Abstract
Cadmium (Cd) exposure in adulthood is associated with nonalcoholic fatty liver disease (NAFLD), characterized by steatosis, inflammation, and fibrosis. The prevalence of NAFLD in children is increasing, suggesting a role for the developmental environment in programming susceptibility. However, the role of developmental Cd exposure in programming NAFLD and the underlying mechanisms remain unclear. We have proposed that imprinted genes are strong candidates for connecting the early life environment and later life disease. In support of this, we previously identified roles for the Imprinted Gene Network (IGN) and its regulator Zac1 in programming NAFLD in response to maternal metabolic dysfunction. Here, we test the hypothesis that developmental Cd exposure is sufficient to program NAFLD, and further, that this process is mediated by Zac1 and the IGN. Using mice, we show that developmental cadmium chloride (CdCl2) exposure leads to histological, biochemical, and molecular signatures of steatosis and fibrosis in juveniles. Transcriptomic analyses comparing livers of CdCl2-exposed and control mice show upregulation of Zac1 and the IGN coincident with disease presentation. Increased hepatic Zac1 expression is independent of promoter methylation and imprinting statuses. Finally, we show that over-expression of Zac1 in cultured hepatocytes is sufficient to induce lipid accumulation in a Ppar-dependent manner and demonstrate direct binding of Zac1 to the Pparγ promoter. Our findings demonstrate that developmental Cd exposure is sufficient to program NAFLD in later life, and with our previous work, establish Zac1 and the IGN as key regulators of prosteatotic and profibrotic pathways, two of the major pathological hallmarks of NAFLD.
Keywords: nonalcoholic fatty liver disease, developmental toxicology, epigenetics, cadmium, genomic imprinting, developmental programming
Anthropogenic activities deposit cadmium (Cd) into the environment at about 8000 tons/year, which is 5 times more than can be expected from natural sources (Nriagu, 1989). As a result, Cd accumulates in drinking water sources and agricultural land (ATSDR, 2012). It is classified as one of the top 10 chemicals of major public health concern by the World Health Organization (W.H.O., 2022). The main route of human exposure is via inhalation of cigarette smoke; however, for the nonsmoking portion of the population, the major route is through ingestion (ATSDR, 2012).
Chronic Cd exposure during adulthood causes renal and hepatotoxicity and is associated with softening of the bones, cancer, cardiovascular disease, and metabolic disorders including nonalcoholic fatty liver disease (NAFLD) (Hyder et al., 2013; Rahimzadeh et al., 2017; Satarug et al., 2010). NAFLD describes a spectrum of liver defects ranging from steatosis and nonalcoholic steatohepatitis to fibrosis and cirrhosis (Bertot and Adams, 2016). In mice, exposure to Cd causes hepatic steatosis, the first major histological hallmark of NAFLD, although a role for Cd in promoting progression of the disease beyond steatosis has not been shown (Go et al., 2015).
NAFLD is the leading type of chronic liver disease, resulting in $103 billion per year in medical costs and affecting 30% of the adult U.S. population (Le et al., 2017; Younossi et al., 2016). Diagnosis of NAFLD is occurring at increasingly younger ages, with up to 20% of adolescents and young adults currently affected, suggesting that susceptibility to NAFLD may be programmed by the environment during early life (Paik et al., 2022). Consistent with this hypothesis, we previously showed that mice nursed by dams with metabolic syndrome develop NAFLD as juveniles, presenting with steatosis and fibrosis (Baptissart et al., 2022). Although developmental exposure to Cd has been shown to program metabolic dysfunction, as well as neurobehavioral, cognitive, cardiovascular, and renal impairments, its contribution to the programming of NAFLD in later life is unclear (Ciesielski et al., 2012; Hudson et al., 2019; Jackson et al., 2020; Swaddiwudhipong et al., 2015).
The pathogenesis of NAFLD includes the activation of pathways that disrupt lipid metabolism, promote inflammation, and drive fibrogenic transformation. The nuclear receptor peroxisome proliferator-activated receptor (PPAR)γ is a major prosteatotic factor that heterodimerizes with retinoid X receptor α to reprogram transcriptional networks involved in lipid storage and metabolism (Morán-Salvador et al., 2011). Although the roles of this and other pathways in NAFLD progression have been extensively studied, the mechanisms through which NAFLD can be programmed by the developmental environment are not well characterized. Epigenetic mechanisms have been proposed as mediators, but few specific examples of causative epigenetic changes have been described (Gallego-Durán and Romero-Gómez, 2015; Sookoian et al., 2013). We and others have proposed that perturbation of imprinted genes, defined by their expression from a single parental allele, is a candidate mechanism for disease programming because of the unique properties and modes of epigenetic regulation of imprinted genes during development. DNA methylation at Imprinting Control Regions (ICRs) is sensitive to environmental perturbation during development, including in response to Cd exposure in humans (Cowley et al., 2018; House et al., 2019; Vidal et al., 2015). Once established, ICR epigenetic states are maintained throughout life, providing an epigenetic memory of the early life environment (Tucci et al., 2019). Imprinted genes also play key roles in liver development and energy homeostasis, suggesting that changes to their expression could influence hepatic metabolism (Pope et al., 2017; Smith et al., 2006). In support of this idea, we previously demonstrated a novel role for imprinted genes in our model of NAFLD programmed by maternal metabolic syndrome (Baptissart et al., 2022).
Many imprinted genes are coordinately expressed as part of an Imprinted Gene Network (IGN) controlled by the transcription factor Zac1 (also called Plagl1), itself encoded by an imprinted gene (Varrault et al., 2017). The IGN, composed of 409 genes, regulates extracellular matrix composition and is implicated in muscle regeneration and adipocyte differentiation (Al Adhami et al., 2015; Varrault et al., 2006). Using gene set enrichment analysis, we previously reported that IGN genes are highly predictive of “Hepatic Fibrosis”, adding further support for imprinted genes playing a role in programming liver disease (Baptissart et al., 2022). We showed that artificial over-expression of Zac1 in cultured hepatoma cells activated the IGN and increased procollagen production, a hallmark of fibrosis. This was associated with transcriptional upregulation of the profibrogenic cytokine Tgf-β1 and activation of Tgf-β1 signaling. Using chromatin immunoprecipitation (ChIP), we demonstrated the binding of Zac1 to the Tgf-β1 promoter, forming a direct link between imprinted genes and a major pathway underlying the pathogenesis of NAFLD-related fibrosis.
In this study, we tested the hypothesis that developmental cadmium chloride (CdCl2) exposure in mice is sufficient to program NAFLD in later life and that this is mediated by the imprinted Zac1 gene. We comprehensively evaluated mice exposed to CdCl2 during development using histological, biochemical, and molecular approaches. Using cultured mouse hepatocytes, we examined the role of Zac1 in lipid accumulation via Pparγ signaling.
Materials and methods
Ethics statement
Animal work was approved by the North Carolina State University Institutional Animal Care and Use Committee, under protocols 15-013-B and 19-049-B. Experiments were conducted in accordance with the Guiding Principles in the Use of Animals in Toxicology.
Animal model and CdCl2 exposure
Female C57Bl/6J and male CAST/EiJ mice were obtained from Jackson Laboratory at 3 and 4 weeks old, respectively, and were allowed an acclimation period of 1 week before experimental manipulations. Mice were maintained on a 14-h/10-h light/dark cycle at 30%–70% humidity, 22 ± 4°C and housed in Green Line IVC Sealsafe cage housing systems (Tecniplast) and fed AIN-93G rodent diet (Research Diets, D10012G) ad libitum for the duration of the study. At 4 weeks of age, female mice were provided unrestricted access to filtered drinking water (Millipore RiOs Essential RO water purification system) containing 0, 1, or 50 ppm CdCl2 (Sigma-Aldrich, 202908). One part per million CdCl2 is comparable to the levels of Cd typically found in the Earth’s crust (ATSDR, 2012). We have previously shown that 50 ppm CdCl2 in this mouse model leads to blood Cd levels in dams of 5.23 ± 0.99 μg/L (mean ± standard error) (Hudson et al., 2019). This is similar to the blood Cd levels of approximately 6 μg/L observed in people in a contaminated area of Japan (Sasaki et al., 2019).
