Abstract
Gastrointestinal infections are a major cause for serious clinical complications in infants. The induction of antibody responses by B cells is critical for protective immunity against infections and requires CXCR5+PD-1++ CD4+ T cells (TFH cells). We investigated the ontogeny of CXCR5+PD-1++ CD4+ T cells in human intestines. While CXCR5+PD-1++ CD4+ T cells were absent in fetal intestines, CXCR5+PD-1++ CD4+ T cells increased after birth and were abundant in infant intestines, resulting in significant higher numbers compared to adults. These findings were supported by scRNAseq analyses, showing increased frequencies of CD4+ T cells with a TFH gene signature in infant intestines compared to blood. Co-cultures of autologous infant intestinal CXCR5+PD-1+/−CD4+ T cells with B cells further demonstrated that infant intestinal TFH cells were able to effectively promote class switching and antibody production by B cells. Taken together, we demonstrate that functional TFH cells are numerous in infant intestines, making them a promising target for oral pediatric vaccine strategies.
Keywords: TFH cells, B cells, Antibodies, Intestine, Pediatrics
Subject terms: Mucosal immunology, Innate lymphoid cells
Introduction
Life expectancy and infant health have improved greatly, however infectious diseases remain the main global cause for infant mortality and morbidity [1, 2]. Almost half a million children under the age of 5 years succumb to gastroenteritis annually [3]. Furthermore, the intestine is the main site of entry for many pathogens causing severe sepsis in young infants, such as Escherichia coli and Klebsiella species [4]. Therefore, oral vaccines that induce antibody-mediated protection could provide critical protection in young infants; however, only few oral vaccines against gastrointestinal pathogens are available [5]. In particular, live-attenuated vaccines have shown to induce relatively efficient long-lasting mucosal antigen-specific humoral and cellular immune responses [5]. However, our understanding of the mechanisms regulating the induction of protective antibody responses at mucosal sites in children remains limited, which impedes the development of novel vaccine strategies against gastrointestinal infections.
In general, induction of antibody responses upon natural infection or vaccination depends on antigen-presenting cells (APCs), such as dendritic cells (DCs), which present antigens to B cells in either an extrafollicular reaction or a germinal center (GC) reaction [6]. The short-lived extrafollicular reaction requires rapid T-cell help, leading to the generation of low-affinity antibodies [6, 7]. In contrast, the GC reaction induces clonal expansion and differentiation of antigen-specific B cells into highly efficient IgG-, IgA- or IgE-producing long-lived plasma cells, requiring help and survival signals from follicular DCs and follicular helper CD4+ T (TFH) cells (CXCR5+PD-1++ CD4+ T cells). The induction of CXCR5+PD-1++ CD4+ T cells by vaccines therefore is a critical denominator for the production of high-affinity antibodies by expanded B cell clones [8, 9].
Enhanced susceptibility to infections and reduced vaccine responses in young children are considered consequences of their relatively immature immune system, exemplified by the presence of mostly naïve CD4+ T cells in cord blood at birth with reduced capacity to stimulate B cells [10–13]. Adaptive immune responses in blood after birth are furthermore known for their tolerance-inducing capacity and regarded as a remnant of the fetal immune system to prevent proinflammatory T cell and antibody responses that could jeopardize the pregnancy outcome [1, 14]. However, recent studies from our group and others have shown that the fetal and infant immune system is highly compartmentalized, and that proinflammatory cytokine-producing effector memory CD4+ T cells are already present in the human intestine at birth [15, 16]. Thus, TFH cell ontogeny may also be compartmentalized. Previous studies have identified CXCR5+ CD4+ T cells in blood and tonsils of older children [9]; however due to limitations obtaining viable intestinal tissues of young infants, the development of TFH responses in human intestines in the first year of life is unknown.
Here, we demonstrate based on analyses of fetal, infant and adult intestines that the numbers of intestinal CXCR5+PD-1++ CD4+ T cells peak in infants and subsequently decline with age. Infant CXCR5+PD-1++ CD4+ T cells exhibited a tissue-resident memory phenotype, allowing retention in intestinal tissues. IL-21, critical for CD4+ T cell help to B cells, was produced by infant CD4+ T cells. Using autologous intestinal CXCR5+PD-1+/− CD4+ T cell-B cell co-cultures, we show that infant intestinal CXCR5+PD-1+/− CD4+ T cells effectively promote B cell maturation and antibody production. Together, our data show that, shortly after birth, functional TFH cells populate infant intestines and are equipped to efficiently promote B cell responses, providing important new rational for the design of oral vaccination strategies in newborn infants.
Materials and methods
Tissue samples
Human intestinal samples were obtained from individuals undergoing abdominal surgery at the Amsterdam University Medical Centre (AUMC) (Amsterdam, the Netherlands), the Altona Children’s Hospital (AKK) (Hamburg, Germany) and the University Medical Centre Hamburg-Eppendorf (UKE) (Hamburg, Germany) after written informed consent by the donors (adult) or by their legal guardians (children). Fetal specimens were obtained by the HIS Mouse (humanized immune system mouse) Facility of the AUMC, Amsterdam. All fetal samples were collected from donors from whom a written informed consent was obtained by the Bloemenhove clinic (Heemstede, the Netherlands) (gestational age: 15–20 weeks, n = 3). Pediatric (median age: 6 months, n = 40) and adult (median age: 56.5 years, n = 27) intestinal samples were collected at surgical intestinal reconstruction. Additional data is provided in Supplementary Table S1. PBMC samples were obtained from donors upon written informed consent. Blood and tissues were obtained in Amsterdam with approval of the ethical committee of the AUMC and tissues and blood were obtained in Hamburg with approval of the ethics committee of the medical association of the Freie und Hansestadt Hamburg (Ärztekammer Hamburg) and in accordance with the Declaration of Helsinki. Due to the limited numbers of cells isolated from the tissues not all samples could be used for all assays.
Lymphocyte isolation from blood and tissue samples
The tissue samples were kept at 4 °C in saline solution or Iscove’s modified Dulbecco’s Medium (IMDM; Thermo Fisher Scientific) supplemented with 5% fetal bovine serum (FBS) and processed within 12 h of collection. The lamina propria and intraepithelial lymphocytes (LPL and IEL, respectively) were isolated as described before [17, 18]. In sum, tissues were washed with phosphate buffered saline (PBS) until cleared of blood and feces and the muscular layer was removed. The intestinal mucosa was spread out in a petri dish and measured to document the surface area and subsequently calculate the cell yield/area. The mucosa was cut into small pieces (0.25–0.5 cm2) and incubated twice for 20 min at 37 °C with IMDM supplemented with 5 mM of ethylenediaminetetraacetic acid (EDTA; Sigma-Aldrich), 2 mM 1,4-dithiothreirol (DTT; Carl Roth GmbH Co. KG) and 1% FBS (Biochrom GmbH). The supernatant was further filtered with a 70 µm cell strainer and the single cell solution of IEL was obtained after a BIOCOLL (Biochrom AG) density gradient centrifugation step. The residual tissue was hashed and incubated twice at 37 °C for 30 min with IMDM supplemented with 1 mg/mL Collagenase D (Sigma-Aldrich), 1% FBS and 1000 U/mL DNAse I (STEMCELL Technologies). The resulting cell suspension was filtered through a 70 µm cell strainer and the single cell suspension of LPL was obtained after a 60% standard isotonic Percoll (Sigma-Aldrich) density gradient centrifugation step. The cell concentration and viability were determined using Trypan Blue (Sigma-Aldrich). Blood samples were collected in heparin-coated tubes. Using a density gradient, the mononuclear cell fraction was isolated. Hank’s Balanced Salt Solution (HBSS; Lonza; Sigma-Aldrich) was used to dilute blood 1:1 followed by layering on top of Lymphoprep (Axis-Shield; Capricorn Scientific) and centrifuged for 22 min at room temperature at 1000 G. Mononuclear cells were collected and washed with PBS.
T cell stimulation
Isolated lamina propria-derived cells from adult and infant samples were resuspended in 500 µL IMDM supplemented with 5% FBS and 1% Penicillin/streptomycin (Pen-Strep, Sigma-Aldrich) and stimulated with 40 ng/mL Phorbol 12-myristate 12-acetate (PMA, Sigma-Aldrich) and 1 µg/mL ionomycin calcium salt (iono, Sigma-Aldrich) at 37 °C and 5% CO2. Unstimulated cells were used as a control. At one hour post-stimulation, Brefeldin A (BFA, Sigma-Aldrich) was added in all conditions at a final concentration of 5 µg/mL. 12–14 h post stimulation, cells were recovered and stained with an antibody panel including antibodies against IL-21, TNF, IL-22, IL-8 and IFNγ (see Supplementary Table S2 for list of antibodies). The cytokine production was measured within the population defined as viable CD45+CD3+CD4+CD8− in the unstimulated cells, and within the viable CD45+CD3+CD4+/−CD8− cells in PMA/iono stimulated cells. As shown previously, the CD3+CD8- populations contained low numbers of non-CD4+ cells in PMA/ionomycin-stimulated samples [16]. The downregulation of CXCR5 upon stimulation prevented to perform analyses on CXCR5+PD-1++ CD4+ T cells. Due to the limited numbers of CXCR5+PD-1++ CD4+ T cells especially in adults, cell sorting of the intestinal CXCR5+PD-1++ CD4+ T population did not result in sufficient numbers of cells for reliable cell stimulations to assess cytokine production in sorted cells.
Flow cytometry analyses
The surface and intracellular expression of molecules by isolated epithelial and lamina propria-derived cells was analyzed by flow cytometry. All antibodies used in the study are listed in Supplementary Table S2. Briefly, cells were collected in a 96-well V plate and washed once with PBS. Cells were resuspended in 30 µL of a master mix containing PBS, the appropriate antibodies and the Zombie Aqua Fixable dye or near infrared (NIR) dye and incubated on an orbital shaker at 4 °C in the dark for 30 min. After a washing step with PBS, cells were fixed with 100 µL BD CellFix (BD Biosciences). For intracellular staining, cells were fixed instead with 150 µL of 1:4 eBioscience IC Fixation Buffer diluted in eBioscience Fixation/Permeabilization Diluent (Thermo Fisher Scientific) at 4 °C for 15 min in the dark. After a washing step with PBS, cells were permeabilized with 30 µL eBioscience Permeabilization Buffer containing the intracellular antibodies for 30 min at 4 °C in the dark. After a washing step, cells were resuspended in 100 µL PBS. Flow cytometry analyses were performed using a LSR Fortessa Flow Cytometer (BD Biosciences) and FACSDIVA software (version 8; BD Biosciences) or using a 5-laser Cytek Aurora Flow Cytometer (Cytek Biosciences) and SpectroFlo software (Cytek Biosciences) within 24 h after staining. The data was analyzed with FlowJo software (version 10.5.0; Treestar).
