Abstract
Machine learning methods have been applied to estimate measures of brain aging from neuroimages. However, only rarely have these measures been examined in the context of biologic age. Here, we investigated associations of an MRI-based measure of dementia risk, the Alzheimer’s disease pattern similarity (AD-PS) scores, with measures used to calculate biological age. Participants were those from visit 5 of the Atherosclerosis Risk in Communities Study with cognitive status adjudication, proteomic data, and AD-PS scores available. The AD-PS score estimation is based on previously reported machine learning methods. We evaluated associations of the AD-PS score with all-cause mortality. Sensitivity analyses using only cognitively normal (CN) individuals were performed treating CNS-related causes of death as competing risk. AD-PS score was examined in association with 32 proteins measured, using a Somalogic platform, previously reported to be associated with age. Finally, associations with a deficit accumulation index (DAI) based on a count of 38 health conditions were investigated. All analyses were adjusted for age, race, sex, education, smoking, hypertension, and diabetes. The AD-PS score was significantly associated with all-cause mortality and with levels of 9 of the 32 proteins. Growth/differentiation factor 15 (GDF-15) and pleiotrophin remained significant after accounting for multiple-testing and when restricting the analysis to CN participants. A linear regression model showed a significant association between DAI and AD-PS scores overall. While the AD-PS scores were created as a measure of dementia risk, our analyses suggest that they could also be capturing brain aging.
Keywords: Machine learning, Aging, Alzheimer’s disease, Proteomics, Deficit accumulation index, Mortality
Introduction
The incidence of Alzheimer’s disease (AD) and related dementias (ADRD) rises exponentially with age. Most etiologic research on ADRD has focused on hallmark pathophysiology of β-amyloid (Aβ) plaques and neurofibrillary tangles (NFT); yet, the role of potentiating age-related brain changes is garnering increased attention [1]. In previous work, we applied machine learning to structural MRIs to develop an index of neuroanatomic risk of dementia called Alzheimer’s Disease Pattern Similarity (AD-PS) score [2]. The algorithm behind the score captures spatial patterns of gray matter tissue able to discriminate between cognitively normal (CN) individuals and those classified as having dementia. The AD-PS scores have been shown to be associated with age, cognitive status, changes in cognitive function, incident cognitive impairment [3], trajectories of global cognitive function [4], and particulate matter air pollution [5, 6]. More recently, using data from the Atherosclerosis Risk in Communities (ARIC) study cohort, we have shown the AD-PS scores to be more predictive of incident cognitive impairment than a volumetric composite measure composed of regions susceptible to AD [7] (e.g., hippocampus, entorhinal, inferior parietal lobule, precuneus). Because the AD-PS score is based on spatial patterns of atrophy detected by a machine learning algorithm and does not directly assess specific brain features (e.g., temporal lobe region volumes) or pathologies (e.g., Aβ, NFT), we considered that the AD-PS score may also be a measure of accelerated brain aging. Therefore, AD-PS may be capable of identifying brains that are relatively “old” compared to others of a similar chronological age.
There is a rapidly growing literature related to the assessment of biologic age using a variety of markers and strategies including telomere length, clinical, laboratory findings, measures of diseases, physical disability, cognitive impairment, proteomics, methylation, and metabolomics. Much of this effort is driven by the emerging field of geroscience, which posits that common biological mechanisms of aging play important roles in the susceptibility of aged persons to multiple chronic diseases [8]. Furthermore, the accumulation of biochemical and cellular features of aging may reciprocally accelerate underlying mechanisms of aging and accelerate development of aged phenotypes [9]. As such, biomarker development in geroscience is focused on biological pillars of aging, molecular and cellular drivers of aging, and accumulated deficits across systems rather than aging in any singular organ system [10, 11]. However, the strategies coming from the geroscience field often do not include any imaging biomarkers and particularly not brain imaging despite relevance to aging [1, 12, 13]. This is a critical gap because brain structural abnormalities are associated with physical function and risk for dementia.
From the neuroscience perspective, several groups have applied machine learning and artificial intelligence methods to brain images to estimate measures of brain aging [14–16]. These estimates are often based on structural MRI because it is more available, less invasive, and cheaper compared to other brain imaging modalities such as Positron Emission Tomography (PET). Accelerated brain aging calculated in this fashion has been shown to be associated with smoking and alcohol consumption [17] and progression to AD [18, 19] among other examples.
In this paper, we evaluated the convergent and predictive validity of the AD-PS score as a measure of biologic age using data from visit 5 of the ARIC cohort study. If the AD-PS score were a measure of biologic age, we would expect it to (a) be associated with non-brain aging biomarkers independent of age and (b) predict age-related outcomes independently of age. We tested these hypotheses by examining (1) cross-sectional associations between AD-PS score and 32 blood-based proteomic markers, which have previously been identified as being strongly associated with age [20], and (2) a deficit accumulation index which is the proportion of prevalent health deficits queried from health record, functional assessment, and clinical laboratories, etc. Deficit accumulation–based frailty indices are strongly associated with mortality outcomes independently of chronological age [21]. With respect to predictive validity, we tested the hypothesis that AD-PS score was associated with all-cause mortality independently of age.
Methods
The ARIC study conducted the baseline exam from 1987 to 1989 among 15,792 White and African American participants aged 45–64 years who were recruited from four field centers located in Forsyth County, NC; Jackson, MS; Minneapolis suburbs, MN; and Washington County, MD. Using probability sampling, each ARIC field center recruited approximately 4000 individuals from their community. Only African Americans were recruited in Jackson, MS; the remaining sites reflected local populations, mostly White in Minneapolis and Washington County and both races in Forsyth County. Due to small sample sizes, we additionally excluded African American participants from the Minneapolis and Washington County centers (N = 8). The Institutional Review Boards from all centers approved ARIC protocols; participants provided written consent. Our analysis included cohort data available through ARIC visit 5 (occurring between 2011 and 2013) who additionally have MRI and cognitive data available (N = 1849).
ARIC data
Cognitive evaluation
The cognitive status (nonimpaired, mild cognitive impairment (MCI), or dementia) of participants who attended visit 5 was determined using a standardized algorithm based on cognitive assessments, verified by expert committee review, using information from in-person cognitive batteries, the Clinical Dementia Rating scale, and Functional Activities Questionnaires completed by participants and/or informants [22].
