Abstract
Self-assembly of macromolecules into higher-order symmetric structures is fundamental for the regulation of biological processes. Higher-order symmetric structure self-assembly by the gene expression machinery, such as bacterial DNA-dependent RNA polymerase (RNAP), has never been reported before. Here, we show that the stress-response σB factor from the human pathogen, Mycobacterium tuberculosis, induces the RNAP holoenzyme oligomerization into a supramolecular complex composed of eight RNAP units. Cryo-electron microscopy revealed a pseudo-symmetric structure of the RNAP octamer in which RNAP protomers are captured in an auto-inhibited state and display an open-clamp conformation. The structure shows that σB is sequestered by the RNAP flap and clamp domains. The transcriptional activator RbpA prevented octamer formation by promoting the initiation-competent RNAP conformation. Our results reveal that a non-conserved region of σ is an allosteric controller of transcription initiation and demonstrate how basal transcription factors can regulate gene expression by modulating the RNAP holoenzyme assembly and hibernation.
Subject terms: Cryoelectron microscopy, Pathogens, Enzyme mechanisms, Molecular biophysics
Biological processes can be regulated via oligomerization of macromolecules into high-order symmetric structures. Here, authors reported high-order structure of RNA polymerase and its role in regulation of gene expression in pathogenic bacterium.
Introduction
Transcription is the first and most highly regulated step in gene expression. In bacteria, transcription is carried out by the RNA polymerase (RNAP) core (E) composed of five subunits (α2ββ‘ω)1. To initiate transcription, the core associates with the promoter-specific initiation factor σ (the σ subunit) to form the RNAP holoenzyme (Eσ)2. Upon holoenzyme assembly, the σ subunit domain 2 (σ2) binds to the β‘ subunit coiled-coil region (β‘-CC) at the RNAP clamp domain while domain 4 (σ4) binds to the β subunit flap (β flap). The σ subunit region 3.2, which connects domains σ2 and σ4 (Fig. 1a), enters deeply into the RNAP active site cleft and RNA exit channel3–5. In the holoenzyme, σ adopts a conformation optimal for recognition of −10 and −35 promoter elements by σ2 and σ4, respectively6,7. However, the molecular mechanisms of σ loading onto RNAP remain obscure. To initiate RNA synthesis, RNAP melts ~13 bp of promoter DNA and forms the transcriptionally competent open promoter complex (RPo). During transcription initiation, the RNAP clamp successively adopts different states, from open (free RNAP) to closed (RPo and transcription elongation complex)8–13. The open-clamp state allows the DNA template entry into the active site, and the closed state is required to hold the DNA template in the active site. Thus, the clamp conformational dynamics drive promoter recognition and melting during transcription initiation10,12. The σ2 and σ4 domains impose restraints on the clamp and β flap relative movements and consequently, should affect the basic RNAP functions in a promoter-dependent manner. Lineage-specific σ factors create gene regulatory networks that ensure the rapid adaptation of bacteria to environmental stress and allow pathogenic bacteria to tolerate antibiotic treatment14,15. For instance, the dormant form of M. tuberculosis (Mtb), the origin of latent tuberculosis, can persist in tissues for decades16. Most Mtb genes are transcribed by RNAP harboring the principal σA subunit and the principal-like σB subunit15 that belong to group I and group II, respectively, and share almost identical promoter binding regions17. It is thought that the σB subunit is responsible for gene transcription during starvation and stress18–20. However, recent findings showed that the mycobacterial σA and σB subunits are present at similar levels and co-transcribe essential genes during exponential growth15. Two general transcription factors, CarD and RbpA, regulate Mtb RNAP activity in the growth phase and in a gene-specific manner21–23. Unlike EσA, the activity of which displays loose dependence on RbpA, EσB is deficient in promoter-dependent transcription initiation in the absence of RbpA17,24. Single-molecule Förster resonance energy transfer (smFRET) analysis showed that RbpA induces the σB conformational change required for the correct assembly of active EσB6. Several solution structures of the Mtb EσΑ holoenzyme and its complexes with CarD, RbpA, and promoter DNA have been solved by cryogenic electron microscopy (cryo-EM)25–27, and have provided the structural basis for understanding σΑ-dependent transcription initiation. However, the lack of EσΒ structure does not allow dissecting the specific roles of the σA and σB subunits in gene regulation. Here, we use single-particle cryo-EM to determine the structural basis of the intrinsically limited EσB transcriptional activity. We find that after σB association with the RNAP core, the EσB holoenzyme remains trapped in an immature conformation in which the σB C-terminus is unloaded from the RNA exit channel. The immature EσB, deficient in promoter recognition, self-assembles into a 3.2 MDa, octamer the size of which exceeds that of the bacterial ribosome. Thus, σB acts as a bona fide RNAP-hibernation factor that can repress transcription.
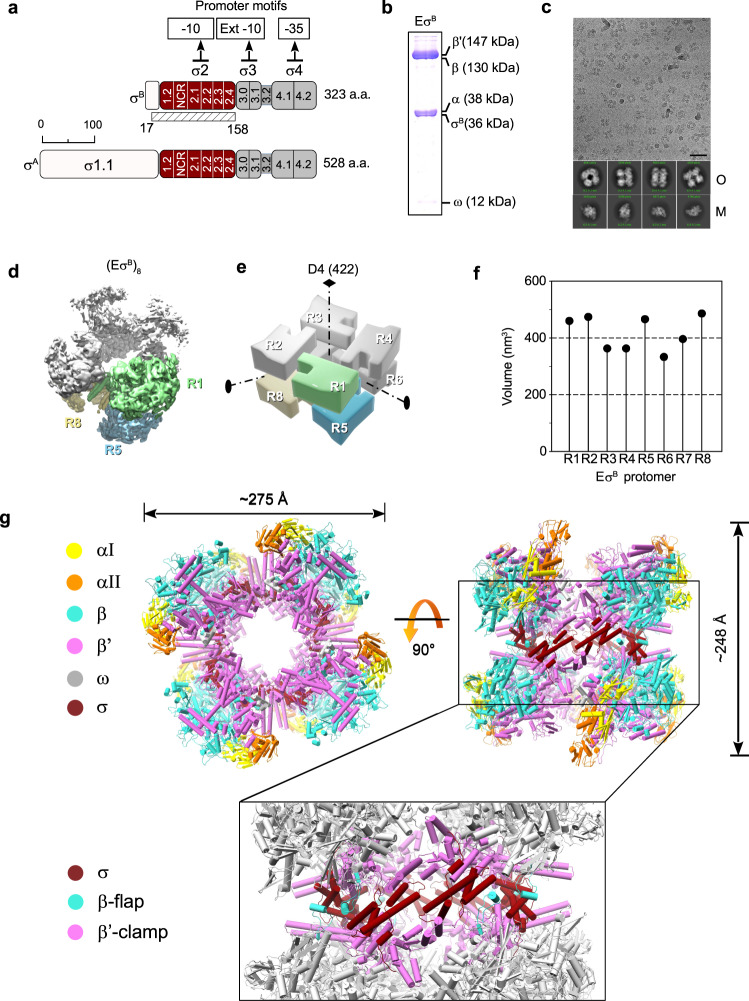
Fig. 1. Cryo-EM structure of the M. tuberculosis EσB octamer.
a Schematic representation of the M. tuberculosis σA and σB subunits with the structural domains σ1.1 in white, σ2 in dark red, σ3 and σ4 in gray. The subregions inside the σ domains are numbered. NCR, non-conserved region. The solved part of σB (residues 17–158) is indicated by a hatched rectangle. b 14% SDS-PAGE of the EσB sample for cryo-EM. c Representative cryo-EM image of the EσB sample. Experiment was repeated independently four times. Scale bar = 50 nm. Bottom, representative 2D class averages of monomers (M) and oligomers (O). d Cryo-EM map of the octamer (EσB)8 refined without imposing symmetry (C1 -map). RNAP protomers with well-defined density are in light green (protomer R1), sky blue (protomer R5), and khaki (protomer R8). The other protomers are in gray. e. 3D-model of the D4 symmetric octamer with the protomers numbered R1 to R8 and the symmetry axes indicated. Color codes are as in d. f Scatter chart shows volumes of the individual protomers in C1-map calculated in UCSF Chimera. g Molecular model of the octamer (EσB)8. Views from the top (protomers R1, R2, R3, R4) and from the side (protomers R2, R3, R7, R8) with the RNAP subunits color-coded as indicated on the left: yellow and orange α, cyan β, magenta β‘, dark red σB. The boxed region shows the junction between the top and bottom rings of RNAP tetramers (EσB)4. Domains holding the RNAP protomers together: β flap (aa 808–832, cyan), β‘ clamp (aa 1–413, magenta) and (dark red). The other regions are in gray. Source data are provided as a Source Data file.
Results
Cryo-EM structure of the M. tuberculosis EσB octamer
For the structural analysis, we first assembled the EσB holoenzyme from the separately purified σB subunit and Mtb RNAP core (Fig. 1b). Analysis of the cryo-EM images revealed two particle populations: RNAP monomers and O-shaped RNAP oligomers (Fig. 1c). We determined the RNAP monomer structure at a nominal resolution of 4.1 Å (Supplementary Figs 1, 2 and Supplementary Table 1). In the cryo-EM map of the monomer, the σB subunit density was absent. Thus, we concluded that monomer particles comprised mainly the RNAP core. Overall, the structure was similar to the published structure of the Mycobacterium smegmatis RNAP core (conformation 2)28 with a ~5 Å bigger distance between the RNAP β lobe 1 (β-L275) and β‘ clamp (β‘-R214) domains. We then refined the RNAP oligomer cryo-EM map without imposing symmetry at a nominal resolution of 6.13 Å (Fig. 1d and Supplementary Fig. 3, C1 -map). Analysis of the C1-map showed that the RNAP-oligomer was formed by eight EσB units assembled into an octamer that exhibited a pseudo dihedral (D4) symmetry (Fig. 1e). For further referencing, we named the RNAP protomers in the C1-map R1 to R8 (Fig. 1d, e). Overall, the C1 -map was non-uniform: the density of the RNAP subunits was well-defined in protomers R1, R2, R5, and R8, (volume of protomers >400 nm3, Fig. 1f) whereas it was incomplete in protomers R3, R4, R6 and R7 (volume of protomers <400 nm3, Fig. 1f). The heterogeneous ab-initio reconstruction with five classes showed that oligomer particles were a mixture of RNAP tetramers that comprised protomers R1, R2, R5 and R8 (~20%) and of RNAP octamers (~80%) (Supplementary Figs 3 and 5a). Refinement of the octamer with the applied D4 symmetry resulted in an improved D4-map at a nominal resolution of 4.39 Å (Supplementary Figs 3, 4a and Supplementary Table 1).
