Abstract
The enteropathogenic Yersinia enterocolitica strains have several systems for scavenging iron from their environment. We have studied the expression of the fyuA gene, which encodes the outer membrane receptor for the siderophore yersiniabactin (Ybt), and the hemR gene, which encodes the receptor for heme, using the reporter genes gfp (encoding green fluorescent protein) and luc (encoding firefly luciferase). To study gene expression in vitro as well as in vivo, we have constructed several translational reporter gene fusions to monitor simultaneously expression of fyuA and hemR or expression of one gene by a gfp-luc tandem reporter. Results of the in vitro expression analysis (liquid media) indicated that fyuA and hemR are strongly derepressed under iron starvation conditions, resulting in strong fluorescence and/or luminescence at 27°C. In the in vivo BALB/C mouse infection model, tissue-specific expression of fyuA and hemR reporter fusions was observed. Surprisingly, fyuA and hemR reporter constructs were weakly expressed by yersiniae located in the liver and intestinal lumen, whereas strong expression was found for yersiniae in the peritoneal cavity and moderate expression was found in the spleen. Strikingly, yersiniae carrying fyuA or hemR reporter fusions exhibited threefold-stronger signals when grown in the peritoneal cavity of mice than those growing under iron derepression in vitro. This hyperexpression suggests that besides Fur derepression, additional activators may be involved in the enhanced expression of fyuA and hemR under peritoneal growth conditions. Differential expression of the fyuA and hemR reporter fusions could not be observed, suggesting similar regulation of fyuA and hemR in the mouse infection model.
Iron is an essential component of many enzymes required for growth and metabolism of bacteria (reviewed in reference 43). In aerobic environments at physiological pH, iron exists predominantly as oxidized ferric iron, Fe(III), which forms insoluble hydroxides. In order to acquire iron under these conditions, gram-negative bacteria usually produce and release high-affinity ferric iron-binding compounds (called siderophores), which capture iron from the environment (reviewed in references 10 and 30). The resulting ferric siderophore complex binds to siderophore-specific surface receptors of bacteria and is subsequently transported into the cytosol in a TonB-dependent pathway. Additionally, many microbes are equipped with low-affinity ferric-iron transport systems, which are usually independent of siderophore and TonB. Under anaerobic conditions, iron exists in the reduced ferrous form Fe(II), which has a higher solubility in aqueous solutions than Fe(III). Accordingly, ferrous iron uptake can be achieved independently of siderophores. Escherichia coli is equipped with an Fe(II) active transport system which is located in the cytoplasmic membrane (Feo system) (24). Many host-adapted microorganisms are also capable of utilizing iron directly from iron-binding proteins (e.g., transferrin) or heme-containing proteins (e.g., hemoglobin), by surface-receptor-mediated release and transport of iron or heme (27, 42, 44). In summary, free-living and host-adapted bacteria have evolved a set of different iron uptake systems, which may be differentially controlled by environmental factors and thus may be used simultaneously or differentially.
Iron overload, on the other hand, can be harmful for bacteria as well as for the host because of its ability to catalyze Fenton reactions. Therefore, bacteria have developed an elaborate regulating system to tightly control iron uptake (19). In the presence of cytosolic ferrous iron surplus, genes involved in iron transport and siderophore production are transcriptionally repressed by the Fe(II)-binding protein, Fur (ferric uptake regulation), which binds to the operator sequence (Fur boxes) of iron-repressible (irp) genes. Ferric iron depletion in the cytosol results in release of the iron-free Fur aporepressor and subsequently in derepression of irp genes (17, 18, 19). There is accumulating evidence that transcription of many irp genes (including fur) can be modulated by additional transcriptional factors to respond to environmental changes, in particular to oxidative stress or siderophore uptake (10, 12, 14, 19, 45). It is reasonable that this regulatory network may enable microorganisms, in particular pathogens, to switch on the most suitable iron uptake system when entering the complex environment of the host organism and to silence less effective iron uptake determinants (4, 41). Thus, one would expect differential expression of irp genes of the pathogen depending on the location or environment in the host.
To study this aspect, we choose mouse virulent Yersinia enterocolitica (biotype IB) and the well-established mouse infection model. Y. enterocolitica and Yersinia pseudotuberculosis are known as the human enteropathogenic species of the genus Yersinia. These enteric pathogens share a virulence plasmid (pYV) with a size of 70 kb (for a review, see reference 9), a chromosomally located high-pathogenicity island of 36 to 43 kb, encoding the siderophore yersiniabactin (Ybt) system with the Fe(III)-Ybt receptor FyuA (22, 29) and the heme uptake system, including the outer membrane receptor for heme, HemR (39). Moreover, several other iron uptake systems have been identified and characterized for Y. enterocolitica: (i) TonB-dependent siderophore uptake systems for ferrioxamine, ferrichrome, and ferri-enterobactin (3, 25, 35, 36), and (ii) the TonB-independent system yfu (33).
It is still unclear whether all these iron uptake systems of Y. enterocolitica are upregulated during infection. Currently, it has only been shown that the Ybt system is absolutely required for mouse virulence and thus should be expressed by yersiniae during infection (22, 29). Yersiniae carrying a functional pYV plasmid are capable of resisting phagocytosis and of replicating extracellularly (16, 20, 38). Presumably, the extracellular concentration of available ferric iron controls the activity of irp genes.
In order to elucidate the expression of iron uptake determinants of Y. enterocolitica during infection of mice, we used reporter gene technology by constructing translational fusions of fyuA (encoding FyuA) and hemR (encoding HemR) with gfp (encoding the green fluorescent protein [GFP]) (6) and luc (encoding the firefly luciferase) (11), respectively. The fyuA and hemR genes both carry a Fur box and thus are repressible by the Fe(II)-Fur repressor. Expression of fyuA (ortholog of psn of Yersinia pestis) is additionally dependent on uptake of Fe(III)-Ybt and the AraC-like transcriptional activator YbtA, which is encoded on the high-pathogenicity island (5, 14, 28). According to this, we anticipated differential regulation of hemR and fyuA, depending on the organs infected by yersiniae.
MATERIALS AND METHODS
Bacterial strains used and plasmid constructions.
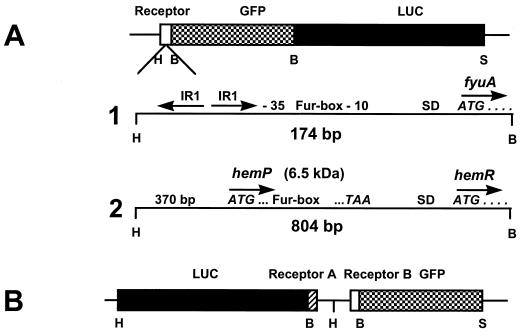
The strains, plasmids, and primers used in this study are listed in Table 1. Escherichia coli DH5α (15) was used as an intermediate host for cloning experiments. The Y. enterocolitica strain WA-C(pYV08), harboring the virulence plasmid pYV08, was used as a host for plasmid constructs (21). We used PCR cloning techniques to construct different fusions between the receptors fyuA and hemR and gfp and/or the luc gene (Fig. 1). We have amplified a 174-bp fragment of the fyuA gene (including the indirect repeats upstream of the −35 region, the Fur box, and the first five codons of the gene (Fig. 1A, line 1) (29) of Y. enterocolitica using the primer pair FyuAHiF and FyuA5BaR (Table 1). We cloned the fragment into the low-copy-number vector pACYC184 (7), resulting in the plasmid pCJFY. Using pGFPmut2 (8) as a template, we have amplified gfpmut2 without its stop codon using the primer pair S65TBaF and S65TBaNstR (pCJFYGNst). The firefly luciferase gene (11) was cloned behind gfp (primers LUCBaF and LUCSaR, resulting in pCJFY-GL). For the hemR reporter gene fusions, we have amplified an 805-bp fragment of the hemR gene including the Fur box and the first five codons of the heme receptor hemR (Fig. 1A, line 2) (primers HemRHiF and HemRBaR). This plasmid (pCJHE-GL) was constructed in a fashion similar to that for pCJFY-GL.