MRI
The ARIC visit 5 (2011–2013) brain MRI scans were performed on four 3 T scanners (Maryland: Siemens Verio; North Carolina: Siemens Skyra; Minnesota: Siemens Trio; Mississippi: Siemens Skyra). The following sequences were obtained: localizer, magnetization-prepared rapid gradient-echo MP-RAGE (1.2-mm slices), axial gradient recalled echo T2-weighted imaging (T2*GRE) (4-mm slices), axial T2 fluid-attenuated inversion recovery (FLAIR) (5-mm slices), field mapping (3-mm slices), and axial diffusion tenor images (2.7-mm slices for Skyra and Verio scanners and 3-mm slices for Trio scanner). T2 FLAIR and T2*GRE sequences were also collected to assess brain lesion burden. The generation of the AD-PS scores was based on the T1 weighted MPRAGE images. Image processing details have been reported previously [7, 23]. A brief description can be found in the supplementary materials.
Mortality information
Ascertainment of mortality was based on medical records and National Death Index searches. Due to lack of access to records at one large Jackson hospital in 2018 and 2019, we excluded from the final datasets any hospitalizations for Jackson participants for 2018 and 2019; thus, the value for the administrative censoring was set to be December 31, 2017, for Jackson participants, instead of the standard value of December 31, 2019, for participants from the other three field centers.
Protein measurements
Full details about proteomic data collection and processing have been previously reported [24]. Briefly, using blood collected at ARIC visit 5, the relative concentration of plasma proteins or protein complexes was measured using a SOMA aptamer-based capture array. This method uses short single strands of DNA with chemically modified nucleotides, called modified aptamers, which act as protein-binding reagents with defined three-dimensional structures and unique nucleotide sequences that are identifiable and quantifiable using DNA detection technology. The SomaScan assay has been described in detail previously, as have the assay’s performance characteristics. Plasma was collected using a standardized protocol at each ARIC site, frozen at − 80 °C and shipped on dry ice to the ARIC central laboratory where it was continuously frozen until aliquoting into barcoded microtiter plates with screw-top lids. The plates were sent to SomaLogic for quantification. In total, 5284 modified aptamers (SOMAmers reagents or “SOMAmers”) were used to measure relative protein concentration. From those 5284 proteins, 4877 passed ARIC quality control. For our analyses, we selected the 32 protein panels (Table S2) which have been consistently associated with chronologic age across several independent data sets [20]. There were 1500 participants with MRI, and proteomic data at visit 5.
Deficit accumulation index
We used 38 measures of physical, cognitive, and emotional function; diagnosed diseases; overall health; and clinical laboratories to estimate a deficit accumulation index (DAI) [21] (see Table S3 in supplementary materials) for data available in ARIC at visit 5. For each one of these items, cutoff values signaling the presence of a deficit in a given individual were defined. The index was computed as the ratio of variables out of defined reference range or health deficits by the total of available variables queried for each individual. If the participant had more than 20% of the items missing, then the index was not computed. For our analyses, we had 1644 participants with DAI, MRI, and cognitive data available at visit 5.
Estimation of the AD-PS scores
The overall approach to estimate the ARIC AD-PS scores has been previously described in detail [7]. Two datasets are involved in the estimation of the AD-PS scores utilized in this study. ARIC comprised the testing cohort while MRI data from the Alzheimer’s Disease Neuroimaging Initiative (ADNI) was used to train machine learning algorithms to generate AD-PS scores when provided with MRI data from ARIC participants. MRI scans from both ARIC and ADNI (Table S1) were aligned to a common template (derived from ADNI images) using image processing tools available in the Advanced Normalization Tools (ANTs) software package [7, 23] (see supplementary materials). Next, we used high-dimensional machine learning methods to estimate the AD-PS scores. Details of the machine learning algorithms were published previously [2, 5, 23, 25, 26]. Briefly, a regularized logistic regression (RLR) classifier was estimated in a voxel-wise manner using the gray matter probability maps from CN and AD participants available in the ADNI training dataset. To estimate the optimal values of the regularization parameters, we combined nested tenfold cross-validations and grid search. Once the RLR classifier was estimated, the conditional probabilities of a given individual having AD-like patterns according to the MRI scan were generated using data from ARIC participants (testing dataset). The final scores were estimated as the mean values of 5 repetitions of the computations, to account for variability due to random partitioning of cross-validation that occurred during model estimation. We refer to these average probabilities as AD-PS scores.
Analyses
To investigate associations of the AD-PS scores with mortality, Cox proportional hazards regression was performed. Participants were stratified by tertiles according to AD-PS values. An additional sensitivity analysis was performed using only the CN individuals and treating causes of death directly related to the central nervous system (CNS) (e.g., strokes, dementia) and not non-CNS causes of death as competing risks [27]. To evaluate associations of the AD-PS scores with proteomics, cross-sectional analyses focused on the 32 proteins reported by Johnson and colleagues [20] were performed. We fitted linear regression models for each protein at a time using the AD-PS scores as the outcome. A Bonferroni correction (α < 0.05) for multiple comparisons was applied. Finally, we fitted a linear regression model to investigate relationships between DAI and the AD-PS scores. All analyses were adjusted for age, sex, center-race, smoking, hypertension, education, and diabetes. The 5-level center-race variable (Forsyth-AA, Forsyth-W, Jackson-AA, Minn-W, Wash Co-W) was created to accommodate a lack of representation of both races in all centers. Analyses were performed using SAS (version 9.4) and R (version 4.0.2).
Results
Table 1 describes the basic demographic characteristics of the ARIC cohort who had AD-PS scores available at visit 5 of the study (N = 1849), stratified by cognitive status at the time of the visit. In comparison with participants classified as having dementia or MCI, the cognitively normal group had lower prevalence of hypertension and was on average younger.
Table 1.
