Abstract
Nucleic acid fragment analysis via separation and detection are routine operations in molecular biology. However, analysis of small single-stranded nucleic acid fragments (<100nt) is challenging and mainly limited to labor-intensive polyacrylamide gel electrophoresis or high-cost capillary electrophoresis methods. Here we report an alternative method, a microfluidic chip electrophoresis system that provides a size resolution of 5nt and a detection time of one minute per sample of fluorescence-labeled DNA/RNA fragments. The feasibility of this system was evaluated by quantifying CRISPR-Cas9 cleavage efficiency and the detection resolution was evaluated by analyzing ssDNA/RNA adenylation and phosphorylation. We employed this system to study the RNA capping efficiency and double-stranded DNA unwinding efficiency in isothermal amplification as two examples for assay design and evaluation. The microfluidic chip electrophoresis system provides a rapid, sensitive, and high-throughput fluorescence fragment analysis (FFA), and can be applied for enzyme characterization, reaction optimization, and product quality control in various molecular biology processes.
INTRODUCTION
Separation and detection of nucleic acid fragment change in size and charge are fundamental to understanding many molecular processes. Large changes in fragment size are easily resolved by regular agarose gel electrophoresis. However, the detection of small changes in size or charge is challenging. For example, nucleic acid phosphorylation, only a change in charge, typically relies on denaturing polyacrylamide gel electrophoresis (PAGE) and autoradiography (1); Adenylation, transferring the adenyl group to the 5'-phosphorylated end of the ‘donor’ strand, is mainly monitored through denaturing PAGE (2); For mRNA capping, adding an N7-methylated guanosine to the first nucleotide of the RNA via a reverse 5′-5′ triphosphate linkage, capillary electrophoresis(CE) is reported as an analytical method (3).
To analyze fragments with such small changes, the following features should be considered in the experimental design and platform selection. First, proper gel chemistry is chosen for better resolution. Second, short fragments are used as the substrates. Shorter fragments are relatively more impacted by charge/size changes in electrophoresis and result in better separation. Third, oligonucleotide substrates are labeled by fluorescence instead of intercalating dye. Fluorescence by intercalating dye results from both the length and quantity of oligonucleotides (4). However, detection from fluorescence-labeled fragments provides more information: its signal intensity reflects the molar ratio of analyzed fragments without calibration; it differentiates the substrate strand from non-labeled fragments. Fourth, the platform can support high-throughput screening through an integrated separation, detection, and data analysis process. Currently, traditional agarose gel electrophoresis cannot meet the separation resolution or streamline data analysis requirements, while PAGE cannot support high-throughput screening or automated data processing (5). Although CE meets both requirements and has been applied for the characterization of nucleic acid modification enzymes (6,7), the high cost of CE instruments limits its availability to core facilities or external resources.
Here we demonstrate a microfluidic chip electrophoresis system for fluorescence fragment analysis (FFA) using LabChip® GX Touch™ Nucleic Acid Analyzer (GX Touch™) with a newly developed fluorescence fragment analysis reagent (FFA evaluation kit) on a microfluidic chip. In this system, sample loading and data acquisition are automated and rapid, allowing ∼1 min processing time per sample with up to 48 samples in a 96-well plate. Substrates of interest are fluorescently labeled for enzymatic reaction setup and sample purification is optional depending on whether reaction solutions are compatible with GX Touch™ (Figure 1A). Subsequent sample denaturing using DMSO is performed to minimize the impact of secondary structures (Figure 1B). Peak size, height, and area of fluorescence-labeled fragments (either initial substrates, intermediates, or end products) are automatically aligned by the lower marker and analyzed using the fluorescence-labeled single-stranded DNA (ssDNA) ladder (Figure 1C). The data can be further processed for peak percentage or peak ratio depending on assay requirements (Figure 1D). This microfluidic system supports high-throughput FFA and integrates sample separation, detection, data visualization, and primary data analysis reports. We apply this microfluidic chip electrophoresis system and demonstrate its capability by RNA capping and dsDNA unwinding efficiency studies. Furthermore, we propose the potential of this system as a powerful tool for a broad range of applications in FFA.