TABLE 1.
Plasmids, bacterial strains, and primers used in this studya
| Plasmid, strain, or primer | Genotype, phenotype, or sequencea | Source or reference |
|---|---|---|
| Plasmids | ||
| pACYC184 | low-copy-number vector; Cm Tc | 7 |
| pGFPmut 2 | pKEN containing cDNA of gfp mutant GFPmut 2 | 8 |
| pT3/T7-LUC | pT3/T7-1 containing cDNA of firefly luciferase | Clontech |
| pCJFY | HindII-BamHI FyuA fragment in pACYC184 | This study |
| pCJFY-GNst | HindIII-BamHI FyuA-gfp fragment in pACYC184 | This study |
| pCJFY-G | HindIII-BamHI FyuA-gfp fragment in pACYC184 | This study |
| pCJFY-L | SalI-HindIII FyuA-luc fragment in pACYC184 | This study |
| pCJHE-G | SalI-HindIII HemR-gfp fragment in pACYC184 | This study |
| pCJHE-L | SalI-HindIII HemR-luc fragment in pACYC184 | This study |
| pCJHE-GL | Like pCJHE-G and luc additionally fused to gfp | This study |
| pCJFY-GL | Like pCJFY-G and luc additionally fused to gfp | This study |
| pCJHE-G/FY-L | FyuA-L cloned in pCJHE-G via HindIII | This study |
| pCJHE-L/FY-G | FyuA-G cloned in pCJHE-L via HindIII | This study |
| Strains | ||
| DH5α (E. coli) | endA1 hsdR17(rk− mk+) supE44 thi-1 recA1 | |
| gyrA relA1Δ(lacZYA-argF)U169 (φ80lacZΔM15) | 15 | |
| WA-C | Plasmidless derivative of Y. enterocolitica | 21 |
| WA-C(pYV08) | WA-C harboring virulence plasmid pYV08 | 21 |
| WA-C(pYV08,pGFPmut2) | WA-C(pYV08) with pGFPmut2 | 23 |
| WA-C(pYV08,pCJHE-GL) | WA-C(pYV08) with pCJHE-GL | This study |
| WA-C(pYV08,pCJFY-GL) | WA-C(pYV08) with pCJFY-GL | This study |
| WA-C(pYV08,pCJHE-G/FY-L) | WA-C(pYV08) with pCJHE-G/FY-L | This study |
| WA-C(pYV08,pCJHE-L/FY-G) | WA-C(pYV08) with pCJHE-L/FY-G | This study |
| Primers | ||
| FyuAHiF | 5′-GCCGAAGCTTGACCGTTATCGGCATTCTG-3′ | |
| FyuA5BaR | 5′-CGCATGGATCCCCGTGTCATTTTCATTGTTG-3′ | |
| HemRHiF | 5′-GCCAGAAGCTTTGAAACGTATGCTTTGGTG-3′ | |
| HemRBaR | 5′-CGCAGGATCCAGTGGAACGCGGCATGTC-3′ | |
| LUCBaF | 5′-GGCAGGATCCATGGAAGACGCCAAAAACATA-3′ | |
| LUCSaR | 5′-GCCCCGTCGACTTACCTGATTTTATTATTCGT-3′ | |
| S65TBaF | 5′-GGCGGATCCATGAGTAAAGGAGAAGAA-3′ | |
| S65TSaR | 5′-AGCCCCGTCGACTTATTTGTATAGTTCATCC-3′ | |
| S65TBaNstR | 5′-AGCATGGATCCGTATAGTTCATCCATGCC-3′ |
Restriction sites are underlined.
FIG. 1.
Construction of translational reporter fusions (vector pACYC184). (A) Translational fusion with GFP and firefly luciferase (LUC) (tandem-reporter). Line 1, enlargement of the 5′ region of fyuA with the ATG codon and the four following codons for the plasmid pCJFY-GL.IR, inverted repeat; SD, Shine-Dalgarno ribosome binding site. Line 2, enlargement of the 5′ region of hemR (including hemP) with ATG and the four following codons for plasmid pCJHE-GL. (B) Bidirectional translational fusion (twin reporter construct). Restriction sites: B, BamHI; H, HindIII; S, SalI.
For the simultaneous detection of expression of fyuA and of hemR reporter gene fusions (Fig. 1B) (twin reporter construct: coexpression), we removed the gfp-luc fragments from pCJFY-GL and pCJHE-GL. Next, we cloned gfpmut2 (with its stop codon; reverse primer S65TSaR) or the luciferase fragment into the restricted vectors, resulting in the plasmids listed in Table 1. In the next step, we restricted plasmids pCJHE-G and pCJHE-L with HindIII and cloned the HindIII-restricted fyuA-luc and fyuA-gfp fragments (amplified with HindIII- containing primers) into the respective plasmids, resulting in pCJHE-G/FY-L and pCJHE-L/FY-G. Each fusion was checked by sequencing. Plasmids were transferred into Y. enterocolitica WA-C(pYV08) by electroporation. Restriction enzyme digests, recovery of DNA from low-melting-point agarose, DNA ligations, and transformations were performed as described previously (1, 34).
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblot analysis of LUC and GFP constructs.
Yersiniae grown overnight in NB (per liter, 8 g of Nutrient broth [Difco], 5 g of NaCl) broth at 27°C were diluted 1:30 with NBD [NB–200 μM dipyridyl for Fe(II) chelation] broth and incubated for 24 h at 27 or 37°C under iron restriction [derepressed state, because of intracellular depletion of Fe(II)]. As a control, yersiniae grown overnight were also diluted 1:30 into NB broth and were incubated in parallel. In some experiments, NB and NBD broth were supplemented with yersiniabactin, heme, 1 mM H2O2, or 0.2 mM paraquat. For direct comparison of the production of reporter fusions, equal amounts of bacterial lysates were used (determined by the optical density at 600 nm [OD600] of 0.1) (10 μl/lane) (Fig. 2). Electrophoretic separation of the proteins was performed as described previously (26) on an 11.5% polyacrylamide gel. For immunoblotting, the proteins were electrophoretically transferred to nitrocellulose sheets (BA85; Schleicher & Schuell) as was described previously (40). The filter sheets were blocked with 5% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) overnight at 4°C. For immunostaining, a polyclonal rabbit anti-LUC antibody (dilution, 1:3,000; Promega) or a polyclonal rabbit anti-GFP antiserum (dilution, 1:200; Clontech) and a peroxidase-conjugated secondary anti-rabbit antibody (dilution, 1:5,000; Dianova) were used. The detection was carried out by using the ECL chemiluminescence detection kit (Amersham Pharmacia).
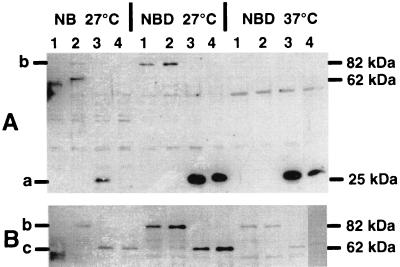
FIG. 2.
Immunoblot of yersiniae cells containing the respective constructs, developed with polyclonal rabbit anti-GFP antiserum (A) or polyclonal rabbit anti-luciferase antibody (B). Yersiniae were grown for 24 h under the conditions indicated at the top. Lane 1, WA-C(pYV08,pCJFY-GL); lane 2, WA-C(pYV08,pCJHE-GL); lane 3: WA-C(pYV08,pCJHE-G/FY-L); lane 4, WA-C(pYV08,pCJHE-L/FY-G). The GFP fusion protein (27 kDa) (a), GFP-LUC fusion protein (89 kDa) (b), and LUC fusion protein (62 kDa) (c) are indicated.
