Abstract
The proposal to use brain connectivity as a biomarker for dementia phenotyping can be potentiated by conducting large-scale multicentric studies using high-density electroencephalography (hd- EEG). Nevertheless, several barriers preclude the development of a systematic “ConnEEGtome” in dementia research. Here we review critical sources of variability in EEG connectivity studies, and provide general guidelines for multicentric protocol harmonization. We describe how results can be impacted by the choice for data acquisition, and signal processing workflows. The implementation of a particular processing pipeline is conditional upon assumptions made by researchers about the nature of EEG. Due to these assumptions, EEG connectivity metrics are typically applicable to restricted scenarios, e.g., to a particular neurocognitive disorder. “Ground truths” for the choice of processing workflow and connectivity analysis are impractical. Consequently, efforts should be directed to harmonizing experimental procedures, data acquisition, and the first steps of the preprocessing pipeline. Conducting multiple analyses of the same data and a proper integration of the results need to be considered in additional processing steps. Furthermore, instead of using a single connectivity measure, using a composite metric combining different connectivity measures brings a powerful strategy to scale up the replicability of multicentric EEG connectivity studies. These composite metrics can boost the predictive strength of diagnostic tools for dementia. Moreover, the implementation of multi-feature machine learning classification systems that include EEG-based connectivity analyses may help to exploit the potential of multicentric studies combining clinical-cognitive, molecular, genetics, and neuroimaging data towards a multi-dimensional characterization of the dementia.
Keywords: EEG, Connectivity, Dementia, Harmonization, Machine learning, Multicentric studies
1. Introduction
The understanding of neurodegeneration as a large-scale network disintegration linked to neuroplasticity has attracted neuroscientists’ and clinicians’ attention (Dorszewska et al., 2020; Schaefers and Teuchert-Noodt, 2016). In recent years, efforts have been directed to develop affordable, scalable and broadly available biomarkers of brain connectivity to help tackle global challenges of dementia (e.g., Mantzavinos and Alexiou, 2017; McKhann et al., 2011; Swift et al., 2021).
Trying to provide the most precise definition of brain connectivity, we revisited a seminal article written by Barry Horwitz (2003), inspired by extensive discussions that had taken place in a previous workshop on functional connectivity. The title of the article, “The elusive concept of brain connectivity”, reflects concerns and opinions that still hold true today. When restricted to EEG, connectivity refers to patterns of statistical dependencies between signals in both sensor- and source-spaces. This type of connectivity, which is termed functional connectivity, is regardless of physical connections between neural assemblies. Also, at the source space, direction of the information flow can be estimated, leading to the concept of effective connectivity (Bowyer, 2016; Sporns, 2014; Friston, 2011).
Brain functional connectivity has provided relevant information for the classification of dementia subtypes and predicting disease severity across dementias (Chen et al., 2019; Moguilner et al., 2021). Examples are Alzheimer’s disease (AD) (Dennis and Thompson, 2014), frontotemporal dementia (Jalilianhasanpour et al., 2019; Sedeño et al., 2017), Huntington’s disease (Johnson and Gregory, 2019; Pini et al., 2020), Parkinson’s disease (Kim et al., 2017; Niethammer et al., 2018), Multiple Sclerosis (Labbe et al., 2020; Pagani et al., 2020), Ataxia (García et al., 2017), and Lewy body dementia (Schumacher et al., 2019), among others neurogenerative disorders. Although this approach to dementia has been mainly developed for magnetic resonance imaging (MRI), the assessment of brain connectivity using high density electroencephalography (hd-EEG) has emerged in the brain network agenda (Babiloni et al., 2021; Horvath et al., 2018; Law et al., 2020; Ibanez and Parra, 2014).
The hd-EEG is a relatively portable, broadly available, and low-cost technology that allows large- scale multisite studies of brain dynamics with high temporal resolution. Over the last two decades, these studies have benefited from the increased sophistication of hd-EEG processing pipelines, allowing researchers to gain more insight into the dynamics and connectivity of the brain (Baş;ar et al., 2013; Larson-Prior et al., 2013; Ibanez et al., 2012). This is illustrated by recent EEG studies incorporating graph theory, nonlinear dynamics, decoding, and whole brain modeling (Ghaderi et al., 2020; Iakovidou, 2017; García et al., 2020; Hesse et al., 2019; Dottori et al., 2017; Josefsson et al., 2019), which bring novel opportunities for the study of dementia. Moreover, the addition of the temporal dimension to these EEG analyses has recently allowed to characterize the switching behavior of EEG microstates, offering a remarkable opportunity for assessing abnormal connectivity fluctuations in neurodegenerative diseases (Khanna et al., 2015; Pal et al., 2021). This can be boosted by connectivity analyses in the EEG source space that incorporate relevant anatomical information provided by MRI.
Further advances in dementia research can result from the multimodal assessment of EEG connectivity, i.e., the “ConEEGtome”. This neologism refers to the multi-feature combination of different connectivity metrics (e.g., tasks vs. rest, linear vs. nonlinear, source vs. electrode spaces, direct connectivity vs. network organization-derived metrics), fueled by machine learning algorithms with feature selection processes. These multimodal metrics may allow for a new multicentric approach to dementia, and may potentiate research in low- and middle-income countries where the access to current mainstream biomarkers (e. g., positron emission tomography) is more restrictive (Ibanez et al., 2021a, 2021b, 2021c; Parra et al., 2018, 2021).
Notwithstanding the evidence endorsing the robustness of the hd-EEG for large-scale multicenter studies, several methodological and analytical caveats preclude the systematic assessment of the dementia ConnEEGtome. EEG datasets present multiple sources of variability (Farzan et al., 2017; Jovicich et al., 2019). They include different experimental and data collection procedures, data monitoring and quality checks, preprocessing pipelines, feature extraction, and statistical approaches. These sources of heterogeneity prevent the development of unified protocols for replication (e.g., Pavlov et al., 2021). Currently, procedures for data harmonization (i.e., signal normalization and standardization), and the subsequent control (e.g., confusion matrices) of multicentric heterogeneity are very scant. The barriers for a ConnEEGtome of dementia have limited the development of standardized multi-feature analyses, in which EEG-based connectivity is integrated with neuroimaging, cognitive, and plasma biomarkers to deliver comprehensive dementia phenotypes.
The present work aims to bring together the first set of guidelines to accelerate the development of a dementia ConnEEGtome. We provide recommendations for a multicentric approach to dementia, which may result in standard procedures for (a) recording and preprocessing, (b) data analysis harmonization, and (c) the development of a multi-feature framework for out-of-sample validation and generalization. In the following sections, we will describe current challenges for the development of diagnosis tools, and intervention strategies based on the analysis of large volumes of EEG data (section 2). Then, we provide considerations for the harmonization of experimental tasks, protocols for data acquisition, and different preprocessing steps (e.g., denoising, artifact removal, and data normalization) (section 3). Subsequently, we analyze tools that could contribute towards the realization of a multifeature framework for EEG connectivity, with special emphasis on those needed for retesting, and the development of integrative measures of connectivity (section 4). We discuss how the use of composite measures of connectivity can promote the development of diagnosis tools based on multi-feature classification (section 5). Lastly, we address the need of metrics to verify the success of EEG data harmonization (section 6).
2. Data staging and harmonization
In recent years, particular emphasis has been placed on the development of EEG-based collaborative multicentric studies assessing predictors of brain disorders and neurodegeneration (Lam et al., 2016; Trivedi et al., 2016). These efforts are set to increase the efficacy of diagnostic tools and intervention strategies based on the analysis of large volumes of data. The current and potential impact of functional connectivity studies based on EEG cannot be ignored, and a great part of this success is owed to the development of affordable and portable EEG systems. Nevertheless, developing EEG collaborative multicentric studies is not an easy road and involves major technical difficulties and organizational needs.
2.1. Resting-state versus task-running EEG
EEG-based functional connectivity can be addressed from either ongoing (resting-state) or task- related activity depending on the question at hand. In a multicentric approach to task-related EEG functional connectivity, the major drawbacks arise from the task design, which needs careful validation. Although task-related connectivity approaches can directly address specific network markers of neurocognitive deficits, they may lack sensitivity to global network trait compromises. Furthermore, multicentric studies involving the acquisition of task-related EEG face several technical challenges, which include compensations for different stimulation timing and EEG synchronization procedures.
A more global view on network dysfunctions is provided by functional connectivity analyses of ongoing (resting-state) EEG. In this case, the thoughts and behavior of the participants are difficult to control, so their cognitive states may vary during the experimental session (van Diessen et al., 2015). Nevertheless, this approach makes multicenter validations easier, since the outcomes of the studies do not largely depend on specific tasks. Furthermore, connectivity analyses of ongoing EEG allow to link intrinsic (spontaneous) neural activity to off-line task performance, different features of diseases, cognitive decline, and other impairments (Bassett and Bullmore, 2009; Supekar et al., 2008). Based on the characterization of ongoing EEG oscillations of individuals with AD, mild cognitive impairment (MCI) and dementia, the Electrophysiology Professional Interest Area (EPIA) and Global Brain Consortium recently endorsed recommendations on EEG measures for clinical trials in AD (Babiloni et al., 2021, see also Babiloni et al., 2020). The EEG power spectrum of these patients shows altered peak frequency, power, and interrelatedness of low and middle EEG frequency-bands (from delta to alpha). Consequently, measures related to power spectrum, including directed transfer function, phase lag index and linear lagged connectivity were suggested for stratification of AD patients, as well as for monitoring the disease progression and the success of interventions (Babiloni et al., 2021).
The above example illustrates that the choice of preprocessing strategy of EEG data for connectivity is conditional on the type of brain activity that is intended to be analyzed, either ongoing or task-related (Fig. 1A). For methodological reasons, the analysis of ongoing EEG connectivity is mainly restricted to the frequency domain and focuses on specific narrow frequency-bands of the EEG. The acquisition of task-related EEG gives the opportunity to expand the connectivity analyses to a variety of time-domain approaches and to establish direct links between brain network dynamics and behavior.
Fig. 1. Flow chart of steps and modules comprising an ideal workflow for multicenter EEG studies of connectivity.
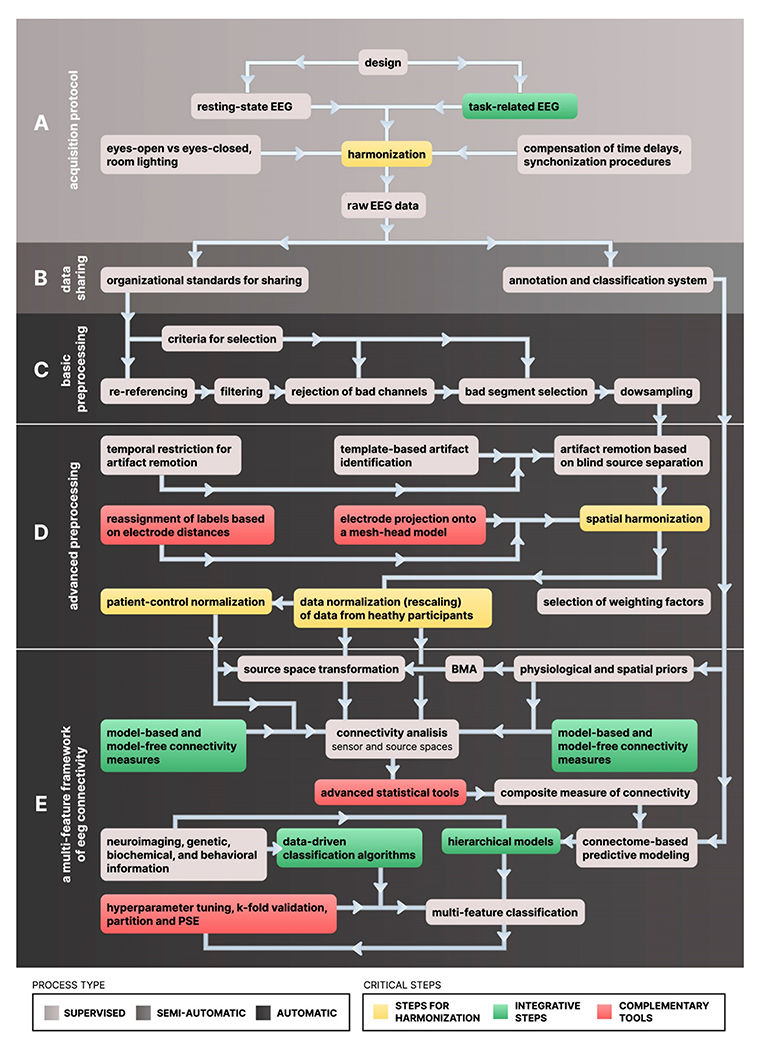
Steps in the workflow are grouped as they belong to the A) Acquisition protocol, B) Data sharing procedures, C) basic preprocessing, D) advanced preprocessing, and E) the final stages of the multi-feature framework for EEG connectivity. Process types are classified as completely supervised, semi- automatic and automatic (grey color code). Cells colors indicate critical steps in the workflow, i.e., where harmonization of datasets is performed (orange cells) or where computation of parameters depend on the integration of several sources of information (green cells). Cells in red denotate additional steps that are favored by processing tools already available. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2.2. Sources of variability in multicentric studies
Multicentric EEG studies benefit greatly when data integration is planned ahead. For many reasons this is often not possible, and variability in task design and acquisition protocols can potentially affect the outcome of the study. The primary sources of variability in multicentric EEG studies span five categories: i) the aim of the primary study for which the data was obtained, ii) the experimental protocol, iii) the acquisition system and acquisition protocol, iv) the procedures to control for the data quality, and v) preprocessing workflows. Within each category, several factors affecting the EEG-data variability need to be considered. Further sources of variability arise from the use of particular feature extraction algorithms, the choice of statistical methods, the format for data storage, and the method of knowledge translation (Farzan et al., 2017).
