Abstract
Adaptive introgression (AI) facilitates local adaptation in a wide range of species. Many state-of-the-art methods detect AI with ad-hoc approaches that identify summary statistic outliers or intersect scans for positive selection with scans for introgressed genomic regions. Although widely used, approaches intersecting outliers are vulnerable to a high false-negative rate as the power of different methods varies, especially for complex introgression events. Moreover, population genetic processes unrelated to AI, such as background selection or heterosis, may create similar genomic signals to AI, compromising the reliability of methods that rely on neutral null distributions. In recent years, machine learning (ML) methods have been increasingly applied to population genetic questions. Here, we present a ML-based method called MaLAdapt for identifying AI loci from genome-wide sequencing data. Using an Extra-Trees Classifier algorithm, our method combines information from a large number of biologically meaningful summary statistics to capture a powerful composite signature of AI across the genome. In contrast to existing methods, MaLAdapt is especially well-powered to detect AI with mild beneficial effects, including selection on standing archaic variation, and is robust to non-AI selective sweeps, heterosis from deleterious mutations, and demographic misspecification. Furthermore, MaLAdapt outperforms existing methods for detecting AI based on the analysis of simulated data and the validation of empirical signals through visual inspection of haplotype patterns. We apply MaLAdapt to the 1000 Genomes Project human genomic data and discover novel AI candidate regions in non-African populations, including genes that are enriched in functionally important biological pathways regulating metabolism and immune responses.
Keywords: adaptive introgression, machine learning, population history, archaic hominins, modern humans
Introduction
The discovery of archaic hominins, such as the Neanderthals in Western Eurasia and the mysterious Denisovans in Asia and Oceania (Browning and Browning 2007; Green et al. 2010; Reich et al. 2010, 2011; Meyer et al. 2012; Prüfer et al. 2013, 2017; Deschamps et al. 2016; Simonti et al. 2016; Slon et al. 2017; Jacobs et al. 2019; Viola et al. 2019; Choin et al. 2021; Larena et al. 2021), is one of the most important scientific findings in human evolution over the last century. The high-quality ancient genomes from both Neanderthals and Denisovans (Meyer et al. 2012; Prüfer et al. 2013, 2017) further revealed that our ancestors not only overlapped with the archaic hominins in space and time during Out-of-Africa migrations, but also interbred with them, through a process known as archaic introgression. Subsequent work has shown that the genomic variants from archaic hominins played a key role in shaping the phenotypic and genotypic landscapes observed in modern humans (Vernot and Akey 2014; Deschamps et al. 2016; Gittelman et al. 2016; Juric et al. 2016; Wall and Brandt 2016; Ahlquist et al. 2021), through adaptive introgression (AI). AI refers to a process by which adaptation occurs via genetic variants that were introgressed into the modern (recipient) population from the archaic (donor) population (Dannemann et al. 2016; Racimo et al. 2017; Burgarella et al. 2019). Currently, there is evidence of AI in modern humans from both Neanderthals and Denisovans in worldwide populations (Browning and Browning 2007; Ding et al. 2013; Sankararaman et al. 2014, 2016; Vernot and Akey 2014; Dannemann and Kelso 2017; Racimo et al. 2017; Xu et al. 2017; Chen et al. 2020), related to the adaptation to UV radiation (Hider et al. 2013; Sankararaman et al. 2014, 2016; Gittelman et al. 2016; Dannemann and Racimo 2018), cold climate (Racimo et al. 2016; Dannemann and Racimo 2018), infectious diseases (Mendez et al. 2012, 2013; Choin et al. 2021), and high altitude environments (Peng et al. 2011; Huerta-Sánchez et al. 2014; Hackinger et al. 2016; Lu et al. 2016; Witt and Huerta-Sánchez 2019; Zhang et al. 2021, 2022). Outside of modern humans, AI also has been observed in a wide range of organisms, including plants (maize, Arabidopsis), invertebrates (Drosophila, butterfly), and vertebrates (mice, fish) (Song et al. 2011; Payseur and Rieseberg 2016; Schumer et al. 2018; Burgarella et al. 2019).
The traditional methodology to detect AI typically relies on the “outlier approach”. Current implementations typically take on one of two flavors. The most commonly used method is to infer genome-wide signals of positive selection and introgressed ancestry separately, and then classify regions that are outliers for both attributes as targets of AI (Browning and Browning 2007; Sankararaman et al. 2014, 2016; Vernot and Akey 2014; Gittelman et al. 2016; Wall and Brandt 2016; Racimo et al. 2017; Chen et al. 2020). Alternatively, one can use standalone summary statistics that capture signatures of AI (Green et al. 2010; Durand et al. 2011; Martin et al. 2014; Racimo et al. 2017). If a genomic region is an outlier for one or two of such summary statistics, it would be identified as an AI candidate region.
Despite their wide use, both implementations of outlier approaches suffer from a series of issues that compromise power and precision. Different methods are typically optimized for application in specific scenarios, and thus, differ in their power. When applied to data, regions that stand out for one statistic may not overlap with the outlier signals from other methods. Therefore, intersecting outliers from different methods can lead to a high false-negative rate. This may particularly be an issue for the inference of archaic AI in modern humans (supplementary Table S1, Supplementary Material online), as the methods for detecting positive selection are generally more powerful at detecting recent sweep events, whereas archaic introgression occurred over more ancient time scales. The standalone statistics, on the other hand, are particularly prone to high false-positive rates due to non-adaptive mechanisms compromising the null distributions for AI (Harris and Nielsen 2016; Kim et al. 2018; Zhang et al. 2020). For example, recessive deleterious variants may accumulate privately in isolated populations. Once admixture occurs, their fitness effects become masked in hybrid individuals, leading to a heterosis effect, where introgressed ancestry increases in frequency in the absence of positive selection. Previous works (Kim et al. 2018; Zhang et al. 2020) suggest that the false positives may particularly be magnified in genomic regions with high exon density and low recombination rate, due to the elevated levels of recessive deleterious mutations leading to heterosis effects in such regions upon introgression.
In addition to challenges related to the population genetic signals of AI, genome-wide scans for selection face several statistical challenges as well. One major challenge with developing genome-wide inference tools is that the genomic regions containing the signature of interest typically represent a small proportion of the genome, compared to the proportion of genomic regions not containing the signatures. Therefore, the highly imbalanced ratio of a few true positives in a background of true negatives can easily lead to a high false discovery rate (FDR) due to multiple testing (Benjamini and Hochberg 1995; Storey and Tibshirani 2003), even if a method has high power and a nominally low false-positive rate (FPR). In addition, genome-wide inference methods to detect selection often have low power due to the presence of various confounding factors, combined with the fact that most of the signatures are mild and hard to distinguish from the genomic background.
With the rapid emergence of genomic data, machine learning (ML) and deep learning-based methods have recently been increasingly applied to the study of population genomics (Schrider and Kern 2018). Recent applications of ML include the inference of selective sweeps (Schrider and Kern 2016, 2018; Sugden and Ramachandran 2016; Schrider et al. 2018), archaic ancestry (Sankararaman et al. 2014; Durvasula and Sankararaman 2019, 2020), population demographic models (Sheehan and Song 2016; Wang et al. 2021), and recombination rates (Chan et al. 2018; Adrion et al. 2020). For the detection of AI, however, the application of ML is still in its infancy. So far, only one study (Gower et al. 2021) has presented a deep learning method called genomatnn. This method is trained using genomic haplotype images and shows high accuracy, but is computationally expensive. Furthermore, a key challenge for ML and deep learning methods is that the underlying model is unknown. Therefore, the deterministic mechanism for the trained model remains a black box. Here, we address this issue by using biologically meaningful features in the model, and use a decision tree-based algorithm so that the importance of all features in making predictions can be retrieved.
In this paper, we present MaLAdapt, a novel ML-based method for detecting AI in whole-genome sequencing data that is generalizable to organisms with a known demographic history. Here, we show its application to modern humans. MaLAdapt utilizes a decision tree-based model called ExtraTreeClassifiers (ETC) (Geurts et al. 2006) as its main algorithm and shows high power and high precision at detecting AI signals at 50kb resolution across the whole genome. MaLAdapt infers AI signatures through a large composite of biologically meaningful population genetic statistics, which addresses a key challenge that it is hard to get mechanistic insights from ML/deep learning predictions. MaLAdapt outperforms existing methods for detecting AI, especially, given highly imbalanced class ratios and their performance, and is robust to demographic misspecifications and other confounding mechanisms such as recessive deleterious mutations and positive selection unrelated to introgression. By applying MaLAdapt to empirical human genetic variation data from the 1000 Genomes Project (1000 Genomes Project Consortium et al. 2015), we discover targets of AI in all non-African human populations by both Neanderthals and Denisovans that were previously undetected. We additionally present a pre-trained version of MaLAdapt optimized for modern human applications, as well as the simulation and ML pipeline scripts that enable the application of MaLAdapt to non-human organisms with different genomic structures and demographic histories.
Results
Overview of MaLAdapt
MaLAdapt is a supervised Machine Learning method for detecting genome-wide Adaptive Introgression, currently optimized at detecting AI from archaic hominins in non-African modern human populations (fig. 1). The goal of MaLAdapt is to predict whether AI has occurred in a given 50 kb genomic window. Essentially, this is a binary classification problem where each window can be classified as “AI” versus “non-AI”. The window length was chosen to capture the mean length of archaic introgressed haplotypes in humans (>44 kb) (Prüfer et al. 2013) (see Material and methods). The underlying ML model for MaLAdapt is a decision tree-based algorithm called the Extra-Tree Classifier (ETC) (Geurts et al. 2006), which creates a hierarchical structure of numerous randomized decision trees that each takes a subset of features computed per 50 kb window. We chose ETC over other commonly used ML algorithms for its highest power and precision (supplementary fig. S1, Supplementary Material online). The model further implements a prediction probability that fits the joint prediction of all decision trees. MaLAdapt relies on the genomic sequence and knowledge of the demographic history of a donor population, a putatively non-introgressed outgroup population, and a recipient population that experienced introgression from the donor population.
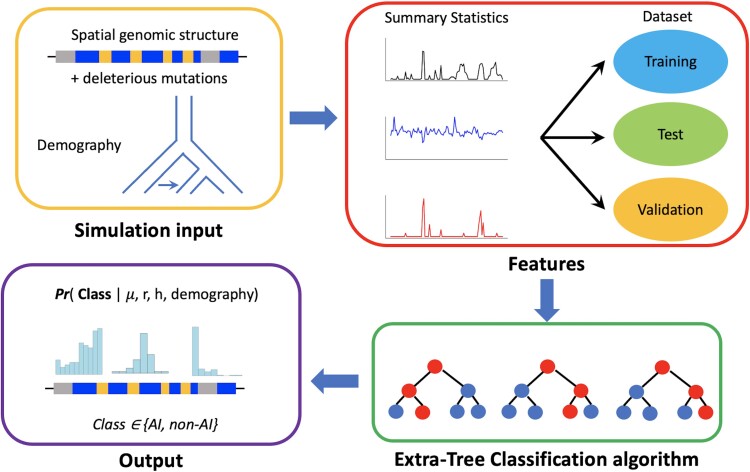
Fig. 1.
Schematic overview of the MaLAdapt workflow. To train MaLAdapt, we simulate 1000 randomly sampled genomic segments of 5MB length with a realistic genic structure, recombination rate, and distribution of deleterious mutations under modern human demography with archaic adaptive introgression (AI). We extract summary statistics in sliding 50kb-windows as features and train a hierarchical decision tree algorithm (ETC) with data labeled with binary AI and non-AI classes. After comprehensive model optimization, testing, and feature selection (supplementary figs. S4–S5, Supplementary Material online), we apply the trained model to empirical modern human genomic data to predict AI candidates.
The ETC model is trained using labeled simulation data obtained from forward-in-time simulations in SLiM (Haller and Messer 2018) of 5MB genomic segments with genic structure and recombination rates sampled from the empirical human genome under a modern Eurasian demographic model that experienced a single pulse of archaic introgression. In each simulation with AI, an adaptive mutation with a selection coefficient drawn from a prior distribution arises and becomes fixed in the archaic population prior to introgression and becomes adaptive in the recipient Eurasian population. We vary the number of generations until the selection starts (See Material and methods, fig. 2, and supplementary Table S2, Supplementary Material online).
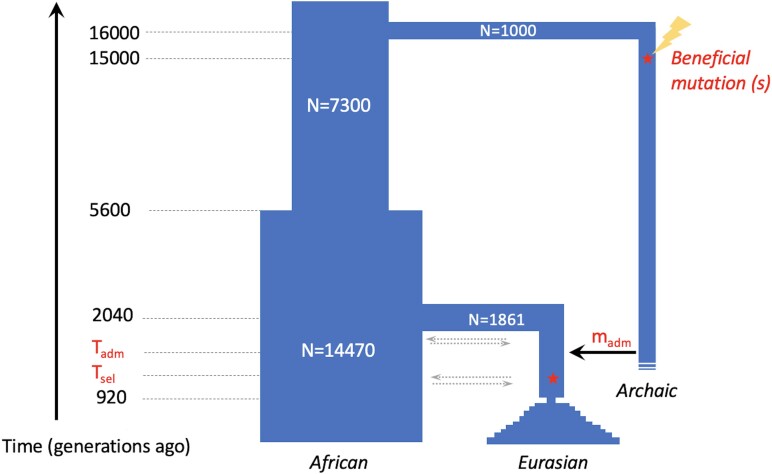
Fig. 2.
