Abstract
Motivation
Adverse outcome pathway (AOP) is a toxicological concept proposed to provide a mechanistic representation of biological perturbation over different layers of biological organization. Although AOPs are by definition chemical-agnostic, many chemical stressors can putatively interfere with one or several AOPs and such information would be relevant for regulatory decision-making.
Results
With the recent development of AOPs networks aiming to facilitate the identification of interactions among AOPs, we developed a stressor-AOP network (sAOP). Using the ‘cytotoxitiy burst’ (CTB) approach, we mapped bioactive compounds from the ToxCast data to a list of AOPs reported in AOP-Wiki database. With this analysis, a variety of relevant connections between chemicals and AOP components can be identified suggesting multiple effects not observed in the simplified ‘one-biological perturbation to one-adverse outcome’ model. The results may assist in the prioritization of chemicals to assess risk-based evaluations in the context of human health.
Availability and implementation
sAOP is available at http://saop.cpr.ku.dk
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Adverse outcome pathway (AOP) is intended to capture existing knowledge, containing empirically based foundations for predicting apical toxicity that are biologically significant and plausible. They proceed through a series of key events (KEs), at different levels of biological organization (cell, tissues and organ), and end in one or more pathological states defined as adverse outcomes AOs (Ankley et al., 2010).
Although one key principle is that AOP represents the chemical-agnostic portion of pathways (independence of any specific chemical) involved in biological perturbation leading to toxicological outcomes, the evidence used to support each KE–KE relationship (KERs) is based on chemical-specific exposure data (Leist et al., 2017). Furthermore, it is observed that some KEs and KERs contribute to several AOPs, and in contrast to the ‘one perturbation-one AO’ model, the characterization of AOP networks has become more suitable to deal with such complex systems (Knapen et al., 2018).
Therefore, we developed an open access chemical stressor-AOP (sAOP) web application (http://saop.cpr.ku.dk) that gives an overview of chemical perturbance of AOP networks.
2 Materials and methods
To develop the sAOP server, we made use of the ToxCast program (Dix et al., 2007). This program builds large collections of in vitro assay data on a diverse set of chemicals. A filtering step using the cytotoxic-associated burst (CTB) was applied, as it was demonstrated that chemicals activate assays at concentration levels also observed for cytotoxicity or cell stress in ToxCast (Judson et al., 2016). The remaining chemicals showing activities on one of the targets in ToxCast were then mapped to AOPs, collected from AOP-wiki (version as of March 2018), a knowledgebase structure of AOP (Villeneuve et al. 2014), resulting to a stressor-AOP network between 4960 chemicals, 369 proteins, 1089 KEs and 207 AOPs. More detailed information is found in the Supplementary Information.
3 Results
Users can query the sAOP database by ‘chemicals’, ‘assay’, ‘protein’, ‘Key Event’ and ‘AOP’. Some parameters can be included in the search such as ‘AC50 score’, ‘z-score’ or ‘degree of separation’ to facilitate the visualization of the network. For example, the search for ‘phenanthrene’, a polycyclic aromatic hydrocarbon, with a degree of separation of 4, results to a network represented in Figure 1, i.e. 13 assays which show activity on three proteins (ESR1, ESR2 and NR1I2) linked to 6 MIEs, 16 KEs and 2 AOs. More examples are provided in the Supplementary Information.
Fig. 1.
