportedly died of old age lived on average 3 years longer than those that succumbed to a specified disease, albeit with substantial variation around this mean value. Studies of human centenarians have suggested that some people are able to live a particularly long and healthy lifespan as “escapers,” individuals that live without chronic disease for their entire lives, while others are “survivors,” managing to not succumb to chronic disease [79]. Ongoing long-term longitudinal studies such as the Dog Aging Project [80] and the Golden Retriever Lifetime Study [81] might tell us whether the same is true in dogs.
Dying of cancer
The lifespan patterns for dogs dying of cancer mirrored that for mean lifespan, showing that cancer deaths contribute to the negative effect of size and the positive effect of genetic diversity on mean lifespan. Just as in humans and other species, canine cancer is typically an age-related disease [41, 82, 83], likely because the aging process and cancer development share several important pathways (reviewed in [63, 82]). Hence, the increased rate of aging in large as well as purebred dogs [9, 11] might be causally linked to the shorter lifespan of dogs of larger and genetically less diverse breeds that die of cancer. One of the proximate mechanisms involved might be the growth hormone or insulin-like growth factor I pathway, since small dogs have lower serum IGF-1 levels than large dogs [64–66]. Some dwarf mice with disruptive mutations in this pathway show increased lifespan and a later onset of cancer (reviewed in [82]). There is also intriguing evidence of cancer protection in a human population with congenital IGF1 deficiency [84]. Our finding that size affects the lifespan of dogs dying of cancer is consistent with earlier evidence suggesting that the onset of cancer might occur later in smaller breeds [85].
Across our breed sample, dogs dying of cancer lived on average 0.9 years longer than those dying of other diseases, consistent with cancer being a disease that typically strikes at an advanced age, but they still lost on average 2.4 years of life compared to those reportedly dying of old age. The evolutionary model of cancer suggests that late onset cancers are often sporadic in origin, i.e., due to somatic mutations alone, while inherited genetic variants are more likely to contribute to the development of early-onset cancers [86]. Due to founder effects, genetic drift and breeding practices, many breeds are at an increased risk for specific types of cancer with variable ages of onset [37, 40, 43]. So, while in some breeds sporadic cancers with little lifespan costs might predominate, in others, these genetic predispositions lead to higher lifetime costs (e.g., histiocytic sarcoma in Flat Coated Retrievers and Bernese Mountain Dogs). This variation might also contribute to the somewhat lower explanatory power of genetic diversity for cancer mortality lifespan compared to mean or old age lifespan.
With respect to the proportional cancer mortality across breeds, we could only identify size as a statistically significant risk factor. Whereas across species, cancer risk is largely independent of size, an observation known as Peto’s Paradox [87–90], the evolutionary model of cancer predicts that within species, cancer risk increases with size, simply because of a larger cell number and higher cell division rate in larger individuals [86, 91, 92]. This should also hold true for dog breeds, since likely not enough time has elapsed on an evolutionary timescale for natural selection to adjust mechanisms of cancer resistance accordingly [88]. Consistent with this, Michell [1] documented higher cancer mortality in large compared to small breeds and Fleming et al. [45] showed that larger size is linked to an increased risk of death due to neoplasia in a large sample of dogs dying at veterinary teaching hospitals (see also [83]). Our findings confirm this relationship for a more representative population of purebred dogs. This within-species effect of size has also been documented for humans for various cancers [93]. Additionally, mice with mutations disrupting the GH/IGF-1 pathway are typically smaller and tend to have a decreased cancer incidence (reviewed in [63, 82]), suggesting that this pathway is also implicated in the decreased cancer mortality risk of small dogs. Increased glycolytic metabolic rates might also contribute to the predisposition of larger dogs to cancer [67, 70]. For a more complete understanding of the positive correlation between size and cancer risk in dogs, it will be necessary to distinguish between cancer types. For example, while size has a major effect on risk of osteosarcoma (reviewed in [94]), we know less about the importance of size for other types of canine cancer. Additionally, analysis of the association between size and cancer risk within breeds could help us to separate the relative effects of size and breed.
