Abstract
Background:
Ex vivo assays of platelet function critically inform mechanistic and clinical hematology studies, where effects of divergent blood processing methods on platelet composition are apparent, but unspecified.
Objective:
Here, we evaluate how different blood anticoagulation options and processing times affect platelet function and protein content ex vivo.
Methods:
Parallel blood samples were collected from healthy human donors into sodium citrate, acid citrate dextrose, EDTA or heparin, and processed over an extended time course for functional and biochemical experiments, including platelet proteome quantification with multiplexed tandem mass tag (TMT) labeling and triple quadrupole mass spectrometry (MS).
Results:
Each anticoagulant had time-dependent effects on platelet function in whole blood. For instance, heparin enhanced platelet agonist reactivity, platelet-monocyte aggregate formation and platelet extracellular vesicle release, while EDTA increased platelet α-granule secretion. Following platelet isolation, TMT-MS quantified 3,357 proteins amongst all prepared platelet samples. Altogether, >400 proteins were differentially abundant in platelets isolated from blood processed at 24 h vs 1 h post-phlebotomy, including proteins pertinent to membrane trafficking and exocytosis. Anticoagulant-specific effects on platelet proteomes included increased complement system and decreased α-granule proteins in platelets from EDTA-anticoagulated blood. Platelets prepared from heparinized blood had higher levels of histone and neutrophil-associated proteins in a manner related to neutrophil extracellular trap (NET) formation and platelet:NET interactions in whole blood ex vivo.
Conclusion:
Our results demonstrate that different anticoagulants routinely used for blood collection have varying effects on platelets ex vivo, where methodology-associated alterations in platelet proteome may influence mechanistic, translational and biomarker studies.
Keywords: Anticoagulants, Biomarkers, Blood Platelets, Platelet Function Tests, Proteomics
1 |. INTRODUCTION
Ex vivo studies of blood platelets have expanded an understanding of mechanisms of hemostasis and thrombosis, and offer promise to uncover diagnostic biomarkers and therapeutic targets in a range of platelet-associated physiological and disease states [1, 2]. As circulating, cellular sentinels of vascular well-being, platelets are exquisitely sensitive to mechanical and chemical stimulation. Accordingly, research laboratories often customize platelet preparation procedures to preserve platelet (ultra)structure and reactivity for downstream biochemical, microscopy, flow cytometry and other ex vivo experiments. While some parameters for quiescent platelet purification such as blood centrifugation forces are largely uniform between research groups [3–6], anticoagulation methods are more variable, especially for blood samples not originally intended for specific platelet studies.
Blood anticoagulation, processing and storage are all known to effect platelet structure, count and function, particularly in clinical diagnostic tests [7–10], and increasingly in basic laboratory investigations of platelet biochemistry and cellular biology [11–13]. However, specific effects of anticoagulation and blood processing on the molecular composition of platelets – determined by genetics and environment, and, posited to be indicative or supportive of disease – remain unknown. Platelet protein content is determined by several factors, including the repertoire of proteins inherited from parent megakaryocytes [14], signal-dependent mRNA splicing and de novo protein synthesis [15], loss of compartment proteins via release of granule contents and extracellular vesicles, and by the sequestration of circulating proteins [16]. However, the effect of blood processing on platelet proteomes remains unknown, where biomarker studies of healthy and “disease-educated” platelets, often involve comparisons of multiple, diverse clinical samples with extended travel and delayed processing times [17, 18]. Many recent biomarker studies find impact of blood processing on plasma proteomes, often pointing to many platelet-derived proteins in plasma preparations [19–24]. However, the reciprocal effects of preanalytical variables on platelet proteomes remain unspecified, where platelets offer promise as circulating drones that continuously survey and reflect the health of the vascular and tissue milieu [1, 25].
We recently established a quantitative mass spectrometry workflow with tandem mass tag (TMT) labeling to measure proteome-level alterations in specific platelet activation or physiological states [1, 26, 27]. In this study, we use these tools to systematically investigate how blood collection into common anticoagulants (sodium citrate, acid citrate dextrose, EDTA, heparin) influences ex vivo platelet protein content across an extended range of post-phlebotomy time points reflective of real-world sample processing situations. We find that significant changes in the platelet proteome can occur over prolonged blood processing times, and we identify protein profiles unique to platelets exposed to EDTA and heparin of relevance to studies of platelet function and biomarkers in health and disease.
2 |. METHODS
2.1 |. Temporal processing of anticoagulated blood
Blood was drawn by venipuncture from a rotating pool of >20 healthy, (equal proportion of male and female) adult volunteers (>18 y, average age 35 y) at Oregon Health & Science University. All samples were collected following institutional review board (IRB) guidelines and after written informed consent from participants. Venous whole blood was collected into vacutainer tubes containing either sodium heparin (158 USP, BD 367874), ethylenediaminetetraacetic acid (K2-EDTA 18 mg, BD #366450), buffered sodium citrate (NaCit, 0.105M, 3.2%, BD #369714), or a combination of sodium citrate and acid citrate dextrose (NaCit+ACD). NaCit+ACD tubes were prepared by addition of 500 μl ACD solution (7.5 g of citric acid, 12.5 g of sodium citrate and 10 g of D-glucose in 500 ml of Milli-Q H2O) into BD NaCit 3.2% vacutainers before blood draw. After gentle mixing, 1 ml anticoagulated blood aliquots were prepared and processed immediately (t = 0 h) or incubated without shaking for t = 1, 2, 3, 5, 9, 24, 48 and 120 h at room temperature (rt). The aliquots were centrifuged at 200 × g for 20 min to obtain platelet-rich plasma (PRP). The upper 2/3 volume of PRP was transferred into a new tube and further centrifuged (for aliquots t = 1–120 h) at 1,000 × g for 10 min, and the resulting supernatant was centrifuged (2,500 × g, 15 min) to obtain platelet poor plasma (PPP) (Fig. 1A).
Figure 1. Progressive effects of anticoagulants on platelet hematologic parameters.
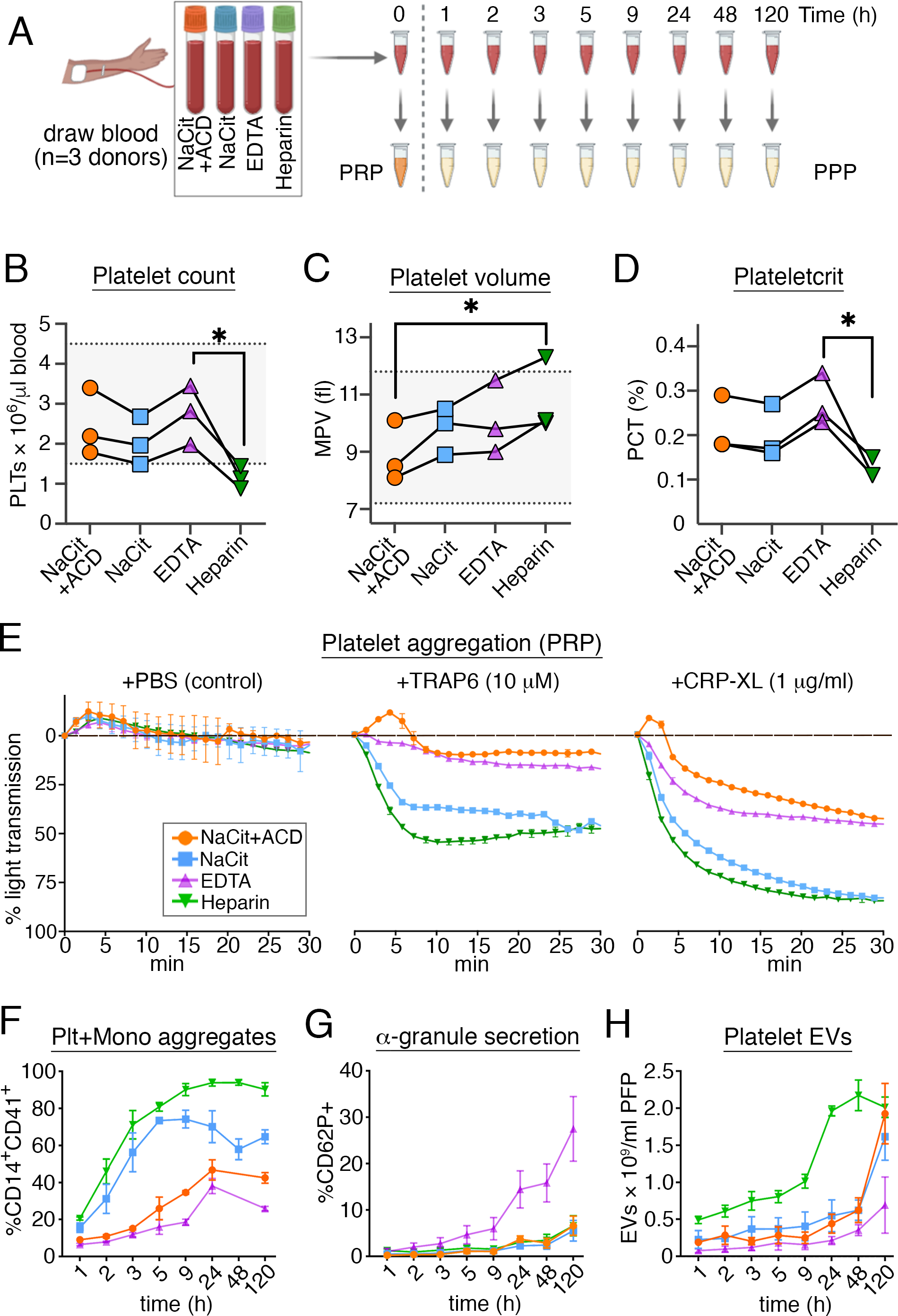
(A) Whole blood was collected from healthy human donors (n = 3) into four different anticoagulants, including sodium citrate (NaCit), a combination of sodium citrate and acid citrate dextrose (NaCit+ACD), EDTA and heparin, and stored for different time intervals (up 120 h) at room temperature (rt) prior to hematological assays of whole blood, platelet rich plasma (PRP) and platelet poor plasma (PPP). Data points in all panels are color-coded to match conventional anticoagulant tube cap colors. (B, C) Platelet concentration and mean platelet volume (MPV) were obtained from complete blood count (CBC) 1 h after blood collection. Counts for citrated blood were adjusted for dilution in liquid anticoagulant. Solid lines connect data points for the same donor. Horizontal dotted lines show normal physiological range of platelet count and MPV values. The Friedman test (non-parametric one-way repeated measures analysis of variance) was used to determine significant differences in platelet count, MPV or PCT across all anticoagulant groups, followed by multiple pair-wise comparisons with Dunn’s correction (*p<0.05). (D) Plateletcrit (PCT) was derived from platelet count and MPV using the formula: PCT (%) = [platelet count (x 109/l) × MPV(fl)]/104. (E) Percent spontaneous and agonist-induced platelet aggregation in PRP prepared immediately after blood draw with different anticoagulants. The absorbance (650 nm) of PRP with normalized platelet counts was tracked in real-time at 37°C. Autologous and anticoagulant-matched PPP samples were used as 100% aggregation controls. (F) The formation of platelet monocyte aggregates in anticoagulated whole blood over storage time was assessed by flow cytometry as %CD14+ (monocyte marker) and CD41a+ (platelet marker) events. (G) Temporal exposure of P-selectin (CD62P) on the surface of CD41a+ platelets in whole blood analyzed by flow cytometry. (H) Platelet EVs, co-expressing CD41a and CD9, were quantified by nanoscale flow cytometry in platelet-depleted plasma prepared after variable storage of whole blood in anticoagulants. Repeated measures two-way analysis of variance (RM-ANOVA) was used to determine interaction between, and contribution of, anticoagulant and storage time variables to variance of PMA, CD62P or platelet EV. See S. Figs. S1–S4 for additional details regarding platelet hematological assays and flow cytometry analyses.
