Abstract
Yersinia pestis, the causative agent of plague, and the enteropathogen Yersinia pseudotuberculosis have nearly identical nucleotide similarity yet cause markedly different diseases. To investigate this conundrum and to study Yersinia pathogenicity, we developed a high-density oligonucleotide array-based modification of signature-tagged mutagenesis (STM). Y. pseudotuberculosis YPIII mutants constructed with the tagged transposons were evaluated in the murine yersiniosis infection model. The DNA tags were amplified using biotinylated primers and hybridized to high-density oligonucleotide arrays containing DNA complementary to the tags. Comparison of the hybridization signals from input pools and output pools identified a mutant whose relative abundance was significantly reduced in the output pool. Sequence data from 31 transposon insertion regions was compared to the complete Y. pestis CO92 genome sequence. The 26 genes present in both species were found to be almost identical, but five Y. pseudotuberculosis genes identified through STM did not have counterparts in the Y. pestis genome and may contribute to the different tropisms in these closely related pathogens. Potential virulence genes identified include those involved in lipopolysaccharide biosynthesis, adhesion, phospholipase activity, iron assimilation, and gene regulation. The phospholipase A (PldA) mutant exhibited reduced phospholipase activity compared to the wild-type strain and in vivo attenuation of the mutant was confirmed. The combination of optimized double tag sequences and high-density array hybridization technology offers improved performance, efficiency, and reliability over classical STM and permits quantitative analysis of data.
Yersinia pseudotuberculosis, a cause of gastroenteritis and mesenteric lymphadenitis in humans (18) has many virulence factors as well as sequences in common with the two other pathogenic representatives of Yersinia, Yersinia pestis and Yersinia enterocolitica (6). In fact, comparison of the available sequences of Y. pseudotuberculosis genes with those from the complete Y. pestis genome (http://www.sanger.ac.uk/Projects/Y_pestis/) reveals that the two species are almost identical at the nucleotide level. Moreover, it has been suggested that Y. pestis is a recently emerged clone of Y. pseudotuberculosis (1). Despite this nucleotide similarity, the mechanisms of pathogenicity of the two species are different: Y. pseudotuberculosis and Y. enterocolitica cause gastrointestinal disease, whereas Y. pestis is the causative agent of plague.
To understand this enigma and to gain further insight into Yersinia pathogenesis, we initiated a signature-tagged mutagenesis (STM) study of Y. pseudotuberculosis and tested tagged mutants in the murine yersiniosis model of infection. STM (20) allows the simultaneous analysis of large numbers of mutants in a complex environment, such as an animal model (reviewed in reference 28). It has been used successfully in the identification of virulence-associated factors in many pathogenic bacteria, such as Staphylococcus aureus (11, 30, 45), Salmonella typhimurium (47), Vibrio cholerae (9), Neisseria meningitidis (10), Streptococcus pneumoniae (35), Legionella pneumophila (17), Y. enterocolitica (13), Proteus mirabilis (55), Mycobacterium tuberculosis (7, 12) Brucella abortus (24), Brucella melitensis (29), and Streptococcus agalactiae (25).
In traditional STM experiments, a transposon (e.g., mini-Tn5) engineered to contain a variable 40-bp region, the signature tag, is used for mutagenesis. The variable region allows different mutants to be distinguished. The library of transposon mutants is passaged through animals, and the relative abundance of each mutant in the input and output pools is monitored by hybridization. A major drawback of the original STM technique is high variation in the signal intensities after hybridization due to variability of the tag sequences, necessitating additional rounds of screening. Mutants are frequently found to contain unamplifiable tags or no tags at all (55). Moreover, the need to remove the invariable arms from the labeled probes before hybridization makes the use of radioactive labels necessary. In addition, colony hybridization often results in high background levels. The use of plasmids carrying transposons with selected tags combined with dot blotting with purified plasmid DNA has helped overcome some of these problems (30), and the technique can be further improved by using membranes containing tags amplified by PCR (35).
A recently developed and powerful tool for analyzing complex mixtures of nucleic acids is hybridization on to high-density oligonucleotide arrays (42). Such arrays can be used to quantitatively probe hundreds of thousands of different DNA or RNA sequences in a single experiment. Using fluorescent detection methods and confocal laser-scanning devices, very low background can be achieved. As the probe sequences on the arrays are selected to have similar hybridization properties, the variability in hybridization intensity often observed when random sequences are used can be reduced. Furthermore, as probes can be selected to be unique, cross-hybridization can be minimized, allowing very complex mixtures to be analyzed.
