Abstract
Objectives
Three-dimensional (3D) genome alterations can dysregulate gene expression by rewiring physical interactions within chromosomes in a tissue-specific or cell-specific manner and lead to diseases. We aimed to elucidate the 3D genome structure and its role in gene expression networks dysregulated in systemic lupus erythematosus (SLE).
Methods
We performed Hi-C experiments using CD4+ T cells from 7 patients with SLE and 5 age-matched and sex-matched healthy controls (HCs) combined with RNA sequencing analysis. Further integrative analyses, including transcription factor motif enrichment, SPI1 knockdown and histone modifications (H3K27ac, H3K4me1, H3K4me3), were performed for altered loop-associated gene loci in SLE.
Results
We deciphered the 3D chromosome organisation in T cells of patients with SLE and found it was clearly distinct from that of HCs and closely associated with the disease activity of SLE. Importantly, we identified loops within chromosomes associated with the disease activity of SLE and differentially expressed genes and found some key histone modifications close to these loops. Moreover, we demonstrated the contribution of the transcription factor SPI1, whose motif is located in the altered loop in SLE, to the overexpression of interferon pathway gene. In addition, we identified the potential influences of genetic variations in 3D genome alterations in SLE.
Conclusions
Our results highlight the 3D genome structure alterations associated with SLE development and provide a foundation for future interrogation of the relationships between chromosome structure and gene expression control in SLE.
Keywords: autoimmune diseases; T-lymphocyte subsets; lupus erythematosus, systemic
WHAT IS ALREADY KNOWN ON THIS TOPIC
Systemic lupus erythematosus (SLE) is a complex autoimmune disease and is widely studied at the epigenetic level, including DNA methylation, histone modification and non-coding RNAs.
Dysregulated expression of some genes is correlated with disease activity in patients with SLE.
Three-dimensional (3D) genomic alterations may be related to the pathogenesis of autoimmune diseases.
WHAT THIS STUDY ADDS
This study showed the alteration of 3D genome organisation in CD4+ T cells of patients with SLE with different disease activities and reveals that both topologically associated domain appearance and loop strength profiles are correlated with the disease activities of patients with SLE.
This study identified the interaction loops within chromosomes associated with SLE and SLE Disease Activity Index scores and revealed the potential relationship among transcription factors, histone modifications, genetic variation and differentially expressed genes in SLE.
HOW THIS STUDY MIGHT AFFECT RESEARCH, PRACTICE OR POLICY
These findings contribute to revealing the epigenetic mechanism and highlighting the relationships among chromosome structure, genetic variations and gene expression control in the pathogenesis of SLE, as well as providing a biomarker for the diagnosis of SLE and a target for disease treatment.
Introduction
To understand the three-dimensional (3D) organisation of the global genome, researchers have applied chromosome conformation capture (3C)1 and its derived methods, such as Hi-C,2 to a variety of organisms from bacteria3 to humans4 for cell ensembles or even single cells.5 6 In the past decade, with increasing sequencing depth, Hi-C data have revealed that the 3D genome architecture comprises a hierarchy of structures7: ‘A’ and ‘B’ compartments, topologically associated domains (TADs), whose boundaries are enriched for architectural proteins such as cohesion and CTCF,8 9 and chromatin loops that promote interaction between regulatory elements and genes.10–12
Accumulating evidence has suggested that the 3D structure has a role in human diseases.13 For example, chromosome translocation frequently occurs in haematological malignancies and influences translocation partner choice.14 15 The disruption of TAD boundaries can lead to aberrant loop interactions that induce oncogene expression in gliomagenesis.16 Enhancer variants associated with Alzheimer’s disease affect gene expression via chromatin looping.17 In short, cell type-specific 3D genome information therefore provides a useful resource to mechanistically dissect disease risk loci by connecting non-coding regions to disease risk genes.18
Systemic lupus erythematosus (SLE) is a complex autoimmune disease characterised by autoreactive antibody production, dysregulation of both T and B lymphocytes and immune complex formation that cause multiorgan damage.19 20 Among many autoimmune diseases, SLE might be the most deeply studied at the epigenetic level,21 including DNA methylation,22 histone modification23 and non-coding RNAs.24 In 3D genomic studies in SLE, Su et al used the combination of promoter-Capture-C and assay for transposase-accessible chromatin using sequencing (ATAC-seq) to map regulatory SLE variants to target genes in follicular helper T cells from healthy children.25 By using circular chromosome conformation capture and high-throughput sequencing (4C-seq), Hou et al found that the genomic region harbouring an SLE susceptibility locus rs2431697 is a cell-type-specific enhancer specifically regulating miR-146a expression.26 However, few 3D genome studies have been performed with samples from patients with SLE, and the association between 3D genome alterations and SLE disease activity and the potential regulatory roles of 3D genome alterations in autoimmune-related gene expression remain unclear.
Here, we used integrative methods to analyse 3D genome alterations and the transcriptome of CD4+ T cells in SLE. We also identified SLE-associated loops with differentially expressed genes (DEGs). We presented a 3D genome landscape of altered loop loci in SLE and demonstrated potential influences of the transcription factor SPI1 at an interferon (IFN)-I pathway-related gene.