2.2 |. Preparation of washed platelets
Washed platelets were prepared for biochemical and proteomics experiments as previously described [26]. Blood was collected into NaCit+ACD, NaCit, EDTA and heparin tubes to prepare washed platelets at 1 h and 24 h post-phlebotomy. For all experimental conditions, 10 ml of anticoagulated blood was centrifuged at 200 × g for 20 min to prepare PRP. Following addition of 10 μl of 1 mg/ml prostacyclin (PGI2) and gentle mixing, PRP was centrifuged (1,000 × g, 10 min) to pellet platelets. The resulting platelet pellet was resuspended and washed in HEPES/Tyrode (H-T) buffer (129 mM NaCl, 0.34 mM Na2HPO4, 2.9 mM KCl, 12 mM NaHCO3, 20 mM HEPES, 5 mM glucose, 1 mM MgCl2; pH 7.3) supplemented with ACD and PGI2. Diluted platelet suspensions were immediately centrifuged at 1,000 × g for 10 min and platelet pellets were was resuspended in H-T buffer.
2.3 |. Platelet lysis
Equal volumes of washed platelet samples H-T buffer were lysed in 2.5% SDS, 150 mM NaCl, 50 mM HEPES pH 8.5, 1× cOmplete mini EDTA-free protease inhibitor (Roche #046933159001), and 1× PhosSTOP (Roche #4906837001) [28]. Lysed platelets were vortexed and immediately flash-frozen in liquid nitrogen before storage at −80°C.
2.4 |. Platelet peptide preparation and quantification
Frozen platelet lysates were thawed, heated at 90°C for 10 min and sonicated using a Bioruptor Pico (Diagenode, # B01060010). Aliquots were analyzed for protein concentration by bicinchoninic acid (BCA) assay. Equal amounts of total protein per sample (50 μg in 5% SDS), were reduced with dithiothreitol (DTT), alkylated with iodoacetamide (IAA) and digested in S-Trap micro columns (ProtiFi, #C02-micro) overnight at 47°C. The Pierce Quantitative Colorimetric Peptide Assay (Thermo, #23275) was then used to determine the concentration of eluted peptides. Briefly, dried S-Trap micro-digests were resuspended in 80 μl of high-performance liquid chromatography (HPLC)-grade water, and 5 μl of the suspension was further diluted to 20 μl. An average of 38 μg of peptides/sample (standard deviation [SD] = 6 μg) was recovered after S-Trap micro-digestion of 50 μg of protein input, that is, 76% (SD = 12%) recovery. Peptide TMT labeling, LC-MS/MS methods, and data analysis are detailed in Supplemental Information. The mass spectrometry proteomics data, Jupyter notebooks, R scripts and other resources are publicly available through the ProteomeXchange Consortium via the PRIDE partner repository with the dataset identifier PXD030225.
2.5 |. Hematologic, Flow Cytometry, Luminex, and Other Assays
Additional methodological details are specified in Supplementary Information.
3 |. RESULTS
3.1 |. Effect of blood anticoagulation on platelet count and function
We first sought to determine how different anticoagulants used for blood collection affect parameters relevant to ex vivo studies of platelet function. As outlined in Fig. 1A, blood was collected from healthy adult human donors directly into four vacutainer tubes with different anticoagulants, including sodium citrate (NaCit), a combination of NaCit together with acid citrate dextrose (hereafter referred to as “NaCit+ACD”), EDTA, and heparin. Anticoagulated blood samples were kept for 0–120 h at rt prior to assay or processing to prepare platelet-rich plasma (PRP). At 1 h after blood draw into NaCit+ACD, NaCit or EDTA, platelet counts were all within the normal range of 150,000–450,000/μl, as determined by CBC analysis (Fig. 1B, S. Table S1);); and, the mean platelet volume (MPV) and the percent of total blood volume occupied by platelets (plateletcrit, PCT) trended higher for blood drawn into EDTA compared to NaCit+ACD, but not in a statistically significant manner (Fig. 1C, D). In contrast, blood that had been collected into heparin had marked decreases in platelet counts to levels below 150,000/μl, in addition to a relative increased MPV and decreased PCT (Fig. 1B–D).
Next, to examine the effects of anticoagulants on ex vivo platelet reactivity, we prepared PRP from t=0 blood samples for platelet aggregation assays. All PRP preparations were confirmed to contain >99.5% platelets and <0.05% leukocytes with flow cytometry analyses (S. Fig. S1). Regardless of anticoagulant, PRP turbidity did not change for over 30 min following the addition of vehicle control (PBS) alone (Fig. 1E). Following addition of TRAP6 or CRP-XL, light transmission (indicative of platelet aggregation) increased through all PRP samples, where PRP from blood collected into heparin or NaCit had the most pronounced platelet aggregation responses (Fig. 1E). As previous studies have determined that acidic pH has an inhibitory effect on platelet aggregation [29], we also measured the pH of citrated PRP and PPP samples, where inclusion of ACD lowered the pH of PRP and PPP preparations relative to NaCit anticoagulation alone (S. Fig. S2). In addition to agonist-induced platelet-platelet aggregation in PRP, we also examined the formation of platelet-monocyte aggregates (CD14+CD41a+) in whole blood over a period of 120 h post phlebotomy (Fig. 1F, S. Fig. S3). Platelet-monocyte aggregation in whole blood increased over time for all anticoagulants, but at a faster rate for blood anticoagulated with NaCit or heparin, reaching >90% beyond 9 h of storage time in heparin (Fig. 1F).
Platelets in whole blood collected into NaCit, NaCit+ACD and heparin all had minimal levels of α-granule secretion, as determined by flow cytometry analysis (CD41a+CD62P+ <7% at 120 h); however, EDTA anticoagulation resulted in an increase in %CD62P+ platelets in whole blood from 1.0% at 1 h, to 4.7% at 5 h, 14.5% at 24 h, and 27.5% at 120 h (Fig. 1G). Likewise, platelet surface expression of CD63 (granulophysin, another granule membrane protein [2, 30]) significantly increased in EDTA-treated blood compared to other anticoagulants, doubling to 9.7% after 24 h (S. Fig. S3C). We also observed a time-dependent decrease in platelet surface CD41a staining intensity exclusively in EDTA-anticoagulated blood (S. Fig. S3D), while detection of CD61 (integrin β3, GPIIIa) did not change (S. Fig. S3E). These observations are consistent with previous studies noting that prolonged exposure of platelets to EDTA alters the confirmation, localization and epitope accessibility of GPIIbIIIa [31]. In addition, with nanoscale flow cytometry (S. Fig. S4), we quantified significantly higher numbers of platelet-derived (CD41a+CD9+) extracellular vesicles (EVs) [32] in platelet depleted plasma prepared from heparinized blood, relative to NaCit+ACD (p = 0.038) or EDTA (p = 0.015) (Fig. 1H).
3.2 |. Platelet proteome composition and variability with anticoagulation
A variety of anticoagulants are commonly used to draw blood in order to prepare peripheral blood cells and plasma for clinical assays as well as mechanistic studies; however, the effects of anticoagulation on platelet composition are not known. To quantify and compare the effects of different anticoagulants on the proteome of platelets prepared for ex vivo studies, we utilized an 11-plex tandem mass tag (TMT) labeling and tandem mass spectrometry (MS) workflow, taking advantage of internal reference samples (IRS) for multiplexing [1, 26, 27, 33]. As outlined in Fig. 2A, blood was drawn into a set of NaCit+ACD, EDTA and heparin vacutainer tubes that were each kept for 1 h and 24 h prior to preparing washed platelets (>99.5% CD41a+; S. Fig. S1; 3 donors × 3 anticoagulants × 2 time points = 18 samples total). These time points were selected, as anticoagulant-specific differences in platelet physiological parameters were less pronounced at 1 h relative to 24 h post phlebotomy (Fig. 1F–H, S. Fig. S3). Following lysis and tryptic digestion, equal amounts of peptides from each test sample were labeled with tandem mass tags, combined into 9-plex mixtures and run as two 11-plex TMT experiments, including two internal reference samples per experiment. A total of 3,746 proteins with quantifiable reporter ion intensities were identified in either of the two plexes, of which, 3,357 proteins representing 99.8% of the sum of reporter ion intensities were identified in both TMT plexes (S. Table S2). After IRS normalization, peptide reporter ion intensities spanned a wide dynamic range of five orders of magnitude, where platelet proteins known to be expressed in high abundance (e.g. filamin A, actin, integrin β3), medium abundance (GPVI, PAR4) and low abundance (e.g. LAMP2, VAMP2) [34], distributed accordingly (Fig. 2B).
Figure 2. Effect of blood anticoagulation and processing time on platelet proteome composition and variability.
