Abstract
We investigated the role of the DDTFR10/A gene of the ethylene response element-binding protein (EREBP) family through the CRISPR/Cas9 genome editing approach. The associated role of this gene in tomato fruit ripening was known. The involvement of ripening-regulatory proteins in plant defense has been documented; therefore, to find the involvement of the DDTFR10/A gene in host susceptibility, we introduced the mutation in DDTFR10/A gene through CRISPR/cas9 in the genome of the tomato plant. The 50% biallelic and 50% homozygous mutations were observed in the T0 generation. The CRISPR/Cas9 edited plants showed 40% reduced symptoms of Fusarium wilt compared to control plants (non-edited). The DDTFR10/A gene expression in tomato plants was evaluated against biotic (Fusarium wilt) and abiotic (salinity) stresses, and the upregulated expression of this gene was found under both challenges. However, a comparative increase in DDTFR10/A gene expression was observed in tomato plants upon inoculation with Fusarium oxysporum f. sp. lycopersici. The phenotypic assay performed on edited tomato plants demonstrated the role of the DDTFR10/A gene in contributing toward susceptibility against Fusarium wilt.
Supplementary Information
The online version contains supplementary material available at 10.1007/s12298-022-01273-6.
Keywords: DDTFR10/A gene, CRISPR/Cas9 genome editing, Salinity, Tomato wilt, Negative regulator
Introduction
Ethylene response factors (ERFs), or ethylene response element-binding protein (EREBP), are unique plant transcriptional factors specified by the integrative node of the ethylene transduction pathway (Guo and Ecker 2004). They are characterized by distinct AP2 (APETALA2)/ERF DNAbinding domain that binds at GCC-box or DRE (/C-repeats) elements of the promoter region of ethylene-responsive genes (Fujimoto et al. 2000). Their major role has been described in ethylene signaling (Cheng et al. 2013). Particularly their role was observed in regulating metabolic pathways, phytohormones signal transduction, response to biotic and abiotic stimuli, and other developmental processes (Gu et al. 2000; Fits and Memelink 2000; Banno et al. 2001; Dubouzet et al. 2003). In response to biotic and abiotic stimuli, signaling or cross-talk among ethylene, salicylic acid, jasmonic acid, and abscisic acid pathways has been documented (Fujita et al. 2006). In Arabidopsis, AtERF1 was studied for its key involvement in the ethylene and jasmonic acid pathway (Lorenzo et al. 2003). The tobacco Tsi1, pepper CaPF1, wheat TaERF1, soybean GmERF3, and rice OsERF71 regulate multiple responses in biotic and abiotic stresses (Park et al. 2001; Yi et al. 2004; Tang et al. 2005; Xu et al. 2007; Zhang et al. 2009; Li et al. 2018a, b, c, d).
In tomato (Solanum lycopersicum), with whole-genome sequence annotation, 77 genes were declared to encode proteins belongings to the ERF subfamily (Pirrello et al. 2012a, b) that might be involved in several physiological and developmental processes. However, most of these were identified with their role in fruit ripening (Pirrello et al. 2012a, b; Fujisawa and Barraclough 2013; Liu et al. 2016). Few ERFs were investigated for their regulatory role in stress response, including Pti4-6, SlPti4, LeERF1-4, JERF3, TSRF1, ERF5, Sl-ERF.B.3, SlERF84, and SlERF2 (Zhou et al. 1997; Gu et al. 2000, 2002; Mysore et al. 2002; Chakravarthy et al. 2003; Tournier et al. 2003; Wang et al. 2004; Zhang et al. 2004; Pan et al. 2012; Klay et al. 2014; Li et al. 2018a, b, c, d; Yu et al. 2018). Multiple aspects of ERF proteins in plant stress responses by interacting with cis-acting elements of the promoter region of downstream genes were studied (Buttner and Singh 1997). The role of ERF proteins, tobacco Tsi1, and tomato JERF3 was underlined in defense pathways related to both salt and pathogenic attack (Park et al. 2001; Wang et al. 2004). Li et al. (2018a, b, c, d) reported the function of SlERF84 in biotic and abiotic stress. The SlERF84 works to enhance tolerance to salt stress but increase susceptibility towards Pseudomonas syringae pv. Tomato DC3000 attack. Similarly, the dual role of several ERFs has also been documented, as a single ERF protein may involve in the developmental process and plant stress response. The expression of the tomato SlPti4 gene is enhanced during the ripening process and upon pathogen attack (Gu et al. 2002; Chakravarthy et al. 2003; Chen et al. 2008). The LeERF3b was studied with high expression in low ethylene fruits, but environmental stimuli induced its expression, i.e., drought, low temperature, and desiccation (Chen et al. 2008). A DDTFR10/A (Differential Display Tomato Fruit Ripening) (synonym, SlERF-B1) gene induced by ethylene and categorized in the ethylene response element-binding protein (EREBP) family. It contains conserved DNA binding AP2/ERF domain. Liu et al. (2016) explained its upregulation at the onset of ripening and suggested it as the best candidate for activating the ripening process. Its continuous expression was also observed in salt stress on tomato plants (Ouyang et al. 2007). However, its differential expression as a negative or positive regulator of transcriptional activity is still unknown. Several ethylene-regulated genes have no assigned particular function; reverse genetics is currently being used to investigate their function (Moin et al. 2018).
Tomato is a vegetable crop grown worldwide due to its great economic value. Although it is native to South America (FAO 2009), some Asian subcontinents contribute to its production. With an average of 9.90 tonnes/ hectare tomato yield, Pakistan occupied the 33rd position in the world (35 tonnes/hectare). The gap between global and Pakistan tomato production is harnessed by producing new cultivars with improved quality, high yielding, or disease resistance. The Ayub Agriculture Research Institute (AARI), Faisalabad, released two tomato hybrids (cv. Sandal and Surkhail) with production from 180 to 190 tonnes/hectare (Soomro et al. 2020). Biotic and abiotic factors are among the leading constraints in obtaining tomato per hectare yield. Yield-limiting tomato wilt is most commonly observed in the field. The vascular wilt caused by Fusarium oxysporum f. sp. lycopersici (Sacc.) W.C. Snyder and H.N. Hans, a soil pathogen, produce heavy economic losses (Hussain et al. 2016). We used two tomato cultivars, cv. Sahel (Fusarium wilt tolerant) (Farooq et al. 2013) and cv. Sandal (Fusarium wilt susceptible) (Syngenta 2015; Soomro et al. 2020). Both hybrids have similar yield potential with good agronomic traits. The CRISPR/Cas9 system has extensive acceptance from the scientific community due to its versatility and easiness among genome editing strategies (Karmakar et al. 2022a). The CRISPR-Cas9 approach enables scientists for developing strategies to induce resistance in plants against pathogens. Scientists have been attempting to develop advanced strategies to leverage the precise nucleic acid cleavage ability of the CRISPR-Cas9 system. CRISPR/cas9 system consists of two components: the Cas9 protein and sgRNA (guided RNA). CRISPR RNA (crRNA) association with trans activating crRNA (tracr RNA) forms sgRNA for the recognition of target sites that direct the endonuclease Cas9 protein for inducing double-stranded breaks (DSBs). When DSBs generate, cellular DNA repair pathways inadvertently induce Insertion–deletion mutations (indels), which causes frameshift mutations (Karmakar et al. 2022a; Molla et al. 2022). Hence, we attempted to edit the DDTFR10/A gene in tomato cv. Sandal (Fusarium wilt susceptible) through CRISPR/Cas9-based genome editing and observed Fusarium wilt tolerance in DDTFR10/A knockout tomato plants. The comparative expression profiling of the DDTFR10/A gene was also performed in biotic and abiotic stresses in tomato plants of cv. Sahel and cv. Sandal. Our study demonstrated a comparative increase in the expression of the DDTFR10/A gene under Fusarium wilt stress.