Factors needed to overcome hardware-related sources of variability in EEG multicentric studies include randomizing the task sequence when EEG is integrated with behavioral and neuroimage datasets, such as MRI images (Farzan et al., 2017). They also include the differences in time delay and jitter among stimulation setups, if task-related EEG is acquired. The use of a dimly lit, sound- attenuated, and electromagnetically-shielded EEG chamber is recommended. Procedures aimed to attenuate the impact of variability linked to the environment (e.g., sources of electromagnetic interference) and movements (i.e., chin rest) need to be considered and implemented. Because of this, it is important to report the amount and nature of lightning, the acoustic background noise, the electromagnetic noise of the EEG chamber and other setting conditions. For consistency, site- specific information about these conditions should be archived.
Noteworthy, researchers are usually encouraged to set the EEG acquisition parameters close to the highest possible boundaries. This includes using the larger possible number of recording electrodes and the utmost possible sampling rate for analog-digital conversion. Nevertheless, the integration of multicentric EEG data will be conditioned by the lowest acquisition capacity among all of the participant institutions. Future efforts should focus on how the proposed ConnEEGtome can inform either ad-hoc or post-hoc theory-driven data harmonization strategies when multicentric collaborations are set up.
3. Preprocessing harmonization
Nowadays, the variety and flexibility of pipelines for the analysis of EEG allow research teams to have their own processing strategy. The choice of algorithms used in different processing steps (artifact removal, filtering, and time-domain transformations) affects the estimation of the power spectral density of different EEG frequency-bands (Alam et al., 2020), with substantial effects on scientific conclusions. The reproducibility of results that are obtained using a single analysis pipeline is hard to estimate, and researchers generally overestimate the likelihood of significant results across hypotheses (Botvinik-Nezer et al., 2020). Consequently, in addition to having standard collection procedures, standardized data preprocessing pipelines are of utmost priority for minimizing raw data heterogeneity across sites, increasing the statistical power and sensitivity of multicentric studies. In this section, we discuss some critical aspects for the standardization of processing workflows in EEG-based studies of connectivity. Additionally, we highlight the need to conduct multiple analyses of the same data, and to have composite metrics that allow the proper integration of the results. These recommendations are armed with an ideal processing workflow (Fig. 1) as the point of departure for developing standard procedures for EEG- based studies of connectivity.
3.1. Control procedures
The adoption of semi-automated quality control procedures (Fig. 1B) ensures that planned acquisition parameters and criteria for data annotation are meet (Farzan et al., 2017). Annotation should not been restricted to technical aspects of the EEG acquisition but should also include tracking experiment- specific events using a standard tag system. This is the case of the Hierarchical Event Descriptors (HED), a tags system used in subsequent processing stages to isolate the effects of events that share a particular feature, e.g., events associated with the same sensory or cognitive phenomena (Bigdely-Shamlo et al., 2016).
Security and organization of the data benefits from organizational standards for EEG data, which also protect patients’ personal data. This is the case of EEG-BIDS, an extension to the brain imaging data structure for EEG, which addresses the heterogeneity of data organization by following the FAIR principles of findability, accessibility, and interoperability (Pernet et al., 2019). The use of EEG-BIDS is closely tied to data repositories that build on BIDS, such as OpenNeuro (https://openneuro.org).
Furthermore, the design of standard pipelines profits from the high number of freely available signal processing software for EEG data. The list of powerful tools is lengthy and includes EEGLab (Delorme and Makeig, 2004), Cartool (Cartool Software FBMLab), Brainstorm (Tadel et al., 2011), MNE-Python (Gramfort et al., 2013) and BioSig, among others. Within each tool, several processing strategies can be designed. Consequently, the selection of tools, the sequence of processing steps, and the analysis parameters need to be planned and carefully reported.
The output of critical processing steps needs to be frequently archived for additional analysis beyond the scope of the original study. Developers of signal processing software for EEG data should also be made aware of data harmonization guidelines as they can greatly contribute to such strategies.
3.2. Denoising
Since multicentric studies typically involve large sample sizes, preprocessing worflow needs to run automatically, and with minimal supervision. An automatic sequence of processing steps provides an efficient workflow for denoising, especially when a large amount of data has been collected (Fig. 1C). Grounded on pre-selected settings, automating EEG pre-processing tools allow for removal of artifact-contaminated EEG segments and channels without subjective judgments precluding standardized signal quality metrics. This is the case of the standardized early-stage EEG processing pipeline (PREP), an open-source tool for automatic removal of line noise, electrode interpolation, and average referencing (Bigdely-Shamlo et al., 2015).
Noteworthy, the preprocessing strategy needs to consider the particular features of the experimental design, the cohort of participants and the expected outcomes of the study. In this regard, pipelines have been developed to address the low signal-to-noise (SNR) ratios of EEG recordings from young children, patients with neurodevelopmental disorders, or individuals with neurodegenerative diseases, where data is relatively more affected by artifacts (Gabard-Durnam et al., 2018). Flexibility of processing pipelines allows users to manage batches of EEG files collected across multiple acquisition setups in multicentric studies (Levin et al., 2018). Furthermore, it permits to implement processing strategies that maximize signal isolation and minimizes data loss using EEG signal quality measures (van Noordt et al., 2020). The emergence of new artificial intelligent solutions (Mashhadi et al., 2020) can significantly contribute not only to remove undesired activity from the EEG but to better characterize it and hopefully contribute in future harmonization strategies.
3.3. Artifact removal
Automatic artifact removal using blind source separation techniques, either alone (Bell and Sejnowski, 1995) or in combination with time-frequency denoising algorithms (e.g., Zima et al., 2012), need to balance the relative contribution of different sources of artifacts (ocular, myographic, cardiac, external) to the EEG signal. Automatic removal of artifacts is facilitated by the identification of eye motions and blinking based on templates (Fig. 1D). For that purpose, processing tools containing a database of manually identified eye-related ICA scalp maps have been developed (Bigdely-Shamlo et al., 2013). This tool (Eyecatch) has been used for the integration of different EEG datasets (Bigdely-Shamlo et al., 2020a) in combination with methods that flag latencies corresponding to different stages (phases) of the blinks (Kleifges et al., 2017) and restrict the corrections to a limited time window.
3.4. Spatial harmonization
To provide general guidelines regarding EEG studies for connectivity, we will restrict the concept of spatial harmonization to the assignment of common labels to channels from recordings that were acquired using different electrode layouts (Fig. 1D). Considerations about spatial transformation (spatial filters) will be given in section 4.1.
The integration of EEG data for connectivity analyses deeply depends on the number of electrodes and the electrode placement system (electrode layout) used for data acquisition. Globally adopted guidelines are not currently available. Recommendations endorsed by international panels and consortiums are critical, since lack of procedures for coregister different electrode layouts may result in distorted EEG spatial representations and inconsistent interpretation of connectivity matrices. Nevertheless, important progress has been achieved. Some multicentric projects have implemented the closest equivalent electrodes between layouts as replacement procedures for integrating different electrode placement systems and electrode configurations (Farzan et al., 2017). A similar assignment procedure, based on electrode distances, has been implemented to reduce a 256-channels electrode layout to standard 10–20 locations (Bigdely-Shamlo et al., 2020b). Alternatively, instead of assigning electrodes, integrating different electrode layouts can be achieved by generating virtual electrodes computed from topographic interpolation transforms either using i) triangulation plus linear interpolation or ii) interpolation by spherical splines over a mesh-head model. This idea has been successfully implemented in studies analyzing the effect of variance across acquisition systems compared to between-subject, and between-session variances (Melnik et al., 2017). The method consists of projecting the electrode positions onto a mesh-head model of 1082 points and interpolating the activity of EEG.
3.5. Data normalization
Dataset variability needs to be compensated when samples across centers are combined. In this sense, efforts have been made to enable the joint connectivity analysis of raw functional MRI (fMRI) data from different scanners (e.g., Legaz et al., 2021; Bachli et al., 2020; Donnelly-Kehoe et al., 2019; Moguilner et al., 2018, 2021; Salamone et al., 2021; Sedeño et al., 2017). More research is needed for EEG multicentric studies to achieve the same standardization. Harmonization of raw EEG data is worthy, since it eliminates technical and methodological sources of variability that impact the interpretation of EEG meta-analysis. Based on previous studies, we propose that between-dataset variability can be reduced by i) multiple normalizations to improve the comparability across recordings, ii) patient-control normalization, and iii) machine learning and confusion matrix approaches (Fig. 1D).
Data normalization (rescaling) can be carried out with methods for data alignment (Bigdely-Shamlo et al., 2018). These methods are linear transformations of the EEG that reduce the between- subject variability by computing statistical properties of the data which in turn are used as a weighting factor for correcting (rescaling) voltage amplitudes. For illustrative purposes, methods for data alignment can be classified as within-electrode and across-electrode transformations. On a much smaller scale, some of these weighting procedures are used to analyze auditory evoked potentials in clinical settings with the objective of reducing the effect of artifacts on the estimated response (e.g., John et al., 2001; Prado-Gutierrez et al., 2019). Across-electrode weighting factors include the mean, Huber mean (Huber, 1964) and the Euclidean (L2) norm (Li et al., 2009). These three methods capture the central tendency of the EEG amplitude. Unlike the mean and Euclidean norm, the Huber mean seems to be more robust to outliers (Bigdely-Shamlo et al., 2020b).
The weighting methods described above are usually implemented in a single dataset, in which different subsets (i.e., different studies) are grouped. This may not be convenient in multicenter studies involving both patients and healthy individuals. In this case, the weighting factor (constant for rescaling, e.g., the Huber mean) can be computed by pooling together the data belonging to healthy participants. Subsequently, both groups can be rescaled using the same weighting factor (Fig. 1D). A similar approach has been implemented in multimodal and multicentric MRI studies on frontotemporal neurodegeneration to avoid MRI-setup bias (e.g., Donnelly-Kehoe et al., 2019). In this study, features (cortical volume and thickness) of participants (both healthy volunteers and patients) who were recruited in a given center were z-scored based on the mean and standard deviation of features of the corresponding healthy participants. Likewise, patient-control normalizations using Z-score have been used for comparing relevant behavioral and electrophysiological measures of patients with MCI and prodromal patients from familial AD carrying the mutation E280A of the presenilin-1 gene (Pietto et al., 2016). In cases where data distribution is not restricted to a narrow range, w-score standardization can be implemented (Chung et al., 2017). This method has been applied to assess neurodegenerative diseases, where correlation of parameters with clinical features has been established (La Joie et al., 2012; Jack et al., 1997; O’Brien and Dyck, 1995; Ossenkoppele et al., 2015).
4. A multi-feature computational framework
Functional and effective connectivity (for robust definitions, see Reid et al., 2019) can be assessed with several methods, which differ in mathematical assumptions about the nature of the signals. Consequently, different measures of dependency and similarity can be used (Ahmadlou et al., 2014; Yuvaraj et al., 2016). An illustrative taxonomic description of methods for quantifying functional connectivity has been provided by Bastos and Schoffelen (2016). These methods can be classified as model-based or model-free, depending on whether they make assumptions about the linear relationship between signals. While model-based metrics of connectivity can be computed from both the EEG envelopes and frequency representations of the signals, popular model-free metrics (e.g., mutual information and transfer entropy) entirely rely on time-domain analyses (Bastos and Schoffelen, 2016).
Studies comparing metrics for functional connectivity typically highlight the advantages and drawbacks of the methods, as well as their applicability to a particular scenario, e.g., to a particular sensory or neurocognitive disorder. Nevertheless, studies comparing the potential of individual connectivity metrics against a pool of alternative measures are scarce. This kind of comparison has been conducted in a recent exploratory study, in which healthy individuals were classified by age (younger versus older) using a binary machine learning classifier based on i) a single functional connectivity measure, and ii) a composite metric, obtained by combining eight measures of functional connectivity (Mohanty et al., 2020b). The result of this study suggests that retesting the data with different analyses and obtaining a composite measure that integrates a pool of connectivity measures adds significant value for EEG multicentric studies (Fig. 1F). This procedure has the potential to increase the consistency of findings and make the results obtained in different multicentric studies more comparable.
4.1. The need for retesting
A critical element behind the aforementioned statement concerns the preprocessing workflow, since a “ground truth” for the choice of methods used in different preprocessing steps is not available. This is the case of the spatial transformations of EEG, which are applied based on theoretical considerations about the physics of the brain and the surrounding tissues (Fig. 1F). These transformations change the voltage values at each electrode according to a weighted combination of voltage values at other (all) electrodes to highlight features that can be difficult to observe in the raw data (Cohen, 2015). They also include adaptive spatial filters (e.g., independent component analysis, ICA; and principal component analysis, PCA) to rescue neural activity that can remains masked when spatial transformations consider that EEG is stationary (Bufacchi et al., 2021). It is hard to know a priori which of several EEG spatial distribution reflects better a particular physiological process. Most of the time correlations with behavioral or clinical measures are needed for validation.
The basic principle of the surface Laplacian transform, as well as the differences between common reference schemes (nose, linked mastoids, average) and the surface Laplacian have been described (Kayser and Tenkem, 2015). These transformations affect the output of EEG connectivity analyses, such that spatial transformation applied to the EEG data needs to be specific to the sensory or cognitive process that is being investigated (Cohen, 2015).
The dependence of EEG connectivity from the spatial transformations implemented in the processing pipeline is show in Fig. 2. We constructed this illustrative example using our own database of auditory steady-state responses (ASSR). Our ASSR datasets comprise recordings from 36 healthy, normal hearing participants (young adults aged 22–36, median age 27) from the region of Valparaíso, Chile. The ASSR were elicited by 1-kHz tones modulated in amplitude at 40 Hz. The EEG was acquired using a 64-channels acquisition system (Biosemi Active Two), such that the EEG was acquired using the zero-reference principle. Spatial transformations of raw data were obtained using different electrode combinations for re-referencing: Cz, linked mastoids (LM), and average reference of the 64 electrodes (AVE). Furthermore, the current source density (CSD) was computed as an estimate of the surface Laplacian based on the EEG voltage values across electrodes. This transformation is considered reference-free, such that the output is independent of any previous choice of reference. The ASSR was computed by the time-domain averaging of synchronized EEG epochs and the subsequent frequency transformation of the averaged signal using the fast Fourier transform. At the sensor space, different coherence-related measures in a narrow-band around 40 Hz (38–42 Hz) were computed between all channel pairs. Furthermore, source generators of the 40-Hz ASSR were estimated using the Bayesian model averaging (BMA) approach of the EEG inverse problem (Trujillo-Barreto et al., 2004). At the source space, pair-wise functional connectivity was estimated in a 20-nodes network comprising temporal, and frontal cortical regions of both hemispheres. Nodes corresponded to the centroid of Broadman areas 20, 21, 22, 37, 38, 41, 42, 44, 45, and 47.