Demographic model used in simulations for MaLAdapt. We simulated an ancestral human population that diverged into an archaic human population and an ancestral African population. The latter population subsequently split into a Eurasian population and experienced two bottleneck events, representing Out-of-Africa migrations and a European–Asian split, followed by an exponential growth. Sometime between the two bottleneck events, the Eurasian population experienced a single pulse of archaic introgression at a varying time and amount, which introduced a mutation that later became beneficial in the Eurasian population. See supplementary Table S2, Supplementary Material online for the full range of simulation parameters.
Features or summary statistics (supplementary Table S3, Supplementary Material online), are computed in 50 kb sliding windows across the 5MB region. Therefore, each genomic variant is predicted five times in sliding windows. Furthermore, given that only 5 of such 50 kb sliding windows would encompass the beneficial mutation, the ratio between the “AI” window and the “non-AI” window across a 5MB segment is approximately 1:100. We divided the simulated segments into training data (90% of all simulations) and testing data (10% of all simulations), so that the testing data are not observed by the model during training, keeping the approximately 1:100 ratio of AI to non-AI segments. In order to optimize the tradeoff between model accuracy and computational efficiency, we downsampled the training data by randomly discarding non-AI windows uniformly across all segments and all replicates, and achieved an approximate 1:2 ratio between the AI and non-AI classes. Using this 1:2 class ratio has little effect on performance (supplementary fig. S2–3, Supplementary Material online), while reducing training time nearly 100-fold compared to the 1:100 class ratio. The non-AI labels in the training data were simulated under the same demographic model that included segments with deleterious mutations and some simulations that included positive selection not related to AI (see Material and methods). The performance of the trained model is evaluated by comparing against other ML algorithms and methods based on the existing AI summary statistics. For standalone statistics-based methods, we used the percentile ranking of the statistic to determine outliers. For methods that yield a prediction probability, we assigned the highest prediction value among all alleles tested within each window.
MaLAdapt Accurately Detects Adaptive Introgression
We first test the accuracy of MaLAdapt on simulated full-5MB genomic segments. The testing dataset is simulated separately using the same range of parameters as the training data (fig. 2), and keeping the 1:100 class ratios (i.e., the proportion of sliding 50 kb windows with and without the introgressed beneficial allele) between AI and non-AI. MaLAdapt predicts the AI class (AI vs. non-AI) for each 50 kb window and returns a prediction probability, which is the mean predicted class probability of all decision trees created by the ETC algorithm. We define true or false positives as whether MaLAdapt predicts AI in a given 50 kb window that contains the beneficial mutation. A window can be predicted as AI if its prediction probability is above a certain threshold. Since various thresholds can be used, we summarize performance using Receiver Operator Characteristic (ROC) and Precision-Recall curves (fig. 3), in which we visualize the True Positive Rate (TPR), FPR, Precision (equivalent to 1-FDR), and recall (equivalent to TPR) at varying thresholds. Figure 3 shows two curves for MaLAdapt in red and blue colors, which represent the accuracy of MaLAdapt at detecting AI and non-AI, respectively.
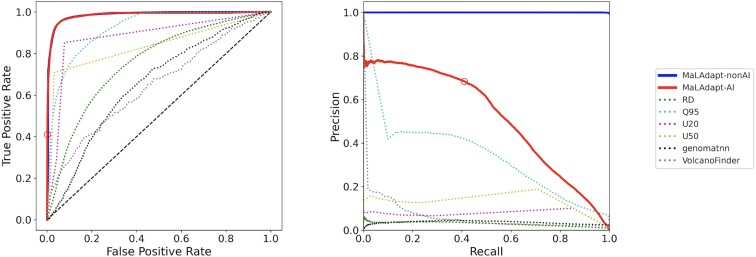
Fig. 3.
Accuracy of MaLAdapt and comparison to related methods. We applied MaLAdapt and other existing AI detection methods to the same set of testing data where the spatial structure of 5MB genomic segments is preserved and the class ratio between AI and non-AI is 1:100, mirroring the highly imbalanced AI class ratio likely present in the empirical human genome. We plot Receiver Operator Characteristic (ROC, left panel) and Precision-Recall (PR, right panel) curves for the prediction probabilities of MaLAdapt AI class (red solid), non-AI class (blue solid), and other AI signature statistics including RD (green dotted), Q95 (turquoise dotted), U20 (pink dotted), U50 (yellow dotted), genomatnn (black dotted), and VolcanoFinder (gray dotted) on the same testing data obtained from figure 2 demography. The red circle corresponds to the MaLAdapt AI prediction threshold of 0.9.
We compare the accuracy of MaLAdapt to other state-of-the-art methods for detecting AI by applying all methods to the same testing dataset we obtained from the three-population archaic AI model. We focus on outlier approaches based on: 1) the RD (average sequence divergence ratio between recipient and donor populations), 2) Q95 (95% quantile of the frequency distribution of uniquely shared derived allele between recipient and donor populations), 3) U20 and 4) U50 (number of uniquely shared derived alleles with a frequency above 20% or 50%, respectively) summary statistics (Racimo et al. 2017). Note that these statistics are themselves used as features in MaLAdapt, and below, we refer to each standalone outlier-based test by the corresponding statistic name. We also compared performance with: 5) the genomatnn (Gower et al. 2021) method, a deep learning-based method for detecting AI leveraging haplotype structure information, as well as 6) VolcanoFinder (Setter et al. 2020), a reference-free method for predicting AI using genomic polymorphic data. We show that across all prediction probability thresholds, MaLAdapt outperforms all other methods by showing the highest power while maintaining the highest precision and the lowest FPR (fig. 3, supplementary Table S4–S5, Supplementary Material online). We reject the null hypothesis that the difference in AUROC between MaLAdapt (when predicting AI) and Q95—the second best-performing method—is zero with a P-value < 2.2e-16 via jackknife, and we reject the null hypothesis that the difference in AUPR between MaLAdapt and Q95 is zero with a P-value = 1.438e-7 via jackknife (Kunsch 1989). Thus, we conclude that MaLAdapt's improvement of power and precision over other methods is statistically significant. We note a substantial reduction of accuracy in both VolcanoFinder and genomatnn when applied to testing data generated under the model in figure 2, while MaLAdapt performs reasonably well on testing data simulated under the model used by genomatnn (supplementary fig. S6, Supplementary Material online). Overall, we believe several key differences between genomatnn, VolcanoFinder, and MaLAdapt may explain their reduced performance on our simulation data, including the complexity of underlying models considered by different methods (See Discussion).
We weigh both the ROC and Precision-Recall curve to determine a prediction probability threshold for calling AI segments that maximizes the power and precision of MaLAdapt. We show in figure 3 that at Pr(AI) = 0.9 (i.e., Pr (non-AI) = 0.1), the precision of MaLAdapt is 0.683 (FDR = 0.317), with a recall (TPR) of 0.410, and FPR at 0.001. At this threshold, MaLAdapt outperforms all other related methods, especially in the precision-recall curve, showing MaLAdapt's outstanding ability to account for the highly imbalanced ratio between AI and non-AI classes, which is 1:100 in testing data. Pr(non-AI) = 0.1 can also be justified as a multiple testing problem: in sliding 50 kb windows, each locus is scanned five times. For the five windows that overlap with a given allele, we treat each window as an independent test. After multiple testing corrections, a significant value for a window being AI (i.e., not being non-AI) should be the default probability threshold, which is 0.5, divided by 5.
Robustness to Misspecification of Model Parameters
Next, we assessed the sensitivity of MaLAdapt to uncertainty and misspecification of the demographic and AI parameters. In the training process, most parameters related to AI, including the time of introgression (Tadm), the time of selection (Tsel), selection coefficient (s), and introgression amount (m), are drawn from uniform distributions (see Material and methods). Additionally, we simulated 1000 randomly sampled genomic segments of 5MB to represent the genic structure and recombination rate distribution for the empirical human genome. The rest of the demography uses a model based on the evolution of modern Eurasians (Gravel et al. 2011) with a pulse of archaic introgression (Prüfer et al. 2017).
To determine the robustness of MaLAdapt to model misspecification, we perturb the key AI-related parameters one at a time. For each alternative parameter, we simulate new testing data of 5MB genomic segments (100 replicates per parameter), and we apply MaLAdapt trained on the original model to the new testing data and evaluate its accuracy. Specifically, we ask how MaLAdapt performs when: 1) The lower bound of Tsel distribution is 200 generations lower (410 generations ago; denoted as “Tsel_low”); 2) The introgression fraction (m) is 2-fold lower than the original lower bound (at 0.5%; denoted as “m_low”); 3) The introgression fraction (m) is 2-fold higher than the original upper bound (at 20%; denoted as “m_high”); 4) the selection coefficient (s) is 10-fold higher than the original upper bound (0.1; denoted as s_high); 5) the genomic segments sampled for generating testing data are different from the ones used in the training process (denoted as “segment”); and 6) the Eurasian population growth rate and Out-of-Africa bottleneck size are different than in the training simulations (denoted as “demo”). We did not explore the selection coefficient (s) being smaller than the original lower bound (1e-4) because, with such weak selection, it would be difficult to generate AI simulations without the beneficial mutation being lost in the recipient population. Nevertheless, MaLAdapt maintains a high precision across all selection strength ranges simulated (supplementary fig. S7a, Supplementary Material online), suggesting robustness to the specific value of s. We also did not perturb the time of introgression (Tadm) because the range of Tadm is bounded by the split time between Eurasians and ancestral Africans, as well as the split time between Europeans and Asians.
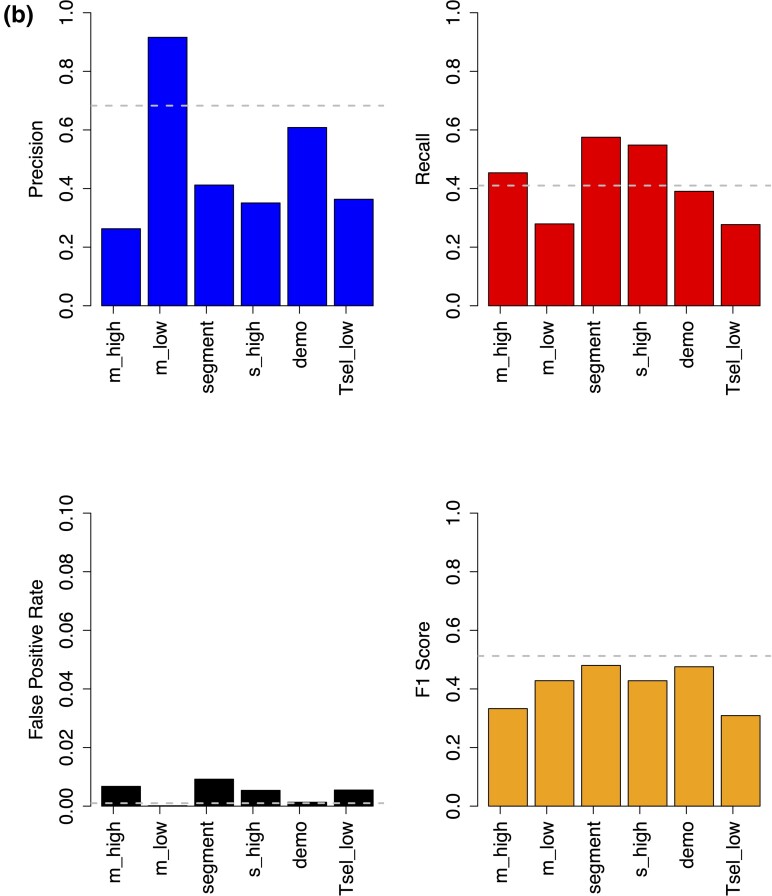
In addition to Precision, Recall (TPR), and FPR, we also computed the F1 score as an accuracy metric. F1 is defined as the weighted average between Precision and Recall (Methods). We evaluate the performance of MaLAdapt at the five alternative parameter combinations listed above by computing the log10-fold change of each accuracy metric when comparing against values obtained from using the original testing data (fig. 4a-b). We find that MaLAdapt remains robust to misspecification of AI model parameters (fig. 4). It is also worth noting that the precision of AI detection was only slightly affected when the selection time is recent (as low as 410 generations/10,000 years ago), representing selection on standing archaic variation in very recent times (Peter et al. 2012; Jagoda et al. 2017; Zhang et al. 2021). Furthermore, performance remained high when the introgression amount is low, representing a low initial frequency of archaic variants. These observations show that MaLAdapt is particularly powerful and reliable at detecting mild, incomplete AI sweeps, which is additionally demonstrated by the fact that MaLAdapt maintains high power when the beneficial allele frequency reaches 0.8 or when the positive selection is weak (s < 0.005) (supplementary fig. S8, Supplementary Material online). MaLAdapt also shows little to moderate precision loss when the demography of the recipient population changes, as well as when the testing genomic segments are different from the training segments.