Materials and methods
Selection of CRISPR-cas9 target sequence in DDTFR10/A gene
The DDTFR10/A gene sequence of tomato was fetched from NCBI (National Center for Biotechnology Information) database and subjected to the web tool CHOPCHOP v2 (Labun et al. 2016) for target selection. It delivers target sequences by perceiving PAM (protospacer adjacent motif) sequence site. RNA Folding Form V2.3 displayed the target-sgRNA secondary structure.
Development of CRISPR/Cas9 constructs for genome editing in tomato.
The CRISPR/Cas9 construct targeting DDTFR10/A was prepared in the pYLCRISPR/Cas9Pubi-B binary vector (Ma et al. 2015). The sgRNA and OsU6a promoter sequence was synthesized in pUC57, an intermediate vector, by taking the services provided by Eurofins, genomics, USA. The target-sgRNA expression cassette was constructed in overlapping PCR using a thermal cycler (peqSTAR), in which the first reaction comprised U-F/UT (-) and gRT ( +)/gR-R primer pairs with the engineered target sequence, and the second reaction included Pps/Pgs primers containing BsaI-cutting site (Supplementary file 1). The nested PCR amplicon (sgRNA expression cassette) proceeded to Golden Gate cloning for BsaI digestion/ligation reaction (NEB Golden Gate assembly Kit; BsaI-HF v2) comprised of pYLCRISPR/Cas9Pubi-B, 10X T4 DNA ligase buffer, and water (nuclease-free). The resultant product was transferred to Escherichia coli cells (Top10 strain) using the heat shock method for bacterial transformation. The transformed bacterial cells selected under kanamycin selection pressure were cultured on an LB medium for plasmid Isolation (GeneJET plasmid Kit, Thermo Scientific). The isolated plasmid was confirmed with Mlu1-mediated restriction digestion and sequencing.
In planta transformation
The pYLCRISPR/Cas9Pubi-B-DDTFR10/A construct was delivered to Agrobacterium tumefaciens strain AGL1 through electroporation. The electroporated cells cultured on LB medium containing streptomycin were incubated at 28 °C. The colonies that appeared under selection pressure were confirmed for harboring the pYLCRISPR/Cas9Pubi-B-DDTFR10/A construct. The transformed cells were cultured and harvested for preparing the cell suspension (1 × 109 concentration ml−1) (Supartana et al. 2006) in sucrose solution (30%). The in planta transformation (floral dip method) was performed following the protocol by Yasmeen et al. (2009). Seeds of tomato varieties, varieties Sandal (Susceptible to wilt), and Sahel (Wilt tolerant) were sown in earthen pots in Belgium compost in the greenhouse of fungal molecular biology laboratory (FMB Lab.), University of Agriculture Faisalabad, Pakistan. They were transfected with A. tumefaciens strain AGL1 containing CRISPR/Cas 9 construct at the floral stage. The sterile syringe (1 ml) filled with cell suspension was injected onto the stigma of unopened flowers (before pollination) without splitting the subtle floral parts. Three consecutive treatments with 24 hourly gaps were made. The Ariel part of treated plants was well irrigated after 48 h of treatment and kept until fruiting. Seeds were taken from harvested fruits and sown in Belgium compost-filled earthen pots. Upon germination, the application of phosphinothricin (80 mg L−1) was executed on juvenile seedlings at 48 h intervals. Plants that survived under selection pressure were put to downstream applications.
Mutation detection
The isolation of gDNA (Genomic DNA) of selected plants (putative targeted plants) was carried out using GeneJET Plant Genomic DNA Purification Kit, Thermo Scientific, USA). PCR analysis was executed using specific primers flanking the target sites (150–250 bp upstream of the target sites) (Forward: 5′TCCACCCTTTATAATCTCCA3′; Reverse: 5′AAACGAGAGACAGATTCAAG3) ′. Target sequence (Ts) includes a diagnostic EcoRI restriction enzyme site (partially overlaps the PAM sequence) spanning the Cas9 cleavage site. Therefore, purified PCR products of putative targeted plants proceeded to restriction digestions with EcoRI endonucleases for edited plant detection. The amplified products of edited plants were eluted (FavorPrep Gel purification kit, Favorgen Biotech Corporation, Taiwan) and cloned into a cloning vector (pTZ57R/T, InsTAclone™ PCR cloning kit) for direct sequencing. The generated chromatograms with superimposed sequences were decoded using the DSDecodeM web tool based on the Degenerate Sequence Decoding method for genotyping targeted mutations.
Phenotypic assay
Diseased parts of wilt-affected tomato plants (lower leaves yellowing, vein clearing, marginal necrosis, and defoliation) were collected. The 3–5 cm tissues were surface sterilized with 5% sodium hypochlorite solution and cultured on PDA (potato dextrose agar) medium containing 250 mg L−1 streptomycin and incubated at 28 ± 2 °C. Fungus culture was purified using the single-spore isolation method (Amini and Sidovich 2010; Baloch et al. 2021). These cultures were morphologically characterized by mycelial growth, spore character, and colony character. Small, hyaline oval to sickle-shaped and single or bi-celled, or multicelled conidia was observed under the microscope. The pinkish-white to light pink colored colonies were formed during incubation. The fluffy mycelial growth was observed on culture plates. Based on morphocultural characteristics, it was characterized as Fusarium oxysporum. The conidial suspension with 1 × 107 ml−1 density was made in distilled water from 5-day-old culture (Catanzariti et al. 2015). Root-dip inoculation of 10 days old tomato seedlings was performed (Catanzariti et al. 2015). The seedlings were carefully uprooted from the compost and dipped into conidial suspension solution for 5 min, and replanted into compost-filled earthen pots. Knockout plants and control plants were inoculated with conidial suspension. Data were recorded daily till the third-week post-inoculation. The disease incidence was recorded with a 0–4 rating scale defined by Song et al. (2004). The scale denoted zero (0) for no infection or symptom, while four (4) indicated whole plant yellowing or dead plants. The one (1), two (2), and three (3) represented slightly wilted or yellowing of 1–2 leaves per plant (about 25% wilt condition), moderately wilted or yellowing of 2–3 leaves per plant (about 50% wilt condition) and heavily wilted, inhibited growth or all plant leaves yellowing ( about 75% wilt condition), respectively.