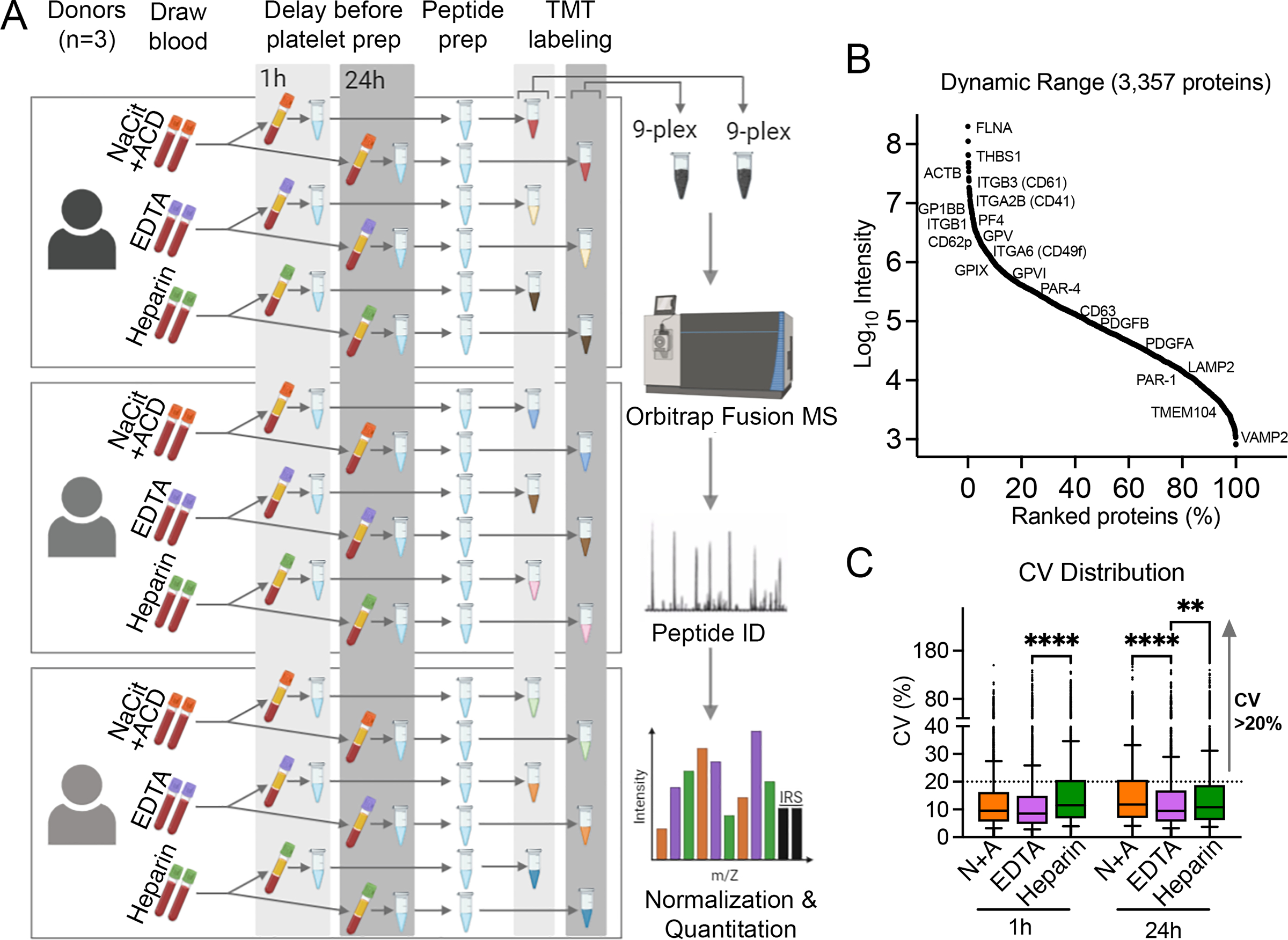
(A) Study design and workflow. A total of 18 washed platelet samples were prepared from blood from n=3 donors × 3 anticoagulants (NaCit+ACD, EDTA and Heparin) × 2 blood storage time points (1 h and 24 h). Following lysis and tryptic digest, peptides from each platelet sample were labeled with isobaric tandem mass tags (TMT) and pooled into two 9-plex mixtures, plus two tagged internal reference samples (IRS) per mixture. Online fractionation (18 fractions) was performed for each 11-plex TMT mixture before mass spectrometry (MS) in an Orbitrap Fusion Tribrid mass spectrometer. (B) Dynamic range of reporter ion intensities of 3,357 proteins identified in both TMT plex 1 and plex 2 after IRS normalization. A selection of high, medium and low abundance platelet proteins are noted with Uniprot gene identifiers. (C) The distribution of inter-individual coefficients of variation (CV) for 3,357 proteins at 1 h and 24 h of blood storage in different anticoagulants. Box and whisker plots represent interquartile range (25th – 75th percentile, lower and upper limit of boxes) and 10th/90th percentiles (whiskers), with median CVs ranging between 8.5 and 12.
To examine how ex vivo anticoagulation influenced global variances in platelet protein expression between healthy individuals, we determined and compared coefficients of variation (CVs) of all 3,357 proteins quantified for each experimental condition. Similar to previous quantitative analyses of platelet proteomes by our group [26, 27] and others [34, 35], the majority (~75%) of platelet proteins had low CVs (≤ 20), irrespective of the anticoagulant or time point (Fig. 2C); nonetheless, there was a general temporal increase in CVs (e.g., NaCit+ACD median CV [25th–75th percentile] = 9.6 [5.6–16.3] at 1 h and 11.7 [6.9–20.6] at 24 h, p <0.0001). The lowest median CVs at both time points were seen for platelets isolated from EDTA-anticoagulated blood (8.5 and 9.5 at 1 h and 24 h, respectively). Not surprising, proteins with the highest inter-subject variance (CV>20) mostly included those expressed at lower abundances (~80% had intensities < median intensity of all proteins), but their distribution was not the same across anticoagulants. For example, 360 proteins had CVs>20 in platelets prepared from heparinized blood, but not in NaCit+ACD- or EDTA-anticoagulated blood at 1 h. Furthermore, only about 25% of all high variance proteins maintained high CVs in all three anticoagulant conditions (265/1,151 at 1 h and 340/1,229 at 24 h).
3.3 |. Systematic effects of blood processing time on platelet protein composition
Unsupervised principal component analysis (PCA) of quantified platelet protein composition markedly clustered samples by blood storage duration more than by anticoagulant type (Fig. 3A, S. Table S2). Accordingly, we first examined the effects of sample processing time (24 h vs 1 h) on platelet proteome content. As summarized in Fig. 3B, of the 3,357 proteins quantified over all samples, 418 proteins were significantly (FDR<0.1) decreased (206) or increased (212) at 24 h vs 1 h. Gene ontology (GO) and Reactome pathway analyses noted systematic relations among many of these proteins differentially abundant at 24 h vs 1 h, where proteins involved in membrane trafficking were downregulated, while other sets of proteins involved in exocytosis were up-regulated (Fig. 3C). STRING and Reactome pathway analysis [36, 37] specifically organized clusters of differentially abundant proteins around mitochondrial respiration and oxidative phosphorylation, ER to Golgi transport, Rab regulation of vesicle trafficking, and ubiquitination and proteasome degradation (Fig. 3D). Altogether, storage of anticoagulated whole blood prior to processing to prepare washed platelets was associated with significant, time-dependent changes in levels of platelet proteins involved in membrane trafficking, energy-dependent metabolism and other biological processes.
Figure 3. Systematic effects of blood anticoagulation time on platelet proteome.
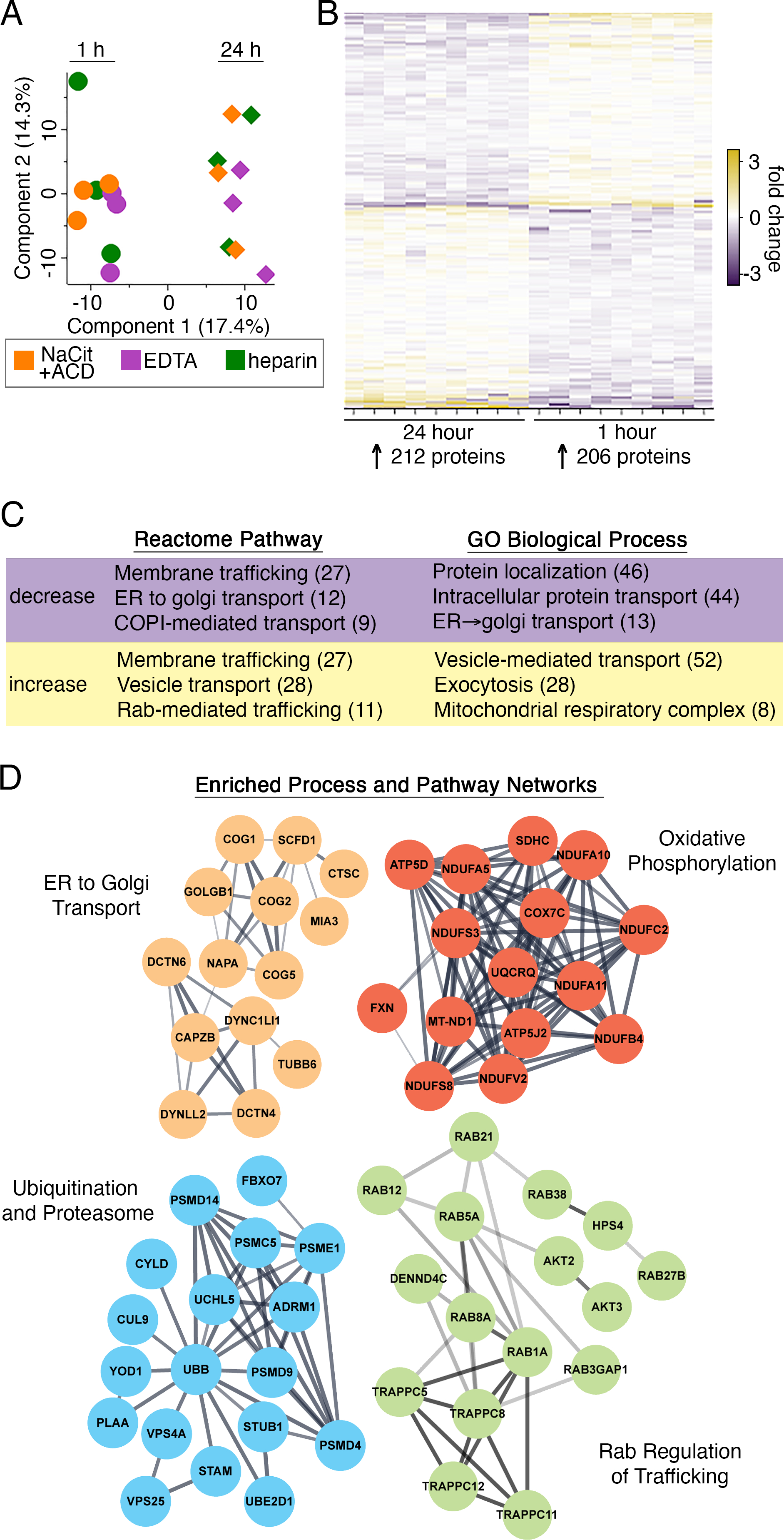
(A) Principal component analysis (PCA) plot of platelet proteome composition clusters samples by storage time (1 h vs 24 h). Circles (1 h samples) and diamond shapes (24 h) are color-coded according to anticoagulant. (B) Heat map of 418 differentially abundant proteins (FDR<0.1, S. Table S2) for 24 h vs 1 h proteomes (n=9, all anticoagulants combined). (C) The top 3 most significant up- and down-regulated Gene Ontology (GO) biological processes and Reactome pathways are noted. Numbers in parentheses indicate number of proteins identified for each corresponding functional annotation. (D) STRING-derived interactomes of 418 DE proteins with functional clusters. Interactive and searchable versions of the proteomics data and analyses above are available via the START app [75] at: https://kcvi.shinyapps.io/STARTapp_846/
3.4 |. Relative effect of EDTA anticoagulation on platelet proteome
To identify platelet proteome alterations specific to EDTA anticoagulation, we compared proteomic data of platelets from blood stored for 1 h and 24 h in EDTA with that of platelets from 1 h storage in NaCit+ACD as reference. Only four differentially abundant proteins were noted between platelet proteomes from 1 h NaCit+ACD and 1 h EDTA-anticoagulated samples (Fig. 4A, S. Table S2). However, a comparison of 24 h EDTA platelets with 1 h NaCit+ACD platelets revealed 28 differentially abundant proteins after adjusting for temporal changes in protein abundance. Of the 28 differentially abundant proteins, 14 were upregulated, especially C1R (complement C1r subcomponent) and FCN3 (ficolin-3) (Fig. 4A–C). The FCN3 protein had the largest fold change of reporter ion intensity (>10-fold) in platelets from EDTA-anticoagulated blood compared to NaCit+ACD or heparin at either time point (S. Table S2). The relatively increased expression of FCN3 and C1R in washed platelets prepared from EDTA anticoagulated blood was orthogonally confirmed by Western blot analysis (Fig. 4D).