Determination of luc and gfp reporter gene activity of yersiniae.
Green fluorescence and luciferase activity of Yersinia cells and lysates (grown under the conditions described above), respectively, were determined as follows. A Beckman Coulter EPICS XL-MCL flow cytometer equipped with an argon 488-nm laser was used for determination of GFP fluorescence of single bacterial cells. In vitro iron-derepressed recombinant yersiniae (grown in NBD broth) were diluted as required, and the bacteria were detected by side scatter as described previously (23, 32). The average intensity of fluorescence was determined (Fig. 3A). The scale was logarithmic, and fluorescence data and scatter data were collected for 10,000 and 50,000 events. For fluorescence determination with Yersinia cells obtained from infected mice (see below), liver, spleen and Peyer's patches were homogenized and diluted as required (1:10 or 1:100). Since homogenates tend to interfere heavily by autofluorescence, the organ homogenates of uninfected mice were used as negative controls. Additionally, a gate was set, allowing only a specific population to be considered in the measurements (Fig. 4A), as has been described previously (23).
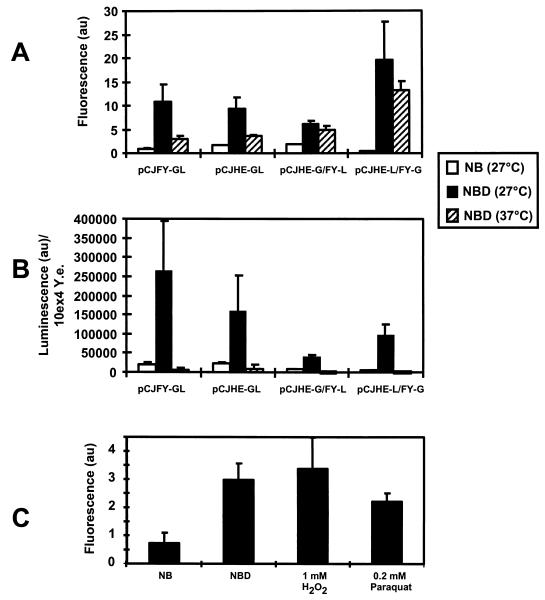
FIG. 3.
In vitro production of FyuA and HemR reporter fusions determined by the fluorescence (GFP) (A) and the luminescence (luciferase) (B) of 104 CFU of Y. enterocolitica (10ex4 Y.e.). The lanes are denoted by names of the recombinant plasmids that had been introduced into WA-C(pYV08); measurements were taken after incubation for 24 h at 27 or 37°C. (C) In vitro production of FyuA-GL of strain WA-C(pYV08,pCJFY-GL) determined by GFP fluorescence in NB (iron-rich) and NBD (iron-restricted) medium at 37°C after 24 h; NBD medium is supplemented with the respective substances. au, arbitrary units for fluorescence (A and C) and luminescence (B).
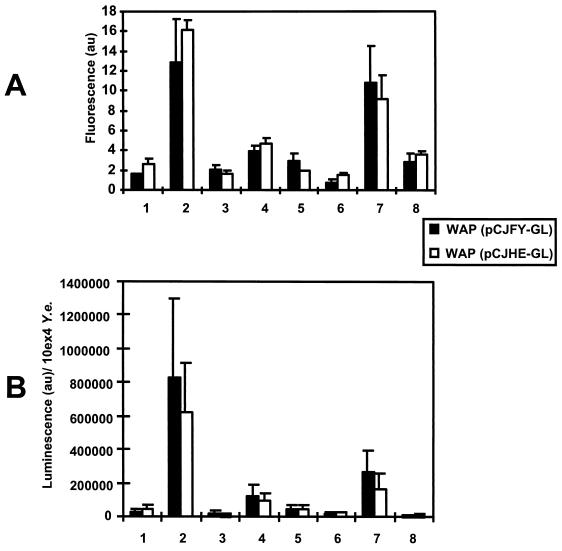
FIG. 4.
In vivo (lanes 1 to 5) and in vitro (lanes 6 to 8) production of FyuA-GL and HemR-GL determined by the GFP fluorescence (A) and luminescence (luciferase) (B) of 104 CFU of Y. enterocolitica (10ex4 Y.e.) after 24 h of in vivo or in vitro incubation. Lanes 1, intestinal lavage sample; lanes 2, peritoneal lavage sample; lanes 3, liver; lanes 4, spleen; lanes 5, Peyer's patches; lanes 6, NB medium (27°C); lanes 7, NBD medium (27°C); lanes 8, NBD medium (37°C). Mice were infected via the i.p., o.g., or i.v. route and were sacrificed 24 h later. Preparation for the samples was done as described in Materials and Methods. au, arbitrary units for fluorescence (A) and luminescence (B).
To measure the luciferase activity, the cell density of the liquid cultures was adjusted to equal an OD600 of 0.1. One milliliter of each sample was sedimented by centrifugation. The corresponding pellets were frozen at −20°C and subsequently resuspended in 1 ml of bacterial lysis buffer (2 mM EDTA, 1% Triton X-100, 5 mg BSA/ml, of 1 mM dithiothreitol, and 5 mg of lysozyme/ml in 100 mM potassium phosphate buffer [pH 7.8]) for 15 min at room temperature, as described by the manufacturer (Boehringer Mannheim). The success of lysis was checked by microscopic examination and by plating the lysate on selective media. Cell debris was removed by centrifugation at 10,000 × g for 15 min, and the supernatant was transferred into a white microtiter plate (if necessary, the supernatants were diluted to measure the luminescence in a linear range). The luciferase activity was measured in a dark box with a microtiter plate chemiluminometer (C 2400-77 charge-coupled-device camera; Hamamatsu Photonics) for 15 min. All experiments were performed independently at least three times. The average and standard deviations were determined, and the luciferase activity of an equivalent of 104 recombinant yersiniae was graphically displayed (Fig. 3B). Luciferase measurements of the in vivo experiments (Fig. 4B) were performed with modifications described below.
Mouse infection model.
Female BALB/c mice (6 to 8 weeks old) were infected (groups of five) with recombinant yersiniae orogastrically (o.g.), intravenously (i.v.), or intraperitoneally (i.p.), respectively. We focused our attention on WA-C(pYV08,pCJFY-GL) and WA-C(pYV08,pCJHE-GL). For i.p. challenge, BALB/c mice were infected with 107 CFU of the respective strains, previously grown in LB broth for 3 h at 27°C. Recombinant yersiniae, 5 × 105 and 5 × 109 CFU, were used for i.v. and o.g. challenge, respectively. Twenty-four hours after infection, mice were sacrificed. Corresponding to the route of infection, a peritoneal (i.p. route) or intestinal (o.g. route) lavage was performed with 4 ml of ice-cold sterile 0.9% NaCl solution; the Peyer's patches of the small intestine were dissected (o.g. route), and the liver and spleen (i.v. route) were recovered. A piece of each organ was prepared for cryosectioning as previously described (13). Briefly, tissues were embedded in Tissue-Tek O.C.T. compound (Miles), frozen in liquid nitrogen, and stored at −80°C. Twenty-five-micrometer cryosections were prepared using a cryomicrotom (Leica). After homogenization of the organs in PBS (with 0.5% Tergitol and 0.5% BSA) (100 mg of spleen tissue in 2 ml and 400 mg liver tissue in 4 ml), aliquots were taken for flow-cytometric measurements and for determination of bacterial numbers (for luciferase measurements). The homogenates and the peritoneal and intestinal lavage samples were centrifuged at 4,000 × g; after the supernatants were decanted, the pellets (mouse cells and yersiniae) were frozen at −20°C. Each pellet was resuspended in lysis buffer, and the lysis procedure was performed as described for luciferase determination (see above). The efficiency of lysis was checked by plating the homogenates before and after the lysis procedure on appropriate media. Aliquots or appropriate dilutions of the samples were transferred into a white microtiter plate (Dynatech), and after addition of the luciferin substrate the luminescence was measured. The mouse experiments were repeated once.
