Abstract
Background and Aims:
The sensitivity of current surveillance methods for detecting early-stage hepatocellular carcinoma (HCC) is suboptimal. Extracellular vesicles (EVs) are promising circulating biomarkers for early cancer detection. In this study, we aim to develop an HCC EV-based surface protein assay for early detection of HCC.
Approach and Results:
Tissue microarray was used to evaluate four potential HCC-associated protein markers. An HCC EV surface protein assay, composed of covalent chemistry-mediated HCC EV purification and real-time immuno-polymerase chain reaction readouts, was developed and optimized for quantifying subpopulations of EVs. An HCC EV ECG score, calculated from the readouts of three HCC EV subpopulations (EpCAM+ CD63+, CD147+ CD63+, and GPC3+ CD63+ HCC EVs), was established for detecting early-stage HCC. A phase 2 biomarker study was conducted to evaluate the performance of ECG score in a training cohort (n = 106) and an independent validation cohort (n = 72).
Overall, 99.7% of tissue microarray stained positive for at least one of the four HCC-associated protein markers (EpCAM, CD147, GPC3, and ASGPR1) that were subsequently validated in HCC EVs. In the training cohort, HCC EV ECG score demonstrated an area under the receiver operating curve (AUROC) of 0.95 (95% confidence interval [CI], 0.90–0.99) for distinguishing early-stage HCC from cirrhosis with a sensitivity of 91% and a specificity of 90%. The AUROCs of the HCC EV ECG score remained excellent in the validation cohort (0.93; 95% CI, 0.87–0.99) and in the subgroups by etiology (viral: 0.95; 95% CI, 0.90–1.00; nonviral: 0.94; 95% CI, 0.88–0.99).
Conclusion:
HCC EV ECG score demonstrated great potential for detecting early-stage HCC. It could augment current surveillance methods and improve patients’ outcomes.
INTRODUCTION
Primary liver cancer is the third-leading cause of cancer-related death worldwide in 2020.[1] In the United States, despite the plateaued incidence and mortality trends over the past decade,[2,3] more than 30,000 deaths from primary liver cancer are estimated in 2021.[4] Hepatocellular carcinoma (HCC) accounts for 80%–85% of primary liver cancers[5,6] and mainly occurs in patients with liver cirrhosis or chronic hepatitis B virus (HBV) infection.[6] Prognosis of HCC is dismal primarily because of advanced stage of disease at diagnosis, and studies have shown early detection of HCC is associated with increased receipt of curative therapy and improved patient prognosis.[3,7] Current clinical practice guidelines recommend HCC surveillance by biannual liver ultrasound with/without serum alpha-fetoprotein (AFP) for at-risk patients to achieve the goal of detecting HCC at a curative stage.[8,9] However, their accuracy remains relevantly low with sensitivity between 60% and 70% at a specificity of 90%.[10-12] As such, novel biomarkers for early detection of HCC are still desperately needed.
Extracellular vesicles (EVs) refer to nanoparticles released by both normal and tumor cells.[13,14] Enclosed by lipid bilayer membranes, EVs contain biomolecules including DNA, RNA, proteins, metabolites, and lipids. Through transferring these cargoes, tumor-derived EVs participate in cell-to-cell communications and cancer progression. Given their early presence in circulation during tumorigenesis, profiling tumor-derived EVs is regarded as a promising liquid biopsy approach for diagnosis of early-stage cancer.[13,14] There have been several studies investigating the role of EVs in detecting HCC among at-risk patients with highly promising results.[13,15,16] For example, von Felden et al. and our team demonstrated the noncoding RNA[15] and mRNA[16] markers in EVs can be used to discriminate Barcelona Clinic Liver Cancer (BCLC) stage 0-A HCC from cirrhosis with area under receiver operator characteristic curves (AUROCs) of 0.93, respectively. Therefore, EVs hold great potential to augment current surveillance strategy and improve the outcomes of patients with HCC.
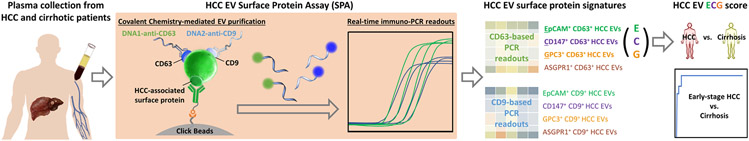
Here, we introduced a streamlined HCC EV surface protein assay (SPA) (Figure 1), capable of dissecting and quantifying eight subpopulations of HCC EVs (i.e., epithelial cellular adhesion molecule [EpCAM]+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, Glypican 3 Protein [GPC3]+ CD63+ HCC EVs, asialoglycoprotein receptor 1 [ASGPR1]+ CD63+ HCC EVs, EpCAM+ CD9+ HCC EVs, CD147+ CD9+ HCC EVs, GPC3+ CD9+ HCC EVs, and ASGPR1+ CD9+ HCC EVs) in 400-μl plasma samples based on the combined use of covalent chemistry(16–18)-mediated EV purification and duplex real-time immuno-polymerase chain reaction (PCR). A logistic regression model, HCC EV ECG score, was established from the resultant HCC EV surface protein signatures (i.e., EpCAM+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, GPC3+ CD63+ HCC EVs) to distinguish early-stage HCC from cirrhosis. We aimed to conduct a phase 2 biomarker study following the International Liver Cancer Association (ILCA) biomarker development guideline[17] to evaluate the performance of HCC EV ECG score for detecting early-stage HCC.
FIGURE 1.
A general workflow of developing HCC EV ECG score for distinguishing early-stage HCC from cirrhosis. Plasma samples from patients with HCC or liver cirrhosis were introduced to HCC EV surface protein assay that combined the use of covalent chemistry-mediated EV purification and duplex real-time immuno-PCR, to quantify eight subpopulations of HCC EVs, i.e., EpCAM+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, GPC3+ CD63+ HCC EVs, ASGPR1+ CD63+ HCC EVs, EpCAM+ CD9+ HCC EVs, CD147+ CD9+ HCC EVs, GPC3+ CD9+ HCC EVs, and ASGPR1+ CD9+ HCC EVs. The resulting HCC EV surface protein signatures was then analyzed to generate HCC EV ECG score (accounted from EpCAM+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, and GPC3+ CD63+ HCC EVs) for distinguishing early-stage HCC from at-risk liver cirrhosis. ASGPR1, Asialoglycoprotein receptor 1; EpCAM, epithelial cellular adhesion molecule; EV, extracellular vesicle; GPC3, Glypican 3 Protein; HCC, hepatocellular carcinoma; PCR, polymerase chain reaction.
MATERIALS AND METHODS
Study population
For this phase 2 biomarker (case–control) study, a total of 106 participants (45 patients with treatment-naïve early-stage HCC and 61 patients with liver cirrhosis) were enrolled between October 2016–August 2021 at Ronald Reagan University of California, Los Angeles (UCLA) Medical Center as a training cohort to develop and optimize HCC EV SPA for distinguishing early-stage HCC from cirrhosis. Additionally, six patients with BCLC stage B-C HCC, seven patients with breast cancer, five patients with prostate cancer, and five patients with thyroid cancer were enrolled as control groups to confirm the specificity of HCC EV ECG score toward HCC. A total of 72 participants (35 patients with treatment-naïve early-stage HCC and 37 patients with liver cirrhosis) were enrolled between October 2019–October 2021 at Cedars-Sinai Medical Center (CSMC) as an independent validation cohort. All participants provided written informed consent for this study according to the institutional review board (IRB) protocols #14-000197, #10-000236-AM-00021, #20-001197 at UCLA and IRB protocols #00000066, #00042197, #00033050 at CSMC. No donor organs were obtained from executed prisons.