The EσB octamer structure was formed by two stacked rings, related by twofold symmetry. Each ring was made of four EσB units, related by fourfold symmetry, in head-to-tail orientation (Fig. 1e, g). The head included the RNAP pincers, formed by β and β‘ subunits (Fig. 1g, in cyan and pink respectively), and the tail comprised two α subunits (Fig. 1g, in orange/yellow). The ring diameter was ~275 Å and the stack height was ~248 Å. The junction between rings was formed by the σΒ subunit domain 2 (, aa 23-158) that interacted with the β‘ clamp (aa 1–413) and β flap (aa 808–832) (Fig. 1g, boxed region). The total buried surface area between in the R1 protomer and all other chains in the R5 and R8 promoters was ~1200 A2, which is similar to that between and the β‘ subunit in the RNAP holoenzyme (~1300 A2). The β clamp contacts were formed by the Actinobacteria-specific insertion in the β‘ subunit (β‘i1, aa 141–230) and invariant residues of the β‘ Zn2+ binding domain (β‘ZBD, aa 60-81). Local resolution calculations showed that the σ2-β‘ clamp module was determined at a resolution between 3.6 and 5.5 Å while the remaining RNAP parts were determined at a resolution between 5 and 10 Å (Supplementary Fig. 4a). Thus we concluded that the σ2/β‘ clamp junction between rings forms a rigid scaffold to hold the whole complex together. The 3D variability analysis29 demonstrated that the RNAP protomers underwent concerted movements relative to the scaffold, which explained the low resolution in the peripheral zones of the EσB octamer (Supplementary Fig. 5 and Supplementary Movie 1). The N-terminal part of the σB subunit, (i.e., domain (Fig. 1a) that comprises residues 17 to 158) was well resolved. Little variation in the density volume between the eight protomers in C1 -map suggested that majority of the octamer molecules contain eight copies of the σΒ subunit (Supplementary Fig. 5d, e). The C-terminal part of σB (i.e., the σ3 and σ4 domains) was poorly delimited, possibly due to its high mobility.
Cryo-EM structure of the EσB protomers
To better characterize the EσB protomer structure, we performed local refinement of the octamer C1 -map with masked R1 and R5 protomers which displayed better defined density for σB (Supplementary Fig. 3). We determined the cryo-EM maps of both protomers at a nominal resolution of 3.8 Å (Fig. 2a, Supplementary Fig. 4b and Supplementary Table 1). Although the cryo-EM map of R1 displayed better resolved electron density for than that of R5, in both maps the σΒ C-terminal domain density was absent. Local resolution calculations of the R1 EσB map showed that the central part of the Mtb RNAP core was determined at a resolution between 3.4 and 3.7 Å and displayed well-defined structural elements (Fig. 2, Supplementary Figs 4b and 6). Domain was determined at resolution between 3.8 and 5 Å. The mobile/flexible peripheral domains: β‘jaw, β lobes, and β‘i1 were determined at a resolution of 4–6 Å.
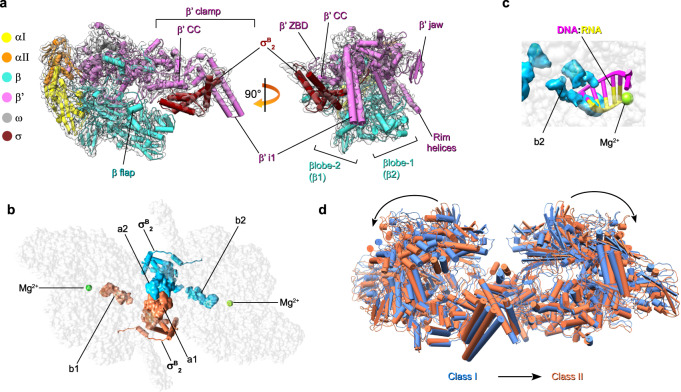
Fig. 2. Cryo-EM structures of the M. tuberculosis EσB protomer and dimer (EσB)2.
a Cryo-EM map and fitted molecular model of EσB (protomer R1): view from the RNA exit channel (left) and from the main channel (right). The RNAP subunits are colored as indicated on the left. The key structural RNAP modules68 are indicated. b Location of σB in the structure of the R1-R5 dimer (EσB)2. The RNAP core is shown as a gray semi-transparent molecular surface. The σB subunits assigned to different protomers are colored in deep sky blue and coral. The solved domain is shown as a cartoon with cylindrical helices. The unmodeled C-terminal segments of σB (i.e. a1, a2, b1, b2) are shown as molecular surfaces. Green spheres indicate catalytic Mg2+ ions. c Steric clash between the C-terminal segment of σB and the RNA:DNA hybrid in the active site of RNAP from the R1-R5 dimer. The 5-nt nascent RNA (yellow) and 8-nt template DNA (pink) (from PDB ID 6KON) are shown as cartoons. The b2 density of σB is shown as a molecular surface colored sky blue. d Superposition of class I (blue) and class II (coral) R1-R5 dimers.
To improve the EσB protomer resolution by including structural information from all eight protomers that constitute EσB octamer, we tested an alternative reconstruction method based on the symmetry-expansion procedure30,31. Briefly, each particle image used to refine the EσB octamer map was replicated and 3D-rotated according to the D4 point group symmetry. Then, we performed alternate cycles of asymmetric 3D classification and local refinement focused on a single EσB protomer. Two selected 3D classes with slightly different (11°) aperture of the β‘ clamp were refined at 3.9 and 4.19 Å resolution respectively (Supplementary Fig. 7). However, we did not observe any improvement in the resolution of the σΒ subunit density map.
Cryo-EM structure of the EσB dimer
To better determine the structure of the interactions holding together the EσB protomers, we performed local refinement of the octamer C1-map with the masked R1-R5 EσB dimer that displayed better-delimited cryo-EM density for the σΒ C-terminal segment (σΒ CTS) (Fig. 2b and Supplementary Fig. 3). We determined the EσB dimer structure at a nominal resolution of 4.36 Å (Supplementary Fig. 4c and Supplementary Table 1). In the RNAP dimer, the σΒ C-terminal domains of the two EσB protomers were stacked together and comprised three disconnected densities, determined at a resolution between 5.5 and 8 Å (Fig. 2b). To improve the σΒ C-terminal segment resolution, we performed a 3D classification using the R1-R5 EσB dimer map as reference. This gave two classes: class 1 (58% of particles; nominal resolution: 4.38 Å) with well-defined density of the σΒ C-terminal segment (resolution between 6.5 and 8 Å), and class 2 (42% of particles; nominal resolution: 6.75 Å) that lacked density of the σΒ C-terminal segment (Supplementary Figs 3, 8 and Supplementary Table 1). The relative orientation and conformation of the RNAP protomers in class 1 were the same as in the reference EσB dimer model. Conversely, the relative orientation of RNAP protomers in class 2 was different. In agreement with the 3D variability analysis of the RNAP octamer, the β lobes and β flaps of the RNAP protomers from class 1 were positioned closer to each other than those in class 2, and thus restrained the movements of the σΒ C-terminal domains (Fig. 2d, Supplementary Fig. 5c and Supplementary Movie 1). Structure analysis showed that the central density of the σΒ C-terminal segment dimer was formed by two copies of tangled polypeptide chains (a1 in R1 and a2 in R5), stacked between the β lobes and β flaps of the neighboring RNAPs (Fig. 2b and Supplementary Fig. 9a, b). Two peripheral densities (b1 in R1 and b2 in R5) were buried deeply in the active site cleft of RNAP and occluded the RNA:DNA hybrid path (Fig. 2b, c). Potentially, the a1-a2 density can comprise domain σΒ4 or domain σΒ3.1. These two domains contact the β flap and β lobe-2, respectively, in the mature form of the RNAP holoenzyme. However, σΒ4 interaction with the β flap is incompatible with the dimer model because the σΒ4 residues 242–260 and 312–323 should clash with the σB region 2 (Supplementary Fig. 9c). Consequently, the atomic model of the σΒ4 dimer could not be fitted into the cryo-EM density without significant rearrangements of the σΒ4 structure. Conversely, the atomic model of the σB3.1 dimer perfectly fitted into the density without significant rearrangements (Supplementary Fig. 9b). However, at the current resolution, we could not unambiguously assign the a1–a2 density to σB3.1.
RNAP conformational flexibility: movements of the β flap
The β flap domain provides an anchoring point for σ4 and paves the RNA exit channel. In the transcriptionally competent RNAP holoenzyme, the β flap tip faced the β‘ dock (β‘a11, aa 440–495) of the RNA exit channel (open flap) and positioned σ4 relative to σ2 at the optimal distance for promoter −10/−35 element recognition (i.e., 60–64 Å in Mtb EσA) (Table 1). In EσB, the β flap tip was turned 111° towards the and β‘ CC that is equal to its 27 Å displacement (closed flap state) (Fig. 3a). The distance between β flap and β‘ CC decreased to 46 Å (Table 1). Thus, σB4 could not be correctly positioned for the −35 element promoter binding. Similarly, Thermus thermophilus EσA in complex with the bacteriophage protein gp39 (inhibited state) displays a closed β flap conformation (rotation angle 80°)32 (Fig. 3a) as well as the terminating elongation complex33. In the RNAP octamer, the β flap was captured in the closed conformation by interactions with the σB subunit subregion 2.3 and σΒ C-terminus of the neighboring protomer (Supplementary Fig. 8a). The β flap density was absent in our Mtb RNAP core structure (Supplementary Fig. 2) and in the published cryo-EM structures of M. smegmatis RNAP core and holoenzyme28. These observations support the notion that the β flap is flexible and can adopt closed/open states if not bound to any partner34. Altogether, these results suggest that the β flap oscillation between two utmost conformations is a target for positive (RbpA) and negative (gp39, ρ termination) regulation of transcription.
Table 1.
Conformational changes in RNAP (distances between Cα atoms in Å)
| Structure | βflap (L820)/β‘CC (L360) | βlobe (T406)/β‘CC (L360) | β1 (G284)/β‘clamp (K123) | PDB ID |
|---|---|---|---|---|
| MtbEσB | 46 | 49 | 33 | This study |
| Msmeg EσA | UNK | 45 | 25 | 6EYD |
| Mtb RbpA/EσA | 64 | 46 | 26 | 6C05 |
| Mtb EσA- Fdx | 60 | 53 | 29 | 6FBV |
| Mtb RbpA/σA-RPo | 63 | 36 | 16 | 6C04 |
Mtb Mycobacterium tuberculosis, Msmeg Mycobacterium smegmatis.