Control experiments were performed to determine whether or not quenching was occurring in the organs: we added luminescing bacterial lysates (bacterial culture at an OD600 of 0.1) to uninfected organ homogenates (100 mg of spleen tissue in 2 ml or 400 mg of liver tissue in PBS–0.5% Tergitol–0.5% BSA) and the peritoneal (4 ml) and intestinal (4 ml) lavage samples and measured the luminescence of these samples (50 μl of organ homogenate plus 50 μl of bacterial lysate), comparing them with the luminescing intensities of the lysate without the organs or lavage samples.
Immunohistochemistry.
Twenty-five-micrometer cryosections of spleen, liver, and Peyer's patches were transferred onto silanized microscopic slides and fixed with 3.7% freshly prepared paraformaldehyde in PBS, pH 7.4, for 20 min at room temperature. After three washes with PBS, the tissue sections were permeabilized by incubation with 2% Triton X-100 for 8 min. The slides were washed again with PBS and then incubated for 30 min in 2% BSA to block unspecific binding, followed by incubation with a polyclonal rabbit anti-yersiniae antiserum for 2 h at 37°C. After the primary antiserum was washed off with PBS, the slides were incubated with a tetramethyl rhodamine isocyanate (TRITC)-labeled secondary antibody (1:100; Dianova) for 60 min (2). After a final wash with PBS, the cryosections were embedded with Fluoprep (BioMerieux), covered carefully with a coverslip, and stored in the dark at 4°C until microscopic examination.
Microscopic examination.
The recombinant, GFP-producing yersiniae obtained from infected mice were visualized using the confocal laser scanning microscope (CLSM) Leica TCS 4D with a standard filter set. A ×63 objective was used for magnification, the laser lines on the krypton/argon laser were 488 (fluorescein isothiocyanate) and 568 nm (TRITC), thus allowing the simultaneous scanning of the two different fluorochromes. Sixteen sections were generated from each image to visualize the three-dimensional structure of the abscesses. The intensity of each section was averaged four times to increase the sharpness of the image and to improve the signal/noise ratio. In addition, a single section in the middle of the “stack” was visualized (Fig. 5C). The settings on the microscope (electronic magnification, offset, and photomultiplier) remained unchanged during the generation of the different images, allowing the comparison (fluorescence intensity and magnification) of each micrograph. The pinhole was set between 100 and 125 μm. Without any modification, the photomicrographs were printed directly on photopaper using a Tektronix Phaser 450 printer.
FIG. 5.
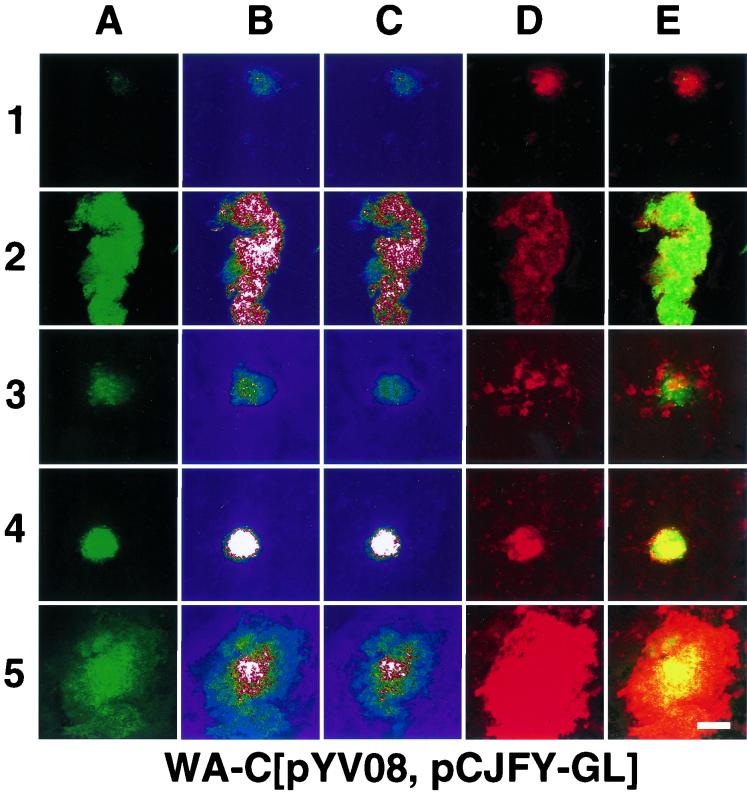
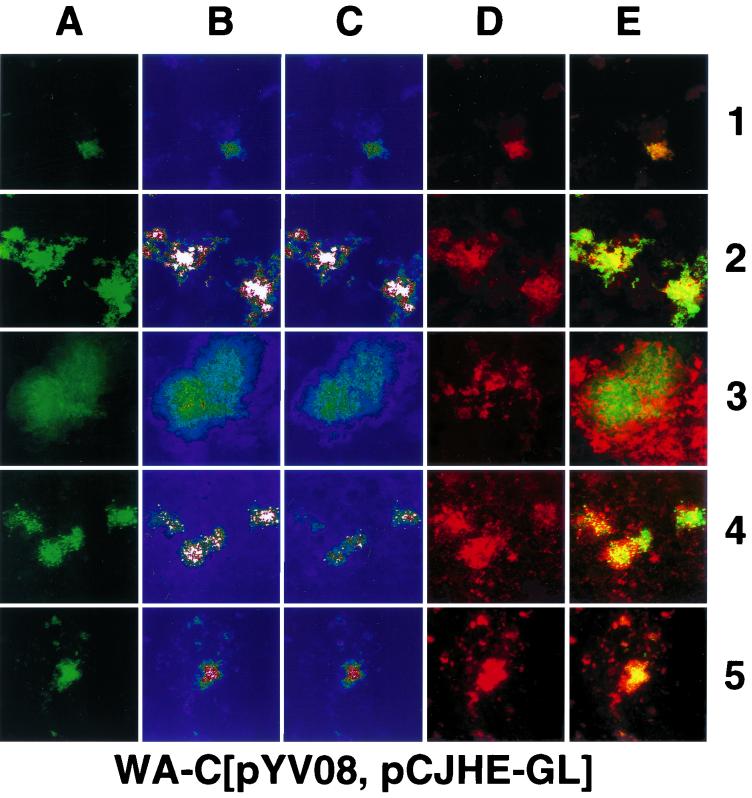
Confocal laser scanning micrographs (overlay of 16 sections) of samples (intestinal [row 1] and peritoneal [row 2] lavage samples, 25-μm cryosections of liver [row 3], spleen [row 4], and Peyer's patches [row 5]) obtained from mouse organs infected with WA-C(pYV08,pCJFY-GL) or WA-C(pYV08,pCJHE-GL), 24 h after of infection (route of infection: i.p. [row 2], o.g. [rows 1 and 5] or i.v. [rows 3 and 4]). (A) GFP fluorescence. (B) Intensity profile (pseudocolor image; white color denotes highest intensity, while blue denotes lowest intensity) of panel A. (C) Single section in the middle with the highest fluorescence intensity (pseudocolor image). (D) Rabbit antiyersiniae antiserum labeled with TRITC. (E) Overlay of panels A and D. Scale bar, 100 μm.
RESULTS
Construction of fyuA and hemR reporter gene fusions.