FACS sorting of lymphocyte subsets
Intestinal lymphocytes were surface stained as described above. Total B cell populations (viable singlets CD45+CD3−CD19+) were isolated using a 5-laser FACS Aria-Fusion (BD Biosciences). Similarly, naive B cells (viable singlets CD45+CD3-CD19+CD20+CD27−IgD+) were obtained. Autologous CXCR5+PD-1+/− CD4+ T cells (viable singlets CD45+CD3+CD4+CXCR5+PD-1+/−) or CXCR5−CD4+ T cells (viable singlets CD45+CD3+CD4+CXCR5-) were isolated from intestinal LPLs after surface-staining and using a 5-laser FACS Aria-Fusion (BD Biosciences). For BCR RNA analyses, cells were sorted directly in 250 µL Buffer RLT (Qiagen) supplemented with 1% β-mercaptoethanol (Sigma-Aldrich) and stored at −80 °C until RNA extraction following the manufacturer’s instructions. For qPCR analyses, cells were sorted in 2 mL of IMDM supplemented with 10% FBS and 1% Pen-Strep, centrifuged, resuspended in Trizol and stored at −80 °C until RNA extraction. For autologous co-cultures, cells were sorted in 2 mL of IMDM supplemented with 10% FBS and 1% Pen-Strep.
Co-culture of B cells and CXCR5+PD-1+/− CD4+ T cells
Sorted CXCR5+PD-1+/− CD4+ T cells and CXCR5−CD4+ T cells from infant samples were activated with a combination of a-CD3 (1.5 µg/mL) and a-CD28 (4 µg/mL) antibodies (Sanquin). They were co-cultured at 37 °C for 6 days at a 1:2 ratio with isolated total intestinal B cells (CD45+CD3-CD19+) or intestinal naive B cells (CD45+CD3−CD19+CD20+CD27−IgD+) from the same infant intestinal tissue, in IMDM supplemented with 10% FBS and 1% Pen-Strep. At day 6, cells were harvested for analyses of surface and intracellular B cell characteristics using flow cytometry.
Enzyme-linked immunosorbent assay (ELISA)
IgG and IgA levels in culture supernatants were measured using Human IgG Total ELISA Kit and Human IgA ELISA Kit, correspondingly (Invitrogen), according to the manufacturer´s instructions. Briefly, 100 μl or 20 μl of culture supernatant were added to the anti-IgG or anti-IgA pre-coated 96-well strip plates, followed by the addition of 50 μl of diluted HRP-conjugated antibody. The plates were covered and incubated at room temperature for 1 h. After washing, 100 μl of TMB substrate was added and the plates were incubated for 30 min at room temperature. The enzyme reaction was stopped by adding 100 μl of the Stop Solution. Absorbance was read at 450 nm with the Safire2™ microplate reader (Tecan Technologies). A standard curve was run on each plate and sample concentrations were interpolated from them.
RNA extraction for qPCR
Trizol-resuspended CXCR5+PD-1++ CD4+ T samples were thawed on ice and chloroform was added to extract the RNA from the cells. To precipitate the RNA, samples were incubated overnight at −20 °C with isopropanol. After a centrifugation step, the supernatant of the precipitate, containing the RNA, was washed first with ethanol at 75% and subsequently resuspended with diethyl pyrocarbonate-treated water (DEPC water). qPCR was performed as previously described [19]. Briefly, cDNA synthesis was performed using the qScriptTM cDNA SuperMix kit (QuantaBiosciences) and qPCR was performed using the Quantifast SYBR Green supermix (Qiagen) in a LightCycler® 96 System (Roche), according to the manufacturer’s instructions. Bcl-6, IL-21, BLIMP-1, STAT3, GAPDH and ACTA expression levels were measured by qPCR. The threshold cycle (Ct) was calculated using the mean of two technical duplicates and normalized to a stable housekeeping mRNA (either ACTA or GAPDH) [20].
B cell receptor (BCR) sequencing
The extracted RNA from B cells was used to synthetize cDNA and the quantitative amplification of the BCR heavy chain repertoire was performed as described earlier [21, 22] using an adaptation for UMI corrected next-generation sequencing on the Illumina MiSeq platform. Bioinformatics analysis of BCR heavy chain repertoire was performed as described [22, 23]. The number of high quality recovered BCR sequences and number of unique BCR clones per sample was a minimum of 16,217 and 1149, respectively (Table S3). A BCR clone was defined as a set of BCR heavy chain sequences with 100% amino acid homology of the CDR3 sequence. A dominant clone was defined as a clone of which the number of BCR reads exceeded 0.5% of the total repertoire [21, 22]. The mutation rate within the V-region was defined as the number of mutated nucleotides divided by the total number of sequenced nucleotides. To determine the mutation rate of a clone, the mean mutation rate of all BCR sequences that belonged to the same clone was taken.
scRNAseq of intestinal and blood-derived CD4+ T cells
CD4+ T cells from infant intestines (n = 3), adult intestines (n = 3), infant blood (n = 2) and adult blood (n = 3) were obtained and isolated as previously described [16]. In short, intestinal and blood-derived mononuclear cells were surface stained and CD4+ T cells (viable CD45+CD3+CD4+), using a FACS Aria IIu SORP (BD Biosciences) and FACSDIVA software (BD Biosciences), were sorted directly into a 384-well plate provided by the single cell sequencing facility (Hubrecht Institute, Utrecht, the Netherlands). Cells were directly dissolved in primer mix. Single cell libraries were prepared using the CEL-Seq2 protocol as previously described [24]. cDNA libraries were sequenced on an Illumina NextSeq500 using 75 bp paired-end sequencing (Utrecht Sequencing Facility).
Analyses of CD4+ T cell scRNAseq data
Building on our previous studies [16] new analyses were performed. Reads obtained from scRNAseq analyses of CD4+ T cells were mapped and deconvoluted into single cell transcriptomes using the zUMIs pipeline (version 2.9.4) [25]. This approach included mapping with STAR 2.6.0c and counting with Rsubread 1.32.4. GRCh38 was used as a reference genome. All further analysis steps were performed in R 4.1.0 and using the Seurat framework (version 4.0.3) [26] and Tidyverse (version 1.3.1) [27]. Cells with less than 100 detected genes were removed. Next, all cells were combined into one Seurat object. Genes were filtered to remove pseudogenes and ribosomal protein encoding genes. Expression count data was normalized using the NormalizeData function and scaled with ScaleData. In total 563 infant intestinal, 635 infant blood, 290 adult intestinal and 817 adult blood CD4+ T cells were part of the final dataset. As a basis to distinguish T helper cell subtypes, geneset enrichment analysis was performed using positive and negative marker gene signatures for T helper type 1 (Th1), T helper type 2 (Th2), T helper type 17 (Th17), T regulatory cells (Treg) and T follicular helper cells (TFH). These signatures are described in Kaufmann et al. [28] and were derived from publicly available bulk RNA sequencing data from T helper cell subtypes sorted from adult blood [29]. Geneset enrichment analysis was performed using AUCell (version 1.14.0) [30]. Signatures tested for enrichment were split in positive and negative marker gene sets. AUC values were then derived separately for positive and negative marker gene sets and the ratio of AUCpositive markers / AUCnegative markers (AUC ratio) was used to determine overall signature enrichment as described previously [28]. To determine the relative position of each cell in the T helper cell differentiation space defined by these signatures, a principal component analysis (PCA) and subsequent clustering using a shared nearest neighbor (SNN) modularity optimization-based algorithm (implemented in the Seurat package) was applied to the AUC ratio matrix with one row for each of the five signatures and one column for each single cell in the dataset. In total, 12 clusters were identified. To visualize the data, uniform manifold approximation and projection (UMAP) was performed. Sufficient integration between groups as a prerequisite for identifying T helper subtypes was validated by projecting sample type identities on the UMAP plot. The relationship between cluster identities and T helper subtype signatures was investigated by projecting the enrichment of each signature on the same UMAP plot. The cluster containing the majority of TFH cells (cluster 7) was further investigated for transcriptomic differences between groups. For this purpose, differential gene expression analysis was performed between the following groups: infant intestine and adult intestine, infant blood and infant intestine, adult blood and adult intestine. Differential gene expression analysis was performed as previously described [28]. In brief, single cell transcriptomes were aggregated as pseudo-bulks for all cells in cluster 7 for each donor. Statistical testing was performed with DEseq2 [31] version 1.32.0 using default settings including FDR-adjustment of p values. Results were visualized as volcano plots contrasting adjusted p values with log2 (fold changes) and boxplots of aggregated expression values for each donor.
Immunohistochemistry (IHC)
Infant and adult tissue samples were fixed with formalin, embedded in paraffin blocks, cut in 5 µm sections and placed on adherent slides. For single antibody stainings, sections were dewaxed, rehydrated and stained in an automatic immunostainer (Benchmark Ultra; Ventana Medical Systems) for Bcl-6, IgA and CD20 (all from DAKO), using a 3,3′-diaminobenzidine detection kit (OptiViewDAB; Ventana Medical Systems). Cell nuclei were counterstained with hematoxylin II and bluing agent (both Ventana Medical Systems). For double antibody stainings, antigen retrieval was performed at 121 °C for 10 min in citrate buffer at pH6. Primary antibody labeling was performed with Bcl-6 (DAKO), followed by secondary antibody labeling with anti-mouse IgG streptavidin alkaline phosphatase (AP). AP activity was visualized with Chromogene Red (DAKO). Red staining was left untouched by washing with TBS-Tween. This step was followed by primary antibody labeling with CD3 (Invitrogen) or CD20 (Abcam). Secondary antibody labeling was performed with biotin-conjugated anti-rabbit IgG and AP activity was visualized with Vector Blue (Biozol). Tissue slides were visualized using a Nikon Eclipse NI microscope equipped with a DS-Ri2 camera and 10×, 20× and 40× objectives, and ImageJ (v1.53 s) was used to analyze the signals.