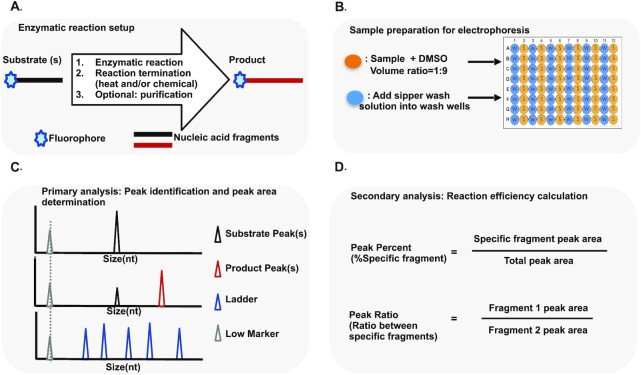
Figure 1.
Microfluidic chip electrophoresis workflow. Microfluidic chip electrophoresis workflow is illustrated schematically in panels A, B, C and D. (A) Enzymatic reaction setup: fluorescence labeled substrates (labeled at 5′ end or 3′ end or internal) are processed through enzymatic reaction based on assay design and followed by reaction termination (heat and/or chemical treatment) and purification (optional). (B) Sample preparation for electrophoresis: samples are mixed with DMSO at a 1:9 volume ratio and added into sample wells of a 96-well plate. Sipper wash solution is added into wash wells of 96 well plate. Then the 96-well sample plate is loaded onto LabChip® GX Touch™ Nucleic Acid Analyzer for electrophoresis. (C) Primary analysis: sample peaks are automatically aligned to lower marker and fluorescently labeled single-stranded DNA (ssDNA) ladder for size calling by GX Touch™ Reviewer software. Peaks (fragments) of interest including substrate, product control, or products with intermediates are identified. GX Touch™ Reviewer software determines the peak height, peak size and peak area and these data can be exported into a .csv file for additional analysis. (D) Secondary analysis: reaction efficiency can be calculated through percentage of specific fragment or ratio between specific fragments.
MATERIALS AND METHODS
Oligonucleotides and fluorescence fragment analysis (FFA) assay
Oligonucleotide sequences are listed in Supplemental Table S1. All microfluidic chip electrophoresis assays were performed using the FFA evaluation kit (for evaluation purpose only, PerkinElmer) on the LabChip® GX Touch™ HT Nucleic Acid Analyzer (PerkinElmer, Catalog# CLS137031) with HT DNA 5K/RNA/CZE LabChip (PerkinElmer, Catalog# 760435).
Assay specifications for FFA including the detection limit of oligonucleotide labeling dyes and size calling precision are listed in Supplemental Tables S2 and S3. This kit is designed for analyzing 20nt to 120nt fragments and includes chip storage buffer, gel matrix, lower marker, and ladder. For assay setup, the chip was rinsed twice before adding gel matrix and lower marker into the designated wells. The ladder tube was prepared by mixing 12μl ladder with 108μl DMSO. 750μl water was added into the buffer tube. Samples were mixed with DMSO at a 1:9 volume ratio and then loaded for electrophoresis. Data analysis was performed using LabChip® GX Reviewer software version 5.8.84.0 (https://www.perkinelmer.com/lab-products-and-services/resources/labchip-and-optimizer-software-downloads.html) following the user manual.
RNA capping
RNA capping was performed using Vaccinia Capping System (NEB, Catalog#: M2080S) (Figure 2A). Various concentrations of Vaccinia virus Capping Enzyme (VCE, 1 nM, 10 nM and 45 nM) were incubated with 0.5μM triphosphate RNA (ppp-RNA1) at 37°C for 30 min in a 20 μl reaction with 1× RNA capping buffer, 100μM S-adenosylmethionine (SAM) and 0.5mM GTP. The reaction was inactivated by the addition of EDTA to 5mM and heated at 70°C for 10 min (3,8). The negative control was uncapped RNA substrate (ppp-RNA1) without VCE, while the positive control was a 1:1 molar ratio mixture of uncapped RNA (ppp-RNA1) and Cap0-mRNA (N-7mGpppRNA1).