First, we compared the suitabilities of the reporter genes luc and gfp for semiquantitative expression studies of yersiniae in vitro and in vivo from different organs of infected mice. Using PCR cloning techniques, we constructed translational fusions as tandem reporter gene constructs (hemR-gfp-luc and fyuA-gfp-luc) and as twin reporter constructs (hemR-gfp/fyuA-luc and hemR-luc/fyuA-gfp), respectively (Fig. 1). The amplified fyuA fragment comprising 174 bp (the 5′ region with the inverted repeats and Fur box and the first five codons) (Fig. 1A, line 1) (29) was ligated into the low-copy-number vector pACYC184 (7), resulting in pCJFY. The gene gfpmut2 (8) was amplified without its stop codon and ligated downstream of the fyuA fragment into pCJFY. The gene encoding luciferase (11) was similarly amplified and fused translationally downstream of gfp, resulting in the plasmid pCJFY-GL. To study the expression of hemR (39), we amplified the 5′ region of the gene plus the first five codons (comprising 805 bp) and ligated this fragment with the gfp-luc hybrid, resulting in the plasmid pCJHE-GL (Fig. 1A, line 2).
In order to study coexpression of fyuA and hemR in yersiniae, we constructed plasmids (twin reporter constructs) carrying both the fyuA and hemR regulatory regions fused to gfp and the luciferase gene, respectively, resulting in the plasmids pCJHE-G/FY-L and pCJHE-L/FY-G (Fig. 1B).
Plasmid constructs were checked by sequencing and transferred into Y. enterocolitica WA-C(pYV08) by electroporation. These recombinant yersiniae strains were used for in vitro and in vivo studies.
Expression of fyuA and hemR reporter gene fusions in liquid medium.
The expression of fyuA and hemR reporter gene fusions by strain WA-C(pYV08) under iron repression (NB medium) and derepression (NBD medium) was studied. After preincubation in NB medium overnight at 27°C, the cultures were diluted 1:30 in NB and NBD medium, respectively, and cultivated for 24 h at 27 or 37°C.
First, production of the FyuA and HemR hybrid proteins at 27 and 37°C in NB and NBD medium was detected by immunoblotting using antiserum and antibody against GFP and LUC, respectively. As shown in Fig. 2A, the hybrid proteins FyuA-GFP and HemR-GFP were visible as strong bands after growth in NBD medium. No protein band or a faint (lane 3) protein band was present after growth in NB medium (not shown). FyuA-GFP-LUC and HemR-GFP-LUC bands were clearly visible only in NBD medium at 27°C. At 27°C in NB medium and at 37°C in both media, the LUC fusion proteins were almost nondetectable. The electrophoretic mobility of these hybrid proteins corresponded to the expected molecular weights (for GFP, 27 kDa; for LUC, 62 kDa; for GFP-LUC, 89 kDa). The LUC immunoblot (Fig. 2B) shows the corresponding LUC hybrid protein bands according to their expected molecular weights. Here, the bands of the respective hybrid proteins corresponded with the bands of the GFP immunoblot, with the exception that at 37°C in NBD no FyuA-LUC or HemR-LUC fusion protein was detectable.
Second, we determined the reporter activities of the produced hybrid proteins using cytofluorometry (GFP fusion proteins) or the charge-coupled device camera (luminescence of LUC fusion proteins). The recombinant yersiniae were grown in NB and NBD medium at 27 and 37°C, respectively.
In accordance with the results obtained by immunoblotting, we were able to detect a strong increase in fluorescence and luminescence (on average about 10- to 20-fold), respectively, when yersiniae were transferred from NB to NBD medium and incubated at 27 or 37°C for 24 h (Fig. 3A and B). With respect to luciferase analysis, the bacterial lysis procedure was more than 99% effective. The fluorescence and luminescence values of the FyuA reporter fusions were on average slightly higher than for the HemR receptor fusions. Incubation of the recombinant bacteria in NBD at 37°C resulted in weak signals of fluorescence and luminescence, respectively, in comparison to results at 27°C. This temperature-dependent decrease of the reporter signals is less prominent in the case of the FyuA- and HemR-GFP fusion proteins (Fig. 3A) (twin reporter constructs pCJHE-G/FY-L and pCJHE-L/FY-G), which is in line with the immunoblot results. This is probably due to the higher stability of the GFP.
When we supplemented NBD medium with yersiniabactin, we observed an increase in expression of about 50% of the fyuA fusions but not of the hemR fusions (results not shown). Similarly, supplementing the NBD medium with 100 μM heme did not result in any increase of expression of the iron receptors. Previously it has been demonstrated that expression of fur is upregulated in E. coli by oxidative stress induced by H2O2 or paraquat treatment (45). Consequently, oxidative stress should result in repression of irp gene expression. As shown in Fig. 3C, 1 mM H2O2 or 0.2 mM paraquat did not significantly influence expression of fyuA-gfp of strain WA-C(pYV08,pCJFY-GL) in NBD medium at 37°C.
In conclusion, these in vitro expression studies with the twin reporter constructs demonstrated that the fyuA and hemR reporter fusions are coexpressed at 27°C under iron-derepressed growth conditions (NBD medium) and are repressed in NB medium. Strikingly, the reporter fusions appeared to be more weakly upregulated in NBD medium at 37°C than at 27°C. An exception is observed with the GFP fusions of the twin reporter constructs: pCJHE-G/FY-L and pCJHE-L/FY-G. Here, the in vitro fluorescence result in NBD medium shows less temperature dependence, probably because of the stability of GFP (Fig. 3A). Obviously, fusion proteins are degraded fairly rapidly in the case of luciferase but not of GFP (unpublished observation). The functional reporter signal intensities were generally in line with the produced reporter protein amounts detected by immunoblotting. To exclude the possibility that the low levels of expression of fyuA and hemR reporter fusions at 37°C are due to the influence of the virulence plasmid, we also performed experiments with the plasmidless strain WAC. Similar results could be obtained (data not shown).
fyuA and hemR reporter gene expression analysis of yersiniae-infected mice.
For analysis of fyuA and hemR reporter gene expression in vivo, mice were infected with strain WA-C(pYV08,pCJFY-GL), expressing the fyuA-gfp-luc tandem construct, or with strain WA-C(pYV08,pCJHE-GL), expressing the hemR-gfp-luc tandem construct. These strains allow the simultaneous determination of GFP fluorescence on a single-cell level and of LUC luminescence of bacterial populations from tissue homogenates. According to infected organs of interest, yersiniae were administered i.v. (spleen and liver), o.g. (intestine, Peyer's patches), or i.p. (peritoneal cavity). After 24 h of infection, the mice were sacrificed, and the organs were processed as described in Materials and Methods. The presence of the respective plasmids was checked, and on average about 90% of bacterial colonies obtained from infected mice carried the reporter plasmids. The numbers differed; while more than 95% of isolated yersiniae from the peritoneal cavity still contained the plasmid, 93 and 90% did so in the liver and spleen, respectively, and about 70% of the isolated colonies from the Peyer's patches still contained the constructs (for each organ more than 100 colonies were checked). Production of bacterial cell-associated fluorescing GFP was determined by employing flow cytometry and CLSM. Figure 4A shows the result of flow cytometry. It is striking that yersiniae obtained from the peritoneal cavity exhibited the highest fluorescence intensity on a single-cell level for both reporter strains, FyuA-GL and HemR-GL. Yersinia cells isolated from spleens showed about 30% of the fluorescence intensity of those from the peritoneal cavity. The weakest fluorescence intensity was determined for yersiniae located within the intestinal lumen and the liver, indicating a relatively iron-rich environment. Surprisingly, the strains producing FyuA-GL and HemR-GL exhibited comparable fluorescence intensities with respect to the location of the pathogen. The cytofluorometric results could be confirmed by CLSM micrographs (Fig. 5) of 25-μm cryosections. The recombinant yersiniae inside the abscesses were fluorescing homogeneously; thus, we did not observe spatial differences of fluorescence intensities within different infected regions. However, within abscesses, the fluorescence is sometimes diffuse, as observed with the CLSM, because of closely packed bacterial clusters or bacterial lysis. Strongly fluorescing bacteria could also be detected in the Peyer's patches (Fig. 5, line 5). As expected, in the intestinal lavage sample, recombinant yersiniae were only weakly fluorescent (Fig. 5, lines 1 and 3). To prove that the fluorescence intensities are not “artifical” due to the overlay of 16 sections, we presented in Fig. 5C single sections from the middle of the “stack,” which verified our observations. Second, to demonstrate that expression of gfp under anaerobic or microaerophilic conditions (like those of the intestinal lumen) results in green-fluorescing bacteria, we administered o.g. strain WA-C(pYV08,pGFPmut2), which expressed constitutively gfp. This strain showed bright fluorescence when isolated from the intestinal lumen (data not shown).