Fig. 2. Effect of EEG spatial transformations on EEG functional connectivity.

Spatial transformations of raw data (a single subject) were obtained using different electrode combinations for re-referencing: Cz, linked mastoids (lm) and average reference of the 64 electrodes (ave). Furthermore, the current source density (csd) was computed as an estimate of the surface Laplacian based on the EEG voltage values across electrodes. Spatial transformations are displayed at the top of the figure. All the results presented in each column were obtained using the same transformation. A) Power spectrum of the 40-ASSR in the electrode Fz are presented in upper panels. The spectral peak corresponding to the 40-Hz frequency bin is denoted by an asterisk (*). The scalp distributions of the ASSR are presented in the lower panels. B) Coherence- related measures were computed in a narrow-band around 40 Hz (38–42 Hz) between all channel pairs. Methods of coherence-related measures were magnitude-squared coherence (msc), phase of cross- spectrum (pcs), and the imaginary part of coherency (ipc). The different coherence-related measures are presented in the box on the left, while contrast between measures derived from different spatial transformations (ave vs. Cz) are presented in the right-hand box. C) Upper panels show neural generators of the 40-Hz ASSR, estimated with BMA. Lower panels display representations of the 40-Hz functional connectivity, estimated in a 20-nodes network including temporal and frontal cortical regions of both hemispheres. Functional connectivity measures were total coherence (totalCoh) and total phase synchronization (totalPhase), as computed by Loreta Key software (Pascual-Marqui et al., 1994) and described in Pascual-Marqui (2007). (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
As expected, spatial transformation of the raw data conditioned the amplitude of the 40-Hz ASSR, and the scalp distribution of the oscillatory response (Fig. 2A). Consequently, results obtained with a given method of coherence varied as a function of the 40-Hz ASS spatial transformation (Fig. 2B, left box). We contrasted the results derived from a pair of spatial transformations to highlight the effect of the processing pipeline on the results (Fig. 1B, right box).
The choice of reference for EEG has been a long-debated methodological topic, due to its impact on the interpretation of the results. Consequently, the potential biases induced by the choice of reference on EEG connectivity patterns have been analyzed at both sensor and source space (Chella et al., 2017). Results consistently show that the choice of reference heavily impacts the connectivity outcome, and that the best reference choice depends on the connectivity analysis that is conducted. In addition to the reference schemes included in the illustrative example presented in Fig. 2 (Cz, LM, AVE and CSD), comparative studies have included an infinity reference estimated via the Reference Electrode Standardization Technique (REST). When cross-frequency connectivity of eye open EEG resting-state is analyzed at the sensor space using the cross-bicoherence and the antisymmetric cross-bicoherence, REST provided superior performance than Cz, LM, and AVE in approximating the ideal neutral reference (Chella et al., 2017). CSD outperforms the rest of aforementioned reference choices, when information-theoretic measures of segregation and integration-interaction complexity are analyzed at the EEG sensor space (Trujillo et al., 2017). Furthermore, changes in the underlying neural source of resting-state EEG were more accurately estimated using CSD (Trujillo et al., 2017), which is likely a consequence of the positive impact of CSD on EEG signal quality, and the reduction of the volume-conduction effect that is obtained when this infinite reference is implemented.
The effect of reference-related bias on the estimation of EEG potentials has been also analyzed in combination with other factors, including the electrode layout, number of channels, SNR at the sensor level and head models (Hu et al., 2018a). By comparing the relative error between the EEG potentials generated by forward calculation using a single dipole in the neural source space (where the reference is the ideal infinity), and the EEG that resulted from reference transformations, authors indicates that REST can be considered as the primary re-reference choice, since REST outperform monopolar references, LM and AVE (Hu et al., 2018a). For solving the inverse solution of the EEG, a regularized version of REST (rREST) has been developed (Hu et al., 2018b). This reference provides the lowest relative error in estimating the EEG potentials, in comparison with a regularized version of AVE and other reference choices. Furthermore, results obtained when using individual and average lead fields are comparable (Hu et al., 2018a), a finding that boost the extended use of rREST. The validation of spatial transformations, as well as any other data transformation, can benefit when methods are tested in a data set stemming from a well-understood experimental manipulation (Kayser and Tenkem, 2015).
4.2. Connectivity at the EEG source space
The different spatial transformations illustrated in Fig. 2 also derived in different current densities maps inside the brain, i.e., different brain activation maps estimated using models to solve the inverse problem of the EEG (Fig 2C, upper panels). Consequently, the output of a particular functional connectivity analysis of the 40-Hz ASSR differed on the basis of the spatial transformation of the EEG at the sensor space (Fig. 2C, lower panels). The spatial transformation particularly affected long-range inter-hemispheric functional connections.
Beside the spatial transformations at the sensor space, the estimation of EEG neural generators heavily depends on the selection of parameters used for modeling the EEG inverse problem. This topic has been addressed by studies in which neural generators of resting-state activity have been estimated using different commonly-used algorithms (based on minimum-norm and beamforming estimates) (Tait et al., 2021). After testing the performance of the source reconstruction methods, based on quantitative metrics that included the explained variance at the sensor level, and resolution properties of the inverse solutions, authors conclude that that there is no “one size fits all” algorithm. Consequently, recommendations for the choice of the inverse solution method were provided, considering available information about the SNR, the spatial resolution requirements, and the properties of the resting-state activity that are statically tested (Tait et al., 2021).
The impact of the choice of EEG source reconstruction algorithm on functional/effective connectivity has been tested as a function of anatomical templates, electrical models of the head, methods of source estimations, and software implementations (Mahjoory et al., 2017). The estimation of EEG sources using weighted minimum-norm estimate (WMNE), exact low resolution brain electromagnetic tomography (eLORETA) and linearly constrained minimum-variance (LCMV) beamformer was more consistent across pipelines than the subsequent estimations of functional connectivity. Although in silico studies suggest that some methods for source-space connectivity can be preferred when a particular density electrode configuration is used (Barzegaran and Knyazeva, 2017), the disparity in the results evidences again that integration of multiple approaches is needed for multisite EEG connectivity settings.
The (lack of) consistency across EEG connectivity analyses in the EEG source-space reinforces the idea that standardized signal processing workflows are necessary for successful multicenter analyses. This is maybe more relevant when EEG is acquired in a resting condition, since the arousal state of the individual represents a major source of inconsistency in source localization and connectivity analyses (Kaufmann et al., 2006; Massimini et al., 2005; Murphy et al., 2009; Ventouras et al., 2010; Moezzi and Goldsworthy, 2018; Tagliazucchi and Laufs, 2014). Variability of the results can be reduced when EEG is integrated with structural or functional MRI images (Ferri et al., 2021), such that physiological information about EEG generators, and anatomical connections are used as priors (Moezzi and Goldsworthy, 2018). Indeed, the validation of EEG connectivity measures at the source space can benefit from electrocorticographic studies in which well-known anatomical connections are probed actively using single pulse electrical stimulation (Hebbink et al., 2019). Likewise, priors for connectivity analysis can be obtained when EEG is combined with transcranial magnetic stimulation (Bortoletto et al., 2015).
4.3. The need of composite measures
As mentioned above, EEG connectivity can be estimated using a large number of methods at both sensor and source spaces (Lee et al., 2017; Sarmukadam et al., 2020). Results are encouraging, since they suggest that EEG connectivity measures can be used as predictors of AD severity (Briels et al., 2020; for a review see delEtoile and Adeli, 2017). Nevertheless, these studies also reveal that different measures that address a particular type of connectivity, e.g., functional connectivity, yield different results for the same dataset. This is also illustrated in Fig. 2, where different EEG connectivity metrics provided different results, even when they were computed on the same scalp distribution of voltages (Fig. 2B and C). Disparity of the results is also exemplified by in silico studies where 42 methods of functional connectivity were benchmarked, using different types of generative models and connectivity structures. No single method was optimal for all types of data (Wang et al., 2014). Likewise, dissimilar results have been obtained when different lag-based effective connectivity measures were tested and benchmarked using simulated data that reflected different scenarios (e.g., different network configurations) with practical interest (Rodrigues and Andrade, 2014). More recently, relevant differences in functional (effective) connectivity estimated with Granger-Geweke causality, directed transfer function, and partial directed coherence methods in real EEG data have been observed (Perera et al., 2020).
The marked disparity in results suggests that a combination of algorithms, and the subsequent integration of the results, e.g., in a composite measure of connectivity, is a promising strategy for the success of multicentric EEG studies (Fig. 1, a multifeatured framework of EEG connectivity). Underlying this approach is the seminal idea that all functional connectivity measures are useful, since they help to constrain the theoretical set of network configurations and causal models that sustain a particular neurocognitive process (Reid et al., 2019). This hypothesis space can undergo progressive refinements when different functional connectivity metrics are integrated (Hyttinen et al., 2016; Ramsey et al., 2010, 2011). This is the case of combining pairwise Pearson correlation and partial correlation analyses, which helps to resolve causal configuration in networks comprising either confounder or colliders nodes, as well as chain interaction (Reid et al., 2019; Sanchez-Romero and Cole, 2021). Likewise, the interpretation of functional connectivity changes can benefit from a “covariance conjunction” method, which combines normalized and non-normalized version of Pearson correlation (Cole et al., 2016). An extension of this approach is to use a relatively large set of functional connectivity metrics to capture different types of dependency between time series, i.e., a composite metrics of connectivity. The usefulness of this metric has been evident in cases where, while time domain Pearson’s correlation indicates low functional connectivity between cortical areas, frequency-domain magnitude-squared coherence suggests otherwise (Mohanty et al., 2020). Noteworthly, classification systems using multiple measures of functional connectivity followed by a feature selection procedure has been proved to provide more consistent results than those in which connectivity metrics are used alone (Mohanty et al., 2020). This result, along with the fact that a particular functional connectivity metric does not typically outperformed others, indicates that a composite metric that integrates information from multiple domains represents an ideal tool for developing the ConnEEGtome for dementia. Nevertheless, further investigation is needed before a composite connectivity metric can be proposed as the standard for neurodegenerative studies. This include assessing the robustness of the connectivity metrics to the intra-subject and inter-subject variability in longitudinal studies (Conti et al., 2019).
Like other aspects of the processing pipeline addressed in this review, the design and implementation of composite metrics of functional connectivity can significantly benefit from computational tools that are already available. This is the case of toolboxes containing combined functional connectivity methods based on correlation, and partial correlation analyses (Sanchez-Romero and Cole, 2021). Another example is SEED-G, a toolbox for the generation of pseudo-EEG data with imposed connectivity patterns, in which ground-truth connectivity models, and different parameters impacting the SNR of the EEG can be controlled (Anzolin et al., 2021).
4.4. Statistical approaches
The standardization of statistical frameworks, particularly methods for correcting multiple comparisons, is a further step to reduce the variability among EEG functional connectivity analyses (Fig. 1F). In this regard, the Unbiased Cluster Estimation (UCE) method, a threshold-free extension to traditional cluster-based analysis (Radhu et al., 2015) has been proposed as a benchmark statistic for multisite initiatives (Farzan et al., 2017). Additionally, promising approaches need to be examined in greater detail, such as permutation-based mass univariate tests in combination with complex factorial designs (Fields and Kuperberg, 2020). Likewise, statistics in multicentric EEG studies can benefit from methods in which hierarchical linear modeling accounts for single trial variability (Pernet et al., 2011). This can be complemented with methods for elucidating nonlinear connectivity (Dottori et al., 2017; Parra et al., 2017), hierarchical representations of higher-order brain areas during complex tasks (Livezey et al., 2019), connectivity-based decoding (Hesse et al., 2019; García et al., 2020) and algorithms that combine Dynamic Causal Modeling and Parametric Empirical Bayes to characterize between-subject variability in effective connectivity (Zeidman et al., 2019). Extensive use of this methodologies is facilitated by freely available computational tools and tutorials with step-by-step instructions (Pernet et al., 2011; Zeidman et al., 2019).
5. Advanced computational tools for diagnosis and classification
The ultimate objective of EEG connectivity studies is to explain (or predict) multi-feature perceptual and cognitive processes, and therefore understand behavior (i.e., Fittipaldi et al., 2020). Consequently, the assessment of EEG connectivity in combination with low-dimensional metrics (like graph theory) can become a powerful tool for diagnosis and classification of neurodegenerative diseases. In this regards, recent studies have analyzed the intra-subject and inter-subject variability of graph-theoretical measures (global and local measures) derived from effective (Granger Causality, and Transfer Entropy), and undirected (Pearson Correlation, and Partial Correlation) connectivity maps, when data is acquired in multiple functional MRI sessions (Conti et al., 2019). Although intersession reproducibility of functional connectivity metrics varies depending on the of graph-theoretical measure selected for the analysis, results like those presented by Conti et al. (2019) are promising and they can be extended to brain connectivity analyses based on EEG. Validation studies can be conducted using well described and publicly available data repositories, such as the Healthy Brain Network (HBN) Biobank, which include a high-density EEG dataset (128 channels, reference Cz) of a diverse sample (1657 individuals) of children and adolescents (aged 5–21) from the New York City area (Alexander et al., 2017).