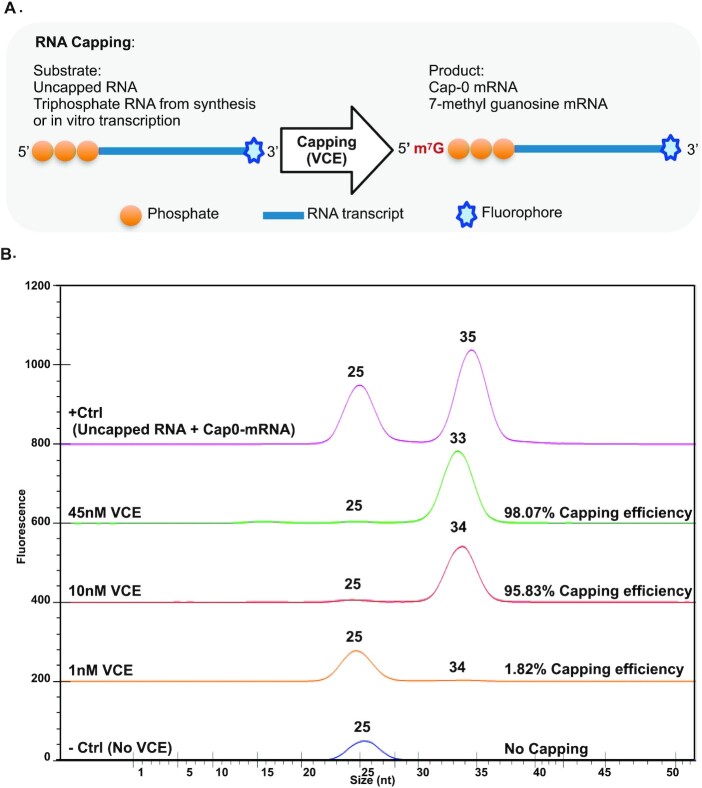
Figure 2.
RNA capping. (A) RNA capping reaction study design using Vaccinia virus Capping Enzyme (VCE). (B) RNA capping efficiency study result. + Ctrl (positive control): a 1:1 ratio mixture of uncapped RNA and Cap0-mRNA. –Ctrl (negative control): uncapped RNA without VCE enzyme mix. Capping efficiency (peak percentage) is automatically calculated by the GX Touch™ software, dividing the peak area of capped RNA by the sum of peak areas of uncapped and capped RNA.
dsDNA unwinding study
A 110-mer pre-annealed duplex was first prepared by a polymerase extension reaction using 2X Phire Hot Start II PCR Master Mix (ThermoFisher, Catalog# F125S) with 3 μM primer (Primer_110) and 2.5 μM template (Template_110mer) in a 20 μl reaction using the following thermal cycler program: 98°C/30 s, 30 cycles (98°C/5 s, 55°C/5 s, 72°C/10 s), 72°C/1 min. The polymerase extension product was tested for purity using the GX Touch™.
Thermophilic helicase-dependent DNA amplification (tHDA)-based isothermal amplification reaction was conducted in a 20 μl reaction mixture with IsoAmp® II Universal tHDA Kit (NEB, Catalog# H0110S) by mixing 4 mM MgSO4, 40 mM NaCl, 1.4 μl IsoAmp® dNTP solution and 1.4 μl IsoAmp® Enzyme Mix with two primer/duplex preparations: 300 nM primer (Primer_80) and 300 nM 110-mer duplex; and 300 nM primer (Primer_80) and 150 nM duplex. The reaction mixture was incubated at 65°C for 20 min and then inactivated at 95°C for 5 min. IsoAmp® Enzyme Mix was substituted with water as the negative control and Bst DNA Polymerase Large Fragment (BstLF) (NEB, Catalog# M0275S) as the polymerase control.