Analyses of IHC images
For quantifying the ratio of cells stained with DAB, a custom written Matlab app was used (see https://github.com/rolandth/Histoquant for source code). Briefly, the app carries out a background subtraction by adaptive image thresholding using local first-order statistics. Then the app allows to choose regions of interest for which the area covered by cells stained with brown and the area covered by all cells (cells stained with brown and blue) is determined. Cells stained with blue only are distinguished from cells stained with brown by having a value larger than 120 for blue in the 8 bit RGB color space. Finally, objects smaller than 100 pixels are removed by an area opening procedure.
Statistical analysis
Statistical analyses were done using GraphPad Prism v7.03 (GraphPad Software). Non-parametric unpaired Mann–Whitney U Test, Wilcoxon matched-pairs signed rank Test, Wilcoxon signed rank Test or Welch’s t-test were used to analyze the data and statistical significance was considered when p < 0.05. In figures and text, median frequencies, interquartile ranges (IQR) are stated and only statistically significant p values are depicted. Samples with CXCR5+PD-1++ CD4+ T cells lower than 10 were excluded from the analysis.
Results
Increased numbers of CXCR5+PD-1++ CD4+ T cells populate infant intestines and decline with age
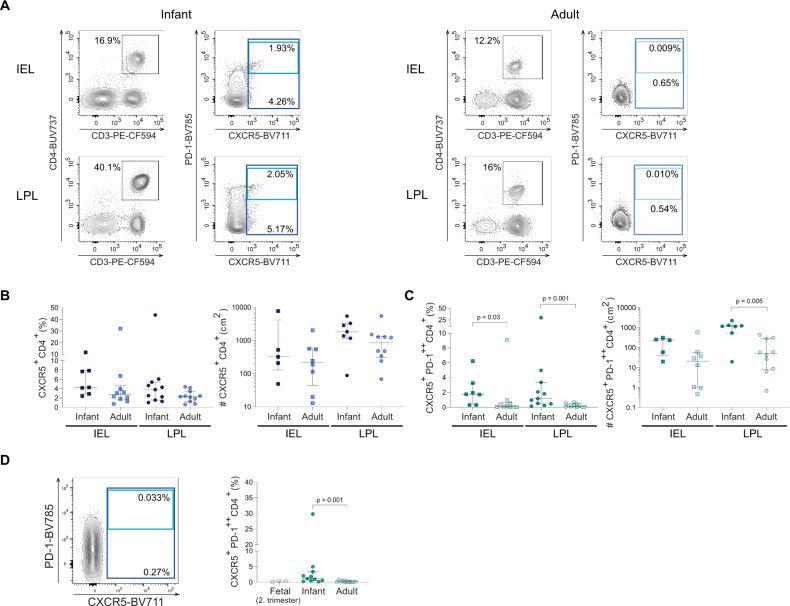
CXCR5 is one of the main chemokine receptors directing B and T cells into close proximity and promoting the interaction between B cells and T cells in tissues [32]. As the numbers of intestinal effector memory CD4+ T cells rapidly change during human development [16], we determined the proportion and absolute numbers of intraepithelial- and lamina propria-derived CXCR5+ CD4+ cells (IEL and LPL respectively) isolated from non-inflamed infant and adult intestinal tissues using flow cytometry (Fig. 1A, Fig. S1A, B). CXCR5+ CD4+ T cells were detected both in infant and adult intestinal tissues (Fig. 1B). Epithelial CXCR5+ CD4+ T cells in infants represented 4.3% (IQR: 2.9–7.9%) of the total CD4+ T cell population in the intestinal epithelium compared to 2.7% (IQR: 1.7-4.6%) of the epithelial CD4+ T cells in adults. A similar trend was observed for CXCR5+ CD4+ T cells derived from the lamina propria, with 3.8% (IQR: 1.6-5.4%) detected in infant and 2.3% (IQR: 1.3–3.5%) in adult intestines (Fig. 1B left). Similarly, the median absolute number of lamina propria-derived CXCR5+ CD4+ cells per cm2 was 1830 cells (IQR: 1167–3275) in infant compared to 851 cells (IQR: 316–1333) in adult intestinal tissues (p = 0.16) (Fig. 1B right).
Fig. 1.
Increased numbers of CXCR5+PD-1++ CD4+ T populate infant intestines and decline with age. A Representative flow cytometric plots of the gating strategies used to identify CXCR5+PD-1++ CD4+T cells in intraepithelial lymphocytes (IEL) and lamina propia lymphocytes (LPL) in infant (dark blue) and adult intestines (light blue). B Frequencies (%) and cell counts/cm2 of epithelial and lamina propria-derived CXCR5+ CD4+ T cells in infant and adult intestines. C Frequencies (%) and cell counts/cm2 of epithelial and lamina propria-derived CXCR5+PD-1++ CD4+ T cells in infant and adult intestines. D Representative flow cytometric plot of fetal intestinal CXCR5+PD-1++ CD4+ T cells and frequencies (%) of lamina propria-derived CXCR5+PD-1++ CD4+ T cells in fetal compared to infant and adult samples. Medians and interquartile ranges (IQRs) are depicted in all figures. (Infant IEL % n = 7; adult IEL % n = 10; infant IEL cell counts/cm2 n = 5; adult IEL cell counts/cm2 n = 8; infant LPL % n = 11; adult LPL % n = 11; infant LPL cell counts/cm2 n = 7; adult LPL cell counts/cm2 n = 10; fetal LPL % n = 3). Mann–Whitney test was used for comparisons. Only significant p values are shown
CXCR5+PD-1++ CD4+ T cells, which are phenotypic CD4+ T follicular helper (FH) cells, were next quantified in human intestines at different ages. Epithelial CXCR5+PD-1++ CD4+ T cells were significantly increased in infant compared to adult intestinal tissues (median: 1.7%, IQR: 0.3–3.3% (infant); median: 0.1%, IQR: 0.03-0.6%, (adults) (p = 0.03)) and in lamina propria tissues (median: 1.2%, IQR: 0.5–3.4%, (infants); median: 0.1%, IQR: 0.03–0.5% (adults) (p = 0.001)) (Fig. 1A, C left). The absolute numbers of lamina propria-derived CXCR5+PD-1++ CD4+ T cells were also significantly increased in infant (median: 1177 cells/cm2) compared to adult intestines (median: 50.3 cells/cm2) (p = 0.005) (Fig. 1C right). The increased numbers of CXCR5+PD-1++ CD4+ T cells in infants led us to quantify CXCR5+PD-1++ CD4+ T cell frequencies during early human developmental stages. Analyses of fetal intestinal tissues showed an absence of CXCR5+PD-1++ CD4+ T cells (Fig. 1D). Moreover, shortly after birth up to one month of age, frequencies of intestinal CXCR5+PD-1++ CD4+ T cells were very low (median: 0.03%; IQR: 0–0.2%), compared to older infants (median: 1.2%, IQR: 0.5–3.4%; p = 0.001) (Fig. S1C, D). Taken together, CXCR5+PD-1++ CD4+ T cells rapidly increased after birth resulting in significantly larger numbers in infant intestines compared to adults.
Infant intestinal CXCR5+PD-1++ CD4+ T cells exhibit a tissue-resident memory phenotype
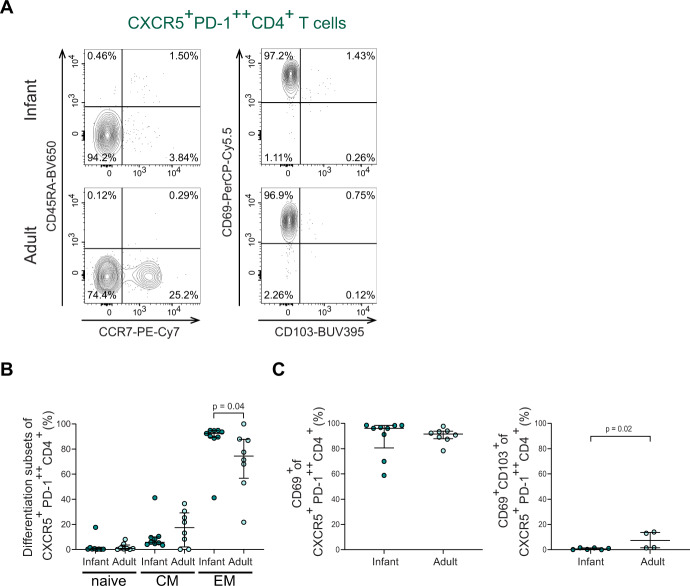
As the intestinal lamina propria lymphocytes (LPL) contained the largest population of CXCR5+PD-1++ CD4+ T cells, and in this layer T cells can have close contact with B cells, the following analyses focused on infant and adult LPL-derived CXCR5+PD-1++ CD4+ T cells. Within the CXCR5+PD-1++ CD4+ T cell population, the majority of CXCR5+PD-1++ CD4+ T cells in infants and adults consisted of TEM (effector memory CCR7-CD45RA-), with the largest frequencies of TEM observed in infant intestines (Fig. 2A, B). Thus, infant and adult intestinal CXCR5+PD-1++ CD4+ T cells exhibit an effector memory phenotype.
Fig. 2.