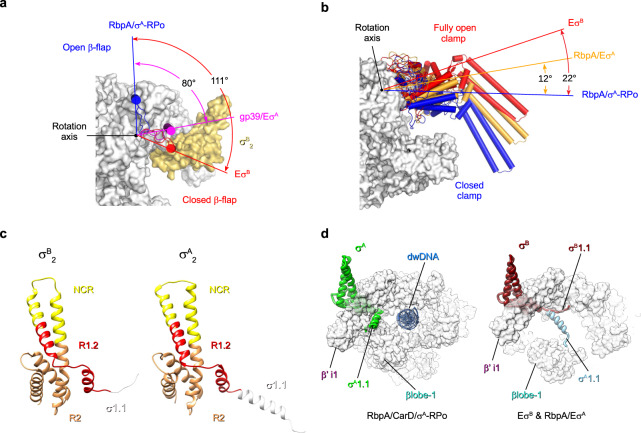
Fig. 3. Conformational flexibility of M. tuberculosis RNAP.
Motions of the RNAP flap (a) and clamp (b). RNAP is shown as a molecular surface with the core subunits in gray and σB in yellow. The flap (β flap, β subunit aa 808–832) and clamp (β‘ subunit aa 1–413, 1219–1245; β subunit aa 1117–1140) domains are shown as cartoons with cylindrical helices. Rotation angles were measured in PyMol as described by26. a The β flap position in EσB (in red) relative to its position in RbpA/σA-RPo (blue, PDB ID 6C04) and in the T. thermophilus gp39/EσA complex (pink, PDB ID 3WOD). b The clamp position in EσB (in red) relative to RbpA/σA-RPo (blue, PDB ID 6C04) and RbpA/EσA (orange, PDB ID 6C05). c Comparison of the σB and σA (PDB ID 6C05) structures. The σ subunits are shown as ribbons with domain σ1.1 in white, subregion R1.2 in red, NCR in yellow, R2 in goldenrod. d Motions of domain σ1.1. Superposition of σ1.1 in EσB (dark red), RbpA/EσA (light blue) and RbpA/CarD/EσA (green, PDB ID 6EDT). The RNAP core is shown as a molecular surface in gray. σA and σB are shown as ribbons; dwDNA (blue): downstream fragment of promoter DNA duplex.
RNAP conformational flexibility: movements of the β‘ clamp
Published cryo-EM structures of Mtb EσA, free or in complex with ligands: RbpA, CarD, promoter DNA, or fidaxomicin (Fdx), have various clamp states, from open in the Fdx/EσA complex to closed in RPo25–27. Superposition of the EσB structure with the published models showed that in EσB, the clamp adopted a “fully open” conformation with a rotation angle of 22° relative to σA-RPo (Fig. 3b) and a clamp-β lobe distance of 28 Å (Table 1). This conformation was different from the open and relaxed clamp conformations observed in EσA, RbpA/EσA, and Fdx/EσA that displayed a clamp rotation angle of 12°−15° (Fig. 3b and Supplementary Fig. 10). In addition, EσB exhibited different positions of the β‘jaw (aa 1037–1116), which moved toward the secondary channel rim-helices (β‘ K742-H792) and of the β‘ dock, which moved toward the clamp (Supplementary Fig. 10). The conformational mobility of these domains may potentially affect promoter binding and RNA synthesis35. A fully open clamp state was the only clamp conformation observed in our Mtb RNAP sample. We did not detect the partially closed or closed clamp states that were observed by smFRET in Escherichia coli Eσ70 10. This discrepancy can be due to the lineage-specific properties of Mtb RNAP or to the different buffer composition (e.g., divalent cation concentration) that affect clamp dynamics12. We concluded that the fully open-clamp state observed in EσB is characteristic of RNAP inactive state in which the σ subunit regions 3.2 and 4 are unloaded from the RNA exit channel. This immature RNAP conformation may represent an assembly intermediate on the pathway to a transcriptionally active mature conformation of the RNAP holoenzyme.
Distinct structural signatures of σB and σA
The overall fold of domain was similar to that of in the published structures of EσA (Fig. 3c). The exception was the N-terminal domain 1.1 that in σB (σΒ1.1) was stacked to the β‘ clamp surface in the downstream DNA (dwDNA) channel (Fig. 3d). Conversely, in σA1.1, the α-helix (a.a. 208–223) was located perpendicular to the clamp inside the dwDNA channel and contacted β lobe 1 (Fig. 3d). Thus, σA1.1 hindered the dwDNA access to the main channel of the RNAP holoenzyme. In the σA-RPo structure, σA1.1 was displaced upstream towards β‘i1, making the main channel accessible for dwDNA (Fig. 3d). In the EσB structure, σΒ1.1 did not block the main channel and thus there was no physical barrier for dwDNA entry. This difference may affect RPo formation that is regulated by domain σ1.136 and may explain the previously observed different behaviors of EσΒ and EσΑ in transcription initiation23.
Structural sequence alignment of and (Fig. 4a) showed high sequence similarity in all regions, but for the σ non-conserved region (NCR). Indeed, in σΑNCR, four residues are inserted between the α-helices 2 and 3, thus making its tip wider (Fig. 3c). An alignment of 250 actinobacterial σΑ and σΒ homologous sequences retrieved in a Blast search showed that σΑ, but not σΒ harbored insertions of various lengths in the σNCR tip. In addition, some σΒ of the genus Pseudonacardia presented 5–7 aa insertions in the NCR (Supplementary Fig. 11a). Alignments revealed the σNCR specific patterns (Fig. 4a). Specifically, 89% of σΑ-NCR harbored conserved tryptophan residue (W283) in α3-helix that was substituted by alanine (A78) in σΒ. Also, σB-NCR harbored an histidine residue at position 60 (94% of σB sequences) and a leucine residue at position 62 (98% of sequences) that were conserved in σΒ, but not in σA. Hydrophobic interaction of σB-L62 in the a2-helix with σB-L76 in the a3-helix created a bridge that can stabilize the helix-turn-helix domain of σΒ NCR.
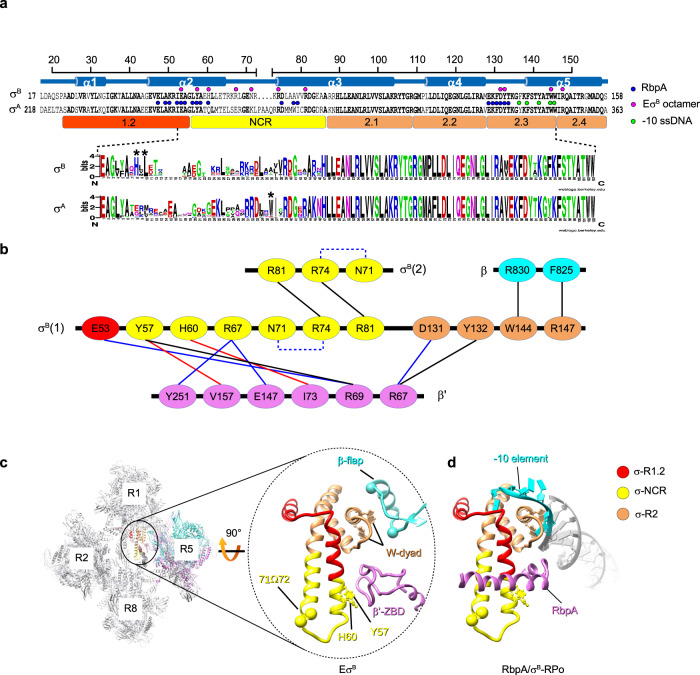
Fig. 4. Structure of the EσB octamer-forming interfaces.
a Schematic representation of the organization of the solved segment of the σB subunit. Top, secondary–structure of σB with the α-helices (α1 to α5) and the structure-based sequence alignment of σB and σA. Dots indicate amino acids that interact with neighboring RNAP protomers in the EσB octamer structure (magenta) or with RbpA (blue) and promoter -10 element ssDNA (light green) in RbpA/σA-RPo. The evolutionarily conserved subregions 1.2, 2.1, 2.2, 2.3, 2.4, and NCR, are depicted by colored rectangles. Bottom, σΒ and σΑ sequence logos generated by Weblogo69 based on the alignment of the 250 Actinobacteria sequences from Uniprot. b Schematic representation of the molecular interactions in the EσB octamer. Residues of the RNAP subunits are presented as ovals and colored according to the color code of a. Interactions between residues are shown by lines: π-stacking in black, Van der Waals in red, ionic in blue. Intra-subunit ionic contacts by dashed lines. c Interactions holding the RNAP protomers together. Left, cartoon presentation of the EσB octamer with the RNAP protomer numbers indicated. The zoomed encircled region (labeled EσB) shows the interactions between σB in the R1 protomer with β‘ ZBD and β flap in the R5 protomer. The W-dyad, (i.e. the invariant W144 and W145 residues), interacts with promoter -10 element ssDNA in RPo. Residues Y57 and H60 that contact β‘ ZBD are shown as ball and stick models. The Cα atoms of residues 71 and 72 in σB NCR (71Ω72) mark the insertion position in the mutant σB71Ω72. The Cα atoms of the β subunit residues 811 and 825 mark the position of the deletion introduced in the β flap. The σB color codes: subregion 1.2 (aa 27–55) in red, NCR (aa 56–86) in yellow, region 2 (aa 87−158) in sandy brown. d Homology model of the RbpA/σB-RPo complex built from the RbpA/σA-RPo model (PDB ID 6C04). Pale green, non-template DNA strand; purple, template DNA strand; magenta, RbpA; gray, DNA; cyan, promoter -10 element.
The σB subunit domain 2 ties RNAP protomers together
Analysis of the EσB octamer and dimer structures revealed four principal intersubunit interfaces to hold the RNAP protomers together: β flap-σB, β‘ZBD-σB, β‘i1-σB and σBNCR-σBNCR (Fig. 4b, c, and Supplementary Fig. 11b). All these interfaces included the residues responsible for binding to the promoter −10 element and RbpA (Fig. 4a). The invariant W144, from the −10 element recognition W-dyad (subregion 2.3), contacted R830 in the β flap. Thus, the W-dyad was sequestered by interactions with the β flap and stabilized in the “edge-on” conformation that is incompatible with −10 element binding9. The residues in the σB subregion 1.2 (E53), σB NCR (Y57, H60, R67) and σB subregion 2.3 (D131, Y132) made contacts with residues R69, R67 and I73 in β‘-ZBD. Of these residues, σB Y57 (σA Y258), σB H60 (σA Q261), σB D131 (σA D336), and σB Y132 (σA Y337) were located at the RbpA-binding interface (Fig. 4a, d). Thus we concluded that RbpA-binding and oligomerization are mutually exclusive events. Residues in σBNCR (Y57, R67) also interacted with residues in β‘i1 (V157, Y251). Finally, R81 (σA R286) and R74 (σA R279) from two adjacent σB NCR faced each other and could make π-stacking interactions37 through guanidinium groups stabilized by N71.