HCC was diagnosed according to the American Association for the Study of Liver Diseases clinical practice guideline[8]: (i) histology or (ii) imaging categorized as Liver Imaging Reporting and Data System 5. Early-stage HCC was defined as BCLC stage 0 or A. Liver cirrhosis was defined according to histology and imaging feature of cirrhosis (nodular surface) or portal hypertension (splenomegaly or portosystemic collaterals). Patients with cirrhosis were ensured the absence of HCC by (i) at least six months of follow-up after blood collection or (ii) a negative contrast-enhanced multiphasic computed tomography or magnetic resonance imaging within two weeks of blood collection. All HCC cases were treatment-naïve at the time of blood collection. Patients were excluded if they had concomitant neoplasms. Etiologies of underlying liver disease were defined in Supplementary Methods.
General information for HCC EV SPA
To conduct HCC EV SPA, we (i) produced Click Beads[18] (i.e., methyltetrazine [mTz]-modified microbeads) according to a chemical modification procedure (Figure S1) and optimized them as described in Supplementary Methods and Figures S2-S4, (ii) prepared the four transcyclooctene (TCO)-conjugated HCC-associated antibodies using the copper-free click chemistry coupling method (Figure S5), and (iii) conjugated DNA barcode onto anti-CD63 and anti-CD9 to target EV markers, followed by validation (Figure S6, the primers and probes for the DNA barcodes were purchased from Integrated DNA Technologies and the sequences are listed in Table S1).
HCC EV SPA for quantification of eight subpopulations of HCC EVs
HCC EV SPA is implemented through a 3-step workflow (Figure 1 and Figure S7). An internal control, fetal bovine serum, was used instead of plasma samples to measure the background binding of the DNA1-anti-CD63 and DNA2-anti-CD9 to Click Beads (Table S2).
Step 1: Sequentially labeling each subpopulation of HCC EVs in plasma
A total of 400 μl of plasma for each patient were evenly aliquoted into four Eppendorf tubes. In the first incubation, these four 100-μ1 plasma aliquots were incubated with the respective TCO-conjugated HCC-associated antibodies: TCO-anti-EpCAM (50 ng), TCO-anti-CD147 (25 ng), TCO-anti-GPC3 (50 ng), and TCO-anti-ASGPR1 (50 ng), respectively. In the second incubation, these aliquots were cocktailed with DNA-conjugated EV-specific antibodies: DNA1-anti-CD63 and DNA2-anti-CD9, at room temperature for 30 min. No TCO-conjugated HCC-associated antibody was added into the internal control that was only incubated with DNA1-anti-CD63 and DNA2-anti-CD9.
Step 2: Covalent chemistry-mediated capture of subpopulations of HCC EVs onto Click Beads
Click Beads[18] were blocked with protein free buffer and then 5% bovine serum albumin (BSA) solution before use. The plasma samples labeled with TCO-antibodies, and DNA barcodes were then incubated with the Click Beads (0.1 mg) for 30 min for isolation of each subpopulation of EVs. Click Beads with isolated EVs were collected by centrifuge at 10,000 g for 2 min. All plasma samples subjected to Click Beads underwent only one freeze-thaw cycle. The internal control was processed with the same protocol.
Step 3: On-bead duplex real-time immuno-PCR for quantifying each subpopulation of HCC EVs
Click Beads with isolated EVs were resuspended in 500 μl phosphate-buffered saline (PBS) with 0.2% BSA and then collected by centrifuge at 10,000 g for 2 min. After 3-time washing step with PBS with 0.2% BSA, Click Beads were then washed by PBS for two times to wash off unbounded DNA1-anti-CD63 and DNA2-anti-CD9. After 5-time washing steps, 90 μl of double-distilled water was added to the sediments of each well and mixed thoroughly. Nine μl of the mixed suspension was loaded into the real-time PCR system (CFX Connect Real-Time PCR Detection System, Bio-Rad). Raw data of real-time PCR runs were evaluated using the corresponding software system (Bio-Rad CFX Manager 3.1, Bio-Rad). The relative signal was calculated using the following equation:
Statistical analysis
For descriptive statistics, continuous variables are reported as median and interquartile range, and categorical variables are reported as numbers and percentages. Comparison of continuous variables and categorical variables between groups were done using Mann–Whitney U test and Fisher exact test or chi-square test, respectively.
In this retrospective phase 2 biomarker (case–control) study, the sample size was calculated for comparing AUROCs between HCC EV SPA and serum AFP,[10] using the paired DeLong test. A sample size of 48 (24 HCC and 24 control) was expected to have 90% power to detect the difference between the AUROCs for our HCC EV SPA versus serum AFP, assuming AUROC = 0.93 for our assay, AUROC = 0.69 for serum AFP[16] for detecting early-stage HCC, when a correlation between the assays of 0.5 was assumed. The power was obtained for a two-sided test at 0.05 significance level.
To identify the HCC EV subpopulations significantly associated with early-stage HCC over cirrhosis (p < 0.05), univariate logistic regression analysis was applied in the training cohort (UCLA cohort; 45 HCC and 61 liver cirrhosis). The HCC EV subpopulations highly associated with HCC at a significance level of 0.005 were included as the final logistic regression model (i.e., HCC EV ECG score) for detecting early-stage HCC from cirrhosis. Youden's index was used to identify the optimal cutoff of HCC EV ECG score. Model calibration indicated the agreement between predicted probabilities from the model and observed event rates was assessed with the calibration plot of probability of actual occurrence versus prediction and the Hosmer–Lemeshow test. Brier score was used to measure the overall accuracy of HCC EV ECG score. Leave-one-out cross-validation was applied to estimate the performance of HCC EV ECG score in the training cohort. Furthermore, classification improvement by adding the serum AFP level to HCC EV ECG score was evaluated with the categorical net reclassification improvement index (NRI) in the training cohort. The categorical NRI was calculated for HCC probability categories <50% and ≥50%. External validation of HCC EV ECG score was performed in the independent validation cohort (CSMC cohort; 35 HCC and 37 liver cirrhosis). The overall study design was summarized in Figure 2. Following the ILCA biomarker development guideline for HCC,[17] sensitivity, specificity, positive predictive value (PPV), negative predictive value (NPV), and AUROC for HCC EV ECG score to discriminate early-stage HCC from at-risk cirrhosis were estimated in both the training and validation cohorts. The AUROC between the HCC EV ECG score and AFP was compared using the paired DeLong test.
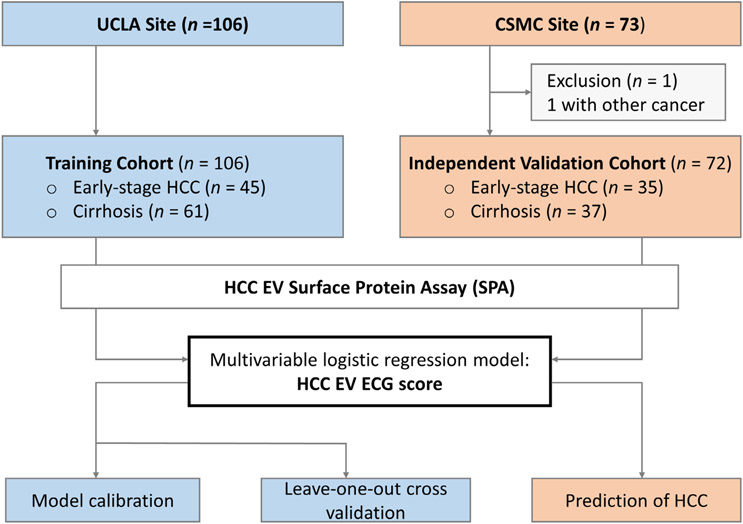
FIGURE 2.
Clinical study design flowchart depicting the recruitment and exclusions from the study cohort. Blood samples from 106 and 73 eligible participants were collected from UCLA (training cohort) and CSMC (independent validation cohort), respectively. In brief, HCC EV surface protein assay was utilized in the training cohort (UCLA cohort, n = 106) to identify the HCC EV subpopulations significantly associated with early-stage HCC over cirrhosis and establish the logistic regression model (i.e., HCC EV ECG score) for detecting early-stage HCC from cirrhosis. Model calibration was performed to evaluate the agreement between predicted probabilities from the model and observed event rates. Leave-one-out cross-validation was applied to estimate the performance of the established HCC EV ECG score in the training cohort. After one participant was excluded because of coexisting with other tumors, external validation of HCC EV ECG score was performed in the independent validation cohort (CSMC cohort, n = 72). CSMC, Cedars-Sinai Medical Center; ECG, EpCAM+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, and GPC3+ CD63+ HCC EVs; EV, extracellular vesicle; GPC3, Glypican 3 Protein; HCC, hepatocellular carcinoma; UCLA, University of California, Los Angeles.