Characteristics of ARIC analytic sample by visit 5 cognitive status. Age and BMI mean and SD values are provided. For categorical variables, sample size and percentages are presented
| Total | Cognitive status at visit 5 | |||
|---|---|---|---|---|
| Normal | MCI | Dementia | ||
| N | 1849 | 1172 | 589 | 88 |
| Gender | ||||
| Female | 1119 (60.5%) | 747 (63.7%) | 320 (54.3%) | 52 (59.1%) |
| Race | ||||
| Black | 538 (29.1%) | 378 (32.3%) | 130 (22.1%) | 30 (34.1%) |
| Education* | ||||
| Basic | 265 (14.4%) | 156 (13.3%) | 82 (13.9%) | 27 (30.7%) |
| Intermediate | 752 (40.7%) | 449 (38.4%) | 270 (45.8%) | 33 (37.5%) |
| Advanced | 830 (44.9%) | 565 (48.3%) | 237 (40.2%) | 28 (31.8%) |
| Smoking status | ||||
| Current | 97 (5.3%) | 55 (4.8%) | 37 (6.4%) | 5 (6.0%) |
| Former | 867 (47.7%) | 565 (48.8%) | 266 (46.3%) | 36 (43.4%) |
| Never | 764 (42.1%) | 488 (42.1%) | 241 (41.9%) | 35 (42.2%) |
| Hypertension | ||||
| No | 449 (24.6%) | 301 (25.8%) | 131 (22.6%) | 17 (20.5%) |
| Age | 76.4 (5.3) | 76.0 (5.3) | 76.7 (5.2) | 79.3 (5.4) |
| Obesity (yes) | 32.4% | 32.6% | 32.4% | 32.2% |
| BMI | 28.5 (5.7) | 28.5 (5.7) | 28.6 (5.7) | 27.7 (5.8) |
| 1849 | 1172 | 589 | 88 | |
*Education categories—Basic is less than completed high school, intermediate is high school or equivalent, and advanced is at least some college
All-cause mortality
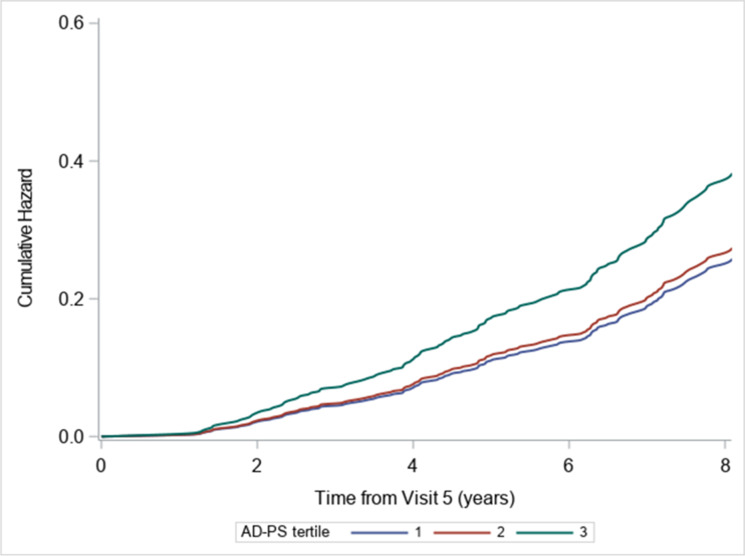
There were 347 deaths among 1808 participants over 8 years of follow-up after visit 5. The AD-PS score was significantly associated (p < 0.001) with all-cause mortality (Table 2). Those in the lowest tertile of the score had a 55% lower all-cause mortality rate compared to those in the highest tertile (hazard rate ratio: 0.45; 95% CI: 0.33–0.62). The association remained strong when restricting the sample to only cognitively normal subjects (hazard rate ratio for lowest tertile: 0.59; 95% CI: 0.39–0.90). When treating CNS causes of death as competing risk among the CN individuals, the association was significant and the hazard rate ratio for the lowest tertile was 0.62; 95%CI: 0.39–0.98 (see Fig. 1).
Table 2.
Survival analyses investigating association between the AD-PS scores and mortality. The analyses were adjusted for age, sex, race-center, smoking, hypertension, education, and diabetes status
| Survival analysis using all participants with MRI at visit 5 (N = 1808, deaths = 347) | ||||
|---|---|---|---|---|
| AD-PS score | p-value | Hazard ratio | 95% hazard ratio confidence limits | |
| Lowest tertile (best) | < 0.0001 | 0.45 | 0.33 | 0.62 |
| Middle tertile | < 0.0001 | 0.59 | 0.45 | 0.78 |
| Restricting to only cognitively normal participants (N = 1148, deaths = 167) | ||||
| Lowest tertile (best) | 0.014 | 0.59 | 0.39 | 0.90 |
| Middle tertile | 0.019 | 0.64 | 0.44 | 0.93 |
| Restricting to cognitively normal and treating CNS-related causes of death as a competing risk (N = 1148, deaths = 144, competing events = 23) | ||||
| Lowest tertile (best) | 0.039 | 0.62 | 0.39 | 0.98 |
| Middle tertile | 0.045 | 0.66 | 0.45 | 0.99 |
Fig. 1.
Cumulative hazard of death by tertile of AD-PS scores for cognitively normal participants at visit 5 treating CNS-related causes of death as competing risk
Age related proteins
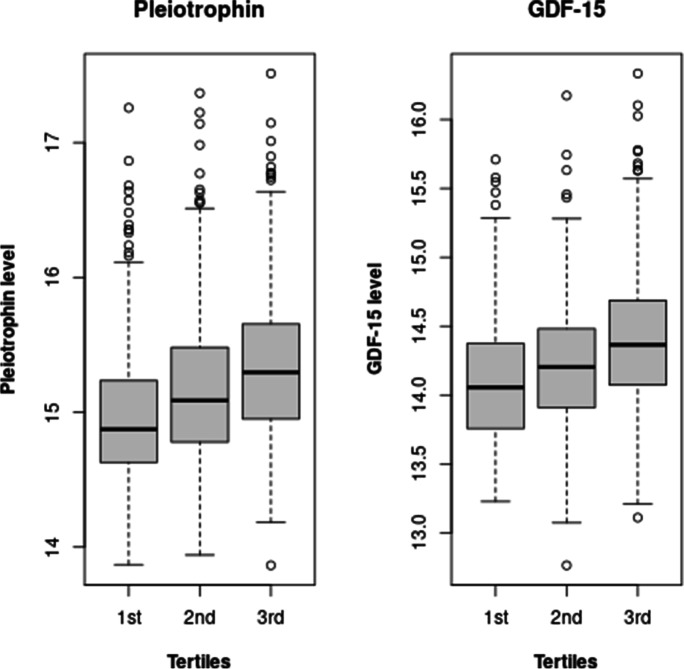
The cross-sectional association analyses between AD-PS scores and age-related proteins revealed the AD-PS scores to be significantly associated with 9 proteins from the above described panel (see Table 3). Growth/differentiation factor 15 (GDF-15) and pleiotrophin remained significant after Bonferroni correction. Figure 2 presents the boxplots of both proteins across AD-PS tertiles. An additional analysis based on cognitively normal participants showed both proteins remaining significant after Bonferroni correction.
Table 3.