Expression profiling of DDTFR10/A gene under biotic and abiotic stresses
For expression profiling of DDTFR10/A, two sets of tomato plants were analyzed. One set of tomato plants of cultivar Sandal (susceptible) was challenged with Fusarium oxysporum f. sp. lycopersici (tomato wilt), and the second set was subjected to salt stress. Tomato plants were inoculated with Fusarium oxysporum f. sp. lycopersici (FOL) by root-dip inoculation of 10-day-old seedlings with conidial spore suspension (1 × 107 ml−1 density). The control plants were irrigated with water (negative control). The wilt-tolerant tomato cultivar (Sahel) plants were also inoculated with Fusarium oxysporum f. sp. lycopersici (FOL). Twenty-one days post-inoculation, leave samples were collected for RNA isolation in liquid nitrogen. In the second set of tomato plants challenged with salt stress, NaCl (8 dS m−1 and 10 dS m−1 electrical conductivity) was applied through irrigation water to tomato seedlings for salinity stress. Seven consecutive irrigations were applied 24 hourly. Control plants were irrigated with tap water. Under salinity stress, leaf samples were taken for RNA isolation.
Total RNA from leave samples were isolated using GeneJET Plant RNA Purification Kit (Thermo Scientific, USA), and cDNA was synthesized using RevertAid First Strand cDNA Synthesis Kit (Thermo Scientific, USA). Comparative expression profiling among control and stress-bearing plants was performed. We used the PrimerQuest™ tool to design qRT-PCR primers for expression profiling of the DDTFR10/A gene. The expression level DDTFR10/A gene was estimated by the 2−ΔCq method, using SlEF1α (S. lycopersicum elongation factor 1-alpha) (Ouyang et al. 2007) as a reference gene under salinity stress and GAPDH (glyceraldehyde 3-phosphate dehydrogenase) reference gene (Catanzariti et al. 2015) for normalizing gene expression under FOL challenge. The reaction mixture consisted of cDNA, primers, Maxima SYBR Green/ROX/qPCR Master Mix (Thermo Scientific, USA), and nuclease-free water. The RT-qPCR analysis was carried out on the CFX96 Touch Real-Time PCR detection system. The reaction was performed with three replicates for each sample. Gene expression level under both conditions was calculated relative to respective reference genes.
Result
Comparative expression profiling of DDTFR10/A gene
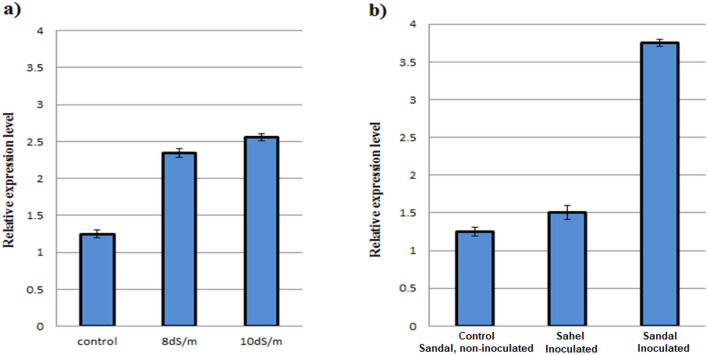
The DDTFR10/A gene expression was investigated under salinity and Fusarium wilt stresses on tomato plants using Rt-qPCR analysis (Fig. 1). DDTFR10/A gene-specific primers were designed by the PrimerQuest tool (Supplementary file 1). Differential expression of the DDTFR10/A gene has been observed in stress conditions. DDTFR10/A gene expression level was determined in Fusarium wilt-challenged plants of cultivars Sandal (susceptible to Fusarium wilt) and Sahel (wilt tolerant) using GAPDH as a reference gene for normalizing gene expression. A comparative increase in DDTFR10/A gene expression was observed in inoculated plants of wilt-susceptible cv. Sandal compared to control plants and inoculated plants of resistant cv. (Sahel). The expression of DDTFR10/A gene in control plants and inoculated plants of resistant cv. (Sahel) was comparatively less. The expression profiling of the DDTFR10/A gene in cultivars Sandal (Susceptible) and Sahel (tolerant) determined the role of this gene as a negative regulator that contributes towards wilt susceptibility. In another set of tomato plants, the expression level of the DDTFR10/A gene was determined under high salt conditions using the SlEF1α gene as a reference gene. The DDTFR10/A gene showed upregulation under a high salinity challenge compared to control plants.
Fig. 1.
Expression level of the DDTFR10/A gene using RT-qPCR analysis was analyzed under wilt and salinity challenges. a The DDTFR10/A gene expression under salinity stress on tomato cv. Sandal was shown to be upregulated compared to control plants. The expression level of the DDTFR10/A gene was increased with an increase in treatment level, 8dS/m and10dS/m, respectively. b Upon inoculation, the expression level of the DDTFR10/A gene in wilt-susceptible tomato cv. Sandal was high compared to the control plants (non-inoculated). However, the expression of the DDTFR10/A gene in control plants and inoculated plants of wilt-tolerant tomato cv. Sahel was similar
The upregulation of DDTFR10/A gene expression under the Fusarium wilt challenge was comparatively higher than the salinity stress. In comparison, the expression level of the DDTFR10/A gene was slightly high in plants treated with NaCl level 10 dSm−1 than plants treated with NaCl level 8 dSm−1. However, the overall expression of the DDTFR10/A gene in NaCl-treated plants was not comparatively high.
CRISPR/cas9 based genome editing in tomato
A DDTFR10/A gene is an ERF family gene that contains DNA binding AP2 domain (https://www.uniprot.org/uniprot/Q9FR33). Some studies revealed it as a ripening-regulated protein; however, up-regulation in the expression of this gene in salt stress was also observed (Ouyang et al. 2007; Liu et al. 2016). Though, the role of the DDTFR10/A gene in plant defense against diseases is not well-explained yet. Therefore, we attempted to introduce the mutation in the DDTFR10/A gene through CRISPR/Cas9 genome editing. The NCBI reference sequence of the DDTFR10/A gene (NM_001319660.1) was retrieved, and the target (5′ TTCTCAAACCTCATCGAATTCGG 3′) was designed. The targeting specificity was confirmed in the Blastn search tool against the S. lycopersicum genome. The specificity criteria of the target to the non-target sequence were a mismatch of > 2 bases at the seed region or PAM-proximal region and differences of > 5 bases at the non-seed region or PAM-distal region (Ma et al. 2015). The sgRNA folding determines the effectiveness of sgRNA-Cas9 working. Therefore, the target-sgRNA folding was also predicted (Supplementary file 2). The predicted folding revealed the pairing of two bases of the target with sgRNA. For efficient targeting, there should be no base pairing between sgRNA and the target. However, the base pairing of fewer than six bases might be tolerated (Ma et al. 2015).