Infant intestinal CXCR5+PD-1++ CD4+ T cells exhibit a tissue-resident memory phenotype. A Representative flow cytometric plots to define i) left: memory subsets based on CCR7 and CD45RA expression in CXCR5+PD-1++ CD4+ T cells and ii) right: CD103 and CD69 expression on CXCR5+PD-1++ CD4+ T cells. B Frequencies (%) of naive (CCR7+CD45RA+), central memory (CCR7+CD45RA−) and effector memory (CCR7-CD45RA−) T cell populations in CXCR5+PD-1++CD4+ T cells comparing infant and adult intestines. C Frequencies (%) of CD69+ cells and CD69+CD103+ cells within CXCR5+PD-1++ CD4+ T cells comparing infant and adult intestines. Medians and IQRs are depicted in all figures. (For B and C left: infant n = 9; adult n = 8. For C right: infant n = 6; adult n = 4). Mann–Whitney test was used to compare infant and adult samples. Only significant p values are shown
Efficient long-term adaptive immune protection in tissues is mediated by tissue-resident T cells (TRM). Retention of CD4+ T cells in intestinal tissues is mediated by the surface molecule CD69 and to a lesser extent by CD103 [33–35]. Most CXCR5+PD-1++ CD4+ T cells expressed CD69 in infant (median: 96.1%; IQR: 80.6–98.4%) and adult intestines (median: 91.6%; IQR: 87.9–93.9%) (Fig. 2C left). CD69 and CD103 co-expression was lower in infant CXCR5+PD-1++ CD4+ T cells compared to adults (Fig. 2C right). To determine whether differences exist within CXCR5+PD-1++ CD4+ TCM and TEM cells, we furthermore analyzed CD69 and CD103 expression within these cell subsets. Most CXCR5+PD-1++ CD4+ TCM (central memory CCR7+CD45RA−) and TEM cells expressed CD69 in infants and adults (Fig. S1E). Thus, infant intestinal CXCR5+PD-1++ CD4+ T cells have an effector memory tissue-resident phenotype, allowing compartmentalization of CXCR5+PD-1++ CD4+ T cell responses in infant intestines.
Infant intestinal CXCR5+PD-1++ CD4+ T cells display a TFH signature
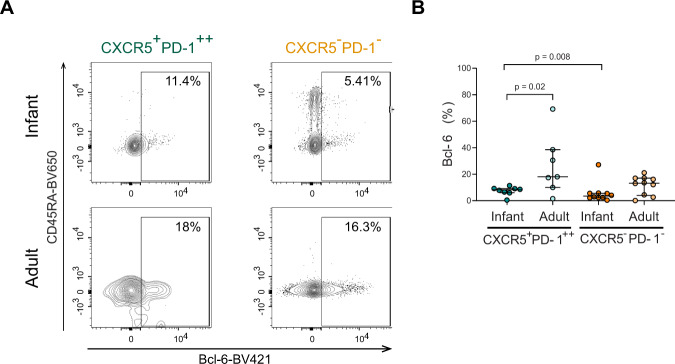
The transcription factor Bcl-6 mediates differentiation into TFH cells [36]. We therefore assessed Bcl-6 expression within the CXCR5+PD-1++ population and compared with expression in CXCR5−PD-1- CD4+ T cells (Fig. 3A). Indeed, infant CXCR5+PD-1++ CD4+ T cells had a higher expression of Bcl-6 compared to infant CXCR5−PD-1- CD4+ T cells (p = 0.008) (Fig. 3B). Compared to adults, Bcl-6 expression in infant CXCR5+PD-1++CD4+T cells was slightly lower (Fig. 3B). mRNA levels of Bcl-6 and of transcription factors involved in TFH differentiation, Blimp-1 and Stat3 [36], did not differ significantly between sorted infant and adult TFH cells (Fig. S2A, B). In sum, infant intestinal CXCR5+PD-1++ CD4+ T cells have higher Bcl-6 expression compared to CXCR5−PD-1− CD4+ T cells, however age-dependent differences may exist. CD4+ T regulatory cells (Tregs) are inhibitory regulators of TFH and B cells [37]. Specifically, TFH regulatory cells have been described, which express CXCR5 and can impact B cell maturation [38]. We measured in intestinal infant and adult CD4+ T cell subsets the expression of CD25, CD127 and FoxP3 (Fig. S2C–F). Overall Treg populations were increased in infant intestines; however, infant and adult CXCR5+PD-1++ CD4+ T cells had similarly low proportions of CD25+CD127− and FoxP3+ cells (Fig. S2C–F). Thus, although Treg responses are increased in infants, this does not translate to increased numbers of infant intestinal TFH regulatory cells.
Fig. 3.
Infant intestinal CXCR5+PD-1++ CD4+ T cells have a TFH phenotype. A Representative flow cytometric plots of the expression levels of Bcl-6 in CXCR5+PD-1++ CD4+ T cells and CXCR5−PD−1−CD4+ T cells in infant and adult intestines. B Frequencies (%) of Bcl-6 expressing intestinal CXCR5+PD-1++ cells (green) and CXCR5−PD-1− CD4+ T cells (orange) in infants and adults. (Infant CXCR5+PD-1++ % n = 8; infant CXCR5−PD-1− % n = 10; adult CXCR5+PD-1++ % n = 7; adult CXCR5−PD-1− % n = 10). Medians and IQRs are depicted in all figures. Mann–Whitney test was used to compare infant and adult samples, and Wilcoxon matched-pairs signed ranked test was used to compare CXCR5+PD-1++ and CXCR5−PD-1− CD4+ T cells. Only significant p values are shown
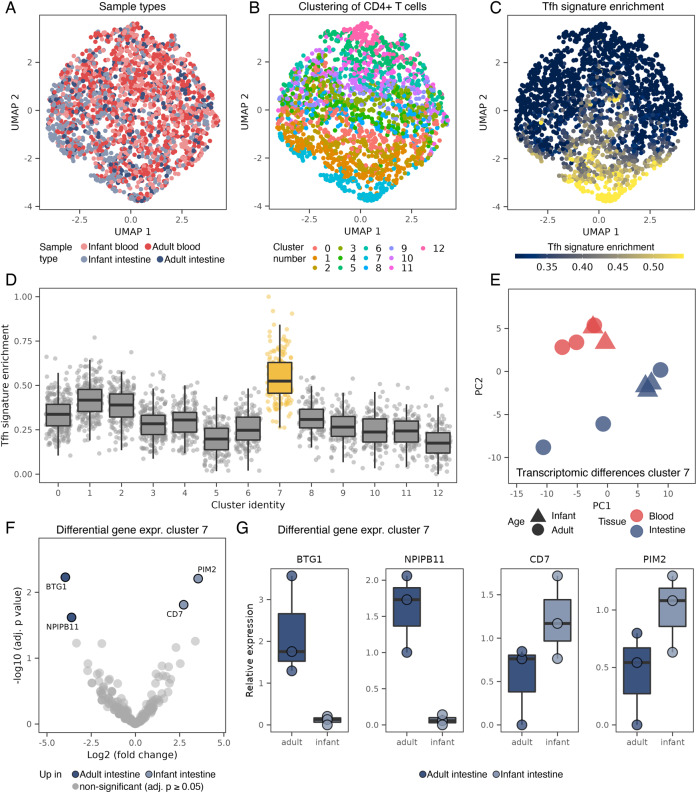
In order to further characterize intestinal TFH cells in infant intestines and compare to populations in adult intestines as well as blood, we performed single cell RNA sequencing (scRNAseq) analyses of infant and adult intestinal and blood-derived CD4+ T helper cells isolated by FACS sorting [16]. A total of 2 305 CD4+ T cells from infant and adult intestines and blood were analyzed (Fig. 4A). To facilitate distinguishing TFH and other T helper cell subtypes in the data, we performed a supervised dimensional reduction based on published transcriptomic signatures of major T helper cell subtypes [28, 29]. As expected after data integration the cells did not cluster by tissue type (Fig. 4A). Instead, 12 groups of T helper cell subtypes were identified using a shared nearest neighbor (SNN) modularity optimization-based clustering algorithm (Fig. 4B). These clusters enriched the different T helper cell subtype signatures to various degrees (Fig. S3B–F). As the only cluster, cluster 7 was highly enriched for the TFH signature (Fig. 4C, D) and did not overlap with other T helper subtypes (Fig. S3B–F), suggesting that the majority of TFH cells in the dataset are detected in this cluster. Cluster 7 contributed 7% to all CD4+ T cells across conditions, which was in the same range as we observed for CXCR5+ CD4+ T cells using flow cytometric analyses shown in Fig. 1. Next the transcriptomic profiles of TFH cells (cluster 7) from the four different sample types (infant and adult intestinal and blood compartments) were compared using differential gene expression analysis. Transcriptomic differences were more pronounced between cells derived from the intestinal and blood compartment than between infant and adult cells from the same compartment (Fig. 4E). The resemblance between transcriptomes of infant and adult intestinal TFH cells was highlighted by the additional observation that only 4 genes passed statistical significance for differential expression between infant and adult intestinal TFH cells in cluster 7 (Fig. 4F). These differentially regulated genes included PIM2, which was increased in infant intestinal TFH cells (Fig. 4F, G). PIM2 has been shown to modulate cytokine production by CD4+ T cells [39]. BTG1 on the other hand was more strongly expressed by adult compared to infant intestinal TFH cells (Fig. 4F, G). BTG1 is recently identified as a critical inhibitor of effector functions of T cells [40]. BTG1 was also decreased in intestinal infant TFH cells compared to blood-derived infant TFH cells potentially indicating reduced effector functions of cells in the blood compared to intestines. SELL, a molecule directing lymphoid tissue homing, was increased on both infant and adult blood-derived TFH cells compared to intestinal TFH cells providing the conditions to promote migration to lymphoid tissues of blood-derived TFH cells (Fig. S3G–I). Taken together, infant intestinal TFH cells display a very similar transcriptomic signature when compared to adult intestinal TFH cells, and to a lesser extend to infant circulating TFH cells.
Fig. 4.