All statistical analyses were performed using MedCalc software (version 20.015; MedCalc Software Ltd), GraphPad Prism (version 9.2.0; GraphPad Software, Inc.), and R statistical software (version 4.0.2; R Foundation) with two-sided tests and a significance level of 0.05.
RESULTS
Selection and validation of the four HCC-associated surface protein markers for HCC EV SPA using TMA and HCC cell line-derived EVs
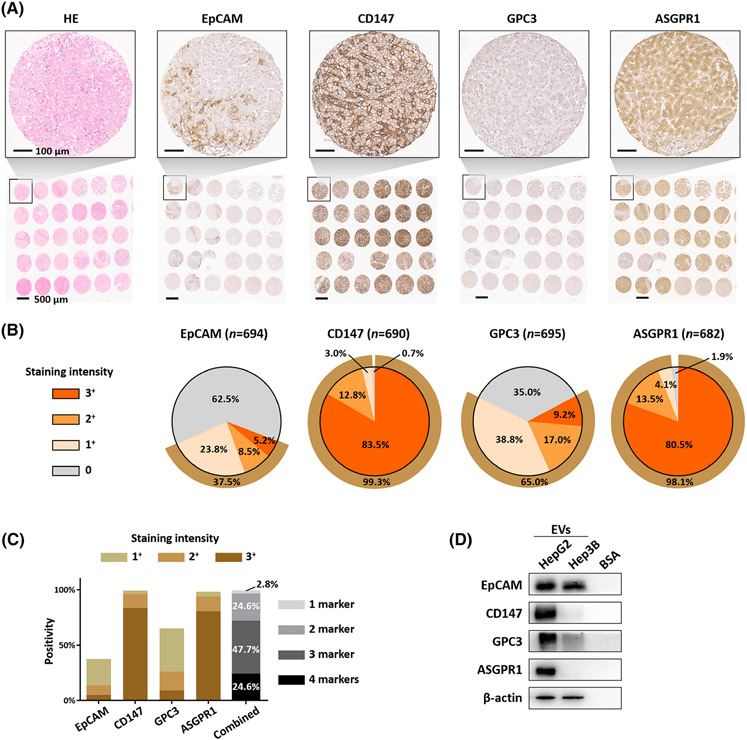
The HCC EV SPA relies on the use of different HCC-associated surface protein markers to target and purify subpopulations of HCC EVs in the plasma samples. Recent studies demonstrated the feasibility of selectively characterizing HCC EVs in plasma samples using HCC-associated surface markers, such as EpCAM[19] and ASGPR1.[20] In particular, one study[20] showed that EVs carrying the surface protein markers AnnexinV, EpCAM, CD147, and ASGPR1 that are significantly increased in the plasma of patients with HCC compared with patients with liver cirrhosis and healthy individuals. Our group also demonstrated that EpCAM, GPC3, and ASGPR1 are HCC-associated surface markers.[21] As such, four candidate HCC EV surface protein markers (i.e., EpCAM, ASGPR1, GPC3, and CD147) were selected for validation. Because the surface proteins on tumor-derived EVs could mirror those of the parental tumor cells, we first validated the expression of the four candidate markers using a 708-sample HCC tissue microarray (TMA) (Supplementary Methods, Supplementary data). The representative TMA hematoxylin and eosin staining and immunohistochemistry (IHC) staining for the four selected HCC-associated surface protein markers were shown in Figure 3A. Figure 3B summarizes the quantification IHC results of the TMA. Among the four selected markers, CD147 exhibited the highest positivity of the 708 HCC tumors (excluding those cases without tumor tissues captured on the TMA slides), 576 (83.5%), 88 (12.8%), and 21 (3.0%) were stained as strong (3+), moderate (2+), and weak (1+), respectively. For the commonly used epithelial marker, EpCAM, only 36 (5.2%), 59 (8.5%), and 165 (23.8%) were stained as strong (3+), moderate (2+), and weak (1+), respectively. In terms of the HCC-specific marker GPC3, 64 (9.2%), 118 (17.0%), and 270 (38.8%) were stained as strong (3+), moderate (2+), and weak (1+), respectively. As expected, the majority of HCC tissues are positive for liver-specific marker ASGPR1 with 549 (80.5%), 92 (13.5%), and 28 (4.1%) stained as strong (3+), moderate (2+), and weak (1+), respectively, demonstrating the primary liver origin of the HCC tissues. Overall, 99.7% (701 out of 703) stained positive for at least one marker (24.6% with all four markers positive, 47.7% with any three markers positive, 24.6% with any two markers positive, 2.8% with any one marker positive, Figure 3C), which underscored the complementary expression of the four surface protein markers in the heterogenous HCC tissues and laid the solid foundation for the use of these HCC-associated antibodies for HCC EV SPA.
FIGURE 3.
Validation of the four selected HCC-associated surface markers using TMA and HCC cells-derived EVs. (A) H&E staining and immunohistochemistry staining of the four selected HCC-associated surface protein markers, EpCAM, CD147, GPC3, and ASGPR1, on a TMA slide containing 708 HCC samples. Scale bar, 100 μm. (B) Percentage of tumors with negative (0), weak (1+), moderate (2+), strong (3+) staining for the four given surface protein markers. (C) The percentage of tumors staining for at least one of the four markers (combined) summarized in the bar chart. (D) Western blotting for the four surface protein markers in HepG2-derived EVs and Hep3B-derived EVs, with BSA as the negative control. ASGPR1, Asialoglycoprotein receptor 1; BSA, bovine serum albumin; EpCAM, epithelial cellular adhesion molecule; EV, extracellular vesicle; GPC3, Glypican 3 Protein; H&E, Hematoxylin and Eosin; HCC, hepatocellular carcinoma; TMA, tissue microarray.
After identifying, selecting, and conducting TMA validation of the four HCC-associated surface proteins, we then validated their presence in HCC cell line-derived EVs by western blotting. As summarized in Figure 3D, all of the four HCC-associated surface protein markers were detected in EVs derived from HepG2, a human liver cancer cell line. In EVs derived from Hep3B, another human liver cancer cell line, both EpCAM and GPC3 were detected. In addition, the expression of these HCC-associated surface protein markers was higher in EVs from HCC cells (Huh-7) than EVs from immortalized human hepatocytes (MIHA, Figure S8A). We also showed low or absent expression of these HCC-associated surface protein markers in EVs from primary human hepatocytes (Figure S8B), providing the rationale for selecting these protein markers as candidates of HCC detection biomarkers.
Quantification of the eight subpopulations of HCC EVs for distinguishing early-stage HCC from at-risk liver cirrhosis in UCLA cohort
After optimizing HCC EV SPA using artificial samples (Figures S3, S6, and S9) and confirming the reproducibility (Figure S10 and Tables S2-S3), 45 patients with early-stage HCC and 61 at-risk patients with liver cirrhosis were enrolled at UCLA as the training cohort for testing HCC EV SPA. The demographic and clinical characteristics of the UCLA cohort are demonstrated in Table 1. Age, sex, and race/ethnicity were similar between patients with HCC and controls with cirrhosis. Approximately three-quarters of patients with HCC (73%) had well-compensated liver disease (Child-Pugh A), compared to nearly half of the controls with cirrhosis (44%). Among the patients with HCC, 82% had liver cirrhosis. Eighty percent of the patients with HCC had BCLC stage A cancer, and the rest had BCLC stage 0.
TABLE 1.