At 9 weeks of age, female mice were mated with unexposed CAST/EiJ males. CdCl2 exposure ended when offspring reached postnatal day (PND) 10, the developmental equivalent of human birth (Semple et al., 2013).
F1 offspring were euthanized via decapitation at PND0 to standardize the litters to 6 pups with a male:female ratio of 1:1 whenever possible and tissues were collected and flash frozen. At PND21, a subset of F1 mice were euthanized after a 6-h fasting period and tissues were flash frozen and stored at −80°C until processing or prepared for histology as described below. The remaining F1 mice were weaned at PND21 and given ad libitum access to AIN-93G diet until euthanasia at PND90 after a 6-h fasting period, at which point tissues were collected and processed as described for PND21. The study was blinded until animals were selected for histological, biochemical, and molecular analyses. The numbers of animals and litters in each treatment group used in the analyses described below are presented in Supplementary Table 1. Animals were selected for inclusion in analyses by determining the 5–8 individuals with liver masses closest to the mean for each treatment group. Same-sex animals from the same litter were avoided whenever possible.
Cell culture
AML12 mouse hepatocytes (ATCC, CRL-2254) were cultured with Dulbecco’s modified Eagle’s medium (DMEM)/F-12 medium (Genesee Scientific) containing 10% fetal bovine serum (Genesee Scientific) and 100 U/mL penicillin, 100 μg/mL streptomycin (HyClone), supplemented with a mixture of 10 µg/mL insulin, 5.5 µg/mL transferrin, 6.7 ng/mL selenium (Thermo Fisher), and 40 ng/mL dexamethasone (Sigma). Cells were maintained in a humidified atmosphere at 5% CO2 and 37°C. All cells used for experiments were under 20 passages.
Transfection
AML12 cells were seeded at 400 000 cells/well in 6-well plates. Cells were starved with serum-free medium for 24 h. Transient expression of EGFP (control) or Zac1 was achieved by transfecting cells with 1 μg of pLenti-CMV-EGFP or pLenti-CMV-Zac1-FLAG using Lipofectamine 3000 according to the manufacturer’s instructions (Thermo Fisher, L3000015). These constructs have been described previously (Baptissart et al., 2022). For mRNA extraction and qRT-PCR, cells were collected 18 h after transfection.
Fatty acid and Pparγ inhibitor exposure
AML12 cells were cultured and transfected as described above. At 18 h post-transfection, cells were exposed to DMEM/F-12 medium containing 50 µg/µL oleic acid-BSA (Sigma-Aldrich, O3008), 100 µg/µL oleic acid-BSA (Sigma-Aldrich, O3008), or comparable levels of BSA (Sigma-Aldrich, A7030) (control). After 24 h, cells were prepared for histology as described below. For experiments involving the Pparγ inhibitor, cells were cultured, transfected, and exposed to fatty acids as described above. At the time of transfection, cells were dosed with 5 or 10 µg/µL of the Pparγ inhibitor T0070907 (Selleck Chemicals, S2871) then prepared for and processed through histology as described below.
Chromatin immunoprecipitation
AML12 cells were transfected with the pLenti-CMV-Zac1-FLAG construct as described above. ChIP was performed as described previously (Baptissart et al., 2022; Johnson et al., 2007) using the FLAG M2 antibody (Sigma-Aldrich, F7425) and an antibody to TBP as a positive control (Abcam, ab220788). IgG (Thermo Fisher, 11203D) was used as a control for nonspecific binding. Binding enrichment was determined by qPCR using primers shown in Supplementary Table 2.
Histology
A portion of F1 median liver lobes was fixed in 4% formaldehyde overnight at 4°C. Lobes were transferred to 70% ethanol and delivered to the NC State University Histology Laboratory for dehydration, paraffin embedding, and sectioning. Sections were stained with hematoxylin and eosin or Sirius red using protocols described previously (Baptissart et al., 2022). A portion of the left lobe for each animal was flash frozen and delivered to the UNC Histology Research Core Facility for 10 µm thick cryosectioning and storage at −80°C. Cryosections were stained with Oil Red O (ORO) using standard protocols (Baptissart et al., 2022). All images were taken at 40× magnification using brightfield microscopy, and a central vein was centered in each image.
AML12 cells were also stained with ORO (Sigma, O-0625). Briefly, cells were fixed in 10% formalin (Midland Scientific, MSI N0019) for 5 min then dehydrated in 1,2-Propanediol (Sigma, 134368) for 1 min. Cells were stained with 0.5% ORO for 10 min at 60°C then rinsed with 1,2-Propanediol for 1 min. The cells were counterstained for 15 s with Harris Hematoxylin (Sigma, HHS16) then placed in 0.5% ammonium water (Ricca Chemical, 633-32) for 1 min to improve contrast. Within 24 h, ORO signal was quantified in Photoshop (Adobe). Briefly, a minimum of 3, discrete images were taken at 40× magnification, imported into Photoshop, and lipid color was selected. A fuzziness setting of 50 was applied to the selection color, all pixels with the color range within the image were selected and then pixel area was calculated using the software.
Biochemical assays
Assays of hepatic triacylglycerides (Thermo Fisher, TR22421) and hydroxyproline (BioVision, K226) were performed according to the manufacturers’ instructions.
Nucleic acid isolation
A portion of the left liver lobe for females and males at PND0, 21, and 90 was homogenized using a microtube homogenizer (Biospec 3110Bx Cell Disrupter 4800, BZ10124883) in Trizol reagent (Invitrogen, 15596026) according to the manufacturer’s instructions. Nucleic acids were quantified on a Nanodrop 2000 and RNA integrity was confirmed using a 1.3% agarose gel.
AML12 cells were processed using the NucleoSpin kit (Macherey Nagel) to obtain RNA, which was quantified and validated as described above for liver tissue samples.
qRT-PCR
According to the manufacturer’s protocol (M-MLV RT enzyme, Promega), 500 ng of total RNA from livers and cells was used to synthesize first-strand cDNA. cDNA was diluted 1/5 or 1/10 for qRT-PCR, which was performed in triplicate on 96-well plates with a QuantStudio 3 system (Thermo Fisher) using SsoAdvanced Universal SYBR Green Supermix (Bio-Rad, 1725271) with standards in sequential dilution 1/5, 1/10, 1/20, 1/40, and 1/80 and water as a no template control (NTC). The cycling conditions were as follows: 95°C for 30 s; 40 cycles of 95°C for 15 s, 60°C for 30 s. The primer sequences are provided in Supplementary Table 2. The dissociation curves showed that there were no products in the NTC and that primers amplified a single PCR product. Amplification efficiencies were calculated. Beta actin was used as a reference gene and was not significantly differentially expressed between treatment groups (data not shown). Gene expression was quantified using the ΔΔCt method (Livak and Schmittgen, 2001).
DNA methylation analysis
Using the Zymo EZ DNA Methylation Kit (Zymo Research), 350 ng of genomic DNA was treated with sodium bisulfite. Bisulfite-converted DNA (bsDNA) was amplified by PCR and products were confirmed on a 1.3% agarose gel. PCR amplicons were then subjected to pyrosequencing using a Pyromark Q96 MD Pyrosequencer (Qiagen). The primer sequences and conditions are provided in Supplementary Table 2. Methylation level was determined for each CpG dinucleotide using Pyromark software (Qiagen).
Allele-specific expression analysis
cDNA was generated as described above and amplified by PCR using gene-specific primers. Forward and reverse primers were designed using Pyromark software to include a single nucleotide polymorphism (SNP) between the C57Bl/6J and CAST/EiJ parental strains within the amplified and sequenced region to assign transcripts to the parental alleles. The primer sequences and conditions are provided in Supplementary Table 2. Products were confirmed on a 1.3% agarose gel and were subject to pyrosequencing using a Pyromark Q96 MD Pyrosequencer (Qiagen). The percent contribution of each allele to total transcript abundance was determined.