EσB octamer formation is hindered by RbpA, but not CarD
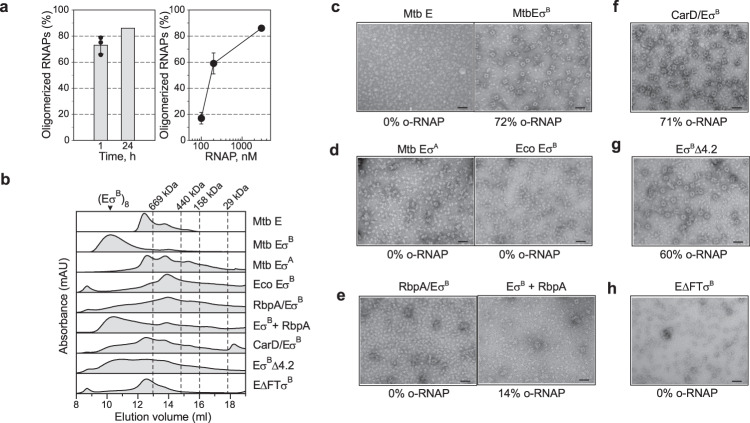
We used negative stain EM and analytical size-exclusion chromatography (SEC) to explore the conditions for EσB oligomerization (Fig. 5). Analysis of negatively stained samples showed that EσB formed octamers spontaneously, within 1 h of mixing the σB subunit with the RNAP core. After 24 h incubation at 4 °C, ~86% of EσB molecules were in oligomers (Fig. 5a) migrating as a single peak during SEC (Fig. 5b, marked as (EσB)8). Octamer formation occurred starting from RNAP concentrations of 0.1 μM which is several orders of magnitude lower than the bulk RNAP concentration in bacterial cells estimated at 5–28 μM38,39. These results, suggest that RNAP octamer assembly may occur in vivo. Next, we explored the capacity to form octamers by the Mtb RNAP core, Mtb EσΑ and a chimeric RNAP holoenzyme assembled from Mtb σB and E. coli RNAP core (Eco EσB). Analysis of the negatively stained images and SEC profiles showed that none of these proteins could form octamers in our experimental conditions (Fig. 5b–d). Although, Mtb RNAP displayed a high propensity for dimer formation (Fig. 5b). Thus, we concluded that the specific structural features of σB and of Mtb RNAP core are essential for spontaneous EσB oligomerization. The specific structural features of σΑ, such as the insertion in the σΑNCR and the bulky side chains of the σA-specific residues (e.g., W283), should interfere with the σANCR-σANCR interactions and hinder oligomerization. Next, we asked whether RbpA and CarD affect EσB oligomerization (Fig. 5b, e, f). As RbpA binds to σNCR, it should interfere with the RNAP-octamer formation (Fig. 4e). Indeed, addition of a 2-fold molar excess of RbpA to the RNAP core before the addition of σB, hindered octamer formation while addition of RbpA to pre-formed EσB octamer induced its dissociation (Fig. 5b, e). As CarD interacts with RNAP β lobe 1, it should not interfere with the octamer-forming interactions of RNAP subunits. Indeed, the negatively stained images show that addition of CarD to the RNAP core before the addition of σB did not prevent octamer formation (Fig. 5b, f). Yet, SEC showed a decrease in the amount of octamers in the presence of CarD, which indicates that CarD affects oligomerization or stability of octamer. These results suggest that RbpA can selectively regulate σB activity by modulating RNAP oligomerization.
Fig. 5. Regulation of the EσB octamer assembly in vitro.
a Time (left panel) and concentration (right panel) dependence of the EσB octamer assembly. Bar graphs show the percentage of RNAP molecules assembled in octamers relative to the total number of RNAP molecules visualized on the EM grid. Values were calculated from the negatively stained images by counting particles using the EMAN2 e2boxer module. When indicated, data are presented as mean values ± SD calculated from n = 3 (bar graph) and n = 4 (line graph) representative images. b Assessment of the RNAP oligomerization state by SEC. Elution profiles of the RNAP core (E) in complex with a different set of transcription factors (indicated on the right) match combinations shown in c–h. The octamer peak is indicated as (EσB)8. Dashed lines indicate the position of the molecular weight markers: 669 kDa, thyroglobulin; 440 kDa, ferritin; 158 kDa, aldolase; 29 kDa, carbonic anhydrase. c Negatively stained images of the Mtb RNAP core (Mtb E) and Mtb RNAP holoenzymes (Mtb EσB). d Negatively stained images of Mtb EσΑ and of the hybrid RNAP holoenzyme assembled from σB and E. coli RNAP core (Eco EσB). e Negatively stained images of Mtb EσB. RbpA was added to E before assembly with σB (RbpA/EσB) and after the octamer formation (EσB + RbpA). f Negatively stained images of Mtb EσB formed in presence of CarD (CarD/EσB) added to E before assembly with σB. Negatively stained images of mutant EσB harboring a deletion in the σΒ subregion 4.2 (EσBΔ4.2) (g) or a deletion in the β flap (EΔFTσB) (h). Scale bar in c–h = 50 nm. Experiments in c–h were repeated independently at least twice with similar results. Source data are provided as a Source Data file.
Interaction of the β flap with σB stabilizes the EσB octamer
In the EσB octamer structure, the σB subregions 2.3 and 2.4 interact with the β flap. To explore whether the β flap and its binding partner σ4 affected octamer assembly, we constructed RNAP mutants in which residues 811-825 in the β flap tip (Mtb EΔFTσB) and residues 252–323 in the σB domain 4.2 (Mtb EσBΔ4.2) were deleted. Analysis of the SEC profiles (Fig. 5b) and the negatively stained images (Fig. 5g) showed that RNAP harboring σBΔ4.2 formed octamers, suggesting that σB4.2 is not essential for oligomerization. Conversely, deletion of the β flap tip (Mtb EΔFTσB) inhibited octamer formation (Fig. 5b, h), suggesting that its interaction with the σB region 2 is critical for oligomerization. Altogether, these results suggest that σB4.2 does not make contacts with the β flap in the octamer.
Role of σB-Y57 and σB-H60 in oligomerization and transcription
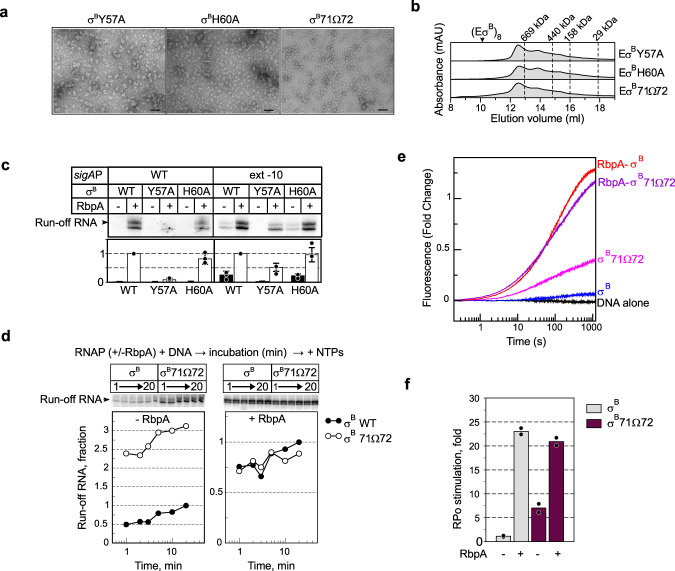
The σBNCR residues Y57 and H60, which make contacts with β‘ZBD in the octamer, also are implicated in RbpA-binding40. To explore their functional role, we constructed two σB mutants harboring the Y57A and H60A substitutions. We used negative stain EM to visualize the RNAP holoenzymes assembled with σB-Y57A and σB-H60A. Analysis of the images demonstrated that both substitutions abolished octamer formation (Fig. 6a), although unstructured oligomers/aggregates were present. SEC also showed no octamer formation by the mutant RNAPs (Fig. 6b).
Fig. 6. Role of σB NCR in octamer assembly and RPo formation.
a Negatively stained images of EσB harboring the following mutant σ subunits: σB-Y57A, σB-H60A, and σB71Ω72. Scale bar = 50 nm. Experiments were repeated independently twice with similar results. b Assessment of the mutant RNAPs oligomerization state by SEC. Elution profiles of the RNAP core (E) in complex with σB-Y57A, σB-H60A, and σB-71Ω72. Expected position of the octamer peak is indicated as (EσB)8. c Transcriptional activity of Mtb RNAP harboring σB (WT) and σB mutants (Y57A, H60A). Transcription was initiated at the wild-type sigAP (WT) and sigAPext-10 promoters either without (-) or with (+) RbpA. Gels show [α32P]-labeled run-off RNA products. Bar graph shows the band quantification. For each panel, the run-off RNA amounts were normalized to the RNA amount synthesized in the presence of RbpA. Data are presented as mean values ± SD of three independent experiments. d Kinetics of promoter binding by the EσB71Ω72 mutant in run-off transcription assays using the sigAPext -10 promoter variant. The experimental scheme is shown at the top. Inserts show run-off RNA products. Graphs show the normalized amounts of run-off RNA (σB71Ω72 vs σB) as a function of time. e Fluorescence fold-change during RPo formation kinetics by EσB and EσB71Ω72 on the Cy3-labeled sigAPext-10 promoter without or with RbpA. f RPo fractions at equilibrium in the time-resolved fluorescence assay shown in d. Values were normalized to the value for EσB. Data points from two technical replicates are shown. Mean values are presented as bar graphs. Source data are provided as a Source Data file.
Run-off transcription assays performed with the mutant EσBY57A and EσBH60A holoenzymes demonstrated that Y57, but not H60, was required for transcription initiation at the RbpA-dependent sigAP promoter and at the RbpA-independent synthetic sigAPext-10 promoter (Fig. 6c). The Y57A substitution abolished stimulation of transcription by RbpA at the sigAP promoter, in agreement with published data on σA40. Unexpectedly, Y57A did not abolish stimulation of transcription by RbpA at the sigAPext-10 promoter, suggesting that Y57 is not essential for RbpA-binding to RNAP. We concluded that extensive contacts between RbpA and the σ subunit can compensate for the lack of the σBY57A-RbpA contact (Fig. 4a). Furthermore, Y57A abolished run-off RNA synthesis from the sigAPext-10 promoter in the absence of RbpA, suggesting that this residue is implicated in transcription initiation also through RbpA-unrelated mechanisms. On the basis of the RbpA/σA-RPo structure, Y57 is positioned too far from promoter DNA (~20 Å) to affect RNAP-DNA binding directly. It may stabilize an overall fold of σ domain 2, and thus stimulates its interaction with the promoter −10 element.