We found only weak, non-robust support for an increase in cancer death risk with decreasing genetic diversity in our data set. This is in line with the lack of a strong support for a mixed breed advantage in cancer death risk ([31, 48], but see [47]). Still, several factors might have masked a more clear-cut effect of genetic diversity on proportional cancer mortality. While suggestive evidence exists that low genetic diversity and inbreeding are linked to an increased cancer risk (reviewed in [95]), the effect does not necessarily have to be linear. Different deleterious alleles rise to high frequency in different populations, with oncogenic mutations appearing to be enriched only in some breeds (reviewed in [37, 40, 42, 44]). Hence, we might expect to see a linear effect of inbreeding level on cancer risk within breeds (e.g., Golden Retrievers [19]), but not necessarily across breeds. Furthermore, the effect of genetic diversity on cancer risk might be tumor specific and not evident in overall cancer risk ([95], e.g., mammary cancer [96], lymphoma [19]). A methodological caveat is that we were not able to account for competing hazards in our analyses, so if an early-onset disease kills many dogs in a breed, a high cancer risk might be masked. Finally, the quality of our cause of death data is not very high. The causes of death in the koiranet database are all owner-reported (i.e., no validation of the diagnosis necessary), and the categories provided are not fully mutually exclusive. In particular, when older dogs have cancer, some owners might not pursue a definitive diagnosis for the actual cause of death and enter their death as due to old age rather than cancer.
Study limitations
There are two main solutions to the problem of biased lifespan estimates due to incomplete birth cohorts. One is using statistical methods developed for right-censored data; the other is to eliminate death data from individuals coming from birth cohorts with a significant percentage of dogs still alive [34]. Urfer et al. [12] used the first method to investigate factors affecting lifespan based on data from primary care US veterinary hospitals. However, if the censoring itself is not random with respect to the outcome, this might lead to biased estimates as well, which might explain the unusually long life-expectancies reported by Urfer et al. [12]. We took the alternative approach and included only completed birth cohorts in our estimates. While this avoids the problem explained above, it is not without potential problems of its own. We inevitably miss more recent changes in breed-specific mortality resulting from changing prevalence of diseases, such as those due to increasing allele frequencies of deleterious mutations because of recent inbreeding or decrease in these same alleles due to better breed management via genetic testing. Another caveat comes from the fact that the population on which our mortality parameter estimates are based is not the same as, but mainly ancestral to, the one that has been genotyped for the genetic diversity estimates used.
We used a large, well-used public database, which allowed us to include more than 40,000 dog deaths while avoiding the problem of case selection bias inherent in earlier large-scale studies on dog mortality based on teaching hospital or insurance data [9, 11, 45, 85, 97]. Because of the large sample size and the permanent accessibility of the database for owners, our study population is likely also more representative than those based on short-term owner surveys. Nevertheless, there are several potential sources of bias that we were not able to control for in our analyses. Desexing has been linked to extended lifespan, especially for females ([31, 98], but see [99, 100]), and to increased cancer risk (reviewed in [101, 102]). However, desexing dogs is not a common practice in Finland [103], rendering it unlikely that this is a major source of error in our analyses. One major drawback shared by many population-based studies on dog mortality is the owner classification of the cause of death data, resulting in a higher degree of uncertainty for this part of our study. Finally, as in most dog mortality studies published so far, most dogs included in our analyses likely were euthanized (estimated at 86% in UK dogs [31]) and euthanasia decisions might partly depend on size.
Conclusions
Our large-scale study shows that size affects lifespan across dog breeds not only via decreasing the age at which dogs die from two major causes of death, but also by affecting the proportional mortality due to “old age” and cancer. Many of the large breeds have become even larger over the last century (compare today’s breed weights to those in, e.g., [104–106]). Our findings suggest that reversing this trend would decrease the probability of these dogs dying prematurely from disease, and hence improve lifespan as well as healthspan. Correcting for right-censor bias, we were able to provide strong evidence that genetic diversity does indeed impact lifespan across dog breeds. As with size, genetic diversity affected the age at death from “old age” and cancer as well as the proportion of old age deaths. These results strongly suggest that many breeds would not only benefit from managing the existing breed-wide genetic diversity, but also from outcross programs to increase genetic diversity [107, 108]. Population genetic simulations by Windig and Doekes [109] emphasize that continuous, low-level outcrossing is needed to minimize inbreeding rate in small populations, typical for many dog breeds. The analyses presented here of two important sources of mortality shed some light on which mortality components behind lifespan are affected by our focal variables, though studies with improved diagnostic validity are clearly needed to confirm our findings and suggest specific underlying mechanisms. The highest quality data on mortality and morbidity come from prospective cohort studies, but only few small-scale ones have been conducted so far in dogs [110, 111]. Large-scale longitudinal studies such as the Golden Retriever Lifetime Study [81] and the Dog Aging Project [80] have the potential to substantially improve our understanding of the determinants and underlying mechanisms shaping mortality and morbidity patterns in our canine companions.
Supplementary Information
Below is the link to the electronic supplementary material.