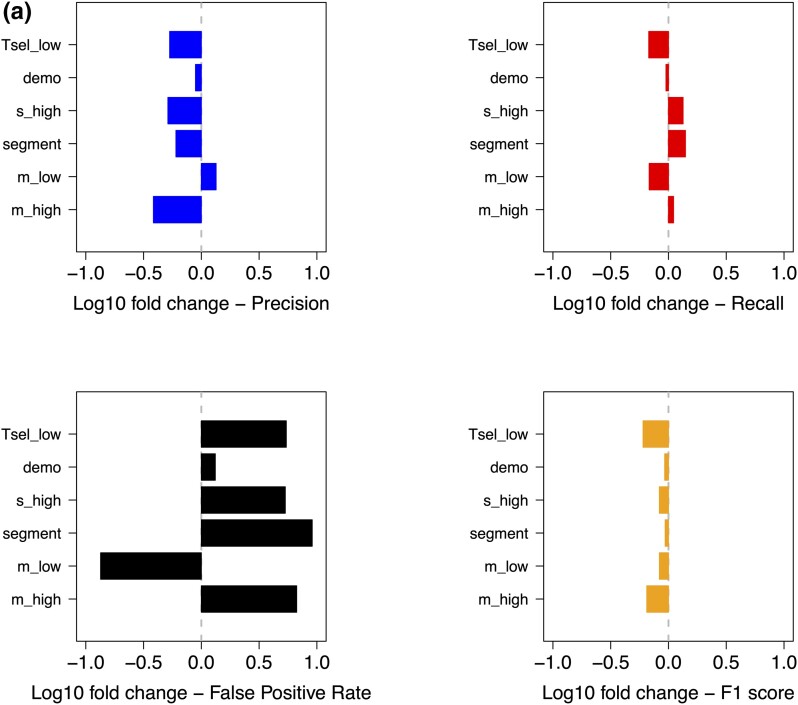
Fig. 4.
MaLAdapt is robust to model misspecification. We evaluated MaLAdapt's robustness by applying the model to testing data with key demographic and AI parameters different from those used in the training. We compute the performance metrics (including Precision, Recall, False-Positive Rate, and F1 score) and compare them against the original data under each testing scenario. Panel (a) shows the log of the value difference (testing scenario minus the original), in which a longer bar indicates a higher fold change for the given metric, and the sign of the bar indicates whether the testing metric value increases (positive) or decreases (negative). Panel (b) shows the absolute value of the performance metric under each testing scenario. Dashed lines indicate performance metrics for the baseline MaLAdapt model.
There are two parameters that, when misspecified, reduce the precision of MaLAdapt by more than 30%. These include large selection coefficients (s = 0.1, 10-fold larger than in training simulations) and high introgression fraction (m = 20%, 2-fold higher than in training simulations). Strong positive selection (s_high) led to a loss in precision since, although both FPR and TPR increased under this scenario, it inflated the FPR more than it did the TPR. A high FPR is potentially caused by falsely classifying windows nearby strong positive selection focal windows as AI. A large amount of introgression, due to a large single pulse or a combination of multiple pulses, reduces precision because it increases the FPR more than it does the TPR. On the contrary, when the amount of introgression is small, MaLAdapt calls fewer windows positive, leading both TPR and FPR to drop, consequently increasing precision. Promisingly, the weighted average of precision and recall (measured by F1) changes little with regard to any of the alternative parameters.
In addition to the AI-related parameters, we further tested MaLAdapt's accuracy on a series of demographic and genomic model-related parameter misspecifications, including back-to-Africa migration (Henn et al. 2012; Chen et al. 2020), possible modern human to Neanderthal introgression (Kuhlwilm et al. 2016; Hubisz and Siepel 2020), a multi-pulse model of archaic introgression (Browning et al. 2018; Jacobs et al. 2019; Yuan et al. 2021), a non-Eurasian demographic model (Malaspinas et al. 2016; Jacobs et al. 2019), and the simulation of genomic segments without a genic structure. For the most part, performance with the misspecified parameters is within 10-fold that of the correctly specified model (supplementary fig. S7, Supplementary Material online), suggesting some robustness to misspecification. Unmodeled migration from Europe back to Africa had the largest effect on performance. Intermediate migration rates (5e-5 to 1e-4), increased the FPR up to 30% more than that in the baseline model. However, given that the baseline model has such a low FPR (~0.001), the increase in FPR is of little practical relevance. For higher migration rates (5e-4 and 1e-3), recall can be up to 100-fold below and precision can be up to 10-fold below that of the correctly specified model. Presumably, power decreases as the migration rate increases due to the increase of sharing of archaic alleles between the outgroup and archaic population. Nevertheless, this model with high migration is likely not relevant for interpreting human data as current evidence suggests that back-to-Africa migration rates are <5e-4 (Chen et al. 2020).
Additionally, we assessed the ability of MaLAdapt to distinguish two non-AI scenarios, including positive selection unrelated to AI, and neutral introgression in lieu of beneficial or deleterious mutations. We simulated both scenarios using 1000 genomic segments that were different from those used in the training data, with the rest of the demography and parameter distributions the same as for the training data. We show in a confusion matrix (supplementary Table S6, Supplementary Material online) that MaLAdapt only mis-assigned 0.13% non-AI sweeps and 0.017% neutral introgressions, both on par or below the 0.1% FPR reported by MaLAdapt.
MaLAdapt Reveals Novel Adaptive Introgression From Neanderthals and Denisovans into Worldwide Human Populations
We computed features in 50 kb sliding windows across the human genome and predicted AI from Neanderthals and Denisovans in 19 non-African populations from the 1000 Genomes Project (1000 Genomes Project Consortium et al. 2015). In all comparisons, we use Yoruba (YRI) as the non-introgressed outgroup. We intersected the 50 kb windows predicted as AI with the GENCODE database to get lists of genes overlapping with the regions, and we merged overlapping AI windows. Here, we show Neanderthal AI in Europeans (CEU) as an example in the main text. Regions with Neanderthal AI in other populations as well as Denisovan AI are in supplementary figs. S9–S10 and supplementary Tables S7–S8, Supplementary Material online. By summarizing previously reported Neanderthal AI candidates from relevant studies and intersecting the findings from MaLAdapt, we identify novel Neanderthal AI candidates in all non-African populations, highlighted in red (fig. 5).
Fig. 5.
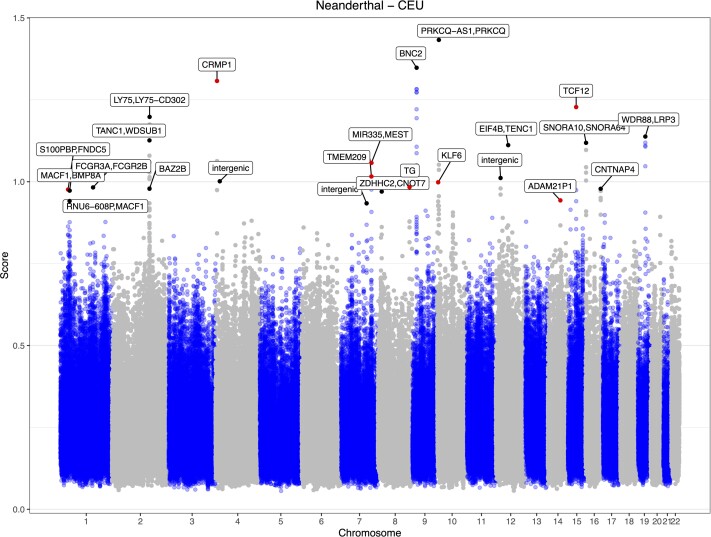
Adaptive introgression from Neanderthals in a European population (CEU). We applied MaLAdapt to predict AI in overlapping 50 kb windows (step size 10 kb) along the genome of non-African populations in the 1000 Genomes Project data. Here, we show the AI prediction results of the European population (CEU), using Yoruba (YRI) as the non-introgressed outgroup and Altai Neanderthal as the introgression donor. The Y-axis shows the AI prediction score, which equals the -Log10 transformed value of [1-Pr(AI)]. Each dot in the plot represents a 50 kb window. The windows that did not reach the MaLAdapt AI threshold are colored in blue or gray depending on the chromosomes, among which the high scoring windows reflect a mix of false negatives and windows in linkage disequilibrium with AI regions. The windows detected as AI are colored in black, if they have been reported by previous studies before, or in red, if they are novel findings from this study. The labels highlight the gene names that overlap with the AI windows.
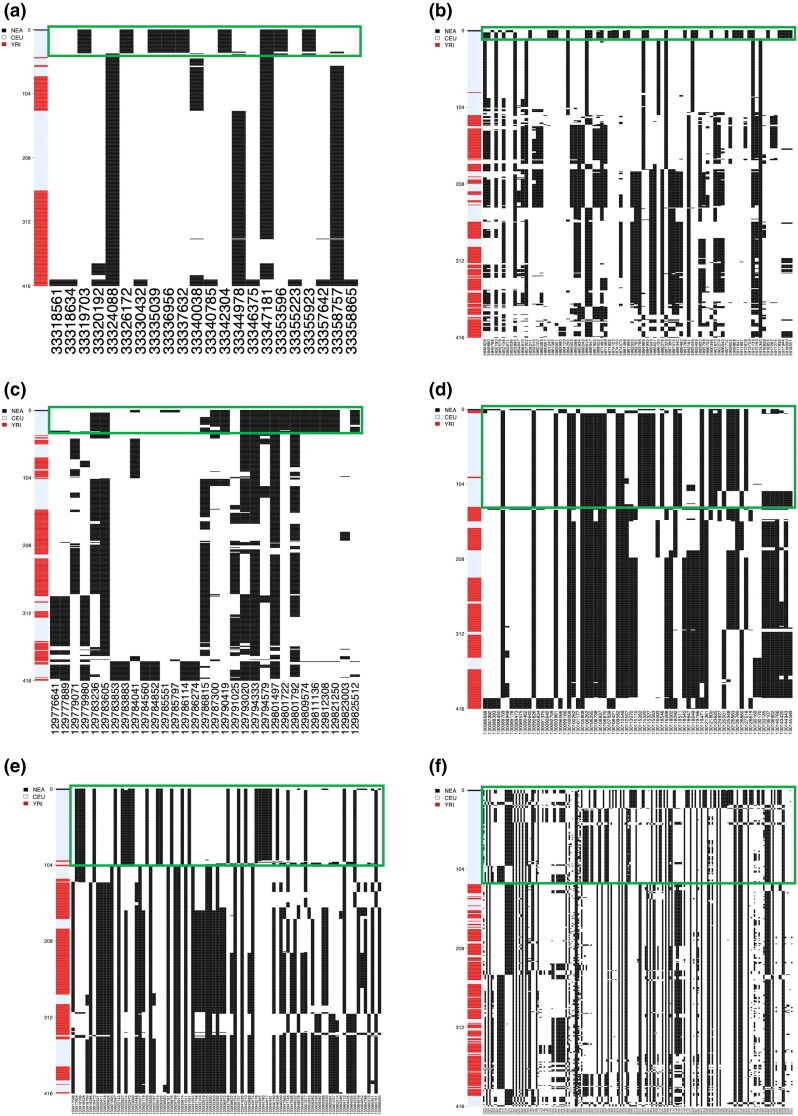
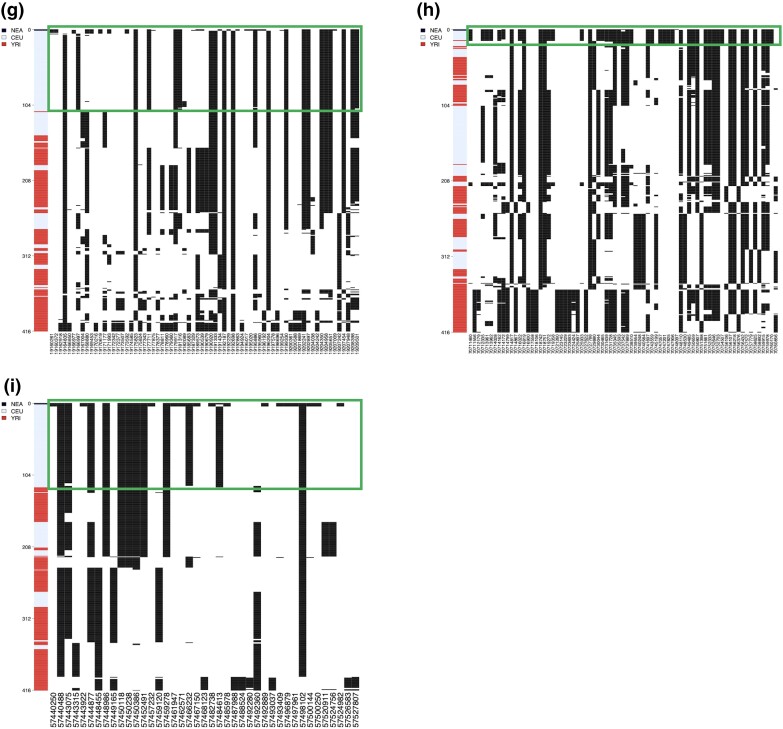
We use a two-step process to evaluate the legitimacy of the novel AI discoveries by MaLAdapt. First, we summarize the canonical hits found by previous studies (Vernot and Akey 2014; Deschamps et al. 2016; Gittelman et al. 2016; Sankararaman et al. 2016; Gouy et al. 2017; Racimo et al. 2017; Browning et al. 2018; Setter et al. 2020; Gower et al. 2021). These are defined as genes that have been reported as a target of Neanderthal AI by more than one study. MaLAdapt found 100% of the most reported hits (those seen by at least five studies). On average, MaLAdapt detected more than 50% of other repeatedly reported Neanderthal AI hits (Table 1). For the repeatedly identified hits that MaLAdapt did not detect as AI, we further examined the prediction probabilities. We found the MaLAdapt predicted Pr(AI) being no less than 0.7, suggesting that MaLAdapt found evidence of AI, despite these genes not making it over the 0.9 cutoff (supplementary fig. S11, Supplementary Material online). Next, we examined the haplotype structure of our AI candidates to visually validate the legitimacy of our hits. Under AI, we expect to see a clear block of haplotypes in the introgressed population (e.g., CEU) that has a close affinity to the archaic genome (e.g., Neanderthal). We do not expect such blocks of haplotypes to be present in the non-introgressed population (e.g., YRI) (Huerta-Sánchez et al. 2014; Marnetto and Huerta-Sánchez 2017). By this criterion, all nine newly discovered gene regions in CEU appear to be legitimate AI candidates (fig. 6, supplementary fig. S12, Supplementary Material online).