Materials and methods
See online supplemental materials and methods.
ard-2022-222653supp001.pdf (2.1MB, pdf)
Results
Widespread 3D genome alterations in SLE CD4+ cells
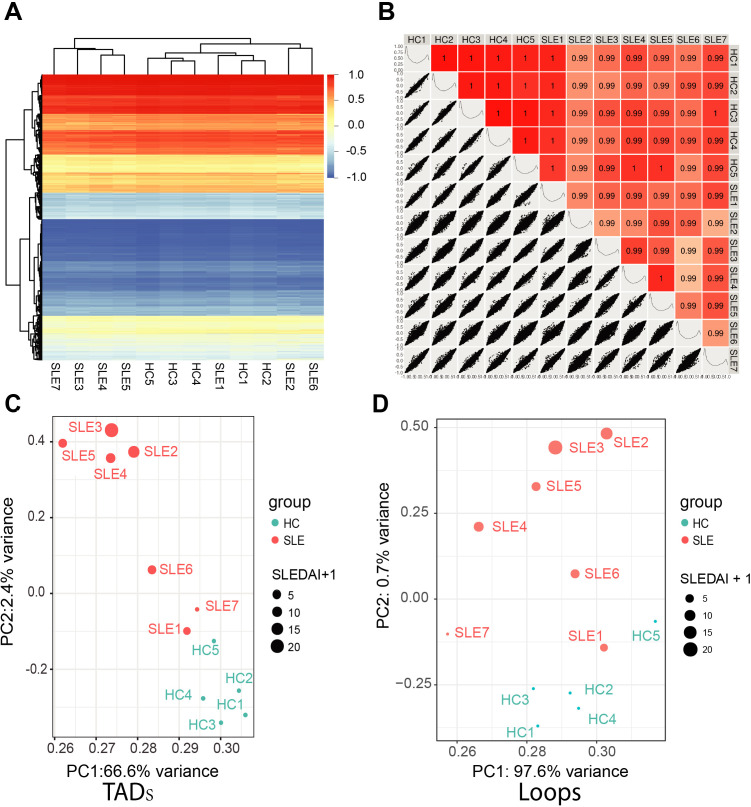
To investigate the 3D genome architecture of patients with SLE, we performed Hi-C with peripheral blood CD4+ T cells from seven patients with SLE and five healthy controls (HCs). The demographic information for both patients with SLE and HCs are described in online supplemental table S1. After Hi-C sequencing, an average of 2 billion raw reads per sample (approximately 610 billion base pairs) were generated from 12 samples (online supplemental table S2). Then, we identified A/B compartments, topologically associating domains (TADs) and loops. As shown in figure 1A and B, there was little change in the compartment structure between the patients with SLE and the HCs. Furthermore, we performed aggregate TAD or loop analysis by principal component analysis (PCA), in which the SLE Disease Activity Index (SLEDAI) score of each patient with SLE was considered. There are certain rules on TAD border appearance and loop strength in 12 individuals. Score plots from PCA displayed clustering trends between the patients with SLE and the HCs (figure 1C and D). Moreover, figure 1C shows an increasing trend of SLEDAI scores from right to left (PC1 from high to low) and from bottom to top (PC2 from low to high) at the TAD appearance level. Figure 1D shows an increasing trend in the SLEDAI score from the bottom to top (PC2 from low to high) at the loop strength level. These findings indicate that genomic 3D features (TADs border and loop strength) of CD4+ T cells between patients with SLE and HCs are distinct, and the higher the activity of SLE, the greater the differences in 3D genomic features.
Figure 1.
Genomic three-dimensional feature difference of CD4+ T cell between patients with systemic lupus erythematosus (SLE) and healthy controls (HCs). (A) Compartments of 12 samples including 7 patients with SLE and 5 HCs. Compartment c score from −1 to 1 are shown with blue to red. (B) Pairwise Pearson’s correlation coefficient and associated scatter plots between each two samples. The upper right displays Pearson’s correlation coefficient. The scores are shown in different degrees of red. The lower left displays scatter plots of compartments c score between each two samples. The c score distributions of each sample are shown on diagonal. (C–D) Principal component analysis of the topologically associated domains (TADs) border appearance (C) and the loop strength (D) of the 12 samples. Patients with SLE are shown as red points and HCs are shown as blue points. The point size reflects the SLE Disease Activity Index (SLEDAI) of patients and we set SLEDAI of HCs as 0.
3D genome alterations associated with differentially expressed genes in SLE CD4+ T cells
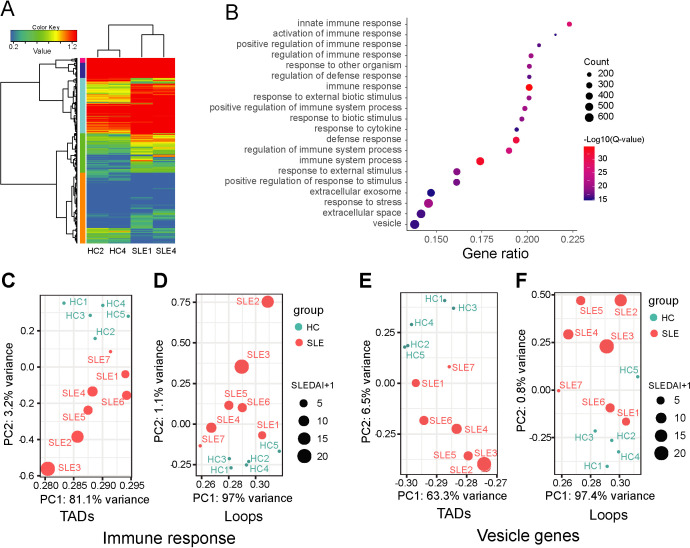
To investigate the role of TADs and chromatin loops in controlling gene expression, we performed RNA sequencing (RNA-seq) to analyse changes in gene expression profiles in samples from three patients with SLE and three HCs tested by Hi-C analysis. We found that the gene expression profiles of the no. 7 patient with SLE (SLE7) who had no apparent clinical phenotype were more similar to HCs (online supplemental table S1 and figure S1). Thus, DEGs were identified by gene expression comparison between two samples from patients with SLE and two samples from HCs (figure 2A). Gene Ontology (GO) enrichment analysis of the DEGs showed that many immune functions were enriched and covered approximately 80% of the DEGs (figure 2B). Two gene sets extracted from the GO enrichments (immune response genes and vesicle genes) were used to profile DEGs overlapping TAD border or loop features using PCA among all samples, in which SLEDAI scores were considered. The PCA revealed that the 3D genomic alterations of these genes can distinguish the patients with SLE from the HCs and were also positively correlated with SLEDAI scores in the patients with SLE (figure 2C-F).
Figure 2.