RNA-sequencing
RNA samples isolated from PND0 and 21 female and male samples as described above were submitted to the NC State University Genomic Sciences Laboratory for Illumina RNA library construction and sequencing. RNA integrity, purity, and concentration were assessed using an Agilent 2100 Bioanalyzer with an RNA 6000 Nano Chip (Agilent Technologies, USA). All samples used for RNA-sequencing (RNA-seq) had an RNA integrity number (RIN) >9.0. cDNA libraries for Illumina sequencing were constructed using the NEBNext Ultra Directional RNA Library Prep Kit (NEB) and NEBNext Mulitplex Oligos for Illumina (NEB) using the manufacturer-specified protocol. The samples were sequenced on one lane of an Illumina NovaSeq S4, utilizing a 150 × 2 bp paired end S4 sequencing reagent kit (Illumina, USA). The software package Real-Time Analysis (RTA) was used to generate raw bcl files, which were then de-multiplexed by sample into fastq files for data submission.
An average of approximately 48.6 million paired-end RNA-seq reads were generated for each replicate. The quality of sequenced data was assessed using fastqc, and 12 poor-quality bases were trimmed from the 5′-end. The remaining reads were aligned to the mouse reference genome (mm38, version 100) downloaded from the Ensembl database using a STAR aligner with a WASP approach. This option allows mapping of one haplotype and supplies the other haplotype variants as VCF files (C57BL_6NJ version 5 SNPs downloaded from dbSNP142) (van de Geijn et al., 2015).
For each replicate, per-gene counts of uniquely mapped reads were calculated using the htseq-count script from the HTSeq python package (Anders et al., 2015). The count matrix was imported to R statistical computing environment for further analysis. Initially, genes that have no count in most replicate samples were discarded. The remaining count data were normalized for sequencing depth and distortion, and dispersion was estimated using the DESeq2 Bioconductor package (Love et al., 2014). A linear model was fitted using the treatment levels, and differentially expressed genes (DEGs) were identified after applying multiple testing corrections using the Benjamini-Hochberg procedure (Benjamini and Hochberg, 1995).
RNA-seq data are deposited in the Gene Expression Omnibus under accession number GSE201906.
Enrichment analysis
To enable comparison, the DEGs identified from the PND0 and 21 RNA-seq and the IGN and imprinted gene lists (Al Adhami et al., 2015) were converted to Ensembl ID v104 using the Database Conversions tool (bioDBnet) and the ID history converter (Ensembl) for consistency prior to enrichment analysis (Supplementary Table 3). After conversion, we retrieved 474/474 (new Ensembl version/old Ensembl version) female PND0 DEGs, 347/347 male PND0 DEGs, 4600/4602 female PND21 DEGs, 4701/4701 male PND21 DEGs, and 408/409 IGN IDs, including 80 imprinted genes. To assess enrichment of the IGN and imprinted genes within DEG datasets (Supplementary Table 3), only IGN and imprinted gene members expressed in the RNA-seq datasets were considered. Hypergeometric calculations were performed as described previously (Baptissart et al., 2022).
Statistical analyses
Unless otherwise stated, statistical analyses were performed using a one-way analysis of variance with Dunnett’s post hoc test comparing 0 ppm to 1 ppm and 50 ppm CdCl2 exposure groups using GraphPad Prism Version 8, or a Student’s t test, two-tailed, comparing 0 and 50 ppm CdCl2 exposure groups using Excel. Organ weight data were analyzed using an analysis of covariance with body weight as a variable. ChIP data were analyzed with a ratio paired t test, comparing FLAG pull down of the Pparγ promoter DNA sequence and a negative control (gene desert) region after normalizing to IgG background.
Unless otherwise stated, data are presented as the mean ± standard error of the mean. *p < .05, **p < .01, ***p < .001, ****p < .0001.
Results
Mouse model of developmental CdCl2 exposure
We established a mouse model of developmental CdCl2 exposure by exposing female C57Bl/6J mice to 0 (control), 1, or 50 ppm CdCl2 through drinking water for 5 weeks prior to mating. Females were bred to unexposed CAST/EiJ male mice to generate F1 hybrid animals that enabled analyses of allele-specific gene expression and DNA methylation (Figure 1A), as we have described previously (Simmers et al., 2022). Exposure continued through gestation until offspring reached PND10 (Figure 1B). The presence of CdCl2 did not significantly alter F0 female water consumption (Supplementary Figure 1A) or body mass (Supplementary Figure 1B) from those of controls. Exposure to CdCl2 did not affect offspring sex ratio (Supplementary Figure 2A) or litter size at PND0 (Supplementary Figure 2B), but exposure to 50 ppm CdCl2 did significantly reduce offspring survival (Supplementary Figure 2C) compared with the control group.
Figure 1.
Mating strategy and Cd exposure model used to produce maternally exposed F1 hybrid offspring. A, Mating strategy in which parental alleles can be discriminated by single nucleotide polymorphisms. B, Animals were exposed until offspring reached PND10, equivalent to human gestation. Abbreviations: Cd, cadmium; PND10, postnatal day 10.
Developmental CdCl2 exposure alters body and liver masses
In utero Cd exposure in humans is associated with fetal growth restriction and smaller body mass at birth (Huang et al., 2019; Wang et al., 2016). Consistent with this, F1 females and males exposed to 50 ppm CdCl2 weighed significantly less than those in the 0 ppm group at PND0 (Figure 2A). This phenotype persisted at PND21 (Figure 2B), when mice presented with significantly reduced masses of brain, gonadal white adipose tissue, kidney, gonad (males only), and leg muscle (males only), and increased masses of retroperitoneal white adipose tissue and heart (Supplementary Table 4). The mass of brown adipose tissue was also reduced for both sexes, albeit not significantly. These outcomes were not observed in the 1 ppm group. At PND90, both sexes showed similar body weights between groups (Figure 2C).
Figure 2.
Effects of CdCl2 exposure on F1 male and female body and liver mass. A-C, Body mass at PND0, PND21, and PND90. D-F, Liver mass at PND0, PND21, and PND90. For (A-C), Data are presented as means ± S.E. *p < .05, ****p < .0001. One-way ANOVA with Dunnett’s post hoc test comparing 1 and 50 ppm CdCl2 exposed mice to 0 ppm CdCl2 controls. For (D-F), Data are presented as means ± S.E. **p < .01. One-way ANCOVA with Fisher’s Least Significant Difference test comparing 1 and 50 ppm CdCl2 exposed mice to 0 ppm CdCl2 controls. Abbreviations: CdCl2, cadmium chloride; PND, postnatal day; ANCOVA, analysis of covariance; S.E., standard error.
At PND0, there were no differences in liver mass between groups for either sex (Figure 2D). However, at PND21, males in the 50 ppm group, but not the 1 ppm group, showed significantly reduced liver mass compared with control mice (Figure 2E). Fifty parts per million exposed males showed significantly increased liver mass at PND90 (Figure 2F).
Overall, these data indicate that 50 ppm developmental CdCl2 exposure induces growth defects, including of the liver, that persist beyond the period of exposure.
Developmental CdCl2 exposure is sufficient to program NAFLD
We next determined if developmental CdCl2 exposure leads to histological and biochemical signatures of NAFLD. At PND0, there was no evidence of differences in hepatic lipid accumulation between groups for either females or males (Supplementary Figures 3A and 3B). At PND21, both female and male F1 mice exposed to 50 ppm CdCl2 during development presented with steatosis, indicated by increased neutral lipid accumulation in hepatocytes. Mice exposed to 50 ppm CdCl2 also presented with fibrosis, indicated by increased collagen deposition (Figures 3A and 3B). The fibrosis phenotype was more pronounced in females than males. No differences in the accumulation of lipids and collagens between the 1 ppm CdCl2 group and controls were observed at this timepoint. At PND90, males and females from all treatment groups showed hepatic lipid accumulation and no differences between the groups were observed (Supplementary Figures 3C and 3D).
Figure 3.