Acknowledgements
NSM and DELP were supported in part by the Dog Aging Project U19 grant AG057377 (PI: Daniel Promislow) from the National Institute on Aging, a part of the National Institutes of Health. We would also like to thank the koiranet team and the Finnish Kennel Club for providing such a valuable resource and making it publicly available. Thanks to Mirka Jantunen and Karina Whittington for breed-specific info. We are grateful for the helpful comments made by the anonymous reviewers on the manuscript.
Declarations
Conflict of interest
Dr. Promislow receives payment for reviewing grants for the Waltham Petcare Science Institute. The other authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Cornelia Kraus, Email: ckraus@posteo.de, http://www.dogagingproject.org.
Noah Snyder-Mackler, Email: nsnyderm@asu.edu, http://www.dogagingproject.org.
Daniel E. L. Promislow, Email: promislo@uw.edu, http://www.dogagingproject.org
References
- 1.Michell AR. Longevity of British breeds of dog and its relationships with sex, size, cardiovascular variables and disease. Vet Rec. 1999;145:625–529. doi: 10.1136/vr.145.22.625. [DOI] [PubMed] [Google Scholar]
- 2.Proschowsky HF, Rugbjerg H, Ersbøll AK. Mortality of purebred and mixed-breed dogs in Denmark. Prev Vet Med. 2003;58:63–74. doi: 10.1016/s0167-5877(03)00010-2. [DOI] [PubMed] [Google Scholar]
- 3.Jimenez AG. Physiological underpinnings in life-history trade-offs in man’s most popular selection experiment: the dog. J Comp Physiol B. 2016;186:813–827. doi: 10.1007/s00360-016-1002-4. [DOI] [PubMed] [Google Scholar]
- 4.Creevy KE, Austad SN, Hoffman JM, O’Neill DG, Promislow DEL. The companion dog as a model for the longevity dividend. Cold Spring Harb Perspect Med. 2016;6:a026633. doi: 10.1101/cshperspect.a026633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoffman JM, Creevy KE, Franks A, O’Neill DG, Promislow DEL. The companion dog as a model for human aging and mortality. Aging Cell. 2018;17:e12737. doi: 10.1111/acel.12737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Creevy KE, Akey JM, Kaeberlein M, Promislow DEL, The Dog Aging Project Consortium. The Dog Aging Project: An open science study of aging in companion dogs. Nature. 2022;in press.
- 7.Patronek GJ, Waters DJ, Glickman LT. Comparative longevity of pet dogs and humans: implications for gerontological research. J Gerontol. 1997;52A:B171–B178. doi: 10.1093/gerona/52a.3.b171. [DOI] [PubMed] [Google Scholar]
- 8.Galis F, Van Der Sluijs I, Van Dooren TJM, Metz JAJ, Nussbaumer M. Do large dogs die young? J Exp Zoolog B Mol Dev Evol. 2007;308B:119–126. doi: 10.1002/jez.b.21116. [DOI] [PubMed] [Google Scholar]
- 9.Kraus C, Pavard S, Promislow DEL. The size–lifespan trade-off decomposed: why large dogs die young. Am Nat. 2013;181:492–505. doi: 10.1086/669665. [DOI] [PubMed] [Google Scholar]
- 10.Adams VJ, Evans KM, Sampson J, Wood JLN. Methods and mortality results of a health survey of purebred dogs in the UK. J Small Anim Pract. 2010;51:512–524. doi: 10.1111/j.1748-5827.2010.00974.x. [DOI] [PubMed] [Google Scholar]
- 11.Yordy J, Kraus C, Hayward JJ, White ME, Shannon LM, Creevy KE, et al. Body size, inbreeding, and lifespan in domestic dogs. Conserv Genet. 2020;21:137–148. doi: 10.1007/s10592-019-01240-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Urfer SR, Kaeberlein M, Promislow DEL, Creevy KE. Lifespan of companion dogs seen in three independent primary care veterinary clinics in the United States. Canine Med Genet. 2020;7:7. doi: 10.1186/s40575-020-00086-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Leroy G, Phocas F, Hedan B, Verrier E, Rognon X. Inbreeding impact on litter size and survival in selected canine breeds. Vet J. 2015;203:74–78. doi: 10.1016/j.tvjl.2014.11.008. [DOI] [PubMed] [Google Scholar]
