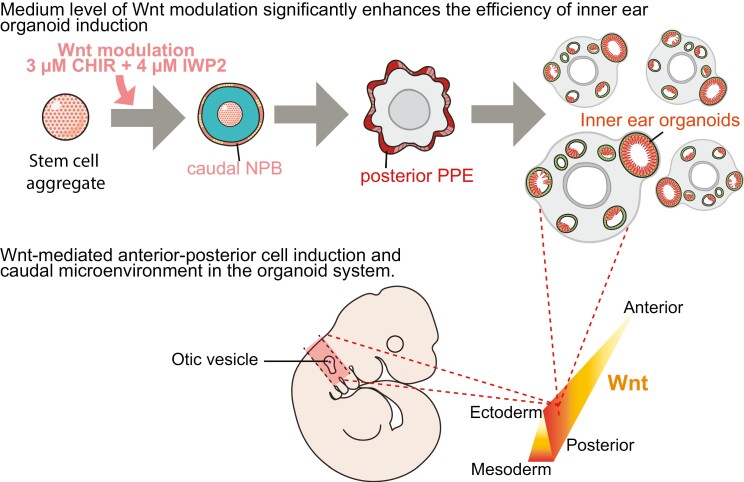
Abstract
The inner ear is derived from the otic placode, one of the numerous cranial sensory placodes that emerges from the pre-placodal ectoderm (PPE) along its anterior-posterior axis. However, the molecular dynamics underlying how the PPE is regionalized are poorly resolved. We used stem cell-derived organoids to investigate the effects of Wnt signaling on early PPE differentiation and found that modulating Wnt signaling significantly increased inner ear organoid induction efficiency and reproducibility. Alongside single-cell RNA sequencing, our data reveal that the canonical Wnt signaling pathway leads to PPE regionalization and, more specifically, medium Wnt levels during the early stage induce (1) expansion of the caudal neural plate border (NPB), which serves as a precursor for the posterior PPE, and (2) a caudal microenvironment that is required for otic specification. Our data further demonstrate Wnt-mediated induction of rostral and caudal cells in organoids and more broadly suggest that Wnt signaling is critical for anterior-posterior patterning in the PPE.
Keywords: inner ear, organoid, otic placode, pre-placodal ectoderm, single-cell-RNA sequencing, stem cell, Wnt
Graphical Abstract
Graphical Abstract.
Significance Statement.
We found that early Wnt activation significantly enhances the induction of hair cell-baring inner ear organoids via cell caudalization. The resulting high yield of otic progenitor cells (and later hair cells) will benefit future inner ear organoid applications, such as high-throughput drug screening and cell therapy. Furthermore, we comprehensively profiled gene expression across various Wnt levels at different times, and these data demonstrate Wnt-mediated induction of cells resembling a rostral and caudal head in organoids and provide valuable information to the fields of stem cell, inner ear, and developmental biology of cranial sensory systems.
Introduction
Stem cell-derived 3D inner ear organoids are derived through sequential modulation of complex signaling pathways based on what we know about in vivo inner ear development.1 Inner ear epithelia and the surrounding neurons are derived from otic vesicles (OVs) and neural crest cells. A key step in the development of the inner ear sensory epithelia is the formation of the OVs that are invaginated from the otic placode, which, along with other cranial sensory placodes, is derived from a common precursor, the pre-placodal ectoderm (PPE), in a region adjacent to the anterior neural plate known as the neural plate border (NPB).2 Specifically, the otic and epibranchial placodes arise from the posterior PPE (pPPE), while the adenohypophysis, trigeminal, lens, and olfactory placodes arise in the anterior PPE (aPPE). Currently, it is unclear how the PPE is regionalized along the anterior-posterior (A-P; ie, rostral-caudal) axis.
Most studies on the early development of sensory placodes have been undertaken in non-mammalian organisms. The small sizes and difficulty in culturing post-gastrulation mammalian embryos contribute to technical obstacles for studying the development of placodal lineages using traditional techniques such as tissue grafting. Alternative models for studying the early development in mammals, especially for scarce tissue types such as placodes, are of need. Stem cell-based systems have shown potential in advancing our understanding of mammalian development,3,4 and we explored the potential of the stem cell-derived inner ear organoid system herein for studying early mammalian placode induction.
One major limitation of the inner ear organoid system is the highly variable and inconsistent yield of organoids. Current organoid derivation protocols5,6 manipulate the TGFβ, BMP, and FGF pathways during early induction. Wnt activation was used in the later differentiation stage to promote the otic placode specification from the pPPE.7 However, Wnt signaling pathways also play dynamic and critical roles earlier in development (ie, prior to otic placode specification). During late gastrulation, Wnt signals reduce rostral gene expression in the NPB.8 Inhibition of Wnt is not initially required but is later involved in PPE induction in birds.9,10 In stem cell-based culture systems, initial Wnt antagonists have been used to enhance the induction of the non-neural ectoderm and PPE.11-13 Herein we focused on the early induction period and explored the effects of Wnt pathways in the induction and regionalization of the PPE using the mouse embryonic stem cell (mESC)-derived inner ear organoid system with the overarching goal of improving the efficiency of the inner ear organoid system.
We hypothesized that Wnt signaling will improve pPPE induction, thereby leading to improved organoid efficiency. We performed stage-specific single-cell RNA-seq (scRNA-seq), and our findings suggest that (1) Wnt activation promotes caudalization of the NPB and (2) Wnt signaling plays an important role in shaping the cellular microenvironment necessary for the specification of the otic placode. Lastly, our data demonstrate that the mESC-derived inner ear organoid system shows similarities with in vivo models in terms of Wnt-mediated induction of rostral-caudal cell types in the head and the early otic development.
Materials and Methods
mESC Culture and Induction of Inner Ear Organoids
Three mESC lines—R1/E, Pax8-tdTomato reporter cell line, and Atoh1-2A-nGFP reporter cell line11—were used; all were maintained under feeder-free conditions using modified LIF-2i media, which was changed every other day. Induction followed5 but with modifications. For details, see Supplementary experimental procedures.
Wnt Modulation
We used the Wnt activator/GSK3 inhibitor CHIR99021 (CHIR) on the R1/E mESC line to activate the canonical Wnt pathway by increasing active β catenin levels in the nuclei, while simultaneously reducing endogenous paracrine Wnt signaling activity by inhibiting the secretion of Wnt ligands with IWP2.14 We refer to this approach as “Wnt modulation” (S1A-C). Because all Wnt modulation treatments include the same concentration of IWP2, we only refer to treatments by their CHIR concentration hereafter. We performed time-course experiments between induction day (D) 0 and D5, but only those treated on D3 for R1/E mESC line and D2 for Pax8-tdTomato reporter cell line subsequently generated inner ear organoids.
OV Productivity
We designed a scoring system to quantify the productivity of OVs. Details of this novel system can be found in the Supplementary experimental procedures. All values have been presented as mean ± SD unless stated otherwise, and all error terms for all other measurements correspond to SD unless stated otherwise.
Generation and Characterization of the Pax8-tdTomato Cell Line
The Pax8-tdTomato reporter cell line was generated using CRISPR/Cas9 technology as described in Supplementary experimental procedures.
Quantitative PCR
Total RNAs were extracted using the PureLink RNA extraction kit following the manufacturer’s instructions (Thermo-Fisher Scientific [TFS]). cDNAs were generated using SuperScript IV (TFS). Reactions were carried out on an ABI QuantStudio 5 (Applied Biosystems). Taqman assays from TFS were used to measure the expression of Six1, Eya1, and Pax8. Expression of Otx2, Gbx2, Pax6, and Pax8 (Supplementary Table S2) was detected using SYBR green I chemistry (mastermix from Applied Biosystems). Tbp (Taqman assay) and L27 (Supplementary Table S2) were used as housekeeping genes in the Taqman and SYBR green assays, respectively.