Effect of developmental CdCl2 exposure on NAFLD programming at PND21. A and B, Representative photomicrographs (40×) of F1 female and male liver sections centered around a central vein. First row: H&E (HE) staining. Second row: Oil Red O (ORO) staining; arrowheads indicate neutral lipid droplets. Third row: Sirius red (SR) staining; arrowheads indicate collagen. Scale bars represent 200 µm. C, Quantification of hepatic triacylglycerides. D, Quantification of hepatic hydroxyproline. For (C and D), Data are presented as means ± S.E. ***p < .001 and ****p < .0001. One-way ANOVA with Dunnett’s post hoc test comparing 1 and 50 ppm CdCl2 exposed mice to 0 ppm CdCl2 controls. Abbreviations: CdCl2, cadmium chloride; NAFLD, nonalcoholic fatty liver disease; PND, postnatal day; H&E, hematoxylin and eosin; S.E., standard error.
Histological evidence for NAFLD at PND21 was supported by biochemical assays on liver tissue. Both females and males in the 50 ppm group showed significantly increased accumulation of hepatic triacylglycerides (Figure 3C), a measure of steatosis, compared with controls. Females but not males showed a significant increase in hepatic hydroxyproline (Figure 3D), a readout for fibrosis, compared with controls. This is consistent with the sex difference observed at the histological level. The 1 ppm CdCl2 group did not show significant increases in either measure when compared with the 0 ppm group.
Together, these histological and biochemical data show that developmental CdCl2 exposure is sufficient to program advanced stages of NAFLD in juvenile mice.
Markers of NAFLD and the IGN are upregulated at the transcriptional level at PND21
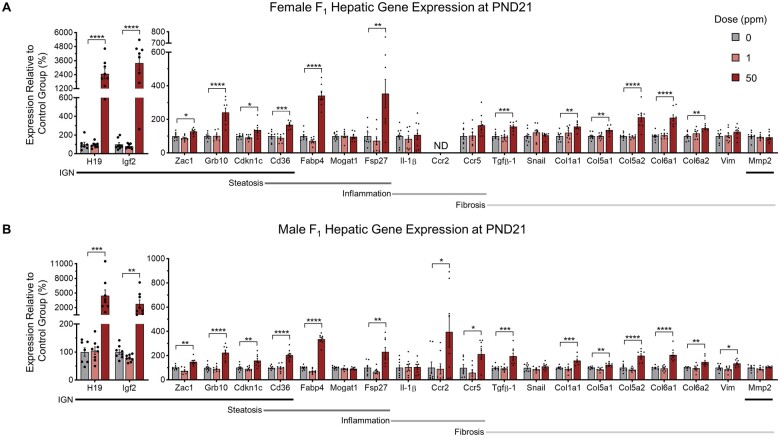
To determine if our observations at the histological and biochemical levels were reflected at the level of gene expression, qRT-PCR was performed on liver tissues. We targeted genes that are canonical markers of steatotic (Cd36, Mogat1, Fsp27, and Fabp4), inflammatory (Il-1β, Ccr2, and Ccr5), and fibrotic (Tgfβ-1, Snail, Col1a1, Col5a1, Col5a2, Col6a1, Col6a2, Vim, and Mmp2) pathways. At PND0, neither female nor male F1 mice showed significant overall differences in the expression of these genes between groups (Supplementary Figures 4A and 4B). However, at PND21, consistent with phenotypic presentation of NAFLD, both sexes in the 50 ppm group showed significant upregulation of genes indicative of these pathologies (Figures 4A and 4B). At PND90, neither female nor male F1 mice showed significant overall differences in the expression of these genes between groups (Supplementary Figures 5A and 5B). These results indicate transcriptional dysregulation of key pathways underlying NAFLD pathogenesis at PND21, consistent with phenotypic presentation at this timepoint.
Figure 4.
Effects of CdCl2 exposure on F1 hepatic gene expression at PND21. A and B, Female and male transcript abundance for genes of the IGN and steatosis, inflammation, and fibrosis pathways. Data are presented as means ± S.E. *p < .05, **p < .01, ***p < .001, ****p < .0001. One-way ANOVA with Dunnett’s post hoc test comparing 1 and 50 ppm CdCl2 exposed mice to 0 ppm CdCl2 controls. Abbreviations: CdCl2, cadmium chloride; PND, postnatal day; IGN, Imprinted Gene Network; S.E., standard error; ND, not detected.
To determine if the IGN was dysregulated in our model of NAFLD, we initially performed qRT-PCR on a subset of IGN genes (Zac1, H19, Grb10, Igf2, Cdkn1c, Cd36, and Mmp2). At PND0, overall, the IGN genes examined were not dysregulated for either sex in response to CdCl2 exposure (Supplementary Figures 4A and 4B). However, at PND21, all representative IGN members except for Mmp2 were upregulated for both sexes in the 50 ppm group compared with controls (Figures 4A and 4B). At PND90, IGN dysregulation did not persist for females or males (Supplementary Figures 5A and 5B). The phenotypic presentation of steatosis and fibrosis is associated with upregulation of IGN members suggesting a role for the IGN in CdCl2-induced juvenile NAFLD.
The IGN is enriched among differentially expressed genes at PND21
To determine the full extent of IGN genes that are dysregulated in response to developmental CdCl2 exposure, RNA-sequencing of F1 liver was performed on both sexes from the 0 ppm and 50 ppm groups at PND0 and 21. There were 474 and 347 DEGs (q-value < 0.05) between the two groups for F1 females and males, respectively, at PND0 (Supplementary Table 3). At PND21, there were 4602 DEGs for F1 females and 4701 DEGs for F1 males between the two groups (Supplementary Table 3). Genes of the IGN were significantly over-represented among PND21 DEGs for both F1 females and males (Figures 5A and 5B), but not among PND0 DEGs (Supplementary Table 3). At PND21, imprinted genes within the IGN were also significantly over-represented among DEGs for both sexes (Figures 5C and 5D), but not among PND0 DEGs (Supplementary Table 3).
Figure 5.
RNA-seq analysis on F1 livers at PND21. A and B, Venn diagrams represent the number of DEGs between the 0 and 50 ppm CdCl2 exposure groups and their intersection with the IGN, for females and males. C and D, Venn diagrams represent the number of DEGs and their intersection with imprinted genes (IGs), for females and males. For (A-D), Enrichment was determined using a hypergeometric test. E and F, Volcano plots showing the directionality of gene expression changes between 0 and 50 ppm CdCl2 exposed female and male mice. Dashed line indicates q-value = 0.05. Abbreviations: PND, postnatal day; DEGs, differentially expressed genes; CdCl2, cadmium chloride; IGN, Imprinted Gene Network.
Most (87% and 89% for females and males, respectively) of the differentially expressed IGN genes at PND21 were upregulated by CdCl2 exposure (Figures 5E and 5F). Consistent with qRT-PCR data, Zac1 was upregulated by CdCl2 exposure in both sexes in RNA-seq. Altogether, these data show that developmental CdCl2 exposure is sufficient to upregulate the IGN at PND21, and provides further support for a role for imprinted genes in the pathogenesis of NAFLD.
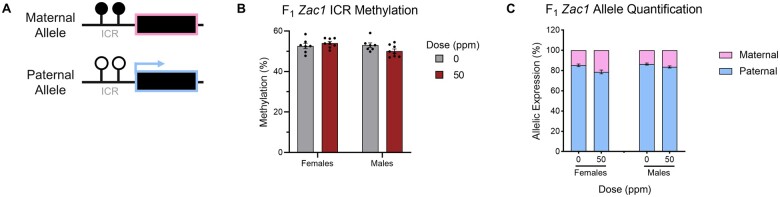
Zac1 upregulation is independent of DNA methylation at its ICR and is not associated with loss of imprinting
Zac1 is a paternally expressed imprinted gene that is silenced on the maternally inherited allele by DNA methylation at its promoter-associated ICR (Figure 6A). Because developmental Cd exposure alters the epigenetic state of imprinted genes in humans (Cowley et al., 2018), we determined whether CdCl2-induced Zac1 upregulation in our mouse model was due to loss of DNA methylation at the ICR and inappropriate transcription from the maternally inherited allele. For both sexes, control mice showed the expected approximately 50% methylation at the ICR, reflecting one methylated and one unmethylated allele (Figure 6B). Mice exposed to 50 ppm CdCl2 showed no significant change in ICR methylation. We then exploited our hybrid mouse model to measure transcription independently from the maternally and paternally inherited alleles using SNPs between the parental genomes. Consistent with no differences in DNA methylation at the ICR, mice from the 50 ppm CdCl2 group showed no significant difference in allele-specific expression compared with controls (Figure 6C). Together, these data show that Zac1 upregulation associated with CdCl2 exposure is not due to a disruption of ICR DNA methylation or a loss of imprinting.