- 14.Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438:803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- 15.Parker HG, Shearin AL, Ostrander EA. Man’s best friend becomes biology’s best in show: genome analyses in the domestic dog. Annu Rev Genet. 2010;44:309–336. doi: 10.1146/annurev-genet-102808-115200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parker HG, Kim LV, Sutter NB, Carlson S, Lorentzen TD, Malek TB, et al. Genetic structure of the purebred domestic dog. 2004;304:6. doi: 10.1126/science.1097406. [DOI] [PubMed] [Google Scholar]
- 17.Dreger DL, Rimbault M, Davis BW, Bhatnagar A, Parker HG, Ostrander EA. Whole genome sequence, SNP chips and pedigree structure: building demographic profiles in domestic dog breeds to optimize genetic trait mapping. Dis Model Mech. 2016;dmm.027037. [DOI] [PMC free article] [PubMed]
- 18.Sams AJ, Boyko AR. Fine-scale resolution of runs of homozygosity reveal patterns of inbreeding and substantial overlap with recessive disease genotypes in domestic dogs. G3 GenesGenomesGenetics. 2019;9:117–23. doi: 10.1534/g3.118.200836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mooney JA, Yohannes A, Lohmueller KE. The impact of identity by descent on fitness and disease in dogs. Proc Natl Acad Sci. 2021;118:e2019116118. doi: 10.1073/pnas.2019116118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tournebize R, Chu G, Moorjani P. Reconstructing the history of founder events using genome-wide patterns of allele sharing across individuals. Lohmueller KE editor. PLOS Genet. 2022;18:e1010243. doi: 10.1371/journal.pgen.1010243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Keller L, Waller D. Inbreeding effects in wild populations. Trends Ecol Evol. 2002;17:230–241. [Google Scholar]
- 22.Leroy G. Inbreeding depression in livestock species: review and meta-analysis. Anim Genet. 2014;45:618–628. doi: 10.1111/age.12178. [DOI] [PubMed] [Google Scholar]
- 23.Boakes EH, Wang J, Amos W. An investigation of inbreeding depression and purging in captive pedigreed populations. Heredity. 2007;98:172–182. doi: 10.1038/sj.hdy.6800923. [DOI] [PubMed] [Google Scholar]
- 24.Hasselgren M, Norén K. Inbreeding in natural mammal populations: historical perspectives and future challenges. Mammal Rev. 2019;49:369–383. [Google Scholar]
- 25.Hedrick PW, Kalinowski ST. Inbreeding depression in conservation biology. Annu Rev Ecol Syst. 2000;3:139–162. [Google Scholar]
- 26.Charlesworth D, Willis JH. The genetics of inbreeding depression. Nat Rev Genet. 2009;10:783–796. doi: 10.1038/nrg2664. [DOI] [PubMed] [Google Scholar]
- 27.Bijlsma Bundgaard, Boerema. Does inbreeding affect the extinction risk of small populations? predictions from Drosophila. J Evol Biol. 2000;13:502–14. [Google Scholar]
- 28.O’Grady JJ, Brook BW, Reed DH, Ballou JD, Tonkyn DW, Frankham R. Realistic levels of inbreeding depression strongly affect extinction risk in wild populations. Biol Conserv. 2006;133:42–51. [Google Scholar]
- 29.Frankham R. Genetics and extinction. Biol Conserv. 2005;126:131–140. [Google Scholar]
- 30.Inoue M, Kwan NCL, Sugiura K. Estimating the life expectancy of companion dogs in Japan using pet cemetery data. J Vet Med Sci. 2018;80:1153–1158. doi: 10.1292/jvms.17-0384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.O’Neill DG, Church DB, McGreevy PD, Thomson PC, Brodbelt DC. Longevity and mortality of owned dogs in England. Vet J. 2013;198:638–643. doi: 10.1016/j.tvjl.2013.09.020. [DOI] [PubMed] [Google Scholar]
- 32.Bannasch D, Famula T, Donner J, Anderson H, Honkanen L, Batcher K, et al. The effect of inbreeding, body size and morphology on health in dog breeds. Canine Med Genet. 2021;8:12. doi: 10.1186/s40575-021-00111-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marsden CD, Ortega-Del Vecchyo D, O’Brien DP, Taylor JF, Ramirez O, Vilà C, et al. Bottlenecks and selective sweeps during domestication have increased deleterious genetic variation in dogs. Proc Natl Acad Sci. 2016;113:152–157. doi: 10.1073/pnas.1512501113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Urfer SR. Right censored data (‘cohort bias’) in veterinary life span studies. Vet Rec. 2008;163:457–458. doi: 10.1136/vr.163.15.457. [DOI] [PubMed] [Google Scholar]