Immunohistochemistry (IHC)
Samples were fixed in 4% paraformaldehyde (PFA) in PBS for a duration that was dependent on the size of the aggregates. The fixed samples were immersed in a sequential gradient of 10%, 20%, and 30% sucrose before being embedded in “tissue freeze media” (Electron Microscopy Sciences [EMS]). Frozen samples were sectioned into 10-12-μm cryosections using a Leica CM1860 cryostat. Sections were incubated in a blocking buffer comprised of 10% goat or horse serum (Rockland) and 0.1% Triton X-100 (Acros) in PBS for 1 h at room temperature (RT). Primary and secondary antibodies (Supplementary Table S3) were diluted in PBS containing 3% goat or horse serum and 0.1% Triton X-100. After blocking, specimens were incubated in primary antibodies for 1 h, washed thrice with PBS, incubated in Alexa Fluor secondary antibody for 1 h, washed thrice with PBS, and embedded in ProLong Gold containing DAPI (Life Technologies). Images were taken using a Leica DMi8 Microscope or a Leica TCS SP8 confocal imaging system.
Western Blots
Two hundred and sixteen organoids from the same experiment were combined to serve as one sample for protein isolation. The nuclear fraction was isolated using the Nuclear/Cytosol Fractionation Kit (BioVision) following the manufacturer’s recommendations. Proteins were stored at −80 °C. Details of SDS-PAGE and Western blots can be found in the Supplementary experimental procedures.
Wnt Reporter Assay
R1/E cells were transfected with the Wnt reporter plasmid 7TFC on D0 of culture; the plasmid was a gift from Roel Nusse (Addgene plasmid # 24307).15 Aggregates were treated with various levels of Wnt modulation. Batches of 8 aggregates were collected 24-h post-Wnt treatment. Luciferase activity was measured using the Firefly Luciferase Assay Kit 2.0 (Biotium) on a Gen5 microplate reader (BioTek).
scRNA-seq Sample Preparation
Organoids were dissociated, and samples were submitted to the Center for Medical Genetics (CMG) at the Indiana University School of Medicine. For details please see the Supplementary experimental procedures.
scRNA-seq Data Analyses
The Cell Ranger pipeline (10× Genomics) was used to perform alignment, filtering, barcode counting, and UMI counting. We then used the R package Seurat (V3; Stuart et al, 201916) to filter out cells that had unique feature counts >6000 or <100, as well as those with >5% mitochondrial counts (calculated from a cell-by-gene matrix). For details, see Supplementary experimental procedures.
Results
Early Modulation of Wnt Signaling Enhances the Induction of Inner Ear Organoids
We first determined the effects of canonical Wnt signaling pathways by the Wnt modulation during the early stage of inner ear organoid induction. Western blotting showed that the active β-catenin ratio in the nuclear fraction increased with increasing levels of CHIR (Supplementary Fig. S1D, S1E). We also implemented the Wnt reporter assay to measure the activation of canonical Wnt pathways15 and found increasing TCF/LEF-dependent luciferase activity with increasing CHIR dosing (Supplementary Fig. S1F). These results indicate activation of canonical Wnt pathways in response to Wnt modulation. There was an effect of Wnt modulation on cell composition, from predominantly OTX2+ ectodermal cells in samples of lower Wnt levels, to a mixture of ectodermal and Brachyury+ mesodermal cells in higher Wnt-modulated samples by D4 (Supplementary Fig. S1G).
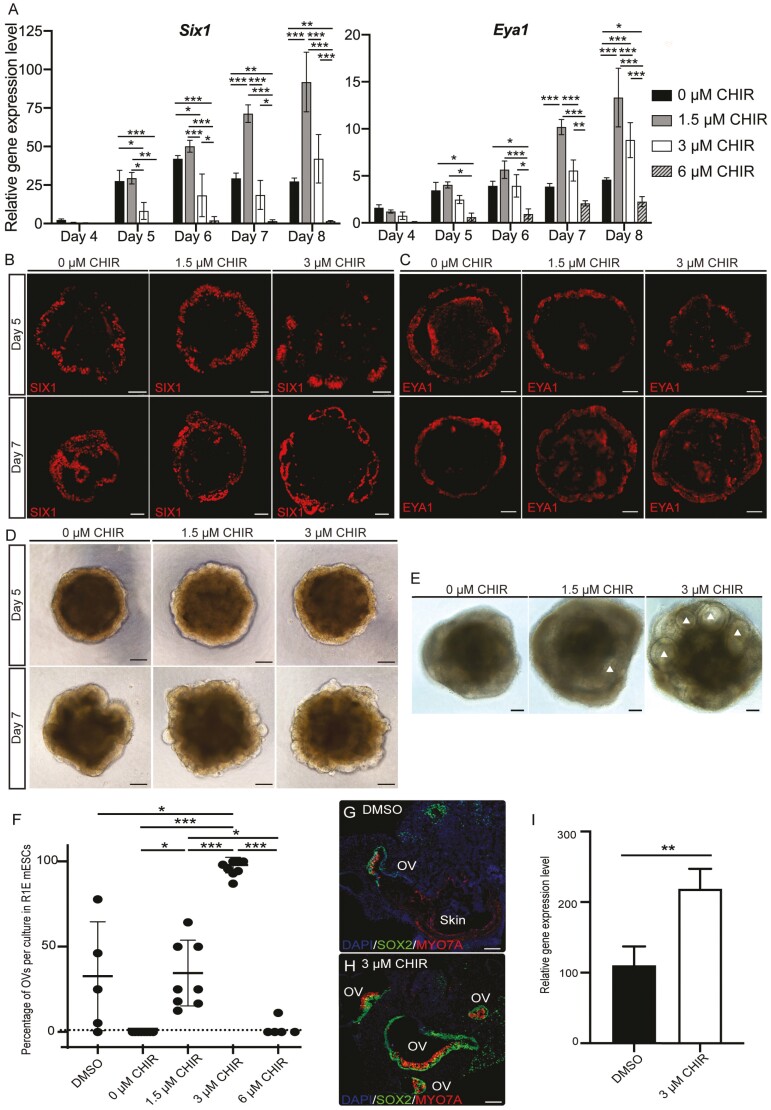
Next, we tracked the induction of placodal progenitors in response to Wnt modulation by quantifying the expression of 2 PPE markers: Six1 and Eya1. We noticed a very low expression of these genes on D4, but significantly increased expression from D5 onwards (Fig. 1A). On D5 and D6, higher expression levels of Six1 were seen in 0 and 1.5 µM CHIR treatments. By D7 and D8, the higher levels of Six1 and Eya1 were observed under 1.5 µM and 3 µM CHIR conditions. High levels of CHIR (6 µM) led to significantly lower expression levels of both genes across all culture days (Fig. 1A). Spatially, both markers labeled the outer layer of aggregates, suggesting that the PPE is located at their outer ruffle borders (Fig. 1B-1D).
Figure 1.
Early Wnt modulation improves inner ear organoid induction. (A) Mean (±SD) expression levels of 2 pre-placodal ectoderm (PPE) marker genes, Six1 and Eya1, under various Wnt levels during the early induction of inner ear organoids (n = 4-6 independent differentiation experiments). Two-way ANOVA: day*** and treatment***. Representative immunohistochemistry (IHC) images of SIX1 (B) and EYA1 (C) signals at the outer layer of samples under various Wnt levels on D5 and 7. (D) Representative bright field (BF) images of aggregates on D5 and D7. (E) Representative BF images of aggregates on D10. More prominent otic vesicles (OVs; white triangles) were seen in 3 µM CHIR samples. (F) Significant increase in the production of OVs in 3 µM CHIR samples. Each dot indicates one 96 well-plate experiment (n = 5-8 independent differentiation experiments). One-way ANOVA of treatment (P = .005) followed by post hoc tests. (G-H) Representative IHC images of D20 OVs in the DMSO control and 3 µM CHIR treatment. (I) Mean (±SD) gene expression levels of the hair cell marker myosin 7A in D20 DMSO controls and the 3 µM CHIR treatment (n = 3 independent differentiation experiments). Scale bars: 100 µm. *, **, and *** denote P < .05, <.001, and <.0001, respectively.