The predictive power of classification systems based on EEG connectivity, and graph-theoretical measures, substantially increases when the estimation of behavior from functional brain connectivity is assessed with machine- learning- based frameworks, such as connectome- based predictive modeling (CPM) (Shen et al., 2017). Using this model, the functional connectivity has been computed with model-based (linear) metrics on the time domain (e.g., Finn et al., 2015; Rosenberg et al.,2018 and model-free approximations (Kumar et al., 2019). So far CPM has been applied to fMRI (Finn et al., 2015), using available dataset such that the Human Connectome Project (HCP) (Van Essen et al., 2013). Nevertheless, CPM can be extended to brain connectivity datasets derived from magnetoencephalography (MEG), and EEG. It is worth to mention that high-quality EMG data for 67 subjects has been released as part of the Young Adult HCP (https://www.humanconnectome.org/study/hcp-young-adult/project-protocol/meg-eeg).
Furthermore, several open-source EEG datasets are available (eg. Valdes-Sosa et al., 2021), and they typically comprise high-density resting-state EEG in different conditions, e.g., eyes closed, eyes open, and hyperventilation. EEG datasets usually belong to multimodal repositories, which also contains MRI datasets, behavioral and psychological data, as well as anthropometries and demographic information. Additionally, physiological measures (e.g., blood pressure, heart rate, and breathing rate), as well as blood and urine samples can be acquired (Babayan et al., 2019). A relatively extensive list of open-source EEG data repositories has been provided (Cavanagh et al., 2017). Furthermore, initiatives to integrate sparse EEG repositories have been launched. This is the case of The Patient Repository for EEG Data + Computational Tools (PRED+CT), an open-source site for gathering, storing, and analyzing clinically relevant data (Cavanagh et al., 2017).
Moving beyond the use of unimodal predictive systems, such as those mentioned above, diagnosis tools based on multi-feature classification can take better advantage of the information provided by functional connectivity maps (Fig. 1F). This type of analysis can exploit the potential of multicentric studies of neurodegeneration in which clinical, molecular, metabolic, and genetic examinations of patients are typically made in combination with neuroimaging. Multimodal assessment of clinical and cognitive process combined with EEG and MRI (Abrevaya et al., 2020; García-Cordero et al., 2016; Ibáñez et al., 2017, Ibáñez, 2018; Melloni et al., 2015a, 2015b, 2017; Salamone et al., 2018, 2021) can improve the characterization of multiple physiopathological and neurocognitive pathways impacted by neurodegeneration. In this sense, a data-driven hierarchical Bayesian model to identify latent factors in AD has been proposed, which performs a joint analysis of atrophy and cognitive deficits. The model outperformed canonical correlation analysis at capturing the heterogeneity of the atrophy-cognitive associations, and revealed atrophy-cognitive factors which had not previously been described (Sun et al., 2019). A comprehensive review of data-driven methods to assess the heterogeneity in neurodegenerative conditions have been recently published (Habes et al., 2020). Another approach is the implementation of models that assess hierarchical associations among sensory processing, neurocognition, clinical symptoms, and functional outcomes, using structural equation modeling (Koshiyama et al., 2021). This approach has been successfully used in other conditions such as schizophrenia, demonstrating that including EEG measures (MMN/P3a and gamma-band ASSR), add explanatory power to traditional models of neuropsychiatric disorders (Koshiyama et al., 2021). This approach can be advanced by multi-feature machine learning classification using hyperparameter tuning and gradient boosting of progressive feature elimination that allows for feature dimensionality reduction and control of overfitting and collinearities.
6. Testing the success of harmonization
Metrics about the success of EEG data harmonization are needed to guide further data analyses and plan future multicentric preclinical studies. Such metrics need to consider the acquisition system and relevant acquisition parameters as covariates in multicentric EEG studies of connectivity. Studies on this matter are scarce, since it is often assumed that the specific choice of EEG system has limited impact on the data and does not add variance to the results (Melnik et al., 2017). Nevertheless, acquisition systems can make a significant contribution to the variance of multisite EEG studies, particularly when they differ in the type of electrodes, and portability- related electronics. This effect is reduced when comparisons are restricted to standard-EEG acquisition systems (Melnik et al., 2017). Since multiple competitive companies participate in the global market of EEG instrumentation, it is necessary to test the equivalence of data acquired with acquisition systems from different companies. We anticipate that such companies should also being informed of sources of variability and suggested guidelines.
Metrics for benchmarking may rely on signal representation approaches, based on the computation of epoch rejection rate, SNR, amplitude variance in particular time windows, the susceptibility of the experimental setup to line noise and the percentage of artifact-contaminated EEG segments (Oliveira et al., 2016; Radüntz, 2018; Hinrichs et al., 2020). When multicentric studies comprise sensory or cognitive tasks with different experimental conditions, the output of algorithms for trial classification can be designed for comparative purposes (Kam et al., 2019). Additionally, metrics about signal stability (Malcolm et al., 2019) based on cross- correlation and auto-correlation analysis can be implemented.
How variations of EEG time series co-vary across headsets have been assessed using distance metrics between recording covariance matrices, e.g., Riemannian distance and Euclidean distance (Bigdely-Shamlo et al., 2020a). For statistical comparisons, random permutations tests are conducted with the output of representational similarity analysis (Kriegeskorte et al., 2008). The latter method computes similarity based on two aspects of the data, for example, the Riemannian distance, and whether EEG was acquired with the same electrode configuration. Using this approach, the effect on any independent parameter that has a potential impact on the integration of multisite EEG data can be tested. This analysis is potentiated when the Hierarchical general modeling (Pernet et al., 2011), which assesses statistically significant ERP and ERSP patterns across subjects, is combined with HED tags (Bigdely-Shamlo et al., 2020b). This method is referred to as the Hierarchical organizational model and allows for the separation of subject, headset, paradigm and cognitive aspects in a manner that is scalable and generalizable across diverse collections of EEG datasets (Bigdely-Shamlo et al., 2020b). Metrics indicating the success of the harmonization can be extended to the EEG source space. This is the case of the pairwise normalized difference/asymmetry (ND), a measure that represent the dimensionless, normalized pairwise asymmetry of functional connectivity matrices and derived graph-based metrics, and that may be used as indicators of inter-subject variability (Conti et al., 2019).
The metrics mentioned above may help to build methodological consensus, which in turn will contribute to establish functional connectivity measures as reliable biomarkers for the early identification of subtypes within a particular neurodegenerative disorder, and facilitate intervention. This early identification system implies that the traditional classification of neurodegenerative disorder based on group analyses shall be complemented with individual-level analyses. The paradigm shift is challenging, and it is not unique to EEG multisite studies. In fact, the need for consensus and harmonization across subtyping methods for AD has been revealed by studies demonstrating a mismatch between the group-level and the individual-level classification of patients, based on MRI images and positron emission tomography (Mohanty et al., 2020). Since this is partially a consequence of the heterogeneity of methodological approaches, metrics comparing the performance of the classification algorithms are needed.
Last but not least, sharing data and processing pipelines is critical for the replicability and cross- validation of multisite studies. These aspects contribute to conducting joint analysis in which two or more multicentric studies are integrated, impacting the power of current diagnosis, monitoring and intervention assessments. Tools for this purpose are available, and some of them have been described in this article. Valuable contributions are initiatives like #EEGManyLabs, a platform for large-scale international collaborative replication studies (Pavlov et al., 2021). The number of peer review journals that seek sharing protocols for result replication is constantly increasing. Many research groups, including us, have participated in these collaborative efforts (e.g., Prado-Gutiérrez et al., 2019). Noteworthy, increasing data sharing and harmonization of methodological procedures is a further step towards the federation of machine learning methods and other computational tools, where model parameters are shared instead of data (Alam et al., 2020). Consensus is needed, and the scientific community is aware of barriers for harmonization of neuroimaging biomarkers for neurodegenerative diseases (Jovicich et al., 2019). Actions to overcome these barriers have been proposed, as well as priorities for funding. They address the creation of a hub of documents to guide the planning and execution of future multicentric studies, and the support of future harmonization challenges (Jovicich et al., 2019).
7. Conclusions
The creation of a global platform for harmonization of EEG-related multi-feature connectivity in neurodegenerative research is challenging, but progress made in the field is encouraging. Several tools for harmonization of preprocessing steps are already available and need to be unified within basic common processing workflows. Summarizing, relevant aspects for data replicability in multicentric EEG studies on connectivity, also relevant for the success of the ConEEGtome, include (a) conducting multiple analyses of the same data to be integrated in subsequent processing steps, and (b) developing composite metrics of connectivity to feed multi-feature classification systems. Undoubtedly, significant advances in these directions will help to revolutionize current EEG approaches in neurodegenerative research, by creating a new generation of objective, computer-based tools for diagnosis, characterization and treatment of neurodegenerative diseases and other disorders.
Funding
This work was supported by Takeda Grant CW2680521; CONICET; FONCYT-PICT (2017-1818, 2017-1820); ANID/FONDECYT Regular (1210195 and 1210176); ANID/FONDAP (15150012); Alzheimer Association GBHI ALZ UK-20-639295; and the Multi-Partner Consortium to Expand Dementia Research in Latin America (ReDLat), funded by the National Institutes of Aging of the National Institutes of Health under award number R01AG057234, an Alzheimer’s Association grant (SG-20-725707-ReDLat), the Rainwater Foundation, and the Global Brain Health Institute. The content is solely the responsibility of the authors and does not represent the official views of these institutions.
Footnotes
Declaration of Competing Interest
Authors declare no competing interests.
Data availability
The data that support analysis presented in Fig. 2 are available from the corresponding author upon reasonable request.
Data availability
Data will be made available on request.
References