In overlapping PCR reactions, the target sequence was introduced into the sgRNA expression cassette. The OsU6a and sgRNA were first amplified from the pUC57 backbone with U-F/UT (-) and gRT ( +) /gR-R primers. In 2nd step, the position-specific primers with an engineered BsaI cutting site were used to develop a sgRNA expression cassette. The expression cassette was introduced into the pYLCRISPR/Cas9Pubi-B binary vector with golden gate assembly. The pYLCRISPR/Cas9Pubi-B- DDTFR10/A construct was delivered to A. tumefaciens for in planta transformation. The floral-dip method was adopted, and transformants were selected under basta selection pressure (80 mg L−1). Ten plants survived under selection pressure, while the remaining died. For full allelic edited plants detection, purified PCR products of putative targeted plants (survived under selection pressure) were treated with EcoRI restriction enzyme. Of the ten selected plants, the restriction digestion assay revealed no restriction digestion in four plants that were a generation of the Sandal variety (Susceptible to wilt).
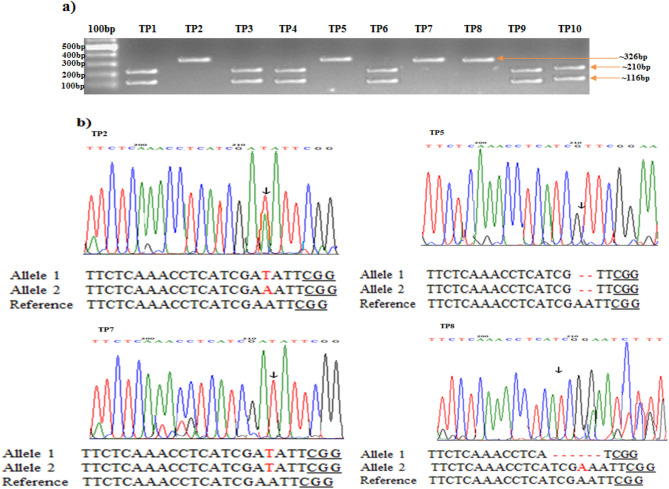
The editing in four tomato plants (targeted generation) survived under the selection regime and was analyzed by direct sequencing of PCR amplicons (~ 326 bp) that were achieved in a PCR analysis using specific primers flanking the target sites. Superimposed chromatograms decoded through DSDecodeM revealed biallelic and homozygous mutations. In 4 sequenced sites, 50% detected mutation was homozygous, and 50% was biallelic (Fig. 2; Table 1). The targeted mutations were due to the addition, deletion, and substitution of nucleotides. In single base insertion or deletion, the proportion for deletion of “A” and insertion of “T” was more significant and corroborated previous reports by Zhang et al. (2014) and Ma et al. (2015). In TP2 and TP7 plants, the “T” nucleotide is inserted in the target sequence that is identical to the nucleotide present at -4 from the protospacer (PAM) sequence, which is in accordance with the findings of Chakrabarti et al. (2019), and Molla et al. (2022). Due to the insertion/deletions of nucleotides, stop codons were formed, and ORF was truncated that led to non-functional protein formation (Supplementary file 3).
Fig. 2.
Detected mutation in targeted DDTFR10/A gene and disease assay of T0 tomato plants a Ten tomato plants, after basta selection, were screened for Indel detection with EcoRI digestion. The targeted plants, TP1, TP3, TP4, TP6, TP9, and TP10, appeared to be un-edited and produced double bands (~ 210 bp and ~ 110 bp), while TP2, TP5, TP7, and TP8 produced single bands (~ 326), indicates fully edited alleles. b Direct sequencing of PCR product containing the targeted site of fully edited tomato T0 plants. The overlapping traces of the sequencing chromatogram were decoded by the DSD method. Arrows indicate the site of mutation
Table 1.
Targeted genomic mutations of DDTFR10/A gene in T0 plants of tomato. Changed nucleotides are highlighted in red color
| Target sequence: TTCTCAAACCTCATCGAATT CGG; Heterozygous: 0, Homozygous: 2, Biallelic: 2, Total: 4 | ||
|---|---|---|
| TP2 |
TTCTCAAACCTCATCGATATTCGG TTCTCAAACCTCATCGAAATTCGG |
Biallelic |
| TP5 | TTCTCAAACCTCATCG ‐ ‐ TTCGG | homozygous |
| TP7 | TTCTCAAACCTCATCGATATTCGG | homozygous |
| TP8 |
TTCTCAAACCTCA ‐ ‐ ‐ ‐ ‐ ‐ TCGG TTCTCAAACCTCATCGAAATTCGG |
Biallelic |
We demonstrated the role of the DDTFR10/A gene in contributing susceptibility by performing the phenotypic assay of edited tomato plants (knock out/mutants) by exerting Fusarium wilt stress. After the first week of inoculation, symptoms started to appear on tomato plants. However, in the third week of post-inoculation, targeted plants were observed with distinct symptoms. The disease incidence percentage of targeted and control tomato plants was measured with observable symptoms, including yellowing, necrotic lesions, stunted growth, and wilt condition. The 25–30% disease severity was recorded on edited plants, showing symptoms like yellowing of 2–3 leaves, vein clearing, and moderately stunted growth (Supplementary file 4). The control plants were heavily wilted (65–70%) with yellowing of all plant leaves, growth inhibition, necrotic lesions formation, and dying of lower leaves.
Discussion
As a robust reverse genetic approach, we used CRISPR/Cas9-based genome editing to evaluate the regulatory role of the DDTFR10/A gene in the Fusarium wilt disease challenge in tomato plants. However, Chen et al. (2013) documented the upregulated expression of the DDTFR10/A gene in response to Tomato Yellow Leaf Curl Virus infection. However, its contribution as a positive or negative regulator of defense system against diseases is unknown and has not yet been attempted to unveil through reverse genetics approaches. Successful targeted mutagenesis using the CRISPR-Cas9 system has already been employed in tomato crops (Ito et al. 2015; Pan et al. 2016; Yu et al. 2017; Li et al. 2018a, b, c, d; Liu et al. 2020). For DDTFR10/A gene targeting, the expression cassette consisted of the OsU6a promoter and sgRNA sequence identified by Ma et al. (2015). Target-sgRNA was transcribed with initiated nucleotide G at the 5' end (Shan et al. 2013; Ma et al. 2015). One of the critical points to determine Cas9/sgRNA effectiveness is sgRNA secondary structure formation (Makarova et al. 2011). Our target-sgRNA sequence folding was shown the pairing of only two bases of the target with sgRNA.