Recombinase Polymerase Amplification (RPA)-based isothermal amplification reaction was conducted in a 50 μl reaction mixture with TwistAmp® Liquid Basic Kit (TwistDx, Catalog# TALQAS01) by mixing 25 μl 2× Reaction Buffer, 5 μl 10× Basic E-Mix, 14 mM MgOAc, 1.8 mM total dNTPs, 2.5 μl 20× Core Reaction Mix with two primer/duplex preparations: 300 nM primer (Primer_80_v2, 35nt) and 300 nM 110-mer duplex; and 300 nM primer (Primer_80_v2) and 150 nM 110-mer duplex. The reaction mixture was incubated at 37°C for 20 min and then inactivated at 95°C for 5 min. 20× Core Reaction Mix was substituted with water as the negative control and BstLF which is reported with amplification activity at 37°C, as the polymerase control. Additionally, the primer length preference of RPA-based amplification was tested by replacing Primer_80_v2 (35nt) with Primer_80 (20nt). RPA products were purified using ZR-96 Oligo Clean & Concentrator kit (ZYMO RESEARCH, Catalog# D4062) before electrophoresis analysis.
The impact of human genomic DNA on RPA was evaluated using Primer_80_v2. 2μl of human genomic DNA (Sigma, Catalog# 11691112001) at 200, 50, 5 and 0.5 ng/μl concentration was added to the RPA reaction with 300 nM primer and 300 nM 110-mer duplex. 2 μl of water substituted human genomic DNA as the positive control. For each human genomic DNA concentration, RPA reaction without a duplex template was prepared as a comparison. The reaction was incubated at 37°C for 20 min, inactivated at 95°C for 5 min, and then purified using ZR-96 Oligo Clean & Concentrator kit before electrophoresis analysis.
RESULTS
Here we demonstrate the use of fluorescent oligonucleotide substrates and high-throughput microfluidic chip electrophoresis to study nucleic acid modifications. The system is applied to fragment analysis (50nt to –120nt) through a CRISPR/Cas9 cleavage efficiency study (Supplemental Figure S1) and provides a good resolution for small ssDNA/RNA fragments with different modifications: one adenylation modification at 25nt (Supplemental Figure S2); one or more phosphorylation modification at 25nt, 40nt and 60nt (Supplemental Figure S3). Detailed design and analysis can be found in supplemental materials. We then applied this system and provided examples of assay development for two research hotspots: RNA capping study; dsDNA unwinding study in isothermal amplification.
Characterization of VCE-mediated mRNA capping
Monitoring the mRNA capping status and characterization of mRNA capping enzymes are important for both in vivo nucleic acid metabolism and in vitro molecular processes. In eukaryotes, 5′ end mRNA capping is an essential modification that protects mature mRNA from degradation and participates in nuclear export and translation initiation (9). In in vitro mRNA vaccine production, the 5′-capping of mRNA is critical to its integrity upon delivery to the target site and overall immunogenicity, which underscores the importance of capping methods (10). Here the mRNA capping system from Vaccinia virus was tested to evaluate the capability of microfluidic chip electrophoresis. First, uncapped RNA (RNA1) is ∼25nt and capped RNA product (N-7mGpppRNA1) is ∼34nt (Figure 2B). Only one peak, the triphosphate RNA substrate (ppp-RNA1), is detected without VCE in the reaction (Figure 2B). The Cap0-mRNA (N-7mGpppRNA1) peak starts to be detected at 1nM VCE and its peak area percentage increases to 95.83% and 98.07% when VCE concentration reaches 10 and 45 nM, which indicates the increment of RNA capping efficiency (Figure 2B). These results show the efficiency of the VCE RNA capping system is enzyme dose dependent. Altogether, microfluidic chip electrophoresis can analyze uncapped RNA and Cap0-mRNA at 25nt with good resolution and be applied to mRNA capping efficiency characterization.