- Abrevaya S, Fittipaldi S, García AM, Dottori M, Santamaria-Garcia H, Birba A, Yoris A, Hildebrandt MK, Salamone P, De la Fuente A, Alarco-Martí S, García-Cordero I, Matorrel-Caro M, Pautassi RM, Serrano C, Sedeño L, Ibáñez A, 2020. At the Heart of Neurological Dimensionality: Cross-Nosological and Multimodal Cardiac Interoceptive Deficits. Psychosom. Med 82 (9), 850–861. 10.1097/PSY.0000000000000868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander LM, Escalera J, Ai L, Andreotti C, Febre K, Mangone A, Vega-Potler N, Langer N, Alexander A, Kovacs M, Litke S, O’Hagan B, Andersen J, Bronstein B, Bui A, Bushey M, Butler H, Castagna V, Camacho N, Chan E, Citera D, Clucas J, Cohen S, Dufek S, Eaves M, Fradera B, Gardner J, Grant-Villegas N, Green G, Gregory C, Hart E, Harris S, Horton M, Kahn D, Kabotyanski K, Karmel B, Kelly SP, Kleinman K, Koo B, Kramer E, Lennon E, Lord C, Mantello G, Margolis A, Merikangas KR, Milham J, Minniti G, Neuhaus R, Levine A, Osman Y, Parra LC, Pugh KR, Racanello A, Restrepo A, Saltzman T, Septimus B, Tobe R, Waltz R, Williams A, Yeo A, Castellanos FX, Klein A, Paus T, Leventhal BL, Craddock RC, Koplewicz HS, Milham MP, 2017. An open resource for transdiagnostic research in pediatric mental health and learning disorders. Sci. Data 4, 170181. 10.1038/sdata.2017.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmadlou M, Adeli A, Bajo R, Adeli H, 2014. Complexity of functional connectivity networks in mild cognitive impairment subjects during a working memory task. Clin. Neurophysiol. 125 (4), 694–702. 10.1016/j.clinph.2013.08.033. [DOI] [PubMed] [Google Scholar]
- Alam RU, Zhao H, Goodwin A, Kavehei O, McEwan A, 2020. Differences in power spectral densities and phase quantities due to processing of EEG signals. Sensors (Basel) 20 (21), 6285. 10.3390/s20216285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anzolin A, Toppi J, Petti M, Cincotti F, Astolfi L, 2021. SEED-G: simulated EEG data generator for testing connectivity algorithms. Sensors (Basel) 21 (11), 3632. 10.3390/s21113632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babayan A, Erbey M, Kumral D, Reinelt JD, Reiter A, Röbbig J, Schaare HL, Uhlig M, Anwander A, Bazin PL, Horstmann A, Lampe L, Nikulin VV, Okon-Singer H, Preusser S, Pampel A, Rohr CS, Sacher J, Thöne-Otto A, Trapp S, Villringer A, 2019. A mind-brain-body dataset of MRI, EEG, cognition, emotion, and peripheral physiology in young and old adults. Sci. Data 6, 180308. 10.1038/sdata.2018.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiloni C, Arakaki X, Azami H, Bennys K, Blinowska K, Bonanni L, Bujan A, Carrillo MC, Cichocki A, de Frutos-Lucas J, Del Percio C, Dubois B, Edelmayer R, Egan G, Epelbaum S, Escudero J, Evans A, Farina F, Fargo K, Fernández A, Ferri R, Frisoni G, Hampel H, Harrington MG, Jelic V, Jeong J, Jiang Y, Kaminski M, Kavcic V, Kilborn K, Kumar S, Lam A, Lim L, Lizio R, Lopez D, Lopez S, Lucey B, Maestú F, McGeown WJ, McKeith I, Moretti DV, Nobili F, Noce G, Olichney J, Onofrj M, Osorio R, Parra-Rodriguez M, Rajji T, Ritter P, Soricelli A, Stocchi F, Tarnanas I, Taylor JP, Teipel S, Tucci F, Valdes-Sosa M, Valdes-Sosa P, Weiergräber M, Yener G, Guntekin B, 2021. Measures of resting-state EEG rhythms for clinical trials in Alzheimer’s disease: recommendations of an expert panel. Alzheimers Dement. 17 (9), 1528–1553. 10.1002/alz.12311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babiloni C, Blinowska K, Bonanni L, Cichocki A, De Haan W, Del Percio C, Dubois B, Escudero J, Fernández A, Frisoni G, Guntekin B, Hajos M, Hampel H, Ifeachor E, Kilborn K, Kumar S, Johnsen K, Johannsson M, Jeong J, LeBeau F, Lizio R, Lopes da Silva F, Maestú F, McGeown WJ, McKeith I, Moretti DV, Nobili F, Olichney J, Onofrj M, Palop JJ, Rowan M, Stocchi F, Struzik ZM, Tanila H, Teipel S, Taylor JP, Weiergräber M, Yener G, Young-Pearse T, Drinkenburg WH, Randall F, 2020. What electrophysiology tells us about Alzheimer’s disease: a window into the synchronization and connectivity of brain neurons. Neurobiol. Aging 85, 58–73. 10.1016/j.neurobiolaging.2019.09.008. [DOI] [PubMed] [Google Scholar]
- Bachli MB, Sedeño L, Ochab JK, Piguet O, Kumfor F, Reyes P, Torralva T, Roca M, Cardona JF, Campo CG, Herrera E, Slachevsky A, Matallana D, Manes F, García AM, Ibáñez A, Chialvo DR, 2020. Evaluating the reliability of neurocognitive biomarkers of neurodegenerative diseases across countries: a machine learning approach. NeuroImage 208, 16456. 10.1016/j.neuroimage.2019.116456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barzegaran E, Knyazeva MG, 2017. Functional connectivity analysis in EEG source space: the choice of method. PLoS One 12 (7), e0181105. 10.1371/journal.pone.0181105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Başar E, Başar-Eroğlu C, Güntekin B, Yener GG, 2013. Brain’s alpha, beta, gamma, delta, and theta oscillations in neuropsychiatric diseases: proposal for biomarker strategies. Suppl. Clin. Neurophysiol 62, 19–54. 10.1016/b978-0-7020-5307-8.00002-8. [DOI] [PubMed] [Google Scholar]
- Bassett DS, Bullmore ET, 2009. Human brain networks in health and disease. Curr. Opin. Neurol 22 (4), 340–347. 10.1097/WCO.0b013e32832d93dd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastos AM, Schoffelen JM, 2016. A tutorial review of functional connectivity analysis methods and their interpretational pitfalls. Front. Syst. Neurosci 9, 175. 10.3389/fnsys.2015.00175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell AJ, Sejnowski TJ, 1995. An information-maximization approach to blind separation and blind deconvolution. Neural Comput. 7 (6), 129–159. 10.1162/neco.l995.7.6.1129. [DOI] [PubMed] [Google Scholar]
- Bigdely-Shamlo N, Cockfield J, Makeig S, Rognon T, La Valle C, Miyakoshi M, Robbins KA, 2016. Hierarchical event descriptors (HED): semi-structured tagging for real-world events in large-scale EEG. Front Neuroinform. 10, 42. 10.3389/fhinf.2016.00042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigdely-Shamlo N, Kreutz-Delgado K, Kothe C, Makeig S, 2013. EyeCatch: data-mining over half a million EEG independent components to construct a fully-automated eye-component detector. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc 2013, 5845–5848. 10.1109/EMBC.2013.6610881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigdely-Shamlo N, Mullen T, Kothe C, Su KM, Robbins KA, 2015. The PREP pipeline: standardized preprocessing for large-scale EEG analysis. Front Neuroinform. 9, 16. 10.3389/fninf.2015.00016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigdely-Shamlo N, Touryan J, Ojeda A, Kothe C, Mullen T, Robbins K, 2020a. Automated EEG mega-analysis I: spectral and amplitude characteristics across studies. NeuroImage 207, 116361. 10.1016/j.neuroimage.2019.116361. [DOI] [PubMed] [Google Scholar]
- Bigdely-Shamlo N, Touryan J, Ojeda A, Kothe C, Mullen T, Robbins K, 2020b. Automated EEG mega-analysis II: cognitive aspects of event related features. NeuroImage 207, 116054. 10.1016/j.neuroimage.2019.116054. [DOI] [PubMed] [Google Scholar]
- Bortoletto M, Veniero D, Thut G, Miniussi C, 2015. The contribution of TMS-EEG coregistration in the exploration of the human cortical connectome. Neurosci. Biobehav. Rev 49, 114–124. 10.1016/j.neubiorev.2014.12.014. [DOI] [PubMed] [Google Scholar]
- Botvinik-Nezer R, Holzmeister F, Camerer CF, Dreber A, Huber J,Johannesson M, Kirchler M, Iwanir R, Mumford JA, Adcock RA, Avesani P, Baczkowski BM, Bajracharya A, Bakst L, Ball S, Barilari M, Bault N, Beaton D, Beitner J, Benoit RG, Bhanji JP, Biswal BB, Bobadilla-Suarez S, Berkers RMWJ, Bortolini T, Bottenhorn KL, Bowring A, Braem S, Brooks HR, Brudner EG, Calderon CB, Camilleri JA, Castrellon JJ, Cecchetti L, Cieslik EC, Cole ZJ, Collignon O, Cox RW, Cunningham WA, Czoschke S, Dadi K, Davis CP, Luca A, Delgado MR, Demetriou L, Dennison JB, Di X, Dickie EW, Dobryakova E, Donnat CL, Dukart J, Duncan NW, Durnez J, Eed A, Eickhoff SB, Erhart A, Fontanesi L, Fricke GM, Fu S, Galván A, Gau R, Genon S, Glatard T, Glerean E, Goeman JJ, Golowin SAE, González-García C, Gorgolewski KJ, Grady CL, Green MA, Guassi, Moreira JF, Guest O, Hakimi S, Hamilton JP, Hancock R, Handjaras G, Harry BB, Hawco C, Herholz P, Herman G, Heunis S, Hoffstaedter F, Hogeveen J, Holmes S, Hu CP, Huettel SA, Hughes ME, Iacovella V, Iordan AD, Isager PM, Isik AI, Jahn A, Johnson MR, Johnstone T, Joseph MJE, Juliano AC, Kable JW, Kassinopoulos M, Koba C, Kong XZ, Koscik TR, Kucukboyaci NE, Kuhl BA, Kupek S, Laird AR, Lamm C, Langner R, Lauharatanahirun N, Lee H, Lee S, Leemans A, Leo A, Lesage E, Li F, Li MYC, Lim PC, Lintz EN, Liphardt SW, Losecaat, Vermeer AB, Love BC, Mack ML, Malpica N, Marins T, Maumet C, McDonald K, McGuire JT, Melero H, Méendez, Leal AS, Meyer B, Meyer KN, Mihai G, Mitsis GD, Moll J, Nielson DM, Nilsonne G, Notter MP, Olivetti E, Onicas AI, Papale P, Patil KR, Peelle JE, Pérez A, Pischedda D, Poline JB, Prystauka Y, Ray S, Reuter-Lorenz PA, Reynolds RC, Ricciardi E, Rieck JR, Rodriguez-Thompson AM, Romyn A, Salo T, Samanez-Larkin GR, Sanz-Morales E, Schlichting ML, Schultz DH, Shen Q, Sheridan MA, Silvers JA, Skagerlund K, Smith A, Smith DV, Sokol, Hessner, Steinkamp SR, Tashjian SM, Thirion B, Thorp JN, Tinghög G, Tisdall L, Tompson SH, Toro-Serey C, Samanez-Larkin GR, Sanz-Morales E, Schlichting ML, Schultz DH, Shen Q, Sheridan MA, Silvers JA, Skagerlund K, Smith A, Smith DV, Sokol, Hessner P, Steinkamp SR, Tashjian SM, Thirion B, Thorp JN, Tinghög G, Tisdall L, Tompson SH, Toro-Serey C, Torre, Tresols JJ, Tozzi L, Truong V, Turella L, Verguts T, van ‘t Veer AE, Vettel JM, Vijayarajah S, Vo K, Wall MB, Weeda WD, Weis S, White DJ, Wisniewski D, Porxas A, Yearling EA, Yoon S, Yuan R, Yuen KSL, Zhang L, Zhang X, Zosky JE, Nichols TE, Poldrack RA, Schonberg T, Xifra-Porxas A, Yearling EA, Yoon S, Yuan R, Yuen KSL, Zhang L, Zhang X, Zosky JE, Nichols TE, Poldrack RA, Schonberg T, 2020. Variability in the analysis of a single neuroimaging dataset by many teams. Nature 582 (7810), 84–88. 10.1038/s41586-020-2314-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bowyer SM, 2016. Coherence a measure of the brain networks: past and present. Neuropsychiatr. Electrophysiol 2 (1) 10.1186/s40810-015-0015-7 [DOI] [Google Scholar]
- Briels CT, Schoonhoven DN, Stam CJ, de Waal H, Scheltens P, Gouw AA, 2020. Reproducibility of EEG functional connectivity in Alzheimer’s disease. Alzheimers Res. Ther 12 (1), 68. 10.1186/sl3195-020-00632-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bufacchi RJ, Magri C, Novembre G, Iannetti GD, 2021. Local spatial analysis: an easy-to-use adaptive spatial EEG filter. J. Neurophysiol 125 (2), 509–521. 10.1152/jn.00560.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanagh JF, Napolitano A, Wu C, Mueen A, 2017. The patient repository for EEG data + computational tools (PRED+CT). Front. Neuroinformatics 11, 67. 10.3389/fninf.2017.00067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chella F, D’Andrea A, Basti A, Pizzella V, Marzetti L, 2017. Non-linear analysis of scalp EEG by using bispectra: the effect of the reference choice. Front. Neurosci 11, 262. 10.3389/ftiins.2017.00262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen D, Jiang J, Lu J, Wu P, Zhang H, Zuo C, Shi K, 2019. Brain network and abnormal hemispheric asymmetry analyses to explore the marginal differences in glucose metabolic distributions among Alzheimer’s disease, Parkinson’s disease dementia, and Lewy body dementia. Front. Neurol 10, 369. 10.3389/fheur.2019.00369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung J, Yoo K, Lee P, Kim CM, Roh JH, Park JE, Kim SJ, Seo SW, Shin JH, Seong JK, Jeong Y, 2017. Normalization of cortical thickness measurements across different T1 magnetic resonance imaging protocols by novel W-score standardization. NeuroImage 159, 224–235. 10.1016/j.neuroimage.2017.07.053. [DOI] [PubMed] [Google Scholar]
- Cohen MX, 2015. Comparison of different spatial transformations applied to EEG data: a case study of error processing. Int. J. Psychophysiol 97 (3), 245–257. 10.1016/j.ijpsycho.2014.09.013. [DOI] [PubMed] [Google Scholar]
- Conti A, Duggento A, Guerrisi M, Passamonti L, Indovina I, Toschi N, 2019. Variability and reproducibility of directed and undirected functional MRI connectomes in the human brain. Entropy (Basel, Switzerland) 21 (7), 661. 10.3390/e21070661 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cole MW, Yang GJ, Murray JD, Repovţ G, Anticevic A, 2016. Functional connectivity change as shared signal dynamics. J. Neurosci. Methods 259, 22–39. 10.1016/j.jneumeth.2015.ll.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- delEtoile J, Adeli H, 2017. Graph theory and brain connectivity in Alzheimer’s disease. Neuroscientist 23 (6), 616–626. 10.1177/1073858417702621. [DOI] [PubMed] [Google Scholar]