We then examined the effects of various Wnt levels on OV formation and observed more OV structures in the 3 µM CHIR group on D10 compared to 1) DMSO controls, which were cultured using published protocols2,5,7 groups with other CHIR levels (Fig. 1E). The 3 µM CHIR group consistently produced the highest yield of OVs by D20 (96.0 ± 4.6%; n= 8 experiments; Fig. 1F and Supplementary Fig. S1H-S1J). To verify that the increased quantity of OVs also contained sensory epithelia, we performed IHC and qPCR for the hair cell marker myosin 7a (Myo7a) and compared the data to those obtained from DMSO controls. Hair cells were seen in most vesicles in the 3 µM CHIR group (Fig. 1G-1I), and the expression of Myo7a was significantly higher in 3 µM CHIR samples compared to controls (Fig. 1I). Additionally, we measured the percentage of hair cell-baring OVs (GFP+ OVs/total vesicles) using the Atoh1-2A-nGFP reporter cell line. We observed both embedded and protruding GFP+ OVs (Supplementary Fig. S1K-S1M) and confirmed that 90.3 ± 8.5% of all vesicles were GFP+ OVs (n = 55 aggregates across 3 experiments). Our data demonstrate that Wnt modulation with 3 µM CHIR significantly increased the productivity of inner ear organoids.
Induction of the aPPE and pPPE is Related to the Level of Wnt Modulation
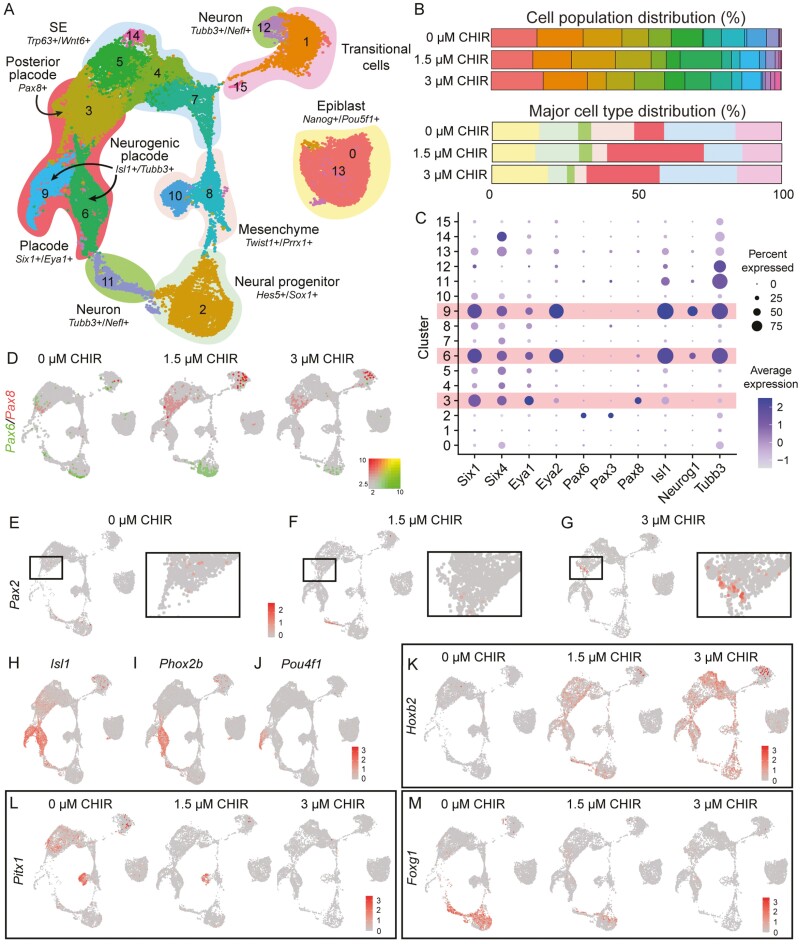
Because there were consistently more OVs in the 3 µM CHIR treatment, despite higher expression levels of Six1 and Eya1 in 0 µM CHIR samples on D5 and 6, we investigated mechanisms underlying this pronounced OV induction. Given (1) the role of Wnt signaling in A-P patterning within the vertebrate head17-19 and (2) that the otic placode arises from the pPPE adjacent to the r5/caudal region,20 we hypothesized that a medium level of Wnt modulation (3 µM CHIR) enhances pPPE induction. A previous study showed that repression between Otx2 and Gbx2 defines the boundary of the aPPE and pPPE.21 OVs arise from the pPPE, as evidenced by the expression of Pax8,22 with the presence of Pax6 signifying aPPE. We, therefore, measured the expression of these 4 genes under various Wnt levels. Given our prior observations that samples of the 6 µM CHIR treatment (1) were composed of more Brachyury+ mesodermal cells (Supplementary Fig. S1F), (2) showed significantly lower levels of PPE gene expression (Fig. 1A), and (3) failed to generate inner ear organoids (Fig. 1F), this condition was excluded from further analysis.
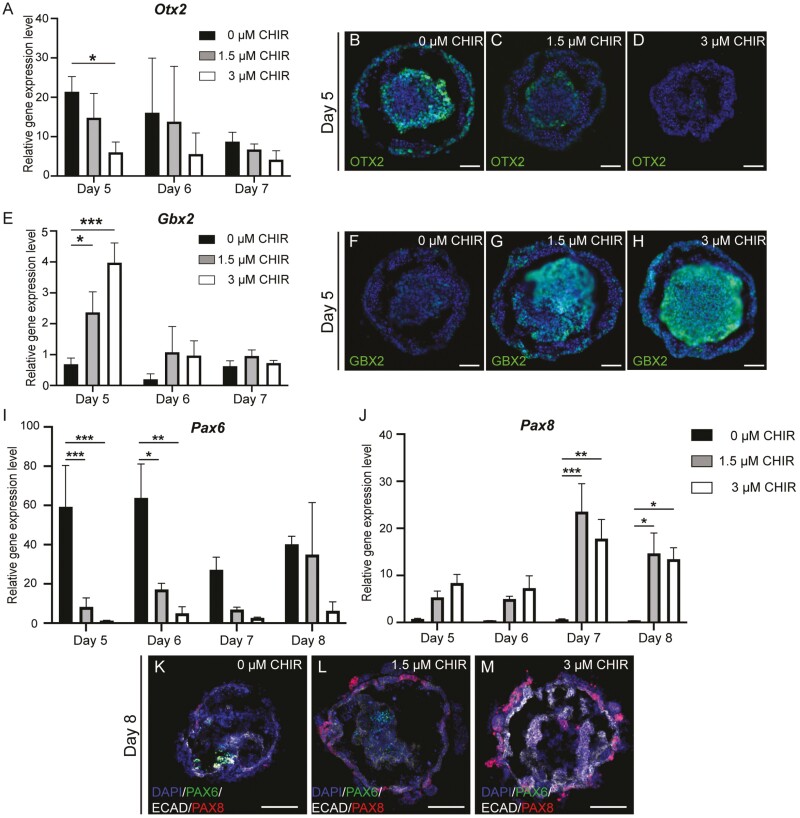
qPCR results revealed significantly higher Otx2 expression in the 0 µM CHIR group (Fig. 2A-2D) alongside significantly higher Gbx2 expression in the 3 µM CHIR group on D5 (Fig. 2E-2H). Suppression of the expression of Pax6 was seen in 3 µM CHIR samples compared to samples with lower Wnt levels (Fig. 2I). In contrast, significantly elevated expression of Pax8 was observed in the 1.5 and 3 µM CHIR treatments, and both peaked on D7 (Fig. 2J). We noticed weak PAX6 signals in the center of the aggregate in 0 and 1.5 µM CHIR samples (Fig. 2K-M). PAX6 was only observed in the outer placodal epithelium in 0 µM CHIR samples when additional FGF8 was supplemented alongside FGF2 and LDN-193189 on D4 (Supplementary Fig. S2A-S2B). In contrast, PAX8 localized to the ruffled outer epithelia in 1.5 and 3 µM CHIR samples (Fig. 2K-2M), where a SIX1 signal was also observed (Fig. 1B). This suggests that Wnt activation elicits induction of the pPPE.
Figure 2.
Wnt modulation regulates the presence of anterior and posterior placodal cells during inner ear organoid induction. Mean (±SD) gene expression and representative IHC images of an anterior marker, Otx2 (A-D), and a posterior marker, Gbx2 (E-H), under various Wnt levels during early inner ear organoid induction. Mean (±SD) gene expression levels of the aPPE marker Pax6 (I) and the pPPE marker Pax8 (J) in samples undergoing various Wnt modulation levels. (K-M) Representative IHC images of PAX6, E-cadherin (ECAD), and PAX8 in D8 samples under various degrees of Wnt modulation. Scale bar=50 µm. n = 3-6 independent differentiation experiments. *, **, and *** denote P < .05, <.001, and <.0001, respectively, and there were significant effects (2-way ANOVA) of day (***) and treatment (***).