Figure 4. EDTA anticoagulation increases complement system and decreases α-granule proteins of washed platelet proteomes.
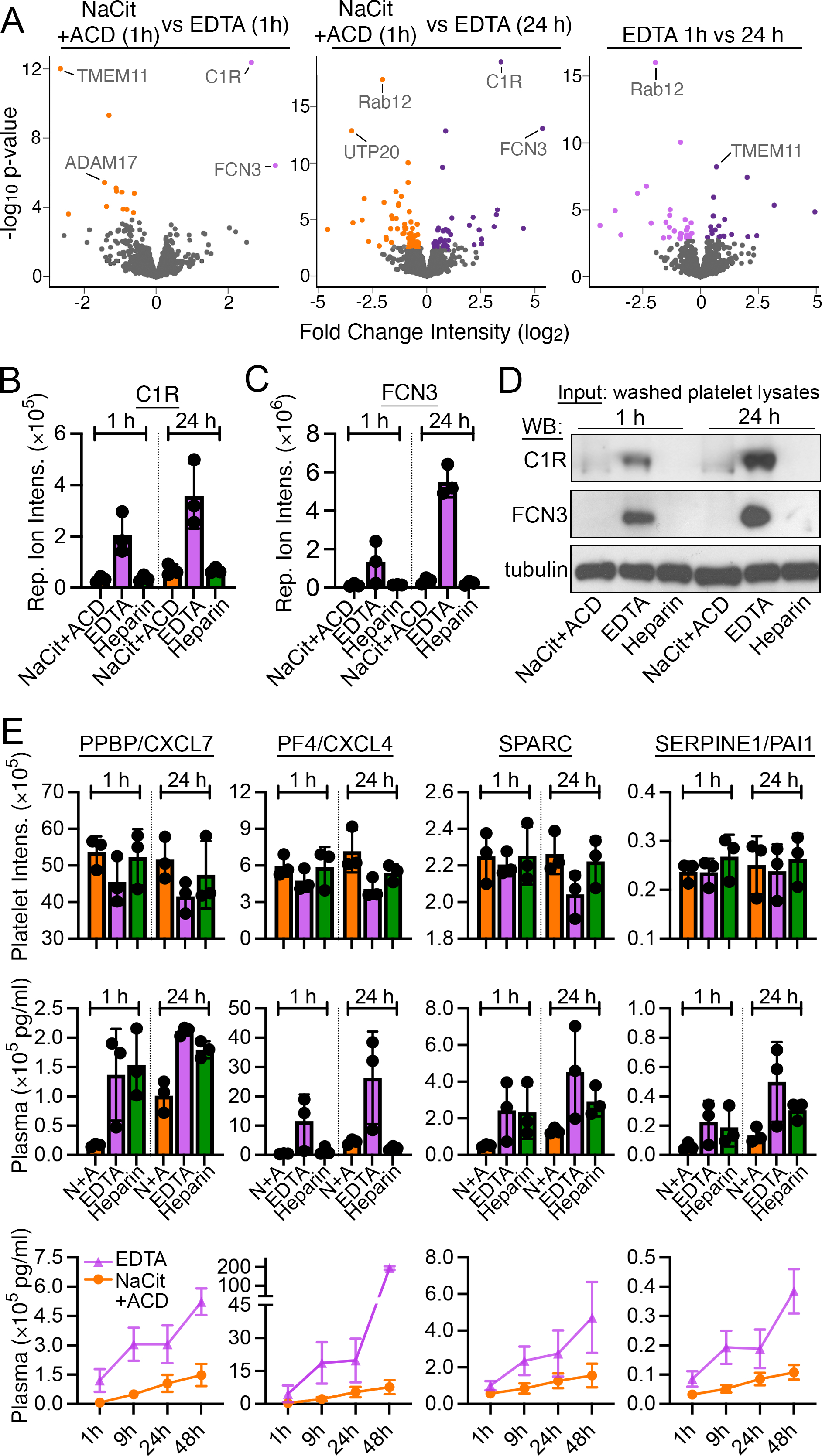
(A) Volcano plots of protein quantity (log2 MS3 reporter ion intensity) vs significance (p values) for washed platelet samples prepared from EDTA-anticoagulated blood. Proteins with significant differential intensities (false discovery rate FDR<0.1) are colored by anticoagulant (S. Table S2). Additional interactive and searchable MS data and statistical analyses above are available via the START app [75] at: https://kcvi.shinyapps.io/STARTapp_846/ (B) Bar plot of MS reporter ion intensity distribution for representative EDTA-upregulated protein C1R and (C) FCN3 (n=3). (D) Undigested platelet lysates from the same samples used for MS studies were analyzed by Western blot (WB) for C1R (~100 kD proenzyme) and FCN3 expression; α-tubulin serves as a control for equal protein amount loading. (E) Platelet MS reporter ion intensity measurements α-granule proteins, including PPBP/CXCL7, PF4/CXCL4, SPARC, and SERPINE1/PAI1 (top row). These same proteins were also quantified by multiplex bead-based Luminex assay in corresponding PPP samples at 1 h and 24 h of blood storage (middle row) and through 48 hours (lower row, only EDTA vs NaCit+ACD). Additional α-granule proteins are analyzed in S. Fig. S5.
Proteins that were significantly under expressed in EDTA (24 h) vs NaCit+ACD (1 h) platelets included proteins involved in clathrin-mediated endocytosis, including CLTA and REPS1, and platelet α-granule proteins THBS1 and TMSB4X. Although reporter ion intensities for other α-granule proteins such as PPBP, PF4, and SPARC [38, 39] were modestly lower in the EDTA platelet proteomes, Luminex assays of matched PPP samples, noted that these proteins were significantly increased in EDTA plasma over time (Fig 4E, S. Fig. S5).
3.5 |. Heparin-associated changes in platelet proteome
Quantitative TMT-MS experiments found that platelets prepared from heparin vs NaCit+ACD anticoagulated blood had significantly increased reporter ion intensities for 9 proteins (Fig. 5A and B, S. Table S2), several of which are commonly found in azurophil granules (MPO, ELANE, LTF, DEFA1) [40, 41] and nucleosomes (HIST1H1B, HIST1H1C, HIST1H4A, HIST1H2AB) [42]. Western blot analysis likewise found higher expression of MPO in lysates of washed platelets prepared from heparin anticoagulated blood (Fig. 5C). As azurophilic granules are typical of neutrophils [40, 41], we hypothesized that heparin induced neutrophil extracellular trap (NET) formation in whole blood, and that NET products (extruded chromatin with histone proteins accompanied by neutrophil granule proteins [43, 44]) were both released into plasma and sequestered by blood platelets before platelet isolation.
Figure 5. Effect of heparin anticoagulation on platelet proteome content.