Age-related proteins that were cross-sectionally associated with the AD-PS scores are presented. GDF-15 and pleiotrophin remained significant after Bonferroni correction. A sensitivity analysis including only cognitively normal participants showed significant associations of both proteins with the AD-PS scores. Both proteins remained significant after the Bonferroni correction in both analyses which is signaled with **. Two more proteins signaled with * remained significant associated with the AD-PS scores. Analyses were adjusted for age, sex, race-center, smoking, education, diabetes, and hypertension
| Proteins | β | SE | t | p-value |
|---|---|---|---|---|
| Growth/differentiation factor 15** | 0.05 | 0.01 | 4.31 | < 0.001 |
| Pleiotrophin** | 0.07 | 0.02 | 3.15 | < 0.001 |
| Laminin subunit alpha-2|Laminin subunit gamma-1|Laminin subunit beta-1 | 0.06 | 0.02 | 2.80 | 0.005 |
| Urokinase plasminogen activator surface receptor* | 0.04 | 0.02 | 2.67 | 0.008 |
| Tumor necrosis factor receptor superfamily member 1A | 0.04 | 0.02 | 2.49 | 0.013 |
| Macrophage metalloelastase | 0.02 | 0.01 | 2.07 | 0.025 |
| Tumor necrosis factor receptor superfamily member 1B | 0.03 | 0.02 | 2.11 | 0.035 |
| Hepatocyte growth factor* | 0.02 | 0.01 | 2.08 | 0.037 |
| Annexin A1 | 0.03 | 0.02 | 2.24 | 0.043 |
Fig. 2.
The levels of GDF-15 and pleiotrophin across AD-PS tertiles are presented for all individuals with MRI and proteomics available at visit 5
Deficit accumulation index
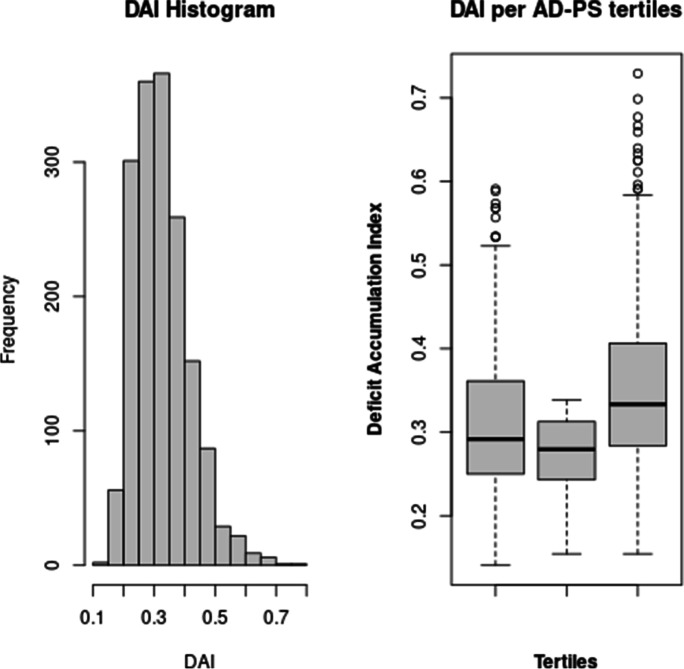
A linear regression model adjusted for age, race-center, sex, education, smoking, hypertension, and diabetes showed a significant association between DAI and AD-PS scores (β = 0.06, SE = 0.008, p < 0.001). For cognitively normal individuals, the association remained significant (β = 0.05, SE = 0.01, p < 0.001). The histogram and boxplots across AD-PS tertiles of the deficit accumulation index are presented in Fig. 3.
Fig. 3.
The histogram of the deficit accumulation index and its distribution across tertiles of AD-PS scores is presented
Discussion
In this paper, we show that a machine-learning derived measure for Alzheimer’s disease risk (AD-PS score) based on structural MRI was associated with three indicators of biologic age. Specifically, AD-PS was associated with all-cause mortality in cognitively normal persons even after censoring deaths from CNS causes; biomarkers of somatic aging, including GDF-15 and pleiotrophin; and a deficit accumulation index. Taken together, these findings suggest the AD-PS score may be a brain-based biomarker of organismal aging.
There is great interest in bringing together geroscience and neuroscience in order to develop a more comprehensive framework to understand brain aging disease, physical function, and cognitive function in the context of biological aging [1]. While there is an increasing body of work based on machine learning to estimate chronological age using neuroimaging data [14–16, 19, 28, 29], it is less common to see these neuroimaging-based measures of brain aging being characterized in the context of biological aging. Cole and colleagues investigated the associations of the gap between chronological age and estimated brain age. They found that accelerated brain aging was associated with weaker grip strength, poorer lung function, slower walking speed, lower fluid intelligence, higher allostatic load, and increased mortality risk [30]. Belsky et al. devised a biomarker called “Pace of Aging” based on tracking declining function in 19 biomarkers indexing the cardiovascular, metabolic, renal, immune, dental, and pulmonary systems across ages 26, 32, 38, and 45 years [31]. In a more recent iteration of the original Pace of Aging biomarkers by Elliot et al. [32], participants aged 45 years with faster Pace of Aging had more cognitive difficulties and signs of accelerated brain aging, including significant reductions of cortical thickness, hippocampal volume, increased brain age, etc. In a follow-up report [33], they were able to produce evidence of accelerated brain aging preceded by age-related degradation of the body and associated with deficit accumulation accelerated by stressors in early life. A different approach based on a machine learning classifier has been used to create an index of advanced brain aging called BA-index [34]. This research group investigated associations of the index with AD risk factors, finding their index to be associated with smoking, anti-hypertensive medication use, and waist circumference for a male cohort after adjusting for age.
Here, we found the AD-PS scores to be strongly associated with all-cause mortality after adjusting for age, race, sex, smoking status, education, diabetes, and hypertension. The association held after the analyses were repeated using only cognitively normal individuals and modeling the CNS related causes of death as competing risk. Previously, several groups have investigated the potential of sMRI-based biomarkers to predict mortality. Kuller et al. reported white matter grade and ventricular volume to be predictors of death [35]. Henneman et al. used MRI scans from 1138 patients to generate visual rating scales for medial temporal lobe atrophy, global cortical atrophy, and white matter hyperintensities (WMH). Number of microbleeds and presence of infarcts were recorded. They found these MRI-derived measures to be predictors of mortality [36]. However, no attempt to link these measures to the biological aging framework was reported.