- Delorme A, Makeig S, 2004. EEGLAB: an open source toolbox for analysis of single-trial EEG dynamics including independent component analysis. J. Neurosci. Methods 134 (1), 9–21. 10.1016/j.jneumeth.2003.10.009. [DOI] [PubMed] [Google Scholar]
- Dennis EL, Thompson PM, 2014. Functional brain connectivity using fMRI in aging and Alzheimer’s disease. Neuropsychol. Rev 24 (1), 49–62. 10.1007/s11065-014-9249-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donnelly-Kehoe PA, Pascariello GO, García AM, Hodges JR, Miller B, Rosen H, Manes F, Landin-Romero R, Matallana D, Serrano C, Herrera E, Reyes P, Santamaria-Garcia H, Kumfor F, Piguet O, Ibanez A, Sedeño L, 2019. Robust automated computational approach for classifying frontotemporal neurodegeneration: multimodal/multicentric neuroimaging. Alzheimers Dement. 11, 588–598. 10.1016/j.dadm.2019.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorszewska J, Kozubski W, Waleszczyk W, Zabel M, Ong K, 2020. Neuroplasticity in the pathology of neurodegenerative diseases. Neural Plast. 2020, 4245821. 10.1155/2020/4245821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dottori M, Sedeño L, Martorell Caro M, Alifano F, Hesse E, Mikulan E, García AM, Ruiz-Tagle A, Lillo P, Slachevsky A, Serrano C, Fraiman D, Ibanez A, Towards affordable biomarkers of frontotemporal dementia: a classification study via network’s information sharing. Sci Rep. 7 (1), 3822. 10.1038/S41598-017-04204-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farzan F, Atluri S, Frehlich M, Dhami P, Kleffner K, Price R, Lam RW, Frey BN, Milev R, Ravindran A, McAndrews MP, Wong W, Blumberger D, Daskalakis ZJ, Vila- Rodriguez F, Alonso E, Brenner CA, Liotti M, Dharsee M, Arnott SR, Evans KR, Rotzinger S, Kennedy SH, 2017. Standardization of electroencephalography for multisite, multi- platform and multi-investigator studies: insights from the Canadian biomarker integration network in depression. Sci. Rep 7 (1), 7473. 10.1038/s41598-017-07613-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferri R, Babiloni C, Karami V, Triggiani AI, Carducci F, Noce G, Lizio R, Pascarelli MT, Soricelli A, Amenta F, Bozzao A, Romano A, Giubilei F, Del Percio C, Stocchi F, Frisoni GB, Nobili F, Patanè L, Arena P, 2021. Stacked autoencoders as new models for an accurate Alzheimer’s disease classification support using resting-state EEG and MRI measurements. Clin Neurophysiol. 132 (1), 232–245. 10.1016/j.clinph.2020.09.015. [DOI] [PubMed] [Google Scholar]
- Fields EC, Kuperberg GR, 2020. Having your cake and eating it too: flexibility and power with mass univariate statistics for ERP data. Psychophysiology 57 (2), e13468. 10.llll/psyp.13468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finn ES, Shen X, Scheinost D, Rosenberg MD, Huang J, Chun MM, Papademetris X, Constable RT, 2015. Functional connectome fingerprinting: identifying individuals using patterns of brain connectivity. Nat. Neurosci 18 (11), 1664–1671. 10.1038/nn.4135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fittipaldi S, Abrevaya S, Fuente A, Pascariello GO, Hesse E, Birba A, Salamone P, Hildebrandt M, Martí SA, Pautassi RM, Huepe D, Martorell MM, Yoris A, Roca M, García AM, Sedeño L, Ibáñez A, 2020. A multidimensional and multi-feature framework for cardiac interoception. NeuroImage 212, 16677. 10.1016/j.neuroimage.2020.116677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friston KJ, 2011. Functional and effective connectivity a review. BrainConnect 1 (1), 13–36. 10.1089/brain.2011.0008. [DOI] [PubMed] [Google Scholar]
- Gabard-Durnam LJ, Mendez Leal AS, Wilkinson CL, Levin AR, 2018. The Harvard automated processing pipeline for electroencephalography (HAPPE) standardized processing software for developmental and high-artifact data. Front. Neurosci 27 (12), 97. 10.3389/fnins.2018.00097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García AM, Abrevaya S, Kozono G, Cordero IG, Cördoba M, Kauffman MA, Pautassi R, Muñoz E, Sedeño L, Ibáñez A, 2017. The, cerebellum, and, embodied, semantics, evidence, from, a, case, of, genetic, ataxia, due, to, STUB1, mutations. J. Med. Genet 54 (2), 114–124. 10.1136/jmedgenet-2016-104148. [DOI] [PubMed] [Google Scholar]
- García AM, Hesse E, Birba A, Adolfi F, Mikulan E, Caro MM, Petroni A, Bekinschtein TA, Del, Carmen, García M, Silva W, Ciraolo C, Vaucheret E, Sedeño L, Ibáñez A, 2020. Time to face language: embodied mechanisms underpin the inception of face-related meanings in the human brain. Cereb. Cortex 30 (11), 6051–6068. 10.1093/cercor/bhaal78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Cordero I, Sedeño L, de la Fuente L, Slachevsky A, Forno G, Klein F, Lillo P, Ferrari J, Rodriguez C, Bustin J, Torralva T, Baez S, Yoris A, Esteves S, Melloni M, Salamone P, Huepe D, Manes F, García AM, Ibañez A, 2016. Feeling, learning from and being aware of inner states: interoceptive dimensions in neurodegeneration and stroke. Philos. Trans. R. Soc. Lond. B Biol. Sci 371 (1708), 20160006. 10.1098/rstb.2016.0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghaderi AH, Baltaretu BR, Andevari MN, Bharmauria V, Balci F, 2020. Synchrony and complexity in state-related EEG networks: an application of spectral graph theory. Neural Comput. 32 (12), 2422–2454. 10.1162/neco_a_01327. [DOI] [PubMed] [Google Scholar]
- Gramfort A, Luessi M, Larson E, Engemann DA, Strohmeier D, Brodbeck C, Goj R, Jas M, Brooks T, Parkkonen L, Hämäläinen M, 2013. MEG and EEG data analysis with MNE-python. Front. Neurosci 7, 267. 10.3389/fnins.2013.00267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Habes M, Grothe MJ, Tunc B, McMillan C, Wolk DA, Davatzikos C, 2020. Disentangling heterogeneity in Alzheimer’s disease and related dementias using data-driven methods. Biol. Psychiatry 88 (1), 70–82. 10.1016/j.biopsych.2020.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebbink J, van Blooijs D, Huiskamp G, Leijten FSS, van Gils SA, Meijer HGE, 2019. A comparison of evoked and non-evoked functional networks. Brain Topogr. 32 (3), 405–417. 10.1007/sl0548-018-0692-l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hesse E, Mikulan E, Sitt JD, Garcia MDC, Silva W, Ciraolo C, Vaucheret E, Raimondo F, Baglivo F, Adolfi F, Herrera E, Bekinschtein TA, Petroni A, Lew S, Sedeno L, Garcia AM, Ibanez A, 2019. Consistent gradient of performance and decoding of stimulus type and valence from local and network activity. IEEE Trans. Neural Syst. Rehabil. Eng. 27 (4), 619–629. 10.1109/TNSRE.2019.2903921 [DOI] [PubMed] [Google Scholar]
- Hinrichs H, Scholz M, Baum AK, Kam JWY, Knight RT, Heinze HJ, 2020. Comparison between a wireless dry electrode EEG system with a conventional wired wet electrode EEG system for clinical applications. Sci. Rep 10 (1), 5218. 10.1038/S41598-020-62154-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath A, Szucs A, Csukly G, Sakovics A, Stefanics G, Kamondi A, 2018. EEG and ERP biomarkers of Alzheimer’s disease: a critical review. Front. Biosci. (Landmark Ed) 23, 183–220. 10.2741/4587. [DOI] [PubMed] [Google Scholar]
- Horwitz B, 2003. The elusive concept of brain connectivity. NeuroImage 19 (2 Pt 1), 466–470. 10.1016/sl053-8119(03)00112-5 [DOI] [PubMed] [Google Scholar]
- Hu S, Lai Y, Valdes-Sosa PA, Bringas-Vega ML, Yao D, 2018. How do reference montage and electrodes setup affect the measured scalp EEG potentials? J. Neural Eng 15 (2), 026013 10.1088/1741-2552/aaal3f. Advance online publication. [DOI] [PubMed] [Google Scholar]
- Hu S, Yao D, Valdes-Sosa PA, 2018b. Unified bayesian estimator of EEG reference at infinity: rREST (regularized reference electrode standardization technique). Front. Neurosci 12, 297. 10.3389/fnins.2018.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber PJ, 1964. Robust estimation of a location parameter. Ann. Math. Stat 35, 73–101. 10.2307/2238020. [DOI] [Google Scholar]
- Hyttinen A, Plis S, Järvisalo M, Eberhardt F, Danks D, 2016. Causal discovery from subsampled time series data by constraint optimization. In: JMLR workshop and conference proceedings, 52, pp. 216–227. [PMC free article] [PubMed] [Google Scholar]
- Ibanez A, Parra MA, Butler C, 2021a. The Latin America and the Caribbean consortium on dementia (LAC-CD): from networking to research to implementation science. J. Alzheimers Dis 82 (s1), S379–S394. 10.3233/JAD-201384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibanez A, Yokoyama JS, Possin KL, Matallana D, Lopera F, Nitrini R, Takada LT, Custodio N, Sosa, Ortiz AL, Avila-Funes JA, Behrens MI, Slachevsky A, Myers RM, Cochran JN, Brusco LI, Bruno MA, Brucki SMD, Pina-Escudero SD, Okada, de Oliveira M, Donnelly, Kehoe P, Garcia AM,Cardona JF, Santamaria-Garcia H, Moguilner S, Duran-Aniotz C,Tagliazucchi E, Maito M, Longoria, Ibarrola EM, Pintado-Caipa M, Godoy ME, Bakman V, Javandel S, Kosik KS, Valcour V, Miller BL, 2021b. The multi-partner consortium to expand dementia research in Latin America (ReDLat): driving multicentric research and implementation science. Front. Neurol 12, 631722 10.3389/fneur.2021.631722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez A, Pina-Escudero SD, Possin KL, Quiroz YT, Peres FA, Slachevsky A, Sosa AL, Brucki SMD, Miller BL, 2021c. Multi-partner consortium to expand dementia research in Latin America. Dementia caregiving across Latin America and the Caribbean and brain health diplomacy. Lancet Healthy Longev. 2 (4), e222–e231. 10.1016/s2666-7568(21)00031-3. Epub 2021 Mar 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibáñez A, 2018. Brain oscillations, inhibition and social inappropriateness in frontotemporal degeneration. Brain 141 (10), e73. 10.1093/brain/awy233. [DOI] [PubMed] [Google Scholar]
- Ibáñez A, Billeke P, de la Fuente L, Salamone P, García AM, Melloni M, 2017. Towards a neurocomputational account of social dysfunction in neurodegenerative disease. Brain 140 (3), e15. 10.1093/brain/aww316. [DOI] [PubMed] [Google Scholar]
- Ibanez A, Melloni M, Huepe D, Helgiu E, Rivera-Rei A, Canales-Johnson A, Baker P, Moya A, 2012. What event-related potentials (ERPs) bring to social neuroscience? Soc. Neurosci 7 (6), 632–649. 10.1080/17470919.2012.691078. [DOI] [PubMed] [Google Scholar]
- Ibanez A, Parra MA, 2014. Mapping memory binding onto the connectome’s temporal dynamics: toward a combined biomarker for Alzheimer’s disease. Front. Hum. Neurosci 22 (8), 237. 10.3389/fnhum.2014.00237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iakovidou ND, 2017. Graph theory at the service of electroencephalograms. Brain Connect. 7 (3), 137–151. 10.1089/brain.2016.0426. [DOI] [PubMed] [Google Scholar]
- Jack CR Jr., Petersen RC, Xu YC, Waring SC, O’Brien PC, Tangalos EG, Smith GE, Ivnik RJ, Kokmen E, 1997. Medial temporal atrophy on MRI in normal aging and very mild Alzheimer’s disease. Neurology 49 (3), 86–94. 10.1212/wnl.49.3.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalilianhasanpour R, Beheshtian E, Sherbaf G, Sahraian S, Sair HI, 2019. Functional connectivity in neurodegenerative disorders: Alzheimer’s disease and frontotemporal dementia. Top. Magn. Reson. Imaging 28 (6), 317–324. 10.1097/RMR.0000000000000223. [DOI] [PubMed] [Google Scholar]
- John MS, Dimitrijevic A, Picton TW, 2001. Weighted averaging of steady-state responses. Clin. Neurophysiol 112 (3), 555–562. 10.1016/sl388-2457(01)00456-4. [DOI] [PubMed] [Google Scholar]
- Johnson EB, Gregory S, 2019. Huntington’s disease: brain imaging in Huntington’s disease. Prog. Mol. Biol. Transl. Sci 165, 321–369. 10.1016/bs.pmbts.2019.04.004. [DOI] [PubMed] [Google Scholar]
- Josefsson A, Ibáñez A, Parra M, Escudero J, 2019. Network analysis through the use of joint-distribution entropy on EEG recordings of MCI patients during a visual short-term memory binding task. Healthc. Technol. Lett 6 (2), 27–31. 10.1049/htl.2018.5060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovicich J, Barkhof F, Babiloni C, Herholz K, Mulert C, van Berckel BNM, Frisoni GB, 2019. SRA-NED JPND working group., harmonization of neuroimaging biomarkers for neurodegenerative diseases: a survey in the imaging community of perceived barriers and suggested actions. Alzheimers Dement. (Amst) 11, 69–73. 10.1016/j.dadm.2018.ll.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kam JWY, Griffin S, Shen A, Patel S, Hinrichs H, Heinze HJ, Deouell LY, Knight RT, 2019. Systematic comparison between a wireless EEG system with dry electrodes and a wired EEG system with wet electrodes. NeuroImage 184, 119–129. 10.1016/j.neuroimage.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaufmann C, Wehrle R, Wetter TC, Holsboer F, Auer DP, Pollmächer T,Czisch M, 2006. Brain activation and hypothalamic functional connectivity during human non-rapid eye movement sleep: an EEG/fMRI study. Brain 129 (Pt 3), 655–667. 10.1093/brain/awh686. [DOI] [PubMed] [Google Scholar]