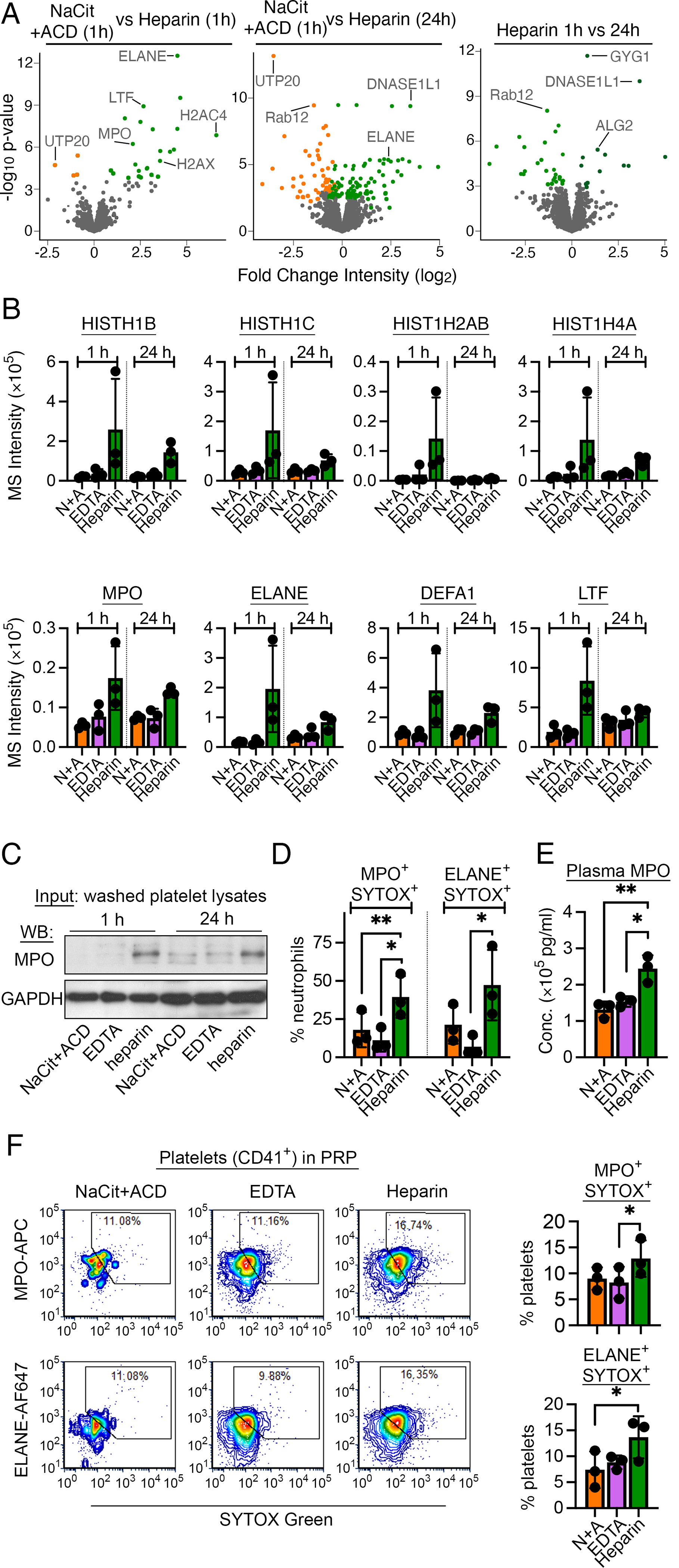
(A) Volcano plots of protein quantity (log2 MS3 reporter ion intensity) vs significance (p values) for washed platelet samples prepared from heparin-anticoagulated blood. Proteins with significant differential intensities (FDR<0.1) are colored by anticoagulant (S. Table S2). Additional interactive and searchable MS data and statistical analyses above are available via the START app [75] at: https://kcvi.shinyapps.io/STARTapp_846/ (B) MS reporter ion intensities are shown for selected histone and neutrophil proteins of interest (n=3). (C) Aliquots of undigested platelet lysates from the same samples used for MS studies were separated by SDS-PAGE, transferred to nitrocellulose membranes and probed by anti-MPO and anti-GAPDH (loading control) antibodies. (D) Anticoagulated whole blood samples were stained for CD45 (pan-leukocyte marker), CD66B (neutrophil marker), SYTOX-Green (membrane-impermeable DNA dye) and MPO or ELANE to examine NETosis in whole blood with flow cytometry (see S. Fig. S6 for additional details). NETs were estimated as the percent of CD45+CD66B+ cells (neutrophils) that co-stained for SYTOX/MPO or SYTOX/ELANE. (E) PPP was prepared 1 h after blood draw and assessed for MPO concentration as part of a multiplex Luminex assay in Fig. 4E above. MPO concentrations in NaCit+ACD plasma were adjusted for dilution in liquid anticoagulant. (F) Differentially anticoagulated PRP preparations (t=1 h) were stained for CD41a (platelet marker), SYTOX-Green and MPO or ELANE to assess platelet:NETs interactions. Left. Flow cytometry fluorescence intensity plots show the degree of co-staining for SYTOX/MPO or SYTOX/ELANE on CD41a+ (platelet) events. Gates show regions deemed double-positive events indicative of platelet:NETs binding. Right. Bar plots of results from n=3 biological replicates are shown. *p<0.05; **p<0.01 (paired t-test).
To examine NETs formation in whole blood samples, we used flow cytometry analysis for measured neutrophils that were also positive for both MPO or ELANE and SYTOX-Green (a membrane-impermeable dye that binds to surface-extruded DNA) (Fig. 5D, S. Fig. S6). Relative to NaCit+ACD and EDTA, heparin anticoagulation significantly increased the percent of neutrophils (CD45+CD66B+) in whole blood that also stained for MPO+SYTOX+ and ELANE+SYTOX+ (Fig. 5D). In addition, Luminex analysis found that MPO concentrations in plasma were significantly higher for heparin anticoagulated samples relative to NaCit+ACD and EDTA anticoagulation (Fig. 5E). Lastly, with flow cytometry analysis, we measured a higher proportion of MPO+SYTOX+ and ELANE+SYTOX+ platelets in heparinized PRP compared to platelets in PRP prepared with NaCit+ACD or EDTA (Fig. 5F). Together, these results suggest that heparin increases NETosis and platelet-NET interactions in whole blood ex vivo in a manner affecting platelet proteome content.
4 |. DISCUSSION
Biochemical and cellular assays of platelets isolated from whole blood are foundational in studies of hemostasis, thrombosis, inflammation, and other active areas of biomedical research, where platelets have causal as well as prognosticative roles in health and disease [1]. The molecular composition of platelets shapes platelet phenotype and is fundamental to understanding platelet function; however, the effects of many common blood processing procedures on platelet composition and associated ex vivo experimental results remain unspecified. Here, we provide the first-ever quantitative proteomics study comparing human platelets prepared from blood collected with different anticoagulants over a time course relevant to real world clinical studies and processing conditions. We find that citrate, EDTA and heparin anticoagulants each have specific effects on platelet function, as well as characteristic changes in platelet proteome in a manner dependent on processing times. Altogether, we show that preanalytical variables around blood collection methods can have systematic effects on platelet proteome content, with potential implications for studies of platelet function, biomarkers and precision medicine.
It has long been recognized that anticoagulants have specific effects on platelet phenotype and function in laboratory experiments as well as in clinical assays. As such, anticoagulants are standardized in clinical hematology tests, such as complete blood count (CBC), where EDTA preserves platelet numbers [7, 45], with the exception of EDTA-dependent pseudothrombocytopenia occurring in 0.1–2% of in-patients [46]. Outside of the clinic, classical electron microscopy studies have detailed that in the presence of EDTA, platelets form pseudopods, become swollen, remodel the open canalicular system (OCS), break down granules [47, 48]. More recent studies continue to note that, relative to citrate anticoagulation, EDTA increases platelet volume, destabilizes protein complexes and alters platelet membrane fluidity and mechanical deformability [9, 11, 13, 45]. In agreement with previous studies [49], we found that platelets in EDTA-anticoagulated blood had a distinct upregulation of α-granule secretion (i.e., CD62P+ platelets), increasing substantially 5 h after blood collection (Fig. 1G, S. Fig. S3). By comparing proteomes of platelets isolated from blood anticoagulated with EDTA vs citrate or heparin, we also noted a broad decrease in levels of several secreted α-granule proteins, together with a corresponding increase of these same proteins in plasma (Fig. 4). Such proteins included PPBP/CXCL7, PF4/CXCL4, SPARC and SERPINE1/PAI1 (Fig. 4E), which appear to be released from platelets into blood in the presence of EDTA. Notably, a number of studies have reported a downregulation of these and other granule proteins in platelets in disease using samples that include EDTA treatments [50–53].
In general, citrate anticoagulants – specifically, a combination of NaCit and ACD – are preferred for preparing blood, PRP and washed platelet samples for laboratory studies of platelet function [12, 54]. However, EDTA is still used in platelet function and biomarker studies, given a lower likelihood of inducing aggregates, decreased technical variability, and the convenience of using residual blood samples from other assays that require EDTA (i.e., studies of platelet nucleic acids with PCR [55, 56]). Several recent proteomics and RNA-sequencing studies of platelet biomarkers in disease, ranging from cancers to bleeding disorders, have included EDTA as an anticoagulant [17, 18, 52, 53, 57]. Our results herein suggest that some platelet profiling studies may benefit from a careful consideration of preanalytical factors, including effects of varying anticoagulants on platelet proteomes. For instance, in this study, we found that EDTA-anticoagulated blood yielded platelets with increased levels of complement system proteins, including complement C1R and ficolin-3 (FCN3) (Fig. 4). It is well established that EDTA inhibits complement-mediated platelet activation, as EDTA disrupts Ca2+-dependent complement protein interactions [58]; but, mechanisms whereby EDTA stabilizes or increases platelet C1R or FCN3 levels remain undescribed. Interestingly, FCN3 and other associated complement system proteins have been reported to be more abundant in platelet-derived vesicles from abdominal aortic aneurysm (AAA) patients using blood samples that were anticoagulated with EDTA [59].
Heparin is not typically used as an anticoagulant for ex vivo human platelet studies, as heparin directly activates platelets and promotes platelet aggregate formation in whole blood; however, heparin is standard to many clotting assays and is also readily available in clinical settings [9, 11]. Heparin is also still often used in mouse platelet studies, where blood collection by retroorbital access into heparinized glass capillary tubes is convenient and standardized in many laboratories, and with only limited effects on mouse platelet function [60]. We found that heparin significantly increased platelet-platelet and platelet monocyte aggregate formation in human whole blood, as well as release of platelet-derived EVs within 1 h of blood draw compared to EDTA and NaCit+ACD (Fig. 1). These effects of heparin occurred relatively rapidly, and more dramatically exacerbated inter-subject platelet proteome variances (Fig. 2C).
Notably, our proteomics analyses found that neutrophil proteins commonly associated with NETs formation were enriched in platelets isolated from blood collected into heparin, including MPO, which was reciprocally enriched in plasma from heparin anticoagulated blood (Fig. 5). Flow cytometry also noted increased neutrophil markers and DNA staining on platelets isolated from heparinized blood (Fig. 5). These findings are congruent with recent imaging flow cytometry studies, noting that heparin directly induces NETs formation ex vivo [61]. Such results may also be related to other developing intersections between platelets and neutrophils, where neutrophil proteins have recently been demonstrated to transfer to platelets in a manner related to cardiovascular pathology [62]. Likewise, histone proteins have been noted to be released into circulation through NETs formation during Dengue infection [63] and to bind to platelets [64] via platelet toll-like (TLRs) and other receptors [65]. Histones generally increase the procoagulant potential of platelets in vitro; as such, heparin-induced NETs formation may provide a means for heparin-enhanced platelet activation in whole blood. NETs and neutrophils also contribute to several features of heparin-induced thrombocytopenia (HIT) and thrombosis in vivo [66–68]. Our findings similarly support the potential of heparin to promote NETs formation – as well as platelet:NETs interactions – in vivo in a manner to be investigated further by future studies with clinical blood samples.