Characterization of dsDNA unwinding efficiency during isothermal amplification
Isothermal amplification-based assays are easy-to-operate processes that rapidly amplify nucleic acids at a constant temperature which is suitable for point-of-care settings or at-home testing. More investigations are underway to further implement and improve these technologies (e.g. protein engineering, new protein characterization) for field-deployable molecular diagnostics. Several isothermal technologies are based on a group of synergistic enzymes that open DNA duplexes without thermocycling, such as helicase-dependent amplification (HDA) and recombinase polymerase amplification (RPA) (11,12). In HDA and RPA technologies, rapid isothermal amplification depends on the synchronization of multiple events (duplex unwinding, primer binding, polymerase extension) engaging multiple enzymes (enzymes for duplex unwinding or strand exchange, polymerases, and accessory proteins). Analyzing duplex opening efficiencies and the rate-limiting factors of synchronization events are essential and fundamental for isothermal amplification optimization. However, traditional unwinding assays are generally performed through gel-shifting or fluorescence/radioactive signal changes to monitor the unwound ssDNA substrates (Supplemental Figure S4). The process of traditional unwinding assays is time-consuming and cannot achieve high-throughput or automation, and the following detection through autoradiography and fluorography image systems only provides qualitative information. Moreover, these assay conditions do not reflect the reaction condition of unwinding events during isothermal amplification. To improve the method of dsDNA unwinding evaluation, here we propose a new assay design (Supplemental Figure S5) and demonstrate the successful application of microfluidic chip electrophoresis on the characterization of double-stranded DNA (dsDNA) unwinding efficiency in HDA and RPA.
Thermophilic helicase-dependent amplification
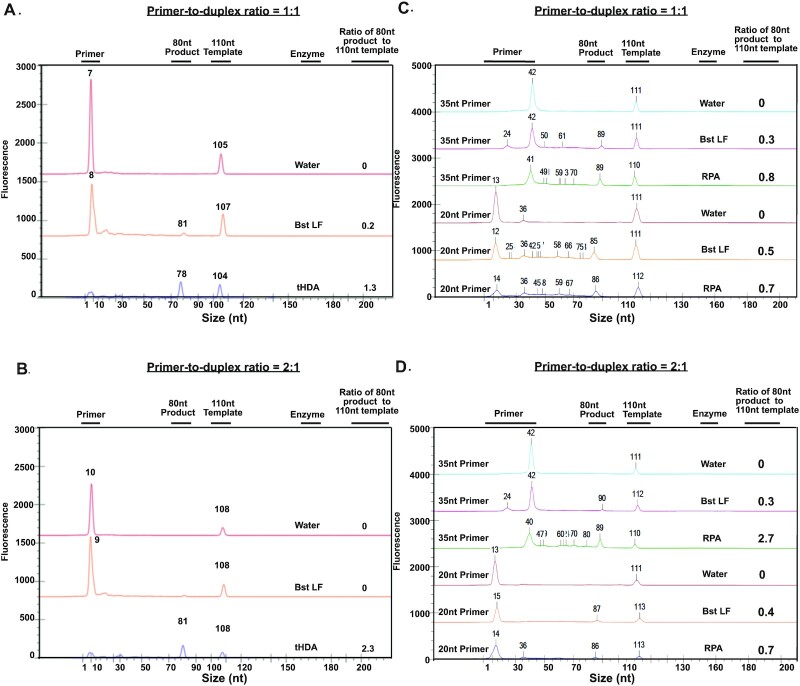
HDA relies on the dsDNA unwinding activity of a helicase to generate single-stranded templates for primer annealing and extension by a strand-displacing DNA polymerase (12). Higher dsDNA unwinding activity of the helicase can promote template-primer binding and contribute to higher amplification efficiency. Thermophilic Helicase-Dependent Amplification (tHDA) is based on a thermostable UvrD helicase (TteUvrD) to selectively amplify target sequences at 60–65°C and shows improved amplification sensitivity compared to the mesophilic form of HDA (mHDA) (12). This study was designed to evaluate dsDNA unwinding efficiency of the tHDA system (Supplemental Figure S5). dsDNA unwinding by tHDA was studied using a short fluorescently labeled primer that was added in excess in the reaction mixture and a pre-annealed duplex where one of the strands are fluorescently labeled. Two peaks are detected when the tHDA reaction system does not contain enzymes (labeled as ‘water’) (Figure 3A, B). The initial primer size is observed at ∼12nt and the 110nt fragment of dsDNA duplex is observed at ∼105nt. With enzymes in the tHDA reaction system, the extended product (80nt) from dsDNA unwinding event can be detected around 78 nt. When the primer-to-duplex ratio is 1:1, the extended-to-initial fragment ratio is 1.3, which indicates that most of the 300nM duplex templates are unwound and most of the 300nM primers can be annealed to the target strand (Figure 3A). When the primer-to-duplex ratio is 2:1, the extended-to-initial fragment ratio increases to 2.3 indicating that more duplex structures (either from the initial templates or extended products) can be unwound with additional primers (Figure 3B). Comparatively, when only BstLF is present in the tHDA reaction system, the extended product (∼80nt peak) is still visible under both conditions: this