We selected the transformants using basta concentration, 80 mgL−1; similarly, Khuong et al. (2013) sprayed basta concentration, 75 mgL−1, on soil-growing tomato plantlets. The selected transformants or putative-targeted plants were screened for mutation detection. Their indel detection was achieved by restriction digestion with the EcoRI enzyme. The sequence analysis revealed two types of mutation, homozygous and biallelic. The wild-type alleles produced a double band, while the mutant ones remained undisrupted and produced a single band. Four plants produced undisrupted alleles. We used a web tool DSDecode, to decode the sequencing chromatogram reads. Li et al. (2018a, b, c, d) also used the DSDecode method to read the overlapped chromatogram of different genes linked to lycopene accumulation in tomato plants.
The knockout plants were assessed for their performance against Fusarium wilt stress. The Fusarium oxysporum was isolated from tomato wilt tissues and morphologically observed to produce hyaline oval to sickle-shaped, single or bicelled or multicelled, conidial spores and pinkish-white to light pink colored colonies. Similar results were observed by Manikandan et al. (2018), who reported single to multicelled, hyaline oval or sickle-shaped micro and macroconidia and whitish or creamy or pinkish white or pinkish fungal colonies of different isolates of Fusarium oxysporum. The isolated Fusarium wilt pathogen was used to inoculate tomato plants. At 21 days of post-inoculation, distinct symptoms in tomato seedlings were observed. However, comparatively lesser disease symptoms were observed in targeted plants to the non-targeted control plants. The disease severity percentage of the control plant was recorded as about 65–70% after three weeks of inoculation. Edited plants survived the stress regime, and disease severity was recorded by nearly 25–30%. Disease incidence on selected control plants (cv. Sahel, known to Fusarium wilt susceptible) was close to the results of Houssien et al. (2010), and Shanmugam and Kanoujia (2011), who studied 69.44% and 78.50% disease incidence on plants in pots experiment, respectively.
The Fusarium oxysporum f. sp. lycopersici is the main contributor to tomato vascular wilt. The penetration and colonization of a fungus in vascular tissues produce discoloration, wilting, collapsing, and ultimately dying. These complex symptoms are physiological disturbance and inactivation of the defense pathway (Srinivas et al. 2019). Disease progression is a complex trait controlled by multipart genetics. It is the first report that indicated the role of the DDTFR10/A gene in Fusarium wilt or vascular wilt. Numerous studies on the involvement of different genes in the Fusarium wilt of tomatoes have been documented in the literature. The NB-LRR protein I (Catanzariti et al. 2017), I-2 (Simons et al. 1998), I-3 (Catanzariti et al. 2015), I-7 (Cendales et al. 2016), and a small transmembrane protein Solyc08g075770 (Prihatna et al. 2018), were proved to be involved in FOL resistance. SlyFRG4 mutants developed through CRISPR-based genome editing displayed enhanced FOL susceptibility (Karmakar et al. 2022b). However, some gene products are documented to facilitate the infection and enhance susceptibility to the disease.
The differential expression of the DDTFR10/A gene was observed under stress conditions (Fusarium wilt and salinity) through expression profiling. The expression level of DDTFR10/A was comparatively higher in Fusarium wilt-susceptible tomato cv. Sandal when inoculated with FOL as compared to control plants. Unlike salt-treatment plants, an increase in DDTFR10/A gene expression was observed in Fusarium wilt-challenged plants. Hence, we attempted the genome-editing strategy using CRISPR-Cas9 tool for finding the role of DDTFR10/A gene in tomato against Fusarium wilt. The findings corroborate the role of DDTFR10/A in disease establishment and indicated its role as a negative regulator to the resistance against Fusarium wilt.
Supplementary Information
Below is the link to the electronic supplementary material.
Author contributions
SI and IUH conceived, designed, and developed the experiment. SI, IH, and HAR experimented. SI, and IH, analyzed and interpreted the data, drafted and revised the manuscript.
Data availability
We declare that the submitted manuscript is our work that has not been published before and is not currently being considered elsewhere.
Declarations
Conflict of interests
Authors declare that they have No Conflict of Interest.
Ethical approval
This article does not contain any studies with human participants or animals performed by any of the authors.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- Amini J, Sidovich D. The effects of fungicides on Fusarium oxysporum f. sp. lycopersici associated with Fusarium wilt of tomato. J Plant Prot Res. 2010;50:172–178. doi: 10.2478/v10045-010-0029-x. [DOI] [Google Scholar]
- Baloch A, Bangulzai BA, Dawood M, Yousaf S (2021) Efficacy of different fungicides against fusarium wilt and their impacts on height and yield of tomato crop under the tunnel farming condition. Pak J Biotechnol 18:1–5. 10.34016/pjbt.2021.18.1.1
- Banno H, Ikeda Y, Niu QW, Chua NH. Overexpression of Arabidopsis ESR1 induces initiation of shoot regeneration. Plant Cell. 2001;13:2609–2618. doi: 10.1105/tpc.010234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buttner M, Singh KB. Arabidopsis thaliana ethylene-responsive element binding protein (AtEBP), an ethylene-inducible, GCC box DNA-binding protein interacts with an ocs element binding protein. Proc Natl Acad Sci USA. 1997;94:5961–5966. doi: 10.1073/pnas.94.11.5961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catanzariti AM, Lim GT, Jones DA. The tomato I-3 gene: a novel gene for resistance to Fusarium wilt disease. New Phytol. 2015;207:106–118. doi: 10.1111/nph.13348. [DOI] [PubMed] [Google Scholar]