Infant and adult intestinal TFH cells share a similar transcriptomic profile. A UMAP plot of CD4+ T cells analyzed by scRNAseq from infant intestinal (n = 3 donors), adult intestinal (n = 3 donors), infant blood (n = 2 donors) and adult blood (n = 3 donors) samples. In total, 2305 cells were analyzed. B Identification of 12 clusters of T helper subtypes using a shared nearest neighbor (SNN) modularity optimization-based clustering algorithm. C Enrichment of published TFH signature [28, 29] (based on sorted CXCR5+CD4+ T cells) projected on the UMAP plot. D Quantification of the TFH signature in single cells, separately for each cluster from B. Cluster 7 stands out as the cluster with the most prominent enrichment of the TFH signature. E Principal component analysis (PCA) of the transcriptome of TFH cells (cluster 7) aggregated for each donor. F Differentially expressed genes between infant and adult intestinal TFH cells (cluster 7), showing only four genes significantly differentially regulated after multi comparison correction (statistical testing using DEseq2). Comparisons between the remaining sample types are shown in Fig. S3G–I. G Expression of significantly differentially expressed genes from F, based on aggregation of single cells in cluster 7 for each donor. Box plots in D and G centered on median, bounds defined between the 25th and 75th percentile with minimum and maximum defined as median ± 1.5 × interquartile range and whiskers extending to the lowest/highest value within this range
Infant intestinal CD4+ T cells produce IL-21 and TNF
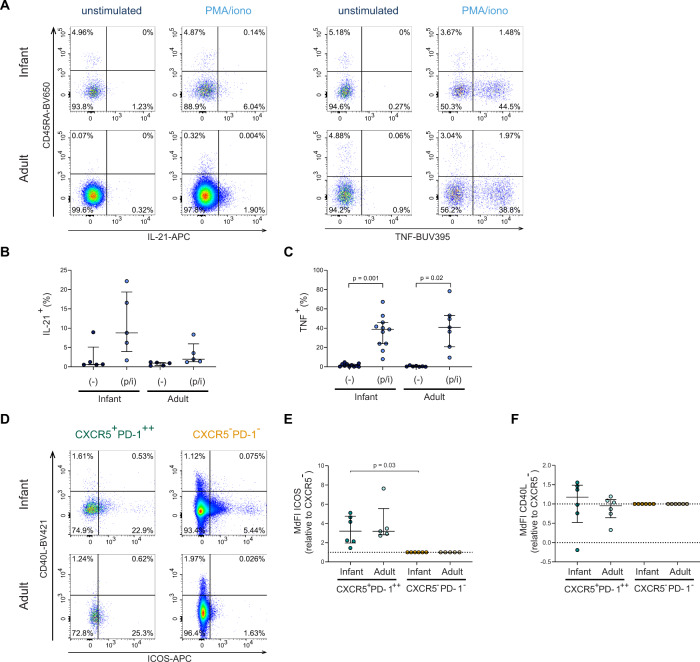
To determine functional capacity, cytokines produced by CD4+ T cells were analyzed after stimulation with phorbol 12-myristate 13-acetate (PMA) and ionomycin (Fig. 5A, S4A). PMA and ionomycin stimulation does not allow for the specific identification of CXCR5+PD-1++ CD4+ T cells due to PD-1 and CXCR5 down-modulation, therefore total infant and adult intestinal CD4+ T cell populations were analyzed. Upon stimulation, IL-21 production, the hallmark cytokine of TFH cells, increased in infant intestinal CD4+ T cells (median unstimulated: 0.7%, median stimulated: 8.8%). Adult intestinal CD4+ T cells also produced IL-21 albeit less (median unstimulated: 0.9%, median stimulated: 1.9%) (Fig. 5B). IL-21 mRNA analyses of isolated unstimulated infant intestinal CXCR5+PD-1++ CD4+ T cells was detectable and showed similar findings compared to adult intestinal CXCR5+PD-1++ CD4+ T cells (Fig. S4C). TNF, which activates B cells, was barely detected at baseline in unstimulated intestinal CD4+ T cells (Fig. 5C), and was significantly increased upon stimulation in infant intestinal CD4+ T cells (median unstimulated: 1.4%, median stimulated: 38.7%; p = 0.001), similar to adult cells. Infant CD4+ T cells have further been shown to produce IL-8 [41], especially in intestines. Indeed IL-8, was significantly higher produced in infant intestinal CD4+ T cells (p = 0.03) (Fig. S4B). In sum, CD4+ T cells in infant intestines were capable to produce cytokines known to activate B cells.
Fig. 5.
Infant intestinal CD4+ T cells produce IL-21 and TNF. A Representative flow cytometric plots of viable infant and adult intestinal CD4+ T cells (CD4+ CD3+, unstimulated; CD3+CD8-, PMA/ionomycin-stimulated) lamina propria-derived lymphocytes showing IL-21 and TNF expression. B Frequencies (%) of infant and adult IL-21+ CD4+ T cells in unstimulated (−) and stimulated (p/i) conditions. C Frequencies (%) of infant and adult TNF+ CD4+ T cells in unstimulated (-) and stimulated (p/i) conditions. D Representative flow cytometry plots showing ICOS expression and CD40L expression on CXCR5+PD-1++ CD4+ T cells (green) and CXCR5−PD-1− CD4+ T cells (orange) in infant and adult intestines. E ICOS expression of CXCR5+PD-1++ CD4+ T cells relative to the sample corresponding CXCR5−PD-1− CD4+ T cells. F CD40L expression of CXCR5+PD-1++ CD4+ T cells relative to the sample corresponding CXCR5−PD-1− CD4+ T cells. Medians and IQRs are depicted in all figures. (For B infant n = 5; adult n = 5. For C infant n = 11; adult n = 7. For E infant n = 6; adult n = 5. For F infant n = 6; adult n = 6). Wilcoxon matched-pairs signed rank test was used to compare unstimulated/stimulated samples, Mann-Whitney test was used to compare cytokine productions by infant and adult cells; and Wilcoxon signed rank test was used to compare ICOS and CD40L expression by CXCR5+PD−1++ and CXCR5−PD-1− CD4+ T cells. Only significant p values are shown
Infant and adult CXCR5+PD-1++ CD4+ T cells express costimulatory molecules
CD4+ TFH cells can also provide B cell help by direct cell contact via ICOS and CD40L receptors [42]. Infant intestinal CXCR5+PD-1++ CD4+ T cells had a higher expression of ICOS compared to CXCR5−PD-1− CD4+ T cells (Fig. 5D, E). CD40L expression did not differ between infant CXCR5+PD-1++ CD4+ T cells and CXCR5−PD-1− CD4+ T cells (Fig. 5F). In sum, CXCR5+PD-1++ CD4+ T cells express ICOS in infant intestines, enhancing the possibilities for infant intestinal CXCR5+PD-1++ CD4+ T cells to provide B cell help.
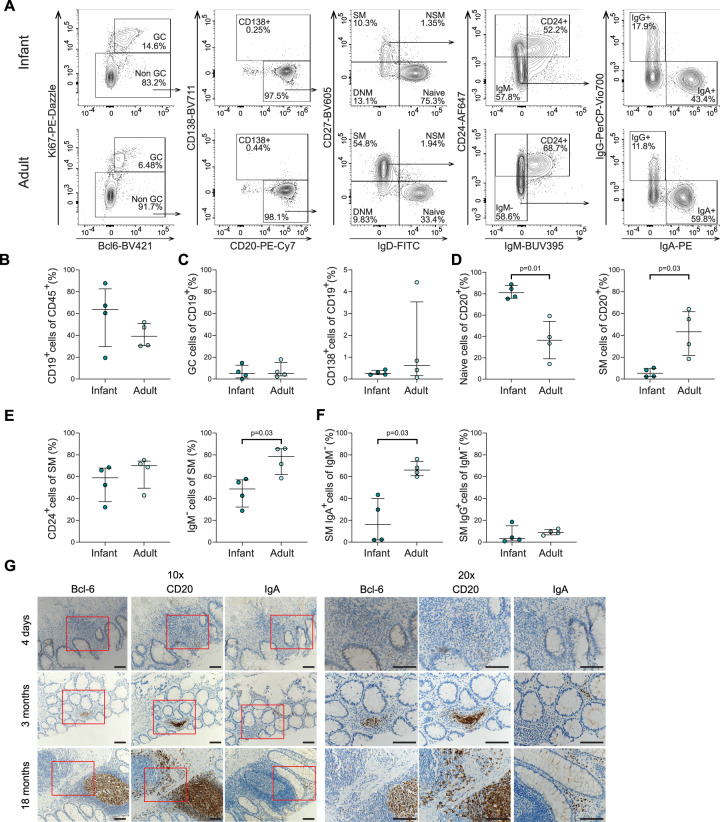
Memory B cells and plasma cells are present in infant intestines
Based on the large population of CXCR5+PD-1++ CD4+ T cells in infant intestines and their documented role in B cell maturation [38, 43], we next performed immune phenotyping of B cells in infant and adult intestines to assess intestinal B cell maturation during human development (Fig. 6A, Fig. S5A). Similar frequencies of CD19+ B cells were observed in infant (median: 64%; IQR: 30–83%) and adult intestines (median: 39%; IQR: 31–51%) (Fig. 6B) as well as frequencies of germinal center (GC) Ki67+Bcl-6+ and CD138+ plasma cells (Fig. 6C). Assessment of B cell differentiation showed that frequencies of naive B cells (CD27−IgD+) were significantly increased in infant intestines compared to adults (p = 0.01) (Fig. 6D left). In line with these observations, switched memory (SM) CD27+IgD− B cells were significantly reduced in infant intestines compared to adults (p = 0.03) (Fig. 6D right) as well as IgM- and IgA+ cells within the SM B cells (Fig. 6E right, 6F left). IgG+ cells within the SM B cells were, however, similar between infants and adults (Fig. 6F right). As antibody secreting plasma cells (CD138+) can lose CD19 expression [44], the expression of CD138, IgM, IgA and IgG cells amongst all CD45+ cells was determined (Fig. S5B). Although no significant differences were observed in the frequencies of IgM+, IgG+ and CD138+ CD45+ cells between infant and adults intestines, IgA+ cells were significantly decreased in infants (p = 0.03) (Fig. S5C). In line, IgA+ and IgG+ cells per cm2 intestine also showed a trend towards lower absolute numbers in infants (IgA median: 2107 cells/cm2; IgG median: 870 cells/cm2) when compared to adults (IgA median: 7506 cells/cm2; IgG median: 1149 cells/cm2) (Fig. S5D). In sum, these findings demonstrate that B cells in infant intestines have an increased naive phenotype; however B cell maturation towards IgG and IgA expressing cells does take place in human intestines early in life.
Fig. 6.