Table 1.
Previously Reported Neanderthal AI Regions Detected by MaLAdapt.
| Number of times reported as Neanderthal AI | Number of genes | Percentage of genes detected by MaLAdapt |
|---|---|---|
| 5 | 4 | 100% |
| 4 | 13 | 76.93% |
| 3 | 25 | 24.00% |
| 2 | 110 | 54.54% |
We summarize gene regions on the human genome by the number of times they have been reported by previous studies as Neanderthal AI candidates (column 1). We count the number of genes in each category (column 2), and examine the percentage of repeatedly reported AI genes that is recovered by MaLAdapt (column 3).
Fig. 6.
Haplotype structure of the novel Neanderthal AI candidate regions in the CEU. Haplotype structure of nine candidate regions predicted by MaLAdapt as AI from Neanderthals in CEU. Haplotypes of the Altai Neanderthal (black), CEU individuals (blue), and YRI individuals (red) are clustered and sorted the haplotypes by increasing distance to the Neanderthal genome. In other words, rows closer to the top of the plot represent haplotypes that are more similar to that of the Neanderthal. In the haplotype structure, each row represents a haplotype, and the column denotes a SNP (black lines indicate the presence of an alternative allele). We highlight the AI candidate haplotype block in green circles.
To examine the biological implications of AI in non-African populations, first, we performed a Gene Ontology (GO) biological process (Ashburner et al. 2000) enrichment analysis of Neanderthal AI candidates using the Enrichr tool (Chen et al. 2013; Xie et al. 2021). We combined the Neanderthal AI candidates identified by MaLAdapt in all 19 non-African populations into four super populations as defined by the 1000 Genomes study. Namely, we grouped the populations as Europeans (EUR), East Asians (EAS), South Asians (SAS), and Americans (AMR). We found that on a global level, introgressed variants from the Neanderthals played a key role in facilitating biological processes involved in metabolism regulation, adaptation to environments, and immune responses (supplementary fig. S13 and, supplementary Table S9, Supplementary Material online). Our findings do not change when population-specific recombination maps (Spence and Song 2019) were used in MaLAdapt applications (supplementary table S10, Supplementary Material online).
We compared the distribution of Neanderthal AI probabilities as predicted by MaLAdapt in genes that code for proteins that interact with RNA viruses (the VIP genes) to other genes and genomic regions. Previous work suggests that RNA viruses drove the AI between Neanderthals and modern humans (Enard and Petrov 2018). Although we find a slight enrichment of AI in VIP genes compared to non-VIP genes (supplementary figs. S14–S15, Supplementary Material online), this difference is not significant (supplementary Table S11, Supplementary Material online, Fisher's exact P-value = 0.846, odds ratio = 1.060). However, VIP genes that were reported as AI candidates (Enard and Petrov 2018) show a substantially higher AI probability in Europeans when compared to the genomic background (P-value < 2.2e–16) and other VIP genes (P-value < 2.2e–16).
Discussion
In this study, we present MaLAdapt—a ML algorithm for detecting signals of AI from genome-wide data. Compared to the existing methods, such as approaches based on standalone summary statistics, MaLAdapt has more power to detect AI, despite the challenges presented by a highly imbalanced class ratio. It is also particularly good at detecting mild, incomplete AI sweeps, and is robust to most model misspecifications and non-AI sweeps. We have applied MaLAdapt to genetic variation data from modern human populations outside of Africa, most of whose ancestral populations experienced at least one archaic introgression event. In doing so, we have discovered AI candidate regions in all non-African populations from both Neanderthals and Denisovans, including novel AI candidates that have not been reported by previous studies.
A key challenge for ML methods is that the deterministic mechanism for the trained model typically remains unknown. Here, we address this issue by using biologically meaningful features in the model, and use a decision tree-based algorithm so that the importance of all features in making predictions can be retrieved. By ranking the features by their importance scores (supplementary fig. S4, Supplementary Material online), we optimize the model by performing feature selection, and in doing so, obtain biological knowledge of AI by examining key features being used in the predictions. We show that the exon density and recombination rates played a critical role in MaLAdapt's underlying prediction mechanism, as both factors jointly determine the extent of heterosis effects (Harris and Nielsen 2016; Kim et al. 2018; Zhang et al. 2020). Additionally, summaries of genetic diversity, such as the number of segregating sites and heterozygosity, are also important factors to distinguish AI from other population genetic processes.
One major challenge in genome-wide studies of AI is that the proportion of the genome undergoing AI is likely to be substantially smaller than the part of the genome not experiencing AI, resulting in imbalanced class ratios. If the class ratio is extremely imbalanced, it can lead to an inflated FDR when performing multiple comparisons. This, of course, is a general statistical challenge in genome-wide studies. Depending on the signature of interest, different types of studies have used different strategies to account for the multiple testing issue. For example, GWAS typically use a Bonferroni correction (Tukey 1977; Bland and Altman 1995; Greenhalgh 1997) to obtain a genome-wide significant P-value threshold of 5e-8 (Risch and Merikangas 1996; International HapMap Consortium 2005; Dudbridge and Gusnanto 2008; Pe’er et al. 2008), which efficiently controls the proportion of false positives in the outstanding signals. However, it may be overly stringent and can lead to a high false-negative rate (Perneger 1998). Other ML or deep learning applications rely on the use of imbalanced datasets in the training process, followed by statistical corrections (e.g., genomatnn uses a beta correction to adjust the class ratio in training and testing data sequentially). However, the main problem with this strategy is that none of the arbitrary ratios used in the training or testing data may be close enough to the empirical ratio. Because MaLAdapt uses a hierarchically structured algorithm with numerous randomly generated decision trees, varying class ratios in the training data led to little change in the TPR and FPR (supplementary figs. S2–S3, Supplementary Material online), so long as the trained model has learned from sufficient observations of both classes as well as the confounders. To best evaluate the performance of methods on highly imbalanced empirical data, we apply MaLAdapt along with other related methods to full 5MB-long genomic segments, the class ratio of which is approximately 1:100 (i.e., 1 true positive window to 100 true negative windows). We also show that at this ratio, MaLAdapt greatly outperforms all existing methods across all thresholds in terms of Precision, Recall, and FPR (fig. 3). Even if the empirical ratio is more extreme than our testing data, all methods including MaLAdapt would suffer from a higher FDR, but MaLAdapt should still retain the highest precision among all.
Another major motivation for developing MaLAdapt is to control for potential false-positive signals due to recessive deleterious mutations in the studies of AI. It is known from multiple previous studies (Harris and Nielsen 2016; Kim et al. 2018; Zhang et al. 2020) that the presence of recessive deleterious mutations can lead to an increase in introgressed ancestry, similar to the manner of AI, and thus, is a confounder of AI detection. This effect is caused by heterosis or heterozygote advantage upon admixture, and is particularly pronounced in genomic regions that have high exon density and low recombination rates. Zhang et al. showed that existing methods for detecting AI, such as the signature summary statistics (Green et al. 2010; Durand et al. 2011; Martin et al. 2014; Racimo et al. 2017), can have exaggerated FPRs in such compact genomic regions when most deleterious mutations are recessive. This effect likely explains the AI signature in HLA and HYAL2, which have been repeatedly discovered as AI candidates in European and Asian populations (Abi-Rached et al. 2011; Ding et al. 2013).
MaLAdapt attempts to control for this potential confounder of recessive deleterious mutations by including them in the simulations used to train the classifier. However, this training process is not without challenges. Similar to the class ratio discussed above, the main challenge for the potential heterosis confounding effect is that the degree of dominance of deleterious mutations in the human genome is poorly known. Most of the studies use models that assume all mutations are either strictly additive or fully recessive, while neither of these extreme assumptions reflects the empirical distribution of dominance. In MaLAdapt, we address the uncertainty in dominance parameters by including three dominance models in the training data, which include an equal ratio of simulations where all deleterious mutations are additive, recessive, or partially recessive.
When applying MaLAdapt to empirical human population data, we do not detect HLA as an AI candidate in any of the populations. This suggests that HLA likely was a falsely identified AI candidate in previous studies (Abi-Rached et al. 2011; Ding et al. 2014; Racimo et al. 2015). However, although we did not detect AI at HYAL2 in all but one Asian population (CHB), we detected AI signatures in the upstream regions of HYAL2 that overlap with multiple genes in nine populations. A possible explanation for this observation is that the earlier reports of HYAL2 being an AI candidate could have been due to linkage to another legitimate AI region upstream of it. However, future studies of the functional changes caused by the archaic variants in this region are needed to test this hypothesis. Furthermore, it is worth noting that the novel discoveries by MaLAdapt show a similar distribution of exon density and recombination rates as previously identified AI candidates (supplementary fig. S16–19, Supplementary Material online), further supporting the conclusion that AI predictions made by MaLAdapt are not likely to be false positives due to heterosis from recessive deleterious mutations.
We show that the accuracy of MaLAdapt is significantly higher than other state-of-the-art AI detection methods. It is unsurprising that MaLAdapt outperforms the outlier methods based on summary statistics such as Q and U, as the limitations of these standalone statistics are complemented by other features incorporated in MaLAdapt. We also noticed that two of the recently developed AI methods—the deep learning-based genomatnn and the polymorphism pattern-based VolcanoFinder—both show lower power when applied to our simulation data (fig. 3). When applied to empirical human genomic data, we noticed that more than half of the candidates predicted by genomatnn as well as VolcanoFinder received low prediction probabilities by MaLAdapt (supplementary fig. S20, Supplementary Material online). There are some essential differences between MaLAdapt, genomatnn, and VolcanoFinder that may explain the differences in their accuracy. For genomatnn, it is trained on simulations of short segments (100 kb) that do not contain genic structure (coding/non-coding regions) similar to what is observed on the empirical human genome. VolcanoFinder, on the other hand, models the volcano shape of heterozygosity around a beneficial allele that is introgressed from a diverged population. This pattern is sensitive to AI but could also be influenced by other non-AI processes and the inherent characteristics of the genome, including the alignability and mappability of sequences. The simulations in our study used a considerable proportion of genomic regions with a high density of exons and low recombination rate due to concerns of the heterosis effect and background selection (Kim et al. 2018; Zhang et al. 2020). In addition, the demographic parameters differ between the methods. For example, both VolcanoFinder and genomatnn assumed a fixed introgression amount and a fixed introgression time in their models. In contrast to MaLAdapt, VolcanoFinder is also optimized to detect AI due to strong selection, whereas MaLAdapt considers weaker and recent sweeps on introgressed variants. Altogether, low power/accuracy could reflect the sensitivity of genomatnn and VolcanoFinder to misspecification of the demographic model and genomic structures used by MaLAdapt.
To further disentangle the potential causes for the discrepancy in accuracy in different methods, we examined the exon density and recombination rates in the AI candidate regions in CEU predicted by MaLAdapt, genomatnn, and VolcanoFinder (supplementary fig. S21, Supplementary Material online). The AI regions predicted by genomatnn tend to have both lower exon density and lower recombination rates than MaLAdapt and VolcanoFinder predictions, which are also lower than the whole-genome distributions. Next, we examined the haplotype structure of the genomatnn candidates using the Haplostrips program (supplementary fig. S22, Supplementary Material online) that ranks European (CEU) and African (YRI) haplotypes by their affinity to the Neanderthal genome. The genomatnn candidates that received low MaLAdapt prediction scores also did not produce a clear AI pattern through this ranking of haplotypes. This could be due to the fact that Haplostrips sorts and ranks the modern human haplotypes by the distance to the archaic reference genome, which is different from the method of haplotype sorting in genomatnn that groups haplotypes by populations. We visually inspected the haplotype structure patterns and annotated them as true positive, false positive, or uncertain labels (supplementary fig. S23, Supplementary Material online). We found that the genomatnn candidates that were not identified by MaLAdapt have strikingly lower exon densities and recombination rates compared to the other two groups. In contrast, the visually false-positive predictions by MaLAdapt are mainly driven by an excess of African (outgroup) haplotypes that also show close affinity to the archaic genome, in which case it is unclear whether they are false-positives or legitimate AI due to back-to-Africa gene flow from Europeans (Chen et al. 2020). Altogether, we believe MaLAdapt is more accurate in predicting AI in regions that contain a small number of mutations and few recombination events.