To validate our findings, we replicated the Wnt dosing experiments in a Pax8-tdTomato reporter line (Supplementary Fig. S3-S4). Consistent with results seen in the R1/E mESCs, the 3 µM CHIR group yielded significantly more hair cell-baring OVs (88.0 ± 7.4%) compared to DMSO controls (39.2 ± 10.1%; Supplementary Fig. S4A-S4C). Pax8 gene expression (qPCR-derived) displayed a similar pattern as for samples derived from R1/E mESCs (Supplementary Fig. S4D). We also documented live images of Pax8 expression in aggregates throughout the induction (Supplementary Fig. S4E-S4P). Spatially, Pax8-tdTomato signals were observed in the outer epithelia of aggregates in 1.5 and 3 µM CHIR treatments but not in the 0 µM CHIR samples. These data are consistent with the results observed in the R1/E mESCs and suggest that the pPPE was only induced in the 1.5 and 3 µM CHIR samples.
Wnt Modulation Promotes Induction of Caudal NPB Cells
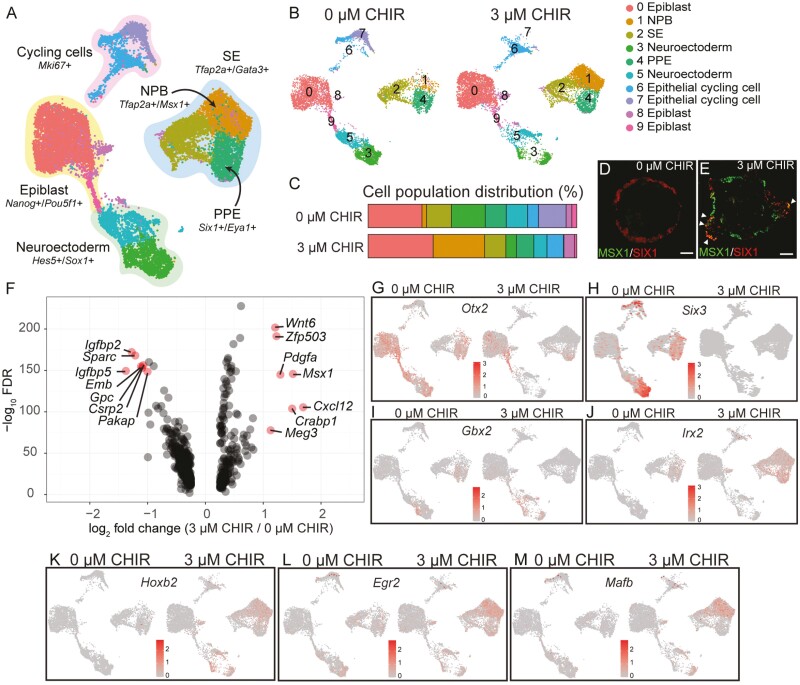
To investigate how Wnt modulation affects the induction of the pPPE, we first conducted scRNA-seq on samples collected on D5, when the PPE first appeared, for both 0 and 3 µM CHIR samples (Fig. 3A-3C). Ten cell subtypes were identified via unbiased clustering, and we annotated the following clusters based on the expression of marker genes (Supplementary Table S1): epiblast (28.5%), surface ectoderm (SE; 11.1%), NPB (13.4%), PPE (9.2%), neuroectoderm (20.7%), and cycling cells (13.3%; Fig. 3A-3C and Supplementary Fig. S5A-S5F). The 3 µM CHIR treatment was associated with a higher percentage of cells within the NPB cluster (24.7% in 3 µM CHIR vs. 2.2% in 0 µM CHIR) along with a decrease in cells within the neuroectoderm clusters (clusters 3 and 5; Fig. 3A-3C). Overlapping IHC signals between MSX1 and SIX1 were only seen in 3 µM CHIR samples on D6 (Fig. 3D-3E), which suggests a transition from MSX1+ NPB to SIX1+ placodal cells.
Figure 3.
WNT modulation increases caudal neural plate border (NPB) cell populations and caudal head region genes in D5 inner ear organoids. (A) Uniform manifold approximation and projection (UMAP) of scRNA-seq data in D5 cells of the 0 and 3 µM CHIR treatments. (B) UMAP of D5 samples of 0 and 3 µM CHIR (plotted individually). Ten clusters were manually annotated in D5 samples from these 2 treatments. (C) Differential cell distribution between 0 and 3 µM CHIR treatments. More NPB cells and fewer neuronal progenitors were seen in 3 µM CHIR samples compared to the 0 µM CHIR group. Representative IHC image of MSX1 and SIX1 staining in 0 µM CHIR (D) and 3 µM CHIR (E) samples on D6. Triangle heads indicate the co-staining of MSX1 and SIX1. (F) Volcano plot of differentially expressed genes (DEGs) between 0 and 3 µM CHIR samples on D5. Seven genes with higher expression in the 3 µM CHIR treatment relative to the 0 µM CHIR group (average log2 fold change > 1) were identified. (G-H) Expression of 2 anterior marker genes: Otx2 and Six3. (I-J) Expression of 2 posterior marker genes: Gbx2 and Irx2. (K-M) Expression of 3 marker genes for the rhombomere 5 region: Hoxb2, Egr2, and Mafb.
In agreement with our qPCR results, more Six1-expressing cells were observed in the 0 µM CHIR samples relative to the 3 µM CHIR samples on D5 (cluster 4 [PPE cluster]: 10.%1 vs. 8.3%, respectively; Fig. 3C and Supplementary S5G-S5H). Eya1 was expressed in both the PPE and NPB clusters in 3 µM CHIR samples (Supplementary Fig. S5H). In the PPE cluster, differentially expressed genes (DEGs; P < .1 & log2 fold change > 1) can be found in Fig. 4F. 0 µM CHIR samples possessed more cells with high expression levels of 2 anterior placodal genes, Otx2 and Six3 (Fig. 3G-3H), while more cells with the posterior markers Gbx2 and Irx2 were present in 3 µM CHIR samples (Fig. 3I-3J). Hoxb2, which is expressed in the hindbrain region (with strong expression adjacent to r3 and r523-25; Trainor, 200326), was observed in 3 µM CHIR samples across several clusters but rarely in 0 µM samples (Fig.3K). We noted the expression of Egr2 (Krox20) and Mafb (Kreisler), which are also expressed around r5, in 3 µM CHIR samples (Fig. 3L-3M). Collectively, our data suggest that Wnt modulation with 3 µM CHIR gives rise to more cells resembling the caudal NPB, from which the pPPE is derived.
Figure 4.
Wnt modulation induces pPPE cells and caudalization in D8 inner ear organoids. (A) UMAP of D8 scRNA-seq samples featuring 16 clusters. Seven major groups- epiblast, surface ectoderm (SE), placode, mesenchyme, transitional cells, neural progenitor, and neuron- were annotated. (B) Differential distribution of cells among the 0, 1.5, and 3 µM CHIR treatments on D8. (C) Dot plot of 4 placodal markers (Six1, Six4, Eya1, and Eya2), 3 A-P placodal markers (Pax6, Pax3, and Pax8), and 3 neurogenic markers (Isl1, Neurog1, and Tubb3). The hypothesized placodal clusters are shaded in pink. (D) Expression of the aPPE marker Pax6 and the pPPE marker Pax8 across 3 Wnt levels. (E-G) Expression of the otic placodal marker, Pax2, in cluster 3, suggesting a transition from the pPPE to the otic placode. (H-J) Representative placode marker genes. Expression of the neurogenic placodal markers Isl1 (H), Phox2b (I), and Pou4f1 in cluster 9 (J). (K-M) Increasing the CHIR concentration leads to 1) increasing number of cells expressing Hox2b across multiple cell clusters (K), 2) decreased expression of the rostral head marker Pitx1 (L), and 3) decreased expression of the forebrain marker Foxg1 (M).