Memory B cells in infant intestines. A Representative flow cytometric plots of the gating strategies showing B cell maturation in infants (upper plots) and adults (lower plots). SM switched memory, NSM non-switched memory, DNM double negative memory. B Frequencies (%) of CD19+ of total CD45+ cells in infants and adults. C Frequencies (%) of GC (germinal center; Bcl-6+Ki67+) and CD138+ cells of total CD19+ cells in infants and adults. D Frequencies (%) of naive and SM cells of total CD20+ cells in infants and adults. E Frequencies (%) of CD24+ and IgM- of total SM cells in infants and adults. F Frequencies (%) of IgA+ and IgG+ of total IgM- cells in infants and adults. G Representative immunohistochemistry (IHC) images of tissue samples from a newborn infant (upper panel), a 3 month-old (middle panel) and 18 month-old infant (lower panel) showing the expression levels of the transcription factor Bcl-6, CD20 and IgA. Images are representative of the analyses of 13 intestinal samples. Scale bars indicate 0.1 mm. Medians and IQRs are depicted in all figures. (For B, C, D, E, F infant n = 4; adult n = 4.). Mann–Whitney test was used for comparisons. Only significant p values are shown
Next to phenotypic changes associated with B cell differentiation, clonal expansion of B cells is a further indication of B cell maturation. To assess whether this occurred in vivo and led to the induction of expanded B cell clones at this time of human development, we performed next-generation sequencing of the BCR repertoire of sorted CD19+ B cells from infant (median age: 5 months; range: 3–9 months) and adult intestinal tissues (median age: 50 years, range: 48-53 years). As a measure for clonal expansion, we compared the number of BCR clones of which the unique BCR sequence exceeded 0.5% of the total BCR repertoire, denoted here as dominant clones [21, 22]. Similar numbers of dominant B cell clones (24–30) (Table S3) were present in infant intestinal tissues compared to adult intestinal tissues (18–58). Collectively, the dominant clones made up 17–35% of the BCR repertoire of the infant intestinal tissue, which was similar to that of adults (13-79%) (Fig. S5E). Somatic hypermutations in B cells are further indicative of TFH help to B cells. Due to the sequencing technique applied, we could not quantify somatic hypermutations in B cell clones, but the overall mutation rate in the V-region was determined. As expected, we observed in all three infant samples that the dominant clones, which contributed >0.5% to the total B cell population, carried more mutations than the non-dominant clones that contributed <0.1% to the total intestinal B cell population (Fig. S5F, G). A similar pattern was observed in adult intestinal B cells, although the overall mutation rate was higher, which is in line with the decreased frequencies of naive B cells in adult intestines. Taken together, infant intestines harbored expanded B cell clones, which carried more mutations in the V-region compared to non-expanded clones.
To assess whether B cell maturation was also reflected by their anatomical distribution in infant intestines, we performed immunohistochemical staining on infant and adult intestinal tissues. B cells (defined as CD20+) were more dispersed in infant tissues and the B cell follicle formation was still under development early after birth (Fig. 6G, Fig. S6A). In line with the flow cytometric analyses described above, IgA+ cells were detected in infant intestinal tissues, further supporting that IgM to IgA class switching of B cells can occur early in life (Fig. 6G). Quantification of the immunohistochemical staining further confirmed the level of maturation of infant and adult intestines, with increasing number of cells expressing CD20 and IgA over life (Fig. S6B).
Next, to determine whether TFH cells (Bcl-6+ and CD3+) are anatomically located in proximity of B cells in infant intestines, double stainings for Bcl-6 and CD3 as well as for Bcl-6 and CD20, respectively were performed. These analyses showed that large numbers of Bcl-6+CD3+ cells were present in infant intestines both in the mucosal and submucosal layers. Furthermore, Bcl-6+CD3+ cells were present near B cell follicles (Fig. S7A), providing the anatomical prerequisites for TFH cell and B cell interactions in infant intestines. In adult intestines, although fewer Bcl-6+CD3+ cells were detected in the mucosa, they were located primarily in proximity of and within B cell follicles in the submucosa. Taken together, these data show that B cell maturation takes place early after birth and that TFH cells in infant intestines are located within B cell follicles where they can provide B cell help.
Infant intestinal CXCR5+PD-1+/– CD4+ T cells promote infant intestinal B cell maturation in vitro
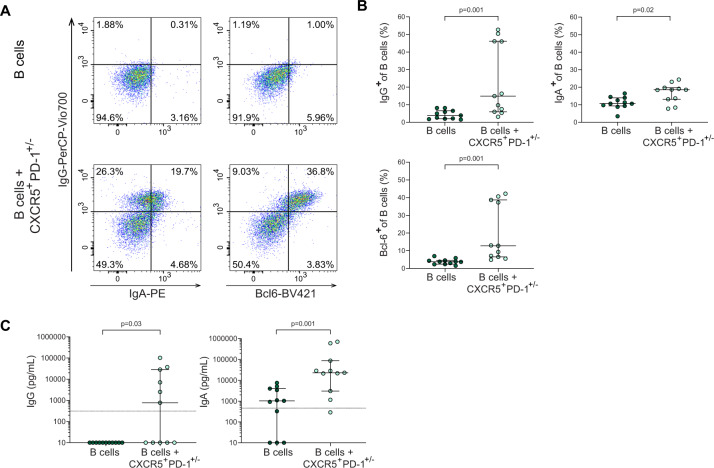
To investigate whether infant intestinal CXCR5+PD-1++ CD4+ T cells are able to provide B cell help, we performed co-cultures of infant intestinal B cells with autologous infant intestinal CXCR5+PD-1+/− CD4+ T cells in an autologous system (Fig. S8A), and assessed B cell maturation after 6 days, similar as previously described [45]. CXCR5+PD-1+/− CD4+ T cells were isolated to optimize cell yield compared to CXCR5+PD-1++ CD4+ T cells. Adult intestinal tissues did not yield sufficient CXCR5+PD-1+/− CD4+ T cells to perform these analyses. Co-cultures of intestinal B cells with intestinal CXCR5+PD-1+/− CD4+ T cells resulted in a significant increase of IgG and IgA expression by B cells (p = 0.001 and p = 0.02, respectively) (Fig. 7A, B). IgG and IgA levels were furthermore significantly increased in the supernatants upon 6 days of culture of B cells together with CXCR5+PD-1+/− CD4+ T cells compared to B cells alone (p = 0.03 and p = 0.001, respectively) (Fig. 7C). Autologous co-cultures with infant naive B cells and CXCR5+PD-1+/– CD4+ T cells showed similar increased expression of especially IgG and to a lesser extend IgA by naive B cells (Fig. S8C–E). To further assess the capacity of CXCR5+PD-1+/– CD4+ T cells to induce B cell differentiation compared to CXCR5− non-TFH cells, we compared co-cultures of B cells with autologous CXCR5+PD-1+/− CD4+ T cells and CXCR5−PD-1+/− CD4+ T cells. CXCR5+PD-1+/– CD4+ T cells induced larger frequencies of IgA+ B cells (p = 0.002) and IgG+ B cells (p = 0.003) compared to CXCR5−PD-1+/− CD4+ T cells, confirming their specialized capacity to induce B cell differentiation (Fig. S8F). Together, these findings demonstrate that infant intestinal CXCR5+PD-1++ CD4+ T cells are functional and can promote B cell maturation.
Fig. 7.
Infant intestinal CXCR5+PD-1+/− CD4+ T cells promote infant intestinal B cell maturation. A Representative flow cytometric plots of B cell analyses of B cell cultures alone or upon co-culture with CXCR5+PD-1+/− CD4+ T cells (lower plots), showing IgG, IgA and Bcl-6 expressions. B Frequencies (%) of IgG+ (top left), IgA+ (top right) and Bcl-6+ (bottom) in B cells cultured alone and in co-culture with autologous CXCR5+PD-1+/− cells. C Levels of IgG (left) and IgA (right) present in culture supernatants of B cells cultured alone and in co-culture with CXCR5+PD-1+/− cells measured by ELISA. Limit of detection for each ELISA experiment is depicted with a horizontal line in the plot. Medians and IQRs are depicted in all figures. N = 5 intestinal samples. For 3 of the donors, 3 replicates were performed. Each dot represents one donor/replicate. Wilcoxon matched-pairs signed rank test was used for comparisons. Only significant p values are shown
Discussion
The gastrointestinal tract is a major site for severe infections in young infants [4, 5]. TFH cells play a critical role to support antibody induction upon vaccination to acquire the required protection [46]. Here, we demonstrate that, although absent during fetal development, human intestines already contained large numbers of CXCR5+PD-1++ CD4+ T cells, phenotypic TFH cells, shortly after birth, which declined with age. Infant intestinal CXCR5+PD-1++ CD4+ T cells expressed costimulatory molecules and were able to promote B cell maturation by enhancing IgG and IgA production. Infant and adult intestinal B cells showed equal numbers of highly expanded clones, providing further in vivo data that infant CXCR5+PD-1++ CD4+ T cells are effective in inducing class switching and clonal B cell expansion in vivo. Taken together, the induction of effective intestinal CD4+ TFH cells is robustly initiated after birth and able to support B cell maturation in infants. The enhanced TFH cell induction in children early after birth may represent a critical time window for vaccine strategies to protect young infants at risk of severe infectious diseases.
The maturation of intestinal CD4+ T cell responses in humans is initiated prior to birth in contrast to mice [10]. Early fetal intestinal CD4+ T cell responses are largely restricted to a Th1 phenotype [15, 16, 47]. Rapid microbial invasion and feeding after birth results in a sudden increase of antigens, promoting enhanced induction and increase of memory CD4+ T cells in humans and in mice [48]. The data presented here demonstrates that CXCR5+PD-1++ CD4+ T cells colonize human intestines within months after birth, which is in line with a recent study in mice, suggesting that early life may represent an unique time window for the enhanced induction of TFH cells by food antigens [49]. Our study demonstrates that during the same developmental time window in humans, there is an enhanced induction of human intestinal CXCR5+PD-1++ CD4+ T cells, and that foreign antigen-exposure is critical, as illustrated by the absence of CXCR5+PD-1++ CD4+ T cells in fetal intestines and their low numbers in intestines of newborn infants.
The induction of TFH cells depends on the regulated expression of specific transcription factors, starting with STAT3 expression that is followed by Bcl-6 expression. Bcl-6 enhances CXCR5 expression in TFH cells, promoting TFH cells to interact with B cells. ICOS-mediated interactions between B cells and TFH cells in GC further enhanced Bcl-6 expression by TFH cells [50–54]. Infant intestinal CXCR5+PD-1++ CD4+ T cells expressed CXCR5; however Bcl-6 expression was slightly lower compared to adult intestinal CXCR5+PD-1++ CD4+ T cells. As we observed that B cell- containing follicles were not yet fully developed, ICOS-mediated upregulation of Bcl-6 may be reduced in infant intestines compared to adults. However, the similar mRNA Bcl-6 levels observed in infants and adults suggest that infant intestinal CXCR5+PD-1++ CD4+ T cells retain their phenotype. This was furthermore supported by a scRNAseq analyses of infant and adult intestinal and blood-derived CD4+ T cells. These analyses showed a population of CD4+ T cells with a TFH signature in infant intestines, their transcriptome however was similar to adult TFH intestinal cells, further supporting the TFH phenotype of CD4+ T cells in infant intestines.