Figure 6.
Determination of F1 hepatic Zac1 ICR methylation and imprinting status. A, Diagram of the imprinted Zac1 locus. Boxes represent the Zac1 gene, closed lollipops represent methylation at the ICR, and the arrow represents transcription from the paternal allele. B, Zac1 ICR methylation levels in male and female 0 and 50 ppm CdCl2 exposed mice. C, Zac1 allele-specific expression in male and female 0 and 50 ppm CdCl2 exposed mice. For (B and C), Data are presented as means ± S.E. Student’s t test comparing 50 ppm CdCl2 exposed mice to 0 ppm CdCl2 controls. Abbreviations: ICR, Imprinting Control Region; CdCl2, cadmium chloride; S.E., standard error.
Zac1 overexpression in cultured mouse hepatocytes is sufficient to promote neutral lipid accumulation
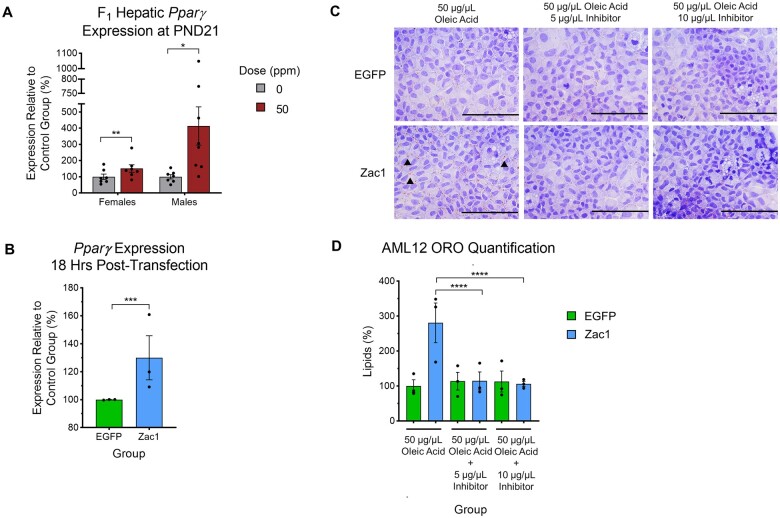
Our lab has previously shown that Zac1 drives fibrosis in NAFLD programming (Baptissart et al., 2022). In our model of developmental CdCl2 exposure, the most striking NAFLD-related phenotype for both females and males is the accumulation of neutral lipids in hepatocytes at PND21. Consistent with this finding, expression of the major prosteatotic factor Pparγ is significantly increased in 50 ppm CdCl2-exposed mice (Figure 7A). A previous study using mouse neuronal cells suggested that Zac1 regulates Pparγ, but whether this signaling pathway is conserved in hepatocytes, and whether Zac1 plays a role in driving steatosis, have not been determined (Barz et al., 2006). To test this, we artificially overexpressed Zac1 in the AML12 mouse hepatocyte cell line (Supplementary Figure 6A). Zac1 overexpression was associated with significant upregulation of Pparγ at 18 h post-transfection compared with control cells over-expressing EGFP (Figure 7B). Exposing cells to 50 or 100 µg/µL oleic acid-BSA led to a significant increase in the accumulation of neutral lipids in Zac1 over-expressing cells compared with controls (Supplementary Figures 6B and 6C). Together, these data suggest a link between Zac1, Pparγ expression, and lipid accumulation in AML12 cells. To test the dependency of Zac1-induced lipid accumulation on Pparγ signaling, we treated Zac1 overexpressing cells with the specific Pparγ inhibitor, T0070907, at 5 and 10 µg/µL in the presence of excess oleic acid-BSA. Both doses of the inhibitor significantly attenuated the Zac1-driven neutral lipid accumulation (Figures 7C and 7D), suggesting that Zac1 drives lipid accumulation via the Pparγ signaling pathway.
Figure 7.
Role of Zac1 in lipid accumulation. A, Pparγ transcript accumulation in F1 female and male livers at PND21. B, Pparγ transcript accumulation in AML12 cells 18 h after transfection. For (A and B), Data are presented as means ± S.E. *p < .05, **p < .01, ***p < .001. Student’s t test comparing 50 ppm CdCl2 to 0 ppm control group and Zac1 overexpressing cells to EGFP controls. C, Representative photomicrographs (40×) of Oil Red O staining of AML12 cells exposed to 50 µg/µL oleic acid with or without 5 or 10 µg/µL T0070907 Pparγ inhibitor. Arrowheads indicate neutral lipids. Scale bars represent 200 µm. D, Quantification of Oil Red O staining. For (D), Data are presented as means ± S.E. ****p < .0001. One-way ANOVA with Dunnett’s post hoc test comparing all conditions. Data represent 3 independent experiments with N = 3 replicates/condition. Three discrete images/slide were taken. Abbreviations: PND, postnatal day; CdCl2, cadmium chloride; S.E., standard error.
Zac1 directly binds to the Pparγ promoter
To further understand the mechanistic link between Zac1 and Pparγ, we performed ChIP on AML12 cells overexpressing Zac1 to test the hypothesis that Zac1 controls Pparγ transcription by binding to its promoter. Zac1 binding was significantly enriched at a predicted binding site (Barz et al., 2006) within the Pparγ promoter region (Figure 8), demonstrating a direct mechanistic link.
Figure 8.

Enrichment of Zac1-FLAG at the Pparγ promoter and a negative control (gene desert, “Neg.”) region relative to IgG. Ratio paired t test, one-tailed, comparing Pparγ to the negative control region. Data represent 3 independent experiments. *p < .05.
Discussion
Cd is naturally present in the soil, but human activities lead to accumulation of it in the environment, presenting a public health concern. Cd is poorly metabolized and excreted. It accumulates in the liver, kidneys, and bone, resulting in negative health effects (Satarug et al., 2010). The Korean National Environmental Health Survey and Third National Health and Nutrition Examination Survey reveal associations between Cd exposure and liver-related mortality (Hyder et al., 2013; Kim et al., 2021). NAFLD is the leading type of chronic liver disease, and observations in humans and model organisms suggest that NAFLD susceptibility can be programmed by the developmental environment. However, the underlying causes and mechanisms remain unclear. We and others have proposed that imprinted genes are strong candidates for bridging the developmental environment with later life disease. In support of this, we previously described a role for the imprinted gene Zac1 in programming NAFLD in response to maternal metabolic syndrome (Baptissart et al., 2022).
To explore the hypothesis that developmental Cd exposure is sufficient to program NAFLD later in life and that this is mediated by Zac1, we exposed hybrid mice to CdCl2 during development for a period equivalent to human gestation. 50 ppm CdCl2 exposure resulted in NAFLD phenotypes in juvenile mice including histological and biochemical evidence of steatosis and fibrosis, and gene expression profiles consistent with diseased liver states. In support of our hypothesis, CdCl2 exposure increased Zac1 expression and upregulated the IGN, a well-characterized transcriptional network of imprinted and biallelically expressed genes. Artificial Zac1 over-expression in vitro promoted lipid accumulation in a Pparγ-dependent manner. Using ChIP, we showed direct binding of Zac1 to the Pparγ promoter. Together, the findings of this study provide evidence that exposure to Cd during early life is sufficient to program NAFLD, and that this process is mediated through Zac1 and the IGN.
Developmental CdCl2 exposure affects offspring growth and development
Mice exposed to 50 ppm CdCl2 through gestation had significantly less body mass than 0 ppm mice at PND0. This is concordant with human studies that have shown an association between in utero Cd exposure, fetal growth restriction, and smaller body size at birth (Huang et al., 2019; Wang et al., 2016).