The σB NCR tip affects oligomerization and RPo formation
We showed that the σΑ subunit cannot induce octamer formation (Fig. 5b, d). As that σΒNCR is implicated in three of four intersubunit interfaces holding RNAPs in the octamer, we hypothesized that the insertion in the σANCR tip (Figs. 3c and 4a) compromises intersubunit interactions and may hinder RNAP oligomerization. To test this hypothesis, we introduced a σA fragment (residues PAAQ) between residues 71 and 72 of σB (σB 71Ω72)(Fig. 4a, c). Analysis of the RNAP holoenzyme harboring the mutant σB 71Ω72 by negative stain EM and SEC showed that the insertion abolished octamer formation (Fig. 6a, b).
Oligomerization captures RNAP in the inactive state and is expected to inhibit transcription, while its suppression should stimulate transcription. To assess the oligomerization effect on transcription initiation, we used σB 71Ω72 and two complementary methods: single-round run-off transcription assay24 and fluorescent assay to follow RPo formation kinetics in real-time41,42. To minimize the RNAP dependence on RbpA, we used as template the sigAPext-10 promoter that forms RPo by EσB without RbpA24. We monitored run-off RNA synthesis at different time points after mixing RNAP with promoter DNA. (Fig. 6d). Without RbpA, the EσB 71Ω72 mutant (monomeric state) displayed ~3-fold higher transcriptional activity than EσB (octameric state). In the presence of RbpA (monomeric state), EσB and EσB 71Ω72 showed similar transcriptional activity. Therefore, we concluded that insertion in σB NCR stimulates transcription initiation possibly by inhibiting octamer formation. As the σB NCR residues 71 and 72 are located too far from the promoter, they cannot affect initiation directly through interaction with promoter DNA, as shown for E. coli σ70 NCR43.
To assess whether the 71Ω72 insertion affected directly RPo formation kinetics, we used the sigAPext-10 promoter with the Cy3 dye tethered to the guanine at position +2 of the non-template DNA strand24. Next, we followed Cy3 fluorescence increase upon RNAP binding to the promoter, which is a characteristic of RPo formation (Fig. 6e, f). In agreement with the previous findings42, the reaction kinetics was best fitted by a triple-exponential equation with three phases: fast, medium, and slow (Table 2). The fast phase was over in the first 15 -20 s and may reflect perturbations in the system during the initial promoter melting step. The next two steps were slow (t1/2 between 0.6 and 2.5 min) and reflected isomerization of the closed promoter complex (RPc) to RPo42. All three kinetics of RPo formation by RNAPs in the monomeric state (RbpA/EσB, EσB71Ω72 and RbpA/EσB 71Ω72) displayed similar fractional amplitudes and rate constants for the intermediate and slow phases. The kinetic constants of RPo formation by RNAP in the octameric state (EσB) were different (Table 2). Specifically, the fractional amplitudes and rate constants for the intermediate and slow phases could not be distinguished probably due to the low signal amplitude. Furthermore, RbpA accelerated by ~3-fold RPo formation by EσB, but not by EσB 71Ω72. Without RbpA, the mutant EσB 71Ω72 formed more RPo (~6-fold) than EσB (Fig. 6f). In the presence of RbpA, the amount of RPo was the same for wild type and mutant RNAPs, in agreement with the results of the transcription assay (Fig. 6d). Thus, we concluded that the insertion in σBNCR stimulates RPo formation by increasing the effective concentration of RNAP monomer available for promoter binding. Mutant EσB 71Ω72 is still susceptible to RbpA activation, in agreement with the dual-mode activation mechanism in which RbpA promotes σ loading to RNAP6 and stabilizes RNAP interaction with promoter DNA40.
Table 2.
Kinetic constants of RPo formation on the sigAPext-10(+2Cy3) promoter
| Condition | Fast k1 (s−1) | Intermediate k2 (s−1) | Slow k3 (s−1) | Fast A1,fraction | Intermediate A2, fraction | Slow A3, fraction | Total ΣAi, FC |
|---|---|---|---|---|---|---|---|
| EσB | 0.049 | 0.005 | 0.005 | 0.253 | 0.373 | 0.373 | 0.064 |
| RbpA/EσB | 0.078 | 0.015 | 0.003 | 0.123 | 0.372 | 0.505 | 1.291 |
| EσB 71Ω72 | 0.083 | 0.018 | 0.002 | 0.212 | 0.348 | 0.440 | 0.405 |
| RbpA/EσB 71Ω72 | 0.052 | 0.011 | 0.002 | 0.262 | 0.303 | 0.436 | 1.177 |
A1, A2, A3 are fractional amplitudes for each phase and ΣAi is the total amplitude of the process at equilibrium.
Discussion
In this study we show that Mtb RNAP harboring the group II σΒ factor can spontaneously oligomerize into symmetric octamers in which RNAP is captured in an inactive conformation with the domain σ4 unloaded from the RNAP core. RNAP oligomerization is reversed by the global transcriptional activator RbpA that, according to smFRET and biochemical studies6,44 induces σ4 domain loading onto RNAP (see model in Fig. 7). In bacterial cells, σ factors compete for a general RNAP core pool38. Our findings suggest that σΒ inhibits its own activity by sequestering the RNAP core and may act as a repressor of σΑ-dependent genes by decreasing the effective concentration of free RNAP. In support to this hypothesis, both σΒ and RbpA are implicated in the regulation of transcription in the M. tuberculosis starvation and dormancy models45,46. Thus, contrary to sigA, the sigB gene expression was shown to be upregulated 6-fold concurrently with RbpA (a 9-fold induction)45–47. We speculate that the EσΒ octamer may serve as a storage of hibernating EσΒ that can be rapidly converted to the active form by RbpA during stress. EσΒ octamer disassembly leads to a burst in the effective RNAP concentration and consequently should boost transcription yields (Fig. 7). From our results, σΒNCR emerges as a major determinant of Mtb EσΒ octamer assembly. Furthermore, our results suggest that small variations in the σNCR structure can have inhibitory and also stimulatory effects on RPo formation.
Fig. 7. Model depicting transcription regulation through EσB holoenzyme assembly and oligomerization.
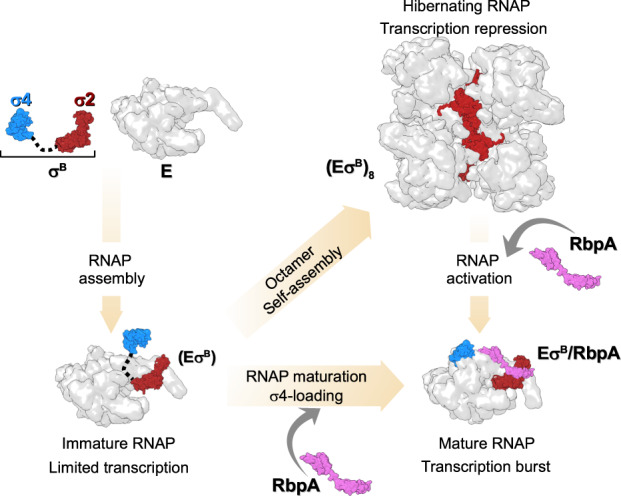
A pathway to Mtb EσB maturation. The pathway starts with the formation of the immature RNAP holoenzyme from free σB [σ2 domain (dark red) and σ4 domain (cyan)] and RNAP core (E, depicted as a gray molecular surface). In the immature EσB, the σB domain 4 remains unloaded from the RNA exit channel, thus making it prone to oligomerization. Binding of RbpA (depicted as a pink molecular surface) to EσB prevents oligomerization and converts EσB to the transcriptionally active, mature form with the σB domain 4 loaded in the RNA exit channel. Binding of RbpA to the (EσB)8 octamer induces its dissociation.
To our knowledge, no structure of octamers formed by RNAPs has been described up to date. Early biochemical studies reported formation of octamers by the E. coli RNAP core in the presence of Mg2+ ions and dimers by the E. coli Eσ70 holoenzyme48,49. The structure of these complexes has not been characterized. Furthermore, neither E. coli nor Mtb RNAP core formed octamers in our experimental conditions. Self-assembly of proteins into high-order structures is essential for regulating all processes in cellular organisms50. For instance, it has been shown that the formation of symmetric tetramers by influenza virus RNA-dependent RNA polymerase (RdRP) is important for the viral life cycle regulation51. Yet, only two cases of regulation that implicate self-assembly of the cellular gene expression machinery into supramolecular structures have been thoroughly described: dimerization of ribosomes in bacteria52,53 and dimerization of RNAP I in yeast54–57. In both cases, dimerization was induced by nutrient deprivation and employed as a regulatory mechanism to control ribosome biogenesis and protein synthesis during cell hibernation. Recently, it has been proposed that dimerization of the Bacillus subtilis RNAP core in complex with the HelD helicase is a hibernation mechanism58. There is an amazing convergence in the mechanisms of RNAP inactivation between the evolutionary distant yeast RNAP I and Mtb EσΒ. Like in Mtb EσΒ, in inactive RNAP I dimers, the clamp adopts an open state56. Furthermore, like RbpA, the eukaryotic initiation factor Rrn3, interacts with the RNAP I regions involved in dimer formation and converts RNAP I dimers to initiation-competent monomers55,59 Thus, RNAP oligomerization controlled by transcription factors emerges as a general regulation mechanism for gene repression in the different kingdoms of life. An exciting direction for future studies will be to explore RNAP octamer formation in live cells and to determine its function.