We reasoned that high-density arrays could be combined with STM. A set of 96 mini-Tn5 transposons labeled with selected tags were constructed and used to generate a library of transposon derivatives in Y. pseudotuberculosis. The input and output pools produced after in vivo experiments were compared by hybridizing the amplified tags to high-density oligonucleotide arrays containing complementary tag sequences. This resulted in the identification of potential virulence factors in Y. pseudotuberculosis, including genes involved in lipopolysaccharide (LPS) biosynthesis and genes with unknown function. A mutant carrying a transposon insertion in a novel phospholipase gene (pldA) was shown to have diminished phospholipase activity and confirmed as a virulence determinant in Y. pseudotuberculosis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
Y. pseudotuberculosis YPIII pIB1 strain (40) was maintained in Luria broth (LB) and LB agar containing nalidixic acid (40 μg ml−1). Escherichia coli XL2 Blue MRF′ (Stratagene), used in cloning experiments, was grown overnight at 37°C on LB agar plates. For selection, agar plates were supplemented with kanamycin (50 μg ml−1), ampicillin (100 μg ml−1), tetracycline (10 μg ml−1), or nalidixic acid. E. coli CC118(λpir) (21) was used as a host strain for maintenance of the pir-dependent pUT mini-Tn5Km2 vector (14) in cloning experiments. The helper strain E. coli S17/pNJ5000 was maintained as described (19).
Construction of double-tagged mini-Tn5 transposon mutants.
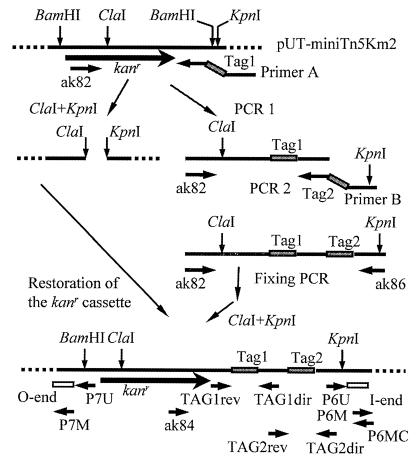
Tag sequences were chosen from those that had been shown to work well in similar experiments with Saccharomyces cerevisiae (53). In addition, common tag priming sites were chosen using the Whitehead Institute primer 3 program (http://www-genome.wi.mit.edu/cgi-bin/primer/primer3.cgi). The sequences of the 192 PCR primers (primer A and primer B) are presented elsewhere (http://www.lshtm.ac.uk/itd/units/pmbbu/karlyshev/tags.htm). Oligonucleotides were synthesized on a 96-well automated multiplex oligonucleotide synthesizer on a 7.5 μM scale (27). Initial amplification was performed in 96-well format on microtiter plates (Hybaid) using primer ak82 (5′-AGC CAT ATT CAA CGG GAA AC), 96 primer A's, and the pUT mini-Tn5Km2 as a template. Second-round amplification was performed in 96-well format on microtiter plates (Hybaid) using primer ak82, 96 primer B's, and 5 μl of the first-round PCR product as template.
A third (fixing) PCR round was performed using primers ak86 (5′-GGG GTA CCG TCG ACC TGC), ak82, and 5 μl of round 2 PCR product as a template. Third-round amplification products were purified on S300 microcolumns (Bio-Rad), digested with KpnI and ClaI, ligated to gel-purified KpnI-ClaI fragment of pUT mini-Tn5Km2 vector, purified using S300, and transformed via electroporation into E. coli CC118(λpir). Kanr transformants were assembled into three microtiter plates. The tag-containing regions of the recombinant plasmids were amplified using primers ak84 (5′-CAC TGG CAG AGC ATT ACG CTG) and p6MC (5′-GTC TCT TGA TCA GAT CTG GC) (Fig. 1). The PCR products were sequenced using primer ak84.
FIG. 1.
Strategy for construction of double-tagged transposons and analysis of transposon derivatives. The 192 different tags were incorporated into the mini-Tn5 vector by means of two sequential sets of PCRs using two sets of long variable primers (primer A and primer B). Also shown are the primers used for amplification and sequencing of the tags (ak84, P6MC, TAG1dir, TAG1rev, TAG2dir, and TAG2rev) and those used for amplification and sequencing of the regions flanking transposon insertion sites (P6M, P7M, P6U, and P7U). The O and I ends of the mini-Tn5 are shown as open boxes. Tag regions are shown as shaded bars.
Conjugation.
Initial triple mating experiments of E. coli CC118 (λpir) donor strain, transformed with the plasmids carrying tagged mini-Tn5, and Y. pseudotuberculosis using a helper strain (E. coli S17/pNJ5000) (19) were performed as described previously (32). Direct mating experiments (without a helper strain) using E. coli 19851 pir+ as the donor strain were performed as described previously (31). Exconjugants were selected for kanamycin and nalidixic acid resistance. Both the recipient strain YPIII pIB1 and the exconjugants were checked for the presence of the virulence plasmid using Congo red magnesium oxalate (CRMOX) plates (39). All attenuated transposon derivatives grew as predominantly small red colonies, confirming that they retained the virulence plasmid.
Tag sequence detection.