peak is detected at 81nt with an extended-to-initial fragment ratio of 0.2 when the primer-to-duplex ratio is 1:1 and this peak is not considered as substantial when the primer-to-duplex ratio is 2:1 since the signal is as low as unspecific products (Figure 3A, B). This result implies that there is a basal level of primer extension from BstLF considering its strand displacement activity and the effect of DNA breathing (13,14), but the amount of the extended product is considerably lower without helicase in the system. Taken together, these data demonstrate that tHDA enzyme mix can unwind blunt-end dsDNA duplexes efficiently and synchronize duplex unwinding, primer binding, and polymerase extension in just 20 minutes at defined tHDA isothermal amplification conditions. More importantly, this dsDNA unwinding process can be monitored by microfluidic chip electrophoresis which can promote high throughput operation on the protein engineering of tHDA and the optimization of tHDA-based amplification systems.
Figure 3.
dsDNA duplex unwinding efficiency study with tHDA and RPA. (A)(B). dsDNA duplex unwinding efficiency study with tHDA. 20nt primer is used at each primer-to-duplex ratio setting. (A) primer-to-duplex ratio is 1:1. (B) primer-to-duplex ratio is 2:1. (C)(D). dsDNA duplex unwinding efficiency study with RPA. 20nt and 35nt primers are used at each primer-to-duplex ratio setting. (C) primer-to-duplex ratio is 1:1. (D) primer-to-duplex ratio is 2:1. Enzymes used in each reaction are labeled on the graph. Water is used as the negative control. Bst LF is used as the no helicase/recombinase control or polymerase only control. dsDNA unwinding efficiency is calculated using the peak area ratio of the 80nt extended product to 110nt duplex template.
Recombinase polymerase amplification
Like the HDA system, the RPA system opens the dsDNA target with a recombinase and has been used in developing many isothermal amplification-based diagnostics products. An efficient method to characterize dsDNA unwinding efficiency in RPA specifically could accelerate the product development of an RPA-based detection system. The RPA process operates at 37–42°C and starts when a recombinase protein uvsX binds to primers forming a recombinase-primer complex which then initiates a strand exchange/invasion process to open the duplex (11). Here we evaluated the strand exchange/invasion efficiency of the RPA system with two different primer designs (Supplemental Figure S5). As shown in Figure 3C and D, mainly two peaks can be detected when no enzyme is present within the RPA system: primers (observed at 41nt for 35nt primer, at 13nt for 20nt primer) and 110nt initial fragments from dsDNA duplex (observed at 111nt). With the RPA enzyme mix, the extended product can be detected at ∼87nt. However, the extended-to-initial fragment ratio is only 0.7 under the 20nt primer condition and with multiple small fragments less than 80nt (Figure 3C, D). Under the 35nt primer condition, the extended-to-initial fragment ratio is 0.8 when the primer-to-duplex ratio is 1:1 and increases to 2.7 when the primer-to-duplex ratio is 2:1 (Figure 3C, D). As a comparison, when BstLF polymerase is applied without the recombinase mixture, the amount of polymerase extension product decreases substantially to an extended-to-initial fragment ratio of 0.3, which does not increase even when primer-to-duplex ratio increases to 2:1. These results suggest that with the strand exchange activity, RPA enzyme mixture provides a higher amplification efficiency compared to BstLF polymerase, and RPA system has a higher strand exchange efficiency with longer primers (35nt) compared to short primers (20nt) in just 20 min at defined RPA isothermal amplification conditions. Compared to the results of the HDA study, more intermediate fragments are observed in RPA isothermal amplification conditions (Figure 3).