Methods
Proteins and DNA templates
The 6xHis-tagged Mtb RNAP core was expressed in BL21 DE3 E. coli cells transformed with the pMR4 plasmid and purified by Ni2+-affinity chromatography17 followed by purification by anion-exchange chromatography on MonoQ 5/50GL (GE Healthcare). In brief, 15 g of bacteria were resuspended in 200 ml of lysis buffer (40 mM Tris-HCl pH 7.9, 500 mM NaCl, 5% Glycerol, 10 μM ZnCl2, 0,2 mM β-mercaptoethanol, 4 tablets of cOmplete™, EDTA-free Protease Inhibitor Cocktail (Roche), 0.1 mg/ml lysozym). Cells were disrupted by sonication and the lysate, cleared by centrifugation at 9,000 g (40 min, 4 °C), was loaded on 5 × 5 ml HisTrap FF columns (GE Heathcare) equilibrated with 40 mM Tris-HCl pH 7.9, 500 mM NaCl, 5% glycerol, 0.2 mM β-mercaptoethanol. Mtb RNAP core was eluted with 300 mM imidazole. Mtb RNAP sample was dialyzed at 4 °C overnight against buffer A (20 mM Tris-HCl pH 7.9, 500 mM NaCl, 5% glycerol, 0,2 mM EDTA, 0.01% Tween20, 10 mM ZnCl2, 0,2 mM β-mercaptoethanol). Next, RNAP sample was dialyzed at 4 °C for 2 h against buffer B (20 mM Tris-HCl pH 7.9, 50 mM NaCl, 5% Glycerol, 0.2 mM EDTA, 0.2 mM β-mercaptoethanol, 0.01% Tween20) and loaded on MonoQ 5/50GL column (GE Heathcare) equilibrated with buffer B. Mtb RNAP core was eluted in linear gradient of NaCl (200–480 mM). Collected peak fractions were pooled, concentrated by Amicon® Ultra-15 Centrifugal Filter Unit (Millipore), supplemented with 20% glycerol, and stored at −80 °C. The 6xHis-tagged E. coli RNAP core was expressed in BL21 DE3 E. coli cells transformed with the pVS10 plasmid and purified as described above for Mtb RNAP. The gene encoding for Mtb CarD (Rv3583c) was PCR-amplified from H37Rv genomic DNA (NR-14867, BEI Resources) and cloned between the NdeI and HindIII sites into the pET28a vector under the N-terminal 6xHis-tag. The 6xHis-tagged Mtb CarD, RbpA, and the σA subunit were expressed and purified by chromatography on HisTrap HP (GE Healthcare) Ni2+-affinity columns as described. The 6xHis-tagged Mtb σB subunit, σB in which residues 252–323 were deleted (σBΔ4.2), and σB with the insertion in NCR (σB71Ω72) were purified by chromatography on HiTrap TALON (GE Healthcare) columns. To construct the mutant σB 71Ω72, residues N71 and R72 of the σB subunit were replaced by six σA residues (segment 272-KLPAAQ-277) using the Quick Change Lightening site-directed mutagenesis kit (Agilent). Residues 811-825 in the Mtb RNAP β flap were deleted using Quick Change II XL site-directed mutagenesis kit (Agilent). Variants of the wild-type sigAP and sigAPext-10 (harboring the T-17G-16T-15G-14 motif) promoters 5’-end-labeled with fluorescein and the sigAPext-10 promoter internally labeled with Cy3 at position +2 of the non-template DNA strand were prepared as described24. Sequences of primers used for CarD cloning and for sigAP promoter fragments preparation are provided in Supplementary Table 2
Cryo-EM sample preparation
To assemble the EσB holoenzyme, 3.4 μM Mtb RNAP core and 3.74 μM σB in transcription buffer (40 mM HEPES pH 8.0, 50 mM NaCl, 5 mM MgCl2, 5% glycerol) were incubated at 37 °C for 5 min. Next, samples were dialyzed in 10 μl drops on 0.025 μm MF-Millipore membrane filters (VSWP) against dialysis buffer (20 mM HEPES pH 8.0, 150 mM NaCl, 5 mM MgSO4) at 22 °C for 45 min. About 3 μl of sample (3 μM EσB final concentration) was spotted on a Quantifoil R2/2 200 Mesh holey carbon grids which were glow-discharged for 10 s using the PELCO easiGlow system (Ted Pella). Grids were flash-frozen in liquid ethane using Vitrobot Mark IV (FEI) at 20 °C and 95–100% of humidity.
Cryo-EM data acquisition
Data were collected using a spherical aberration (Cs) - corrected Titan Krios S-FEG instrument (FEI) operating at 300 kV acceleration voltage and equipped with a Gatan K2 Summit direct electron detector (Gatan, Warrendale, PA). Automatic image acquisition was carried out using the EPU software (FEI) in a super-resolution mode at a nominal magnification of ×105,000 with a pixel size of 0.55 Å. Movies (31 frames) were collected at an exposure rate of 6.2 e−/Å−2/s and a total electron dose of 49.6 e−/Å−2 over a nominal defocus range from −0.5 to −5.0 µm.
Cryo-EM data processing
Movie frames were aligned, dose-weighted, binned by two, and averaged using Motioncor260. Movie sums were used for contrast transfer function (CTF) estimation with Gctf61. A 3064 dose-weighted movie sums were used in the subsequent image-processing steps. About 100 particles comprising monomers and oligomers were manually picked in cryoSPARC62 and subjected to 2D classification to create templates for automatic picking.
Mtb RNAP core reconstruction using cryoSPARC
A total set of 721,752 particles that included RNAP monomers underwent several 2D classification rounds. A cleaned dataset of 153,953 particles was used in the ab-initio reconstruction to compute the initial model. The ab-initio model was used as reference for the 3D heterogeneous refinement and classification. Two 3D classification rounds produced a cleaned set of 55,008 particles that were used for the homogeneous refinement at 4.19 Å. Then, the local non-uniform refinement resulted in an improved cryo-EM map refined at 4.08 Å.
Mtb EσB octamer and protomer reconstructions using cryoSPARC
A dataset of 254,380 particles that included RNAP oligomers underwent several 2D classification rounds to produce a clean dataset of 115,112 particles. The best class averages that included 82,893 particles were used in the reference-free ab-initio heterogeneous reconstruction to produce the initial model. The homogeneous non-uniform refinement of the initial map using 115,112 particles without applying symmetry resulted in a C1-map at a nominal resolution of 6.13 Å. The dataset of 115,457 particles was used for the non-uniform refinement with imposed D4 symmetry to compute the D4-map at a nominal resolution of 4.39 Å. The dataset of 115,112 subtracted particles and the masked C1-map were used for the local non-uniform refinement to calculate the maps of the RNAP monomer and RNAP dimer at a nominal resolution of 3.84 Å and 4.36 Å, respectively The dataset of 115,112 subtracted particles underwent 3D heterogeneous refinement using the R1-R5 RNAP dimer map as reference. The 3D classification produced two RNAP dimer classes with different conformations: class 1, which included 66,519 particles, resolved at 4.38 Å, and class 2, which included 48,593 particles, resolved at 6.75 Å.
Mtb EσB protomer reconstruction using RELION
A dataset of 94,191 particles was used in RELION (version 1.4) 3D refinement to compute the D4-map of the EσB octamer at resolution of 6.3 Å. Then, each particle 2D image was replicated and 3D-rotated according to the D4 point group symmetry and the re-projected density of all but one EσB protomer was subtracted. The resulting expanded dataset of 753,528 subtracted particles was used in alternate cycles of asymmetric 3D classification and 3D local refinement focused on the region occupied by a single EσB protomer. Refinement was first focused on the RNAP core density devoid of the β‘ clamp. A D4-map of the octamer, from which the focused region was masked out, was used for the signal subtraction. The focused region of the RNAP core, which represented the least resolved peripheral zones in the D4-map of the EσB octamer (Supplementary Fig. 4a), was reconstructed at 3.6 Å resolution (Supplementary Fig. 7). Next, the β‘ clamp and σB subunit were included in the focused refinement. The signal of seven of the eight β‘ clamp/σB re-projected densities was subtracted from the particles projections based on the initial D4-map of the EσB octamer and using an appropriate mask. For each particle projection, the signal of the remaining RNAP core subunits outside the focused region was subtracted using seven successive re-projections based on the 3.6 Å RNAP core reconstruction and the angular parameters obtained in the preceding 3D refinement. Two out of six 3D classes were selected and locally refined at a resolution up to 3.9 Å and 4.19 Å, respectively (Supplementary Fig. 7).
Model building and refinement
The coordinates of the Mtb EσA holoenzyme (PDB ID 6FBV) were used as starting model. The σΑ subunit was replaced by σB using homology modeling in Modeller63. The model was fitted to cryo-EM maps in UCSF Chimera64 and was manually modified in Coot65. Several cycles of real-space refinement, using secondary-structure restrains and geometry optimization, were performed in Phenix66 using the R1-R5 RNAP dimer and R1 RNAP protomer maps. To improve the σΒ subunit model, it was real-space refined separately in Phenix with the RNAP octamer D4-map. The final EσΒ model was assembled and modified in Coot. The R1-R5 RNAP dimer model was assembled from two copies of EσΒ rigid body refined in Phenix. The EσΒ octamer model was built by applying NCS operators to the EσB protomer model in Phenix followed by modification in Coot.
Negative stain EM sample preparation and data acquisition
100–400 nM RNAP core was mixed with threefold molar excess of the σ subunit in the transcription buffer described above. When indicated, RbpA and CarD (3-fold molar excess) were added to the RNAP core. Samples were incubated at 22 °C for 10 min. The reaction mixtures were dialyzed for 1 h, as described for the cryo-EM samples. Then, 3 μl of mixture was spotted on a Formvar/Carbon copper 200 mesh grids (Electron Microscopy Sciences) glow-discharged for 10 s. Grids were stained with uranyl acetate (1% w/v). Images were collected using an 120 kV JEOL 1200 EX II EM equipped with an EMSIS Quemesa 11Mpixels camera with a nominal magnification of ×50,000 and pixel size 2.86 Å. Particles were counted using the EMAN2 e2boxer module67.
Analytical size-exclusion chromatography
To assess oligomeric states of RNAP, about 200 pmole of E and Eσ holoenzymes formed with twofold molar excess of σΑ, σB variants, RbpA and CarD in 120 μl of transcription buffer were incubated at 24 °C for 1 h. Mtb E was first mixed with RbpA and CarD and then supplemented with the σB subunit. To test the effect of RbpA on the octamer dissociation, Mtb E was incubated with σB overnight at +4 °C, then supplemented with RbpA and incubated at 24 °C for 1 h. Next, a 100 μl of the sample was applied to Superose® 6 Increase 10/300 GL column (Cytiva) equilibrated with 10 mM Tris-HCl pH 8.0, 150 mM NaCl, 5 mM MgCl2, 5% glycerol and run at 0.5 ml/min flow rate.