Gene expression difference between patients with systemic lupus erythematosus (SLE) and healthy controls (HCs). (A) Heat map represent differentially expressed genes (DEGs) between patients with SLE (SLE1, SLE4) and HCs (HC2, HC4). (B) Gene Ontology (GO) functional enrichment of the DEGs. (C–F) Principal component analysis of the topologically associated domains (TADs) border appearance and loop strength from which the TADs and loops overlap with genes from DEGs and annotated as GO function ‘immune response’ and ‘vesicle genes’. SLEDAI, SLE Disease Activity Index.
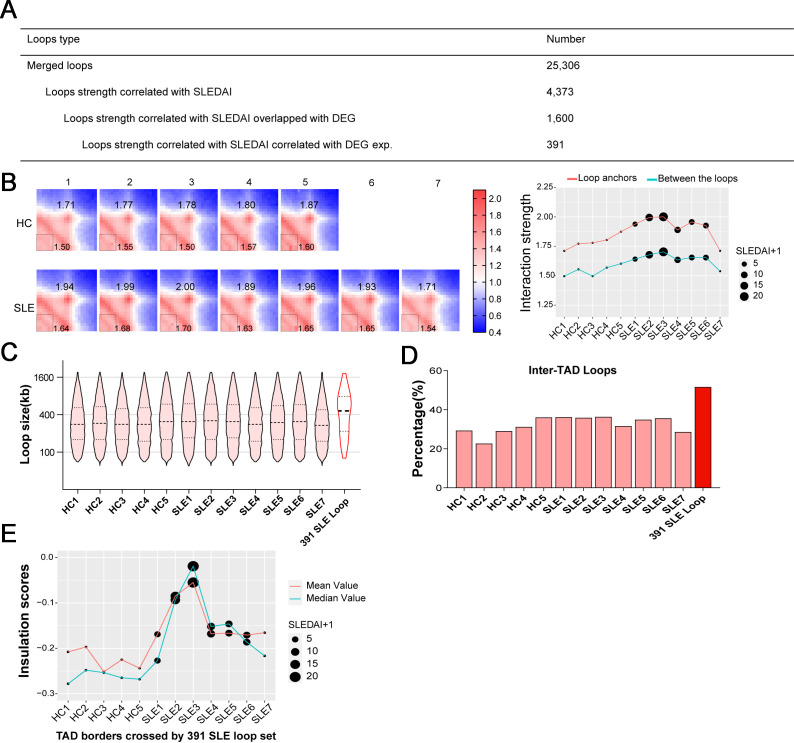
Three hundred ninety-one loops highly correlated with SLEDAI and distinct gene expression
To better understand the relationship between loop strength and SLEDAI score, we performed Pearson’s correlation analysis. More loops were positively correlated with SLEDAI scores (online supplemental figure S2A). Recently, gene signature expression was shown to be correlated with SLE disease activity.27 We quantified the relationship between SLEDAI-associated loops and gene expression by intersection analysis (figure 3A). In the merged loop set, 4373 loops that were highly correlated with the SLEDAI score were identified. Then, loop anchors without overlap with DEGs in SLE were removed. Finally, 391 SLEDAI-associated loops were identified as SLE-associated loops (online supplemental table S3). In addition, we also analysed the relationships between loop strength and other clinical parameters of patients with SLE, respectively, including the levels of complement 3 and complement 4, the number of red blood cell, white blood cell and platelet, haemoglobin concentration. The loops that correlated with these clinical parameters are shown in online supplemental table S4. For display and visual consistency with the SLE-associated loops that we screened, scatter plots were used to show the relationship between the SLEDAI range and changes in the SLE-associated loop strength in all samples (figure 3B). In addition, functional enrichment and pathway analysis revealed that the 391 genes with SLE-associated loops were mainly related to inflammatory and immune-related biological processes (online supplemental figure S2B and online supplemental table S5). In addition, some immune-related pathways including influenza infection, TNF Receptor Associated Factor 6 (TRAF6)-mediated IRF7 activation in toll-like receptor 7/8 or 9 signalling and the NLRP3 inflammasome were enriched in 391 genes (online supplemental table S5). To further investigate the SLE-associated loop features, we compared the length distributions of all loops in 12 samples and the SLE-associated loop set. A longer length distribution was observed in the SLE-associated loop set (figure 3C), which has the potential to confer long-range interactions over the TAD border. We characterised inter-TAD loop differences between loops set of 12 individual samples and SLE-associated loop sets and found that more SLE-associated loops tended to be long-range inter-TAD loops (figure 3D). Further analysis was performed on TAD border insulation strength. We selected the TAD borders overlapping with SLE-associated inter-TAD loops set in each sample and calculated the mean and median insulation scores around selected TAD borders. A decreasing trend of insulation strength was observed in the patients with SLE and indicated that SLE-associated inter-TADs loops were determined by weak insulation scores around TAD borders, and SLEDAI scores were involved in this trend (figure 3E).
Figure 3.
Systemic lupus erythematosus (SLE) associated loops are identified by SLE Disease Activity Index (SLEDAI) scores and different gene expression. (A) Number of loops that correlated with SLEDAI scores and gene expression. (B) Aggregate peak analysis result on 391 SLE loops that associated with SLEDAI scores and differentially expressed genes (DEGs) expression in each sample. With normalised APA matrix (left), interaction strengths are indicated at central pixel (loop anchors) and the lower left pixels (between the loop anchors). SLEDAI are indicated in line plot (right). (C) Length distribution of loops in all the samples and 391 SLE loops. (D) Percentages of intertopologically associated domain (inter-TAD) loops in all the samples and 391 SLE loops. (E) The mean value and median value of insulation scores from the TAD border. In each sample, the TAD border that are crossed by 391 SLE-associated loops are selected. SLEDAI are indicated in all samples. HC, healthy control.
ard-2022-222653supp002.xlsx (38.6KB, xlsx)
ard-2022-222653supp003.xlsx (2.1MB, xlsx)
ard-2022-222653supp004.xlsx (79.3KB, xlsx)
Additionally, to confirm the changes in SLE loop-associated gene expression observed by RNA-seq, we compared the relative expression of SLE loop-associated genes between our RNA-seq data and the published CD4+ T RNA-seq dataset (GSE97263). And a heat map revealed the similarities in the differential gene expression profile between them (online supplemental figure S2D). We also performed RT-quantitative PCR (qPCR) to detect some DEGs identified by RNA-seq (online supplemental table S6). The results showed that the expression of DDX60L, LTBP1, CXCL13, SLC8A1, ATG5 and IFIT1 was significantly upregulated and the expression of FCER1A and HDC was downregulated in CD4+ T cells of patients with SLE compared with HCs (online supplemental figure S2E), which was consistent with the results of our RNA-seq. Thus, comparable different interactions, especially SLE-associated loops, were identified in CD4+ T cells from patients with SLE.