Wnt Modulation Induces the pPPE and a Caudal Head-Like Microenvironment in D8 Inner Ear Organoids
The 3 µM CHIR condition generated significantly more OVs than the 1.5 µM CHIR group (Fig. 1F) despite no significant difference in the expression of Pax8 genes. We explored the cause of this discrepancy by comparing scRNA-seq data among the 3 Wnt modulation levels on D8. Specifically, we investigated the cell constituents in aggregates among the Wnt treatments. Sixteen subtypes were annotated into 7 major cell groups based on commonly expressed gene signatures (Fig. 4A and Supplementary Fig. S6A-S6G): undifferentiated epiblast (16.9%), neural progenitors (16.9%), differentiating neurons (3.9%), mesenchyme (8%), sensory placode (23%), SE (21.5%), and transitional cells (15%). The distributions of these major cell types varied according to Wnt modulation level (Fig. 4B). Specifically, 25%-33% of cells were of the placodal lineage in the 1.5 and 3 µM CHIR groups compared to only 10% of cells in 0 µM CHIR condition (Fig. 4B). On the other hand, 18%-20% of cells were of the neuronal lineage in the 0 and 1.5 µM CHIR groups, but only 9% of neural cells were seen in the 3 µM CHIR condition (Fig. 4B). In sum, similar levels of placodal cells were seen in the 1.5 and 3 µM CHIR treatments while more neuronal-like cells were present in the 0 and 1.5 µM CHIR samples.
According to the expression of the Six and Eya genes, 3 clusters-3, -6, and -9 were of the placodal lineage (Fig. 4C). Among these, the pPPE marker Pax8 was more prevalent in cluster 3 of the 1.5 and 3 µM CHIR samples compared to the 0 µM CHIR group, whereas the aPPE marker Pax6 was mainly present in cluster 3 of the 0 µM CHIR group (Fig. 4D). Noticeable expression of Pax6 was also seen in neuronal progenitors (cluster 2). Interestingly, the early otic placodal marker Pax2 was observed in cluster 3 of the 3 µM CHIR group, but rarely in the other treatments (Fig. 4E-4G). Placodal clusters 6 and 9 featured the expression of several neurogenic markers, including Isl1, Neurog1, Neurod1, and Tubb3, suggesting the presence of neurogenic placodes (Fig. 4C, 4H; Supplementary Fig. S6H). Within the presumed neurogenic placode regions, 2 epibranchial placodal genes, Phox2b and Neurog2, were seen in cluster 6 (Fig. 4I and Supplementary Fig. S6I). The expression of Pou4f1, a sensory neuron marker, was only documented in cluster 9 (Fig. 4J). These data suggest the induction of 2 pPPE derivatives, the otic and epibranchial placodes.
Hoxb2 was the most highly expressed anterior Hox gene in the 3 µM CHIR group (Fig. 4K and Supplementary Fig. S6J-S6N). We, therefore, explored whether any rostral head markers were expressed in response to Wnt modulation and documented decreased expression of Pitx1, Foxg1, Six3, and Lhx2 at higher Wnt levels (Fig. 4L-4M, Supplementary S6O-S6P). Taken together, our results indicate that the cell composition shifted from the anterior neuronal lineage to the posterior placodal lineage with increasing Wnt levels.
A Specific Level of Wnt Modulation (3 µM CHIR) is Required to Create the Optimal Microenvironment for Derivation of the Otic Placode from the pPPE
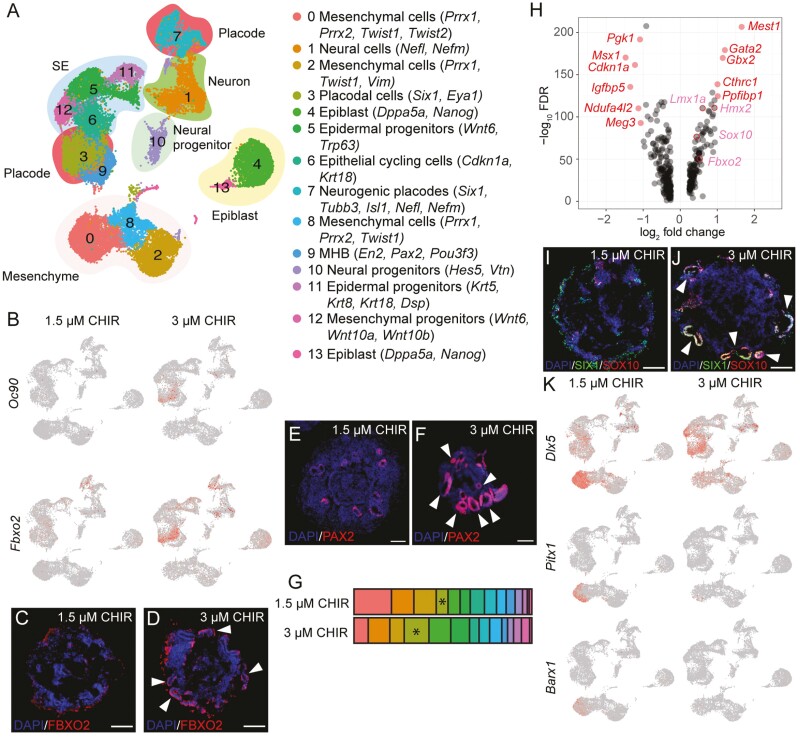
Because we noticed that Pax2 was only present in 3 µM CHIR on D8, we hypothesized that the derivation of the otic placode from the pPPE only occurs under 3 µM CHIR conditions. We compared the scRNA-seq data between 1.5 and 3 µM CHIR samples on D10. Fourteen subgroups were identified and annotated according to key marker genes (Fig. 5A). The otic markers Oc90 and Fbxo2 were mainly seen in the placodal cluster (cluster 3) of the 3 µM CHIR samples but not in the 1.5 µM CHIR group (Fig. 5B-5D). More PAX2+ OVs were observed in 3 µM CHIR samples than in the 1.5 µM CHIR group (Fig. 5E-5F). The percentage of cluster 3 in the 3 µM CHIR group (13.9%) was double that of the 1.5 µM CHIR group (6.6%; Fig. 5G). This suggests that despite the presence of Pax8+ pPPE in both 1.5 and 3 µM CHIR samples on D8, subsequent differentiation from the pPPE to the otic placode only occurred in the 3 µM CHIR treatment.
Figure 5.
A caudal microenvironment in aggregates is required for otic placode induction from the pPPE. (A) UMAP of D10 samples in 1.5 and 3 µM CHIR treatments featuring 14 annotated clusters. (B) Expression of 2 otic placodal markers, Oc90 and Fbxo2, in 3 µM CHIR samples. Representative IHC images of D10 OV staining with FBXO2 (C-D) and PAX2 (E-F). (G) Differential distribution of cells between the 1.5 and 3 µM CHIR treatments. Asterisks indicate cluster 3 (placodal cluster). (H) Volcano plot of differential gene expression in cluster 3 between 1.5 and 3 µM CHIR samples on D10. DEGs (average log2 fold change>1) are labeled in red, and pink labels indicate upregulated otic genes (log2 fold change<1) in 3 µM samples. (I-J) Representative IHC images of D10 OV staining with the placodal marker SIX1 and the otic marker SOX10. (K) Decreased expression of the rostral oral markers Dlx5, Pitx1, and Barx1 in the mesenchymal cluster of the 3 µM CHIR samples. White arrowheads indicate OVs. Scale bars = 50 µm.
Eleven DEGs were identified between the 1.5 and 3 µM CHIR treatments within placodal cluster 3 alone (Fig. 5H). Three of the upregulated genes—Gata2, Gbx2, and Cthrc1—in 3 µM CHIR were annotated as “inner ear morphogenesis-related.” Furthermore, we noticed that several otic genes uncovered in prior analyses of mouse otocysts—Lmx1a, Hmx2, Sox10, and Fbxo227- were upregulated in 3 µM CHIR samples, although the average log2 fold change was <1 and so they were not deemed DEGs (Fig. 5H). Indeed, some of these otic-related proteins are labeled OVs (Fig. 5C-5D, 5I-5J). In the 1.5 µM CHIR group, we documented more mesenchymal cells expressing higher levels of 3 rostral genes- Dlx5, Pitx1, and Barx1- related to oral development28,29; this implies that the 1.5 µM CHIR condition leads to more rostral cells compared to the 3 µM CHIR treatment (Fig. 5K). We conclude that, despite the presence of pPPE under both 1.5 and 3 µM conditions, the microenvironment required for the transition from the pPPE to the otic placode was only present in 3 µM CHIR samples.