Genomic DNA was extracted using the PureGen kit (Flowgen). Tags were amplified using two pairs of primers: BTAG1dir (biotin-5′-AGA GAC CTC GTG GAC ATC A) and TAG1rev (5′-GTG GCA AAG CAA AAG TTC AA) for tag1 and BTAG2dir (biotin-5′-GTC GAC CTG CAG CGT ACG) and TAG2rev (5′-GAT GTC CAC GAG GTC TCT) for tag2 (Fig. 1) using Taq polymerase Gold (Perkin Elmer). Biotinylated PCR products (10 μl) were hybridized for 1 h at 42°C to high-density oligonucleotide arrays (Affymetrix) in a buffer (200 μl total) containing 6× SSPE (BioWhittaker), 0.005% Triton X-100, 0.5 nM control oligonucleotide 213B (which hybridizes to the corners of the array, for alignment purposes), 200 pmol of TAG2rev, 100 pmol of TAG1rev, and 200 pmol of FTAGMX4COMP (5′-CGT ACG CTG CAG GTC GAC). The addition of the unlabeled primers has been shown to reduce background (47a).
Following hybridization, the arrays were washed as previously described (53). Hybridization was visualized by staining with a streptavidin-phycoerythrin conjugate (Molecular Probes) and scanning with a modified confocal scanning device as previously described (54). The relative signal intensities were determined using the histogram method with Affymetrix Genechip III software. FilemakerPro program was used in background subtraction and to associate the hybridization intensity data with attenuated mutants.
In vivo experiments.
Three input pools containing 60, 40, and 33 transposon mutants, respectively, were constructed and stored at −70°C. Aliquots (0.1 ml) containing approximately 107 CFU were inoculated into 10 ml of LB and incubated with shaking overnight at 30°C. The overnight culture (2 ml) was used to inoculate 20 ml of fresh warmed LB and further incubated at 37°C for 3 h with shaking. Genomic DNA was isolated from approximately 108 cells and stored (input pool). Bacteria were pelleted at 3,000 × g and diluted in phosphate-buffered saline (PBS; Oxoid) for infection and viable count determination. Pairs of 8-week-old female BALB/c mice (Charles River Labs) were challenged intravenously (i.v.) via the tail vein with 105 or 5 × 105 CFU. After 3 days, the surviving mice were culled, and spleens were removed and homogenized in 3 ml of LB using a stomacher (Seward Medical Ltd.) on maximum setting for 5 min. Dilutions of the extract were plated on LB agar containing kanamycin (LB-Kan) and nalidixic acid (LB-Nal). Plates containing approximately 104 colonies were washed with saline and mixed, and aliquots were taken for making lysates (for PCR) or for total DNA preparation
The median lethal dose (MLD) (the expected median dose required to produce morbidity or death based on previous experimental data) was assessed by i.v. injection of groups of five female 6-week-old BALB/c mice with serial dilutions in PBS of exponential-phase cultures grown at 37°C in LB-Nal (wild type) or LB-Nal-Kan (mutant) (37). Humane endpoints were strictly observed, and animals deemed incapable of survival (unable to right themselves or to respond to a pinch on the foot or tail) were killed by cervical dislocation.
For in vivo competition studies, mutant and wild-type strains were grown separately to exponential phase in LB with appropriate antibiotics. Bacteria were washed with LB, and concentration was adjusted to 5 × 106 CFU/ml. Equal volumes of each bacterial suspension were mixed together, and 0.1-ml volumes were injected i.v. into four mice as above. Viable counts on LB, LB-Nal, and LB-Nal-Kan allowed the exact input ratio to be calculated. After 3 days, spleens were recovered and passed through sieves (70 μm; Becton Dickinson) to produce a cell suspension in 3 ml of LB. Homogenates were plated on selective medium to determine the output ratio. The competitive index (CI) is defined as the output ratio (mutant/wild type) divided by the input ratio (mutant/wild type). In vitro CI was determined as described (9). Briefly, mixtures containing a mutant and the wild-type strain were inoculated into LB medium supplemented with nalidixic acid at approximately 104 CFU/ml. The cultures were grown overnight at 28°C, and the mutant-to-wild type ratios were determined by plating on medium with and without the selective marker (kanamycin).
Detection of transposon insertion sites and yplA gene.
The genomic regions of DNA flanking the transposon were amplified using single-primer PCR (26) with primers P6U (5′-CGA GCT CGA ATT CGG CCT AG) and P7U (5′-CTG CAG GCA TGC AAG CTT CG), and sequenced with internal primers P6M (5′-GCC AGA TCT GAT CAA GAG AC) and P7M (5′-GCC GAA CTT GTG TAT AAG AGT C), respectively (Fig. 1). Primers designed from the Y. enterocolitica yplA gene, yplA-dir (5′-CAG TGA TAA TCA ACA GTA TGT C) and yplA-rev (5′-CAT ACC GCC GCT CCC AAG G), were used for detection of the gene in Y. pseudotuberculosis (43).
Sequence analysis.
DNA sequencing was performed on an ABI 373 automatic sequencer using an ABI PRISM Dye Terminator cycle sequencing kit (Perkin Elmer). The sequences were analyzed using BLASTn and BLASTp software (http://www.ncbi.nlm.nih.gov/BLAST/) with a nonredundant database and with the Y. pestis genome sequence (http://www.sanger.ac.uk/Projects/Y_pestis/). The entire sequence of the Y. pseudotuberculosis pldA gene was deposited at EMBL under accession no. AJ245393.