3D chromatin changes around SLE-related gene loci
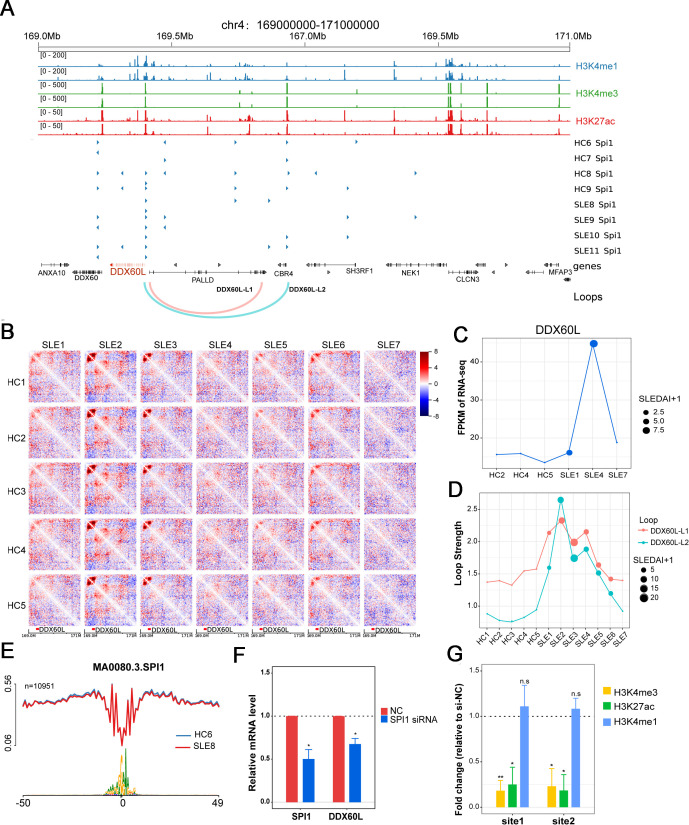
As over 90% of SLE-associated loops were enhancer or promoter-based interactions (online supplemental figure S2C), we focused on SLE-associated loop-associated gene loci. There were two SLE-associated loops overlapping with the DDX60L gene promoter (figure 4A). DDX60L is an ATP-dependent helicase gene involved in the IFN antiviral response, which is an innate immune response.28 At the DDX60L locus, the Hi-C interaction matrix indicated that patients with SLE with higher SLEDAI scores had a stronger interaction frequency than HCs (figure 4B). Moreover, the SLEDAI scores of the patients were higher, and the loop strength and gene expression at the locus were stronger (figure 4C and D). To assess the interaction between the SLE loop-associated promoter in DDX60L and long-range interacting region, we cloned the DDX60L gene promoter with or without promoter interacting region into the luciferase reporter vector, respectively (online supplemental table S7). The results showed that the group containing DDX60L promoter and the promoter interacting region, an SLE loop interaction, had greater luciferase activity than the group containing only the DDX60L promoter (online supplemental figure S3A), suggesting the SLE loop interaction promoted the transcription of DDX60L gene. In addition, according to previous study,25 we used CRISPR-Cas9 methodology to delete the promoter interacting regions of DDX60L gene, and observed that the respective deletion of two fragments in the region downregulated DDX60L expression in Jurkat cell line (online supplemental figure S3B).
Figure 4.
Chromatin loop and SPI1 regulate DDX60L expression. (A) Genomic features of localised region of gene DDX60L. From the top, genomic coordinates; enrichment peaks of H3K4me1, H3K4me3 and H3K27ac of activate CD4+ T cell from public data, transcription factor SPI1 ATAC footprint of sample HC6~HC9 and SLE8~SLE11, Gene location (DDX60L is marked with red), the two loops that strength correlated with SLEDAI score and DDX60L gene expression. (B) The different chromatin interaction map of DDX60L localised region. Shown are the 20 kb binned heatmap depicting the Z-score difference between HCs (different rows) and patients with SLE (different columns). DDX60L gene is marked at the bottom. (C) DDX60L gene expression of sample HC2, HC4, HC5 and SLE1, SLE4, SLE7. The points size reflects the SLEDAI scores. (D) The strengths of two loops in all the samples. (E) Nucleotide resolution average chromatin accessibility (y-axis) along the binding site of SPI1 in the sample HC6 and SLE8. (F) Changes of gene expression levels of SPI1 and DDX60L in CD4+ T cells with SPI1 knockdown. (G) Changes of enrichment levels of H3K4me1, H3K4me3 and H3K27ac in CD4+ T cells with SPI1 knockdown. HC, healthy controls; mRNA, messenger RNA; siRNA, small interfering RNA; SLEDAI, SLE Disease Activity Index; SLE, systemic lupus erythematosus.