TFH cells provide B cell help through interactions with costimulatory molecules and cytokines. The signaling pathway of ICOS, through PI3 kinase (PI3K), further enhances production of TFH-associated cytokines, such as IL-21 [36, 55–57], that promote B cell maturation and immunoglobulin production [58–60]. Autologous co-cultures of intestinal B cells and CXCR5+PD-1+/− CD4+ T cells showed increased differentiation of B cells and IgG and IgA production compared to B cells cultured alone. Furthermore, TFH cells were anatomically located near and within B cell follicles in infant intestines, allowing cellular interactions and promotion of B cell maturation. This was in line with the detection of IgG+ and IgA+ B cells in infant intestines, as well as comparable numbers of highly expanded B cell clones in infants and adults.
In conclusion, our study demonstrates that, although absent during fetal development, functional effector memory CXCR5+PD-1++ CD4+ T cells are induced after birth, with the appropriate molecular tools to shape B cell immunity and promote highly expanded B cell clones in human intestines [61]. This feature of infant intestines to support antibody responses can be harnessed for oral vaccines.
Supplementary information
Acknowledgements
The authors would like to thank all donors for participating in this study; Dr. Weijer, Mrs. Voordouw, Mrs. Siteur-van Rijnstra, and the Bloemenhove Clinic (Heemstede, the Netherlands) for collecting and providing fetal tissues. The authors also thank the FACS Core facility members of the Leibniz Institute of Virology (LIV) for their support, Mr. Arne Düsedau and Mrs. Jana Henessen. This study is supported by the Innovative Antiviral Therapy Program, Leibniz Institute of Virology (LIV), the German Center for Infection Research (DZIF), EFRE 2014-2020 REACT-EU, Dutch Digestive Fund (MLDS CDG 15-02), Deutsche Forschungsgemeinschaft (BU 3630/2-1) and Hüet Roëll Foundation. MK is supported by a Walter Benjamin Fellowship of the Deutsche Forschungsgemeinschaft (KA5554/1-1, KA5554/1-2). The Leibniz Institute of Virology is supported by the Free and Hanseatic City of Hamburg and the Federal Ministry of Health.
Author contributions
AJP, GM, FLS, and MJB designed the experiments. AJP, GM, FLS, MK, AFS, RRCES, AR, MEB, CG, RT, JSS, JBG, TBH, MA, US, STP, MAF and PLK contributed to experiments and interpretation of data. AJP, FLS, AFS, RRCES, JMJ, KJM, LW, STP, IK, DP, KR, CT, GS and NM contributed to collection of tissue samples. LR supported the statistical analysis. AJP, GM, FLS, LR, MAF, MK and PLK performed the data analyses. AJP, GM and MJB wrote the manuscript with input from all authors, who revised the manuscript. MJB supervised the study.
Competing interests
The authors declare no competing interests.
Footnotes
These authors contributed equally: Ana Jordan-Paiz, Glòria Martrus, Fenja L. Steinert.
Supplementary information
The online version contains supplementary material available at 10.1038/s41423-022-00944-4.
References
- 1.Prendergast AJ, Klenerman P, Goulder PJ. The impact of differential antiviral immunity in children and adults. Nat Rev Immunol. 2012;12:636–48. doi: 10.1038/nri3277. [DOI] [PubMed] [Google Scholar]
- 2.Liu L, Oza S, Hogan D, Perin J, Rudan I, Lawn JE, et al. Global, regional, and national causes of child mortality in 2000-13, with projections to inform post-2015 priorities: an updated systematic analysis. Lancet. 2015;385:430–40. doi: 10.1016/S0140-6736(14)61698-6. [DOI] [PubMed] [Google Scholar]
- 3.GBD 2016 Diarrhoeal Disease Collaborators. Estimates of the global, regional, and national morbidity, mortality, and aetiologies of diarrhoea in 195 countries: a systematic analysis for the Global Burden of Disease Study 2016. Lancet Infect Dis. 2018;18:1211–28. [DOI] [PMC free article] [PubMed]
- 4.Okomo U, Akpalu ENK, Le Doare K, Roca A, Cousens S, Jarde A, et al. Aetiology of invasive bacterial infection and antimicrobial resistance in neonates in sub-Saharan Africa: a systematic review and meta-analysis in line with the STROBE-NI reporting guidelines. Lancet Infect Dis. 2019;19:1219–34. doi: 10.1016/S1473-3099(19)30414-1. [DOI] [PubMed] [Google Scholar]
- 5.Zhu Q, Berzofsky JA. Oral vaccines: directed safe passage to the front line of defense. Gut Microbes. 2013;4:246–52. doi: 10.4161/gmic.24197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Plotkin S, Orenstein W, Offit P, Edwards KM. Plotkin’s vaccines - 7th ed. Elsevier. 2017.
- 7.MacLennan IC, Toellner KM, Cunningham AF, Serre K, Sze DM, Zúñiga E, et al. Extrafollicular antibody responses. Immunol Rev. 2003;194:8–18. doi: 10.1034/j.1600-065x.2003.00058.x. [DOI] [PubMed] [Google Scholar]
- 8.Linterman MA, Hill DL. Can follicular helper T cells be targeted to improve vaccine efficacy? F1000Res. 2016;5:F1000 Faculty Rev-1088. [DOI] [PMC free article] [PubMed]
- 9.Roider J, Maehara T, Ngoepe A, Ramsuran D, Muenchhoff M, Adland E, et al. High-frequency, functional HIV-specific T-follicular helper and regulatory cells are present within germinal centers in children but not adults. Front Immunol. 2018;9:1975. doi: 10.3389/fimmu.2018.01975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bunders MJ, Van Der Loos CM, Klarenbeek PL, Van Hamme JL, Boer K, Wilde JC, et al. Memory CD4(+)CCR5(+) T cells are abundantly present in the gut of newborn infants to facilitate mother-to-child transmission of HIV-1. Blood. 2012;120:4383–90. doi: 10.1182/blood-2012-06-437566. [DOI] [PubMed] [Google Scholar]
- 11.Bunders MJ, Van Hamme JL, Jansen MH, Boer K, Kootstra NA, Kuijpers TW. Fetal exposure to HIV-1 alters chemokine receptor expression by CD4+T cells and increases susceptibility to HIV-1. Sci Rep. 2014;4:6690. doi: 10.1038/srep06690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Brugnoni D, Airò P, Graf D, Marconi M, Lebowitz M, Plebani A, et al. Ineffective expression of CD40 ligand on cord blood T cells may contribute to poor immunoglobulin production in the newborn. Eur J Immunol. 1994;24:1919–24. doi: 10.1002/eji.1830240831. [DOI] [PubMed] [Google Scholar]
- 13.Nonoyama S, Penix LA, Edwards CP, Lewis DB, Ito S, Aruffo A, et al. Diminished expression of CD40 ligand by activated neonatal T cells. J Clin Investig. 1995;95:66–75. doi: 10.1172/JCI117677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang X, Zhivaki D, Lo-Man R. Unique aspects of the perinatal immune system. Nat Rev Immunol. 2017;17:495–507. doi: 10.1038/nri.2017.54. [DOI] [PubMed] [Google Scholar]
- 15.Li N, van Unen V, Abdelaal T, Guo N, Kasatskaya SA, Ladell K, et al. Memory CD4 + T cells are generated in the human fetal intestine. Nat Immunol. 2019;20:301–12. doi: 10.1038/s41590-018-0294-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schreurs RRCE, Baumdick ME, Sagebiel AF, Kaufmann M, Mokry M, Klarenbeek PL, et al. Human fetal TNF-α-cytokine-producing CD4+ effector memory T cells promote intestinal development and mediate inflammation early in life. Immunity. 2019;50:462–76.e8. doi: 10.1016/j.immuni.2018.12.010. [DOI] [PubMed] [Google Scholar]
- 17.Schreurs RRCE, Drewniak A, Bakx R, Corpeleijn WE, Geijtenbeek THB, van Goudoever JB, et al. Quantitative comparison of human intestinal mononuclear leukocyte isolation techniques for flow cytometric analyses. J Immunol Methods. 2017;445:45–52. doi: 10.1016/j.jim.2017.03.006. [DOI] [PubMed] [Google Scholar]
- 18.Sagebiel AF, Steinert F, Lunemann S, Körner C, Schreurs RRCE, Altfeld M, et al. Tissue-resident Eomes (+) NK cells are the major innate lymphoid cell population in human infant intestine. Nat Commun. 2019;10:975. doi: 10.1038/s41467-018-08267-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ziegler SM, Beisel C, Sutter K, Griesbeck M, Hildebrandt H, Hagen SH, et al. Human pDCs display sex-specific differences in type I interferon subtypes and interferon α/β receptor expression. Eur J Immunol. 2017;47:251–6. doi: 10.1002/eji.201646725. [DOI] [PubMed] [Google Scholar]
- 20.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 21.Pollastro S, Klarenbeek PL, Doorenspleet ME, Van Schaik BDC, Esveldt REE, Thurlings RM, et al. Non-response to rituximab therapy in rheumatoid arthritis is associated with incomplete disruption of the B cell receptor repertoire. Ann Rheum Dis. 2019;78:1339–45. doi: 10.1136/annrheumdis-2018-214898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Doorenspleet ME, Klarenbeek PL, De Hair MJ, Van Schaik BD, Esveldt RE, Van Kampen AH, et al. Rheumatoid arthritis synovial tissue harbours dominant B cell and plasma-cell clones associated with autoreactivity. Ann Rheum Dis. 2014;73:756–62. doi: 10.1136/annrheumdis-2012-202861. [DOI] [PubMed] [Google Scholar]
- 23.Klarenbeek PL, Tak PP, van Schaik BDC, Zwinderman AH, Jakobs ME, Zhang Z, et al. Human T-cell memory consists mainly of unexpanded clones. Immunol Lett. 2010;133:42–8. doi: 10.1016/j.imlet.2010.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Hashimshony T, Senderovich N, Avital G, Klochendler A, de Leeuw Y, Anavy L, et al. CEL-Seq2: sensitive highly-multiplexed single-cell RNA-Seq. Genome Biol. 2016;17:77. doi: 10.1186/s13059-016-0938-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parekh S, Ziegenhain C, Vieth B, Enard W, Hellmann I. zUMIs - a fast and flexible pipeline to process RNA sequencing data with UMIs. Gigascience. 2018;7:giy059. [DOI] [PMC free article] [PubMed]