Phospholipase detection assay.
Bacterial strains were grown overnight in LB supplemented with appropriate antibiotics. Cells were washed in PBS, resuspended in 1 ml of sonication buffer (0.6 M NaCl, 0.1 NaPO4 [pH 8]), and lysed by sonication (five 30-s bursts). Cell debris was centrifuged for 10 min at 13,000 rpm, and the supernatant was retained. Protein concentration was determined using the BCA protein assay kit (Pierce, Rockford, Ill.), and phospholipase activity was determined using the PLA2 detection kit (Assay Designs, Ann Arbor, Mich.). Each strain was tested in triplicate, and results are expressed as units of PLA2 per milligram of protein.
Phospholipase activity assay.
Phospholipase activity towards liposomes was measured using carboxyfluoroscein (CF)-loaded liposomes composed of either egg yolk phosphatidylcholine (Lipoid) or sphingomyelin (Sigma) in a 1:1 molar ratio with cholesterol. Cell lysates (100 μl) from wild-type or mutant strains were incubated with liposomes (100 μl) for 15 min at 37°C in a 96-well Microfluor plate (Dynex). Fluorescent intensity was measured (excitation at 485 nm and emission at 520 nm) immediately after addition of liposomes (0 min) and after 15 min of incubation using the Fluoroskan Ascent fluorimeter (Life Sciences). A rate of activity was calculated by taking the average change in fluorescent intensity of the replicates over time (minutes) and total protein concentration (milligrams). Activity was expressed as units per milligram of total protein per minute.
RESULTS
Library construction and mutagenesis.
Initial experiments using the random tagging technique resulted in variable hybridization signals in the input pools, causing difficulties in data interpretation. Sequencing the tags from different clones revealed that weak signals were due to large deletions in the variable regions, possibly a result of errors in primer synthesis, whereas exceptionally strong signals were due to the presence of an above-average GC/AT ratio in the variable regions (data not shown). To overcome these difficulties, we introduced selected tag sequences into a transposon. Tags were chosen from a yeast mutagenesis project (48) that are 20 bases long, lack sequences known to form secondary structure, have similar melting temperatures, and have a minimum of 5 bp differences between each tag. From this set of 6,000, 192 tag sequences (two per transposon) that had demonstrated strong hybridization potential were chosen. The use of two tags provided more easily interpreted results. After three amplification reactions for each of the 96, 96 purifications, 96 restriction-ligation reactions, and 96 transformations, a library of E. coli CC118(λpir) carrying 96 double-tagged mini-Tn5 transposons was constructed (Fig. 1).
To check whether the hybridization signals for different clones containing the same tags were similar in strength, the tags were amplified using biotinylated primers from three clones from each transformation and hybridized to high-density oligonucleotide arrays containing the tag complements. The arrays contain approximately 4,000 different 20-mer sequences synthesized in a spatially addressable format using photolithography and photosensitive oligonucleotide chemistry. Analysis of the scanned images for the three initial sets revealed that a large proportion of tags gave very similar signal intensities (data not shown). However, some clonal variation was also observed. Sequencing of the tags showed deletions of single bases in the tags derived from the clones with reduced signals. Such microdeletions may have resulted from errors during chemical synthesis of the long PCR primers used for the library construction. The master library of 96 donor E. coli strains containing double-tagged transposons was assembled via selection of the clones producing the strongest signals from the three sets.
During mating experiments, we encountered problems caused by frequent lysogenization of the recipient strain by the spontaneous release of λpir prophage in the donor strain. Similar problems during conjugation have been described previously (46). The frequency of ampicillin-resistant (Ampr) exconjugants carrying markers of the delivery plasmid can be highly variable depending on the recipient strain. For example, as many as 52% of exconjugants in Vibrio cholerae were found to be ampicillin resistant (9). For different E. coli donor strains, the frequency was shown to vary between 10 and 90% (14). We attempted to use sodium citrate to inhibit possible phage adsorption during mating, but this completely inhibited conjugation. The addition of sodium citrate to the selective medium at a later stage increased the relative proportion of Amps colonies but resulted in a significant decrease in the conjugation efficiency. These results imply that divalent cations are possibly required for both phage infection and conjugation.
To avoid the complications caused by potential prophage induction, we employed an E. coli 19851 donor strain with a stable pir+ gene (31). Use of the donor library based on this strain resulted in a sufficient number of exconjugants. Almost all of the exconjugants in this case were Amps, which implies that prophage induction was the reason for the observed high frequency of Ampr colonies obtained in the previous experiments. Southern hybridization and PCR experiments with the Ampr clones confirmed random insertion of the delivery plasmid, possibly due to illegitimate recombination (data not shown).