MaLAdapt can be used for the study of AI in other populations and organisms with different demographic histories and genomic structures. The simulation and training of MaLAdapt are easy to implement, computationally efficient, and modifiable for other organisms. We provide all necessary scripts to replicate our results, and users can adapt any component of our pipeline for AI applications in other organisms or other similar population genetics questions. It is important to note that MaLAdapt is the most suitable for applications in study systems where a reliable demographic model, genomic annotation, and recombination map are available. It is possible to use the MaLAdapt pipeline without one or more pieces of such information, but the robustness to non-AI processes (e.g., heterosis) may be compromised. For AI detection in humans, MaLAdapt currently relies on a well-understood Eurasian population history as its demographic model backbone. This model may not accurately describe the evolutionary history of human populations distantly related to Eurasians, such as those in the Americas. Furthermore, the current model does not account for the complex demography in some of the regional populations, especially in Asia and Oceania, where populations are known to have experienced complex archaic introgression and admixture patterns (Reich et al. 2011; Jacobs et al. 2019; Choin et al. 2021; Larena et al. 2021). However, since MaLAdapt can be easily retrained, we expect to continually revisit and revise our model when better-fitting demographic models become available. Despite the possible deficiencies of the demographic model in training simulations, MaLAdapt demonstrates its power and robustness by recovering most of the canonical AI candidates that have been reported by previous studies.
Another requirement for the use of MaLAdapt is an archaic reference genome. The empirical findings reported in this study are based on using the Altai Neanderthal individual (Prüfer et al. 2013) as the Neanderthal reference genome, and the Altai Denisovan (Meyer et al. 2012) as the Denisovan reference genome. Without a further discovery of more high-quality archaic hominin genomes, we do not have the power to detect AI from unknown, “ghost” introgressions (Chen et al. 2020; Durvasula and Sankararaman 2020) from archaic hominin groups that are distantly related to either Neanderthals or Denisovans. Nevertheless, we discovered numerous novel AI candidates in all non-African populations from Neanderthals and/or Denisovans which were undetected in previous studies, and have been verified by visual inspection of the haplotype structure (fig. 6). These genes are enriched in a range of biological pathways, shedding light on the functional influence of archaic introgression to the phenotype spectrum, local adaptation, and health in our species. We provide a comprehensive summary of AI candidates in all non-African populations, with informative annotations of studies that reported them. We hope this can serve as a useful resource for future studies to investigate open questions related to AI in humans. For example, the function and selection history of novel AI genes discovered by this study should be characterized in follow-up studies. We also observe that AI candidate loci overlap across populations. Whether this reflects a shared population history of independent AI events requires further investigation.
In conclusion, MaLAdapt provides an example of how ML, especially feature-based algorithms, can help solve complex population genetics and human genomics problems. Such ML models can particularly be powerful at tackling questions with highly imbalanced classes, mild signals, and various confounding factors. We make available the complete software and development pipeline of MaLAdapt to enable customization and improvement for future studies. We look forward to integrating new knowledge of archaic genomes and human evolutionary history into the MaLAdapt model, and to seeing novel methods for detecting AI in other biological systems inspired by MaLAdapt.
Materials and Methods
Simulation Settings
We used the software SLiM (version 3.2.0) (Haller and Messer 2018) throughout this work for simulations. We simulated introgression between archaic humans and modern humans under a three-population demographic model, shown in figure 2 and supplementary Table S2, Supplementary Material online. This demographic model is adapted from Gravel et al. 2011 and Prüfer et al. 2017. In this demography, an archaic hominin population (Narc = 1,000) splits from the ancestral African population (Nanc = 7,300) 16,000 generations ago. The ancestral African population further splits into a modern African population 5,600 generations ago (Nafr = 14,470) and a modern Eurasian population 2,040 generations ago (Neur_OoA = 1,861). The Eurasian population further experiences a population bottleneck 920 generations ago (Neur_split = 550), representing the split of European and East Asian populations, followed by a population expansion at an exponential rate of 0.55% per generation, until the end of the simulation. In simulations with AI, a beneficial mutation with a selection coefficient (s ∊[1e-4, 1e-2]) arises in an exon of the simulated genomic region 15,000 generations ago and is simulated as fixed in the archaic population by introducing mutation to all haplotypes. A single pulse of introgression occurs at a random time (Tadm ∊ in [1530, 2030]) at a random proportion (m ∊ {1%, 2%, 5%, 10%}). The introgressed beneficial mutation does not necessarily become immediately beneficial in the Eurasian population, depending on the selection time (Tsel ∊ [610, Tadm−1]). All simulations are conditioned on the introgressed beneficial mutation not being lost in the recipient Eurasian population by the end of the simulation. It is worth noting that the fixation of the beneficial allele in the donor population was strategic for computational efficiency to ensure that beneficial mutation is present in the pulse of introgression. Although no real selective sweep occurred in the donor population, this lack of diversity in the sweep region in the donor population should not affect the signature of AI in the recipient population, as only a small number of lineages from the donor was sampled.
We simulated 1,000 randomly sampled genomic regions from the modern human genome build GRCh37/hg19 with a length of 5MB. As such, the simulated segments represent the empirical distribution of exon density and recombination rates on the human genome so that the inference of MaLAdapt accounts for the confounding effects of heterosis due to recessive deleterious mutations (Zhang et al. 2020). Specifically, we use the exon ranges defined by the GENCODE v.14 annotations (Harrow et al. 2012) and the sex-averaged recombination map by Kong et al. (Kong et al. 2010) averaged over a 10 kb scale. The per base pair mutation rate was fixed at 1.08e-8. Deleterious mutations can only occur in exonic regions of the segment with fitness effects drawn from a distribution estimated from modern humans (Kim et al. 2017), with a shape parameter of 0.186 and an average selection coefficient of −0.01315, as well as a 2.31:1 ratio of nonsynonymous to synonymous mutations (Huber et al. 2017). Additionally, to account for the heterosis effect in the inference of AI while accounting for the fact that the dominance distribution of mutations in the human genome is poorly understood, we simulated three models of dominance effects. In the first model, all deleterious mutations were fully additive (h = 0.5). In the second, all were fully recessive (h = 0). In the third model, all were partially recessive (hs relationship) (Henn et al. 2016), where more strongly deleterious mutations were more likely to be recessive. For each of the sampled genomic segments, we repeated simulations 1000 times under the demography shown in figure 2 for each dominance model (deleterious mutations being additive, recessive, or partially recessive). Because there are three dominance models and 1000 sampled segments in total, this exercise resulted in 3 × 1000 × 1000 = 3 million simulation replicates.
For the computational efficiency of simulations, we scale the simulation parameters by a scaling factor of c (c = 10). In all simulations, the population size is rescaled to N/c, generation times to t/c, selection coefficient to s*c, mutation rate to μ*c, and the recombination rate to 0.5(1-(1-2r)c). Other evolutionary parameters remained the same.
Features Used by MaLAdapt
We consider biologically meaningful summary statistics that are likely informative of archaic AI (supplementary Table S3, Supplementary Material online). The untrained MaLAdapt model learns which features are most important. All statistics are calculated in Python3. For each simulation replicate, we computed features in sliding 50 kb windows (step size 10 kb) throughout the simulated segments. We used 50 kb as the prediction window size because it encompasses the average archaic introgressed haplotype length in modern humans, which is approximately 44 kb (Prüfer et al. 2013). We define “AI” as genomic windows in the admixed Eurasian population that contain beneficial mutations originating from archaic introgression. In contrast, windows with the label “non-AI” do not contain the beneficial mutation, even if such windows are on the same genomic segment as the “AI” windows. Therefore, at most, only 5 out of 496 windows per segment contain beneficial mutations.
A full list of features used by the MaLAdapt can be found in supplementary Table S3, Supplementary Material online, which includes summary statistics that are informative about archaic introgression (Green et al. 2010; Durand et al. 2011; Martin et al. 2014), positive selection (Garud et al. 2015; Racimo et al. 2017), linkage disequilibrium (Hill and Robertson 1968; Kelly 1997; Pritchard and Przeworski 2001; Slatkin 2008), genetic diversity (Crow and Kimura 1970; Nei 1973; Watterson 1975; Saitou and Nei 1987), and the genic structure and recombination rates (Kong et al. 2010; Harrow et al. 2012).
Training MaLAdapt and the Choice of the ETC Algorithm
Using features computed from all windows in all simulated replicates, we further divided the dataset into training and testing datasets at a 9:1 ratio. For the training dataset, we added additional segments containing selective sweeps due to de novo beneficial mutations. As these windows were not due to AI, these simulations were added to the “non-AI” labels. Up to 10% of the training dataset was comprised of these particular windows. In these selective sweep simulations, the beneficial mutations are de novo mutations in the Eurasian populations (arising at Tsel), rather than introduced by archaic introgression. We also shuffle the training dataset to break down the genomic structure of the segments, and we further evaluate the influence of class ratios on the performance of MaLAdapt (supplementary figs. S2–S3, Supplementary Material online). We show that in the training data, a relatively balanced class ratio optimizes the performance of MaLAdapt, as the model is trained by observing sufficient examples of both classes. Therefore, we downsize the “non-AI” labeled windows to be twice the amount of the “AI” labeled windows. The final training data contain “AI” and “non-AI” windows at approximately a 1:2 ratio. In the testing data, on the other hand, the original simulation class ratio (AI:non-AI ∼ 1:100) and genomic segment structures are preserved, because AI likely is a rare event on the human genome.
We compared the performance of five ML algorithms to be used in MaLAdapt including Logistic Regression, LASSO, Ridge, traditional Random Forest, and ETC. The algorithms are trained and tested using the same datasets as each other and are evaluated in terms of different performance metrics including the True positive rate (TPR), False-positive rate (FPR), Precision (1-False Discovery Rates), Recall (TPR), and F1 Score at different prediction probability thresholds (supplementary fig. S1, Supplementary Material online). We show that ETC is the best-performing algorithm at detecting genome-wide AI, as its hierarchical structure is optimized to detect mild AI signatures, especially when the class ratio is highly imbalanced. Therefore, we chose to use the ETC algorithm. We additionally carried out feature selection for model optimization by looking at the feature importance score ranking from the original ETC-based MaLAdapt (see Supplementary Material online text for details).
MaLAdapt Robustness and Model Misspecification Analysis
To evaluate the robustness of MaLAdapt to model misspecification, we obtained different sets of testing data that include 6 independent scenarios where one of the key parameters in the simulation model is perturbed (supplementary Table S2, Supplementary Material online). Specifically, we define 1) “Tsel_low” as the selection time being 200 generations lower than the original lower bound, 2) “m_low” as the introgression fraction (m) being 2-fold lower than the original lower bound, 3) “m_high” as the introgression fraction (m) being 2-fold higher than the original upper bound, 4) “s_high” as the selection coefficient (s) being 10-fold higher than the original upper bound, 5) “segment” as the genomic segments in simulations being different from the training data, and 6) “demo” as the Eurasian population growth rate and Out-of-Africa bottleneck size being different than the training simulations. We performed a series of additional parameter misspecifications to evaluate MaLAdapt's robustness (supplementary Methods and supplementary fig. S7, Supplementary Material online).
We applied MaLAdapt to each of the above 6 perturbed testing datasets, and computed accuracy metrics including FPR, Precision, TPR, Recall, and F1 Score with a prediction probability threshold of 0.9. We compared the metrics with the values obtained from applying MaLAdapt to the original testing dataset (without parameter perturbation), and computed the log10-fold change of the metrics to the original values.
Analysis of AI in the 1000 Genomes Data
For the application of trained MaLAdapt on empirical modern human genetic variation data, we scanned the autosomes from Phase 3 of the 1000 Genomes Project and computed the features used in supplementary Table S3, Supplementary Material online in 50 kb sliding windows (step size = 10 kb). Specifically, we first defined the genomic coordinates of the sliding 50 kb windows throughout each of the autosomes (excluding the telomere and centromere regions). Within each window, we use the start and end positions to extract the genotypes from Yoruba (YRI, phased) as the non-introgressed population/outgroup, one of the 19 non-African populations (phased) as the introgressed population/recipient group, and one of the high-quality archaic genomes (Altai Neanderthal (Prüfer et al. 2013) or Altai Denisovan ((Meyer et al. 2012), unphased) as the introgressing population/donor group. We join the genotypes together as a matrix, and additionally removed sites in the archaic genomes having potential quality issues (quality score < 40 and/or mapping quality < 30). We computed all summary statistics included in the feature set in MaLAdapt, and repeated the process across all windows across all autosomes. We computed features for Neanderthal introgression and Denisovan introgression separately for all populations. We applied the trained model to all 19 non-African populations and obtained prediction probabilities in all windows across the whole genome for Neanderthal or Denisovan AI, respectively. For windows predicted as AI that overlap with each other, we joined them as one AI region and used the boundary of the region to determine overlapping genes. We further converted the prediction probability of Pr(AI) to a prediction score, which equals -log10(1-Pr(AI)). We plot the prediction scores of all windows for each population, and label the gene names in the AI regions.