Beyond the specific preanalytical effects of anticoagulants noted above, we found that extended lapses between blood collection and sample processing times had even more pronounced effects on platelet protein composition, where significant changes in platelet proteomes were apparent for samples with 24 h vs 1 h delays in processing (Fig. 3, S. Table S2). While data on the effects of preanalytical variables on platelet proteomes is sparse, a number of related studies of plasma proteomes find that proteins in (platelet poor) plasma preparations vary in a manner related to blood processing conditions, where many such proteins of interest in plasma likely originate from platelets [19, 20, 22, 24, 69]. These and other prior studies have already indirectly suggested that blood processing compromises the stability platelet proteomes. Moreover, a recent study of EDTA anticoagulated blood found that processing time increased total protein content in plasma likely due to cellular leakage, in a processing time-dependent manner [70]. Similarly, we find that delays in blood processing decreased levels of several α-granule proteins in platelets, which reciprocally increased in plasma, suggesting that platelets continuously release (or fail to retain) α-granule contents following blood collection (Fig. 4). We also noted changes in platelet proteasome and mitochondrial respiratory complex proteins (Fig. 3), suggestive of increasing physiological stress during sample processing delays. Collectively, sample processing considerations are likely of strong interest to analyses of disease educated platelets as potential biomarker troves. Our results may also inform other translational areas, including efforts to limit platelet storage lesions for transfusion, where granule proteins in proteomes of platelet concentrates have been reported to decrease as a result of pathogen reduction technology [71]. Indeed, data on the relative effects of different anticoagulants on platelets after prolonged blood storage is sparse, but crucial for better design of platelet clinical studies that are constrained by long storage times in non-citrate anticoagulants [17, 18, 57]. Other ongoing clinical studies of ex vivo and in vivo anticoagulation with citrate vs heparin also note phenotypic differences in platelets in contexts such as kidney replacement therapy [72].
While our study provides a deep perspective on the consequences of anticoagulation and delayed processing on human platelet proteomes ex vivo, there are limitations to our work. For instance, due to technical limitations, we only compare platelet proteomes from a small set of healthy individuals at two different delayed processing time points in the context of a single research group. Future efforts will aim to determine how more specific delays between 1 h and 24 h temporally affect features of platelet proteomes, where ongoing studies from our group suggest minimal variations in platelet proteomes from citrate-anticoagulated blood with 1–3.5 h delays in processing (S. Fig. S7). Analyses in this study also focus on platelets prepared from blood anticoagulated with citrate under acidic conditions (NaCit+ACD) without extensively characterizing effects of sodium citrate (NaCit) anticoagulation alone on platelet proteomes. To address these types of limitations, during the course of our study, we carried out an additional TMT 11-plex experiment with a different set of donors to examine any differential effects of NaCit (alone) vs NaCit+ACD anticoagulation on platelet proteomes. As detailed in S. Fig. S8, platelets from NaCit vs NaCit+ACD samples had no significant differences in proteome composition, and NaCit vs EDTA or heparin samples showed similar proteome alterations as NaCit+ACD experiments above. Many other sample handling variables are not investigated in our study, including temperatures, centrifugation forces, as well as specific proteomics methodologies (i.e., mass spectrometry, 2D-DIGE [35], aptamer array capture [73], high-throughput imaging [74]). Beyond sample collection and processing effects, other preanalytical variables such as choice of lysis buffer, protease, and sample fractionation, are not specifically considered within the scope of the current study. Moreover, we only measure differences in abundance of entire proteins in platelets, and we do not examine how preanalytical factors affect protein phosphorylation (or any other protein posttranslational modifications) to inform platelet activation state. Nonetheless, the present work brings to light a need for standardization, or a recognition that some results of platelet studies may be related to sample processing, as well as in vivo physiology. Ideally, with further studies, it will become apparent how to best distinguish sample quality markers and process-related artifacts from clinically-relevant biomarkers and in vivo molecular changes in biochemical studies of platelet phenotype and function.
5 |. CONCLUSION
In conclusion, our study provides data on effects of blood sample anticoagulation and processing time on the results of ex vivo platelet proteome analyses. Previous studies of platelet proteomes have systematically assessed platelet protein composition in disease and other contexts, where some methodological steps vary widely. We find that differences in anticoagulation agents and preanalytical processing times affect platelet function and platelet protein content ex vivo. Future studies and concerted efforts will determine how to best approach platelet omics studies, and, provide a means to understand physiological and pathological mechanisms underlying platelet phenotypes independent of any impact from preanalytical processing variabilities.
Supplementary Material
ESSENTIALS.
Variations in blood processing protocols affect platelet function and protein content ex vivo.
Effects of preanalytical processing on platelet proteomes are measured with mass spectrometry.
Delays in blood processing contribute to platelet proteome variability ex vivo.
EDTA and heparin anticoagulation distinctly effect ex vivo platelet and plasma protein content.
AKNOWLEDGEMENTS
This work was supported by the Medical Research Foundation of Oregon, a Scholar Award from the American Society of Hematology (to J.E.A.) and the National Institutes of Health (R01HL146549 to J.E.A. and R01HL101972 to O.J.T.M.). Mass spectrometric analysis was partially supported by NIH core Grants P30 EY010572 and P30 CA069533 and shared instrument Grant S10OD-012246. Support for this work also comes from the Cancer Early Detection Advanced Research Center (CEDAR3500918) and Knight Cancer Institute at Oregon Health & Science University.
Footnotes
CONFLICT OF INTEREST DISCLOSURES
The authors do not declare any conflicts of interest.
REFERENCES
- 1.Aslan JE. Platelet Proteomes, Pathways, and Phenotypes as Informants of Vascular Wellness and Disease. Arterioscler Thromb Vasc Biol. 2021; 41: 999–1011. 10.1161/ATVBAHA.120.314647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gremmel T, Frelinger AL 3rd, Michelson AD. Platelet Physiology. Semin Thromb Hemost. 2016; 42: 191–204. 10.1055/s-0035-1564835. [DOI] [PubMed] [Google Scholar]
- 3.Gomez LA, Escobar M, Penuela O. Standardization of a Protocol for Obtaining Platelet Rich Plasma from blood Donors; a Tool for Tissue Regeneration Procedures. Clin Lab. 2015; 61: 973–80. [PubMed] [Google Scholar]
- 4.Dagur PK, McCoy JP, Jr. Collection, Storage, and Preparation of Human Blood Cells. Curr Protoc Cytom. 2015; 73: 5 1–5 1 16. 10.1002/0471142956.cy0501s73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Burzynski LC, Pugh N, Clarke MCH. Platelet Isolation and Activation Assays. Bio Protoc. 2019; 9: e3405. 10.21769/BioProtoc.3405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Roos B, Duthie SJ, Polley AC, Mulholland F, Bouwman FG, Heim C, Rucklidge GJ, Johnson IT, Mariman EC, Daniel H, Elliott RM. Proteomic methodological recommendations for studies involving human plasma, platelets, and peripheral blood mononuclear cells. J Proteome Res. 2008; 7: 2280–90. 10.1021/pr700714x. [DOI] [PubMed] [Google Scholar]
- 7.Hardy M, Lessire S, Kasikci S, Baudar J, Guldenpfennig M, Collard A, Dogne JM, Chatelain B, Jacqmin H, Lecompte T, Mullier F. Effects of Time-Interval since Blood Draw and of Anticoagulation on Platelet Testing (Count, Indices and Impedance Aggregometry): A Systematic Study with Blood from Healthy Volunteers. J Clin Med. 2020; 9. 10.3390/jcm9082515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salzman EW, Rosenberg RD, Smith MH, Lindon JN, Favreau L. Effect of heparin and heparin fractions on platelet aggregation. J Clin Invest. 1980; 65: 64–73. 10.1172/JCI109661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Spezia J, Hermann PB, Comar SR, Picheth G, Henneberg R, Utiyama SRR. Anticoagulant Choices Affect the Mean Platelet Volume Measurement by Impedance. Clin Lab. 2018; 64: 217–20. 10.7754/Clin.Lab.2017.170806. [DOI] [PubMed] [Google Scholar]
- 10.Weber D, Nakashima MO. Platelet count in sodium citrate-anticoagulated whole blood: Comparison to EDTA-anticoagulated results and stability over time. Int J Lab Hematol. 2021; 43: e35–e7. 10.1111/ijlh.13350. [DOI] [PubMed] [Google Scholar]
- 11.Golanski J, Pietrucha T, Baj Z, Greger J, Watala C. Molecular insights into the anticoagulant-induced spontaneous activation of platelets in whole blood-various anticoagulants are not equal. Thromb Res. 1996; 83: 199–216. 10.1016/0049-3848(96)00129-6. [DOI] [PubMed] [Google Scholar]