Amps Kanr Nalr exconjugants were assembled on microtiter plates. Using CRMOX plates and agarose gel analysis of the extracted DNA, no loss of the virulence plasmid in the randomly selected exconjugants was observed. Total DNA extracted from arbitrarily selected exconjugants was used in Southern blot experiments, and the random integration of the transposon into the Yersinia chromosome was confirmed (data not shown).
Selection of attenuated mutants.
Determination of the appropriate infectious dose and selection of the complexity of the input pool are critical to obtain meaningful STM data (22). The optimal infection dose and pool complexity depend on the route of infection, the animal model, and the microorganism used (9). To determine the optimum infection dose and the number of mutants in each dose, we varied the pool complexity between 30 and 60 clones and tested two infection doses, 105 and 5 × 105 CFU. The choice of infection doses was based on the following considerations. According to the published data, the 50% lethal dose (LD50) of Y. pseudotuberculosis via the i.v. route in mice is between 20 CFU (52) and 100 CFU (49). Our figure in a similar test for strain YPIII pIB1 was 51.5 CFU. In Y. enterocolitica STM experiments, the optimum infectious dose leading, to reproducible results via the intraperitoneal infection route was 100 times greater than the LD50 (13). We therefore chose infection doses significantly higher (varied between approximately 30 and 300 times the LD50 per mutant in the pools) than the LD50 in order to select mutants with severely impaired virulence properties.
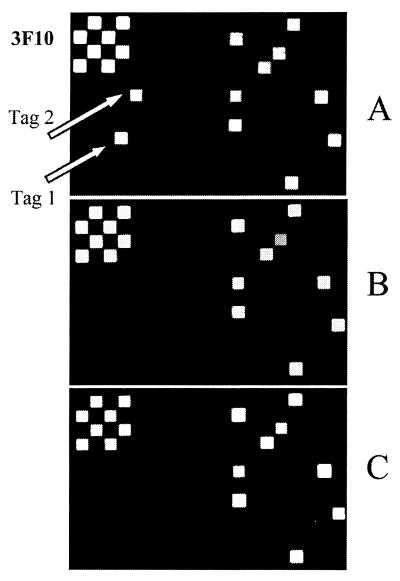
Genomic DNA was isolated from an aliquot of the bacterial culture used for infection (input pool) and from bacteria recovered from spleens (output pools). Mutants with reduced survival in vivo were visually identified by comparing the scanned images from arrays that had been hybridized with tags amplified from the input pools with images obtained from two independent output pools (Fig. 2). In addition, the hybridization intensities of different oligonucleotide sequences on the array were quantitated.
FIG. 2.
Fragments of images of arrays hybridized with tags from three pools. The infection dose used is 105 CFU. (A) Input pool; (B and C) output pools for two animals. The arrows point to the signals for both tags of mutant 3F10 in the input pool. No signals for these tags could be detected in output pools B and C.
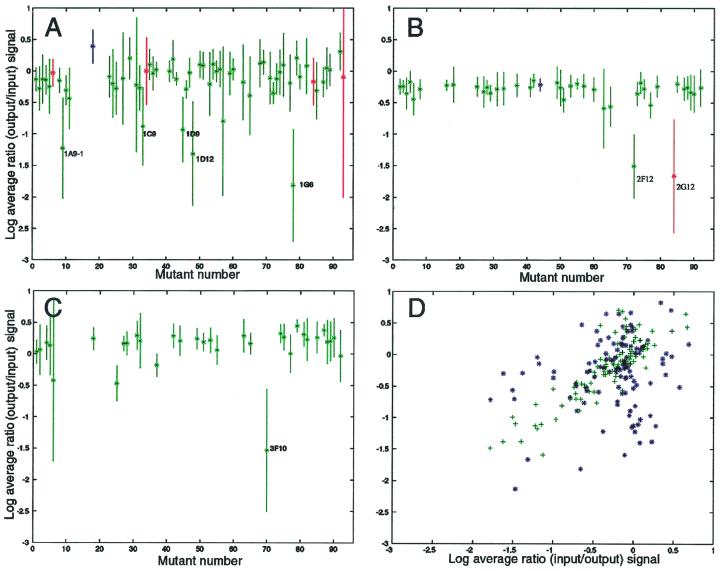
The input and output pools of the mutants were compared by hybridizing the labeled amplified tags to high-density oligonucleotide arrays (Affymetrix) containing complementary DNA sequences (Fig. 2). The hybridization patterns were found to be reproducible (Fig. 3A to C) regardless of the infection dose (105 or 5 × 105 CFU) or the number of colonies in the output pool (104 or 5 × 104). However, less variability was found in smaller (lower complexity) input pools (compare Fig. 3A and 3C). For larger (or higher complexity) input pools, animal-to-animal variability was greater than tag-to-tag variability (Fig. 3D), suggesting that there are biological limits on pool size. Mutants that showed reduced signals in the output pool for both tags in duplicate mice were selected for further analysis. Authenticity of the mutants was verified via direct sequencing of both tags after PCR amplification (data not shown).
FIG. 3.