In addition, within the DDX60L gene promoter, we observed the following: (1) the active histone marks H3K27ac, H3K4me1 and H3K4me3 were enriched at two loop anchors based on the public data of normal activated CD4+ T cells;29 (2) SPI1 is the transcription factor binding footprint most frequently enriched at the two loop anchors according to our ATAC-seq data from other patients with SLE (NODE: https://www.biosino.org/node/index, accession number: OEP003228) (figure 4A and E). SPI1 is an ETS-domain transcription factor that activates gene expression during lymphoid and myeloid cell development.30 The chromatin immunoprecipitation (ChIP) atlas database also shows SPI1 binding to the DDX60L promoter in lymphoid and myeloid cells.31 When SPI1 gene expression was suppressed by small interfering RNA (siRNA), moderate inhibition of DDX60L gene expression was observed, and the levels of H3K4me3 and H3K27ac were also decreased at the two loci of the DDX60L p rinomoter (figure 4F and G). To show the role of SPI1 in gene regulation and looping in T cells, we performed ChIP-qPCR and 3C-qPCR detections in CD4+ T cells with SPI1 knockdown. The ChIP experiment was performed using SPI1 antibody and the enriched DNA fragments were detected by qPCR to determine the binding of SPI1 in DDX60L promoter and/or distal looping region. The result showed that silencing SPI1 decreased the binding of SPI1 in the DDX60L promoter (200 bp upstream of transcription starting site) compared with negative control (online supplemental figure S3C). The results suggested that SPI1 was able to bind the promoter region of DDX60L. Moreover, we performed the 3C assay and detected the change of DDX60L gene-associated looping by multiple primers in CD4+ T cells with SPI1 knockdown. The results of 3C-qPCR showed that the interaction between the DDX60L promoter and distal looping region was decreased in CD4+ T cells transfected with SPI1 siRNA compared with negative control (online supplemental figure S3D, E). Overall, these results suggest that the transcriptional regulation of the DDX60L locus was controlled by SPI1, histone modifications and chromatin interactions in CD4+ T cells from patients with SLE.
Recent studies have demonstrated that SPI1 was associated with the Th9 phenotype in CD4+ T cells.32 T helper (Th)9 cell and interleukin (IL)-9 are involved in the pathogenesis of SLE,33 so we performed RT-qPCR analysis on IL9 and other genes associated with Th9 cell in HCs and patients with SLE. The expression of Th9 cell-associated genes (IL9, BATF, IRF1, ETV5, NFATC2 and FOXO1) was significantly upregulated in CD4+ T cells of patients with SLE compared with HCs (online supplemental figure S3F). In addition, we also detected the regulation of SPI1 on other T cell genes expression such as Th9 cell-associated genes IL9 and BATF, which have SPI1 binding motif within their promoter regions, when we knocked down SPI1 gene with siRNA in induced Th9 cells in vitro. We observed a significant decrease in IL9 and BATF expression in Th9 cells with SPI1 knockdown compared with negative control (online supplemental figure S3G). Taken together, these data indicated that SPI1 might play an important role in regulating T cell-related genes in patients with SLE.
Moreover, we found that other SLE-associated loop-associated genes, such as CXCL13, SLC8A1-AS1 and DNAJB4, exhibited similar patterns in that both gene expression and their associated loop strengths were positively correlated with SLEDAI scores. The 3D genomic features and gene expression levels of these gene loci in patients with SLE and HCs are shown in online supplemental figures S4–S6. We also determined the function of SLE-associated loop in regulating CXCL13 gene transcription (online supplemental file S4 A-D). The luciferase reporter gene assay showed a stronger luciferase activity in group containing CXCL13 promoter and the promoter interacting region compared with control group with only promoter (online supplemental figure S4E). In addition, deletion the CXCL13 promoter interacting region by CRISPR-Cas9 system in Jurkat cell line could lead to a decreased messenger RNA expression of CXCL13 gene (online supplemental figure S4F). However, we also found a few SLE-associated loops in which the SLEDAI score negatively correlated with loop strength and positively correlated with gene expression, such as a loop near the CASP5 locus (online supplemental figure S7).
3D chromatin changes around SLE-associated single-nucleotide polymorphisms
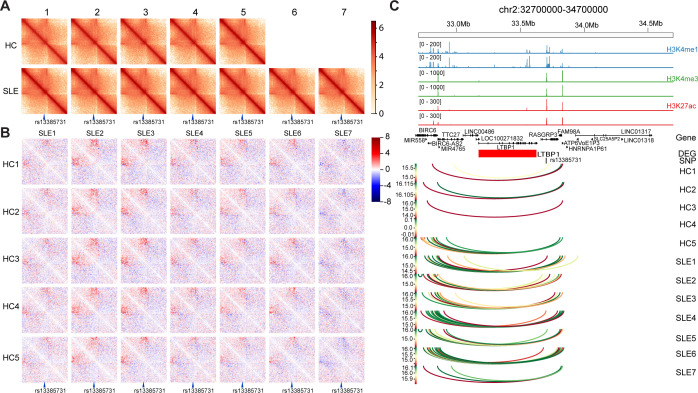
Recently, disease-associated single-nucleotide polymorphisms (SNPs) as well as variations were shown to affect the 3D genomic structure.34 35 To further elucidate the roles of SLE-associated SNPs in 3D genomic structure, we compared the difference in loop strength at SNP loci between the patients with SLE and the HCs without focusing on the 391 SLE-associated loop set. An SLE-associated SNP, rs13385731, was located in the region separated by contact depletion zones (figure 5A). Compared with that of the HCs, the interaction in the rs13385731 region was increased in the patients with SLE, except in SLE7 (figure 5B). Interestingly, we found that the SNP locus overlapped with an activated enhancer according to the enrichments of the histone markers H3K4me1, H3K4me3 and H3K27ac indicated by ChIP-seq data from ENCODE (figure 5C).29 Consistent with the Hi-C interaction heatmap results, loop interactions over a longer range occurred in the enhancer region with rs13385731 in SLE samples, which may affect the local chromatin configuration and gene expression of LTBP1 (figure 5C). LTBP1 is a component of the transforming growth factor beta (TGF-β) pathway that binds to fibrillin-1 in the extracellular matrix to inactivate TGF-β.36
Figure 5.