In vitro transcription assay
In multiple-round transcription assays, 100 nM RNAP core was mixed with 300 nM σB and 300 nM RbpA in 10μl of transcription buffer and incubated at 37 °C for 5 min. 50 nM of promoter DNA was added and incubated at 37 °C for 10 min. Transcription was initiated by adding 50 μM/each of ATP, GTP, CTP, 5 μM of UTP, and 0.5 μM of [32P]-UTP, and performed at 37 °C for 5 min. In single-round kinetics run-off assays, 200 nM RNAP core was mixed with 600 nM σB with or without 600 nM RbpA in 10μl of transcription buffer and incubated at 37 °C for 5 min. Samples prepared without RbpA were incubated at +4 °C overnight to reach the maximum yield of RNAP octamer. After the addition of 100 nM of sigAPext-10 promoter, DNA samples were incubated at 37 °C for 1, 2, 3, 5, 10, or 20 min. Transcription was initiated by adding 100 μM/each of CTP, GTP, ATP, 10 μM of UTP, 0.5 μM of [32P]-UTP, and 0.1 mg/ml poly(dI-dC). Reactions were incubated at 37 °C for 3 min. RNA transcripts were analyzed by denaturing 18% PAGE/7 M urea gels. Gels were scanned with a Molecular Dynamics STORM Imager and quantified by ImageQuant software.
Real-time fluorescent assay of RPo formation
Data were acquired using a SF-61 DX2 stopped flow spectrophotometer (TgK Scientific UK) with a shot volume of 100 µl, excitation at 535 nm and emission at 570 nm. 200 nM RNAP core was mixed with 1 μM of σ subunit with or without 1 μM RbpA in 100 μl of transcription buffer and incubated at 37 °C for 5 min. Samples prepared without RbpA were incubated at +4 °C overnight. Protein samples were diluted 4-fold in transcription buffer containing 0.1 mg/ml BSA immediately before mixing with promoter DNA. Experiments were initiated by mixing equal volumes of 50 nM RNAP and 10 nM Cy3-labeled sigAPext-10 promoter in a transcription buffer containing 0.1 mg/ml BSA. The final RNAP concentration was 25 nM and the DNA concentration was 5 nM. Data were collected at 30 °C for 20 min. Two consecutive shots were performed and averaged. Each dataset was normalized to the fluorescence signal value at equilibrium and plotted as the fluorescence fold change (FC), where FC = (F–Fo)/Fo. Fo is the signal for DNA alone and F is the signal for RNAP-bound DNA. Values from two experiments were averaged and fitted using the Grace software (v. 5.1.25) with the triple-exponential equation FCt = A0 + Α1 · exp(-k1 · t)+ Α2 · exp(-k2 · t) + Α3· exp(-k3 · t) where FCt is the total fluorescence change.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.
Supplementary information
Description of Additional Supplementary Files
Acknowledgements
We thank Irina Artsimovitch and Mickael Blaise for critical reading of the manuscript and discussion. We thank Jean-Paul Leonetti for help with figure preparation and discussion. We thank Franck Godiard for the negative stained EM data collection at the MEA platform, University of Montpellier, Montpellier, France. We thank Alexander Myasnikov, Jean-Francois Menetret, Julio Ortiz for assistance with grid preparation and cryo-EM data collection at the Center for Integrative Biology, IGBMC, Strasbourg, France. Funding from the French National Research Agency [MycoMaster, ANR-16-CE11-0025] to K.B. and P.B. Support from Instruct-ERIC (PID: 1309) to K.B. Support from CNRS (Frozen, Mission pour l’Interdisciplinarité, AAP Interne 2017) to C.L. The CBS is a member of the French Infrastructure for Integrated Structural Biology (FRISBI), a national infrastructure supported by the French National Research Agency (ANR-10-INBS-05).
Source data
Author contributions
Conceptualization, K.B.; methodology, K.B., P.B., and S.T.; software, S.T.; investigation, Z.M., R.K.V., C.L., J.L.K.H., P.B., and K.B; formal Analysis, C.L., Z.M., L.C., S.T., and K.B.; validation, K.B., and P.B. writing, K.B.; supervision, K.B., and P.B.; funding acquisition, K.B., P.B., and C.L.
Peer review
Peer review information
Nature Communications thanks Elizabeth Campbell, Yu Zhang and the other, anonymous, reviewer(s) for their contribution to the peer review of this work. Peer reviewer reports are available.
Data availability
The data that support this study are available from the corresponding authors upon reasonable request. Cryo-EM density maps reported in this paper have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-13579 (Mtb EσB protomer), EMD-14696 (Mtb EσB protomer), EMD-13817 (Mtb EσB octamer D4-map), EMD-14697 (Mtb EσB octamer D4-map), EMD-13829 (Mtb EσB dimer), EMD-14378 (Mtb EσB dimer class 1), EMD-14974 (Mtb EσB dimer class 2), EMD-14560 (Mtb RNAP core). Model coordinates have been deposited in the Protein Data Bank (PDB) with accession codes 7PP4 (Mtb EσB protomer), 7ZF2 (Mtb EσB protomer), 7Q4U (Mtb EσB octamer), 7Q59 (Mtb EσB dimer), 7Z8Q (Mtb RNAP core). The publicly available datasets with PDB accession codes 6FBV, 6EYD, 6C04, 6C05, 6KON, 3WOD, 6EDT were used in this study for figure preparation and data analysis. Source data are provided with this paper.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Patrick Bron, Email: patrick.bron@cbs.cnrs.fr.
Konstantin Brodolin, Email: konstantin.brodolin@inserm.fr.
Supplementary information
The online version contains supplementary material available at 10.1038/s41467-023-36113-y.
References
- 1.Burgess RR. Separation and characterization of the subunits of ribonucleic acid polymerase. J. Biol. Chem. 1969;244:6168–6176. [PubMed] [Google Scholar]
- 2.Burgess RR, Travers AA, Dunn JJ, Bautz EK. Factor stimulating transcription by RNA polymerase. Nature. 1969;221:43–46. doi: 10.1038/221043a0. [DOI] [PubMed] [Google Scholar]
- 3.Murakami KS, Masuda S, Darst SA. Structural basis of transcription initiation: RNA polymerase holoenzyme at 4 A resolution. Science. 2002;296:1280–1284. doi: 10.1126/science.1069594. [DOI] [PubMed] [Google Scholar]
- 4.Vassylyev DG, et al. Crystal structure of a bacterial RNA polymerase holoenzyme at 2.6 A resolution. Nature. 2002;417:712–719. doi: 10.1038/nature752. [DOI] [PubMed] [Google Scholar]
- 5.Bae B, Feklistov A, Lass-Napiorkowska A, Landick R, Darst SA. Structure of a bacterial RNA polymerase holoenzyme open promoter complex. eLife. 2015;4:08504. doi: 10.7554/eLife.08504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vishwakarma RK, et al. Single-molecule analysis reveals the mechanism of transcription activation in M. tuberculosis. Sci. Adv. 2018;4:eaao5498. doi: 10.1126/sciadv.aao5498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zuo Y, Steitz TA. Crystal structures of the E. coli transcription initiation complexes with a complete bubble. Mol. Cell. 2015;58:534–540. doi: 10.1016/j.molcel.2015.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schulz S, et al. TFE and Spt4/5 open and close the RNA polymerase clamp during the transcription cycle. Proc. Natl Acad. Sci. USA. 2016;113:1816–1825. doi: 10.1073/pnas.1515817113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen J, et al. Stepwise promoter melting by bacterial RNA Polymerase. Mol. Cell. 2020;78:275–288. doi: 10.1016/j.molcel.2020.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chakraborty A, et al. Opening and closing of the bacterial RNA polymerase clamp. Science. 2012;337:591–595. doi: 10.1126/science.1218716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Feklistov A, et al. RNA polymerase motions during promoter melting. Science. 2017;356:863–866. doi: 10.1126/science.aam7858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mazumder A, Wang A, Uhm H, Ebright RH, Kapanidis AN. RNA polymerase clamp conformational dynamics: Long-lived states and modulation by crowding, cations, and nonspecific DNA binding. Nucleic Acids Res. 2021;49:2790–2802. doi: 10.1093/nar/gkab074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Duchi D, Mazumder A, Malinen AM, Ebright RH, Kapanidis AN. The RNA polymerase clamp interconverts dynamically among three states and is stabilized in a partly closed state by ppGpp. Nucleic Acids Res. 2018;46:7284–7295. doi: 10.1093/nar/gky482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodrigue S, Provvedi R, Jacques PÉ, Gaudreau L, Manganelli R. The σ factors of Mycobacterium tuberculosis. FEMS Microbiol. Rev. 2006;30:926–941. doi: 10.1111/j.1574-6976.2006.00040.x. [DOI] [PubMed] [Google Scholar]
- 15.Hurst-Hess K, et al. Mycobacterial SigA and SigB cotranscribe essential housekeeping genes during exponential growth. mBio. 2019;10:e00273–19. doi: 10.1128/mBio.00273-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gengenbacher M, Kaufmann SHE. Mycobacterium tuberculosis: success through dormancy. FEMS Microbiol. Rev. 2012;36:514–532. doi: 10.1111/j.1574-6976.2012.00331.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hu Y, Morichaud Z, Perumal AS, Roquet-Baneres F, Brodolin K. Mycobacterium RbpA cooperates with the stress-response σB subunit of RNA polymerase in promoter DNA unwinding. Nucleic Acids Res. 2014;42:10399–10408. doi: 10.1093/nar/gku742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lee JH, Karakousis PC, Bishai WR. Roles of SigB and SigF in the Mycobacterium tuberculosis sigma factor network. J. Bacteriol. 2008;190:699–707. doi: 10.1128/JB.01273-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fontán PA, et al. The Mycobacterium tuberculosis sigma factor σB is required for full response to cell envelope stress and hypoxia in vitro, but it is dispensable for in vivo growth. J. Bacteriol. 2009;191:5628–5633. doi: 10.1128/JB.00510-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Manganelli R, Dubnau E, Tyagi S, Kramer FR, Smith I. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 1999;31:715–724. doi: 10.1046/j.1365-2958.1999.01212.x. [DOI] [PubMed] [Google Scholar]
- 21.Stallings CL, et al. CarD is an essential regulator of rRNA transcription required for Mycobacterium tuberculosis persistence. Cell. 2009;138:146–159. doi: 10.1016/j.cell.2009.04.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Newell KV, Thomas DP, Brekasis D, Paget MSB. The RNA polymerase-binding protein RbpA confers basal levels of rifampicin resistance on Streptomyces coelicolor. Mol. Microbiol. 2006;60:687–696. doi: 10.1111/j.1365-2958.2006.05116.x. [DOI] [PubMed] [Google Scholar]