Relative attenuation for different mutants from three pools. Each point represents the mean of the log of the ratio of the hybridization signal for the output pool relative to the input pool. Up to 12 measurements were collected for each mutant (tag1, tag2, etc.) for three (C), four (B), or six (A) different animals. Tags whose hybridization intensities were less than 500 (about threefold above background) in the input pool for each tag were not used in calculations. When signal was detectable in the input pool but below background in the output pool, the ratio was set to 0.001. Error bars indicate one standard deviation above and below the mean. Mixtures of 59 (A), 39 (B), or 31 (C) different mutants were used. Mutants from Table 1 are indicated. Points are colored green if data from both tag1 and tag2 were used in the calculations, red for tag1 only, or blue for tag2 only. (D) Comparison of reproducibility of hybridization and infection dose. The log of the ratio of the output to input hybridization signal for tag1 versus tag2 for the same animal (+) and tag1 versus tag1 and tag2 versus tag2 for different animals (∗) is shown. An infection dose of 105 CFU was used for 60 different mutants. Two mutants were not included in Table 1: 2F12 could not be sequenced, and 2G12 was not analyzed because data for one tag only were available.
Characterization of attenuated mutants and identification of transposon insertion sequences.
Approximately 5% of 603 exconjugants exhibited a statistically valid reduction in signal intensity. Transposon insertion sites in 31 mutants were sequenced using a single-primer PCR sequencing procedure (28) The results are summarized in Table 1. The Y. pestis genome sequence database (http://www.sanger.ac.uk/Projects/Y_pestis/) was used for identification of the corresponding genes in that pathogen. Almost 100% identity of the Y. pseudotuberculosis sequences to the Y. pestis DNA sequences implies similarity in their function.
TABLE 1.
Analysis of transposon derivatives selected by STMa
| No. | Mutant | In vivo CI | In vitro CI | Similarity to Y. pestis (%) | Y. pestis ORF | Similarity to NRDB (%) | Product | Microorganism | Putative function |
|---|---|---|---|---|---|---|---|---|---|
| 1 | 4H9 | 0.03 | 0.06 | 97 | YPO0054 | 85 (256) | Glycosyltransferase | S. marcescens | LPS core biosynthesis |
| 2 | 2B3 | 1.02 | 0.85 | 94 | YPO0337 | NS | UK | ||
| 3 | 5E4 | NT | NT | 99 | YPO0702 | 46 (87) | Putative lipoprotein | Mycobacterium tuberculosis | UK |
| 4 | 3E1 | 0.48 | 0.31 | 98 | YPO1108 | 91 (427) | Citrate synthase | S. enterica | Citrate synthesis |
| 5 | 2G8 | 0.53 | 1.34 | 96 | YPO1174 | 39 (222) | Adhesin | E. coli | UK |
| 6 | 3C10 | 0.055 | 1.02 | 98 | YPO1186 | 66 (150) | Hypothetical | E. coli | Amino acid transport |
| 7 | 3H10 | 0.08 | 0.07 | 97 | YPO1382 (promoter region) | 43 | LpsA, glycosyltransferase | Pasteurella haemolytica | LPS core biosynthesis |
| 8 | 5G6 | 0.021 | 0.88 | 95 | YPO1987 | NS | UK | ||
| 9 | 1D12 | 0.084 | 1.34 | 97 | YPO1994 | NS | UK | ||
| 10 | 1H10 | NT | NT | 98 | YPO2174 | 62 (442) | UDP-glucose 6-dehydrogenase | Rhizobium meliloti | LPS/capsular polysaccharide biosynthesis |
| 11 | 4H10 | 0.0036 | 1.95 | 96 | YPO2287a | 65 (153) | ABC transporter | Methanococcus jannaschii | Amino acid transport |
| 12 | 5G7 | 0.25 | 1.09 | 98 | YPO2440 | 82 (275) | Iron(III) dicitrate ATP binding protein | Haemophilus influenzae | Iron transport |
| 13 | 1A9 | NT | NT | 100 | YPO2532 | NS | UK | ||
| 14 | 4H2 | NT | NT | 98 | YPO2712 | 77 (218) | RseA, negative regulator of RpoE (sigma 24) | E. coli | Transcription regulation of RpoE |
| 15 | 5E6 | 0.27 | 1.24 | 97 | YPO3004 | 47 (268) | Prodipeptidase | B. subtilis | UK |
| 16 | 1D2 | 0.43 | 0.08 | 96 | YPO3099 | 100 (468) | ManC, mannose-1-P guanylyltransferase | Y. pseudotuberculosis | LPS biosynthesis |
| 17 | 1D12-1 | NT | 0.16 | 95 | YPO3099 | 100 (468) | ManC, mannose-1-phosphate guanyltransferase | Y. pseudotuberculosis | LPS biosynthesis |
| 18 | 3G2 | 0.13 | NT | 98 | YPO3100 | 100 (237) | Fcl, fucose synthetase | Y. pseudotuberculosis | LPS biosynthesis |