Demographic and clinical characteristics of UCLA cohort and CSMC cohort
| Characteristic | UCLA cohort (Training cohort) |
CSMC cohort (Validation cohort) |
p value HCC: UCLA versus CSMC |
p value Cirrhosis: UCLA versus CSMC |
||||
|---|---|---|---|---|---|---|---|---|
| Early-stage HCC (n = 45) |
Cirrhosis (n = 61) |
p value | Early-stage HCC (n = 35) |
Cirrhosis (n = 37) |
p value | |||
| Age, y (IQR) | 64 (59–70) | 62 (54–70) | 0.12 | 67 (62–70) | 59 (50.5–65) | <0.001 | 0.48 | 0.13 |
| Sex, n (%) | – | – | 0.23 | – | – | 0.15 | >0.99 | >0.99 |
| Female | 14 (31.1) | 27 (44.3) | – | 10 (28.6) | 17 (45.9) | – | – | – |
| Male | 31 (68.9) | 34 (55.7) | – | 25 (71.4) | 20 (54.1) | – | – | – |
| Race/ethnicity, n (%) | – | – | 0.33 | – | – | 0.01 | 0.27 | 0.13 |
| White | 18 (40.0) | 18 (29.5) | – | 8 (22.8) | 13 (35.1) | – | – | – |
| Black | 1 (2.2) | 1 (1.6) | – | 3 (8.6) | 0 (0) | – | – | – |
| Asian | 8 (17.8) | 12 (19.7) | – | 10 (28.6) | 2 (5.4) | – | – | – |
| Hispanic | 17 (37.8) | 22 (36.1) | – | 14 (40.0) | 20 (54.1) | – | – | – |
| Others/Unknown | 1 (2.2) | 8 (13.1) | – | 0 (0) | 2 (5.4) | – | – | – |
| Child-Pugh Score, n (%) | – | – | 0.003 | – | – | <0.001 | >0.99 | 0.13 |
| A | 33 (73.3) | 27 (44.3) | – | 25 (71.4) | 10 (27.0) | – | – | – |
| B-C | 12 (26.7) | 34 (55.7) | – | 10 (28.6) | 27 (73.0) | – | – | – |
| MELD score (IQR) | 11 (7–14) | 13 (9–17) | 0.06 | 10 (7–18) | 23 (13–27.8) | <0.001 | 0.87 | <0.001 |
| Etiology, n (%) | – | – | 0.50 | – | – | 0.001 | 0.54 | 0.03 |
| HBV | 7 (15.6) | 7 (11.5) | – | 6 (17.1) | 2 (5.4) | – | – | – |
| HCV | 14 (31.1) | 13 (21.3) | – | 13 (37.1) | 2 (5.4) | – | – | – |
| ALD | 12 (26.7) | 21 (34.4) | – | 4 (11.5) | 15 (40.6) | – | – | – |
| NAFLD | 9 (20.0) | 18 (29.5) | – | 8 (22.8) | 11 (29.7) | – | – | – |
| Others | 3 (6.7) | 2 (3.3) | – | 4 (11.5) | 7 (18.9) | – | – | – |
| Cirrhosis, n (%) | 37 (82.2) | 61 (100.0) | <0.001 | 30 (85.7) | 37 (100) | 0.02 | 0.77 | >0.99 |
| ALT, median (IQR) | 22 (29–43) | 26 (18.5–37) | 0.44 | 27 (18–43) | 26 (16–47.5) | 0.74 | 0.54 | 0.56 |
| AFP, median (IQR) | 9.9 (3.1–43.0) | 3.5 (2.3–5.1) | <0.001 | 29.5 (4.3–188.4) | 2.5 (1.9–4.3) | <0.001 | 0.12 | 0.07 |
| AFP <20, n (%) | 27 (60.0) | 57 (93.4) | – | 16 (45.7) | 36 (97.3) | – | – | – |
| AFP ≥20, n (%) | 17 (37.8) | 1 (1.6) | – | 18 (51.4) | 0 (0) | – | – | – |
| NA, n (%) | 1 (2.2) | 3 (4.9) | – | 1 (2.9) | 1 (2.7) | – | – | – |
| Tumor stage | – | – | NA | – | – | NA | 0.79 | NA |
| Very early (BCLC 0), n (%) | 9 (20.0) | NA | – | 8 (22.8) | NA | – | – | NA |
| Early (BCLC A), n (%) | 36 (80.0) | NA | – | 27 (77.2) | NA | – | – | NA |
| Single tumor, n (%) | 43 (95.6) | NA | NA | 29 (82.9) | NA | NA | 0.13 | NA |
| Largest tumor, cm (IQR) | 3.0 (2.2–4.5) | NA | NA | 3.0 (2.0–4.0) | NA | NA | 0.64 | NA |
| Within Milan criteria, n (%) | 37 (82.2) | NA | NA | 31 (88.6) | NA | NA | 0.54 | NA |
Abbreviations: AFP, alpha-fetoprotein; ALD, alcoholic liver disease; ALT, alanine aminotransferase; BCLC, Barcelona clinic liver cancer; CSMC, Cedars-Sinai Medical Center; HBV, hepatitis B virus; HCC, hepatocellular carcinoma; HCV, hepatitis C virus; IQR, interquartile range; MELD, Model for End-Stage Liver Disease; NA, not available; UCLA, University of California, Los Angeles.
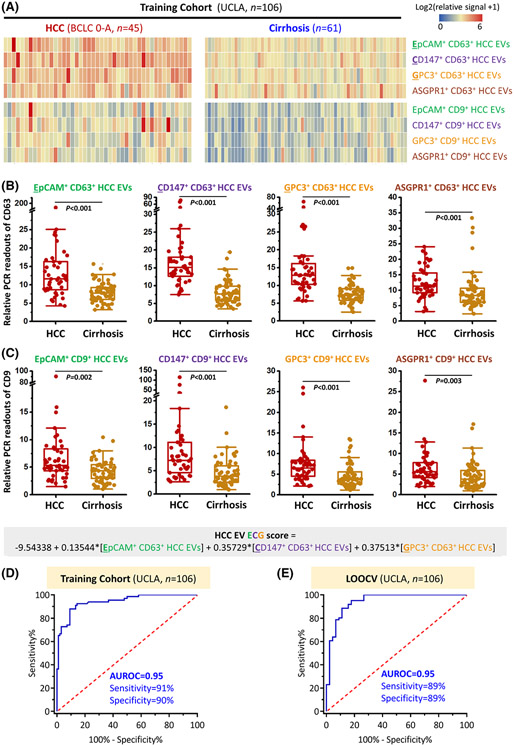
The optimized HCC EV SPA was utilized to obtain the HCC EV surface protein signatures of the UCLA cohort (Figure 1). In brief, the identified four HCC-associated antibodies (i.e., anti-EpCAM, anti-CD147, anti-GPC3, and anti-ASGPR1) were applied to purify the EVs and two conjugated DNA-antibodies (i.e., DNA1-anti-CD63 and DNA2-anti-CD9) targeting EV markers were applied to report the presence of EVs. The resulting readouts of eight subpopulations of HCC EVs (i.e., EpCAM+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, GPC3+ CD63+ HCC EVs, ASGPR1+ CD63+ HCC EVs, EpCAM+ CD9+ HCC EVs, CD147+ CD9+ HCC EVs, GPC3+ CD9+ HCC EVs, and ASGPR1+ CD9+ HCC EVs) were summarized in Figure 4A. Overall, significantly higher signals were observed in the HCC group compared to those found in the liver cirrhosis group (p <0.005 in all subpopulations; Figure 4B,C).
FIGURE 4.