- 12.Hechler B, Dupuis A, Mangin PH, Gachet C. Platelet preparation for function testing in the laboratory and clinic: Historical and practical aspects. Res Pract Thromb Haemost. 2019; 3: 615–25. 10.1002/rth2.12240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sachs L, Wesche J, Lenkeit L, Greinacher A, Bender M, Otto O, Palankar R. Ex vivo anticoagulants affect human blood platelet biomechanics with implications for high-throughput functional mechanophenotyping. Commun Biol. 2022; 5: 86. 10.1038/s42003-021-02982-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fisher MH, Di Paola J. Genomics and transcriptomics of megakaryocytes and platelets: Implications for health and disease. Res Pract Thromb Haemost. 2018; 2: 630–9. 10.1002/rth2.12129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zimmerman GA, Weyrich AS. Signal-dependent protein synthesis by activated platelets: new pathways to altered phenotype and function. Arterioscler Thromb Vasc Biol. 2008; 28: s17–24. 10.1161/ATVBAHA.107.160218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klement GL, Yip TT, Cassiola F, Kikuchi L, Cervi D, Podust V, Italiano JE, Wheatley E, Abou-Slaybi A, Bender E, Almog N, Kieran MW, Folkman J. Platelets actively sequester angiogenesis regulators. Blood. 2009; 113: 2835–42. 10.1182/blood-2008-06-159541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mantini G, Meijer LL, Glogovitis I, In ‘t Veld S, Paleckyte R, Capula M, Le Large TYS, Morelli L, Pham TV, Piersma SR, Frampton AE, Jimenez CR, Kazemier G, Koppers-Lalic D, Wurdinger T, Giovannetti E. Omics Analysis of Educated Platelets in Cancer and Benign Disease of the Pancreas. Cancers (Basel). 2020; 13. 10.3390/cancers13010066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Best MG, Sol N, Kooi I, Tannous J, Westerman BA, Rustenburg F, Schellen P, Verschueren H, Post E, Koster J, Ylstra B, Ameziane N, Dorsman J, Smit EF, Verheul HM, Noske DP, Reijneveld JC, Nilsson RJA, Tannous BA, Wesseling P, Wurdinger T. RNA-Seq of Tumor-Educated Platelets Enables Blood-Based Pan-Cancer, Multiclass, and Molecular Pathway Cancer Diagnostics. Cancer Cell. 2015; 28: 666–76. 10.1016/j.ccell.2015.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Geyer PE, Voytik E, Treit PV, Doll S, Kleinhempel A, Niu L, Muller JB, Buchholtz ML, Bader JM, Teupser D, Holdt LM, Mann M. Plasma Proteome Profiling to detect and avoid sample-related biases in biomarker studies. EMBO Mol Med. 2019; 11: e10427. 10.15252/emmm.201910427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Halvey P, Farutin V, Koppes L, Gunay NS, Pappas DA, Manning AM, Capila I. Variable blood processing procedures contribute to plasma proteomic variability. Clin Proteomics. 2021; 18: 5. 10.1186/s12014-021-09311-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kaisar M, van Dullemen LFA, Thezenas ML, Zeeshan Akhtar M, Huang H, Rendel S, Charles PD, Fischer R, Ploeg RJ, Kessler BM. Plasma degradome affected by variable storage of human blood. Clin Proteomics. 2016; 13: 26. 10.1186/s12014-016-9126-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rai AJ, Gelfand CA, Haywood BC, Warunek DJ, Yi J, Schuchard MD, Mehigh RJ, Cockrill SL, Scott GB, Tammen H, Schulz-Knappe P, Speicher DW, Vitzthum F, Haab BB, Siest G, Chan DW. HUPO Plasma Proteome Project specimen collection and handling: towards the standardization of parameters for plasma proteome samples. Proteomics. 2005; 5: 3262–77. 10.1002/pmic.200401245. [DOI] [PubMed] [Google Scholar]
- 23.Savage AK, Gutschow MV, Chiang T, Henderson K, Green R, Chaudhari M, Swanson E, Heubeck AT, Kondza N, Burley KC, Genge PC, Lord C, Smith T, Thomson Z, Beaubien A, Johnson E, Goldy J, Bolouri H, Buckner JH, Meijer P, Coffey EM, Skene PJ, Torgerson TR, Li XJ, Bumol TF. Multimodal analysis for human ex vivo studies shows extensive molecular changes from delays in blood processing. iScience. 2021; 24: 102404. 10.1016/j.isci.2021.102404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Melani RD, Gerbasi VR, Anderson LC, Sikora JW, Toby TK, Hutton JE, Butcher DS, Negrao F, Seckler HS, Srzentic K, Fornelli L, Camarillo JM, LeDuc RD, Cesnik AJ, Lundberg E, Greer JB, Fellers RT, Robey MT, DeHart CJ, Forte E, Hendrickson CL, Abbatiello SE, Thomas PM, Kokaji AI, Levitsky J, Kelleher NL. The Blood Proteoform Atlas: A reference map of proteoforms in human hematopoietic cells. Science. 2022; 375: 411–8. 10.1126/science.aaz5284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li JL, Zarbock A, Hidalgo A. Platelets as autonomous drones for hemostatic and immune surveillance. J Exp Med. 2017; 214: 2193–204. 10.1084/jem.20170879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Babur O, Melrose AR, Cunliffe JM, Klimek J, Pang J, Sepp AI, Zilberman-Rudenko J, Tassi Yunga S, Zheng T, Parra-Izquierdo I, Minnier J, McCarty OJT, Demir E, Reddy AP, Wilmarth PA, David LL, Aslan JE. Phosphoproteomic quantitation and causal analysis reveal pathways in GPVI/ITAM-mediated platelet activation programs. Blood. 2020; 136: 2346–58. 10.1182/blood.2020005496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cooper ST, Wilmarth PA, Cunliffe JM, Klimek JE, Pang J, Tassi Yunga S, Minnier J, Reddy A, David LL, Aslan JE. Platelet proteome dynamics in hibernating 13-lined ground squirrels. Physiol Genomics. 2021. 10.1152/physiolgenomics.00078.2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Paulo JA, McAllister FE, Everley RA, Beausoleil SA, Banks AS, Gygi SP. Effects of MEK inhibitors GSK1120212 and PD0325901 in vivo using 10-plex quantitative proteomics and phosphoproteomics. Proteomics. 2015; 15: 462–73. 10.1002/pmic.201400154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shape Patscheke H. and functional properties of human platelets washed with acid citrate. Haemostasis. 1981; 10: 14–27. 10.1159/000214383. [DOI] [PubMed] [Google Scholar]
- 30.Hatskelzon L, Dalal BI, Shalev A, Robertson C, Gerrard JM. Wide distribution of granulophysin epitopes in granules of human tissues. Lab Invest. 1993; 68: 509–19. [PubMed] [Google Scholar]
- 31.Nomura S, Nagata H, Oda K, Kokawa T, Yasunaga K. Effects of EDTA on the membrane glycoproteins IIb-IIIa complex--analysis using flow cytometry. Thromb Res. 1987; 47: 47–58. 10.1016/0049-3848(87)90239-8. [DOI] [PubMed] [Google Scholar]
- 32.Aatonen MT, Ohman T, Nyman TA, Laitinen S, Gronholm M, Siljander PR. Isolation and characterization of platelet-derived extracellular vesicles. J Extracell Vesicles. 2014; 3. 10.3402/jev.v3.24692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Plubell DL, Wilmarth PA, Zhao Y, Fenton AM, Minnier J, Reddy AP, Klimek J, Yang X, David LL, Pamir N. Extended Multiplexing of Tandem Mass Tags (TMT) Labeling Reveals Age and High Fat Diet Specific Proteome Changes in Mouse Epididymal Adipose Tissue. Mol Cell Proteomics. 2017; 16: 873–90. 10.1074/mcp.M116.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Burkhart JM, Vaudel M, Gambaryan S, Radau S, Walter U, Martens L, Geiger J, Sickmann A, Zahedi RP. The first comprehensive and quantitative analysis of human platelet protein composition allows the comparative analysis of structural and functional pathways. Blood. 2012; 120. 10.1182/blood-2012-04-416594. [DOI] [PubMed] [Google Scholar]
- 35.Winkler W, Zellner M, Diestinger M, Babeluk R, Marchetti M, Goll A, Zehetmayer S, Bauer P, Rappold E, Miller I, Roth E, Allmaier G, Oehler R. Biological variation of the platelet proteome in the elderly population and its implication for biomarker research. Mol Cell Proteomics. 2008; 7: 193–203. 10.1074/mcp.M700137-MCP200. [DOI] [PubMed] [Google Scholar]
- 36.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021; 49: D605–D12. 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, Doncheva NT, Legeay M, Fang T, Bork P, Jensen LJ, von Mering C. Correction to ‘The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets’. Nucleic Acids Res. 2021; 49: 10800. 10.1093/nar/gkab835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wijten P, van Holten T, Woo LL, Bleijerveld OB, Roest M, Heck AJ, Scholten A. High precision platelet releasate definition by quantitative reversed protein profiling--brief report. Arterioscler Thromb Vasc Biol. 2013; 33: 1635–8. 10.1161/ATVBAHA.113.301147. [DOI] [PubMed] [Google Scholar]
- 39.Maynard DM, Heijnen HF, Gahl WA, Gunay-Aygun M. The alpha-granule proteome: novel proteins in normal and ghost granules in gray platelet syndrome. J Thromb Haemost. 2010; 8: 1786–96. 10.1111/j.1538-7836.2010.03932.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rorvig S, Ostergaard O, Heegaard NH, Borregaard N. Proteome profiling of human neutrophil granule subsets, secretory vesicles, and cell membrane: correlation with transcriptome profiling of neutrophil precursors. J Leukoc Biol. 2013; 94: 711–21. 10.1189/jlb.1212619. [DOI] [PubMed] [Google Scholar]
- 41.Lominadze G, Powell DW, Luerman GC, Link AJ, Ward RA, McLeish KR. Proteomic analysis of human neutrophil granules. Mol Cell Proteomics. 2005; 4: 1503–21. 10.1074/mcp.M500143-MCP200. [DOI] [PubMed] [Google Scholar]
- 42.Kurumizaka H, Kujirai T, Takizawa Y. Contributions of Histone Variants in Nucleosome Structure and Function. J Mol Biol. 2021; 433: 166678. 10.1016/j.jmb.2020.10.012. [DOI] [PubMed] [Google Scholar]
- 43.Masuda S, Shimizu S, Matsuo J, Nishibata Y, Kusunoki Y, Hattanda F, Shida H, Nakazawa D, Tomaru U, Atsumi T, Ishizu A. Measurement of NET formation in vitro and in vivo by flow cytometry. Cytometry A. 2017; 91: 822–9. 10.1002/cyto.a.23169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rada B Neutrophil Extracellular Traps. Methods Mol Biol. 2019; 1982: 517–28. 10.1007/978-1-4939-9424-3_31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McShine RL, Sibinga S, Brozovic B. Differences between the effects of EDTA and citrate anticoagulants on platelet count and mean platelet volume. Clin Lab Haematol. 1990; 12: 277–85. 10.1111/j.1365-2257.1990.tb00038.x. [DOI] [PubMed] [Google Scholar]
- 46.Mant MJ, Doery JC, Gauldie J, Sims H. Pseudothrombocytopenia due to platelet aggregation and degranulation in blood collected in EDTA. Scand J Haematol. 1975; 15: 161–70. 10.1111/j.1600-0609.1975.tb01070.x. [DOI] [PubMed] [Google Scholar]
- 47.White JG, Krumwiede MD, Escolar G. EDTA induced changes in platelet structure and function: influence on particle uptake. Platelets. 1999; 10: 327–37. 10.1080/09537109975979. [DOI] [PubMed] [Google Scholar]
- 48.White JG. Effects of ethylenediamine tetracetic acid (EDTA) on platelet structure. Scand J Haematol. 1968; 5: 241–54. 10.1111/j.1600-0609.1968.tb01743.x. [DOI] [PubMed] [Google Scholar]
- 49.Ritchie JL, Alexander HD, Rea IM. Flow cytometry analysis of platelet P-selectin expression in whole blood--methodological considerations. Clin Lab Haematol. 2000; 22: 359–63. 10.1046/j.1365-2257.2000.00339.x. [DOI] [PubMed] [Google Scholar]