- Kayser J, Tenkem CE, 2015. Hemifield-dependent N1 and event-related theta/delta oscillations: an unbiased comparison of surface laplacian and common EEG reference choices. Int. J. Psychophysiol 97 (3), 258–270. 10.1016/j.ijpsycho.2014.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna A, Pascual-Leone A, Michel CM, Farzan F, 2015. Microstates in resting-state EEG: current status and future directions. Neurosci. Biobehav. Rev 49, 105–113. 10.1016/j.neubiorev.2014.12.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J, Criaud M, Cho SS, Díez-Cirarda M, Mihaescu A, Coakeley S, Ghadery C, Valli M, Jacobs MF, Houle S, Strafella AP, 2017. Abnormal intrinsic brain functional network dynamics in Parkinson’s disease. Brain 140 (11), 2955–2967. 10.1093/brain/awx233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kleifges K, Bigdely-Shamlo N, Kerick SE, Robbins KA, 2017. BLINKER: automated extraction of ocular indices from EEG enabling large-scale analysis. Front. Neurosci 11, 12. 10.3389/fnins.2017.00012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koshiyama D, Thomas ML, Miyakoshi M, Joshi YB, Molina JL, Tanaka-Koshiyama K, Sprock J, Braff DL, Swerdlow NR, Light GA, 2021. Hierarchical pathways from sensory processing to cognitive, clinical, and functional impairments in schizophrenia. Schizophr. Bull 47 (2), 373–385. 10.1093/schbul/sbaal16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kriegeskorte N, Mur M, Bandettini P, 2008. Representational similarity analysis - connecting the branches of systems neuroscience. Front. Syst. Neurosci 24 (2), 4. 10.3389/neuro.06.004.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar S, Yoo K, Rosenberg MD, Scheinost D, Constable RT, Zhang S, Li CR, Chun MM, 2019. An information network flow approach for measuring functional connectivity and predicting behavior. Brain Behav. 9 (8), e01346 10.1002/brb3.1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- La Joie R, Perrotin A, Barré L, Hommet C, Mézenge F, Ibazizene M, Camus V, Abbas A, Landeau B, Guilloteau D, de La Sayette V, Eustache F, Desgranges B, Chételat G, 2012. Region-specific hierarchy between atrophy, hypometabolism, and β-amyloid (Aβ) load in Alzheimer’s disease dementia. J. Neurosci 32 (46), 16265–16273. 10.1523/JNEUROSCI.2170-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Labbe TP, Zurita M, Montalba C, Ciampi EL, Cruz JP, Vasquez M, Uribe S, Crossley N, Cárcamo C, 2020. Social cognition in multiple sclerosis is associated to changes in brain connectivity: a resting- state fMRI study. Mult. Scler. Relat. Disord 45, 102333 10.1016/j.msard.2020.102333. [DOI] [PubMed] [Google Scholar]
- Lam RW, Milev R, Rotzinger S, Andreazza AC, Blier P, Brenner C, Daskalakis ZJ, Dharsee M, Downar J, Evans KR, Farzan F, Foster JA, Frey BN,Geraci J, Giacobbe P, Feilotter HE, Hall GB, Harkness KL, Hassel S,Ismail Z, Leri F, Liotti M, MacQueen GM, McAndrews MP, Minuzzi L, Müller DJ, Parikh SV, Placenza FM, Quilty LC, Ravindran AV, Salomons TV, Soares CN, Strother SC, Turecki G, Vaccarino AL, Vila-Rodriguez F, Kennedy SH, 2016. CAN-BIND Investigator Team. Discovering biomarkers for antidepressant response: protocol from the Canadian biomarker integration network in depression (CAN-BIND) and clinical characteristics of the first patient cohort. BMC Psychiatry 16, 105. 10.1186/sl2888-016-0785-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson-Prior LJ, Oostenveld R, Della Penna S, Michalareas G, Prior F, Babajani-Feremi A, Schoffelen JM, Marzetti L, de Pasquale F, Di Pompeo F, Stout J, Woolrich M, Luo Q, Bucholz R, Fries P, Pizzella V, Romani GL, Corbetta M, Snyder AZ, WU-Minn HCP, 2013. Consortium. Adding dynamics to the human connectome project with MEG. Neuroimage 80, 190–201. 10.1016/j.neuroimage.2013.05.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law ZK, Todd C, Mehraram R, Schumacher J, Baker MR, LeBeau FEN,Yarnall A, Onofrj M, Bonanni L, Thomas A, Taylor JP, 2020. The role of EEG in the diagnosis, prognosis and clinical correlations of dementia with lewy bodies-a systematic review. Diagnostics (Basel) 10 (9), 616. 10.3390/diagnosticsl0090616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M, Sehatpour P, Hoptman MJ, Lakatos P, Dias EC, Kantrowitz JT, Martinez AM, Javitt DC, 2017. Neural mechanisms of mismatch negativity dysfunction in schizophrenia. Mol. Psychiatry 22 (11), 1585–1593. 10.1038/mp.2017.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Legaz A, Abrevaya S, Dottori M, Campo CG, Birba A, Caro MM, Aguirre J, Slachevsky A, Aranguiz R, Serrano C, Gillan CM, Leroi I, García AM, Fittipaldi S, Ibañez A, 2021. Multimodal mechanisms of human socially reinforced learning across neurodegenerative diseases. Brain. 10.1093/brain/awab345. Sep 16:awab345 Epub ahead of print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levin AR, Méndez, Leal AS, Gabard-Durnam LJ, O’Leary HM, 2018. BEAPP: the batch electroencephalography automated processing platform. Front. Neurosci 12, 513. 10.3389/fhins.2018.00513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li SZ, Jain A, L2 norm, 2009. Encyclopedia of Biometrics. Springer, Boston, MA. 10.1007/978-0-387-73003-5_1070 [DOI] [Google Scholar]
- Livezey JA, Bouchard KE, Chang EF, 2019. Deep learning as a tool for neural data analysis: speech classification and cross-frequency coupling in human sensorimotor cortex. PLoS Comput. Biol 15 (9), e1007091 10.1371/journal.pcbi.1007091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahjoory K, Nikulin VV, Botrel L, Linkenkaer-Hansen K, Fato MM, Haufe S, 2017. Consistency of EEG source localization and connectivity estimates. NeuroImage 152, 590–601. 10.1016/j.neuroimage.2017.02.076. [DOI] [PubMed] [Google Scholar]
- Malcolm BR, Foxe JJ, Butler JS, Mowrey WB, Molholm S, De Sanctis P, 2019. Long-term test-retest reliability of event-related potential (ERP) recordings during treadmill walking using the mobile brain/body imaging (MoBI) approach. Brain Res. 1716, 62–69. 10.1016/j.brainres.2017.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mantzavinos V, Alexiou A, 2017. Biomarkers for Alzheimer’s disease diagnosis. Curr. Alzheimer Res 14 (11), 1149–1154. 10.2174/1567205014666170203125942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashhadi N, Khuzani AZ, Heidari M, Khaledyan D, 2020. Deep learning denoising for EOG artifacts removal from EEG signals. In: Paper Presented at the 2020 IEEE Global Humanitarian Technology Conference (GHTC). [Google Scholar]
- Massimini M, Ferrarelli F, Huber R, Esser SK, Singh H, Tononi G, 2005. Breakdown of cortical effective connectivity during sleep. Science 309 (5744), 2228–2232. 10.1126/science.Ill7256. [DOI] [PubMed] [Google Scholar]
- McKhann GM, Knopman DS, Chertkow H, Hyman BT, Jack CR Jr., Kawas CH, Klunk WE, Koroshetz WJ, Manly JJ, Mayeux R, Mohs RC, Morris JC, Rossor MN, Scheltens P, Carrillo MC, Thies B, Weintraub S, Phelps CH, 2011. The diagnosis of dementia due to Alzheimer’s disease: recommendations from the National Institute on Aging-Alzheimer’s Association workgroups on diagnostic guidelines for Alzheimer’s disease. Alzheimers Dement. 7 (3), 263–269. 10.1016/j.jalz.2011.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloni M, Sedeño L, Hesse E, García-Cordero I, Mikulan E, Plastino A, Marcotti A, López JD, Bustamante C, Lopera F, Pineda D, García AM, Manes F, Trujillo N, Ibáñez A, 2015a. Cortical dynamics and subcortical signatures of motor-language coupling in Parkinson’s disease. Sci. Rep 5, 11899. 10.1038/srepl1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloni M, Sedeño L, Hesse E, García-Cordero I, Mikulan E, Plastino A, Marcotti A, López JD, Bustamante C, Lopera F, Pineda D, García AM, Manes F, Trujillo N, Ibáñez A, 2015b. Cortical dynamics and subcortical signatures of motor-language coupling in Parkinson’s disease. Sci. Rep 5, 11899. 10.1038/srepl1899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloni M, Billeke P, Baez S, Hesse E, Forno G, Birba A, García-Cordero I, Serrano C, Plastino A, Slachevsky A, Huepe D, Sigman M, Manes F, García AM, Sedeño L, Ibáñez A, 2017. Your perspective and my benefit: multiple lesion models of self-other integration strategies during social bargaining. Brain 139 (11), 3022–3040. 10.1093/brain/aww231. [DOI] [PubMed] [Google Scholar]
- Melnik A, Legkov P, Izdebski K, Kärcher SM, Hairston WD, Ferris DP, König P, 2017. Systems, subjects, sessions: to what extent do these factors influence EEG Data? Front. Hum. Neurosci 11, 150. 10.3389/fhhum.2017.00150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moezzi B, Goldsworthy MR, 2018. Commentary: consistency of EEG source localization and connectivity estimates. Front. Neurosci 12, 147. 10.3389/fhins.2018.00147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moguilner S, García AM, Perl YS, Tagliazucchi E, Piguet O, Kumfor F, Reyes P, Matallana D, Sedeño L, Ibáñez A, 2021. Dynamic brain fluctuations outperform connectivity measures and mirror pathophysiological profiles across dementia subtypes: a multicentric study. NeuroImage 225, 117522. 10.1016/j.neuroimage.2020.117522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moguilner S, García AM, Mikulan E, Hesse E, García-Cordero I, Melloni M, Cervetto S, Serrano C, Herrera E, Reyes P, Matallana D, Manes F, Ibáñez A, Sedeño L, 2018. Weighted symbolic dependence metric (wSDM) for fMRI resting-state connectivity: a multicentric validation for frontotemporal dementia. Sci. Rep 8 (1), 11181. 10.1038/s41598-018-29538-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohanty R, Sethares WA, Nair VA, Prabhakaran V, 2020b. Rethinking measures of functional connectivity via feature extraction. Sci. Rep 10 (1), 1298. 10.1038/S41598-020-57915-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy M, Riedner BA, Huber R, Massimini M, Ferrarelli F, Tononi G, 2009. Source modeling sleep slow waves. Proc. Natl. Acad. Sci. U. S. A 106 (5), 1608–1613. 10.1073/pnas.0807933106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bigdely-Shamlo N, Ibagon G, Kothe C, Mullen T, 2018. Finding the optimal cross-subject EEG data alignment method for analysis and BCI. In: 2018 IEEE International Conference on Systems, Man, and Cybernetics (SMC), pp. 1110–1115. 10.1109/SMC.2018.00196 [DOI] [Google Scholar]
- Niethammer M, Tang CC, Vo A, Nguyen N, Spetsieris P, Dhawan V, Ma Y, Small M, Feigin A, During MJ, Kaplitt MG, Eidelberg D, 2018. Gene therapy reduces Parkinson’s disease symptoms by reorganizing functional brain connectivity. Sci. Transl. Med 10 (469), eaau0713 10.1126/scitranslmed.aau0713, 28. [DOI] [PubMed] [Google Scholar]
- O’Brien PC, Dyck PJ, 1995. Procedures for setting normal values. Neurology 45 (1), 17–23. 10.1212/wnl.45.1.17. [DOI] [PubMed] [Google Scholar]
- Oliveira AS, Schlink BR, Hairston WD, König P, Ferris DP, 2016. Proposing metrics for benchmarking novel EEG technologies towards real-world measurements. Front. Hum. Neurosci 10 (10), 188. 10.3389/fnhum.2016.00188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ossenkoppele R, Cohn-Sheehy BI, La Joie R, Vogel JW, Möller C, Lehmann M, van Berckel BN, Seeley WW, Pijnenburg YA, Gorno-Tempini ML, Kramer JH, Barkhof F, Rosen HJ, van der Flier WM, Jagust WJ, Miller BL, Scheltens P, Rabinovici GD, 2015. Atrophy patterns in early clinical stages across distinct phenotypes of Alzheimer’s disease. Hum. Brain Mapp 36 (11), 4421–4437. 10.1002/hbm.22927 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pagani E, Rocca MA, De Meo E, Horsfield MA, Colombo B, Rodegher M, Comi G, Filippi M, 2020. Structural connectivity in multiple sclerosis and modeling of disconnection. Mult. Scler 26 (2), 220–232. 10.1177/1352458518820759. [DOI] [PubMed] [Google Scholar]
- Pal A, Behari M, Goyal V, Sharma R, 2021. Study of EEG microstates in Parkinson’s disease: a potential biomarker? Cogn. Neurodyn 15 (3), 463–471. 10.1007/sl1571-020-09643-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pascual-Marqui R, 2007. Instantaneous and lagged measurements of linear and nonlinear dependence between groups of multivariate time series: frequency decomposition. In: arXiv: Methodology n. pag.