Mice in the 50 ppm group also showed a significantly lower rate of survival prior to weaning than animals in the 0 ppm group. Although the reason for this is unknown, we observed that most deaths occurred around PND14. This timepoint is consistent with the timing at which mice undergo key metabolic changes including a second phase of brown adipose tissue recruitment to enable proper thermoregulation outside the nest and transition to a solid food diet (Charalambous et al., 2012; Rugh, 1968). At PND21, we observed that offspring in the 50 ppm group showed a trend toward decreased brown adipose tissue mass when compared with the 0 ppm group, although the difference was not statistically significant. Our working hypothesis is that developmental CdCl2 exposure disrupts brown adipose tissue recruitment and metabolism, affecting offspring survival. Further studies are required to test this empirically, including collecting brown adipose tissue at PND14 for molecular and histological analyses.
Exposure to developmental CdCl2 programs NAFLD in juvenile mice
Exposure to Cd in adulthood has been associated with NAFLD (Hyder et al., 2013). However, increasingly younger individuals are being diagnosed with NAFLD each year. A recent study found an association between epigenetic alterations and liver disease in children, potentially indicating a role of the developmental environment in programming NAFLD (Moylan et al., 2022). In our study, mice exposed to 50 ppm CdCl2 during gestation and early postnatal life presented with histological, biochemical, and molecular signatures of NAFLD at PND21, a timepoint corresponding to the onset of adolescence in rodents (Laviola et al., 2003). These data suggest that early life Cd exposure could be contributing to the increasing rates of juvenile NAFLD in humans.
A recent study, which examined the effects of life-long Cd exposure on high-fat diet-induced NAFLD, reported that 5 ppm CdCl2 exposure exacerbated high-fat diet-induced NAFLD but CdCl2-exposed mice on a normal diet did not develop the disease (Young et al., 2022). The different outcomes between this study and our own are most likely due to differences in CdCl2 dose. In support of this, mice exposed to 1 ppm CdCl2 in our own study did not present with NAFLD, indicating that the disease is induced in mice only by exposure to higher levels of Cd.
Our study identified sex-specific differences in Cd-induced NAFLD phenotypes at PND21. At the level of gene transcription, males showed more DEGs in RNA-seq than females. Both females and males presented with steatosis, the first major hallmark of NAFLD, whereas only females presented with the more advanced stage of fibrosis. This observation is consistent with previous studies reporting sex-specific differences in the metabolic response to developmental Cd exposure, which revealed female mice to be more susceptible (Jackson et al., 2020). This could be explained by Cd having a longer half-life in females, both in mice and humans, and by the increased permeability of the female placenta to Cd (Jackson et al., 2022; Suwazono et al., 2009; Taguchi and Suzuki, 1981). The significance of this for human NAFLD is unclear because our understanding of sex-specific differences in susceptibility to this disease requires further study (Lonardo et al., 2019).
At PND90, only males in the 50 ppm group showed a significant increase in liver mass. There were no differences between the Cd exposure groups at the histological level. At this timepoint, control mice exhibited considerable accumulation of neutral lipids in hepatocytes, consistent with the propensity of C57Bl/6J mice to develop metabolic deficiencies in adulthood (Stahl et al., 2020). This result may also be linked to the different gene signatures associated with lipid metabolism and immune response at different liver developmental phases (Gong et al., 2020). Genes relevant to steatosis and fibrosis overall were no longer upregulated at PND90 in Cd-exposed mice compared with controls. Our results indicate that developmental CdCl2 exposure is sufficient to program juvenile NAFLD in a sex-specific manner, but that differences are no longer discernible between groups in the absence of continued exposure. We and others have proposed a multiple-hit hypothesis for NAFLD whereby predisposed individuals require another insult to maintain NAFLD phenotypes in later life (Buzzetti et al., 2016). Our future work will test whether developmental Cd-induced NAFLD can persist into later life in the presence of a second environmental insult, a model that is potentially more consistent with the life-long exposures experienced by humans (Wild, 2005).
CdCl2 upregulates Zac1 and the IGN
Our lab previously showed that Zac1 and the IGN are upregulated in NAFLD programming after exposure to maternal metabolic syndrome, and that exposure specifically during postnatal development drives the phenotype (Baptissart et al., 2022). Consistent with those findings, developmental exposure to 50 ppm CdCl2 upregulated Zac1 and the IGN in female and male livers at PND21, suggesting that these genes form a conserved mechanism that responds to different environmental stressors. Leveraging samples from the Newborn Epigenetics Study (NEST) cohort, we previously showed that DNA methylation at ICRs in humans is sensitive to Cd exposure in maternal and newborn cord blood, providing a potential mechanism for imprinted gene dysregulation (Cowley et al., 2018). However, Zac1 upregulation in our mouse model was independent of methylation changes at its ICR, a finding that is mirrored at additional imprinted loci in other mouse models of developmental CdCl2 exposure developed by our group (Simmers et al., 2022). The differences in the epigenetic response to Cd between mice and humans could be explained by interaction effects from the diverse environmental stressors that humans are exposed to, which can be more carefully controlled in mouse studies. An alternative explanation is that our mouse model examines only the impact of maternal CdCl2 exposure, whereas in human populations, paternal exposure to environmental insults influences offspring epigenetics. A recent study showed that exposure to Cd can impact the sperm methylome, although the impact this has on imprinted genes in offspring still needs to be determined (Saintilnord et al., 2021). Furthermore, the differences could be explained by species-specific variation in ICR sensitivity. Cd-induced Zac1 upregulation could occur via alternative epigenetic mechanisms. Imprinted genes including Zac1 are highly expressed in prenatal development; however, they are progressively downregulated after birth by chromatin remodeling, a process mediated by the α-thalassemia mental retardation X-linked (Atrx) protein (Kernohan et al., 2010; Lui et al., 2008; Voon et al., 2015). Our future work will test the hypothesis that Cd exposure during this critical time-period disrupts Atrx function and the placement of repressive histone variants, using ChIP.
Zac1 regulates Pparγ signaling to drive steatosis
Fibrosis is dependent upon interactions between hepatocytes and hepatic stellate cells (HSCs) (Zeisberg et al., 2007; Tsuchida and Friedman, 2017). Although we previously showed that hepatocyte-specific overexpression of Zac1 is sufficient to induce fibrosis, the role of HSCs in this context requires further study (Baptissart et al., 2022). However, hepatocytes are the primary drivers of liver steatosis (Sovaila et al., 2019). Another study in nonhepatic cell types indicated a link between Zac1 and the prosteatotic gene Pparγ (Barz et al., 2006). Given our observations in vivo, we tested whether Zac1 could be responsible for Cd-induced hepatic lipid accumulation through this pathway. Overexpressing Zac1 in the presence of fatty acids in a murine hepatocyte cell line leads to significantly increased lipid accumulation, which can be abrogated by a Pparγ-specific antagonist. Furthermore, using ChIP, we confirmed that Zac1 directly controls Pparγ expression by binding to its promoter region.
Our results support the hypothesis that imprinted genes play a central role in mediating between Cd exposure in development and juvenile NAFLD and, together with our previous findings, identify the imprinted gene Zac1 as a novel regulator connecting prosteatotic and profibrotic pathways in hepatocytes. Future work in our lab will exploit genetic models of Zac1 manipulation to determine whether Zac1 is required for the programming of NAFLD in response to developmental CdCl2 exposure.
Supplementary data
Supplementary data are available at Toxicological Sciences online.
Supplementary Material
Acknowledgments
We are grateful to our collaborators and the NCSU Toxicology Animal Facility staff for their contributions, which enabled the successful completion of this work. We thank members of the Cowley Lab and faculty at NC State University for helpful comments on the manuscript.
Funding
National Institute of Environmental Health Sciences (R01ES031596, K22ES027510, P30ES025128, and T32ES007046).