It has been reported that RPA amplification is inhibited by background genomic DNA (15). However, it is still not clear what event(s) during RPA amplification are affected by background genomic DNA. To understand the impact of background genomic DNA on RPA strand exchange efficiency, additional studies were performed. Five different amounts of human genomic DNA from 0, 1, 10, 100 to 400 ng were tested in the RPA system where the primer-to-duplex ratio is 1:1, and reactions without duplex template were set up as the comparison. The results show that the ratio of 80nt product to 110nt template gradually drops from 0.68 to 0.35 along with the increase of background human genomic DNA (Supplemental Figure S6). Above data indicate that more background DNA does interfere with the RPA strand exchange process. Overall, the studies have demonstrated that the strand exchange efficiency study is helpful to understand and further optimize RPA reactions.
DISCUSSION
Analysis of fluorescent oligonucleotide fragments using microfluidic chip electrophoresis provides a good resolution and a rapid method to characterize nucleic acid modification enzymes. The examples presented here demonstrate the utility and capability of microfluidic chip fluorescence fragment analysis for a broad analysis of nucleic acid modification enzymes. In addition, we have developed a novel approach to understanding dsDNA unwinding efficiency in isothermal amplification reaction as an example of assay design for potential applications.
Microfluidic chip fluorescence fragment analysis can be applied to study many biological processes by monitoring fluorescently labeled single-stranded DNAs or RNAs beyond the applications reported in this work. For example, different features of DNA/RNA polymerase can be evaluated using fluorescently labeled oligos to study polymerase extension activity, 3′-5′ exonuclease activity and strand displacement activity, hot-start technology efficiency, etc. Fluorescent substrates can be designed to study DNA/RNA cleavage specificity and efficiency of various DNA/RNA nucleases such as RNase H, RNase A, Restriction enzymes, and DNase. Furthermore, this system can also aid in optimizing Cas12a-based DETECTR, Cas13-based SHERLOCK, and SHERLOCKv2 nucleic acid diagnostics systems (16,17,18). Unlike CRISPR-Cas9 which is RNA-guided on-target DNA cleavage, when Cas12a or Cas13 recognizes its target in vitro, it becomes activated and promiscuously cleaves DNAs, and RNAs respectively (19). This promiscuous and rapid cleavage activity can be used to amplify signals from fluorescently labeled reporter fragments, making Cas12a and Cas13 well-suited for nucleic acid detection. Using similar assay design principles as shown in CRIPSR-Cas9 application (Supplemental Figure S1), fluorescence substrates can be DNA/RNA targets, reporter fragments, or CRISPR-guide RNAs. Assessing the specificity and efficiency of CRIPSR-Cas systems by fluorescence fragment analysis on the microfluidic chip will accelerate the development of rapid molecular diagnostics products with better sensitivity and specificity.
DATA AVAILABILITY
The data underlying this article are available in the article and in its online supplementary material.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Nidhi Nandu, Eleanore Dougherty, James White, and Lloyd Bwanali for their support during preparing this manuscript.
Contributor Information
Yali Sun, PerkinElmer Health Sciences Division, Waltham, MA 02451, USA.
Zhi-xiang Lu, PerkinElmer Health Sciences Division, Waltham, MA 02451, USA.
Michael Miller, PerkinElmer Health Sciences Division, Waltham, MA 02451, USA.
Thomas Perroud, PerkinElmer Health Sciences Division, Waltham, MA 02451, USA.
Yanhong Tong, PerkinElmer Health Sciences Division, Waltham, MA 02451, USA.
SUPPLEMENTARY DATA
Supplementary Data are available at NARGAB Online.
FUNDING
Funding for open access charge: PerkinElmer Health Sciences, Inc.
Conflict of interest statement. Although the authors are employed and funded by PerkinElmer Health Sciences, Inc., this should not detract from the objectivity of data generation or interpretation.