Our analyses using proteomics found significant associations between the AD-PS scores and 9 proteins from 32 that have consistently been linked to mammalian aging. The proteins with stronger associations with AD-PS scores (after correction for multiple comparisons) were GDF-15 and pleiotrophin. GDF-15 has been linked to regulation of obesity, cancer, nervous system disease, metabolism, and cardiovascular disease. It has been shown to be associated with mortality in different conditions [37–41]. Serum GDF-15 levels have been proposed as potential diagnostic markers for aging-related diseases, such as cognitive impairment, frailty, and cardiovascular disease, and more recently has been explored as therapeutic target [42–47]. Pleiotrophin is another protein that has been widely linked to aging [1]. Importantly, pleiotrophin is a secreted cell signaling cytokine that is involved in numerous biological pathways, including cell growth, differentiation, and proliferation. Pleiotrophin has both systemic (e.g., bone, vascular) and brain-related actions, including neuromodulation and regulation of neuroinflammation [40, 48–50]. Both proteins remained significantly associated with the AD-PS scores after correction for multiple comparisons and sub-setting the analyses to cognitively normal individuals. Several studies have investigated associations between MRI and proteomics [24, 51, 52] in the context of identifying biomarkers for AD and not aging. Our results do not necessarily rule out the relevance of the other 23 proteins in brain aging. We employed a conservative criteria to protect against Type I errors, and so false negative results are possible. Certainly, the findings reported here should be replicated, since some aspects of the patterns observed may reflect features unique to the population included in this analysis.
Finally, we found the AD-PS scores to be significantly correlated with the DAI independent of covariates. Kant and colleagues investigated associations between frailty, based on the Fried frailty phenotype, and MRI features of cerebral small vessel disease [53], finding that white matter hyperintensity volumes were associated with frailty, with similar finding reported by Siejka and colleagues who used frailty index in a cross-sectional study [54].
Overall, we have gathered evidence of convergent and predictive validity of the AD-PS scores as a brain-based measure of biologic aging based on three different and independent measures of aging selected a priori. While the AD-PS scores were created as a measure of AD risk by using a machine learning algorithm to discriminate between cognitively normal individuals and AD patients, our analyses suggest that they are also capturing brain aging. Franke and colleagues have reported that individuals with dementia and ADRDs have accelerated brain age [55]. Similarly, cognitive decline [33] and diagnoses known to impair cognitive ability and accelerate aging, including schizophrenia and Down syndrome [16], were reported to be associated to accelerated aging.
Our work is not without limitations: Participants in both ADNI and ARIC are older adults. In particular, ages in ARIC vary between 67 and 90. It is unclear how the AD-PS scores will perform for younger cohorts. The MRI participants were not selected randomly from all V5 participants; thus, caution should be taken in generalizing the result. Part of the analyses are cross-sectional, implying the possibility of reverse causation. The most important limitation is that there is no gold-standard measure of biologic aging and so we cannot say with confidence that an interpretation in this direction is appropriate. The ARIC cohort is basically composed of White and African American participants. No Hispanic, Asian, or Native American individuals were available for our study.
Our study also has important strengths. The ARIC cohort is deeply phenotyped with a wealth of biomedical information (imaging, omics, cognition, etc.) collected over decades. Also, it is a cohort racially diverse. We adjusted for multiple comparisons and used results from other studies and tested specific hypotheses based on these as opposed to a broad non-hypothesis-based approach. The AD-PS scores are based on T1 weighted images, one of the more common brain imaging modalities across studies. The fact that structural MRI is not specific to AD and that the AD-PS scores are based on global patterns detected by a machine learning algorithm increases their potential as a measure of brain aging.
The MRI sequence used to derive the AD-PS score is based on a sequence that is commonly used clinically. Thus, it is possible that with appropriate acquisition protocols and processing, this or similarly derived measures might be deployable to clinical settings more quickly than some other kinds of measures like blood-based measures of epigenetic modifications. But, this is just a first step in that direction and more work is needed to understand how brain age/aging is related to aging of the rest of the body.
Conclusion
We have investigated the potential of the AD-PS scores, an MRI-based measure of dementia risk, as a measure of accelerated aging. The AD-PS scores were strongly associated with three different types of biological aging measures: (1) all-cause mortality; (2) proteins previously linked to aging; and (3) accumulation of deficits which suggests that the AD-PS scores could be capturing patterns of brain aging.
Acknowledgements
The Atherosclerosis Risk in Communities Study is carried out as a collaborative study supported by National Heart, Lung, and Blood Institute contracts (HHSN268201700001I, HHSN268201700002I, HHSN268201700003I, HHSN268201700005I, HHSN268201700004I). Neurocognitive data is collected by U01 2U01HL096812, 2U01HL096814, 2U01HL096899, 2U01HL096902, and 2U01HL096917 from the NIH (NHLBI, NINDS, NIA, and NIDCD), and with previous brain MRI examinations funded by R01-HL70825 from the NHLBI. The authors thank the staff and participants of the ARIC study for their important contributions. SomaLogic Inc. conducted the SomaScan assays in exchange for use of ARIC data. This work was supported in part by NIH/NHLBI grant R01 HL134320. RC and TH receive funding from the Wake Forest Alzheimer’s Disease Core Center (P30AG049638-01A1). We thank the ARIC Neurocognitive Study and the grant P30 AG021332 for funding to develop these analyses. RC and TH receive funding from the P30AG072947. Data collection and sharing for this project were also funded by the Alzheimer’s Disease Neuroimaging Initiative (ADNI) (National Institutes of Health Grant U01 AG024904) and DOD ADNI (Department of Defense award number W81XWH-12-2-0012). ADNI is funded by the National Institute on Aging, the National Institute of Biomedical Imaging and Bioengineering, and through generous contributions from the following: AbbVie, Alzheimer’s Association; Alzheimer’s Drug Discovery Foundation; Araclon Biotech; BioClinica, Inc.; Biogen; Bristol-Myers Squibb Company; CereSpir, Inc.; Cogstate; Eisai Inc.; Elan Pharmaceuticals, Inc.; Eli Lilly and Company; EuroImmun; F. Hoffmann-La Roche Ltd and its affiliated company Genentech, Inc.; Fujirebio; GE Healthcare; IXICO Ltd.; Janssen Alzheimer Immunotherapy Research & Development, LLC.; Johnson & Johnson Pharmaceutical Research & Development LLC.; Lumosity; Lundbeck; Merck & Co., Inc.; Meso Scale Diagnostics, LLC.; NeuroRx Research; Neurotrack Technologies; Novartis Pharmaceuticals Corporation; Pfizer Inc.; Piramal Imaging; Servier; Takeda Pharmaceutical Company; and Transition Therapeutics. The Canadian Institutes of Health Research is providing funds to support ADNI clinical sites in Canada. Private sector contributions are facilitated by the Foundation for the National Institutes of Health (www.fnih.org). The grantee organization is the Northern California Institute for Research and Education, and the study is coordinated by the Alzheimer’s Therapeutic Research Institute at the University of Southern California. ADNI data are disseminated by the Laboratory for Neuro Imaging at the University of Southern California.