Long-term interactions change at systemic lupus erythematosus (SLE) risk single-nucleotide polymorphism (SNP) rs13385731 between patients with SLE and healthy controls (HCs). (A) Chromatin interaction map of rs13385731 localised region of all the samples. rs13385731 is indicated at the bottom. (B) Difference in chromatin interaction map of rs13385731 localised region. The 20 kb binned heatmap depicting the Z-score difference between HCs (different rows) and patients with SLE (different columns) are shown. rs13385731 is indicated at the bottom. Long-term interaction enhanced in sample SLE1, SLE2, SLE3, SLE4, SLE5, SLE6 compared with HC1~HC5 could be viewed. (C) Genomic features of localised region of rs13385731. From the top, genomic coordinates, ChIP-seq peaks of H3K4me1, H3K4me3 and H3K27ac in activate CD4+ T cell from public data, gene location, gene expression difference between patients with SLE and HCs. Red means upregulation in SLE sample, blue means downregulation in SLE sample. The colour reflects the log base 10 of interaction’s p value. DEG, differentially expressed gene.
The 3D chromatin changes in two SLE-associated SNPs, rs2732549 and rs2245214, are also shown in online supplemental figures S8–9. The rs2732549 SNP is located in a condensed TAD domain with two gene loci, APIP and SLC1A2, which are DEGs in SLE CD4+ T cell (online supplemental figure S8A, B). We observed many interactions in the TAD and strong enhancer markers (H3K4me1 and H3K27ac) near the two gene loci (online supplemental figure S8C). According to the Enhancer-Atlas2.0 database, the region was annotated as a super enhancer in CD4+ T/CD8+ T cells.37 Another SLE-associated SNP, rs2245214, is located at loop anchors, including short-distance interactions, long-distance interactions and longer-distance interactions (online supplemental figure S9A). The longer distance interactions were increased in most of the patients with SLE compared with the HCs (online supplemental figure S9B, C). As a result, we found that the ATG5 locus, a gene involved in autophagic vesicle formation,38 overlapped with SNP rs2245214, which may be related to its overexpression in patients with SLE.
Discussion
In recent decades, increasing evidence has illustrated the importance of the 3D genomic structure in autoimmune diseases. Human islet 3D genome maps have been developed to identify the target genes of type 2 diabetes-relevant regulatory elements.39 Burren et al identified common candidate genes for five immune-mediated diseases in activated CD4+ T cells, highlighting an interaction-mediated regulation of IL2RA expression.40 In this study, we generated comprehensive and high-resolution 3D genome maps in CD4+ T cells of seven patients with SLE and five HCs, and revealed the difference in 3D genome maps between patients with SLE and HCs, especially in TAD appearance and loop strength profiles. Interestingly, our Hi-C analysis provided the first evidence that both TAD appearance and loop strength profiles in CD4+ T cells were correlated with the disease activities of patients with SLE. Furthermore, we identified 391 SLE-associated loops by Hi-C and RNA-seq data, which will help understand the epigenetic mechanisms underlying the aberrant activation and functions of CD4+ T cells in patients with SLE.
Our analysis indicated the increased loop interactions and gene expression at several SLE-associated loops and gene loci, such as DDX60L, CXCL13, SLC8A-AS1, DNJB4 and CASP5 locus. DDX60L belongs to the IFN pathway genes, which are considered as important factors in the development of SLE. Thus, in this study we investigated the potential epigenetic regulatory mechanism in DDX60L loci and found the binding motif of SPI1 within the increased interaction loop around DDX60L loci. Then, we demonstrated that SPI1 regulated histone modifications and interaction loop in DDX60L promoter to induce DDX60L overexpression in SLE CD4+ T cells. Previous study showed that SPI1, as an important transcription factor in the Ets family, was significantly hypomethylated at promoter regions, leading to upregulated gene expression in SLE CD4+ T cells.41 Recently, PU.1, a SPI1 coding protein, was indicated to regulate the local chromatin state, enhancer-promoter contacts and downstream gene expression and genetic variants associated with differential PU.1 binding were associated with neutrophil transcriptional control and immune disease susceptibility.42 Therefore, we speculate that SPI1 may play an important role in 3D chromatin alteration and gene expression regulation in SLE CD4+ T cells.
Much evidence has confirmed that genetic factors and environmental factors are involved in the pathogenesis of SLE. Previous genome-wide association studies identified many SLE-associated SNPs, most of which located in the non-coding regions of the genome.43 44 The functions of these SNPs remain unclear. In this study, we found that some SLE-associated SNPs were located in chromatin looping in patients with SLE, where enhancers were identified according to the enrichments of H3K27ac and H3K4me1. Moreover, we observed long-range controls of disease-related gene expression by looping interactions of 0.5~1 Mb and even longer in patients with SLE. Therefore, the regulatory elements harbouring the SLE-associated SNPs have more long-range or more frequent looping interactions, which lead to spatial proximity between specific genomic regions and cis-regulatory regions for the regulation of genes expression. These findings suggested that SLE-associated SNPs may be related to 3D chromatin alterations in CD4+ T cells of patients with SLE, leading to gene expression changes and aberrant immune responses.
Together, integrating the data from Hi-C and gene expression and SLEDAI scores in patients with SLE, we established the 3D genomic interaction landscape of SLE and identified potential SLE-associated loops, which will help elucidate the genetic and epigenetic mechanisms underlying SLE. In addition, we found that the alterations in chromatin interactions were associated with the disease activity of SLE, which suggests an important role of chromatin interactions in the pathogenesis of SLE and may provide potential markers for the diagnosis of SLE. However, there are some limitations in this study. Blood samples from some of the patients with SLE failed to isolate sufficient numbers of CD4+ T cells for both Hi-C and RNA-seq simultaneously. In this study, these sorted CD4+ T cells were preferentially used for Hi-C, and the rest were used for RNA-seq, which resulted in only three SLE and three HCs being included in RNA-seq. Next, we will validate the chromatin interaction alterations and gene expression regulation in a larger SLE cohort and uncover the changes in histone modification patterns and CTCF binding around the SLE-associated loops in CD4+ T cells of patients with SLE with different SLEDAI scores. Importantly, we will investigate the roles of SLE-associated loops and potential regulated genes in the pathogenesis of SLE.