Supplementary Material
Acknowledgments
X.Z. was supported by NIH Grant K99GM143466. B.K. was supported by NIH grant F32GM135998. K.E.L. was supported by NIH Grant R35GM119856. S.S. was supported by R35GM125055 and an Alfred P. Sloan Research Fellowship. We thank Dr Graham Gower and Dr Xiaoheng Cheng for sharing scripts of genomatnn and VolcanoFinder, and providing insightful comments related to the comparison of adaptive introgression inference results from different methods. We also thank Dr David Enard for providing the VIP-related datasets for analyses in this study.
Contributor Information
Xinjun Zhang, Department of Ecology and Evolutionary Biology, UCLA, Los Angeles, CA.
Bernard Kim, Department of Biology, Stanford University, Palo Alto, CA.
Armaan Singh, Department of Computer Science, UCLA, Los Angeles, CA.
Sriram Sankararaman, Department of Computer Science, UCLA, Los Angeles, CA; Department of Computational Medicine, UCLA, Los Angeles, CA; Department of Human Genetics, David Geffen School of Medicine, UCLA, Los Angeles, CA.
Arun Durvasula, Department of Genetics, Harvard Medical School, Boston, MA.
Kirk E Lohmueller, Department of Ecology and Evolutionary Biology, UCLA, Los Angeles, CA; Department of Human Genetics, David Geffen School of Medicine, UCLA, Los Angeles, CA.
Author Contribution
X.Z., B.K., and A.D. conceived the study. X.Z. designed the study, carried out the simulations, ML implementation, empirical data analyses, and wrote the manuscript. B.K., A.D., S.S., and K.E.L. contributed to the design of the study, data analysis, and participated in manuscript writing. B.K. designed the simulation framework. A.S. participated in code optimization and ML data analysis. All authors read and approved the manuscript.
Supplementary Material
Supplementary data are available at Molecular Biology and Evolution online.
Data Availability Statement
All scripts necessary to recreate the simulations, ML training and testing, robustness analysis, and empirical predictions can be found at GitHub: https://github.com/xzhang-popgen/maladapt
Reference
- 1000 Genomes Project Consortium, Auton A, Abecasis GR, Altshuler DM, Durbin RM, Bentley DR, Chakravarti A, Clark AG, Donnelly P, Eichler EE, et al. 2015. A global reference for human genetic variation. Nature. 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, Gragert L, Babrzadeh F, Gharizadeh B, Luo M, Plummer FA, et al. 2011. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 334:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrion JR, Galloway JG, Kern AD. 2020. Predicting the landscape of recombination using deep learning. Mol Biol Evol. 37:1790–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist KD, Bañuelos MM, Funk A, Lai J, Rong S, Villanea FA, Witt KE. 2021. Our tangled family tree: new genomic methods offer insight into the legacy of archaic admixture. Genome Biol Evol. 13:evab115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 57:289–300. [Google Scholar]
- Bland JM, Altman DG. 1995. Multiple significance tests: the Bonferroni method. BMJ. 310:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 81:1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL, Zhou Y, Tucci S, Akey JM. 2018. Analysis of human sequence data reveals two pulses of archaic Denisovan admixture. Cell. 173:53–61.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgarella C, Barnaud A, Kane NA, Jankowski F, Scarcelli N, Billot C, Vigouroux Y, Berthouly-Salazar C. 2019. Adaptive introgression: an untapped evolutionary mechanism for crop adaptation. Front Plant Sci. 10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Perrone V, Spence JP, Jenkins PA, Mathieson S, Song YS. 2018. A likelihood-free inference framework for population genetic data using exchangeable neural networks. Adv Neural Inf Process Syst. 31:8594–8605. [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wolf AB, Fu W, Li L, Akey JM. 2020. Identifying and interpreting apparent Neanderthal ancestry in African individuals. Cell. 180:677–687.e16. [DOI] [PubMed] [Google Scholar]
- Choin J, Mendoza-Revilla J, Arauna LR, Cuadros-Espinoza S, Cassar O, Larena M, Ko AM-S, Harmant C, Laurent R, Verdu P, et al. 2021. Genomic insights into population history and biological adaptation in Oceania. Nature. 592:583–589. [DOI] [PubMed] [Google Scholar]
- Crow JF, Kimura M. 1970. An introduction to population genetics theory. New York, Evanston and London: Harper & Row, Publishers. [Google Scholar]
- Dannemann M, Andrés AM, Kelso J. 2016. Introgression of Neandertal- and Denisovan-like haplotypes contributes to adaptive variation in human toll-like receptors. Am J Hum Genet. 98:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemann M, Kelso J. 2017. The contribution of Neanderthals to phenotypic variation in modern humans. Am J Hum Genet. 101:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemann M, Racimo F. 2018. Something old, something borrowed: admixture and adaptation in human evolution. Curr Opin Genet Dev. 53:1–8. [DOI] [PubMed] [Google Scholar]
- Deschamps M, Laval G, Fagny M, Itan Y, Abel L, Casanova J-L, Patin E, Quintana-Murci L. 2016. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am J Hum Genet. 98:5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Hu Y, Jin L. 2014. Non-Neanderthal origin of the HLA-DPB1*0401. J Biol Chem. 289:10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Hu Y, Xu S, Wang J, Jin L. 2013. Neanderthal introgression at chromosome 3p21.31 was under positive natural selection in East Asians. Mol Biol Evol. 31:683–695. [DOI] [PubMed] [Google Scholar]
- Dudbridge F, Gusnanto A. 2008. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 32:227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 28:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durvasula A, Sankararaman S. 2019. A statistical model for reference-free inference of archaic local ancestry. PLoS Genet. 15:e1008175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durvasula A, Sankararaman S. 2020. Recovering signals of ghost archaic introgression in African populations. Sci Adv. 6:eaax5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard D, Petrov DA. 2018. Evidence that RNA viruses drove adaptive introgression between Neanderthals and modern humans. Cell. 175:360–371.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud NR, Messer PW, Buzbas EO, Petrov DA. 2015. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 11:e1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts P, Ernst D, Wehenkel L. 2006. Extremely randomized trees. Mach Learn. 63:3–42. [Google Scholar]
- Gittelman RM, Schraiber JG, Vernot B, Mikacenic C, Wurfel MM, Akey JM. 2016. Archaic hominin admixture facilitated adaptation to out-of-Africa environments. Curr Biol. 26:3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy A, Daub JT, Excoffier L. 2017. Detecting gene subnetworks under selection in biological pathways. Nucleic Acids Res. 45:e149–e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower G, Picazo PI, Fumagalli M, Racimo F. 2021. Detecting adaptive introgression in human evolution using convolutional neural networks. Elife. 10:e64669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG, Yu F, Gibbs RA, Bustamante CD. 2011. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci. 108:11983–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MHY, et al. 2010. A draft sequence of the Neandertal genome. Science. 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh T. 1997. How to read a paper. Statistics for the non-statistician. I: different types of data need different statistical tests. BMJ. 315:364–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackinger S, Kraaijenbrink T, Xue Y, Mezzavilla M, Asan van Driem G, Jobling MA, de Knijff P, Tyler-Smith C, Ayub Q. 2016. Wide distribution and altitude correlation of an archaic high-altitude-adaptive EPAS1 haplotype in the Himalayas. Hum Genet. 135:393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller BC, Messer PW. 2019. SLim 3: forward genetic simulations beyond the Wright–Fisher model. Mol Biol Evol. 36:632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K, Nielsen R. 2016. The genetic cost of Neanderthal introgression. Genetics. 203:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Gravel S, Wang W, Brisbin A, Byrnes JK, Fadhlaoui-Zid K, Zalloua PA, Moreno-Estrada A, Bertranpetit J, et al. 2012. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 8:e1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Peischl S, Dupanloup I, Lipatov M, Maples BK, Martin AR, Musharoff S, Cann H, Snyder MP, et al. 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci. 113:E440–E449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hider JL, Gittelman RM, Shah T, Edwards M, Rosenbloom A, Akey JM, Parra EJ. 2013. Exploring signatures of positive selection in pigmentation candidate genes in populations of East Asian ancestry. BMC Evol Biol. 13:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. 1968. Linkage disequilibrium in finite populations. Theor Appl Genet. 38:226–231. [DOI] [PubMed] [Google Scholar]
- Huber CD, Kim BY, Marsden CD, Lohmueller KE. 2017. Determining the factors driving selective effects of new nonsynonymous mutations. Proc Natl Acad Sci. 114:4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz M, Siepel A. 2020. Inference of ancestral recombination graphs using ARGweaver. Methods Mol Biol. 2090:231–266. [DOI] [PubMed] [Google Scholar]
- Huerta-Sánchez E, Jin X, Asan BZ, Peter BM, Vinckenbosch N, Liang Y, Yi X, He M, Somel M, et al. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 512:194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium . 2005. A haplotype map of the human genome. Nature. 437:1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs GS, Hudjashov G, Saag L, Kusuma P, Darusallam CC, Lawson DJ, Mondal M, Pagani L, Ricaut F-X, Stoneking M, et al. 2019. Multiple deeply divergent Denisovan ancestries in Papuans. Cell. 177:1010–1021. [DOI] [PubMed] [Google Scholar]
- Jagoda E, Lawson DJ, Wall JD, Lambert D, Muller C, Westaway M, Leavesley M, Capellini TD, Mirazón Lahr M, Gerbault P, et al. 2017. Disentangling immediate adaptive introgression from selection on standing introgressed variation in humans. Mol Biol Evol. 35:623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric I, Aeschbacher S, Coop G. 2016. The strength of selection against Neanderthal introgression. PLoS Genet. 12:e1006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK. 1997. A test of neutrality based on interlocus associations. Genetics. 146:1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Huber CD, Lohmueller KE. 2017. Inference of the distribution of selection coefficients for new nonsynonymous mutations using large samples. Genetics. 206:345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Huber CD, Lohmueller KE. 2018. Deleterious variation shapes the genomic landscape of introgression. PLoS Genet. 14:e1007741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB, Jonasdottir A, Gylfason A, Kristinsson KT, et al. 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 467:1099–1103. [DOI] [PubMed] [Google Scholar]
- Kuhlwilm M, Gronau I, Hubisz MJ, de Filippo C, Prado-Martinez J, Kircher M, Fu Q, Burbano HA, Lalueza-Fox C, de la Rasilla M, et al. 2016. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature. 530:429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunsch HR. 1989. The jackknife and the bootstrap for general stationary observations. Ann Stat. 17:1217–1241. [Google Scholar]
- Larena M, McKenna J, Sanchez-Quinto F, Bernhardsson C, Ebeo C, Reyes R, Casel O, Huang J-Y, Hagada KP, Guilay D, et al. 2021. Philippine Ayta possess the highest level of Denisovan ancestry in the world. Curr Biol. 31:4219–4230.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lou H, Yuan K, Wang X, Wang Y, Zhang C, Lu Y, Yang X, Deng L, Zhou Y, et al. 2016. Ancestral origins and genetic history of Tibetan highlanders. Am J Hum Genet. 99:580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspinas A-S, Westaway MC, Muller C, Sousa VC, Lao O, Alves I, Bergström A, Athanasiadis G, Cheng JY, Crawford JE, et al. 2016. A genomic history of Aboriginal Australia. Nature. 538:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnetto D, Huerta-Sánchez E. 2017. Haplostrips: revealing population structure through haplotype visualization. Methods Ecol Evol. 8:1389–1392. [Google Scholar]
- Martin SH, Davey JW, Jiggins CD. 2014. Evaluating the use of ABBA–BABA statistics to locate introgressed loci. Mol Biol Evol. 32:244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez FL, Watkins JC, Hammer MF. 2012. A haplotype at STAT2 introgressed from Neanderthals and serves as a candidate of positive selection in Papua New Guinea. Am J Hum Genet. 91:265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez FL, Watkins JC, Hammer MF. 2013. Neandertal origin of genetic variation at the cluster of OAS immunity genes. Mol Biol Evol. 30:798–801. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science. 338:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci. 70:3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur BA, Rieseberg LH. 2016. A genomic perspective on hybridization and speciation. Mol Ecol. 25:2337–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe’er I, Yelensky R, Altshuler D, Daly MJ. 2008. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 32:381–385. [DOI] [PubMed] [Google Scholar]
- Peng Y, Yang Z, Zhang H, Cui C, Qi X, Luo X, Tao X, Wu T, Ouzhuluobu B, et al. 2011. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 28:1075–1081. [DOI] [PubMed] [Google Scholar]