Declarations
Conflict of interest
The authors declare no competing interest.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Higgins-Chen AT, Thrush KL, Levine ME. Aging biomarkers and the brain. Semin Cell Dev Biol. 2021;116:180–193. doi: 10.1016/j.semcdb.2021.01.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Casanova R, Hsu FC, Sink KM, Rapp SR, Williamson JD, Resnick SM, Espeland MA, Alzheimer's Disease Neuroimaging I. Alzheimer’s disease risk assessment using large-scale machine learning methods. PLoS ONE. 2013;8:e77949. doi: 10.1371/journal.pone.0077949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Espeland MA, Chen JC, Weitlauf J, Hayden KM, Rapp SR, Resnick SM, Garcia L, Cannell B, Baker LD, Sachs BC, Tindle HA, Wallace R, Casanova R, Women's Health Initiative Memory Study Magnetic Resonance Imaging Study G Trajectories of relative performance with 2 measures of global cognitive function. J Am Geriatr Soc. 2018;66:1575–1580. doi: 10.1111/jgs.15431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Espeland MA, Luchsinger JA, Neiberg RH, Carmichael O, Laurienti PJ, Pi-Sunyer X, Wing RR, Cook D, Horton E, Casanova R, Erickson K, Nick Bryan R, Action for Health in Diabetes Brain Magnetic Resonance Imaging Research G Long term effect of intensive lifestyle intervention on cerebral blood flow. J Am Geriatr Soc. 2018;66:120–126. doi: 10.1111/jgs.15159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Younan D, Petkus AJ, Widaman KF, Wang X, Casanova R, Espeland MA, Gatz M, Henderson VW, Manson JE, Rapp SR, Sachs BC, Serre ML, Gaussoin SA, Barnard R, Saldana S, Vizuete W, Beavers DP, Salinas JA, Chui HC, Resnick SM, Shumaker SA, Chen JC. Particulate matter and episodic memory decline mediated by early neuroanatomic biomarkers of Alzheimer's disease. Brain. 2020;143:289–302. doi: 10.1093/brain/awz348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Younan D, Wang X, Casanova R, Barnard R, Gaussoin SA, Saldana S, Petkus AJ, Beavers DP, Resnick SM, Manson JE, Serre ML, Vizuete W, Henderson VW, Sachs BC, Salinas JA, Gatz M, Espeland MA, Chui HC, Shumaker SA, Rapp SR, Chen JC PM2.5 associated with gray matter atrophy reflecting increased Alzheimers risk in older women. Neurology. 2020. [DOI] [PMC free article] [PubMed]
- 7.Casanova R, Hsu FC, Barnard RT, Anderson AM, Talluri R, Whitlow CT, Hughes TM, Griswold M, Hayden KM, Gottesman RF, Wagenknecht LE. Comparing data-driven and hypothesis-driven MRI-based predictors of cognitive impairment in individuals from the Atherosclerosis Risk in Communities (ARIC) study. Alzheimers Dement. 2022;18:561–571. doi: 10.1002/alz.12427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kennedy BK, Berger SL, Brunet A, Campisi J, Cuervo AM, Epel ES, Franceschi C, Lithgow GJ, Morimoto RI, Pessin JE, Rando TA, Richardson A, Schadt EE, Wyss-Coray T, Sierra F. Geroscience: linking aging to chronic disease. Cell. 2014;159:709–713. doi: 10.1016/j.cell.2014.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kohanski RA, Deeks SG, Gravekamp C, Halter JB, High K, Hurria A, Fuldner R, Green P, Huebner R, Macchiarini F, Sierra F. Reverse geroscience: how does exposure to early diseases accelerate the age-related decline in health? Ann N Y Acad Sci. 2016;1386:30–44. doi: 10.1111/nyas.13297. [DOI] [PubMed] [Google Scholar]
- 10.Justice JN, Ferrucci L, Newman AB, Aroda VR, Bahnson JL, Divers J, Espeland MA, Marcovina S, Pollak MN, Kritchevsky SB, Barzilai N, Kuchel GA. A framework for selection of blood-based biomarkers for geroscience-guided clinical trials: report from the TAME Biomarkers Workgroup. Geroscience. 2018;40:419–436. doi: 10.1007/s11357-018-0042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.LeBrasseur NK, de Cabo R, Fielding R, Ferrucci L, Rodriguez-Manas L, Vina J, Vellas B. Identifying biomarkers for biological age: geroscience and the ICFSR task force. J Frailty Aging. 2021;10:196–201. doi: 10.14283/jfa.2021.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hoffman JM, Hernandez CM, Hernandez AR, Bizon JL, Burke SN, Carter CS, Buford TW. Bridging the gap: a geroscience primer for neuroscientists with potential collaborative applications. J Gerontol A Biol Sci Med Sci. 2021. [DOI] [PMC free article] [PubMed]
- 13.Hernandez CM, Hernandez AR, Hoffman JM, King PH, McMahon LL, Buford TW, Carter C, Bizon JL, Burke SN. A neuroscience primer for integrating geroscience with the neurobiology of aging. J Gerontol A Biol Sci Med Sci. 2021. [DOI] [PMC free article] [PubMed]
- 14.Franke K, Gaser C. Ten years of BrainAGE as a neuroimaging biomarker of brain aging: what insights have we gained? Front Neurol. 2019;10:789. doi: 10.3389/fneur.2019.00789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cole JH. Neuroimaging-derived brain-age: an ageing biomarker? Aging (Albany NY) 2017;9:1861–1862. doi: 10.18632/aging.101286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cole JH, Franke K. Predicting age using neuroimaging: innovative brain ageing biomarkers. Trends Neurosci. 2017;40:681–690. doi: 10.1016/j.tins.2017.10.001. [DOI] [PubMed] [Google Scholar]
- 17.Ning K, Zhao L, Matloff W, Sun F, Toga AW. Association of relative brain age with tobacco smoking, alcohol consumption, and genetic variants. Sci Rep. 2020;10:10. doi: 10.1038/s41598-019-56089-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gaser C, Franke K, Kloppel S, Koutsouleris N, Sauer H. BrainAGE in mild cognitive impaired patients: predicting the conversion to Alzheimer’s disease. PLoS ONE. 2013;8:e67346. doi: 10.1371/journal.pone.0067346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gaser C, Franke K, Kloppel S, Koutsouleris N, Sauer H, Alzheimer's Disease Neuroimaging I. BrainAGE in mild cognitive impaired patients: predicting the conversion to Alzheimer’s disease. PLoS ONE. 2013;8:e67346. doi: 10.1371/journal.pone.0067346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Johnson AA, Shokhirev MN, Wyss-Coray T, Lehallier B. Systematic review and analysis of human proteomics aging studies unveils a novel proteomic aging clock and identifies key processes that change with age. Ageing Res Rev. 2020;60:101070. doi: 10.1016/j.arr.2020.101070. [DOI] [PubMed] [Google Scholar]