The application of gfp-luc tandem reporter fusions enabled us to determine the luciferase activity of yersiniae of the same samples used for GFP fluorescence determination. In order to compare and quantify the luminescence of the recombinant yersiniae obtained from the different organs, the numbers of bacteria (CFU) present in each organ and in the intestinal lumen as well as in the peritoneal cavity were determined by plating aliquots of each sample on solid media (the numbers of CFU were in the following ranges: for the spleen, ∼5 × 106 CFU; for the liver, ∼106 CFU; for the peritoneal cavity, ∼106 CFU; for the intestinal lumen, ∼107 CFU; and for Peyer's patch, ∼105 CFU). The luminescence data were calculated for 104 CFU as a reference value (CFU obtained from solid media, selective for the reporter plasmid) (Fig. 4B). The luciferase activities of the strains producing FyuA-GL or HemR-GL were strikingly high for yersiniae from the peritoneal cavity, whereas yersiniae from spleens showed only 20% of the luminescence intensity with respect to the peritoneal cavity. The luciferase activity of yersiniae from the intestinal lumen and liver was found to be low.
From comparison of the GFP fluorescence and luciferase data, respectively, obtained from yersiniae of infected mice, it is evident that yersiniae in the peritoneal cavity are producing large amounts of FyuA and HemR reporter proteins. To exclude the possibility that this high level of fluorescence of yersiniae in the peritoneal cavity is due to, e.g., upregulation of the copy number of the reporter plasmids, we infected mice i.p. or i.v. with yersiniae, producing GFP constitutively, carrying gfpmut2 on pACYC184. After 24 h of infection, we performed confocal laser microscopic analysis of yersiniae from the peritoneal cavity, the spleen, and the liver. The fluorescence intensities of all samples were equal, suggesting that the differences in fluorescence of the different organs are not due to copy number effects of the plasmid.
Control experiments were performed to rule out the possibility that differences in the intensities of luminescence are due to the effect of quenching substances. We seeded equal amounts of luminescent yersiniae, pregrown in NBD medium at 27°C, in homogenized liver, spleen, or Peyer's patches and additionally into the intestinal and peritoneal lavage samples from uninfected mice and determined the luminescence intensities, in comparison to the intensities without the homogenates (see Materials and Methods). The luminescence intensities with and without the homogenates were similar, indicating that mouse tissue and the lavage liquid do not contain quenching activities for luciferase. Another control experiment was performed with peritoneal exudate as a growth medium. First, we injected sterile NB medium into the peritoneal cavity of each mouse, sacrificed the mice 5 h later, and removed the peritoneal NB-exudate mixture from each cavity. Next, we inoculated WA-C(pYV08,pCJFY-GL) into this mixture for 24 h. Most interestingly, we measured fluorescence and luminescence values comparable to those from NBD medium (which are about 20-fold higher than in NB medium but threefold less than in peritoneal infections).
DISCUSSION
This study is focused on monitoring the expression of the iron-repressible genes fyuA and hemR of Y. enterocolitica under different in vitro and in vivo (mouse infection model) growth conditions. As a first step, we constructed various fyuA and hemR translational reporter fusions by using gfp and luc for simultaneous monitoring (twin reporter construct) of fyuA and hemR expression or monitoring expression of one gene by immunofluorescence and luminescence using tandem gfp-luc reporter constructs (Fig. 1).
As shown in Fig. 2, all protein fusions were produced in vitro under iron starvation (NBD medium) at 27°C. The amounts of the fusion proteins were not equal, as judged by the intensities of the bands detected by immunoblotting. However, the results from immunoblotting corresponded well with the GFP fluorescence and luciferase luminescence intensities, respectively, as shown in Fig. 3. While the FyuA-GFP fluorescence of WA-C(pYV08,pCJHE-L/FY-G) was higher than the FyuA-GFP-LUC fluorescence of WA-C(pYV08,pCJFY-GL) at 27°C with NBD (which was also observed on the protein level) (Fig. 2A), the level of luciferase activity of the FyuA-LUC was lower than that of the FyuA-GFP-LUC construct. This is in line with the protein level shown by immunoblots in Fig. 2B. Since GFP is known to be very stable (6), the GFP might not only stabilize itself but might also protect the luciferase, resulting in a stronger luminescence than the luminescence of the single LUC fusion. On the other hand, the GFP fluorescence intensity was higher for FyuA-GFP or HemR-GFP than for the tandem constructs. This may be attributed to correct folding and thus a higher level of stability of the unfused GFP molecule. At 27°C under iron starvation conditions, the FyuA-GFP strain exhibited higher fluorescence than the HemR-GFP strain, which was, however, not observed on a protein level (Fig. 2A). Surprisingly, the reporter fusions were weakly expressed at 37°C in NBD (iron-derepressed condition) in comparison to results at a 27°C growth temperature. This temperature effect was not associated with the presence of the virulence plasmid. This unusual temperature-dependent expression pattern of fyuA and hemR in vitro has also been observed with other yersiniae virulence genes, such as inv (encoding invasin) and yst (encoding heat-stable enterotoxin) (31). As previously demonstrated, a psn-lacZ translational reporter fusion (psn encodes the FyuA homolog of Y. pestis and is nearly identical to the fyuA sequence) was significantly expressed by a virulence plasmidless Y. pestis strain at 37°C in deferrated PMH medium (14). However, expression studies at 27°C were not reported. Thus, these results cannot be compared with our results with Y. enterocolitica because of different parameters (growth medium, different species, different reporter).
We also tested the influence of oxidative stress on the expression of fyuA reporter fusions under in vitro growth conditions to simulate conditions close to those of the mouse infection model. As shown in Fig. 3C, under iron starvation conditions and with added 1 mM H2O2 or 0.2 mM paraquat (superoxide-generating compound) we observed no significant change of fyuA reporter expression at 37°C with the fyuA-gfp-luc fusion. Recently it has been demonstrated with E. coli that the concentration of Fur molecules increases at least twofold in the cytoplasm under oxidative stress (45). However, whether this Fur increase leads to repression of irp genes has not been shown. Increased Fur production might be compensated for by oxidative damage of Fur and thus might be without influence on irp gene expression (45). This could explain our in vitro results.
Next we examined the regulation of the fyuA and hemR reporter fusions in the BALB/c mouse infection model. BALB/c mice were infected with the recombinant yersiniae by different routes. Twenty-four hours later, yersiniae and tissue specimens were taken to analyze the reporter activities. Comparison of luminescence intensities indicated that fyuA and hemR reporter fusions showed similar organ-specific expression profiles. The highest level of expression was found with yersiniae obtained from the peritoneal cavity, whereas yersiniae recovered from the intestinal lumen and the liver showed weak reporter signals and those from the Peyer's patches and spleen expressed moderate signals. The intestinal lumen represents an anaerobic environment; thus, iron is expected to be in its reduced (Fe2+) state. We can assume that Y. enterocolitica, like E. coli, is equipped with a feo system for ferrous iron uptake, which may lead to repression of fyuA and hemR in the intestinal lumen. When yersiniae enter the Peyer's patches, fyuA and hemR reporter expression are obviously induced, as seen by an increase in luminescence and fluorescence, indicating iron starvation. From the Peyer's patches, yersiniae typically disseminate into spleen and liver. Surprisingly, yersiniae from the liver showed much weaker reporter signals than those from spleen. Presumably, iron is less available for yersiniae in the spleen (derepressed state) than in the liver, in spite of the fact that the amount of tissue iron in the liver is around 250 μg per g of dry weight and in the is spleen around 900 μg per g of dry weight for mice on a standard diet (46). This assumption is also supported by the observation that Ybt-negative Y. enterocolitica strains (non-biotype IB strains) occasionally cause liver abscesses, probably because of sufficient iron accessibility in the liver (37).