Acknowledgments
The authors thank Dr Xieling He for providing the 293T cell line.
Footnotes
Handling editor: Josef S Smolen
MZ and DF contributed equally.
Contributors: MZ and QL designed and supervised the experiments. MZ and LH collected samples and isolated cells. MZ and DF performed Hi-C, RNA-seq, 3C and RT-qPCR and analysed data. LH performed cell transfection, CRISPR-Cas9 gene editing and ChIP-qPCR detection. LL performed 3C and data analysis. QK performed luciferase reporter assay. QL, JW, ZH, HL and LO helped collect samples. MZ, DF and QL prepared the manuscript. MZ and QL are responsible for the overall content as the guarantors.
Funding: This work was supported by the National Natural Science Foundation of China (No. 82030097 and No. 32141004), CAMS Innovation Fund for Medical Sciences (CIFMS) (2019-I2M-5-033), the Key project for international and regional cooperation in science and technology innovation of Hunan province (2019WK2081) and the Project for leading talents in science and technology in Hunan province (2019RS3003).
Competing interests: None declared.
Patient and public involvement: Patients and/or the public were not involved in the design, or conduct, or reporting, or dissemination plans of this research.
Provenance and peer review: Not commissioned; externally peer reviewed.
Supplemental material: This content has been supplied by the author(s). It has not been vetted by BMJ Publishing Group Limited (BMJ) and may not have been peer-reviewed. Any opinions or recommendations discussed are solely those of the author(s) and are not endorsed by BMJ. BMJ disclaims all liability and responsibility arising from any reliance placed on the content. Where the content includes any translated material, BMJ does not warrant the accuracy and reliability of the translations (including but not limited to local regulations, clinical guidelines, terminology, drug names and drug dosages), and is not responsible for any error and/or omissions arising from translation and adaptation or otherwise.
Data availability statement
Data are available on reasonable request.
Ethics statements
Patient consent for publication
Not applicable.
Ethics approval
This study was approved by the ethics committees and institutional review board of the Second Xiangya Hospital of Central South University (No. 20193044). Participants gave informed consent to participate in the study before taking part.
References
- 1. Dekker J, Rippe K, Dekker M, et al. Capturing chromosome conformation. Science 2002;295:1306–11. 10.1126/science.1067799 [DOI] [PubMed] [Google Scholar]
- 2. Lieberman-Aiden E, van Berkum NL, Williams L, et al. Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009;326:289–93. 10.1126/science.1181369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Le TBK, Imakaev MV, Mirny LA, et al. High-resolution mapping of the spatial organization of a bacterial chromosome. Science 2013;342:731–4. 10.1126/science.1242059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Jin F, Li Y, Dixon JR, et al. A high-resolution map of the three-dimensional chromatin interactome in human cells. Nature 2013;503:290–4. 10.1038/nature12644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nagano T, Lubling Y, Stevens TJ, et al. Single-cell Hi-C reveals cell-to-cell variability in chromosome structure. Nature 2013;502:59–64. 10.1038/nature12593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ramani V, Deng X, Qiu R, et al. Sci-Hi-C: a single-cell Hi-C method for mapping 3D genome organization in large number of single cells. Methods 2020;170:61–8. 10.1016/j.ymeth.2019.09.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Gibcus JH, Dekker J. The hierarchy of the 3D genome. Mol Cell 2013;49:773–82. 10.1016/j.molcel.2013.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dixon JR, Selvaraj S, Yue F, et al. Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 2012;485:376–80. 10.1038/nature11082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Beagan JA, Phillips-Cremins JE. On the existence and functionality of topologically associating domains. Nat Genet 2020;52:8–16. 10.1038/s41588-019-0561-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Nora EP, Goloborodko A, Valton A-L, et al. Targeted degradation of CTCF decouples local insulation of chromosome domains from genomic compartmentalization. Cell 2017;169:930–44. 10.1016/j.cell.2017.05.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Krietenstein N, Abraham S, Venev SV, et al. Ultrastructural details of mammalian chromosome architecture. Mol Cell 2020;78:554–65. 10.1016/j.molcel.2020.03.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chen Y, Zhang Y, Wang Y, et al. Mapping 3D genome organization relative to nuclear compartments using TSA-Seq as a cytological ruler. J Cell Biol 2018;217:4025–48. 10.1083/jcb.201807108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Krumm A, Duan Z. Understanding the 3D genome: emerging impacts on human disease. Semin Cell Dev Biol 2019;90:62–77. 10.1016/j.semcdb.2018.07.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hakim O, Resch W, Yamane A, et al. DNA damage defines sites of recurrent chromosomal translocations in B lymphocytes. Nature 2012;484:69–74. 10.1038/nature10909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Roix JJ, McQueen PG, Munson PJ, et al. Spatial proximity of translocation-prone gene loci in human lymphomas. Nat Genet 2003;34:287–91. 10.1038/ng1177 [DOI] [PubMed] [Google Scholar]
- 16. Flavahan WA, Drier Y, Liau BB, et al. Insulator dysfunction and oncogene activation in IDH mutant gliomas. Nature 2016;529:110–4. 10.1038/nature16490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kikuchi M, Hara N, Hasegawa M, et al. Enhancer variants associated with Alzheimer's disease affect gene expression via chromatin looping. BMC Med Genomics 2019;12:128. 10.1186/s12920-019-0574-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Armand EJ, Li J, Xie F, et al. Single-cell sequencing of brain cell transcriptomes and Epigenomes. Neuron 2021;109:11–26. 10.1016/j.neuron.2020.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Durcan L, O'Dwyer T, Petri M. Management strategies and future directions for systemic lupus erythematosus in adults. Lancet 2019;393:2332–43. 10.1016/S0140-6736(19)30237-5 [DOI] [PubMed] [Google Scholar]