- Catanzariti A-M, Do HT, Bru P, de Sain M, Thatcher LF, Rep M, et al. The tomato I gene for Fusarium wilt resistance encodes an atypical leucine-rich repeat receptor-like protein whose function is nevertheless dependent on SOBIR1 and SERK3/BAK1. Plant J. 2017;89:1195–1209. doi: 10.1111/tpj.13458. [DOI] [PubMed] [Google Scholar]
- Chakrabarti AM, Henser-Brownhill T, Monserrat J, Poetsch AR, Luscombe NM, Scaffidi P. Target-specific precision of CRISPR-mediated genome editing. Mol Cel. 2019;73:699–713. doi: 10.1016/j.molcel.2018.11.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakravarthy S, Tuori RP, D’Ascenzo MD, Fobert PR, Despres C, Martin GB. The tomato transcription factor Pti4 regulates defense-related gene expression via GCC box and non-GCC box cis elements. Plant Cell. 2003;15:3033–3050. doi: 10.1105/tpc.017574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen G, Hu Z, Grierson D. Differential regulation of tomato ethylene responsive factor LeERF3b, a putative repressor, and the activator Pti4 in ripening mutants and in response to environmental stresses. J Plant Physiol. 2008;165:662–670. doi: 10.1016/j.jplph.2007.03.006. [DOI] [PubMed] [Google Scholar]
- Chen T, Lv Y, Zhao T, Li N, Yang Y, Yu W, He X, Liu T, Zhang, B (2013) Comparative transcriptome profiling of a resistant vs. susceptible tomato (Solanum lycopersicum) cultivar in response to infection by tomato yellow leaf curl virus. PloS one, 8(11), e80816. [DOI] [PMC free article] [PubMed]
- Cheng MC, Liao PM, Kuo WW, Lin TP. The Arabidopsis ETHYLENE RESPONSE FACTOR1 regulates abiotic stress-responsive gene expression by binding to different cis-acting elements in response to different stress signals. Plant Physiol. 2013;162:1566–1582. doi: 10.1104/pp.113.221911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dubouzet JG, Sakuma Y, Ito Y, Kasuga M, Dubouzet EG, Miura S, Seki M, Shinozaki K, Yamaguchi-Shinozaki K. OsDREB genes in rice, Oryza sativa L., encode transcription activators that function in drought-, high-salt- and cold-responsive gene expression. Plant J. 2003;33:751–763. doi: 10.1046/j.1365-313X.2003.01661.x. [DOI] [PubMed] [Google Scholar]
- Farooq M, Ullah H, Nawab NN, Qureshi KM. Evaluation of indigenous tomato hybrids under plastic tunnel. Pak J Agric Res. 2013;26(2):97–103. [Google Scholar]
- Flowers TJ, Yeo AR. Breeding for salinity resistance in crop plants: Where next? Aust J Plant Physiol. 1995;22:875–884. doi: 10.1071/PP9950875. [DOI] [Google Scholar]
- Food and agriculture organization corporate statistical database (2009). Available from: http://en.wikipedia.org/wiki/Food_and_Agriculture_Organization_Corporate_Statistical_Databasee
- Fujimoto SY, Ohta M, Usui A, Shinshi H, Ohme-Takagi M. Arabidopsis ethylene-responsive element binding factors act as transcriptional activators or repressors of GCC box–mediated gene expression. Plant Cell. 2000;12:393–404. doi: 10.1105/tpc.12.3.393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujisawa T, Barraclough TG. Delimiting species using single-locus data and the generalized mixed yule coalescent approach: a revised method and evaluation on simulated data sets. Syst Biol. 2013;62:707–724. doi: 10.1093/sysbio/syt033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujita M, Fujita Y, Noutoshi Y, Takahashi F, Narusaka Y, Yamaguchi-Shinozaki K, Shinozaki K. Cross-talk between abiotic and biotic stress responses: a current view from the points of convergence in the stress signaling networks. Curr Opin Plant Biol. 2006;9:436–442. doi: 10.1016/j.pbi.2006.05.014. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Cendales Y, Catanzariti AM, Baker B, Mcgrath DJ, Jones DA. Identification of I-7 expands the repertoire of genes for resistance to Fusarium wilt in tomato to three resistance gene classes. Mol Plant Pathol. 2016;17:448–463. doi: 10.1111/mpp.12294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Yang C, Thara VK, Zhou J, Martin GB. Pti4 is induced by ethylene and salicylic acid, and its product is phosphorylated by the Pto kinase. Plant Cell. 2000;12:771–786. doi: 10.1105/tpc.12.5.771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu YQ, Wildermuth MC, Chakravarthy S, Loh YT, Yang C, He X, Han Y, Martin GB. Tomato transcription factors pti4, pti5, and pti6 activate defense responses when expressed in Arabidopsis. Plant Cell. 2002;14:817–831. doi: 10.1105/tpc.000794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo H, Ecker JR. The ethylene signaling pathway: new insights. Curr Opin Plant Biol. 2004;7:40–49. doi: 10.1016/j.pbi.2003.11.011. [DOI] [PubMed] [Google Scholar]
- Houssien AA, Ahmed SM, Ismail AA. Activation of tomato plant defense response against Fusarium wilt disease using Trichoderma harzianum and salicylic acid under greenhouse conditions. Res J Agric Biol Sci. 2010;6:328–338. [Google Scholar]
- Hussain I, Alam SS, Khan I, Shah B, Naeem A, Khan N, Ullah W, Iqbal B, Adnan M, Junaid K, Shah SR. Study on the biological control of fusarium wilt of tomato. J Entomol Zool Studies. 2016;6:328–338. [Google Scholar]
- Ito Y, Nishizawa-Yokoi A, Endo M, Mikami M, Toki S. CRISPR/Cas9-mediated mutagenesis of the RIN locus that regulates tomato fruit ripening. Biochem Biophys Res Commun. 2015;467:76–82. doi: 10.1016/j.bbrc.2015.09.117. [DOI] [PubMed] [Google Scholar]
- Karmakar S, Das P, Panda D, Xie K, Baig MJ, Molla KA. A detailed landscape of CRISPR-Cas-mediated plant disease and pest management. Plant Sci. 2022;323:111376. doi: 10.1016/j.plantsci.2022.111376. [DOI] [PubMed] [Google Scholar]
- Karmakar A, Taufiqa S, Baig MJ, Molla KA (2022a) Increasing disease resistance in host plants through genome editing. In Proceedings of the Indian National Science Academy. pp 1–13
- Khuong TT, Crété P, Robaglia C, Caffarri S. Optimisation of tomato Micro-tom regeneration and selection on glufosinate/Basta and dependency of gene silencing on transgene copy number. Plant Cell Rep. 2013;32:1441–1454. doi: 10.1007/s00299-013-1456-8. [DOI] [PubMed] [Google Scholar]