- 21.Mitnitski AB, Mogilner AJ, Rockwood K. Accumulation of deficits as a proxy measure of aging. ScientificWorldJournal. 2001;1:323–336. doi: 10.1100/tsw.2001.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Knopman DS, Gottesman RF, Sharrett AR, Wruck LM, Windham BG, Coker L, Schneider AL, Hengrui S, Alonso A, Coresh J, Albert MS, Mosley TH., Jr Mild cognitive impairment and dementia prevalence: the Atherosclerosis Risk in Communities Neurocognitive Study (ARIC-NCS) Alzheimers Dement (Amst) 2016;2:1–11. doi: 10.1016/j.dadm.2015.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Casanova R, Barnard RT, Gaussoin SA, Saldana S, Hayden KM, Manson JE, Wallace RB, Rapp SR, Resnick SM, Espeland MA, Chen JC, Group W-MS, the Alzheimer's disease Neuroimaging I Using high-dimensional machine learning methods to estimate an anatomical risk factor for Alzheimer’s disease across imaging databases. Neuroimage. 2018;183:401–411. doi: 10.1016/j.neuroimage.2018.08.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Walker KA, Chen J, Zhang J, Fornage M, Yang Y, Zhou L, Grams ME, Tin A, Daya N, Hoogeveen RC, Aozhou Wu, Sullivan KJ, Ganz P, Zeger SL, Gudmundsson EF, Emilsson V, Launer LJ, Jennings LL, Gudnason V, Chatterjee N, Gottesman RF, Mosley TH, Boerwinkle E, Ballantyne CM, Coresh J. Large-scale plasma proteomic analysis identifies proteins and pathways associated with dementia risk. Nature Aging. 2021;1:473–489. doi: 10.1038/s43587-021-00064-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Casanova R, Hsu FC, Espeland MA. Classification of structural MRI images in Alzheimer’s disease from the perspective of ill-posed problems. PLoS ONE. 2012;7:e44877. doi: 10.1371/journal.pone.0044877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Casanova R, Maldjian JA, Espeland MA. Evaluating the impact of different factors on voxel-wise classification methods of ADNI structural MRI brain images. International Journal of Biomedical Datamining. 2011;1:11. [Google Scholar]
- 27.Fine JP, Gray RJ. A proportional hazards model for the subdistribution of a competing risk. J Am Stat Assoc. 1999;94:496–509. [Google Scholar]
- 28.Franke K, Gaser C, Manor B, Novak V. Advanced BrainAGE in older adults with type 2 diabetes mellitus. Front Aging Neurosci. 2013;5:90. doi: 10.3389/fnagi.2013.00090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cole JH, Poudel RPK, Tsagkrasoulis D, Caan MWA, Steves C, Spector TD, Montana G. Predicting brain age with deep learning from raw imaging data results in a reliable and heritable biomarker. Neuroimage. 2017;163:115–124. doi: 10.1016/j.neuroimage.2017.07.059. [DOI] [PubMed] [Google Scholar]
- 30.Cole JH, Ritchie SJ, Bastin ME, Valdes Hernandez MC, Munoz Maniega S, Royle N, Corley J, Pattie A, Harris SE, Zhang Q, Wray NR, Redmond P, Marioni RE, Starr JM, Cox SR, Wardlaw JM, Sharp DJ, Deary IJ. Brain age predicts mortality. Mol Psychiatry. 2018;23:1385–1392. doi: 10.1038/mp.2017.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Belsky DW, Caspi A, Houts R, Cohen HJ, Corcoran DL, Danese A, Harrington H, Israel S, Levine ME, Schaefer JD, Sugden K, Williams B, Yashin AI, Poulton R, Moffitt TE. Quantification of biological aging in young adults. Proc Natl Acad Sci U S A. 2015;112:E4104–4110. doi: 10.1073/pnas.1506264112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Elliott ML, Caspi A, Houts RM, Ambler A, Broadbent JM, Hancox RJ, Harrington H, Hogan S, Keenan R, Knodt A, Leung JH, Melzer TR, Purdy SC, Ramrakha S, Richmond-Rakerd LS, Righarts A, Sugden K, Thomson WM, Thorne PR, Williams BS, Wilson G, Hariri AR, Poulton R, Moffitt TE. Disparities in the pace of biological aging among midlife adults of the same chronological age have implications for future frailty risk and policy. Nat Aging. 2021;1:295–308. doi: 10.1038/s43587-021-00044-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elliott ML, Belsky DW, Knodt AR, Ireland D, Melzer TR, Poulton R, Ramrakha S, Caspi A, Moffitt TE, Hariri AR. Brain-age in midlife is associated with accelerated biological aging and cognitive decline in a longitudinal birth cohort. Mol Psychiatry. 2021;26:3829–3838. doi: 10.1038/s41380-019-0626-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Habes M, Janowitz D, Erus G, Toledo JB, Resnick SM, Doshi J, Van der Auwera S, Wittfeld K, Hegenscheid K, Hosten N, Biffar R, Homuth G, Volzke H, Grabe HJ, Hoffmann W, Davatzikos C. Advanced brain aging: relationship with epidemiologic and genetic risk factors, and overlap with Alzheimer disease atrophy patterns. Transl Psychiatry. 2016;6:e775. doi: 10.1038/tp.2016.39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kuller LH, Arnold AM, Longstreth WT, Jr, Manolio TA, O'Leary DH, Burke GL, Fried LP, Newman AB. White matter grade and ventricular volume on brain MRI as markers of longevity in the cardiovascular health study. Neurobiol Aging. 2007;28:1307–1315. doi: 10.1016/j.neurobiolaging.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 36.Henneman WJ, Sluimer JD, Cordonnier C, Baak MM, Scheltens P, Barkhof F, van der Flier WM. MRI biomarkers of vascular damage and atrophy predicting mortality in a memory clinic population. Stroke. 2009;40:492–498. doi: 10.1161/STROKEAHA.108.516286. [DOI] [PubMed] [Google Scholar]