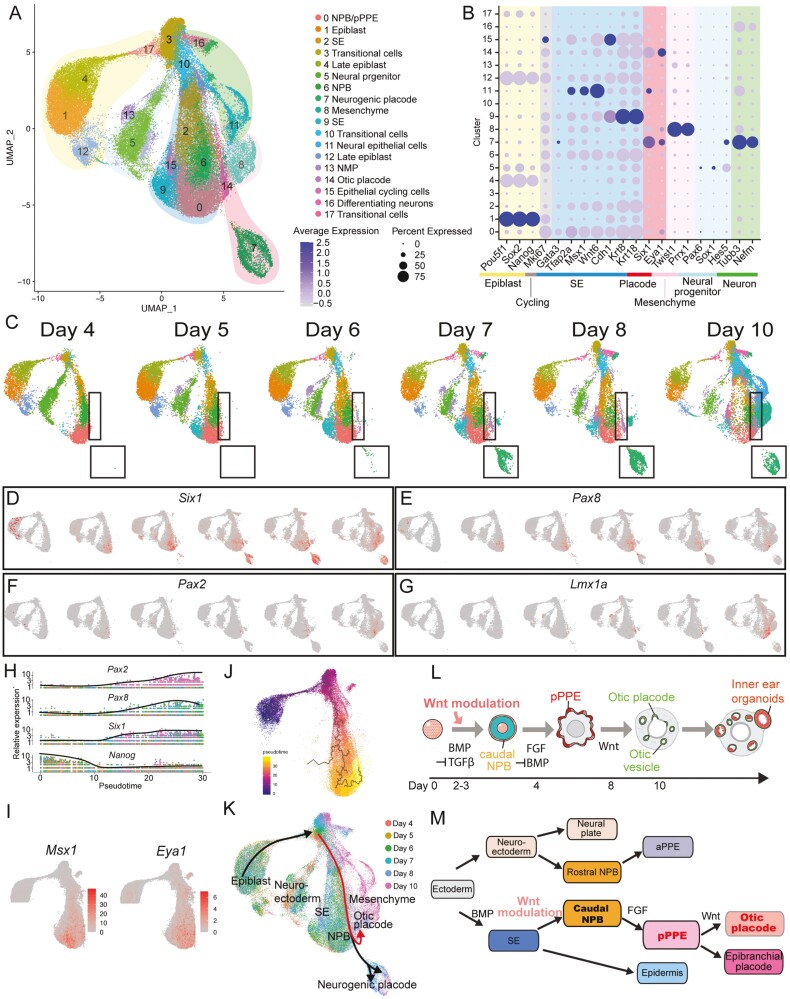
Modeling the Otic Trajectory in the mESC-Derived Organoid System
We reconstructed the differentiation trajectory of inner ear organoids. To examine the otic trajectory of cells induced by the optimal Wnt level (3 µM CHIR) from epiblasts to OVs, samples were collected every 24-48 h between D4 and 10 for scRNA-seq analyses. Seventeen cell subtypes were identified, which were annotated and sorted into 7 major groups based on the expression of conventional marker genes (Supplementary Table S1): epiblast, cycling cells, SE, placodal cells, mesenchymal cells, neural progenitors, and differentiating neurons (Fig. 6A-6B). Cell composition shifted over time, and cells exhibiting gene signatures of otic and neurogenic placodes arose around D6-7 (Fig. 6C). Six1 expression represented the distribution of PPE over time (Fig. 6D). Consistent with the qPCR results, the expression of Pax8 increased from D5 and peaked at D7 before decreasing by D8 and 10 (Fig. 6E). While Pax8 expression decreased, the expression of Pax2 began increasing (Fig. 6E-6F). Interestingly, a potential regulator of Pax2, Lmx1a, presented a similar expression pattern as Pax2 (Fig. 6F-6G).
Figure 6.
Wnt modulation drives the development of the otic placode from the caudal neural plate border (NPB) to the posterior PPE (pPPE). (A) UMAP visualization of 3 µM CHIR samples across induction days (D4 through D10). (B) Dot plot of marker genes of different lineages. (C) Feature plots of samples from each induction day demonstrating that cell composition shifted over time. The black boxes indicate the placodal clusters. The expression of critical otic lineage genes Six1 (D), Pax8 (E), Pax2 (F), and Lmx1a (G) across induction days. (H) Kinetic plot showing relative gene expression levels of otic placode and pluripotency (Nanog) markers across developmental pseudotime. (I) The expression of select genes associated with the NBP (Msx1) and PPE (Eya1). (J) Trajectory of the otic lineage colored by inferred pseudotime. (K) Putative placodal trajectories (black lines with arrowheads). The red lines indicate the otic trajectory. Schematics of inner ear organoid induction (L) and hypothetical ectodermal cell differentiation (M).
We then focused on the otic lineage and performed pseudo-temporal ordering. Along pseudo-time, we observed a simple, early-to-late transition from the epiblast (Nanog) to the pPPE (Six1 & Pax8) followed by a transition to the otic placode (Pax2; Fig. 6H). Msx1, an early-stage NPB marker, was expressed broadly with higher expression at the lower left side of cluster 0 (Fig. 6I). Eya1 was expressed broadly and overlapped with both Msx1-and Six1-expressing cells (Fig. 6D, 6I). The progression from cells expressing Msx1 (lower left) to Six1 (lower right) indicates a trajectory from the NPB to the pPPE, as also evidenced by the Pax8 data (Fig. 6D-6E, 6I). Finally, we reconstructed the lineage trajectories followed by extracting the otic trajectory using Monocle3 (Fig. 6J). This analysis revealed that the otic trajectory undergoes differentiation from (1) pluripotent stem cells through (2) the caudal NPB, and (3) the pPPE, and then to the (4) otic placodes in the inner ear organoid system (Fig. 6K).
Discussion
Previously, Wnt activation on D8 was reported to improve the inner ear organoid induction from ~20 to ~50% efficiency, although the variability was nevertheless high.5,7,30 Our study showed that exogenous Wnt modulation during the early induction stage facilitates the formation of hair cell-baring OVs through caudalization of the NBP, which subsequently gives rise to pPPE and otic placode (Fig. 6I-6J). Among the various Wnt modulation treatments, we identified that the combination of 3 µM CHIR and 4 µM IWP2 produced inner ear organoids in ~90% of cultures. Reproducibility was improved, as the SD dropped from 31.9% in the DMSO controls to only 4.6% in 3 µM CHIR samples. Wnt modulation during early induction therefore drastically improved the consistency and productivity of the inner ear organoid culture protocol.
Consistent with previous studies,13 we showed that, under lower Wnt levels, relatively more PPE cells were present. On D5, the presence of the early anterior marker gene Otx2 in the PPE cluster suggests that 0 µM CHIR samples primarily consisted of aPPE. Furthermore, by supplementing with additional factors, i.e., Fgf8, this induction protocol generates Pax6+ anterior placodal epithelium by D8. Though the 1.5 µM CHIR condition generated an equivalent number of, or more, Pax8+Six1+ cells, these cells failed to give rise to the Pax2+ otic lineage. We observed that cells exhibited more rostral characteristics in 1.5 µM CHIR samples based on the scRNA-seq data. The microenvironment critically influences the induction trajectories. Overall, we demonstrated subsequent differentiation from the PPE into each specific placodal derivative requires the manipulation of specific signaling pathways and/or a suitable microenvironment.
Our IHC images and scRNA-seq data indicate that Gbx2 was expressed in neuroectodermal rather than PPE clusters. Gbx2 plays an important role in defining the mid-hindbrain boundary31; expression of Gbx2 in the neuroectodermal clusters, therefore, implies hindbrain cell fate in the 3 µM CHIR treatment. Furthermore, more cells expressing Hoxb2, Egr2, and Mafb, which are known to be expressed in the hindbrain r3-r5 region,32-34 were present in the 3 µM CHIR group. Samples of the lower Wnt treatments (0 and 1.5 µM CHIR) instead expressed rostral genes. For instance, the expression patterns of Otx2 and Six3 in D5 0 µM CHIR neural lineages suggest a forebrain identity. Moreover, more rostral cell types, ie, putative oral mandibular mesenchyme (Pitx1+ and Barx1+) and forebrain (Foxg1+, Six3+, and Lhx2+) cells,35-38 increased in abundance at lower Wnt levels.