- Klay I, Pirrello J, Riahi L, Bernadac A, Cherif A, Bouzayen M, Bouzid S. Ethylene response factor Sl-ERFB3 is responsive to abiotic stresses and mediates salt and cold stress response regulation in tomato. Sci World J. 2014;2014:167681. doi: 10.1155/2014/167681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J, Guo X, Zhang M, Wang X, Zhao Y, Yin Z, Zhang Z, Wang Y, Xiong H, Zhang H, Todorovska E. OsERF71 confers drought tolerance via modulating ABA signaling and proline biosynthesis. Plant Sci. 2018;270:131–139. doi: 10.1016/j.plantsci.2018.01.017. [DOI] [PubMed] [Google Scholar]
- Li R, Fu D, Zhu B, Luo Y, Zhu H. CRISPR/Cas9-mediated mutagenesis of lncRNA1459 alters tomato fruit ripening. Plant J. 2018;94:513–524. doi: 10.1111/tpj.13872. [DOI] [PubMed] [Google Scholar]
- Li X, Wang Y, Chen S, Tian H, Fu D, Zhu B, Luo Y, Zhu H. Lycopene is enriched in tomato fruit by CRISPR/Cas9-mediated multiplex genome editing. Front Plant Sci. 2018;9:559. doi: 10.3389/fpls.2018.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Tian Y, Xu J, Fu X, Gao J, Wang BO, Han H, Wang L, Peng R, Yao Q. A tomato ERF transcription factor, SlERF84, confers enhanced tolerance to drought and salt stress but negatively regulates immunity against Pseudomonas syringae pv. tomato DC3000. Plant Physiol Biochem. 2018;132:683–695. doi: 10.1016/j.plaphy.2018.08.022. [DOI] [PubMed] [Google Scholar]
- Liu M, Gomes BL, Mila I, Purgatto E, Peres LE, Frasse P, Maza E, Zouine M, Roustan JP, Bouzayen M, Pirrello J. Comprehensive profiling of ethylene response factor expression identifies ripening-associated ERF genes and their link to key regulators of fruit ripening in tomato. Plant Physiol. 2016;170:1732–1744. doi: 10.1104/pp.15.01859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu L, Zhang J, Xu J, Li Y, Guo L, Wang Z, Zhang X, Zhao B, Guo YD, Zhang N. CRISPR/Cas9 targeted mutagenesis of SlLBD40, a lateral organ boundaries domain transcription factor, enhances drought tolerance in tomato. Plant Sci. 2020;301:110683. doi: 10.1016/j.plantsci.2020.110683. [DOI] [PubMed] [Google Scholar]
- Lorenzo O, Piqueras R, Sánchez-Serrano JJ, Solano R. Ethylene response factor1 integrates signals from ethylene and jasmonate pathways in plant defense. Plant Cell. 2003;15:165–178. doi: 10.1105/tpc.007468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma X, Zhang Q, Zhu Q, Liu W, Chen Y, Qiu R, Wang B, Yang Z, Li H, Lin Y, Xie Y, Shen R, Chen S, Wang Z, Chen Y, Guo J, Chen L, Zhao X, Dong Z, Liu YG. A robust CRISPR/Cas9 system forconvenient, high-efficiency multiplex genome editing in monocot and dicot plants. Mol Plant. 2015;8:1274–1284. doi: 10.1016/j.molp.2015.04.007. [DOI] [PubMed] [Google Scholar]
- Makarova KS, Haft DH, Barrangou R, Brouns SJ, Charpentier E, Horvath P, Moineau S, Mojica FJ, Wolf YI, Yakunin AF, Van Der Oost J. Evolution and classification of the CRISPR-Cas systems. Nat Rev Microbiol. 2011;9:467–477. doi: 10.1038/nrmicro2577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manikandan R, Harish S, Karthikeyan G, Raguchander T. Comparative proteomic analysis of different isolates of Fusarium oxysporum f sp lycopersici to exploit the differentially expressed proteins responsible for virulence on tomato plants. Front Microbial. 2018;9:420. doi: 10.3389/fmicb.2018.00420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moin M, Bakshi A, Madhav MS, Kirti PB. Cas9/sgRNA-based genome editing and other reverse genetic approaches for functional genomic studies in rice. Brief Funct Genomics. 2018;17:339–351. doi: 10.1093/bfgp/ely010. [DOI] [PubMed] [Google Scholar]
- Molla KA, Shih J, Wheatley MS, Yang Y. Predictable NHEJ insertion and assessment of HDR editing strategies in plants. Front Genome Ed. 2022;4:825236. doi: 10.3389/fgeed.2022.825236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mysore KS, Crasta OR, Tuori RP, Folkerts O, Swirsky PB, Martin GB. Comprehensive transcript profiling of Pto-and Prf-mediated host defense responses to infection by Pseudomonas syringae pv. tomato. Plant J. 2002;32:299–315. doi: 10.1046/j.1365-313X.2002.01424.x. [DOI] [PubMed] [Google Scholar]
- Ouyang B, Yang T, Li H, Zhang L, Zhang Y, Zhang J, Fei Z, Ye Z. Identification of early salt stress response genes in tomato root by suppression subtractive hybridization and microarray analysis. J Exp Bot. 2007;58:507–520. doi: 10.1093/jxb/erl258. [DOI] [PubMed] [Google Scholar]
- Pan Y, Seymour GB, Lu C, Hu Z, Chen X, Chen G. An ethylene response factor (ERF5) promoting adaptation to drought and salt tolerance in tomato. Plant Cell Rep. 2012;31:349–360. doi: 10.1007/s00299-011-1170-3. [DOI] [PubMed] [Google Scholar]
- Pan C, Ye L, Qin L, Liu X, He Y, Wang J, Chen L, Lu G. CRISPR/Cas9-mediated efficient and heritable targeted mutagenesis in tomato plants in the first and later generations. Sci Rep. 2016;6:1–9. doi: 10.1038/srep24765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park JM, Park CJ, Lee SB, Ham BK, Shin R, Paek KH. Overexpression of the tobacco Tsi1 gene encoding an EREBP/AP2–type transcription factor enhances resistance against pathogen attack and osmotic stress in tobacco. Plant Cell. 2001;13:1035–1046. doi: 10.1105/tpc.13.5.1035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrello J, Prasad BN, Zhang W, Chen K, Mila I, Zouine M, Latche A, Pech JC, Ohme-Takagi M, Regad F, Bouzayen M. Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol. 2012;12:1–15. doi: 10.1186/1471-2229-12-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pirrello J, Prasad BN, Zhang W, Chen K, Mila I, Zouine M, Latché A, Pech JC, Ohme-Takagi M, Regad F, Bouzayen M. Functional analysis and binding affinity of tomato ethylene response factors provide insight on the molecular bases of plant differential responses to ethylene. BMC Plant Biol. 2012;12:190. doi: 10.1186/1471-2229-12-190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prihatna C, Barbetti MJ, Barker SJ. A novel tomato fusarium wilt tolerance gene. Front Microbiol. 2018;9:1226. doi: 10.3389/fmicb.2018.01226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman MM, Hossain M, Hossain KF, Sikder MT, Shammi M, Rasheduzzaman M, Hossain MA, Alam AM, Uddin MK. Effects of NaCl-salinity on tomato (Lycopersicon esculentum Mill.) plants in a pot experiment. Open Agric. 2018;3:578–585. doi: 10.1515/opag-2018-0061. [DOI] [Google Scholar]