- Pascual-Marqui RD, Michel CM, Lehmann D, 1994. Low resolution electromagnetic tomography: a new method for localizing electrical activity in the brain. Int. J. Psychophysiol 18 (1), 49–65. 10.1016/0167-8760(84)90014-x. [DOI] [PubMed] [Google Scholar]
- Parra MA, Baez S, Sedeño L, Gonzalez, Campo C, Santamaría-García H, Aprahamian I, Bertolucci PH, Bustin J, Camargos, Bicalho MA, Cano-Gutierrez C, Caramelli P, Cogram P, Beber BC, MLF, Chaves, Court FA, de Souza LC, Custodio N, Damian A, de la Cruz M, Diehl, Rodriguez R, Brucki SMD, Fajersztajn L, Farías GA, De Felice FG, Ferrari R, de Oliveira FF, Ferreira ST, Ferretti C, Figueredo, Balthazar ML, Ferreira, Frota NA,Fuentes P, García AM, Garcia PJ, de Gobbi, Porto FH, Duque, Peñailillo L, Engler HW, Maier I, Mata IF, Gonzalez-Billault C, Lopez OL, Morelli L, Nitrini R, Quiroz YT, Guerrero, Barragan A, Huepe D, Pio FJ, Suemoto CK, Kochhann R, Kochen S, Kumfor F, Lanata S, Miller B, Mansur LL, Hosogi ML, Lillo P, Llibre, Guerra J, Lira D, Lopera F, Comas A, Avila-Funes JA, Sosa AL, Ramos C, Snyder HM, Tarnanas I, Yokoyama J, Llibre J,Cardona JF, Possin K, Kosik KS, Montesinos R, Moguilner S, Resende EPF, Solis PCL, Ferretti-Rebustini REL, Ramirez JM, Matallana D, Mbakile-Mahlanza L, Marques, Ton AM, Tavares RM, Miotto EC, Muniz-Terrera G, Muñoz-Nevárez LA, Orozco D, Okada, de Oliveira M, Piguet O, Pintado Caipa, Piña Escudero, Schilling LP, de Oliveira M, Piguet O, Pintado, Caipa M, Piña, Escudero SD, Schilling LP, Rodrigues, Palmeira AL, Yassuda MS, Santacruz-Escudero JM, Serafim RB, Smid J, Slachevsky A, Serrano C, Soto-Añari M, Takada LT, Grinberg LT, Teixeira AL, Barbosa MT, Trépel D, Ibanez A, 2021. Dementia in Latin America: paving the way toward a regional action plan. Alzheimers Dement. 17 (2), 295–313. 10.1002/alz.12202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra MA, Baez S, Allegri R, Nitrini R, Lopera F, Slachevsky A, Custodio N, Lira D, Piguet O, Kumfor F, Huepe D, Cogram P, Bak T, Manes F, Ibanez A, 2018. Dementia in Latin America: assessing the present and envisioning the future. Neurology 90 (5), 222–231. 10.1212/WNL.0000000000004897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parra MA, Mikulan E, Trujillo N, Sala SD, Lopera F, Manes F, Starr J,Ibanez A, 2017. Brain information sharing during visual short-term memory binding yields a memory biomarker for familial Alzheimer’s disease. Curr. Alzheimer Res 14(12), 1335–1347. 10.2174/1567205014666170614163316. [DOI] [PubMed] [Google Scholar]
- Pavlov YG, Adamian N, Appelhoff S, Arvaneh M, Beste C, Bland AR,Bradford DE, Bublatzky F, Busch NA, Clayson PE, Cruse D, Czeszumski A, Dreber A, Dumas G, Ehinger B, Ganis G, He X, Benwell CSY, Hinojosa JA, Huber-Huber C, Inzlicht M, Jack BN, Johannesson M, Jones R,Kalenkovich E, Kaltwasser L, Karimi-Rouzbahani H, Keil A, König P,Kouara L, Kulke L, Ladouceur CD, Langer N, Liesefeld HR, Luque D, MacNamara A, Mudrik L, Muthuraman M, Neal LB, Nilsonne G, Niso G, Ocklenburg S, Oostenveld R, Pernet CR, Pourtois G, Ruzzoli M, Sass SM, Schaefer A, Senderecka M, Snyder JS, Tamnes CK, Tognoli E, van Vugt MK, Verona E, Vloeberghs R, Welke D, Wessel JR, Zakharov I, Mushtaq F, 2021. #EEGManyLabs: Investigating the replicability of influential EEG experiments. Cortex 2. 10.1016/j.cortex.2021.03.013.S0010-9452(21)00110-6. [DOI] [PubMed] [Google Scholar]
- Perera D, Wang YK, Lin CT, Zheng J, Nguyen HT, Chai R, 2020. Statistical analysis of brain connectivity estimators during distracted driving. Annu. Int. Conf. IEEE Eng. Med. Biol. Soc 2020, 3208–3211. 10.1109/EMBC44109.2020.9176240. [DOI] [PubMed] [Google Scholar]
- Pernet CR, Appelhoff S, Gorgolewski KJ, Flandin G, Phillips C, Delorme A, Oostenveld R, 2019. EEG- BIDS, an extension to the brain imaging data structure for electroencephalography. Sci. Data 6 (1), 103. 10.1038/s41597-019-0104-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pernet CR, Chauveau N, Gaspar C, Rousselet GA, 2011. LIMO EEG: a toolbox for hierarchical Linear Modeling of ElectroEncephaloGraphic data. Comput. Intell. Neurosci, 831409 10.1155/2011/831409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietto M, Parra MA, Trujillo N, Flores F, García AM, Bustin J, Richly P,Manes F, Lopera F, Ibáñez A, Baez S, 2016. Behavioral and electrophysiological correlates of memory binding deficits in patients at different risk levels for Alzheimer’s disease. J. Alzheimers Dis 53 (4), 1325–1340. 10.3233/JAD-160056. [DOI] [PubMed] [Google Scholar]
- Pini L, Jacquemot C, Cagnin A, Meneghello F, Semenza C, Mantini D, Vallesi A, 2020. Aberrant brain network connectivity in presymptomatic and manifest Huntington’s disease: a systematic review. Hum. Brain Mapp 41 (1), 256–269. 10.1002/hbm.24790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Gutierrez P, Martínez-Montes E, Weinstein A, Zañartu M, 2019a. Estimation of auditory steady- state responses based on the averaging of independent EEG epochs. PLoS One 14 (1), e0206018. 10.1371/journal.pone.0206018, 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prado-Gutiérrez P, Otero M, Marténez-Montes E, Weinstein A, Escobar MJ, El-Deredy W, Zañartu M, 2019b. A method for tracking the time evolution of steady-state evoked potentials. J. Vis. Exp 147 10.3791/59898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radhu N, Garcia, Dominguez L, Farzan F, Richter MA, Semeralul MO, Chen R, Fitzgerald PB, Daskalakis ZJ, 2015. Evidence for inhibitory deficits in the prefrontal cortex in schizophrenia. Brain 138 (Pt 2), 483–497. 10.1093/brain/awu360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radüntz T, 2018. Signal quality evaluation of emerging EEG devices. Front. Physiol 14, 9–98. 10.3389/fphys.2018.00098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramsey JD, Hanson SJ, Hanson C, Halchenko YO, Poldrack RA, Glymour C, 2010. Six problems for causal inference from fMRI. NeuroImage 49, 1545–1558. 10.1016/j.neuroimage.2009.08.065. [DOI] [PubMed] [Google Scholar]
- Ramsey JD, Hanson SJ, Glymour C, 2011. Multi-subject search correctly identifies causal connections and most causal directions in the DCM models of the Smith et al. simulation study. Neuroimage 58, 838–848. [DOI] [PubMed] [Google Scholar]
- Reid AT, Headley DB, Mill RD, Sanchez-Romero R, Uddin LQ, Marinazzo D, Lurie DJ, Valdés-Sosa PA, Hanson SJ, Biswal BB, Calhoun V, Poldrack RA, Cole MW, 2019. Advancing functional connectivity research from association to causation. Nat. Neurosci 22 (11), 1751–1760. 10.1038/s41593-019-0510-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues J, Andrade A, 2014. Lag-based effective connectivity applied to fMRI: a simulation study highlighting dependence on experimental parameters and formulation. NeuroImage 89 (358), 377. 10.1016/j.neuroimage.2013.10.029. [DOI] [PubMed] [Google Scholar]
- Rosenberg MD, Hsu WT, Scheinost D, Todd, Constable R, Chun MM, 2018. Connectome-based models predict separable components of attention in novel individuals. J. Cogn. Neurosci 30 (2), 160–173. 10.1162/jocn_a_01197. [DOI] [PubMed] [Google Scholar]
- Salamone PC, Legaz A, Sedeño L, Moguilner S, Fraile-Vazquez M, Campo CG, Fittipaldi S, Yoris A, Miranda M, Birba A, Galiani A, Abrevaya S, Neely A, Caro MM, Alifano F, Villagra R, Anunziata F, Okada, de Oliveira M, Pautassi RM, Slachevsky A, Serrano C, García AM, Ibañez A, 2021. Interoception primes emotional processing: multimodal evidence from neurodegeneration. J. Neurosci 41 (19), 4276–4292. 10.1523/JNEUROSCI.2578-20.2021, 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salamone PC, Esteves S, Sinay VJ, García-Cordero I, Abrevaya S, Couto B, Adolfi F, Martorell M, Petroni A, Yoris A, Torquati K, Alifano F, Legaz A, Cassará FP, Bruno D, Kemp AH, Herrera E, García AM, Ibáñez A,Sedeño L, 2018. Altered neural signatures of interoception in multiple sclerosis. Hum. Brain Mapp 39 (12), 4743–4754. 10.1002/hbm.24319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanchez-Romero R, Cole MW, 2021. Combining multiple functional connectivity methods to improve causal inferences. J. Cogn. Neurosci 33 (2), 180–194. 10.1162/jocn_a_01580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedeño L, Piguet O, Abrevaya S, Desmaras H, García-Cordero I, Baez S, Alethia, de la Fuente L, Reyes P, Tu S, Moguilner S, Lori N, Landin-Romero R, Matallana D, Slachevsky A, Torralva T, Chialvo D, Kumfor F, García AM, Manes F, Hodges JR, Ibanez A, 2017. Tackling variability: a multicentric study to provide a gold-standard network approach for frontotemporal dementia. Hum. Brain Mapp 38 (8), 3804–3822. 10.1002/hbm.23627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarmukadam K, Bitsika V, Sharpley CF, McMillan MME, Agnew LL, 2020. Comparing different EEG connectivity methods in young males with ASD. Behav. Brain Res 383, 112482 10.1016/j.bbr.2020.112482. [DOI] [PubMed] [Google Scholar]
- Schaefers AT, Teuchert-Noodt G, 2016. Developmental neuroplasticity and the origin of neurodegenerative diseases. World J. Biol. Psychiatry 17 (8), 587–599. 10.3109/15622975.2013.797104. [DOI] [PubMed] [Google Scholar]
- Schumacher J, Peraza LR, Firbank M, Thomas AJ, Kaiser M, Gallagher P, O’Brien JT, Blamire AM, Taylor JP, 2019. Dysfunctional brain dynamics and their origin in lewy body dementia. Brain 142 (6), 1767–1782. 10.1093/brain/awz069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen X, Finn ES, Scheinost D, Rosenberg MD, Chun MM, Papademetris X, Constable RT, 2017. Using connectome-based predictive modeling to predict individual behavior from brain connectivity. Nat. Protoc 12 (3), 506–518. 10.1038/nprot.2016.178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sporns O, 2014. Towards network substrates of brain disorders. Brain 137 (Pt 8), 2117–2118. 10.1093/brain/awul48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun N, Mormino EC, Chen J, Sabuncu MR, Yeo BTT, 2019. Alzheimer’s disease neuroimaging initiative. Multmodal latent factor exploration of atrophy, cognitive and tau heterogeneity in Alzheimer’s disease. NeuroImage 201, 116043. 10.1016/j.neuroimage.2019.116043. [DOI] [PubMed] [Google Scholar]
- Supekar K, Menon V, Rubin D, Musen M, Greicius MD, 2008. Network analysis of intrinsic functional brain connectivity in Alzheimer’s disease. PLoS Comput. Biol 4 (6), 1000100. 10.1371/journal.pcbi.1000100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swift IJ, Sogorb-Esteve A, Heller C, Synofzik M, Otto M, Graff C, Galimberti D, Todd E, Heslegrave AJ, van der Ende EL, Van Swieten JC, Zetterberg H, Rohrer JD, 2021. Fluid biomarkers in frontotemporal dementia: past, present and future. J. Neurol. Neurosurg. Psychiatry 92 (2), 204–215. 10.1136/jnnp-2020-323520. [DOI] [PubMed] [Google Scholar]
- Tadel F, Baillet S, Mosher JC, Pantazis D, 2011. Leahy RM. Brainstorm: a user-friendly application for MEG/EEG analysis. Comput. Intell. Neurosci, 879716 10.1155/2011/879716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tagliazucchi E, Laufs H, 2014. Decoding wakefulness levels from typical fMRI resting-state data reveals reliable drifts between wakefulness and sleep. Neuron 82 (3), 695–708. 10.1016/j.neuron.2014.03.020. [DOI] [PubMed] [Google Scholar]
- Tait L, Özkan A, Szul MJ, Zhang J, 2021. A systematic evaluation of source reconstruction of resting MEG of the human brain with a new high-resolution atlas: performance, precision, and parcellation. Hum. Brain Mapp 42 (14), 4685–4707. 10.1002/hbm.25578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trivedi MH, McGrath PJ, Fava M, Parsey RV, Kurian BT, Phillips ML, Oquendo MA, Bruder G, Pizzagalli D, Toups M, Cooper C, Adams P, Weyandt S, Morris DW, Grannemann BD, Ogden RT, Buckner R,McInnis M, Kraemer HC, Petkova E, Carmody TJ, Weissman MM, 2016. Establishing moderators and biosignatures of antidepressant response in clinical care (EMBARC): rationale and design. J. Psychiatr. Res 78, 11–23. 10.1016/j.jpsychires.2016.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo-Barreto NJ, Aubert-Vázquez E, Valdés-Sosa PA, 2004. Bayesian model averaging in EEG/MEG imaging. NeuroImage 21 (4), 1300–1319. 10.1016/j.neuroimage.2003.11.008. [DOI] [PubMed] [Google Scholar]
- Trujillo LT, Stanfield CT, Vela RD, 2017. The effect of electroencephalogram (EEG) reference choice on information-theoretic measures of the complexity and integration of EEG signals. Front. Neurosci 11, 425. 10.3389/fnins.2017.00425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valdes-Sosa PA, Galan-Garcia L, Bosch-Bayard J, Bringas-Vega ML, Aubert-Vazquez E, Rodriguez-Gil I, Das S, Madjar C, Virues-Alba T, Mohades Z, MacIntyre LC, Rogers C, Brown S, Valdes-Urrutia L, Evans AC, Valdes-Sosa MJ, 2021. The Cuban human brain mapping project, a young and middle age population-based EEG, MRI, and cognition dataset. Sci. Data 8 (1), 45. 10.1038/s41597-021-00829-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Diessen E, Numan T, van Dellen E, van der Kooi AW, Boersma M, Hofman D, van Lutterveld R, van Dijk BW, van Straaten EC, Hillebrand A, Stam CJ, 2015. Opportunities and methodological challenges in EEG and MEG resting-state functional brain network research. Clin. Neurophysiol 126 (8), 1468–1481. 10.1016/j.clinph.2014.11.018 [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, 2013. WU-Minn HCP consortium. The WU-Minn human connectome project: an overview. NeuroImage 80, 62–79. 10.1016/j.neuroimage.2013.05.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Noordt S, Desjardins JA, Huberty S, Abou-Abbas L, Webb SJ, Levin AR, Segalowitz SJ, Evans AC, Elsabbagh M, 2020. EEG-IP: an international infant EEG data integration platform for the study of risk and resilience in autism and related conditions. Mol. Med 26 (1), 40. 10.1186/sl0020-020-00149-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventouras EM, Ktonas PY, Tsekou H, Paparrigopoulos T, Kalatzis I, Soldatos CR, 2010. Independent component analysis for source localization of EEG sleep spindle components. Comput. Intell. Neurosci 2010, 329436 10.1155/2010/329436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang HE, Bénar CG, Quilichini PP, Friston KJ, Jirsa VK, Bernard C, 2014. A systematic framework for functional connectivity measures. Front. Neurosci 8, 405. 10.3389/ftiins.2014.00405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuvaraj R, Murugappan M, Acharya UR, Adeli H, Ibrahim NM, Mesquita E, 2016. Brain functional connectivity patterns for emotional state classification in Parkinson’s disease patients without dementia. Behav. Brain Res 298 (Pt B), 248–260. 10.1016/j.bbr.2015.10.036. [DOI] [PubMed] [Google Scholar]
- Zeidman P, Jafarian A, Seghier ML, Litvak V, Cagnan H, Price CJ, Friston KJ, 2019. A guide to group effective connectivity analysis, part 2: second level analysis with PEB. NeuroImage 200, 12–25. 10.1016/j.neuroimage.2019.06.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zima M, Tichavský P, Paul K, Krajča V, 2012. Robust removal of short-duration artifacts in long neonatal EEG recordings using wavelet-enhanced ICA and adaptive combining of tentative reconstructions. Physiol. Meas 33 (8), N39–N49. 10.1088/0967-3334/33/8/N39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support analysis presented in Fig. 2 are available from the corresponding author upon reasonable request.
Data will be made available on request.