- 35.O’Neill DG, Packer RMA, Lobb M, Church DB, Brodbelt DC, Pegram C. Demography and commonly recorded clinical conditions of Chihuahuas under primary veterinary care in the UK in 2016. BMC Vet Res. 2020;16:42. doi: 10.1186/s12917-020-2258-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gladyshev TV, Gladyshev VN. A disease or not a disease? Aging as a pathology. Trends Mol Med. 2016;22:995–996. doi: 10.1016/j.molmed.2016.09.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dobson JM. Breed-predispositions to cancer in pedigree dogs. ISRN Vet Sci. 2013;2013:1–23. doi: 10.1155/2013/941275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Paoloni M, Khanna C. Translation of new cancer treatments from pet dogs to humans. Nat Rev Cancer. 2008;8:147–156. doi: 10.1038/nrc2273. [DOI] [PubMed] [Google Scholar]
- 39.Alvarez CE. Naturally occurring cancers in dogs: insights for translational genetics and medicine. ILAR J. 2014;55:16–45. doi: 10.1093/ilar/ilu010. [DOI] [PubMed] [Google Scholar]
- 40.Ostrander EA, Dreger DL, Evans JM. Canine cancer genomics: lessons for canine and human health. Annu Rev Anim Biosci. 2019;7:449–472. doi: 10.1146/annurev-animal-030117-014523. [DOI] [PubMed] [Google Scholar]
- 41.Davis BW, Ostrander EA. Domestic dogs and cancer research: a breed-based genomics approach. ILAR J. 2014;55:59–68. doi: 10.1093/ilar/ilu017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gardner HL, Fenger JM, London CA. Dogs as a model for cancer. Annu Rev Anim Biosci. 2016;4:199–222. doi: 10.1146/annurev-animal-022114-110911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Rowell JL, McCarthy DO, Alvarez CE. Dog models of naturally occurring cancer. Trends Mol Med. 2011;17:380–388. doi: 10.1016/j.molmed.2011.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schiffman JD, Breen M. Comparative oncology: what dogs and other species can teach us about humans with cancer. Philos Trans R Soc B Biol Sci. 2015;370:20140231. doi: 10.1098/rstb.2014.0231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fleming JM, Creevy KE, Promislow DEL. Mortality in North American dogs from 1984 to 2004: an investigation into age-, size-, and breed-related causes of death: mortality of dogs in North America. J Vet Intern Med. 2011;25:187–198. doi: 10.1111/j.1939-1676.2011.0695.x. [DOI] [PubMed] [Google Scholar]
- 46.Dorn CR, Taylor DON, Schneider R, Hibbard HH, Klauber MR. Survey of animal neoplasms in Alameda and Contra Costa Counties, California. II. Cancer morbidity in dogs and cats from Alameda County. J Natl Cancer Inst. 1968;40:307–18. [PubMed] [Google Scholar]
- 47.Vascellari M, Baioni E, Ru G, Carminato A, Mutinelli F. Animal tumour registry of two provinces in northern Italy: incidence of spontaneous tumours in dogs and cats. BMC Vet Res. 2009;5:39. doi: 10.1186/1746-6148-5-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bellumori TP, Famula TR, Bannasch DL, Belanger JM, Oberbauer AM. Prevalence of inherited disorders among mixed-breed and purebred dogs: 27,254 cases (1995–2010) J Am Vet Med Assoc. 2013;242:1549–1555. doi: 10.2460/javma.242.11.1549. [DOI] [PubMed] [Google Scholar]
- 49.Parker HG, Dreger DL, Rimbault M, Davis BW, Mullen AB, Carpintero-Ramirez G, et al. Genomic analyses reveal the influence of geographic origin, migration, and hybridization on modern dog breed development. Cell Rep. 2017;19:697–708. doi: 10.1016/j.celrep.2017.03.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Faraway JJ. Linear Models with R. 2nd ed. Chapman & Hall; 2015.