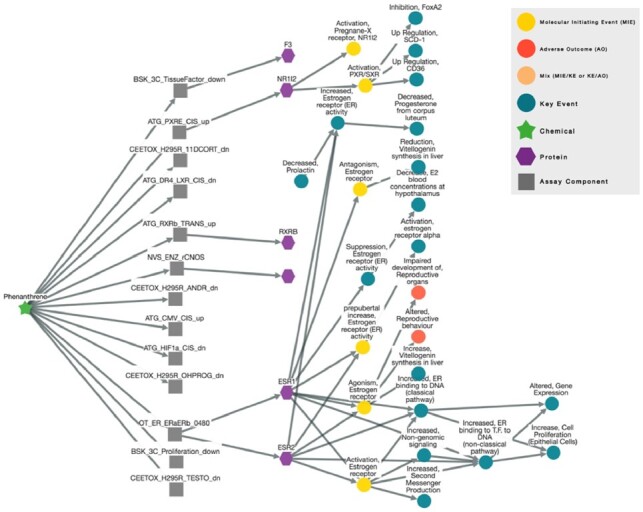
Results of the sAOP network for the chemical ‘phenanthrene’. A color scheme is defined for each element of the network. The arrows show the edges directionality
4 Conclusion
The proposed stressor-AOP network described here allows to explore the knowledge ‘space’ regarding chemicals and adverse effects from a mechanistic point of view. It can assist with the identification of chemicals involved in an AO and in the prioritization of biological assay endpoints associated to known AOPs. Finally, it suggests how the combination of various toxic compounds can, by affecting the same pathway or different modules of the same AOP, be a sufficient perturbation for the appearance of the AO (Miller et al., 2017). With the interest of regulatory agencies to consider new approach methodologies (NAMs) in risk assessment, such integration could be suitable for regulatory decision-making.
Supplementary Material
Acknowledgement
We would like to acknowledge Catherine Bjerre Collin’s help on the revision of English.
Funding
This work was supported by the European Union’s Horizon 2020 program [681002, EUtoxRisk], the Novo Nordisk Foundation under [NNF14CC0001], the University of Paris Descartes-USPC, the university of Paris Diderot and INSERM.
Conflict of Interest: none declared.
Contributor Information
Alejandro Aguayo-Orozco, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, DK-2200, Denmark.
Karine Audouze, Environmental Toxicity, Therapeutic Targets, Cellular Signaling and Biomarkers (T3S) Unit, Université de Paris, INSERM UMR-S 1124, Paris, France.
Troels Siggaard, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, DK-2200, Denmark.
Robert Barouki, Environmental Toxicity, Therapeutic Targets, Cellular Signaling and Biomarkers (T3S) Unit, Université de Paris, INSERM UMR-S 1124, Paris, France.
Søren Brunak, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, DK-2200, Denmark.
Olivier Taboureau, Novo Nordisk Foundation Center for Protein Research, Faculty of Health and Medical Sciences, University of Copenhagen, Copenhagen, DK-2200, Denmark; Université de Paris, INSERM U1133, Computational Modeling of Protein-Ligand Interactions group, CNRS UMR 8251, Unit of Functional and adaptive Biology, Paris, France.
References
- Ankley G.T. et al. (2010) Adverse outcome pathways: a conceptual framework to support ecotoxicology research and risk assessment. Environ. Toxicol. Chem., 29, 730–741. [DOI] [PubMed] [Google Scholar]
- Dix D.J. et al. (2007) The ToxCast program for prioritizing toxicity testing of environmental chemicals. Toxicol. Sci., 95, 5–12. [DOI] [PubMed] [Google Scholar]
- Judson R.S. et al. (2016) Analysis of the effects of cell stress and cytotoxicity on in vitro assay activity across a diverse chemical and assay space. Toxicol. Sci., 152, 323–339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knapen D. et al. (2018) Adverse outcome pathway networks I: development and applications. Environ. Toxicol. Chem., 37, 1723–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leist M. et al. (2017) Adverse outcome pathways: opportunities, limitations and open questions. Arch. Toxicol., 91, 3477–3505. [DOI] [PubMed] [Google Scholar]
- Miller M.F. et al. (2017) Low-dose mixture hypothesis of carcinogenesis workshop: scientific underpinning and research recommendations. Environ. Health Perspect., 125, 163–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve D.L. et al. (2014) Adverse outcome pathway (AOP) development I: strategies and principles. Toxicol. Sci., 142, 312–320. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