Wnt signaling pathways have been known to be critical for A-P patterning, even within the head region.39,40 The rostral portion of the head is formed upon the negative regulation of the Wnt pathway,41 which could explain our observation of gene signatures of the forebrain in samples with lower levels of Wnt activity. Collectively, lower Wnt levels appear to provide a more rostral environment, whereas 3 µM CHIR samples were composed of cells more characteristic of those of the r5 region (near where OVs are found in vivo). Our results signify that the 3 µM CHIR treatment gives rise to a microenvironment that is critical for proper cell derivation toward the otic placode and demonstrates that the induction of rostral–caudal cell types in this organoid system are Wnt-dependent.
Significantly more Irx2+ NPB cells were seen in 3 µM CHIR samples compared to 0 µM CHIR samples on D5, highlighting that Wnt activity is required for the caudalization of the NPB during late gastrulation8; Fig. 6J). These caudal NPB cells likely serve as precursors for the pPPE; this could explain the continuously increased pPPE presence in 3 µM CHIR samples on D7 and 8 (Fig. 6I-6J). In mouse embryos, Six1 around the PPE region has been seen as early as embryonic day (E) 8.42 In addition, the expression of Pax8 has been detected near the location of the future otic placode around E8-8.5 in mice.43 We first detected the expression of Six1 on D5. We also detected the expression of Pax8 from D5 onwards, with a peak on D7. This suggests that D5-7 in our in vitro system corresponds to ~E8-8.5 in vivo. The otic markers, Oc90, Fbxo2, and Sox10, were seen in D10, which is consistent with findings in mouse E10.5.44 In addition to the otic placode, there was evidence for induction of neurogenic placodes (clusters 6 and 9 on D8 and cluster 7 on D10) during inner ear organoid induction. The epibranchial placode is also derived from the pPPE45; therefore, it is unsurprising that Phox2b+ epibranchial placodal cells (cluster 6 on D8) were present in our cultures. Additionally, we observed that the potential Pou4f1+ otic neuron46 was exclusively expressed in the neurogenic placodal cluster 9 on D8. This suggests the likely existence of otic neurons in the inner ear organoid systems.
Conclusion
Our data revealed that Wnt levels coordinate the induction of anterior and posterior PPE. In addition, this study showed that the inner ear organoid system exhibits (1) multiple ectodermal derivatives, including 2 pPPE derivatives—epibranchial and otic placodes, and (2) similarities in early otic placode development with those steps documented in vivo. Finally, our results show that caudalization of placodal cells is Wnt-dependent and is a critical step for inner ear development.
Supplementary Material
Acknowledgments
This work was supported by the National Institutes of Health (NIH; K08-DC016034 to R.F.N.; R01-DC013072 to A.K.G.; R35-GM142701 to L.C.; R01-DC005575 to X.L; and R21-DC019450 to P.T.), the Hearing Health Foundation (Emerging Research Grants to P.T.), the Indiana Clinical and Translational Sciences Institute (which is funded in part by grant UL1TR002529 from NIH’s National Center for Advancing Translational Sciences) through a Clinical and Translational Sciences Award to P.T., and the Department of Otolaryngology—Head & Neck Surgery at Indiana University (to R.F.N.).
Contributor Information
Pei-Ciao Tang, Department of Otolaryngology—Head and Neck Surgery, Indiana University School of Medicine, Indianapolis, IN, USA; Department of Otolaryngology—Head and Neck Surgery, University of Miami Miller School of Medicine, Miami, FL, USA.
Li Chen, Department of Biostatistics, University of Florida, Gainesville, FL, USA.
Sunita Singh, Department of Neuroscience, Baylor College of Medicine, Houston, TX, USA.
Andrew K Groves, Department of Neuroscience, Baylor College of Medicine, Houston, TX, USA; Department of Molecular and Human Genetics, Baylor College of Medicine, Houston, TX, USA.
Karl R Koehler, Department of Otolaryngology, Boston Children’s Hospital, Boston, MA, USA; Department of Otolaryngology– Head and Neck Surgery, Harvard Medical School, Boston, MA, USA.
Xue Zhong Liu, Department of Otolaryngology—Head and Neck Surgery, University of Miami Miller School of Medicine, Miami, FL, USA.
Rick F Nelson, Department of Otolaryngology—Head and Neck Surgery, Indiana University School of Medicine, Indianapolis, IN, USA.
Conflict of Interest
The authors declared no potential conflicts of interest.
Author Contributions
P-C.T.: conception and design, financial support, collection and/or assembly of data, data analysis and interpretation, manuscript writing, and final approval of the manuscript. L.C.: data analysis, financial support, and final approval of the manuscript. S.S.: provision of study material and final approval of the manuscript. A.K.G.: financial support, provision of study material, and final approval of the manuscript. K.R.K.: conception and design and final approval of the manuscript. X.Z.L., R.F.N.: financial support and final approval of the manuscript.
Data Availability
scRNA-seq data have been deposited on NCBI’s Gene Expression Omnibus database under accession code GSE180062. Scripts used for scRNA-seq analyses are available at https://github.com/lichen-lab/PeiCiao/blob/main/codeforpaper.R
References
- 1. Koehler KR, Mikosz AM, Molosh AI, Patel D, Hashino E.. Generation of inner ear sensory epithelia from pluripotent stem cells in 3D culture. Nature. 2013;500:217-221. 10.1038/nature12298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Baker CVH, Bronner-Fraser M.. Vertebrate cranial placodes I. Embryonic induction. Dev Biol. 2001;232:1-61. 10.1006/dbio.2001.0156. [DOI] [PubMed] [Google Scholar]
- 3. Baillie-Benson P, Moris N, Martinez Arias A.. Pluripotent stem cell models of early mammalian development. Curr Opin Cell Biol. 2020;66:89-96. 10.1016/j.ceb.2020.05.010. [DOI] [PubMed] [Google Scholar]
- 4. Kretzschmar K, Clevers H.. Organoids: modeling development and the stem cell niche in a dish. Dev Cell. 2016;38:590-600. 10.1016/j.devcel.2016.08.014. [DOI] [PubMed] [Google Scholar]
- 5. Koehler KR, Hashino E.. 3D mouse embryonic stem cell culture for generating inner ear organoids. Nat Protoc. 2014;9:1229-1244. 10.1038/nprot.2014.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Koehler KR, Nie J, Longworth-Mills E, et al. Generation of inner ear organoids containing functional hair cells from human pluripotent stem cells. Nat Biotechnol. 2017;35:583-589. 10.1038/nbt.3840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. DeJonge RE, Liu X, Deig CR, et al. Modulation of Wnt signaling enhances inner ear organoid development in 3D culture. PLoS One. 2016;11:e0162508. 10.1371/journal.pone.0162508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Patthey C, Gunhaga L, Edlund T.. Early development of the central and peripheral nervous systems is coordinated by Wnt and BMP signals. PLoS One. 2008;3:e1625. 10.1371/journal.pone.0001625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Litsiou A, Hanson S, Streit A.. A balance of FGF, BMP and WNT signalling positions the future placode territory in the head. Development. 2005;132:4051-4062. 10.1242/dev.01964. [DOI] [PubMed] [Google Scholar]