| 19 | 1B3 | 0.29 | 0.98 | 90 | YPO3104 | 100 (337) | O-antigen polymerase | Y. pseudotuberculosis | LPS biosynthesis |
| 20 | 1D9 | NT | 0.07 | 98 | YPO3114 | 100 (314) | DdhB, CDP-D-glucose-dehydratase | Y. pseudotuberculosis | LPS biosynthesis |
| 21 | 3F3 | 0.04 | 0.44 | 95 | YPO3116 | 100 (328) | AscD, ascarylose biosynthesis | Y. pseudotuberculosis | LPS biosynthesis |
| 22 | 3G1 | 0.21 | 1.25 | 97 | YPO3144 | 89 (587) | MdlB, multidrug resistance protein | E. coli | UK |
| 23 | 5A5 | 0.44 | 1.16 | 98 | YPO3572 | 88 (209) | Hypothetical transcription factor | E. coli | Transcription regulation |
| 24 | 1G6 | NT | NT | 97 | YPO3657/8 (intergenic) | NS | UK | UK | |
| 25 | 3F10 | 0.017 | 0.46 | 99 | YPO3834 | 87 (284) | PldA, phospholipase A | E. coli | UK |
| 26 | 1G6-1 | NT | 0.41 | 96 | YPO3965 | 47 (397) | VirA, His kinase | Agrobacterium tumefaciens | Sensory transduction |
| 27 | 5D12 | 0.003 | 0.98 | NS | 80 (66) | Wzx | S. enterica | O-antigen transporter | |
| 28 | 5H10 | NT | NT | NS | 70 (30) | Phage-related transcription activator | Xylella fastidiosa | Transcription activation | |
| 29 | 5B12 | 0.89 | 1.32 | NS | 59 (98) | Hypothetical | Phage HP1 | UK | |
| 30 | 1A9-1 | NT | NT | NS | NS | UK | UK | ||
| 31 | 1C9 | NT | 0.36 | NS | NS | UK | UK |
Analysis of Y. pseudotuberculosis transposon derivatives selected by STM. NS, no similarity; UK, unknown; NT, not tested; Similarity to Y. pestis determined using BLASTn at http://www.sanger.ac.uk/Projects/Y_pestis; similarity of the corresponding Y. pestis gene product (length in amino acids shown parentheses) to an orthologue in the NRDB using BLASTp at http://www.blast.genome.ad.jp.
Genes involved in polysaccharide biosynthesis.
One third of the sequenced mutants had transposon insertions in genes related to polysaccharide biosynthesis (mainly LPS core or O-antigen biosynthesis). In six cases (1B3, 1D2, 1D9, 1D12-1, 3G2, and 3F3; Table 1), the genes disrupted belonged to a single characterized O-antigen biosynthesis locus of Y. pseudotuberculosis. The disrupted genes encode mannose-phosphate-guanylyl transferase YPO3099 (1D12-1 and 1D2), fucose synthetase YPO3100 (3G2), LPS core biosynthesis protein YPO3104 (1B3), sugar dehydratase YPO3114 (1D9), and ascarylose biosynthesis protein YPO3116 (3F3). These genes are also present in Y. pestis. Other genes related to polysaccharide biosynthesis, such as those encoding UDP-glucose 6-dehydrogenase (1H10) and glycosyltransferase (4H9), were also identified. One mutant, 3H10, contained an insert in a putative promoter region of a single-gene operon encoding glycosyltransferase. The only mutation in an LPS-related gene which was absent in Y. pestis encoded an O-antigen transporter (5D12).
Putative virulence-related genes with orthologues in other bacteria.
Putative interrupted virulence genes include those encoding phospholipase A (3F10), sensory transducer His kinase VirA (1G6-1), a putative adhesin (2G8), a Pro-dipeptidase (5E6), RseA, a negative regulator of sigma 24 transcription factor (4H2), a transcription activator (5H10), and a transcription regulator (5A5) flanked by a vspC gene essential for secretion of virulence factors. The genes found in 4H10 and 3G1 are related to ABC transporters, a large class of proteins involved in export-import of a range of molecules. A clue to their possible function can be found from the analysis of corresponding regions of Y. pestis. In the case of 3G1, other genes of the operon are involved in ammonia assimilation. These genes are assumed to be essential in vivo, when bacteria have a depleted source of nitrogen. Similarly, the Y. pestis orthologue of the gene disrupted in the ABC transporter (4H10) is also located in the region involved in nitrate as well as amino acid transport. An unusual feature of this region in Y. pestis is that this gene overlaps by 270 nucleotides with another gene (AMP nucleosidase) transcribed in the opposite direction. Disrupted genes in the iron(III) dicitrate ATP-binding protein 5G7 are likely to be involved in iron transport. The importance of this system is not surprising, since iron ions are normally in limited supply in vivo.
Unknown and hypothetical genes.