- Shan Q, Wang Y, Li J, Zhang Y, Chen K, Liang Z, Zhang K, Liu J, Xi JJ, Qiu JL, Gao C. Targeted genome modification of crop plants using a CRISPR-Cas system. Nat Biotechnol. 2013;31:686–688. doi: 10.1038/nbt.2650. [DOI] [PubMed] [Google Scholar]
- Shanmugam V, Kanoujia N. Biological management of vascular wilt of tomato caused by Fusarium oxsporum f.sp. lycopersici by plant growth promoting rhizobacterial mixture. Biol Control. 2011;57:85–93. doi: 10.1016/j.biocontrol.2011.02.001. [DOI] [Google Scholar]
- Simons G, Groenendijk J, Wijbrandi J, Reijans M, Groenen J, Diergaarde P, Van der Lee T, Bleeker M, Onstenk J, de Both M, Haring M. Dissection of the Fusarium I2 gene cluster in tomato reveals six homologs and one active gene copy. Plant Cell. 1998;10:1055–1068. doi: 10.1105/tpc.10.6.1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song W, Zhou L, Yang C, Cao X, Zhang L, Liu X. Tomato Fusarium wilt and its chemical control strategies in a hydroponic system. Crop Prot. 2004;23:243–247. doi: 10.1016/j.cropro.2003.08.007. [DOI] [Google Scholar]
- Soomro AF, Ali M, Yasin A (2020) Tomato cluster feasibility and transformation study. In Ali Mubarik, (ed.). (2020). Cluster development based agriculture transformation plan vision-2025. Project No. 131(434)PC/AGR/CDBAT-120/2018. Planning commission of Pakistan, Islamabad, Pakistan and centre for agriculture and biosciences international (CABI), Rawalpindi, Pakistan
- Srinivas C, Devi DN, Murthy KN, Mohan CD, Lakshmeesha TR, Singh B, Kalagatur NK, Niranjana SR, Hashem A, Alqarawi AA, Tabassum B. Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: biology to diversity–a review. Saudi J Biol Sci. 2019;26:1315–1324. doi: 10.1016/j.sjbs.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Supartana P, Shimizu T, Nogawa M, Shioiri H, Nakajima T, Haramoto N, Nozue M, Kojima M. Development of simple and efficient in planta transformation method for wheat (Triticum aestivum L.) using Agrobacterium tumefaciens. J Biosci Bioeng. 2006;102(3):162–170. doi: 10.1263/jbb.102.162. [DOI] [PubMed] [Google Scholar]
- Syngenta (2015) Tomato sahel: crops & products. Available on http://www3.syngenta.com/country/za/en/cropsandproducts/productbrands/Vegetable%20Seeds/tomatofreshmarket/ Pages/Sahel.aspx.
- Tang W, Charles TM, Newton RJ. Overexpression of the pepper transcription factor CaPF1 in transgenic Virginia pine (Pinus virginiana Mill.) confers multiple stress tolerance and enhances organ growth. Plant Mol Biol. 2005;59:603–617. doi: 10.1007/s11103-005-0451-z. [DOI] [PubMed] [Google Scholar]
- Tournier B, Sanchez-Ballesta MT, Jones B, Pesquet E, Regad F, Latché A, Pech JC, Bouzayen M. New members of the tomato ERF family show specific expression pattern and diverse DNA-binding capacity to the GCC box element. FEBS Lett. 2003;550:149–154. doi: 10.1016/S0014-5793(03)00757-9. [DOI] [PubMed] [Google Scholar]
- van der Fits L, Memelink J. ORCA3, a jasmonate-responsive transcriptional regulator of plant primary and secondary metabolism. Science. 2000;289:295–297. doi: 10.1126/science.289.5477.295. [DOI] [PubMed] [Google Scholar]
- Wang H, Huang Z, Chen Q, Zhang Z, Zhang H, Wu Y, Huang D, Huang R. Ectopic overexpression of tomato JERF3 in tobacco activates downstream gene expression and enhances salt tolerance. Plant Mol Biol. 2004;55:183–192. doi: 10.1007/s11103-004-0113-6. [DOI] [PubMed] [Google Scholar]
- Xu ZS, Xia LQ, Chen M, Cheng XG, Zhang RY, Li LC, Zhao YX, Lu Y, Ni ZY, Liu L, Qiu ZG (2007) Isolation and molecular characterization of the Triticum aestivum L. ethylene-responsive factor 1 (TaERF1) that increases multiple stress tolerance. Plant Mol Biol 65(6):719–732. 10.1007/s11103-007-9237-9 [DOI] [PubMed]
- Yasmeen A, Mirza B, Inayatullah S, Safdar N, Jamil M, Ali S, Choudhry MF. In planta transformation of tomato. Plant Mol Biol Rep. 2009;27:20–28. doi: 10.1007/s11105-008-0044-5. [DOI] [Google Scholar]
- Yi SY, Kim JH, Joung YH, Lee S, Kim WT, Yu SH, Choi D. The pepper transcription factor CaPF1 confers pathogen and freezing tolerance in Arabidopsis. Plant Physiol. 2004;136:2862–2874. doi: 10.1104/pp.104.042903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu QH, Wang B, Li N, Tang Y, Yang S, Yang T, Xu J, Guo C, Yan P, Wang Q, Asmutola P. CRISPR/Cas9-induced targeted mutagenesis and gene replacement to generate long-shelf life tomato lines. Sci Rep. 2017;7(1):1–9. doi: 10.1038/s41598-017-12262-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu W, Zhao R, Sheng J, Shen L. SlERF2 is associated with methyl jasmonate-mediated defense response against Botrytis cinerea in tomato fruit. J Agric Food Chem. 2018;66:9923–9932. doi: 10.1021/acs.jafc.8b03971. [DOI] [PubMed] [Google Scholar]
- Zhang H, Zhang D, Chen J, Yang Y, Huang Z, Huang D, Wang XC, Huang R. Tomato stress-responsive factor TSRF1 interacts with ethylene responsive element GCC box and regulates pathogen resistance to Ralstonia solanacearum. Plant Mol Biol. 2004;55:825–834. doi: 10.1007/s11103-005-2140-3. [DOI] [PubMed] [Google Scholar]
- Zhang G, Chen M, Li L, Xu Z, Chen X, Guo J, Ma Y. Overexpression of the soybean GmERF3 gene, an AP2/ERF type transcription factor for increased tolerances to salt, drought, and diseases in transgenic tobacco. J Exp Bot. 2009;60:3781–3796. doi: 10.1093/jxb/erp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang H, Zhang J, Wei P, Zhang B, Gou F, Feng Z, et al. The CRISPR/Cas9 system produces specific and homozygous targeted gene editing in rice in one generation. Plant Biotechnol J. 2014;12:797–807. doi: 10.1111/pbi.12200. [DOI] [PubMed] [Google Scholar]
- Zhou J, Tang X, Martin GB. The Pto kinase conferring resistance to tomato bacterial speck disease interacts with proteins that bind a cis-element of pathogenesis-related genes. EMBO J. 1997;16:3207–3218. doi: 10.1093/emboj/16.11.3207. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
We declare that the submitted manuscript is our work that has not been published before and is not currently being considered elsewhere.