The highest level of fyuA and hemR reporter expression was observed in the peritoneal cavity. It is known that i.p. injection of yersiniae leads to rapid infiltration of the peritoneal cavity by polymorphonuclear leukocytes within 10 to 20 h and thus should be associated with release of oxygen intermediates and the iron-sequestering protein lactoferrin. However, we could not show in vitro that H2O2 or paraquat (known as oxidative stress inducers) significantly induce fyuA reporter expression at 37°C in NBD.
As a control experiment, we have tested fyuA reporter activity after incubation in peritoneal exudate at 37°C. Also, although we obtained 20-fold-higher reporter activities than in NB medium, they did not approach the in vivo data of the peritoneal infection. Obviously, other still-unknown factors are involved in fyuA upregulation of yersiniae in the peritoneal cavity.
The in vivo GFP fluorescence results (Fig. 5) corresponded well with the luciferase luminescence, as was also seen in vitro. Using the CLSM in connection with cryosections and intestinal lavage samples, we observed weak fluorescence of yersiniae in the liver and in the intestinal lumen, respectively. Again, the fluorescence intensity of yersiniae from the peritoneal lavage sample was the highest, followed by that of the spleen. Analyzing the fluorescing abscesses closely, we did not observe any spatially dependent expression of fyuA or hemR within a single abscess after 24 h of infection.
Infecting mice by different routes did not give conflicting results in the respective reporter gene activities. However, since we have tested only a single time point after infection, nothing can be said about the dynamics of the expression of these receptors. We are also aware of the problem of the high stability of the GFP, which can accumulate in the cytoplasm and thus may not reflect the reporter fusion expression rate at the time point of the experiment.
While the impact of the yersiniabactin uptake system is well known, the role of the heme uptake system in the pathogenesis of yersiniosis is still unresolved. Preliminary experiments with a spontaneous hemR mutant of Y. enterocolitica indicated that the heme uptake has no significant effect on mouse virulence compared to the results with the wild type (39). However, more experiments with genetically defined mutants in the heme uptake operon are needed for analysis of organ-specific infection in the mouse model.
In summary, we were able to detect organ-specific expression of the fyuA and the hemR receptor gene in the mouse model, which probably reflects the availability of iron for yersiniae. A striking result was the extraordinary upregulation of fyuA and hemR in the peritoneal cavity, which may indicate a specific activation mechanism. Moreover, we demonstrated that gfp-luc tandem reporter construct is a useful hybrid reporter system for in vivo expression studies and that regulation of two genes can be studied simultaneously in single bacterial cells using gfp and luc.
ACKNOWLEDGMENTS
C.A.J. is supported by the Graduiertenkolleg “Infektion und Immunität (GRK 303)” of the Deutsche Forschungsgemeinschaft.
We thank D. Brem and A. M. Geiger for critical reading of the manuscript.
REFERENCES
- 1.Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K. Current protocols in molecular biology. New York, N.Y: John Wiley & Sons, Inc.; 1989. [Google Scholar]
- 2.Autenrieth I B, Hantchmann P, Heymer B, Heesemann J. Immunohistological characterisation of the cellular immune response against Yersinia enterocolitica in mice: evidence for the involvement of T-lymphocytes. Immunobiology. 1993;187:1–16. doi: 10.1016/S0171-2985(11)80241-X. [DOI] [PubMed] [Google Scholar]
- 3.Baumler A J, Hantke K. Ferrioxamine uptake in Yersinia enterocolitica: characterization of the receptor protein FoxA. Mol Microbiol. 1992;6:1309–1321. doi: 10.1111/j.1365-2958.1992.tb00852.x. [DOI] [PubMed] [Google Scholar]
- 4.Bearden S W, Perry R D. The Yfe system of Yersinia pestis transports iron and manganese and is required for full virulence of plague. Mol Microbiol. 1999;32:403–414. doi: 10.1046/j.1365-2958.1999.01360.x. [DOI] [PubMed] [Google Scholar]
- 5.Brem D, Pelludat C, Rakin A, Jacobi C A, Heesemann J. Functional analysis of yersiniabactin transport genes of Yersinia enterocolitica. Microbiology. 2001;147:1115–1127. doi: 10.1099/00221287-147-5-1115. [DOI] [PubMed] [Google Scholar]
- 6.Chalfie M, Tu Y, Euskirchen G, Ward W W, Prasher D C. Green fluorescent protein as a marker for gene expression. Science. 1994;263:802–805. doi: 10.1126/science.8303295. [DOI] [PubMed] [Google Scholar]
- 7.Chang A C Y, Cohen S N. Construction and characterization of amplifiable multicopy DNA cloning vehicles derived from the P15A cryptic miniplasmid. J Bacteriol. 1978;134:1141–1156. doi: 10.1128/jb.134.3.1141-1156.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cormack B P, Valdivia R H, Falkow S. FACS-optimized mutants of green fluorescent protein (GFP) Gene. 1996;173:33–38. doi: 10.1016/0378-1119(95)00685-0. [DOI] [PubMed] [Google Scholar]
- 9.Cornelis G R, Boland A, Boyd A P, Geuijen C, Iriarte M, Neyt C, Sory M P, Stanier E. The virulence plasmid of Yersinia, an antihost genome. Microbiol Mol Biol Rev. 1998;62:1315–1352. doi: 10.1128/mmbr.62.4.1315-1352.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Crosa J H. Signal transduction and transcriptional and posttranscriptional control of iron-regulated genes in bacteria. Microbiol Mol Biol Rev. 1997;61:319–336. doi: 10.1128/mmbr.61.3.319-336.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.DeWet J R, Wood K V, DeLuca M, Helinski D R, Subramani S. Firefly luciferase gene: structure and expression in mammalian cells. Mol Cell Biol. 1987;7:725–737. doi: 10.1128/mcb.7.2.725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escolar L, Perez-Martin J, de Lorenzo V. Opening the iron box: transcriptional metalloregulation by the Fur protein. J Bacteriol. 1999;181:6223–6229. doi: 10.1128/jb.181.20.6223-6229.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ewald J H, Heesemann J, Rüdiger H, Autenrieth I B. Interaction of polymorphonuclear leucocytes with Yersinia enterocolitica: role of the Yersinia virulence plasmid and modulation by the iron-chelator desferrioxamine B. Infect Dis. 1994;170:140–150. doi: 10.1093/infdis/170.1.140. [DOI] [PubMed] [Google Scholar]
- 14.Fetherston J D, Bearden S W, Perry R D. YbtA, an AraC-type regulator of the Yersinia pestis pesticin/yersiniabactin receptor. Mol Microbiol. 1996;22:315–325. doi: 10.1046/j.1365-2958.1996.00118.x. [DOI] [PubMed] [Google Scholar]
- 15.Hanahan D. Studies on transformation of Escherichia coli with plasmids. J Mol Biol. 1983;166:557–580. doi: 10.1016/s0022-2836(83)80284-8. [DOI] [PubMed] [Google Scholar]
- 16.Hanski C, Kutschka U, Schmoranzer H P, Naumann M, Stallmach A, Hahn H, Menge H, Riecken E O. Immunohistochemical and electron microscopic study of interaction of Yersinia enterocolitica serotype O8 with intestinal mucosa during experimental enteritis. Infect Immun. 1989;57:673–678. doi: 10.1128/iai.57.3.673-678.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hantke K. Cloning of a repressor protein gene of iron-regulated systems in Escherichia coli K12. Mol Gen Genet. 1984;197:337–341. doi: 10.1007/BF00330982. [DOI] [PubMed] [Google Scholar]