- 23.Hu Y, Morichaud Z, Chen S, Leonetti J-P, Brodolin K. Mycobacterium tuberculosis RbpA protein is a new type of transcriptional activator that stabilizes the σ A-containing RNA polymerase holoenzyme. Nucleic Acids Res. 2012;40:6547–6557. doi: 10.1093/nar/gks346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Perumal AS, Vishwakarma RK, Hu Y, Morichaud Z, Brodolin K. RbpA relaxes promoter selectivity of M. tuberculosis RNA polymerase. Nucleic Acids Res. 2018;46:10106–10118. doi: 10.1093/nar/gky714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lin W, et al. Structural basis of transcription inhibition by fidaxomicin (Lipiarmycin A3) Mol. Cell. 2018;70:60–71.e15. doi: 10.1016/j.molcel.2018.02.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Boyaci H, et al. Fidaxomicin jams Mycobacterium tuberculosis RNA polymerase motions needed for initiation via RbpA contacts. eLife. 2018;7:e34823. doi: 10.7554/eLife.34823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boyaci H, Chen J, Jansen R, Darst SA, Campbell EA. Structures of an RNA polymerase promoter melting intermediate elucidate DNA unwinding. Nature. 2019;565:382–385. doi: 10.1038/s41586-018-0840-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kouba T, et al. The core and holoenzyme forms of RNA polymerase from mycobacterium smegmatis. J. Bacteriol. 2019;201:e00583–18. doi: 10.1128/JB.00583-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Punjani A, Fleet DJ. 3D variability analysis: resolving continuous flexibility and discrete heterogeneity from single particle cryo-EM. J. Struct. Biol. 2021;213:107702. doi: 10.1016/j.jsb.2021.107702. [DOI] [PubMed] [Google Scholar]
- 30.Zhou M, et al. Atomic structure of the apoptosome: mechanism of cytochrome c- and dATP-mediated activation of Apaf-1. Genes Dev. 2015;29:2349–2361. doi: 10.1101/gad.272278.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Scheres SHW. Processing of structurally heterogeneous cryo-EM Data in RELION. Methods Enzymol. 2016;579:125–157. doi: 10.1016/bs.mie.2016.04.012. [DOI] [PubMed] [Google Scholar]
- 32.Tagami S, et al. Structural basis for promoter specificity switching of RNA polymerase by a phage factor. Genes Dev. 2014;28:521–531. doi: 10.1101/gad.233916.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Said N, et al. Steps toward translocation-independent RNA polymerase inactivation by terminator ATPase ρ. Science. 2021;371:eabd1673. doi: 10.1126/science.abd1673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang G, et al. Crystal structure of thermus aquaticus core RNA polymerase at 3.3 å resolution. Cell. 1999;98:811–824. doi: 10.1016/s0092-8674(00)81515-9. [DOI] [PubMed] [Google Scholar]
- 35.Kang JY, et al. RNA polymerase accommodates a pause RNA hairpin by global conformational rearrangements that prolong pausing. Mol. Cell. 2018;69:802–815.e5. doi: 10.1016/j.molcel.2018.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vuthoori S, Bowers CW, McCracken A, Dombroski AJ, Hinton DM. Domain 1.1 of the sigma(70) subunit of Escherichia coli RNA polymerase modulates the formation of stable polymerase/promoter complexes. J. Mol. Biol. 2001;309:561–572. doi: 10.1006/jmbi.2001.4690. [DOI] [PubMed] [Google Scholar]
- 37.Lee D, Lee J, Seok C. What stabilizes close arginine pairing in proteins? Phys. Chem. Chem. Phys. 2013;15:5844–5853. doi: 10.1039/c3cp00160a. [DOI] [PubMed] [Google Scholar]
- 38.Grigorova IL, Phleger NJ, Mutalik VK, Gross CA. Insights into transcriptional regulation and σ competition from an equilibrium model of RNA polymerase binding to DNA. Proc. Natl Acad. Sci. USA. 2006;103:5332–5337. doi: 10.1073/pnas.0600828103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Klumpp S, Hwa T. Growth-rate-dependent partitioning of RNA polymerases in bacteria. Proc. Natl Acad. Sci. USA. 2008;105:20245–20250. doi: 10.1073/pnas.0804953105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hubin EA, et al. Structural, functional, and genetic analyses of the actinobacterial transcription factor RbpA. Proc. Natl Acad. Sci. 2015;112:7171–7176. doi: 10.1073/pnas.1504942112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Matlock DL, Heyduk T. A real-time fluorescence method to monitor the melting of duplex DNA during transcription initiation by RNA polymerase. Anal. Biochem. 1999;270:140–147. doi: 10.1006/abio.1999.4078. [DOI] [PubMed] [Google Scholar]
- 42.Rammohan J, Ruiz Manzano A, Garner AL, Stallings CL, Galburt EA. CarD stabilizes mycobacterial open complexes via a two-tiered kinetic mechanism. Nucleic Acids Res. 2015;43:3272–3285. doi: 10.1093/nar/gkv078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Narayanan A, et al. Cryo-EM structure of Escherichia coli σ70 RNA polymerase and promoter DNA complex revealed a role of σ non-conserved region during the open complex formation. J. Biol. Chem. 2018;293:7367–7375. doi: 10.1074/jbc.RA118.002161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brodolin K, Morichaud Z. Region 4 of the RNA polymerase σ subunit counteracts pausing during initial transcription. J. Biol. Chem. 2021;296:100253. doi: 10.1074/jbc.RA120.016299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Betts JC, Lukey PT, Robb LC, McAdam RA, Duncan K. Evaluation of a nutrient starvation model of Mycobacterium tuberculosis persistence by gene and protein expression profiling. Mol. Microbiol. 2002;43:717–731. doi: 10.1046/j.1365-2958.2002.02779.x. [DOI] [PubMed] [Google Scholar]
- 46.Voskuil MI, Visconti KC, Schoolnik GK. Mycobacterium tuberculosis gene expression during adaptation to stationary phase and low-oxygen dormancy. Tuberculosis. 2004;84:218–227. doi: 10.1016/j.tube.2004.02.003. [DOI] [PubMed] [Google Scholar]
- 47.Pettersson BMF, et al. Comparative sigma factor-mRNA levels in mycobacterium marinum under stress conditions and during host infection. PloS One. 2015;10:e0139823. doi: 10.1371/journal.pone.0139823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaner SL, et al. Aggregation equilibria of Escherichia coli RNA polymerase: evidence for anion-linked conformational transitions in the protomers of core and holoenzyme. Biochemistry. 1982;21:5539–5551. doi: 10.1021/bi00265a025. [DOI] [PubMed] [Google Scholar]
- 49.Kansara SG, Sukhodolets MV. Oligomerization of the E. coli core RNA polymerase: formation of (α2ββ’ω)2-DNA complexes and regulation of the oligomerization by auxiliary subunits. PloS One. 2011;6:e18990. doi: 10.1371/journal.pone.0018990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marsh JA, Teichmann SA. Structure, dynamics, assembly, and evolution of protein complexes. Annu. Rev. Biochem. 2015;84:551–575. doi: 10.1146/annurev-biochem-060614-034142. [DOI] [PubMed] [Google Scholar]
- 51.Chang S, et al. Cryo-EM structure of influenza virus rna polymerase complex at 4.3Å resolution. Mol. Cell. 2015;57:925–935. doi: 10.1016/j.molcel.2014.12.031. [DOI] [PubMed] [Google Scholar]
- 52.Beckert B, et al. Structure of the Bacillus subtilis hibernating 100S ribosome reveals the basis for 70S dimerization. EMBO J. 2017;36:2061–2072. doi: 10.15252/embj.201696189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Beckert B, et al. Structure of a hibernating 100S ribosome reveals an inactive conformation of the ribosomal protein S1. Nat. Microbiol. 2018;3:1115–1121. doi: 10.1038/s41564-018-0237-0. [DOI] [PubMed] [Google Scholar]
- 54.Torreira E, et al. The dynamic assembly of distinct RNA polymerase I complexes modulates rDNA transcription. eLife. 2017;6:e20832. doi: 10.7554/eLife.20832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pilsl M, et al. Structure of the initiation-competent RNA polymerase I and its implication for transcription. Nat. Commun. 2016;7:12126. doi: 10.1038/ncomms12126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Heiss FB, Daiß JL, Becker P, Engel C. Conserved strategies of RNA polymerase I hibernation and activation. Nat. Commun. 2021;12:758. doi: 10.1038/s41467-021-21031-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Engel C, Sainsbury S, Cheung AC, Kostrewa D, Cramer P. RNA polymerase i structure and transcription regulation. Nature. 2013;502:650–655. doi: 10.1038/nature12712. [DOI] [PubMed] [Google Scholar]
- 58.Pei H-H, et al. The δ subunit and NTPase HelD institute a two-pronged mechanism for RNA polymerase recycling. Nat. Commun. 2020;11:6418. doi: 10.1038/s41467-020-20159-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Engel C, Plitzko J, Cramer P. RNA polymerase I-Rrn3 complex at 4.8 Å resolution. Nat. Commun. 2016;7:12129. doi: 10.1038/ncomms12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zheng SQ, et al. MotionCor2: anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat. Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang K. Gctf: real-time CTF determination and correction. J. Struct. Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Punjani A, Rubinstein JL, Fleet DJ, Brubaker MA. cryoSPARC: algorithms for rapid unsupervised cryo-EM structure determination. Nat. Methods. 2017;14:290–296. doi: 10.1038/nmeth.4169. [DOI] [PubMed] [Google Scholar]
- 63.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 64.Pettersen EF, et al. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 65.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D. Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 66.Adams PD, et al. PHENIX: a comprehensive python-based system for macromolecular structure solution. Acta Crystallogr. D. Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tang G, et al. EMAN2: an extensible image processing suite for electron microscopy. J. Struct. Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 68.Lane WJ, Darst SA. Molecular evolution of multisubunit RNA polymerases: structural analysis. J. Mol. Biol. 2010;395:686–704. doi: 10.1016/j.jmb.2009.10.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Description of Additional Supplementary Files
Data Availability Statement
The data that support this study are available from the corresponding authors upon reasonable request. Cryo-EM density maps reported in this paper have been deposited in the Electron Microscopy Data Bank (EMDB) with accession codes EMD-13579 (Mtb EσB protomer), EMD-14696 (Mtb EσB protomer), EMD-13817 (Mtb EσB octamer D4-map), EMD-14697 (Mtb EσB octamer D4-map), EMD-13829 (Mtb EσB dimer), EMD-14378 (Mtb EσB dimer class 1), EMD-14974 (Mtb EσB dimer class 2), EMD-14560 (Mtb RNAP core). Model coordinates have been deposited in the Protein Data Bank (PDB) with accession codes 7PP4 (Mtb EσB protomer), 7ZF2 (Mtb EσB protomer), 7Q4U (Mtb EσB octamer), 7Q59 (Mtb EσB dimer), 7Z8Q (Mtb RNAP core). The publicly available datasets with PDB accession codes 6FBV, 6EYD, 6C04, 6C05, 6KON, 3WOD, 6EDT were used in this study for figure preparation and data analysis. Source data are provided with this paper.