HCC EV SPA for measuring subpopulations of HCC EVs and detecting early-stage HCC in the UCLA training cohort. (A) Heatmaps summarize relative duplex real-time immuno-PCR readouts of plasma samples from patients with early-stage HCC (BCLC Stage 0-A, n = 45) and patients with liver cirrhosis (n = 61). Significantly higher immuno-PCR signals of both (B) CD63+ and (C) CD9+ HCC EV subpopulations were observed in patients with HCC compared to those with cirrhosis. (D) ROC curve of HCC EV ECG score that was calculated by the signals from EpCAM+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, and GPC3+ CD63+HCC EVs using HCC EV SPA, for detecting early-stage HCC from cirrhosis in the UCLA training cohort. (E) ROC curve of HCC EV ECG score after leave-one-out cross-validation for detecting early-stage HCC from cirrhosis in the UCLA training cohort. ASGPR1, Asialoglycoprotein receptor 1; AUROC, area under the receiver operating characteristic curve; BCLC, Barcelona Clinic Liver Cancer; EpCAM, epithelial cellular adhesion molecule; EV, extracellular vesicle; GPC3, Glypican 3 Protein; HCC, hepatocellular carcinoma; LOOCV, leave-one-out cross-validation; PCR, polymerase chain reaction; ROC, receiver operating characteristic; SPA, Surface Protein Assay; UCLA, University of California, Los Angeles.
Development of HCC EV ECG score for distinguishing early-stage HCC from at-risk cirrhosis
To evaluate the ability of the HCC EV surface protein signatures (based on quantification of eight subpopulations of HCC EVs) for distinguishing early-stage HCC from at-risk cirrhosis, the UCLA cohort was employed as the training cohort, and the diagnostic performance of each HCC EV subpopulation was summarized in Figure S11. Among the eight HCC EV subpopulations, the AUROCs of CD147+ CD63+ HCC EVs and GPC3+ CD63+ HCC EVs are the highest at 0.91 (95% confidence interval [CI], 0.86–0.96) and 0.86 (95% CI, 0.79–0.94), respectively. In addition, univariate logistic regression analysis was performed to identify the HCC EV subpopulations significantly associated with early-stage HCC over cirrhosis (Table S3). Three HCC EV subpopulations highly associated with HCC at a significance level of 0.005, EpCAM+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, and GPC3+ CD63+ HCC EVs, were selected in the final logistic regression model for detecting early-stage HCC from cirrhosis, named HCC EV ECG score.
HCC EV ECG score is defined as:
This HCC EV ECG score exhibited excellent accuracy for discriminating patients with early-stage HCC from controls with cirrhosis in the training cohort with an AUROC of 0.95 (95% CI, 0.90–0.99; Figure 4D). At the optimal cutoff of −0.40, the sensitivity and specificity of HCC EV ECG score for early-stage HCC detection were 91% (95% CI, 79%–96%) and 90% (95% CI, 80%–95%), respectively, with the PPV of 87% (95% CI, 76%–94%) and NPV of 93% (95% CI, 84%–97%) (Table S5). The model, HCC EV ECG score, was well calibrated with a low Brier score (0.08) and a low mean absolute probability error (0.015) to predict early-stage HCC after 1000 bootstrap resampling (Figure S12). The p value of HCC EV ECG score for the Hosmer-Lemeshow test was also higher than 0.05. Leave-one-out cross-validation of the training cohort confirmed the accuracy of the model with the AUROC of 0.95 (95% CI, 0.88–0.98; Figure 4E), sensitivity of 89% (95% CI, 76%–96%), and specificity of 89% (95% CI, 78%–95%).
Next, we examined if adding serum AFP level to HCC EV ECG score would improve its performance for distinguishing early-stage HCC from cirrhosis. However, as shown in Table S6 and Figure S13, addition of serum AFP level did not make any contributions to reclassification improvement (NRI, −0.06 [95% CI, −0.06–0.16]; DeLong test: p = 0.31).
Finally, we compared the HCC EV ECG scores from patients with early-stage HCC (n = 45) to those from patients with BCLC stage B-C HCC (n = 6), breast cancer (n = 7), prostate cancer (n = 5), or thyroid cancer (n = 5) (the demographic and clinical characteristics are shown in Table S7). As shown in Figure S14, HCC EV ECG scores from patients with HCC were significantly higher than patients with other cancers (p <0.001, respectively), confirming the specificity of HCC EV ECG score to HCC rather than cancers in general.
Performance of HCC EV ECG score for detecting early-stage HCC in an independent validation cohort
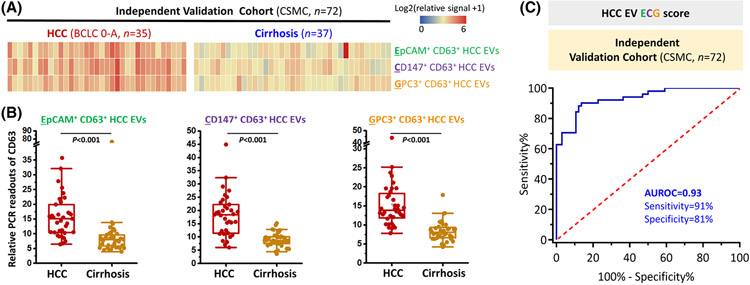
To further validate HCC EV ECG score for detecting early-stage HCC, a total of 35 patients with early-stage HCC and 37 patients with at-risk liver cirrhosis recruited at CSMC were utilized as an independent validation cohort and analyzed. The clinical characteristics of the CSMC cohort are shown in Table 1 and the HCC EV surface protein signatures of the CMSC cohort were summarized in Figure 5A. There was no significant difference between CSMC and UCLA cohorts in terms of the patient characteristics. The readouts of EpCAM+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, and GPC3+ CD63+ HCC EVs were significantly higher in patients with HCC than patients with liver cirrhosis as expected (p < 0.001; Figure 5B).
FIGURE 5.
HCC EV ECG score for detecting early-stage HCC in the CSMC independent validation cohort. (A) Heatmaps summarize relative duplex real-time immuno-PCR signals of plasma samples from patients with early-stage HCC (BCLC Stage 0-A, n = 35) and patients with liver cirrhosis (n = 37). Significantly higher immuno-PCR signals of (B) EpCAM+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, and GPC3+ CD63+ HCC EVs were observed in patients with HCC compared to those with cirrhosis. (C) ROC curve of HCC EV ECG score for detecting early-stage HCC from cirrhosis in the CSMC independent validation cohort. ASGPR1, Asialoglycoprotein receptor 1; AUROC, area under the receiver operating characteristic curve; BCLC, Barcelona Clinic Liver Cancer; CSMC, Cedars-Sinai Medical Center; EpCAM, epithelial cellular adhesion molecule; EV, extracellular vesicle; GPC3, Glypican 3 Protein; HCC, hepatocellular carcinoma; PCR, polymerase chain reaction; ROC, receiver operating characteristic.
In the CSMC validation cohort, the AUROC of HCC EV ECG score for detecting early-stage HCC remained excellent as 0.93 (95% CI, 0.87–0.99; Figure 5C). At the cutoff of −0.65, the sensitivity was 94% (95% CI, 81%–99%) and the specificity was 81% (95% CI, 65%–92%). When setting the cutoff value at −0.40, the same optimal cutoff value identified in the training cohort, HCC EV ECG score still had great accuracy of detecting early-stage HCC with sensitivity of 91% (95% CI, 77%–98%), specificity of 81% (95% CI, 65%–92%), PPV of 82% (95% CI, 70%–90%) and NPV of 91% (95% CI, 76%–93%) (Table S8).