- 52.Jimenez AG, Downs CJ. Untangling life span and body mass discrepancies in canids: phylogenetic comparison of oxidative stress in blood from domestic dogs and wild canids. Am J Physiol-Regul Integr Comp Physiol. 2020;319:R203–R210. doi: 10.1152/ajpregu.00067.2020. [DOI] [PubMed] [Google Scholar]
- 53.Watowich MM, MacLean EL, Hare B, Call J, Kaminski J, Miklósi Á, et al. Age influences domestic dog cognitive performance independent of average breed lifespan. Anim Cogn. 2020;23:795–805. doi: 10.1007/s10071-020-01385-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.MacLean EL, Snyder-Mackler N, vonHoldt BM, Serpell JA. Highly heritable and functionally relevant breed differences in dog behaviour. Proc R Soc B Biol Sci. 2019;286:20190716. doi: 10.1098/rspb.2019.0716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Akdemir D, Godfrey OU. 2015 EMMREML: fitting mixed models with known covariance structures. R Package Version 31 HttpsCRANR-Proj
- 56.Bürkner P-C. An R package for Bayesian multilevel models using Stan. J Stat Softw. 2017;80:1–28. [Google Scholar]
- 57.Bürkner P-C. Advanced Bayesian multilevel modeling with the R Package brms. R J. 2018;10:395–411. [Google Scholar]
- 58.R Core Team. R: a language and environment for statistical computing. R Found Stat Comput Vienna Austria URL HttpswwwR-Proj [Internet]. 2020; Available from: https://www.R-project.org/. Accessed Dec 2020.
- 59.Long JA. Jtools: analysis and presentation of social scientific data. R Package Version 2020; 210 URL Httpscranr-Proj
- 60.Teng KT. Life tables of annual life expectancy and mortality for companion dogs in the United Kingdom. Sci Rep. 2022 ;11 [DOI] [PMC free article] [PubMed]
- 61.O’Neill DG, Baral L, Church DB, Brodbelt DC, Packer RMA. Demography and disorders of the French Bulldog population under primary veterinary care in the UK in 2013. Canine Genet Epidemiol. 2018;5:3. doi: 10.1186/s40575-018-0057-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Greer KA, Canterberry SC, Murphy KE. Statistical analysis regarding the effects of height and weight on life span of the domestic dog. Res Vet Sci. 2007;82:208–214. doi: 10.1016/j.rvsc.2006.06.005. [DOI] [PubMed] [Google Scholar]
- 63.Tian X, Seluanov A, Gorbunova V. Molecular mechanisms determining lifespan in short- and long-lived species. Trends Endocrinol Metab. 2017;28:722–734. doi: 10.1016/j.tem.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Eigenmann JE, Patterson DF, Froesch ER. Body size parallels insulin-like growth factor I levels but not growth hormone secretory capacity. Acta Endocrinol (Copenh). 1984;106:448–453. doi: 10.1530/acta.0.1060448. [DOI] [PubMed] [Google Scholar]
- 65.Eigenmann JE, Amador A, Patterson DF. Insulin-like growth factor I levels in proportionate dogs, chondrodystrophic dogs and in giant dogs. Acta Endocrinol (Copenh). 1988;118:105–108. doi: 10.1530/acta.0.1180105. [DOI] [PubMed] [Google Scholar]
- 66.Greer KA, Hughes LM, Masternak MM. Connecting serum IGF-1, body size, and age in the domestic dog. Age. 2011;33:475–483. doi: 10.1007/s11357-010-9182-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Jimenez AG, Winward J, Beattie U, Cipolli W. Cellular metabolism and oxidative stress as a possible determinant for longevity in small breed and large breed dogs. Ling F editor. PLOS ONE. 2018;13:e0195832. doi: 10.1371/journal.pone.0195832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Nicholatos JW, Robinette TM, Tata SVP, Yordy JD, Francisco AB, Platov M, et al. Cellular energetics and mitochondrial uncoupling in canine aging. GeroScience. 2019;41:229–242. doi: 10.1007/s11357-019-00062-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Hoffman JM, Kiklevich JV, Austad M, Tran V, Jones DP, Royal A, et al. Tryptophan metabolism is differently regulated between large and small dogs. GeroScience. 2020;42:881–896. doi: 10.1007/s11357-019-00114-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Jimenez AG. The physiological conundrum that is the domestic dog. Integr Comp 2021; Biol.icab005 [DOI] [PubMed]
- 71.Urfer SR. Beyond inbreeding depression? A case study of the Irish Wolfhound. J Vet Behav. 2011;6:99. doi: 10.1016/j.jveb.2010.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lewis TW, Wiles BM, Llewellyn-Zaidi AM, Evans KM, O’Neill DG. Longevity and mortality in Kennel Club registered dog breeds in the UK in 2014. Canine Genet Epidemiol. 2018;5:10. doi: 10.1186/s40575-018-0066-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Blanckenhorn WU. The evolution of body size: what keeps organisms small? Q Rev Biol. 2000;75:385–407. doi: 10.1086/393620. [DOI] [PubMed] [Google Scholar]