Declaration of conflicting interests
The authors declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Contributor Information
Sierra D Riegl, Center for Human Health and the Environment, North Carolina State University, Raleigh, North Carolina 27695, USA; Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
Cassie Starnes, Center for Human Health and the Environment, North Carolina State University, Raleigh, North Carolina 27695, USA; Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
Dereje D Jima, Center for Human Health and the Environment, North Carolina State University, Raleigh, North Carolina 27695, USA; Bioinformatics Research Center, North Carolina State University, Raleigh, North Carolina 27695, USA.
Marine Baptissart, Center for Human Health and the Environment, North Carolina State University, Raleigh, North Carolina 27695, USA; Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
Anna Mae Diehl, Department of Medicine, Duke University, Durham, North Carolina 27710, USA.
Scott M Belcher, Center for Human Health and the Environment, North Carolina State University, Raleigh, North Carolina 27695, USA; Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
Michael Cowley, Center for Human Health and the Environment, North Carolina State University, Raleigh, North Carolina 27695, USA; Department of Biological Sciences, North Carolina State University, Raleigh, North Carolina 27695, USA.
References
- Agency for Toxic Substances and Disease Registry (ATSDR). (2012). Toxicological Profile for Cadmium. U.S. Department of Health and Human Services, Public Health Service, Atlanta, GA. [PubMed] [Google Scholar]
- Al Adhami H., Evano B., Le Digarcher A., Gueydan C., Dubois E., Parrinello H., Dantec C., Bouschet T., Varrault A., Journot L., et al. (2015). A systems-level approach to parental genomic imprinting: The imprinted gene network includes extracellular matrix genes and regulates cell cycle exit and differentiation. Genome Res. 25, 353–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anders S., Pyl P. T., Huber W. (2015). HTSeq-A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baptissart M., Bradish C. M., Jones B. S., Walsh E., Tehrani J., Marrero-Colon V., Mehta S., Jima D. D., Oh S. H., Diehl A. M., et al. (2022). Zac1 and the Imprinted Gene Network program juvenile NAFLD in response to maternal metabolic syndrome. Hepatology 76, 1090–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barz T., Hoffmann A., Panhuysen M., Spengler D. (2006). Peroxisome proliferator-activated receptor gamma is a Zac target gene mediating Zac antiproliferation. Cancer Res. 66, 11975–11982. [DOI] [PubMed] [Google Scholar]
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B 57, 289–300. [Google Scholar]
- Bertot L. C., Adams L. A. (2016). The natural course of non-alcoholic fatty liver disease. IJMS 17, 774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buzzetti E., Pinzani M., Tsochatzis E. A. (2016). The multiple-hit pathogenesis of non-alcoholic fatty liver disease (NAFLD). Metabolism 65, 1038–1048. [DOI] [PubMed] [Google Scholar]
- Charalambous M., Ferron S. R., da Rocha S. T., Murray A. J., Rowland T., Ito M., Schuster-Gossler K., Hernandez A., Ferguson-Smith A. C. (2012). Imprinted gene dosage is critical for the transition to independent life. Cell Metab. 15, 209–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciesielski T., Weuve J., Bellinger D. C., Schwartz J., Lanphear B., Wright R. O. (2012). Cadmium exposure and neurodevelopmental outcomes in U.S. children. Environ. Health Perspect. 120, 758–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cowley M., Skaar D. A., Jima D. D., Maguire R. L., Hudson K. M., Park S. S., Sorrow P., Hoyo C. (2018). Effects of cadmium exposure on DNA methylation at imprinting control regions and genome-wide in mothers and newborn children. Environ. Health Perspect. 126, 037003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallego-Durán R., Romero-Gómez M. (2015). Epigenetic mechanisms in non-alcoholic fatty liver disease: An emerging field. World J. Hepatol. 7, 2497–2502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Go Y.-M., Sutliff R. L., Chandler J. D., Khalidur R., Kang B.-Y., Anania F. A., Orr M., Hao L., Fowler B. A., Jones D. P., et al. (2015). Low-dose cadmium causes metabolic and genetic dysregulation associated with fatty liver disease in mice. Toxicol. Sci. 147, 524–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gong T., Zhang C., Ni X., Li X., Li J., Liu M., Zhan D., Xia X., Song L., Zhou Q., et al. (2020). A time-resolved multi-omic atlas of the developing mouse liver. Genome Res. 30, 263–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- House J. S., Hall J., Park S. S., Planchart A., Money E., Maguire R. L., Huang Z., Mattingly C. J., Skaar D., Tzeng J. Y., et al. (2019). Cadmium exposure and MEG3 methylation differences between Whites and African Americans in the NEST Cohort. Environ. Epigenet. 5, dvz014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang S., Kuang J., Zhou F., Jia Q., Lu Q., Feng C., Yang W., Fan G. (2019). The association between prenatal cadmium exposure and birth weight: A systematic review and meta-analysis of available evidence. Environ. Pollut. 251, 699–707. [DOI] [PubMed] [Google Scholar]
- Hudson K. M., Belcher S. M., Cowley M. (2019). Maternal cadmium exposure in the mouse leads to increased heart weight at birth and programs susceptibility to hypertension in adulthood. Sci. Rep. 9, 13553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyder O., Chung M., Cosgrove D., Herman J. M., Li Z., Firoozmand A., Gurakar A., Koteish A., Pawlik T. M. (2013). Cadmium exposure and liver disease among US adults. J. Gastrointest. Surg. 17, 1265–1273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T. W., Baars O., Belcher S. M. (2022). Gestational Cd exposure in the CD-1 mouse sex-specifically disrupts essential metal ion homeostasis. Toxicol. Sci. 187, 254–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson T. W., Ryherd G. L., Scheibly C. M., Sasser A. L., Guillette T. C., Belcher S. M. (2020). Gestational Cd exposure in the CD-1 mouse induces sex-specific hepatic insulin insensitivity, obesity, and metabolic syndrome in adult female offspring. Toxicol. Sci. 178, 264–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson D. S., Mortazavi A., Myers R. M., Wold B. (2007). Genome-wide mapping of in vivo protein-DNA interactions. Science 316, 1497–1502. [DOI] [PubMed] [Google Scholar]
- Kernohan K. D., Jiang Y., Tremblay D. C., Bonvissuto A. C., Eubanks J. H., Mann M. R. W., Bérubé N. G. (2010). ATRX partners with cohesin and MeCP2 and contributes to developmental silencing of imprinted genes in the brain. Dev. Cell. 18, 191–202. [DOI] [PubMed] [Google Scholar]
- Kim D. W., Ock J., Moon K. W., Park C. H. (2021). Association between Pb, Cd, and Hg exposure and liver injury among Korean adults. IJERPH 18, 6783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laviola G., Macrì S., Morley-Fletcher S., Adriani W. (2003). Risk-taking behavior in adolescent mice: Psychobiological determinants and early epigenetic influence. Neurosci. Biobehav. Rev. 27, 19–31. [DOI] [PubMed] [Google Scholar]
- Le M. H., Devaki P., Ha N. B., Jun D. W., Te H. S., Cheung R. C., Nguyen M. (2017). Prevalence of non-alcoholic fatty liver disease and risk factors for advanced fibrosis and mortality in the United States. PLoS One 12, e0173499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lonardo A., Nascimbeni F., Ballestri S., Fairweather D., Win S., Than T. A., Abdelmalek M. F., Suzuki A. (2019). Sex differences in nonalcoholic fatty liver disease: State of the art and identification of research gaps. Hepatology 70, 1457–1469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Love M. I., Huber W., Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lui J. C., Finkielstain G. P., Barnes K. M., Baron J. (2008). An imprinted gene network that controls mammalian somatic growth is down-regulated during postnatal growth deceleration in multiple organs. Am. J. Physiol. Regul. Integr. Comp. Physiol. 295, R189–R196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morán-Salvador E., López-Parra M., García-Alonso V., Titos E., Martínez-Clemente M., González-Périz A., López-Vicario C., Barak Y., Arroyo V., Clària J. (2011). Role for PPARγ in obesity-induced hepatic steatosis as determined by hepatocyte- and macrophage-specific conditional knockouts. FASEB J. 25, 2538–2550. [DOI] [PubMed] [Google Scholar]