Comparison between HCC EV ECG score and serum AFP for detecting early-stage HCC and subgroup analyses
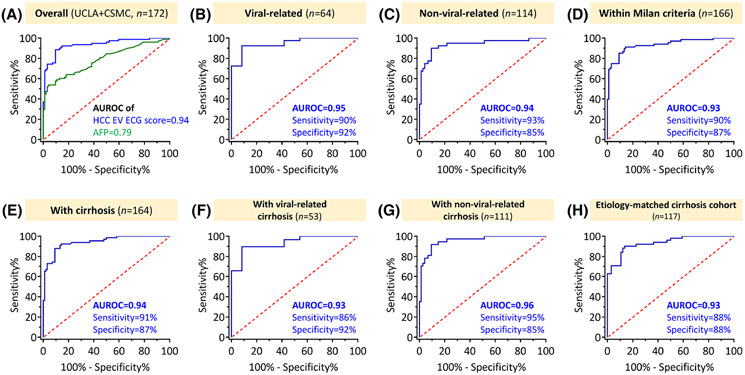
After validating HCC EV ECG score in an independent cohort, we then compared the performance of HCC EV ECG score with serum AFP for detecting early-stage HCC in all the participants in this study (UCLA+CSMC cohorts, n = 172; Table S9). Individuals without serum AFP records (n = 6) were excluded in the analyses. As demonstrated in Figure 6A, HCC EV ECG score outperformed serum AFP in distinguishing early-stage HCC from cirrhosis (AUROC, 0.94; 95% CI, 0.90–0.97 vs. AUROC, 0.79; 95% CI, 0.72–0.86; p < 0.001). At the identified cutoff of −0.40, the sensitivity of HCC EV ECG score was 91% (95% CI, 82%–96%) and the specificity was 86% (95% CI, 78%–92%). For serum AFP, when using the standard cutoff of 20 ng/ml, it had the sensitivity of 45% (95% CI, 34%–57%) and the specificity of 99% (95% CI, 94%–100%). The HCC EV ECG score was not correlated with serum AFP level, serum alanine aminotransferase level, and model for end-stage liver disease (MELD) score (Figure S15), that suggested our results were not biased by these factors.
FIGURE 6.
Comparison between HCC EV ECG score and serum AFP for detecting early-stage HCC among all the participants and subgroup analyses. (A) Comparison of the performance of HCC EV ECG score and serum AFP for detecting early-stage HCC among all the participants (n = 172). (B) ROC curve of HCC EV ECG score for detecting early-stage HCC from cirrhosis among patients with viral etiology (n = 64). (C) ROC curve of HCC EV ECG score for detecting early-stage HCC from cirrhosis among patients with nonviral etiology (n = 114). (D) ROC curve of HCC EV ECG score for detecting early-stage HCC within Milan criteria (n = 68) from cirrhosis (n = 98). (E) ROC curve of HCC EV ECG score for detecting early-stage HCC from cirrhosis among patients with cirrhosis (n = 164). (F) ROC curve of HCC EV ECG score for detecting early-stage HCC from cirrhosis among patients with viral-related cirrhosis (n = 53). (G) ROC curve of HCC EV ECG score for detecting early-stage HCC from cirrhosis among patients with nonviral-related cirrhosis (n = 111). (H) ROC curve of HCC EV ECG score for detecting early-stage HCC from cirrhosis among a cohort with cirrhosis after frequency matching of the etiology (n = 117). AUROC with the sensitivity and specificity of the assays at the optimal cutoffs are listed for each graph. AFP, alpha-fetoprotein; AUROC, area under the receiver operating characteristic curve; ECG, EpCAM+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, and GPC3+ CD63+ HCC EVs; EV, extracellular vesicle; HCC, hepatocellular carcinoma; ROC, receiver operating characteristic.
The performance of HCC EV ECG score remained excellent in the subpopulations of patients stratified by etiology (viral vs. nonviral) and Milan criteria. Among patients with chronic viral hepatitis (n = 64), the AUROC was 0.95 (95% CI, 0.90–1.00; Figure 6B); on the other hand, the AUROC was 0.94 (95% CI, 0.88–0.99; Figure 6C) for patients with nonviral etiology (n = 114). Among patients with nonviral-related disease (Figure S16A-C), we demonstrated consistent performance of HCC EV ECG score in metabolic-related subgroup (i.e., NAFLD-related and alcoholic liver disease [ALD]-related; AUROC, 0.96; 95% CI, 0.93–1.00), NAFLD-related subgroup (AUROC, 0.97; 95% CI, 0.91–1.00), and ALD-related subgroup (AUROC, 0.95; 95% CI, 0.90–1.00). When restricting the patients with HCC to those with tumor within Milan criteria (n = 166), HCC EV ECG score still had excellent performance with AUROC of 0.93 (95% CI, 0.89–0.97; Figure 6D), sensitivity of 90% (95% CI, 80%–96%), and a specificity of 87% (95% CI, 78%–93%) for HCC detection.
HCC EV ECG score for detecting early-stage HCC among subgroups with cirrhosis
HCC EV ECG score was further evaluated in the subgroup of patients with HCC and cirrhosis (n = 164). At the identified cutoff of −0.40, the sensitivity was 91% (95% CI, 81%–97%) and the specificity was 87% (95% CI, 78%–93%) in the subgroup with cirrhosis with an AUROC of 0.94 (Figure 6E). The sensitivity and specificity were comparable among patients with cirrhosis with different etiologies (Figure 6F,G and Figure S16D-F). In addition, HCC EV ECG score had similar performance in a cohort with cirrhosis after frequency matching of the etiology between cases and controls (Figure 6H, n = 117; the cohort's characteristics were provided in Table S10).
DISCUSSION
In the current study, we developed a streamlined HCC EV SPA capable of quantifying eight subpopulations of HCC EVs in 400-μl plasma samples based on the combined use of four HCC-associated surface protein markers and two EV markers. Biostatistical analysis of the resulting HCC EV surface protein signatures established HCC EV ECG score, allowing for distinguishing early-stage HCC from at-risk cirrhosis. We conducted a phase 2 biomarker study and demonstrated a potential of HCC EV ECG score for detecting early-stage HCC with an AUROC of 0.95 and 0.93 in the UCLA training cohort and independent CSMC validation cohort, respectively. Its diagnostic ability remained excellent among the subpopulations by etiology and those with tumors within Milan criteria.
Current HCC surveillance is based on ultrasound with/without AFP, despite their suboptimal diagnostic performance. A statistical model, the GALAD score (derived from gender, age, and three serum biomarkers [AFP-L3, AFP, and Des-carboxyprothrombin]), demonstrated high sensitivity in detecting HCC in phase II biomarker studies.[22,23] However, the more recent phase III study showed overall lower accuracy, emphasizing the need for better biomarkers for HCC surveillance.[24] Liquid biopsy has emerged as another promising strategy for HCC surveillance.[13,25,26] For example, several circulating tumor DNA-based assays measuring the molecular characteristics have been proposed for detecting HCC and received Food and Drug Administration breakthrough device designation.[27-29] In addition, as shown in previous studies, the biomolecular cargoes such as microRNA (miRNA),[13] noncoding RNA,[15] mRNA,[16] and proteins in EVs are also regarded as potential biomarkers for early detection of HCC.
HCC heterogeneity includes spatial intratumor heterogeneity within each tumor, and interpatient heterogeneity with genetic and molecular diversity among tumors from different patients.[30] Similar to the parental tumor cells, HCC EVs are highly heterogeneous, underscoring the need for integration of different surface protein markers for developing liquid biopsy-based diagnostic assay to detect early-stage HCC. Our results from HCC tissue microarray showed that the selected four surface protein markers (i.e., EpCAM, CD147, GPC3, and ASGPR1) are complementarily expressed on the tumor cell surfaces. The complementary expression pattern of surface protein markers prompted us to profile and integrate different subpopulations of HCC EVs for detecting highly heterogeneous HCC. These surface protein markers were included in the final ECG score except ASGPR1, although ASGPR1 showed strong positivity in HCC TMA. It's well known that ASGPR1 is a liver-specific marker that will also be expressed in liver cells of patients with liver cirrhosis.