- 18.Hantke K. Selection procedure for deregulated iron transport mutants (fur) in Escherichia coli K12: Fur not only affects iron metabolism. Mol Gen Genet. 1987;210:135–139. doi: 10.1007/BF00337769. [DOI] [PubMed] [Google Scholar]
- 19.Hantke K. Iron and metal regulation in bacteria. Curr Opin Microbiol. 2001;4:172–177. doi: 10.1016/s1369-5274(00)00184-3. [DOI] [PubMed] [Google Scholar]
- 20.Heesemann J, Laufs R. Double immunofluorescence microscopic technique for accurate differentiation of extracellularly and intracellularly located bacteria in cell culture. J Clin Microbiol. 1985;22:168–175. doi: 10.1128/jcm.22.2.168-175.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Heesemann J. Chromosomal-encoded siderophores are required for mouse virulence of enterophathogenic Yersinia species. FEMS Microbiol Lett. 1987;48:229–233. [Google Scholar]
- 22.Heesemann J, Hantke K, Vocke T, Saken E, Rakin A, Stojiljkovic I, Berner R. Virulence of Yersinia enterocolitica is closely associated with siderophore production, expression of an iron repressible outer membrane protein of 65000 Da and pesticin sensitivity. Mol Microbiol. 1993;8:397–408. doi: 10.1111/j.1365-2958.1993.tb01583.x. [DOI] [PubMed] [Google Scholar]
- 23.Jacobi C A, Roggenkamp A, Rakin A, Zumbihl R, Leitriz L, Heesemann J. In vitro and in vivo expression studies of yopE from Yersinia enterocolitica using the gfp reporter gene. Mol Microbiol. 1998;30:865–882. doi: 10.1046/j.1365-2958.1998.01128.x. [DOI] [PubMed] [Google Scholar]
- 24.Kammler M, Schon C, Hantke K. Characterization of the ferrous iron uptake system of Escherichia coli. J Bacteriol. 1993;175:6212–6219. doi: 10.1128/jb.175.19.6212-6219.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Koebnik R, Hantke K, Braun V. The TonB-dependent ferrichrome receptor FcuA of Yersinia enterocolitica: evidence against a strict co-evolution of receptor structure and substrate specificity. Mol Microbiol. 1993;7:383–393. doi: 10.1111/j.1365-2958.1993.tb01130.x. [DOI] [PubMed] [Google Scholar]
- 26.Laemmli U K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- 27.Otto B R, Verweijj-van Vught A M J, Mac Laren D M. Transferrins and heme-compounds as iron sources for pathogenic bacteria. Crit Rev Microbiol. 1992;18:217–233. doi: 10.3109/10408419209114559. [DOI] [PubMed] [Google Scholar]
- 28.Pelludat C, Rakin A, Jacobi C A, Schubert S, Heesemann J. The yersiniabactin biosynthetic gene cluster of Yersinia enterocolitica: organization and siderophore-dependent regulation. J Bacteriol. 1998;180:538–546. doi: 10.1128/jb.180.3.538-546.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rakin A, Saken E, Harmsen D, Heesemann J. The pesticin receptor of Yersinia enterocolitica: a novel virulence factor with dual function. Mol Microbiol. 1994;13:253–263. doi: 10.1111/j.1365-2958.1994.tb00420.x. [DOI] [PubMed] [Google Scholar]
- 30.Ratledge C, Dover L G. Iron metabolism in pathogenic bacteria. Annu Rev Microbiol. 2000;54:881–941. doi: 10.1146/annurev.micro.54.1.881. [DOI] [PubMed] [Google Scholar]
- 31.Revell P A, Miller V L. A chromosomally encoded regulator is required for expression of the Yersinia enterocolitica inv gene and for virulence. Mol Microbiol. 2000;35:677–685. doi: 10.1046/j.1365-2958.2000.01740.x. [DOI] [PubMed] [Google Scholar]
- 32.Russo-Marie F, Roederer M, Sager B, Herzenberg L A, Kaiser D. β-Galactosidase activity in single differentiating bacterial cells. Proc Natl Acad Sci USA. 1993;90:8194–8198. doi: 10.1073/pnas.90.17.8194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Saken E, Rakin A, Heesemann J. Molecular characterization of a novel siderophore-independent iron transport system in Yersinia. Int J Med Microbiol. 2000;290:51–60. doi: 10.1016/S1438-4221(00)80106-X. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch E F, Maniatis T. Molecular cloning: a laboratory manual. Cold Spring Harbor, N.Y: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Schubert S, Fischer D, Heesemann J. Ferric enterochelin transport in Yersinia enterocolitica: molecular and evolutionary aspects. J Bacteriol. 1999;181:6387–6395. doi: 10.1128/jb.181.20.6387-6395.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schubert S, Sorsa J L, Cuenca S, Fischer D, Jacobi C A, Heesemann J. HPI of high-virulent Yersinia is found in E. coli strains causing urinary tract infection. Structural, functional aspects, and distribution. Adv Exp Med Biol. 2000;485:69–73. doi: 10.1007/0-306-46840-9_9. [DOI] [PubMed] [Google Scholar]
- 37.Schuchmann M, Gerbes A L, Heesemann J, Sauter G. Multiple liver abscesses caused by Yersinia enterocolitica in a patient receiving long-term transfusion therapy for osteomyelosclerosis. Dig Dis Sci. 1997;42:2501–2504. doi: 10.1023/a:1018808527632. [DOI] [PubMed] [Google Scholar]
- 38.Simonet M, Richard S, Berche P. Electron microscopic evidence for in vivo extracellular localization of Yersinia pseudotuberculosis harboring the pYV plasmid. Infect Immun. 1990;58:841–845. doi: 10.1128/iai.58.3.841-845.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stojiljkovic I, Hantke K. Hemin uptake system of Yersinia enterocolitica: similarities with other TonB-dependent systems in Gram-negative bacteria. EMBO J. 1992;12:4359–4367. doi: 10.1002/j.1460-2075.1992.tb05535.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Towbin H, Staehelin T, Gordon J. Electrophoretic transfer of proteins from polyacrylamide gels to nitrocellulose sheets: procedure and some applications. Proc Natl Acad Sci USA. 1979;76:4350–4354. doi: 10.1073/pnas.76.9.4350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tsolis R M, Baumler A J, Heffron F, Stojiljkovic I. Contribution of TonB- and Feo-mediated iron uptake to growth of Salmonella typhimurium in the mouse. Infect Immun. 1996;64:4549–4556. doi: 10.1128/iai.64.11.4549-4556.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wandersman C, Stojiljkovic I. Bacterial heme sources: the role of heme, hemoprotein receptors and hemophores. Curr Opin Microbiol. 2000;3:215–220. doi: 10.1016/s1369-5274(00)00078-3. [DOI] [PubMed] [Google Scholar]
- 43.Weinberg E D. Iron and infection. Microbiol Rev. 1978;42:45–66. doi: 10.1128/mr.42.1.45-66.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Weinberg E D. Iron withholding: a defense against infection and neoplasia. Physiol Rev. 1984;64:65–107. doi: 10.1152/physrev.1984.64.1.65. [DOI] [PubMed] [Google Scholar]
- 45.Zheng M, Doan B, Schneider T D, Storz G. OxyR and SoxRS regulation of fur. J Bacteriol. 1999;181:4639–4643. doi: 10.1128/jb.181.15.4639-4643.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhou X Y, Tomatsu S, Fleming R E, Parkilla S, Waheed A, Jiang J, Fei Y, Brunt E M, Ruddy D A, Prass C E, Schatzman R C, O'Neil R, Britton R S, Bacon B R, Sly W S. HFE gene knockout produces mouse model of hereditary hemochromatosis. Proc Natl Acad Sci USA. 1998;95:2492–2497. doi: 10.1073/pnas.95.5.2492. [DOI] [PMC free article] [PubMed] [Google Scholar]