REFERENCES
- 1. Jain R., Shuman S. Characterization of a thermostable archaeal polynucleotide kinase homologous to human Clp1. RNA. 2009; 15:923–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Zhelkovsky A.M., McReynolds L.A. Simple and efficient synthesis of 5′ pre-adenylated DNA using thermostable RNA ligase. Nucleic Acids Res. 2011; 39:e117–e117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chan S.-H., Whipple J.M., Dai N., Kelley T.M., Withers K., Tzertzinis G., Corrêa I.R., Robb G.B. RNase H-based analysis of synthetic mRNA 5′ cap incorporation. RNA. 2022; 28:1144–1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Wong K.H., Campbell K.L. Factors that affect use of intercalating dyes such as PicoGreen, SYBR Green I, Syto-13 and Syto-82 in DNA assays. Biol. Reprod. 2011; 85:725–725. [Google Scholar]
- 5. Barril P., Nates S. Introduction to agarose and polyacrylamide gel electrophoresis matrices with respect to their detection sensitivities. Gel Electrophoresis - Principles and Basics. 2012; InTech. [Google Scholar]
- 6. Greenough L., Schermerhorn K.M., Mazzola L., Bybee J., Rivizzigno D., Cantin E., Slatko B.E., Gardner A.F. Adapting capillary gel electrophoresis as a sensitive, high-throughput method to accelerate characterization of nucleic acid metabolic enzymes. Nucleic Acids Res. 2016; 44:e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Durney B.C., Crihfield C.L., Holland L.A. Capillary electrophoresis applied to DNA: determining and harnessing sequence and structure to advance bioanalyses (2009–2014). Anal Bioanal. Chem. 2015; 407:6923–6938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Robb G.B., Chan S.-H., Roy B. Enzymatic RNA capping method. 2020; U.S. Patent Application No. 17/001,236.
- 9. Ramanathan A., Robb G.B., Chan S.-H. mRNA capping: biological functions and applications. Nucleic Acids Res. 2016; 44:7511–7526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rouf N.Z., Biswas S., Tarannum N., Oishee L.M., Muna M.M. Demystifying mRNA vaccines: an emerging platform at the forefront of cryptic diseases. RNA Biol. 2022; 19:386–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Piepenburg O., Williams C.H., Stemple D.L., Armes N.A. DNA detection using recombination proteins. PLoS Biol. 2006; 4:e204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. An L., Tang W., Ranalli T.A., Kim H.-J., Wytiaz J., Kong H. Characterization of a thermostable UvrD helicase and its participation in helicase-dependent amplification. J. Biol. Chem. 2005; 280:28952–28958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Roychoudhury R., Tu C.-P.D., Wu R. Influence of nucleotide sequence adjacent to duplex DNA termini on 3′ terminal labeling by terminal transferase. Nucleic Acids Res. 1979; 6:1323–1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Putnam B.F., van Zandt L.L., Prohofsky E.W., Mei W.N. Resonant and localized breathing modes in terminal regions of the DNA double helix. Biophys. J. 1981; 35:271–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Rohrman B., Richards-Kortum R. Inhibition of recombinase polymerase amplification by background DNA: a lateral flow-based method for enriching target DNA. Anal. Chem. 2015; 87:1963–1967. [DOI] [PubMed] [Google Scholar]
- 16. Chen J.S., Ma E., Harrington L.B., da Costa M., Tian X., Palefsky J.M., Doudna J.A. CRISPR-Cas12a target binding unleashes indiscriminate single-stranded dnase activity. Science. 2018; 360:436–439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gootenberg J.S., Abudayyeh O.O., Kellner M.J., Joung J., Collins J.J., Zhang F. Multiplexed and portable nucleic acid detection platform with Cas13, Cas12a, and Csm6. Science. 2018; 360:439–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gootenberg J.S., Abudayyeh O.O., Lee J.W., Essletzbichler P., Dy A.J., Joung J., Verdine V., Donghia N., Daringer N.M., Freije C.A. et al. Nucleic acid detection with CRISPR-Cas13a/C2c2. Science. 2017; 356:438–442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Mustafa M.I., Makhawi A.M. SHERLOCK and DETECTR: cRISPR-cas systems as potential rapid diagnostic tools for emerging infectious diseases. J. Clin. Microbiol. 2021; 59:e00745-20. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are available in the article and in its online supplementary material.