- 74.Metcalfe N. Growth versus lifespan: perspectives from evolutionary ecology. Exp Gerontol. 2003;38:935–940. doi: 10.1016/s0531-5565(03)00159-1. [DOI] [PubMed] [Google Scholar]
- 75.Sargan DR. IDID: Inherited Diseases in Dogs: web-based information for canine inherited disease genetics. Mamm Genome. 2004;15:503–506. doi: 10.1007/s00335-004-3047-z. [DOI] [PubMed] [Google Scholar]
- 76.Karlsson EK, Lindblad-Toh K. Leader of the pack: gene mapping in dogs and other model organisms. Nat Rev Genet. 2008;9:713–725. doi: 10.1038/nrg2382. [DOI] [PubMed] [Google Scholar]
- 77.Donner J, Anderson H, Davison S, Hughes AM, Bouirmane J, Lindqvist J, et al. Frequency and distribution of 152 genetic disease variants in over 100,000 mixed breed and purebred dogs. Leeb T editor. PLOS Genet. 2018;14:e1007361. doi: 10.1371/journal.pgen.1007361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Wallis LJ, Szabó D, Erdélyi-Belle B, Kubinyi E. Demographic change across the lifespan of pet dogs and their impact on health status. Front Vet Sci. 2018;5:200. doi: 10.3389/fvets.2018.00200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Evert J, Lawler E, Bogan H, Perls T. Morbidity profiles of centenarians: survivors, delayers, and escapers. J Gerontol A Biol Sci Med Sci. 2003;58:M232–M237. doi: 10.1093/gerona/58.3.m232. [DOI] [PubMed] [Google Scholar]
- 80.Kaeberlein M, Creevy KE, Promislow DEL. The dog aging project: translational geroscience in companion animals. Mamm Genome. 2016;27:279–288. doi: 10.1007/s00335-016-9638-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Guy MK, Page RL, Jensen WA, Olson PN, Haworth JD, Searfoss EE, et al. The Golden Retriever Lifetime Study: establishing an observational cohort study with translational relevance for human health. Philos Trans R Soc B Biol Sci. 2015;370:20140230. doi: 10.1098/rstb.2014.0230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.de Magalhães JP. How ageing processes influence cancer. Nat Rev Cancer. 2013;13:357–365. doi: 10.1038/nrc3497. [DOI] [PubMed] [Google Scholar]
- 83.Doherty A, Lopes I, Ford CT, Monaco G, Guest P, de Magalhães JP. A scan for genes associated with cancer mortality and longevity in pedigree dog breeds. Mamm Genome. 2020;31:215–227. doi: 10.1007/s00335-020-09845-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Steuerman R, Shevah O, Laron Z. Congenital IGF1 deficiency tends to confer protection against post-natal development of malignancies. Eur J Endocrinol. 2011;164:485–489. doi: 10.1530/EJE-10-0859. [DOI] [PubMed] [Google Scholar]
- 85.Egenvall A, Bonnett BN, Hedhammar Å, Olson P. Mortality in over 350,000 insured Swedish dogs from 1995–2000: II. Breed-specific age and survival patterns and relative risk for causes of death. 2005;46:16. doi: 10.1186/1751-0147-46-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Nunney L. The real war on cancer: the evolutionary dynamics of cancer suppression. Evol Appl. 2013;6:11–19. doi: 10.1111/eva.12018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Peto R, Roe FJ, Lee PN, Levy L, Clack J. Cancer and aging in mice and men. Br J Cancer. 1975;32:411–426. doi: 10.1038/bjc.1975.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Caulin AF, Maley CC. Peto’s Paradox: evolution’s prescription for cancer prevention. Trends Ecol Evol. 2011;26:175–182. doi: 10.1016/j.tree.2011.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nunney L. Resolving Peto’s paradox: Modeling the potential effects of size-related metabolic changes, and of the evolution of immune policing and cancer suppression. Evol Appl. 2020;13:1581–1592. doi: 10.1111/eva.12993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Vincze O, Colchero F, Lemaître J-F, Conde DA, Pavard S, Bieuville M, et al. Cancer risk across mammals. Nature. 2022;601:263–267. doi: 10.1038/s41586-021-04224-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Armitage P, Doll R. The age distribution of cancer and a multi-stage theory of carcinogenesis. Br J Cancer. 1954;8:1–12. doi: 10.1038/bjc.1954.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Albanes D, Winick M. Are cell number and cell proliferation risk factors for cancer? J Natl Cancer Inst. 1988;80:772–774. doi: 10.1093/jnci/80.10.772. [DOI] [PubMed] [Google Scholar]