- Perneger T V. 1998. What's wrong with Bonferroni adjustments. BMJ. 316:1236–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BM, Huerta-Sanchez E, Nielsen R. 2012. Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLoS Genet. 8:e1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M. 2001. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 69:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, de Filippo C, Grote S, Mafessoni F, Korlević P, Hajdinjak M, Vernot B, Skov L, Hsieh P, Peyrégne S, et al. 2017. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science. 358:655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2013. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 505:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F, Gokhman D, Fumagalli M, Ko A, Hansen T, Moltke I, Albrechtsen A, Carmel L, Huerta-Sánchez E, Nielsen R. 2016. Archaic adaptive introgression in TBX15/WARS2. Mol Biol Evol. 34:509–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F, Marnetto D, Huerta-Sánchez E. 2017. Signatures of archaic adaptive introgression in present-day human populations. Mol Biol Evol. 34:296–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F, Sankararaman S, Nielsen R, Huerta-Sánchez E. 2015. Evidence for archaic adaptive introgression in humans. Nat Rev Genet. 16:359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PLF, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 468:1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N, Kircher M, Delfin F, Nandineni MR, Pugach I, Ko AM-S, Ko Y-C, Jinam TA, Phipps ME, et al. 2011. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 89:516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. 1996. The future of genetic studies of complex human diseases. Science. 273:1516–1517. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D. 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 507:354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Patterson N, Reich D. 2016. The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr Biol. 26:1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Ayroles J, Matute DR, Kern AD. 2018. Supervised machine learning reveals introgressed loci in the genomes of Drosophila simulans and D. sechellia. PLoS Genet. 14:e1007341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Kern AD. 2016. S/HIC: robust identification of soft and hard sweeps using machine learning. PLoS Genet. 12:e1005928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Kern AD. 2018. Supervised machine learning for population genetics: a new paradigm. Trends Genet. 34:301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumer M, Xu C, Powell DL, Durvasula A, Skov L, Holland C, Blazier JC, Sankararaman S, Andolfatto P, Rosenthal GG, et al. 2018. Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science. 360:656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setter D, Mousset S, Cheng X, Nielsen R, DeGiorgio M, Hermisson J. 2020. Volcanofinder: genomic scans for adaptive introgression. PLoS Genet. 16:e1008867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan S, Song YS. 2016. Deep learning for population genetic inference. PLoS Comput Biol. 12:e1004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonti CN, Vernot B, Bastarache L, Bottinger E, Carrell DS, Chisholm RL, Crosslin DR, Hebbring SJ, Jarvik GP, Kullo IJ, et al. 2016. The phenotypic legacy of admixture between modern humans and Neandertals. Science. 351:737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. 2008. Linkage disequilibrium–understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 9:477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slon V, Viola B, Renaud G, Gansauge MT, Benazzi S, Sawyer S, Hublin JJ, Shunkov MV, Derevianko AP, Kelso J, et al. 2017. A fourth Denisovan individual. Sci Adv. 3:e1700186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Endepols S, Klemann N, Richter D, Matuschka F-R, Shih C-H, Nachman MW, Kohn MH. 2011. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between Old World mice. Curr Biol. 21:1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JP, Song YS. 2019. Inference and analysis of population-specific fine-scale recombination maps across 26 diverse human populations. Sci Adv . 5:eaaw9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci. 100:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden LA, Ramachandran S. 2016. Integrating the signatures of demic expansion and archaic introgression in studies of human population genomics. Curr Opin Genet Dev. 41:140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey JW. 1977. Some thoughts on clinical trials, especially problems of multiplicity. Science. 198:679–684. [DOI] [PubMed] [Google Scholar]
- Vernot B, Akey JM. 2014. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 343:1017–1021. [DOI] [PubMed] [Google Scholar]
- Viola BT, Gunz P, Neubauer S, Slon V, Kozlikin MB, Shunkov MV, Meyer M, Paabo S, Derevianko AP. 2019. A parietal fragment from Denisova Cave. Am J Phys Anthropol. 168:258–258. [Google Scholar]
- Wall JD, Brandt DYC. 2016. Archaic admixture in human history. Curr Opin Genet Dev. 41:93–97. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang J, Kourakos M, Hoang N, Lee HH, Mathieson I, Mathieson S. 2021. Automatic inference of demographic parameters using generative adversarial networks. Mol Ecol Resour. 21:2689–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. 1975. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 7:256–276. [DOI] [PubMed] [Google Scholar]
- Witt KE, Huerta-Sánchez E. 2019. Convergent evolution in human and domesticate adaptation to high-altitude environments. Philos Trans R Soc B Biol Sci. 374:20180235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Bailey A, Kuleshov M V, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, et al. 2021. Gene set knowledge discovery with enrichr. Curr Protoc. 1:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Pavlidis P, Taskent RO, Alachiotis N, Flanagan C, DeGiorgio M, Blekhman R, Ruhl S, Gokcumen O. 2017. Archaic hominin introgression in Africa contributes to functional salivary MUC7 genetic variation. Mol Biol Evol. 34:2704–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, Ni X, Liu C, Pan Y, Deng L, Zhang R, Gao Y, Ge X, Liu J, Ma X, et al. 2021. Refining models of archaic admixture in Eurasia with ArchaicSeeker 2.0. Nat Commun. 12:6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhang X, Zhang X, Gao X, Huerta-Sanchez E, Zwyns N. 2022. Denisovans and Homo sapiens on the Tibetan Plateau: dispersals and adaptations. Trends Ecol Evol. 37:257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kim B, Lohmueller KE, Huerta-Sánchez E. 2020. The impact of recessive deleterious variation on signals of adaptive introgression in human populations. Genetics. 215(3):799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Witt KE, Bañuelos MM, Ko A, Yuan K, Xu S, Nielsen R, Huerta-Sanchez E. 2021. The history and evolution of the Denisovan-EPAS1 haplotype in Tibetans. Proc Natl Acad Sci. 118:e2020803118. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All scripts necessary to recreate the simulations, ML training and testing, robustness analysis, and empirical predictions can be found at GitHub: https://github.com/xzhang-popgen/maladapt
Reference
- 1000 Genomes Project Consortium, Auton A, Abecasis GR, Altshuler DM, Durbin RM, Bentley DR, Chakravarti A, Clark AG, Donnelly P, Eichler EE, et al. 2015. A global reference for human genetic variation. Nature. 526:68–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abi-Rached L, Jobin MJ, Kulkarni S, McWhinnie A, Dalva K, Gragert L, Babrzadeh F, Gharizadeh B, Luo M, Plummer FA, et al. 2011. The shaping of modern human immune systems by multiregional admixture with archaic humans. Science. 334:89–94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adrion JR, Galloway JG, Kern AD. 2020. Predicting the landscape of recombination using deep learning. Mol Biol Evol. 37:1790–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahlquist KD, Bañuelos MM, Funk A, Lai J, Rong S, Villanea FA, Witt KE. 2021. Our tangled family tree: new genomic methods offer insight into the legacy of archaic admixture. Genome Biol Evol. 13:evab115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. 2000. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 25:25–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 57:289–300. [Google Scholar]
- Bland JM, Altman DG. 1995. Multiple significance tests: the Bonferroni method. BMJ. 310:170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL. 2007. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 81:1084–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browning SR, Browning BL, Zhou Y, Tucci S, Akey JM. 2018. Analysis of human sequence data reveals two pulses of archaic Denisovan admixture. Cell. 173:53–61.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgarella C, Barnaud A, Kane NA, Jankowski F, Scarcelli N, Billot C, Vigouroux Y, Berthouly-Salazar C. 2019. Adaptive introgression: an untapped evolutionary mechanism for crop adaptation. Front Plant Sci. 10:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J, Perrone V, Spence JP, Jenkins PA, Mathieson S, Song YS. 2018. A likelihood-free inference framework for population genetic data using exchangeable neural networks. Adv Neural Inf Process Syst. 31:8594–8605. [PMC free article] [PubMed] [Google Scholar]
- Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. 2013. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 14:128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen L, Wolf AB, Fu W, Li L, Akey JM. 2020. Identifying and interpreting apparent Neanderthal ancestry in African individuals. Cell. 180:677–687.e16. [DOI] [PubMed] [Google Scholar]
- Choin J, Mendoza-Revilla J, Arauna LR, Cuadros-Espinoza S, Cassar O, Larena M, Ko AM-S, Harmant C, Laurent R, Verdu P, et al. 2021. Genomic insights into population history and biological adaptation in Oceania. Nature. 592:583–589. [DOI] [PubMed] [Google Scholar]
- Crow JF, Kimura M. 1970. An introduction to population genetics theory. New York, Evanston and London: Harper & Row, Publishers. [Google Scholar]
- Dannemann M, Andrés AM, Kelso J. 2016. Introgression of Neandertal- and Denisovan-like haplotypes contributes to adaptive variation in human toll-like receptors. Am J Hum Genet. 98:22–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemann M, Kelso J. 2017. The contribution of Neanderthals to phenotypic variation in modern humans. Am J Hum Genet. 101:578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dannemann M, Racimo F. 2018. Something old, something borrowed: admixture and adaptation in human evolution. Curr Opin Genet Dev. 53:1–8. [DOI] [PubMed] [Google Scholar]
- Deschamps M, Laval G, Fagny M, Itan Y, Abel L, Casanova J-L, Patin E, Quintana-Murci L. 2016. Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am J Hum Genet. 98:5–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Hu Y, Jin L. 2014. Non-Neanderthal origin of the HLA-DPB1*0401. J Biol Chem. 289:10252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Q, Hu Y, Xu S, Wang J, Jin L. 2013. Neanderthal introgression at chromosome 3p21.31 was under positive natural selection in East Asians. Mol Biol Evol. 31:683–695. [DOI] [PubMed] [Google Scholar]
- Dudbridge F, Gusnanto A. 2008. Estimation of significance thresholds for genomewide association scans. Genet Epidemiol. 32:227–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand EY, Patterson N, Reich D, Slatkin M. 2011. Testing for ancient admixture between closely related populations. Mol Biol Evol. 28:2239–2252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durvasula A, Sankararaman S. 2019. A statistical model for reference-free inference of archaic local ancestry. PLoS Genet. 15:e1008175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durvasula A, Sankararaman S. 2020. Recovering signals of ghost archaic introgression in African populations. Sci Adv. 6:eaax5097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enard D, Petrov DA. 2018. Evidence that RNA viruses drove adaptive introgression between Neanderthals and modern humans. Cell. 175:360–371.e13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garud NR, Messer PW, Buzbas EO, Petrov DA. 2015. Recent selective sweeps in North American Drosophila melanogaster show signatures of soft sweeps. PLoS Genet. 11:e1005004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geurts P, Ernst D, Wehenkel L. 2006. Extremely randomized trees. Mach Learn. 63:3–42. [Google Scholar]
- Gittelman RM, Schraiber JG, Vernot B, Mikacenic C, Wurfel MM, Akey JM. 2016. Archaic hominin admixture facilitated adaptation to out-of-Africa environments. Curr Biol. 26:3375–3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gouy A, Daub JT, Excoffier L. 2017. Detecting gene subnetworks under selection in biological pathways. Nucleic Acids Res. 45:e149–e149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gower G, Picazo PI, Fumagalli M, Racimo F. 2021. Detecting adaptive introgression in human evolution using convolutional neural networks. Elife. 10:e64669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gravel S, Henn BM, Gutenkunst RN, Indap AR, Marth GT, Clark AG, Yu F, Gibbs RA, Bustamante CD. 2011. Demographic history and rare allele sharing among human populations. Proc Natl Acad Sci. 108:11983–11988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Green RE, Krause J, Briggs AW, Maricic T, Stenzel U, Kircher M, Patterson N, Li H, Zhai W, Fritz MHY, et al. 2010. A draft sequence of the Neandertal genome. Science. 328:710–722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenhalgh T. 1997. How to read a paper. Statistics for the non-statistician. I: different types of data need different statistical tests. BMJ. 315:364–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hackinger S, Kraaijenbrink T, Xue Y, Mezzavilla M, Asan van Driem G, Jobling MA, de Knijff P, Tyler-Smith C, Ayub Q. 2016. Wide distribution and altitude correlation of an archaic high-altitude-adaptive EPAS1 haplotype in the Himalayas. Hum Genet. 135:393–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haller BC, Messer PW. 2019. SLim 3: forward genetic simulations beyond the Wright–Fisher model. Mol Biol Evol. 36:632–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris K, Nielsen R. 2016. The genetic cost of Neanderthal introgression. Genetics. 203:881–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrow J, Frankish A, Gonzalez JM, Tapanari E, Diekhans M, Kokocinski F, Aken BL, Barrell D, Zadissa A, Searle S, et al. 2012. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 22:1760–1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Gravel S, Wang W, Brisbin A, Byrnes JK, Fadhlaoui-Zid K, Zalloua PA, Moreno-Estrada A, Bertranpetit J, et al. 2012. Genomic ancestry of North Africans supports back-to-Africa migrations. PLoS Genet. 8:e1002397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henn BM, Botigué LR, Peischl S, Dupanloup I, Lipatov M, Maples BK, Martin AR, Musharoff S, Cann H, Snyder MP, et al. 2016. Distance from sub-Saharan Africa predicts mutational load in diverse human genomes. Proc Natl Acad Sci. 113:E440–E449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hider JL, Gittelman RM, Shah T, Edwards M, Rosenbloom A, Akey JM, Parra EJ. 2013. Exploring signatures of positive selection in pigmentation candidate genes in populations of East Asian ancestry. BMC Evol Biol. 13:150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill WG, Robertson A. 1968. Linkage disequilibrium in finite populations. Theor Appl Genet. 38:226–231. [DOI] [PubMed] [Google Scholar]