- Moylan C. A., Mavis A. M., Jima D., Maguire R., Bashir M., Hyun J., Cabezas M. N., Parish A., Niedzwiecki D., Diehl A. M., et al. (2022). Alterations in DNA methylation associate with fatty liver and metabolic abnormalities in a multi-ethnic cohort of pre-teenage children. Epigenetics 17, 1446–1461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nriagu J. O. (1989). A global assessment of natural sources of atmospheric trace metals. Nature 338, 47–49. [Google Scholar]
- Paik J. M., Kabbara K., Eberly K. E., Younossi Y., Henry L., Younossi Z. M. (2022). Global burden of NAFLD and chronic liver disease among adolescents and young adults. Hepatology 75, 1204–1217. [DOI] [PubMed] [Google Scholar]
- Pope C., Mishra S., Russell J., Zhou Q., Zhong X. B. (2017). Targeting H19, an imprinted long non-coding RNA, in hepatic functions and liver diseases. Diseases 5, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahimzadeh M. R., Rahimzadeh M. R., Kazemi S., Moghadamnia A. A. (2017). Cadmium toxicity and treatment: An update. Caspian J. Intern. Med. 8, 135–145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rugh R. (Ed.) (1968). The Mouse; Its Reproduction and Development. Burgess Pub. Co., Minneapolis, MN. [Google Scholar]
- Saintilnord W. N., Tenlep S. Y. N., Preston J. D., Duregon E., DeRouchey J. E., Unrine J. M., de Cabo R., Pearson K. J., Fondufe-Mittendorf Y. N. (2021). Chronic exposure to cadmium induces differential methylation in mice spermatozoa. Toxicol. Sci. 180, 262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki T., Horiguchi H., Arakawa A., Oguma E., Komatsuda A., Sawada K., Murata K., Yokoyama K., Matsukawa T., Chiba M., et al. (2019). Hospital-based screening to detect patients with cadmium nephropathy in cadmium-polluted areas in Japan. Environ. Health Prev. Med. 24, 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Satarug S., Garrett S. H., Sens M. A., Sens D. A. (2010). Cadmium, environmental exposure, and health outcomes. Environ. Health Perspect. 118, 182–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semple B. D., Blomgren K., Gimlin K., Ferriero D. M., Noble-Haeusslein L. J. (2013). Brain development in rodents and humans: Identifying benchmarks of maturation and vulnerability to injury across species. Prog. Neurobiol. 106–107, 1–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simmers M. D., Hudson K. M., Baptissart M., Cowley M. (2022). Epigenetic control of the imprinted growth regulator Cdkn1c in cadmium-induced placental dysfunction. Epigenetics 1–17. doi: 10.1080/15592294.2022.2088173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith F. M., Garfield A. S., Ward A. (2006). Regulation of growth and metabolism by imprinted genes. Cytogenet. Genome Res. 113, 279–291. [DOI] [PubMed] [Google Scholar]
- Sookoian S., Gianotti T. F., Burgueño A. L., Pirola C. J. (2013). Fetal metabolic programming and epigenetic modifications: A systems biology approach. Pediatr. Res. 73, 531–542. [DOI] [PubMed] [Google Scholar]
- Sovaila S., Purcarea A., Gheonea D., Ionescu S., Ciurea T. (2019). Cellular interactions in the human fatty liver. J. Med. Life. 12, 338–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahl E. C., Delgado E. R., Alencastro F., LoPresti S. T., Wilkinson P. D., Roy N., Haschak M. J., Skillen C. D., Monga S. P., Duncan A. W., et al. (2020). Inflammation and ectopic fat deposition in the aging murine liver is influenced by CCR2. Am. J. Pathol. 190, 372–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suwazono Y., Kido T., Nakagawa H., Nishijo M., Honda R., Kobayashi E., Dochi M., Nogawa K. (2009). Biological half-life of cadmium in the urine of inhabitants after cessation of cadmium exposure. Biomarkers 14, 77–81. [DOI] [PubMed] [Google Scholar]
- Swaddiwudhipong W., Mahasakpan P., Jeekeeree W., Funkhiew T., Sanjum R., Apiwatpaiboon T., Phopueng I. (2015). Renal and blood pressure effects from environmental cadmium exposure in Thai children. Environ. Res. 136, 82–87. [DOI] [PubMed] [Google Scholar]
- Taguchi T., Suzuki S. (1981). Influence of sex and age on the biological half-life of cadmium in mice. J. Toxicol. Environ. Health. 7, 239–249. [DOI] [PubMed] [Google Scholar]
- Tsuchida T., Friedman S. L. (2017). Mechanisms of hepatic stellate cell activation. Nat. Rev. Gastroenterol. Hepatol. 14, 397–411. [DOI] [PubMed] [Google Scholar]
- Tucci V., Isles A. R., Kelsey G., Ferguson-Smith A. C., Bartolomei M. S., Benvenisty N.; Erice Imprinting Group. (2019). Genomic imprinting and physiological processes in mammals. Cell 176, 952–965. [DOI] [PubMed] [Google Scholar]
- van de Geijn B., McVicker G., Gilad Y., Pritchard J. K. (2015). WASP: Allele-specific software for robust molecular quantitative trait locus discovery. Nat. Methods. 12, 1061–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varrault A., Gueydan C., Delalbre A., Bellmann A., Houssami S., Aknin C., Severac D., Chotard L., Kahli M., Le Digarcher A., et al. (2006). Zac1 regulates an imprinted gene network critically involved in the control of embryonic growth. Dev. Cell. 11, 711–722. [DOI] [PubMed] [Google Scholar]
- Varrault A., Dantec C., Le Digarcher A., Chotard L., Bilanges B., Parrinello H., Dubois E., Rialle S., Severac D., Bouschet T., et al. (2017). Identification of Plagl1/Zac1 binding sites and target genes establishes its role in the regulation of extracellular matrix genes and the imprinted gene network. Nucleic Acids Res. 45, 10466–10480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vidal A. C., Semenova V., Darrah T., Vengosh A., Huang Z., King K., Nye M. D., Fry R., Skaar D., Maguire R., et al. (2015). Maternal cadmium, iron and zinc levels, DNA methylation and birth weight. BMC Pharmacol. Toxicol. 16, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Voon H. P. J., Hughes J. R., Rode C., De La Rosa-Velázquez I. A., Jenuwein T., Feil R., Higgs D. R., Gibbons R. J. (2015). ATRX plays a key role in maintaining silencing at interstitial heterochromatic loci and imprinted genes. Cell Rep. 11, 405–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Liu L., Hu Y. F., Hao J. H., Chen Y. H., Su P. Y., Fu L., Yu Z., Zhang G. B., Wang L., et al. (2016). Maternal serum cadmium level during pregnancy and its association with small for gestational age infants: A population-based birth cohort study. Sci. Rep. 6, 22631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- W.H.O. (2022). Ten Chemicals of Major Public Health Concern. https://www.who.int/news-room/photo-story/photo-story-detail/10-chemicals-of-public-health-concern. Accessed April 24, 2022.
- Wild C. P. (2005). Complementing the genome with an “exposome”: The outstanding challenge of environmental exposure measurement in molecular epidemiology. Cancer Epidemiol. Biomarkers Prev. 14, 1847–1850. [DOI] [PubMed] [Google Scholar]
- Young J. L., Cave M. C., Xu Q., Kong M., Xu J., Lin Q., Tan Y., Cai L. (2022). Whole life exposure to low dose cadmium alters diet-induced NAFLD. Toxicol. Appl. Pharmacol. 436, 115855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Younossi Z. M., Blissett D., Blissett R., Henry L., Stepanova M., Younossi Y., Racila A., Hunt S., Beckerman R. (2016). The economic and clinical burden of nonalcoholic fatty liver disease in the United States and Europe. Hepatology 64, 1577–1586. [DOI] [PubMed] [Google Scholar]
- Zeisberg M., Yang C., Martino M., Duncan M. B., Rieder F., Tanjore H., Kalluri R. (2007). Fibroblasts derive from hepatocytes in liver fibrosis via epithelial to mesenchymal transition. J. Biol. Chem. 282, 23337–23347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.