- 26.Butler A, Hoffman P, Smibert P, Papalexi E, Satija R. Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol. 2018;36:411–20. doi: 10.1038/nbt.4096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wickham H, Averick M, Bryan J, Chang W, McGowan L, François R, et al. Welcome to the Tidyverse. J Open Source Softw. 2019;4:1686. [Google Scholar]
- 28.Kaufmann M, Evans H, Schaupp AL, Engler JB, Kaur G, Willing A, et al. Identifying CNS-colonizing T cells as potential therapeutic targets to prevent progression of multiple sclerosis. Med (N Y) 2021;2:296–312.e8. doi: 10.1016/j.medj.2021.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Monaco G, Lee B, Xu W, Mustafah S, Hwang YY, Carré C, et al. RNA-Seq signatures normalized by mRNA abundance allow absolute deconvolution of human immune cell types. Cell Rep. 2019;26:1627–40.e7. doi: 10.1016/j.celrep.2019.01.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Aibar S, González-Blas CB, Moerman T, Huynh-Thu VA, Imrichova H, Hulselmans G, et al. SCENIC: Single-cell regulatory network inference and clustering. Nat Methods. 2017;14:1083–6. doi: 10.1038/nmeth.4463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550. doi: 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vinuesa CG, Linterman MA, Yu D, Maclennan ICM. Follicular helper T cells. Annu Rev Immunol. 2016;34:335–68. doi: 10.1146/annurev-immunol-041015-055605. [DOI] [PubMed] [Google Scholar]
- 33.Mackay LK, Braun A, Macleod BL, Collins N, Tebartz C, Bedoui S, et al. Cutting edge: CD69 interference with sphingosine-1-phosphate receptor function regulates peripheral T cell retention. J Immunol. 2015;194:2059–63. doi: 10.4049/jimmunol.1402256. [DOI] [PubMed] [Google Scholar]
- 34.Thome JJC, Farber DL. Emerging concepts in tissue-resident T cells: lessons from humans. Trends Immunol. 2015;36:428–35. doi: 10.1016/j.it.2015.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.López-Cabrera M, Santis AG, Fernández-Ruiz E, Blacher R, Esch F, Sánchez-Mateos P, et al. Molecular cloning, expression, and chromosomal localization of the human earliest lymphocyte activation antigen AIM/CD69, a new member of the C-Type animal lectin superfamily of signal-transmitting receptors. J Exp Med. 1993;178:537–47. doi: 10.1084/jem.178.2.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Crotty S. Follicular helper CD4 T cells (TFH) Annu Rev Immunol. 2011;29:621–63. doi: 10.1146/annurev-immunol-031210-101400. [DOI] [PubMed] [Google Scholar]
- 37.Yu N, Li X, Song W, Li D, Yu D, Zeng X, et al. CD4(+)CD25(+)CD127(low/-) T cells: a more specific treg population in human peripheral blood. Inflammation. 2012;35:1773–80. doi: 10.1007/s10753-012-9496-8. [DOI] [PubMed] [Google Scholar]
- 38.Sage PT, Sharpe AH. T follicular regulatory cells. Immunol Rev. 2016;271:246–59. doi: 10.1111/imr.12411. [DOI] [PubMed] [Google Scholar]
- 39.Daenthanasanmak A, Wu Y, Iamsawat S, Nguyen HD, Bastian D, Zhang M, et al. PIM-2 protein kinase negatively regulates T cell responses in transplantation and tumor immunity. J Clin Invest. 2018;128:2787–801. doi: 10.1172/JCI95407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hwang SS, Lim J, Yu Z, Kong P, Sefik E, Xu H, et al. mRNA destabilization by BTG1 and BTG2 maintains T cell quiescence. Science. 2020;367:1255–60. doi: 10.1126/science.aax0194. [DOI] [PubMed] [Google Scholar]
- 41.Gibbons D, Fleming P, Virasami A, Michel ML, Sebire NJ, Costeloe K, et al. Interleukin-8 (CXCL8) production is a signatory T cell effector function of human newborn infants. Nat Med. 2014;20:1206–10. doi: 10.1038/nm.3670. [DOI] [PubMed] [Google Scholar]
- 42.Choi YS, Kageyama R, Eto D, Escobar TC, Johnston RJ, Monticelli L, et al. ICOS receptor instructs T follicular helper cell versus effector cell differentiation via induction of the transcriptional repressor Bcl6. Immunity. 2011;34:932–46. doi: 10.1016/j.immuni.2011.03.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Vu Van D, Beier KC, Pietzke LJ, Al Baz MS, Feist RK, Gurka S, et al. Local T/B cooperation in inflamed tissues is supported by T follicular helper-like cells. Nat Commun. 2016;7:10875. [DOI] [PMC free article] [PubMed]
- 44.Malkiel S, Barlev AN, Atisha-Fregoso Y, Suurmond J, Diamond B. Plasma cell differentiation pathways in systemic lupus erythematosus. Front Immunol. 2018;9:427. [DOI] [PMC free article] [PubMed]
- 45.Havenith SHC, Remmerswaal EBM, Idu MM, van Donselaar-van der Pant KAMI, Van der Bom N, Bemelman FJ, et al. CXCR5+CD4+ follicular helper T cells accumulate in resting human lymph nodes and have superior B cell helper activity. Int Immunol. 2014;26:183–92. doi: 10.1093/intimm/dxt058. [DOI] [PubMed] [Google Scholar]
- 46.Crotty S. T follicular helper cell differentiation, function, and roles in disease. Immunity. 2014;41:529–42. doi: 10.1016/j.immuni.2014.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Halkias J, Rackaityte E, Hillman SL, Aran D, Mendoza VF, Marshall LR, et al. CD161 contributes to prenatal immune suppression of IFN-γ-producing PLZF+ T cells. J Clin Investig. 2019;129:3562–77. doi: 10.1172/JCI125957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ganal-Vonarburg SC, Hornef MW, Macpherson AJ. Microbial–host molecular exchange and its functional consequences in early mammalian life. Science. 2020;368:604–7. doi: 10.1126/science.aba0478. [DOI] [PubMed] [Google Scholar]
- 49.Hong SW, Eunju O, Lee JY, Lee M, Han D, Ko HJ, et al. Food antigens drive spontaneous IgE elevation in the absence of commensal microbiota. Sci Adv. 2019;5:eaaw1507. doi: 10.1126/sciadv.aaw1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vinuesa CG, Linterman MA, Goodnow CC, Randall KL. T cells and follicular dendritic cells in germinal center B-cell formation and selection. Immunol Rev. 2010;237:72–89. doi: 10.1111/j.1600-065X.2010.00937.x. [DOI] [PubMed] [Google Scholar]
- 51.Deenick EK, Chan A, Ma CS, Gatto D, Schwartzberg PL, Brink R, et al. Follicular helper T Cell differentiation requires continuous antigen presentation that is independent of unique B cell signaling. Immunity. 2010;33:241–53. doi: 10.1016/j.immuni.2010.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Poholek AC, Hansen K, Hernandez SG, Eto D, Chandele A, Weinstein JS, et al. In vivo regulation of Bcl6 and T follicular helper cell development. J Immunol. 2010;185:313–26. doi: 10.4049/jimmunol.0904023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Crotty S, Johnston RJ, Schoenberger SP. Effectors and memories: Bcl-6 and Blimp-1 in T and B lymphocyte differentiation. Nat Immunol. 2010;11:114–20. doi: 10.1038/ni.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kitano M, Moriyama S, Ando Y, Hikida M, Mori Y, Kurosaki T, et al. Bcl6 protein expression shapes pre-germinal center B cell dynamics and follicular helper T cell heterogeneity. Immunity. 2011;34:961–72. doi: 10.1016/j.immuni.2011.03.025. [DOI] [PubMed] [Google Scholar]
- 55.Bauquet AT, Jin H, Paterson AM, Mitsdoerffer M, Ho IC, Sharpe AH, et al. The costimulatory molecule ICOS regulates the expression of c-Maf and IL-21 in the development of follicular T helper cells and TH -17 cells. Nat Immunol. 2009;10:167–75. doi: 10.1038/ni.1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gigoux M, Shang J, Pak Y, Xu M, Choe J, Mak TW, et al. Inducible costimulator promotes helper T-cell differentiation through phosphoinositide 3-kinase. Proc Natl Acad Sci USA. 2009;106:20371–6. doi: 10.1073/pnas.0911573106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rolf J, Bell SE, Kovesdi D, Janas ML, Soond DR, Webb LMC, et al. Phosphoinositide 3-kinase activity in T cells regulates the magnitude of the germinal center reaction. J Immunol. 2010;185:4042–52. doi: 10.4049/jimmunol.1001730. [DOI] [PubMed] [Google Scholar]
- 58.Good KL, Bryant VL, Tangye SG. Kinetics of human B cell behavior and amplification of proliferative responses following stimulation with IL-21. J Immunol. 2006;177:5236–47. doi: 10.4049/jimmunol.177.8.5236. [DOI] [PubMed] [Google Scholar]
- 59.Kuchen S, Robbins R, Sims GP, Sheng C, Phillips TM, Lipsky PE, et al. Essential role of IL-21 in B cell activation, expansion, and plasma cell generation during CD4+ T cell-B cell collaboration. J Immunol. 2007;179:5886–96. doi: 10.4049/jimmunol.179.9.5886. [DOI] [PubMed] [Google Scholar]
- 60.Bryant VL, Ma CS, Avery DT, Li Y, Good KL, Corcoran LM, et al. Cytokine-mediated regulation of human B cell differentiation into Ig-secreting cells: predominant role of IL-21 produced by CXCR5+ T follicular helper cells. J Immunol. 2007;179:8180–90. doi: 10.4049/jimmunol.179.12.8180. [DOI] [PubMed] [Google Scholar]
- 61.Lavelle EC, Ward RW. Mucosal vaccines — fortifying the frontiers. Nat Rev Immunol. 2022;22;236–50. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.