- Huber CD, Kim BY, Marsden CD, Lohmueller KE. 2017. Determining the factors driving selective effects of new nonsynonymous mutations. Proc Natl Acad Sci. 114:4465–4470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubisz M, Siepel A. 2020. Inference of ancestral recombination graphs using ARGweaver. Methods Mol Biol. 2090:231–266. [DOI] [PubMed] [Google Scholar]
- Huerta-Sánchez E, Jin X, Asan BZ, Peter BM, Vinckenbosch N, Liang Y, Yi X, He M, Somel M, et al. 2014. Altitude adaptation in Tibetans caused by introgression of Denisovan-like DNA. Nature. 512:194–197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- International HapMap Consortium . 2005. A haplotype map of the human genome. Nature. 437:1299–1320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs GS, Hudjashov G, Saag L, Kusuma P, Darusallam CC, Lawson DJ, Mondal M, Pagani L, Ricaut F-X, Stoneking M, et al. 2019. Multiple deeply divergent Denisovan ancestries in Papuans. Cell. 177:1010–1021. [DOI] [PubMed] [Google Scholar]
- Jagoda E, Lawson DJ, Wall JD, Lambert D, Muller C, Westaway M, Leavesley M, Capellini TD, Mirazón Lahr M, Gerbault P, et al. 2017. Disentangling immediate adaptive introgression from selection on standing introgressed variation in humans. Mol Biol Evol. 35:623–630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juric I, Aeschbacher S, Coop G. 2016. The strength of selection against Neanderthal introgression. PLoS Genet. 12:e1006340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly JK. 1997. A test of neutrality based on interlocus associations. Genetics. 146:1197–1206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Huber CD, Lohmueller KE. 2017. Inference of the distribution of selection coefficients for new nonsynonymous mutations using large samples. Genetics. 206:345–361. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim BY, Huber CD, Lohmueller KE. 2018. Deleterious variation shapes the genomic landscape of introgression. PLoS Genet. 14:e1007741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong A, Thorleifsson G, Gudbjartsson DF, Masson G, Sigurdsson A, Jonasdottir A, Walters GB, Jonasdottir A, Gylfason A, Kristinsson KT, et al. 2010. Fine-scale recombination rate differences between sexes, populations and individuals. Nature. 467:1099–1103. [DOI] [PubMed] [Google Scholar]
- Kuhlwilm M, Gronau I, Hubisz MJ, de Filippo C, Prado-Martinez J, Kircher M, Fu Q, Burbano HA, Lalueza-Fox C, de la Rasilla M, et al. 2016. Ancient gene flow from early modern humans into Eastern Neanderthals. Nature. 530:429–433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunsch HR. 1989. The jackknife and the bootstrap for general stationary observations. Ann Stat. 17:1217–1241. [Google Scholar]
- Larena M, McKenna J, Sanchez-Quinto F, Bernhardsson C, Ebeo C, Reyes R, Casel O, Huang J-Y, Hagada KP, Guilay D, et al. 2021. Philippine Ayta possess the highest level of Denisovan ancestry in the world. Curr Biol. 31:4219–4230.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu D, Lou H, Yuan K, Wang X, Wang Y, Zhang C, Lu Y, Yang X, Deng L, Zhou Y, et al. 2016. Ancestral origins and genetic history of Tibetan highlanders. Am J Hum Genet. 99:580–594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malaspinas A-S, Westaway MC, Muller C, Sousa VC, Lao O, Alves I, Bergström A, Athanasiadis G, Cheng JY, Crawford JE, et al. 2016. A genomic history of Aboriginal Australia. Nature. 538:207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnetto D, Huerta-Sánchez E. 2017. Haplostrips: revealing population structure through haplotype visualization. Methods Ecol Evol. 8:1389–1392. [Google Scholar]
- Martin SH, Davey JW, Jiggins CD. 2014. Evaluating the use of ABBA–BABA statistics to locate introgressed loci. Mol Biol Evol. 32:244–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez FL, Watkins JC, Hammer MF. 2012. A haplotype at STAT2 introgressed from Neanderthals and serves as a candidate of positive selection in Papua New Guinea. Am J Hum Genet. 91:265–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendez FL, Watkins JC, Hammer MF. 2013. Neandertal origin of genetic variation at the cluster of OAS immunity genes. Mol Biol Evol. 30:798–801. [DOI] [PubMed] [Google Scholar]
- Meyer M, Kircher M, Gansauge M-T, Li H, Racimo F, Mallick S, Schraiber JG, Jay F, Prüfer K, de Filippo C, et al. 2012. A high-coverage genome sequence from an archaic Denisovan individual. Science. 338:222–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nei M. 1973. Analysis of gene diversity in subdivided populations. Proc Natl Acad Sci. 70:3321–3323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Payseur BA, Rieseberg LH. 2016. A genomic perspective on hybridization and speciation. Mol Ecol. 25:2337–2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pe’er I, Yelensky R, Altshuler D, Daly MJ. 2008. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 32:381–385. [DOI] [PubMed] [Google Scholar]
- Peng Y, Yang Z, Zhang H, Cui C, Qi X, Luo X, Tao X, Wu T, Ouzhuluobu B, et al. 2011. Genetic variations in Tibetan populations and high-altitude adaptation at the Himalayas. Mol Biol Evol. 28:1075–1081. [DOI] [PubMed] [Google Scholar]
- Perneger T V. 1998. What's wrong with Bonferroni adjustments. BMJ. 316:1236–1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peter BM, Huerta-Sanchez E, Nielsen R. 2012. Distinguishing between selective sweeps from standing variation and from a de novo mutation. PLoS Genet. 8:e1003011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pritchard JK, Przeworski M. 2001. Linkage disequilibrium in humans: models and data. Am J Hum Genet. 69:1–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, de Filippo C, Grote S, Mafessoni F, Korlević P, Hajdinjak M, Vernot B, Skov L, Hsieh P, Peyrégne S, et al. 2017. A high-coverage Neandertal genome from Vindija Cave in Croatia. Science. 358:655–658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prüfer K, Racimo F, Patterson N, Jay F, Sankararaman S, Sawyer S, Heinze A, Renaud G, Sudmant PH, de Filippo C, et al. 2013. The complete genome sequence of a Neanderthal from the Altai Mountains. Nature. 505:43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F, Gokhman D, Fumagalli M, Ko A, Hansen T, Moltke I, Albrechtsen A, Carmel L, Huerta-Sánchez E, Nielsen R. 2016. Archaic adaptive introgression in TBX15/WARS2. Mol Biol Evol. 34:509–524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F, Marnetto D, Huerta-Sánchez E. 2017. Signatures of archaic adaptive introgression in present-day human populations. Mol Biol Evol. 34:296–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Racimo F, Sankararaman S, Nielsen R, Huerta-Sánchez E. 2015. Evidence for archaic adaptive introgression in humans. Nat Rev Genet. 16:359–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Green RE, Kircher M, Krause J, Patterson N, Durand EY, Viola B, Briggs AW, Stenzel U, Johnson PLF, et al. 2010. Genetic history of an archaic hominin group from Denisova Cave in Siberia. Nature. 468:1053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich D, Patterson N, Kircher M, Delfin F, Nandineni MR, Pugach I, Ko AM-S, Ko Y-C, Jinam TA, Phipps ME, et al. 2011. Denisova admixture and the first modern human dispersals into Southeast Asia and Oceania. Am J Hum Genet. 89:516–528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Risch N, Merikangas K. 1996. The future of genetic studies of complex human diseases. Science. 273:1516–1517. [DOI] [PubMed] [Google Scholar]
- Saitou N, Nei M. 1987. The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425. [DOI] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Dannemann M, Prüfer K, Kelso J, Pääbo S, Patterson N, Reich D. 2014. The genomic landscape of Neanderthal ancestry in present-day humans. Nature. 507:354–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sankararaman S, Mallick S, Patterson N, Reich D. 2016. The combined landscape of Denisovan and Neanderthal ancestry in present-day humans. Curr Biol. 26:1241–1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Ayroles J, Matute DR, Kern AD. 2018. Supervised machine learning reveals introgressed loci in the genomes of Drosophila simulans and D. sechellia. PLoS Genet. 14:e1007341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Kern AD. 2016. S/HIC: robust identification of soft and hard sweeps using machine learning. PLoS Genet. 12:e1005928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrider DR, Kern AD. 2018. Supervised machine learning for population genetics: a new paradigm. Trends Genet. 34:301–312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumer M, Xu C, Powell DL, Durvasula A, Skov L, Holland C, Blazier JC, Sankararaman S, Andolfatto P, Rosenthal GG, et al. 2018. Natural selection interacts with recombination to shape the evolution of hybrid genomes. Science. 360:656–660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Setter D, Mousset S, Cheng X, Nielsen R, DeGiorgio M, Hermisson J. 2020. Volcanofinder: genomic scans for adaptive introgression. PLoS Genet. 16:e1008867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheehan S, Song YS. 2016. Deep learning for population genetic inference. PLoS Comput Biol. 12:e1004845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simonti CN, Vernot B, Bastarache L, Bottinger E, Carrell DS, Chisholm RL, Crosslin DR, Hebbring SJ, Jarvik GP, Kullo IJ, et al. 2016. The phenotypic legacy of admixture between modern humans and Neandertals. Science. 351:737–741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slatkin M. 2008. Linkage disequilibrium–understanding the evolutionary past and mapping the medical future. Nat Rev Genet. 9:477–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slon V, Viola B, Renaud G, Gansauge MT, Benazzi S, Sawyer S, Hublin JJ, Shunkov MV, Derevianko AP, Kelso J, et al. 2017. A fourth Denisovan individual. Sci Adv. 3:e1700186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song Y, Endepols S, Klemann N, Richter D, Matuschka F-R, Shih C-H, Nachman MW, Kohn MH. 2011. Adaptive introgression of anticoagulant rodent poison resistance by hybridization between Old World mice. Curr Biol. 21:1296–1301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spence JP, Song YS. 2019. Inference and analysis of population-specific fine-scale recombination maps across 26 diverse human populations. Sci Adv . 5:eaaw9206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storey JD, Tibshirani R. 2003. Statistical significance for genomewide studies. Proc Natl Acad Sci. 100:9440–9445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugden LA, Ramachandran S. 2016. Integrating the signatures of demic expansion and archaic introgression in studies of human population genomics. Curr Opin Genet Dev. 41:140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tukey JW. 1977. Some thoughts on clinical trials, especially problems of multiplicity. Science. 198:679–684. [DOI] [PubMed] [Google Scholar]
- Vernot B, Akey JM. 2014. Resurrecting surviving Neandertal lineages from modern human genomes. Science. 343:1017–1021. [DOI] [PubMed] [Google Scholar]
- Viola BT, Gunz P, Neubauer S, Slon V, Kozlikin MB, Shunkov MV, Meyer M, Paabo S, Derevianko AP. 2019. A parietal fragment from Denisova Cave. Am J Phys Anthropol. 168:258–258. [Google Scholar]
- Wall JD, Brandt DYC. 2016. Archaic admixture in human history. Curr Opin Genet Dev. 41:93–97. [DOI] [PubMed] [Google Scholar]
- Wang Z, Wang J, Kourakos M, Hoang N, Lee HH, Mathieson I, Mathieson S. 2021. Automatic inference of demographic parameters using generative adversarial networks. Mol Ecol Resour. 21:2689–2705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watterson GA. 1975. On the number of segregating sites in genetical models without recombination. Theor Popul Biol. 7:256–276. [DOI] [PubMed] [Google Scholar]
- Witt KE, Huerta-Sánchez E. 2019. Convergent evolution in human and domesticate adaptation to high-altitude environments. Philos Trans R Soc B Biol Sci. 374:20180235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie Z, Bailey A, Kuleshov M V, Clarke DJB, Evangelista JE, Jenkins SL, Lachmann A, Wojciechowicz ML, Kropiwnicki E, Jagodnik KM, et al. 2021. Gene set knowledge discovery with enrichr. Curr Protoc. 1:e90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu D, Pavlidis P, Taskent RO, Alachiotis N, Flanagan C, DeGiorgio M, Blekhman R, Ruhl S, Gokcumen O. 2017. Archaic hominin introgression in Africa contributes to functional salivary MUC7 genetic variation. Mol Biol Evol. 34:2704–2715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan K, Ni X, Liu C, Pan Y, Deng L, Zhang R, Gao Y, Ge X, Liu J, Ma X, et al. 2021. Refining models of archaic admixture in Eurasia with ArchaicSeeker 2.0. Nat Commun. 12:6232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Zhang X, Zhang X, Gao X, Huerta-Sanchez E, Zwyns N. 2022. Denisovans and Homo sapiens on the Tibetan Plateau: dispersals and adaptations. Trends Ecol Evol. 37:257–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kim B, Lohmueller KE, Huerta-Sánchez E. 2020. The impact of recessive deleterious variation on signals of adaptive introgression in human populations. Genetics. 215(3):799–812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Witt KE, Bañuelos MM, Ko A, Yuan K, Xu S, Nielsen R, Huerta-Sanchez E. 2021. The history and evolution of the Denisovan-EPAS1 haplotype in Tibetans. Proc Natl Acad Sci. 118:e2020803118. [DOI] [PMC free article] [PubMed] [Google Scholar]