- 93.Nunney L. Size matters: height, cell number and a person’s risk of cancer. Proc R Soc B Biol Sci. 2018;285:20181743. doi: 10.1098/rspb.2018.1743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Makielski KM, Mills LJ, Sarver AL, Henson MS, Spector LG, Naik S, et al. Risk factors for development of canine and human osteosarcoma: a comparative review. Vet Sci. 2019;6:48. doi: 10.3390/vetsci6020048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Ujvari B, Klaassen M, Raven N, Russell T, Vittecoq M, Hamede R, et al. Genetic diversity, inbreeding and cancer. Proc R Soc B Biol Sci. 2018;285:20172589. doi: 10.1098/rspb.2017.2589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Dorn CR, Schneider R. Inbreeding and canine mammary ancer: a retrospective study. JNCI J Natl Cancer Inst. 1976;57:545–548. doi: 10.1093/jnci/57.3.545. [DOI] [PubMed] [Google Scholar]
- 97.Bonnett BN, Egenvall A, Hedhammar Å, Olson P. Mortality in over 350,000 insured Swedish dogs from 1995–2000: I. Breed-, gender-, age- and cause-specific rates. 2005;46:16. doi: 10.1186/1751-0147-46-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Hoffman JM, Creevy KE, Promislow DEL. Reproductive capability is associated with lifespan and cause of death in companion dogs. Helle S editor. PLoS ONE. 2013;8:e61082. doi: 10.1371/journal.pone.0061082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Waters DJ, Kengeri SS, Clever B, Booth JA, Maras AH, Schlittler DL, et al. Exploring mechanisms of sex differences in longevity: lifetime ovary exposure and exceptional longevity in dogs. Aging Cell. 2009;8:752–755. doi: 10.1111/j.1474-9726.2009.00513.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Waters DJ, Kengeri SS, Maras AH, Chiang EC. Probing the perils of dichotomous binning: how categorizing female dogs as spayed or intact can misinform our assumptions about the lifelong health consequences of ovariohysterectomy. Theriogenology. 2011;76:1496–1500. doi: 10.1016/j.theriogenology.2011.06.017. [DOI] [PubMed] [Google Scholar]
- 101.Oberbauer AM, Belanger JM, Famula TR. A review of the impact of neuter status on expression of inherited conditions in dogs. Front Vet Sci. 2019;6:397. doi: 10.3389/fvets.2019.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Urfer SR, Kaeberlein M. Desexing dogs: a review of the current literature. Animals. 2019;9:1086. doi: 10.3390/ani9121086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Niskanen M, Thrusfield MV. Associations between age, parity, hormonal therapy and breed, and pyometra in Finnish dogs. Vet Rec. 1998;143:493–498. doi: 10.1136/vr.143.18.493. [DOI] [PubMed] [Google Scholar]
- 104.Walsh (Stonehenge) JH, Armatage G. The dog: its varieties and management in health [Internet]. London, New York: F. Warne and Co; 1896. Available from: https://archive.org/details/dogitsvarietiesm00wals2. Accessed Dec 2021.
- 105.Huntington HW. The show dog [Internet]. Providence, R.I.: For the author, by the Remington print. co. 1901. Available from: https://archive.org/details/showdogbeingabo00huntgoog/mode/2up. Accessed Dec 2021.
- 106.Speelman SR, Williams JO. 1926. Breeds of dogs. U.S. Department of Agriculture
- 107.Frankham R. Genetic rescue of small inbred populations: meta-analysis reveals large and consistent benefits of gene flow. Mol Ecol. 2015;24:2610–2618. doi: 10.1111/mec.13139. [DOI] [PubMed] [Google Scholar]
- 108.Melis C, Pertoldi C, Ludington WB, Beuchat C, Qvigstad G, Stronen AV. Genetic rescue of the highly inbred Norwegian Lundehund. Genes. 2022;13:163. doi: 10.3390/genes13010163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Windig JJ, Doekes HP. Limits to genetic rescue by outcross in pedigree dogs. J Anim Breed Genet. 2018;135:238–248. doi: 10.1111/jbg.12330. [DOI] [PubMed] [Google Scholar]
- 110.van Hagen MAE, Ducro BJ, van den Broek J, Knol BW. Life expectancy in a birth cohort of Boxers followed up from weaning to 10 years of age. Am J Vet Res. 2005;66:1646–1650. doi: 10.2460/ajvr.2005.66.1646. [DOI] [PubMed] [Google Scholar]
- 111.Dobson J, Hoather T, McKinley TJ, Wood JLN. Mortality in a cohort of flat-coated retrievers in the UK. Vet Comp Oncol. 2009;2:115–21. doi: 10.1111/j.1476-5829.2009.00181.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.