To dissect and quantify the subpopulations of HCC EVs, our team proposed an innovative assay, HCC EV SPA, by integrating two powerful technologies: Click Beads[18] for purification of HCC EVs and duplex real-time immuno-PCR for quantification of the purified subpopulations of HCC EVs. Compared with conventional immunoaffinity-based EV capture approaches, Click Beads achieve a more rapid and irreversible purification of HCC EVs by leveraging the click chemistry-reaction[31,32] between beads and targeted EVs. As demonstrated in our previous study,[16,18,33] the ligation between mTz-grafted devices and TCO-grafted EVs not only improved capture efficiency and but also reduced the nonspecific EV capture from the background.[16,33] Duplex real-time immuno-PCR technology combines the advantages of (i) flexibility and robustness of immunoassays and (ii) sensitivity of PCR. Immuno-PCR is capable of multiplex detection of several antigens using specific antibodies grafted with DNA barcodes. Real-time immuno-PCR typically exhibits a 10- to 1000-fold increase in sensitivity compared to an analogous enzyme-amplified immunoassay.[34] As such, the integration of Click Beads and duplex real-time immuno-PCR enables HCC EV SPA to quantify the HCC EV subpopulations sensitively and specifically from each individual. Perhaps most important of all, this assay only requires a very small amount of plasma (ca. 400 μl) to obtain patients’ HCC EV surface protein signatures.
Leveraging the optimized HCC EV SPA, the HCC EV surface protein signatures of patients with liver cirrhosis and HCC were analyzed. Three HCC EV subpopulations (i.e., EpCAM+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, and GPC3+ CD63+ HCC EVs) highly associated with HCC diagnosis were selected to establish HCC EV ECG score, allowing for distinguishing early-stage HCC from at-risk cirrhosis. With the guidance of the ILCA White Paper on HCC biomarker development,[17] a phase 2 case–control biomarker study was conducted to evaluate the performance of HCC EV ECG score. It is noteworthy that we restricted the patients with HCC to BCLC stage 0-A and the controls to at-risk cirrhosis to better assess the score's ability as a surveillance test. Importantly, HCC EV ECG score had excellent performance among the subgroups stratified by the underlying liver disease. Currently, viral hepatitis-related cirrhosis remains the most common cause of HCC, and cirrhosis resulting from ALD or NAFLD are rapidly growing contributors to HCC.[6,35] Verifying the performance of HCC EV ECG score in these subpopulations lays the solid foundation for future phase 3 biomarker study. Lastly, we showed the tumors within Milan criteria could also be identified by the assay with a high sensitivity of 90%, emphasizing the potential of HCC EV ECG score for timely detection of curable cancer. In addition to its excellent performance, HCC EV SPA has the advantages of fast turnaround time (within six hours from sample to result), convenient and user-friendly protocol, cost-efficiency, and requires only 400 μl of plasma to calculate HCC EV ECG score. Furthermore, after the COVID-19 era, real-time PCR system is ubiquitously deployed in clinical settings. Once validated in the subsequent phases of biomarker studies, we envision HCC EV SPA can be conveniently adopted in existing PCR facilities, paving the way for wide dissemination.
We acknowledge there are some limitations in the current study. First, the sample sizes of both cohorts are small, making it difficult to conduct granular subgroup analyses. In response, we dichotomized participants into viral-related and nonviral-related subgroups and investigated the assay's performance in each subpopulation. Despite small sample size, excellent performance of HCC EV ECG score was validated in an independent cohort of patients with different clinical characteristics (e.g., cirrhosis etiology, MELD score), highlighting the external validity of our test. Second, given the retrospective design of a phase 2 biomarker study, patients with HCC were detected by the current surveillance tool, liver ultrasound with/without serum AFP, and diagnosed before enrollment. As such, it is not feasible to directly compare the performance of HCC EV ECG score with liver ultrasound with/without serum AFP for detecting early-stage HCC. Instead, we reported the comparison between HCC EV ECG score and serum AFP in the current study. Finally, despite adherence to the ILCA guideline, there could be some potential biases in a phase 2 case–control study. Subsequent phase 3 and 4 biomarker studies will address this concern and confirm the ability of HCC EV SPA and HCC EV ECG score for detecting early-stage HCC.
In conclusion, HCC EV SPA and HCC EV ECG score were developed and validated for accurately detecting early-stage HCC from cirrhosis. The sensitivity and specificity of this assay significantly outperformed serum AFP and remained consistently excellent through the subgroup analyses. This approach holds great promise to help identify HCC at a curable stage and improve patients’ long-term outcomes. Further validation in a larger phase 2 multicenter biomarker study and a subsequent phase 3 study are essential to confirm its utility in clinical settings.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge all the patients and healthy donors who participated in this study.
FUNDING INFORMATION
This work is supported by American College of Gastroenterology Junior Faculty Development Award, Department of Defense Peer Reviewed Cancer Research Program Career Development Award (CA191051), and National Institutes of Health (R01 CA218356, U01 CA198900, P01 CA233452, R01 CA255727, R01 CA253651, R01 CA246304, U01 CA230705, U01 EB026421, U54 CA14391, R21 CA240887, and R21 CA235340). The funders had no role in the collection of data; the design and conduct of the study; management, analysis, and interpretation of the data; preparation, review, or approval of the manuscript; and decision to submit the manuscript for publication.
Abbreviations:
- ALD
alcoholic liver disease
- AFP
alpha-fetoprotein
- ASGPR1
asialoglycoprotein receptor 1
- AUROC
area under the receiver operating characteristic curve
- BCLC
Barcelona Clinic Liver Cancer
- BSA
bovine serum albumin
- CI
confidence interval
- CSMC
Cedars-Sinai Medical Center
- ECG score
the score calculated from the readouts of EpCAM+ CD63+ HCC EVs, CD147+ CD63+ HCC EVs, and GPC3+ CD63+ HCC EVs
- EpCAM
epithelial cellular adhesion molecule
- EV
extracellular vesicle
- GPC3
Glypican 3 Protein
- HBV
hepatitis B virus
- HCC
hepatocellular carcinoma
- HCV
hepatitis C virus
- ILCA
International Liver Cancer Association
- MELD
Model for End-stage Liver Disease
- NPV
negative predictive value
- NRI
net reclassification improvement index
- PBS
phosphate-buffered saline
- PCR
polymerase chain reaction
- PPV
positive predictive value
- ROC
receiver operating characteristic curve
- SPA
Surface Protein Assay
- TCO
transcyclooctene
- TMA
tissue microarray
- mTz
methyltetrazine
- UCLA
University of California, Los Angeles
Footnotes
CONFLICT OF INTEREST
Hsian-Rong Tseng owns stock in CytoLumina and Pulsar. Vinay Sundaram consults for Saol. He is on the speakers’ bureau for Gilead, AbbVie, and Intercept. Mazen Noureddin owns stock in, consults for, and received grants from Viking. He consults for and received grants from Gilead, Pfizer, and Madrigal. He consults for 89 Bio, Altimmune, CoBar, Cytodyn, Intercept, Novo Nordisk, Blade, EchoSens, Fractyl, NorthSea, Perspecturm, Terns, Sami-Sabina Group, Siemens, and Roche. He received grants from Allergan, BMS, Galmed, Galectin, Genfit, Conatus, Enanta, Novartis, Shire, and Zydus. Richard S. Finn consults for AstraZeneca, Bayer, BMS, Exelixis, CStone, Eisai, Eli Lilly, Pfizer, Merck, and Roche/Genentech. His institution receives grants from Bayer, Eisai, Bristol Myers Squibb, Eli Lilly, Roche/Genentech, and Pfizer. Saeed Sadeghi consults for and is in the speakers’ bureau for Eisai. Wenyuan Li is a co-founder and shareholder in Early Diagnostics Inc.
SUPPORTING INFORMATION
Additional supporting information can be found online in the Supporting Information section at the end of this article.