- 20. Bialas AR, Presumey J, Das A, et al. Microglia-dependent synapse loss in type I interferon-mediated lupus. Nature 2017;546:539–43. 10.1038/nature22821 [DOI] [PubMed] [Google Scholar]
- 21. Kraig E, Linehan LA, Liang H, et al. A randomized control trial to establish the feasibility and safety of rapamycin treatment in an older human cohort: immunological, physical performance, and cognitive effects. Exp Gerontol 2018;105:53–69. 10.1016/j.exger.2017.12.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Imgenberg-Kreuz J, Carlsson Almlöf J, Leonard D, et al. DNA methylation mapping identifies gene regulatory effects in patients with systemic lupus erythematosus. Ann Rheum Dis 2018;77:736–43. 10.1136/annrheumdis-2017-212379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hedrich CM. Epigenetics in SLE. Curr Rheumatol Rep 2017;19:58. 10.1007/s11926-017-0685-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ding S, Zhang Q, Luo S, et al. BCL-6 suppresses miR-142-3p/5p expression in SLE CD4+ T cells by modulating histone methylation and acetylation of the miR-142 promoter. Cell Mol Immunol 2020;17:474–82. 10.1038/s41423-019-0268-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Su C, Johnson ME, Torres A, et al. Mapping effector genes at lupus GWAS loci using promoter Capture-C in follicular helper T cells. Nat Commun 2020;11:3294. 10.1038/s41467-020-17089-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hou G, Harley ITW, Lu X, et al. SLE non-coding genetic risk variant determines the epigenetic dysfunction of an immune cell specific enhancer that controls disease-critical microRNA expression. Nat Commun 2021;12:135. 10.1038/s41467-020-20460-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Petri M, Fu W, Ranger A, et al. Association between changes in gene signatures expression and disease activity among patients with systemic lupus erythematosus. BMC Med Genomics 2019;12:4. 10.1186/s12920-018-0468-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Grünvogel O, Esser-Nobis K, Reustle A, et al. DDX60L is an interferon-stimulated gene product restricting hepatitis C virus replication in cell culture. J Virol 2015;89:10548–68. 10.1128/JVI.01297-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. ENCODE Project Consortium . An integrated encyclopedia of DNA elements in the human genome. Nature 2012;489:57–74. 10.1038/nature11247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Ungerbäck J, Hosokawa H, Wang X, et al. Pioneering, chromatin remodeling, and epigenetic constraint in early T-cell gene regulation by SPI1 (PU.1). Genome Res 2018;28:1508–19. 10.1101/gr.231423.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Oki S, Ohta T, Shioi G, et al. ChIP-Atlas: a data-mining suite powered by full integration of public ChIP-Seq data. EMBO Rep 2018;19. 10.15252/embr.201846255. [Epub ahead of print: 09 11 2018]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Gerlach K, Hwang Y, Nikolaev A, et al. Th9 cells that express the transcription factor PU.1 drive T cell-mediated colitis via IL-9 receptor signaling in intestinal epithelial cells. Nat Immunol 2014;15:676–86. 10.1038/ni.2920 [DOI] [PubMed] [Google Scholar]
- 33. Leng R-X, Pan H-F, Ye D-Q, et al. Potential roles of IL-9 in the pathogenesis of systemic lupus erythematosus. Am J Clin Exp Immunol 2012;1:28–32. [PMC free article] [PubMed] [Google Scholar]
- 34. Tsuchiya H, Ota M, Sumitomo S, et al. Parsing multiomics landscape of activated synovial fibroblasts highlights drug targets linked to genetic risk of rheumatoid arthritis. Ann Rheum Dis 2021;80:440–50. 10.1136/annrheumdis-2020-218189 [DOI] [PubMed] [Google Scholar]
- 35. Jablonski KP, Carron L, Mozziconacci J, et al. Contribution of 3D genome topological domains to genetic risk of cancers: a genome-wide computational study. Hum Genomics 2022;16:2. 10.1186/s40246-022-00375-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Zhuang X, Zhang H, Li X, et al. Differential effects on lung and bone metastasis of breast cancer by Wnt signalling inhibitor DKK1. Nat Cell Biol 2017;19:1274–85. 10.1038/ncb3613 [DOI] [PubMed] [Google Scholar]
- 37. Gao T, Qian J. EnhancerAtlas 2.0: an updated resource with enhancer annotation in 586 tissue/cell types across nine species. Nucleic Acids Res 2020;48:D58–64. 10.1093/nar/gkz980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nishida Y, Arakawa S, Fujitani K, et al. Discovery of Atg5/Atg7-independent alternative macroautophagy. Nature 2009;461:654–8. 10.1038/nature08455 [DOI] [PubMed] [Google Scholar]
- 39. Miguel-Escalada I, Bonàs-Guarch S, Cebola I, et al. Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes. Nat Genet 2019;51:1137–48. 10.1038/s41588-019-0457-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Burren OS, Rubio García A, Javierre B-M, et al. Chromosome contacts in activated T cells identify autoimmune disease candidate genes. Genome Biol 2017;18:165. 10.1186/s13059-017-1285-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Xiang N, Fang X, Sun X-G, et al. Expression profile of PU.1 in CD4+T cells from patients with systemic lupus erythematosus. Clin Exp Med 2021;21:621–32. 10.1007/s10238-021-00717-9 [DOI] [PubMed] [Google Scholar]
- 42. Watt S, Vasquez L, Walter K, et al. Genetic perturbation of PU.1 binding and chromatin looping at neutrophil enhancers associates with autoimmune disease. Nat Commun 2021;12:2298. 10.1038/s41467-021-22548-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science 2012;337:1190–5. 10.1126/science.1222794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Catalina MD, Owen KA, Labonte AC, et al. The pathogenesis of systemic lupus erythematosus: harnessing big data to understand the molecular basis of lupus. J Autoimmun 2020;110:102359. 10.1016/j.jaut.2019.102359 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
ard-2022-222653supp001.pdf (2.1MB, pdf)
ard-2022-222653supp002.xlsx (38.6KB, xlsx)
ard-2022-222653supp003.xlsx (2.1MB, xlsx)
ard-2022-222653supp004.xlsx (79.3KB, xlsx)
Data Availability Statement
Data are available on reasonable request.