- 10. Hintze M, Prajapati RS, Tambalo M, et al. Cell interactions, signals and transcriptional hierarchy governing placode progenitor induction. Development. 2017;144:2810-2823. 10.1242/dev.147942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Oshima K, Shin K, Diensthuber M, et al. Mechanosensitive hair cell-like cells from embryonic and induced pluripotent stem cells. Cell. 2010;141:704-716. 10.1016/j.cell.2010.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Lahlou H, Nivet E, Lopez-Juarez A, et al. Enriched differentiation of human otic sensory progenitor cells derived from induced pluripotent stem cells. Front Mol Neurosci. 2018;11:452. 10.3389/fnmol.2018.00452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Britton G, Heemskerk I, Hodge R, Qutub AA, Warmflash A.. A novel self-organizing embryonic stem cell system reveals signaling logic underlying the patterning of human ectoderm. Development. 2019;146:dev179093. 10.1242/dev.179093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Chen B, Dodge ME, Tang W, et al. Small molecule-mediated disruption of Wnt-dependent signaling in tissue regeneration and cancer. Nat Chem Biol. 2009;5:100-107. 10.1038/nchembio.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Fuerer C, Nusse R.. Lentiviral vectors to probe and manipulate the Wnt signaling pathway. PLoS One. 2010;5:e9370. 10.1371/journal.pone.0009370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Stuart T, Butler A, Hoffman P, et al. Comprehensive integration of single-cell data. Cell. 2019;17:1888-1902.e21. 10.1016/j.cell.2019.05.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Feinberg S, Roure A, Piron J, Darras S.. Antero-posterior ectoderm patterning by canonical Wnt signaling during ascidian development. PLoS Genet. 2019;15:e1008054. 10.1371/journal.pgen.1008054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Massey J, Liu Y, Alvarenga O, et al. Synergy with TGFβ ligands switches WNT pathway dynamics from transient to sustained during human pluripotent cell differentiation. PNAS. 2019;116:4989-4998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Schilling TF. Anterior-posterior patterning and segmentation of the vertebrate head. Integr Comp Bio. 2008;48:658-667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Singh S, Groves AK.. The molecular basis of craniofacial placode development. Wiley Interdiscip Rev Dev Biol. 2016;5:363-376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Steventon B, Mayor R, Streit A.. Mutual repression between Gbx2 and Otx2 in sensory placodes reveals a general mechanism for ectodermal patterning. Dev Biol. 2012;367:55-65. 10.1016/j.ydbio.2012.04.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Pfeffer PL, Gerster T, Lun K, Brand M, Busslinger M.. Characterization of three novel members of the zebrafish Pax2/5/8 family: dependency of Pax5 and Pax8 expression on the Pax2.1 (noi) function. Development. 1998;125:3063-3074. 10.1242/dev.125.16.3063. [DOI] [PubMed] [Google Scholar]
- 23. Bosse A, Zülch A, Becker MB, et al. Identification of the vertebrate Iroquois homeobox gene family with overlapping expression during early development of the nervous system. Mech Dev. 1997;69:169-181. 10.1016/s0925-4773(97)00165-2. [DOI] [PubMed] [Google Scholar]
- 24. Matsumoto K, Nishihara S, Kamimura M, et al. The prepattern transcription factor Irx2, a target of the FGF8/MAP kinase cascade, is involved in cerebellum formation. Nat Neurosci. 2004;7:605-612. 10.1038/nn1249. [DOI] [PubMed] [Google Scholar]
- 25. Minoux M, Antonarakis GS, Kmita M, Duboule D, Rijli FM.. Rostral and caudal pharyngeal arches share a common neural crest ground pattern. Development. 2009;136:637-645. 10.1242/dev.028621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Trainor PA. Making headway: The roles of hox genes and neural crest cells in craniofacial development. Sci World J. 2003;3:240-264. 10.1100/tsw.2003.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Durruthy-Durruthy R, Gottlieb A, Hartman BH, et al. Reconstruction of the mouse otocyst and early neuroblast lineage at single-cell resolution. Cell. 2014;157:964-978. 10.1016/j.cell.2014.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Balic A, Adams D, Mina M.. Prx1 and Prx2 cooperatively regulate the morphogenesis of the medial region of the mandibular process. Dev Dyn. 2009;238:2599-2613. 10.1002/dvdy.22092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mitsiadis TA, Drouin J.. Deletion of the Pitx1 genomic locus affects mandibular tooth morphogenesis and expression of the Barx1 and Tbx1 genes. Dev Biol. 2008;313:887-896. 10.1016/j.ydbio.2007.10.055. [DOI] [PubMed] [Google Scholar]
- 30. Schaefer SA, Higashi AY, Loomis B, et al. From otic induction to hair cell production: Pax2(EGFP) cell line illuminates key stages of development in mouse inner ear organoid model. Stem Cell Dev. 2018;27:237-251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hidalgo-Sánchez M, Simeone A, Alvarado-Mallart RM.. Fgf8 and Gbx2 induction concomitant with Otx2 repression is correlated with midbrain-hindbrain fate of caudal prosencephalon. Development. 1999;126:3191-3203. 10.1242/dev.126.14.3191. [DOI] [PubMed] [Google Scholar]
- 32. Cordes SP, Barsh GS.. The mouse segmentation gene kr encodes a novel basic domain-leucine zipper transcription factor. Cell. 1994;79:1025-1034. 10.1016/0092-8674(94)90033-7. [DOI] [PubMed] [Google Scholar]
- 33. Hunt P, Krumlauf R.. Hox codes and positional specification in vertebrate embryonic axes. Annu Rev Cell Biol. 1992;8:227-256. 10.1146/annurev.cb.08.110192.001303. [DOI] [PubMed] [Google Scholar]
- 34. Seitanidou T, Schneider-Maunoury S, Desmarquet C, Wilkinson DG, Charnay P.. Krox-20 is a key regulator of rhombomere-specific gene expression in the developing hindbrain. Mech Dev. 1997;65:31-42. 10.1016/s0925-4773(97)00051-8. [DOI] [PubMed] [Google Scholar]
- 35. Godbole G, Shetty AS, Roy A, et al. Hierarchical genetic interactions between FOXG1 and LHX2 regulate the formation of the cortical hem in the developing telencephalon. Development. 2018;145:dev154583. 10.1242/dev.154583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Lanctôt C, Lamolet B, Drouin J.. The bicoid-related homeoprotein Ptx1 defines the most anterior domain of the embryo and differentiates posterior from anterior lateral mesoderm. Development. 1997;124:2807-2817. 10.1242/dev.124.14.2807. [DOI] [PubMed] [Google Scholar]
- 37. Oliver G, Mailhos A, Wehr R, et al. Six3, a murine homologue of the sine oculis gene, demarcates the most anterior border of the developing neural plate and is expressed during eye development. Development. 1995;121:4045-4055. 10.1242/dev.121.12.4045. [DOI] [PubMed] [Google Scholar]
- 38. Xuan S, Baptista CA, Balas G, et al. Winged helix transcription factor BF-1 is essential for the development of the cerebral hemispheres. Neuron. 1995;14:1141-1152. 10.1016/0896-6273(95)90262-7. [DOI] [PubMed] [Google Scholar]
- 39. Newman EA, Wu D, Taketo MM, Zhang J, Blackshaw S.. Canonical Wnt signaling regulates patterning, differentiation and nucleogenesis in mouse hypothalamus and prethalamus. Dev Biol. 2018;442:236-248. 10.1016/j.ydbio.2018.07.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Yamaguchi TP. Heads or tails: Wnts and anterior–posterior patterning. Curr. 2001;11:R713-R724. [DOI] [PubMed] [Google Scholar]
- 41. Fossat N, Jones V, Khoo P-L, et al. Stringent requirement of a proper level of canonical WNT signalling activity for head formation in mouse embryo. Development. 2011;138:667-676. 10.1242/dev.052803. [DOI] [PubMed] [Google Scholar]
- 42. Sato S, Ikeda K, Shioi G, et al. Conserved expression of mouse Six1 in the pre-placodal region (PPR) and identification of an enhancer for the rostral PPR. Dev Biol. 2010;344:158-171. 10.1016/j.ydbio.2010.04.029. [DOI] [PubMed] [Google Scholar]
- 43. Bouchard M, de Caprona D, Busslinger M, Xu P, Fritzsch B.. Pax2 and Pax8 cooperate in mouse inner ear morphogenesis and innervation. BMC Dev Biol. 2010;10:89-89. 10.1186/1471-213X-10-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hartman BH, Durruthy-Durruthy R, Laske RD, Losorelli S, Heller S.. Identification and characterization of mouse otic sensory lineage genes. Front Cell Neurosci. 2015;9:79-79. 10.3389/fncel.2015.00079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Schlosser G. Induction and specification of cranial placodes. Dev Biol. 2006;294:303-351. 10.1016/j.ydbio.2006.03.009. [DOI] [PubMed] [Google Scholar]
- 46. Deng M, Yang H, Xie X, Liang G, Gan L.. Comparative expression analysis of POU4F1, POU4F2 and ISL1 in developing mouse cochleovestibular ganglion neurons. GEP. 2014;15:31-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
scRNA-seq data have been deposited on NCBI’s Gene Expression Omnibus database under accession code GSE180062. Scripts used for scRNA-seq analyses are available at https://github.com/lichen-lab/PeiCiao/blob/main/codeforpaper.R