REFERENCES
- 1.Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71:209–49. [DOI] [PubMed] [Google Scholar]
- 2.Lee YT, Wang JJ, Luu M, Tseng HR, Rich NE, Lu SC, et al. State-level HCC incidence and association with obesity and physical activity in the United States. Hepatology. 2021;74:1384–94. [DOI] [PubMed] [Google Scholar]
- 3.Lee YT, Wang JJ, Luu M, Noureddin M, Kosari K, Agopian VG, et al. The mortality and overall survival trends of primary liver cancer in the United States. J Natl Cancer Inst. 2021;113:1531–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Siegel RL, Miller KD, Fuchs HE, Jemal A. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71:7–33. [DOI] [PubMed] [Google Scholar]
- 5.Lee YT, Wang JJ, Luu M, Noureddin M, Nissen NN, Patel TC, et al. Comparison of clinical features and outcomes between intrahepatic cholangiocarcinoma and hepatocellular carcinoma in the United States. Hepatology. 2021;74:2622–32. [DOI] [PubMed] [Google Scholar]
- 6.Yang JD, Hainaut P, Gores GJ, Amadou A, Plymoth A, Roberts LR. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16:589–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Toni EN, Schlesinger-Raab A, Fuchs M, Schepp W, Ehmer U, Geisler F, et al. Age independent survival benefit for patients with hepatocellular carcinoma (HCC) without metastases at diagnosis: a population-based study. Gut. 2020;69:168–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marrero JA, Kulik LM, Sirlin CB, Zhu AX, Finn RS, Abecassis MM, et al. Diagnosis, staging, and management of hepatocellular carcinoma: 2018 practice guidance by the American Association for the Study of Liver Diseases. Hepatology. 2018;68:723–50. [DOI] [PubMed] [Google Scholar]
- 9.European Association for the Study of the Liver. EASL Clinical Practice Guidelines: Management of hepatocellular carcinoma. J Hepatol. 2018;69:182–236. [DOI] [PubMed] [Google Scholar]
- 10.Tzartzeva K, Obi J, Rich NE, Parikh ND, Marrero JA, Yopp A, et al. Surveillance imaging and alpha fetoprotein for early detection of hepatocellular carcinoma in patients with cirrhosis: a meta-analysis. Gastroenterology. 2018;154:1706–18.e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Singal AG, Conjeevaram HS, Volk ML, Fu S, Fontana RJ, Askari F, et al. Effectiveness of hepatocellular carcinoma surveillance in patients with cirrhosis. Cancer Epidemiol Biomarkers Prev. 2012;21:793–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Trinchet JC, Chaffaut C, Bourcier V, Degos F, Henrion J, Fontaine H, et al. Ultrasonographic surveillance of hepatocellular carcinoma in cirrhosis: a randomized trial comparing 3- and 6-month periodicities. Hepatology. 2011;54:1987–97. [DOI] [PubMed] [Google Scholar]
- 13.Lee YT, Tran BV, Wang JJ, Liang IY, You S, Zhu Y, et al. The role of extracellular vesicles in disease progression and detection of hepatocellular carcinoma. Cancers (Basel). 2021;13:3076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Niel G, D'Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat Rev Mol Cell Biol. 2018;19:213–28. [DOI] [PubMed] [Google Scholar]
- 15.von Felden J, Garcia-Lezana T, Dogra N, Gonzalez-Kozlova E, Ahsen ME, Craig A, et al. Unannotated small RNA clusters associated with circulating extracellular vesicles detect early stage liver cancer. Gut. 2021. 10.1136/gutjnl-2021-325036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sun N, Lee YT, Zhang RY, Kao R, Teng PC, Yang Y, et al. Purification of HCC-specific extracellular vesicles on nanosubstrates for early HCC detection by digital scoring. Nat Commun. 2020;11:4489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Singal AG, Hoshida Y, Pinato DJ, Marrero J, Nault JC, Paradis V, et al. International Liver Cancer Association (ILCA) white paper on biomarker development for hepatocellular carcinoma. Gastroenterology. 2021;160:2572–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sun N, Tran BV, Peng Z, Wang J, Zhang C, Yang P, et al. Coupling lipid labeling and click chemistry enables isolation of extracellular vesicles for noninvasive detection of oncogenic gene alterations. Adv Sci (Weinh). 2022;9:e2015853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Willms A, Müller C, Julich H, Klein N, Schwab R, Güsgen C, et al. Tumour-associated circulating microparticles: a novel liquid biopsy tool for screening and therapy monitoring of colorectal carcinoma and other epithelial neoplasia. Oncotarget. 2016;7:30867–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Julich-Haertel H, Urban SK, Krawczyk M, Willms A, Jankowski K, Patkowski W, et al. Cancer-associated circulating large extracellular vesicles in cholangiocarcinoma and hepatocellular carcinoma. J Hepatol. 2017;67:282–92. [DOI] [PubMed] [Google Scholar]
- 21.Court CM, Hou S, Winograd P, Segel NH, Li QW, Zhu Y, et al. A novel multimarker assay for the phenotypic profiling of circulating tumor cells in hepatocellular carcinoma. Liver Transpl. 2018;24:946–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Johnson PJ, Pirrie SJ, Cox TF, Berhane S, Teng M, Palmer D, et al. The detection of hepatocellular carcinoma using a prospectively developed and validated model based on serological biomarkers. Cancer Epidemiol Biomarkers Prev. 2014;23:144–53. [DOI] [PubMed] [Google Scholar]
- 23.Yang JD, Addissie BD, Mara KC, Harmsen WS, Dai J, Zhang N, et al. GALAD score for hepatocellular carcinoma detection in comparison with liver ultrasound and proposal of GALADUS score. Cancer Epidemiol Biomarkers Prev. 2019;28:531–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singal AG, Nabihah T, Mehta A, Marrero JA, El-Serag H, Jin Q, et al. GALAD demonstrates high sensitivity for HCC surveillance in a cohort of patients with cirrhosis. Hepatology. 2022;75:541–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ahn JC, Teng PC, Chen PJ, Posadas E, Tseng HR, Lu SC, et al. Detection of circulating tumor cells and their implications as a biomarker for diagnosis, prognostication, and therapeutic monitoring in hepatocellular carcinoma. Hepatology. 2021;73:422–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tran NH, Kisiel J, Roberts LR. Using cell-free DNA for HCC surveillance and prognosis. JHEP Rep. 2021;3:100304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oussalah A, Rischer S, Bensenane M, Conroy G, Filhine-Tresarrieu P, Debard R, et al. Plasma mSEPT9: a novel circulating cell-free DNA-based epigenetic biomarker to diagnose hepatocellular carcinoma. EBioMedicine. 2018;30:138–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chalasani NP, Ramasubramanian TS, Bhattacharya A, Olson MC, Edwards VD, Roberts LR, et al. A novel blood-based panel of methylated DNA and protein markers for detection of early-stage hepatocellular carcinoma. Clin Gastroenterol Hepatol. 2021;19:2597–605.e4. [DOI] [PubMed] [Google Scholar]
- 29.Xu RH, Wei W, Krawczyk M, Wang W, Luo H, Flagg K, et al. Circulating tumour DNA methylation markers for diagnosis and prognosis of hepatocellular carcinoma. Nat Mater. 2017;16:1155–61. [DOI] [PubMed] [Google Scholar]
- 30.Caruso S, Nault JC. A dive into the deep heterogeneity of hepatocellular carcinoma. Gastroenterology. 2019;157:1477–9. [DOI] [PubMed] [Google Scholar]
- 31.Kolb HC, Finn MG, Sharpless KB. Click chemistry: diverse chemical function from a few good reactions. Angew Chem Int Ed. 2001;40:2004–21. [DOI] [PubMed] [Google Scholar]
- 32.Karver MR, Weissleder R, Hilderbrand SA. Synthesis and evaluation of a series of 1,2,4,5-tetrazines for bioorthogonal conjugation. Bioconjug Chem. 2011;22:2263–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dong J, Zhang RY, Sun N, Hu J, Smalley MD, Zhou A, et al. Coupling nanostructured microchips with covalent chemistry enables purification of sarcoma-derived extracellular vesicles for downstream functional studies. Adv Funct Mater. 2020;30:2003237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.He X, Patfield SA. Immuno-PCR assay for sensitive detection of proteins in real time. Methods Mol Biol. 2015;1318:139–48. [DOI] [PubMed] [Google Scholar]
- 35.Huang DQ, El-Serag HB, Loomba R. Global epidemiology of NAFLD-related HCC: trends, predictions, risk factors and prevention. Nat Rev Gastroenterol Hepatol. 2021;18:223–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.