- 37.Doerstling S, Hedberg P, Ohrvik J, Leppert J, Henriksen E. Growth differentiation factor 15 in a community-based sample: age-dependent reference limits and prognostic impact. Ups J Med Sci. 2018;123:86–93. doi: 10.1080/03009734.2018.1460427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim JH, Jeon Y, Ahn JS, Kim S, Kim DK, Lee JP, Ryu DR, Seong EY, Ahn SY, Baek SH, Jung HY, Choi JY, Park SH, Kim CD, Kim YL, Cho JH. GDF-15 predicts in-hospital mortality of critically ill patients with acute kidney injury requiring continuous renal replacement therapy: a multicenter prospective study. J Clin Med 10. 2021 [DOI] [PMC free article] [PubMed]
- 39.Meyer SL, Wolff D, Ridderbos FS, Eshuis G, Hillege H, Willems TP, Ebels T, van Melle JP, Berger RMF. GDF-15 (growth differentiation factor 15) is associated with hospitalization and mortality in patients with a fontan circulation. J Am Heart Assoc. 2020;9:e015521. doi: 10.1161/JAHA.119.015521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Sathyan S, Ayers E, Gao T, Weiss EF, Milman S, Verghese J, Barzilai N. Plasma proteomic profile of age, health span, and all-cause mortality in older adults. Aging Cell. 2020;19:e13250. doi: 10.1111/acel.13250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Breniere C, Meloux A, Pedard M, Marie C, Thouant P, Vergely C, Bejot Y. Growth differentiation factor-15 (GDF-15) is associated with mortality in ischemic stroke patients treated with acute revascularization therapy. Front Neurol. 2019;10:611. doi: 10.3389/fneur.2019.00611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chai YL, Hilal S, Chong JPC, Ng YX, Liew OW, Xu X, Ikram MK, Venketasubramanian N, Richards AM, Lai MKP, Chen CP. Growth differentiation factor-15 and white matter hyperintensities in cognitive impairment and dementia. Medicine (Baltimore) 2016;95:e4566. doi: 10.1097/MD.0000000000004566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.He L, Barreto PS, Sanchez Sanchez JL, Rolland Y, Guyonnet S, Parini A, Lucas A, Vellas B, Group MD. Prospective associations of plasma growth differentiation factor 15 with physical performance and cognitive functions in older adults. J Gerontol A Biol Sci Med Sci. 2022. [DOI] [PubMed]
- 44.Alcazar J, Frandsen U, Prokhorova T, Kamper RS, Haddock B, Aagaard P, Suetta C. Changes in systemic GDF15 across the adult lifespan and their impact on maximal muscle power: the Copenhagen Sarcopenia Study. J Cachexia Sarcopenia Muscle. 2021;12:1418–1427. doi: 10.1002/jcsm.12823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sarkar S, Melchior JT, Henry HR, Syed F, Mirmira RG, Nakayasu ES, Metz TO. GDF15: a potential therapeutic target for type 1 diabetes. Expert Opin Ther Targets. 2022;26:57–67. doi: 10.1080/14728222.2022.2029410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchis J, Ruiz V, Bonanad C, Sastre C, Ruescas A, Diaz M, Rodriguez E, Valero E, Garcia-Blas S, Carratala A, Nunez E, Nunez J. Growth differentiation factor 15 and geriatric conditions in acute coronary syndrome. Int J Cardiol. 2019;290:15–20. doi: 10.1016/j.ijcard.2019.05.034. [DOI] [PubMed] [Google Scholar]
- 47.Wischhusen J, Melero I, Fridman WH. Growth/differentiation factor-15 (GDF-15): from biomarker to novel targetable immune checkpoint. Front Immunol. 2020;11:951. doi: 10.3389/fimmu.2020.00951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gonzalez-Castillo C, Ortuno-Sahagun D, Guzman-Brambila C, Pallas M, Rojas-Mayorquin AE. Pleiotrophin as a central nervous system neuromodulator, evidences from the hippocampus. Front Cell Neurosci. 2014;8:443. doi: 10.3389/fncel.2014.00443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamprou M, Kaspiris A, Panagiotopoulos E, Giannoudis PV, Papadimitriou E. The role of pleiotrophin in bone repair. Injury. 2014;45:1816–1823. doi: 10.1016/j.injury.2014.10.013. [DOI] [PubMed] [Google Scholar]
- 50.Fernandez-Calle R, Vicente-Rodriguez M, Gramage E, Pita J, Perez-Garcia C, Ferrer-Alcon M, Uribarri M, Ramos MP, Herradon G. Pleiotrophin regulates microglia-mediated neuroinflammation. J Neuroinflammation. 2017;14:46. doi: 10.1186/s12974-017-0823-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toledo JB, Da X, Bhatt P, Wolk DA, Arnold SE, Shaw LM, Trojanowski JQ, Davatzikos C, Alzheimer's Disease Neuroimaging I. Relationship between plasma analytes and SPARE-AD defined brain atrophy patterns in ADNI. PLoS ONE. 2013;8:e55531. doi: 10.1371/journal.pone.0055531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nazeri A, Ganjgahi H, Roostaei T, Nichols T, Zarei M, Alzheimer's Disease Neuroimaging I. Imaging proteomics for diagnosis, monitoring and prediction of Alzheimer’s disease. Neuroimage. 2014;102(Pt 2):657–665. doi: 10.1016/j.neuroimage.2014.08.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kant IMJ, Mutsaerts H, van Montfort SJT, Jaarsma-Coes MG, Witkamp TD, Winterer G, Spies CD, Hendrikse J, Slooter AJC, de Bresser J. The association between frailty and MRI features of cerebral small vessel disease. Sci Rep. 2019;9:11343. doi: 10.1038/s41598-019-47731-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Siejka TP, Srikanth VK, Hubbard RE, Moran C, Beare R, Wood A, Phan T, Callisaya ML. Frailty and cerebral small vessel disease: a cross-sectional analysis of the Tasmanian Study of Cognition and Gait (TASCOG) J Gerontol A Biol Sci Med Sci. 2018;73:255–260. doi: 10.1093/gerona/glx145. [DOI] [PubMed] [Google Scholar]
- 55.Franke K, Gaser C. Longitudinal changes in individual BrainAGE in healthy aging, mild cognitive impairment, and Alzheimer’s disease. GeroPsych J Gerontopsychology Geriatr Psychiatry. 2012;25:235–245. [Google Scholar]