- 50.Sims MC, Mayer L, Collins JH, Bariana TK, Megy K, Lavenu-Bombled C, Seyres D, Kollipara L, Burden FS, Greene D, Lee D, Rodriguez-Romera A, Alessi MC, Astle WJ, Bahou WF, Bury L, Chalmers E, Da Silva R, De Candia E, Deevi SVV, Farrow S, Gomez K, Grassi L, Greinacher A, Gresele P, Hart D, Hurtaud MF, Kelly AM, Kerr R, Le Quellec S, Leblanc T, Leinoe EB, Mapeta R, McKinney H, Michelson AD, Morais S, Nugent D, Papadia S, Park SJ, Pasi J, Podda GM, Poon MC, Reed R, Sekhar M, Shalev H, Sivapalaratnam S, Steinberg-Shemer O, Stephens JC, Tait RC, Turro E, Wu JKM, Zieger B, BioResource N, Kuijpers TW, Whetton AD, Sickmann A, Freson K, Downes K, Erber WN, Frontini M, Nurden P, Ouwehand WH, Favier R, Guerrero JA. Novel manifestations of immune dysregulation and granule defects in gray platelet syndrome. Blood. 2020; 136: 1956–67. 10.1182/blood.2019004776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Bergemalm D, Ramstrom S, Kardeby C, Hultenby K, Eremo AG, Sihlbom C, Bergstrom J, Palmblad J, Astrom M. Platelet proteome and function in X-linked thrombocytopenia with thalassemia and in silico comparisons with gray platelet syndrome. Haematologica. 2021; 106: 2947–59. 10.3324/haematol.2020.249805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van Bergen M, Marneth AE, Hoogendijk AJ, Van Alphen FPJ, Van den Akker E, Laros-Van Gorkom BAP, Hoeks M, Simons A, De Munnik SA, Janssen J, Martens JHA, Jansen JH, Meijer AB, Van der Reijden BA. Specific proteome changes in platelets from individuals with GATA1-, GFI1B-, and RUNX1-linked bleeding disorders. Blood. 2021; 138: 86–90. 10.1182/blood.2020008118. [DOI] [PubMed] [Google Scholar]
- 53.van Oorschot R, Hansen M, Koornneef JM, Marneth AE, Bergevoet SM, van Bergen M, van Alphen FPJ, van der Zwaan C, Martens JHA, Vermeulen M, Jansen P, Baltissen MPA, Gorkom B, Janssen H, Jansen JH, von Lindern M, Meijer AB, van den Akker E, van der Reijden BA. Molecular mechanisms of bleeding disorderassociated GFI1B(Q287*) mutation and its affected pathways in megakaryocytes and platelets. Haematologica. 2019; 104: 1460–72. 10.3324/haematol.2018.194555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aslan JE, Itakura A, Gertz JM, McCarty OJ. Platelet shape change and spreading. Methods Mol Biol. 2012; 788: 91–100. 10.1007/978-1-61779-307-3_7. [DOI] [PubMed] [Google Scholar]
- 55.Garcia ME, Blanco JL, Caballero J, Gargallo-Viola D. Anticoagulants interfere with PCR used to diagnose invasive aspergillosis. J Clin Microbiol. 2002; 40: 1567–8. 10.1128/JCM.40.4.1567-1568.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Djordjevic V, Stankovic M, Nikolic A, Antonijevic N, Rakicevic LJ, Divac A, Radojkovic M. PCR amplification on whole blood samples treated with different commonly used anticoagulants. Pediatr Hematol Oncol. 2006; 23: 517–21. 10.1080/08880010600751900. [DOI] [PubMed] [Google Scholar]
- 57.Best MG, Sol N, In ‘t Veld S, Vancura A, Muller M, Niemeijer AN, Fejes AV, Tjon Kon Fat LA, In Huis, ‘t Veld AE, Leurs C, Le Large TY, Meijer LL, Kooi IE, Rustenburg F, Schellen P, Verschueren H, Post E, Wedekind LE, Bracht J, Esenkbrink M, Wils L, Favaro F, Schoonhoven JD, Tannous J, Meijers-Heijboer H, Kazemier G, Giovannetti E, Reijneveld JC, Idema S, Killestein J, Heger M, de Jager SC, Urbanus RT, Hoefer IE, Pasterkamp G, Mannhalter C, Gomez-Arroyo J, Bogaard HJ, Noske DP, Vandertop WP, van den Broek D, Ylstra B, Nilsson RJA, Wesseling P, Karachaliou N, Rosell R, Lee-Lewandrowski E, Lewandrowski KB, Tannous BA, de Langen AJ, Smit EF, van den Heuvel MM, Wurdinger T. Swarm Intelligence-Enhanced Detection of Non-Small-Cell Lung Cancer Using Tumor-Educated Platelets. Cancer Cell. 2017; 32: 238–52 e9. 10.1016/j.ccell.2017.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Thielens NM, Illy C, Bally IM, Arlaud GJ. Activation of human complement serine-proteinase C1r is down-regulated by a Ca(2+)-dependent intramolecular control that is released in the C1 complex through a signal transmitted by C1q. Biochem J. 1994; 301 (Pt 2): 509–16. 10.1042/bj3010509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fernandez-Garcia CE, Burillo E, Lindholt JS, Martinez-Lopez D, Pilely K, Mazzeo C, Michel JB, Egido J, Garred P, Blanco-Colio LM, Martin-Ventura JL. Association of ficolin-3 with abdominal aortic aneurysm presence and progression. J Thromb Haemost. 2017; 15: 575–85. 10.1111/jth.13608. [DOI] [PubMed] [Google Scholar]
- 60.Grill A, Kiouptsi K, Karwot C, Jurk K, Reinhardt C. Evaluation of blood collection methods and anticoagulants for platelet function analyses on C57BL/6J laboratory mice. Platelets. 2020; 31: 981–8. 10.1080/09537104.2019.1701185. [DOI] [PubMed] [Google Scholar]
- 61.Lelliott PM, Momota M, Shibahara T, Lee MSJ, Smith NI, Ishii KJ, Coban C. Heparin induces neutrophil elastase-dependent vital and lytic NET formation. Int Immunol. 2020; 32: 359–68. 10.1093/intimm/dxz084. [DOI] [PubMed] [Google Scholar]
- 62.Joshi A, Schmidt LE, Burnap SA, Lu R, Chan MV, Armstrong PC, Baig F, Gutmann C, Willeit P, Santer P, Barwari T, Theofilatos K, Kiechl S, Willeit J, Warner T, Mathur A, Mayr M. Neutrophil-Derived Protein S100A8/A9 Alters the Platelet Proteome in Acute Myocardial Infarction and Is Associated With Changes in Platelet Reactivity. Arterioscler Thromb Vasc Biol. 2021: ATVBAHA121317113. 10.1161/ATVBAHA.121.317113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Opasawatchai A, Amornsupawat P, Jiravejchakul N, Chan-In W, Spoerk NJ, Manopwisedjaroen K, Singhasivanon P, Yingtaweesak T, Suraamornkul S, Mongkolsapaya J, Sakuntabhai A, Matangkasombut P, Loison F. Neutrophil Activation and Early Features of NET Formation Are Associated With Dengue Virus Infection in Human. Front Immunol. 2018; 9: 3007. 10.3389/fimmu.2018.03007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Trugilho MRO, Hottz ED, Brunoro GVF, Teixeira-Ferreira A, Carvalho PC, Salazar GA, Zimmerman GA, Bozza FA, Bozza PT, Perales J. Platelet proteome reveals novel pathways of platelet activation and platelet-mediated immunoregulation in dengue. PLoS Pathog. 2017; 13: e1006385. 10.1371/journal.ppat.1006385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Semeraro F, Ammollo CT, Morrissey JH, Dale GL, Friese P, Esmon NL, Esmon CT. Extracellular histones promote thrombin generation through platelet-dependent mechanisms: involvement of platelet TLR2 and TLR4. Blood. 2011; 118: 1952–61. 10.1182/blood-2011-03-343061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Perdomo J, Leung HHL, Ahmadi Z, Yan F, Chong JJH, Passam FH, Chong BH. Neutrophil activation and NETosis are the major drivers of thrombosis in heparin-induced thrombocytopenia. Nat Commun. 2019; 10: 1322. 10.1038/s41467-019-09160-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Gollomp K, Kim M, Johnston I, Hayes V, Welsh J, Arepally GM, Kahn M, Lambert MP, Cuker A, Cines DB, Rauova L, Kowalska MA, Poncz M. Neutrophil accumulation and NET release contribute to thrombosis in HIT. JCI Insight. 2018; 3. 10.1172/jci.insight.99445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zhou P, Li T, Jin J, Liu Y, Li B, Sun Q, Tian J, Zhao H, Liu Z, Ma S, Zhang S, Novakovic VA, Shi J, Hu S. Interactions between neutrophil extracellular traps and activated platelets enhance procoagulant activity in acute stroke patients with ICA occlusion. EBioMedicine. 2020; 53: 102671. 10.1016/j.ebiom.2020.102671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Kim HJ, Rames MJ, Tassi Yunga S, Armstrong R, Morita M, Ngo ATP, McCarty OJT, Civitci F, Morgan TK, Ngo TTM. Irreversible alteration of extracellular vesicle and cell-free messenger RNA profiles in human plasma associated with blood processing and storage. Sci Rep. 2022; 12: 2099. 10.1038/s41598-022-06088-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Huang J, Khademi M, Lindhe O, Jonsson G, Piehl F, Olsson T, Kockum I. Assessing the Preanalytical Variability of Plasma and Cerebrospinal Fluid Processing and Its Effects on Inflammation-Related Protein Biomarkers. Mol Cell Proteomics. 2021; 20: 100157. 10.1016/j.mcpro.2021.100157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Salunkhe V, De Cuyper IM, Papadopoulos P, van der Meer PF, Daal BB, Villa-Fajardo M, de Korte D, van den Berg TK, Gutierrez L. A comprehensive proteomics study on platelet concentrates: Platelet proteome, storage time and Mirasol pathogen reduction technology. Platelets. 2019; 30: 368–79. 10.1080/09537104.2018.1447658. [DOI] [PubMed] [Google Scholar]
- 72.Zarbock A, Kullmar M, Kindgen-Milles D, Wempe C, Gerss J, Brandenburger T, Dimski T, Tyczynski B, Jahn M, Mulling N, Mehrlander M, Rosenberger P, Marx G, Simon TP, Jaschinski U, Deetjen P, Putensen C, Schewe JC, Kluge S, Jarczak D, Slowinski T, Bodenstein M, Meybohm P, Wirtz S, Moerer O, Kortgen A, Simon P, Bagshaw SM, Kellum JA, Meersch M, Investigators R, the Sepnet Trial G. Effect of Regional Citrate Anticoagulation vs Systemic Heparin Anticoagulation During Continuous Kidney Replacement Therapy on Dialysis Filter Life Span and Mortality Among Critically Ill Patients With Acute Kidney Injury: A Randomized Clinical Trial. JAMA. 2020; 324: 1629–39. 10.1001/jama.2020.18618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Daniels JR, Cao Z, Maisha M, Schnackenberg LK, Sun J, Pence L, Schmitt TC, Kamlage B, Rogstad S, Beger RD, Yu LR. Stability of the Human Plasma Proteome to Pre-analytical Variability as Assessed by an Aptamer-Based Approach. J Proteome Res. 2019; 18: 3661–70. 10.1021/acs.jproteome.9b00320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Backstrom A, Kugel L, Gnann C, Xu H, Aslan JE, Lundberg E, Stadler C. A Sample Preparation Protocol for High Throughput Immunofluorescence of Suspension Cells on an Adherent Surface. J Histochem Cytochem. 2020; 68: 473–89. 10.1369/0022155420935403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Nelson JW, Sklenar J, Barnes AP, Minnier J. The START App: a web-based RNAseq analysis and visualization resource. Bioinformatics. 2017; 33: 447–9. 10.1093/bioinformatics/btw624. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