Six mutants (1G6, 5D12, 5H10, 5B12, 1A9-1, and 1C9) contained inserts in genes with unknown function. Two of them (1C9 and 1A9-1) did not have counterparts in Y. pestis. Both of these mutants contained inserts in the region identical to an E. coli integrase-recombinase pseudogene, but actually had inserts into an adjacent region similar to an E. coli gene with unknown function. Transposon integration sites in these mutants were separated by approximately 200 nucleotides. A region flanking the insertion site in the 1G6 appeared to be unique for Yersinia, as it had 97% identity in the Y. pestis database, whereas no similarity to other bacteria was found. The genes in mutants 3C10 and 5B12 have similarities in a nonredundant database (NRDB) and also have counterparts in Y. pestis. The genes inactivated in mutants 2B3, 5G6, 1D12, and 1A9 appear to be unique to Yersinia, as no homologues could be identified in an NRDB. Genes inactivated in five mutants (5D12, 5H10, 5B12, 1C9, 1A9-1, and 1D12) do not have Y. pestis orthologues. One of these mutants (5D12) contains an insert in the gene encoding a putative O-antigen transferase (see above).
In vivo and in vitro CIs.
The majority of the selected mutants with reduced output signals did not reveal significant reduction in in vitro growth properties and were confirmed to be attenuated (Table 1). Five mutants (1D9, 1D2, 1D12-1, 4H9, and 3H10) revealed substantial (more than six times) reduction in CI. In all cases, the genes affected are related to LPS or LPS core biosynthesis. For example, an orthologue to a gene inactivated in 4H10 is located in an operon containing a number of other genes, such as kdtB, waaA, rfaC, rfaD, and rfaF, all related to core biosynthesis. The insert in mutant 3H10 would also inactivate a gene encoding an LPS core-related glycosyltransferase. Inactivation of genes in other mutants of this class may have a dramatic effect on outer membrane stability due to an affect on LPS biosynthesis. In vivo CI figures of less than 0.3 were demonstrated in 14 of 20 cases, confirming the attenuated properties of these mutants.
Characterization of phospholipase A mutant 3F10.
Mutant 3F10 contained an insertion into a pldA gene encoding a member of a family of bacterial phospholipases A (4). Given that phospholipases have been identified as virulence determinants in other pathogens but have not been detected in Y. pseudotuberculosis previously, mutant 3F10 was selected for more detailed characterization. The entire pldA gene was sequenced by primer walking. The gene encoded a protein of 282 amino acid residues and was preceded by an AGG Shine-Dalgarno sequence 6 nucleotides from the translation start site. A perfect inverted repeat was found after the termination codon of the pldA gene, indicating the presence of the rho-independent transcription terminator found in other bacteria (5). The sequence of the Y. pseudotuberculosis PldA protein also contained a Ca2+ binding motif (Gly-X-Gly-Gly) present in other bacterial PldAs (4, 5). The determined 1,468-bp sequence, including the pldA gene with flanking regions, was found to be 100% identical to the corresponding sequence of Y. pestis (http://www.sanger.ac.uk/Projects/Y_pestis/).
To test whether mutant 3F10 had reduced levels of PldA, we measured phospholipase activity and found that it was significantly reduced: 172 ± 9 U of phospholipase A2/mg of protein compared to 449 ± 27 for the wild-type strain (P < 0.05). Furthermore, using liposomes to assay phospholipase activity, total cell sonicates of the wild-type strain of Y. pseudotuberculosis caused the lysis of phosphatidylcholine or sphingomyelin liposomes. By contrast, cell sonicates from the pldA mutant showed only 28% of the activity of the wild-type extract towards either phosphatidylcholine or sphingomyelin liposomes. The remaining activity in both assays is probably attributable to other phospholipases present in the bacteria. Indeed, another phospholipase A, YplA, was recently discovered and shown to be essential for virulence in Y. enterocolitica (43).
In contrast to YplA (an extracellular protein), PldA is likely to be located in the outer membrane. A BLAST search of the Y. pestis sequence database indicated that both the yplA and pldA genes are present in this species. To check the possibility that the residual phospholipase activity in the 3F10 mutant could be the result of the presence of another phospholipase, a PCR was performed using Y. enterocolitica yplA-derived primers. Both Y. pestis (strain CO92) and Y. pseudotuberculosis DNAs gave identical products of the expected size, indicating the presence of a second phospholipase gene in these two Yersinia species (data not shown).
In vivo competition studies revealed significant attenuation of the 3F10 derivative. The CI (CI = 0.01655) obtained in the mixed-infection experiment confirmed that the strain was severely attenuated. However, an in vitro CI of 0.46 ± 0.04 indicated that growth of the mutant was not significantly impaired. Attenuation was also confirmed by the expected MLD determination. The expected MLD of the mutant (1.04 × 104 CFU per animal) was approximately 200-fold higher than that of the wild-type strain (51.5 CFU per animal). The expected MLD of the wild-type strain was in good agreement with published data (20 CFU per animal [52]). These data suggest that PldA is an essential virulence factor in the murine yersiniosis model of infection.
DISCUSSION
Despite being a powerful approach for identification of virulence-related genes, the output of STM generally depends on many different parameters, such as complexity of infection dose, efficiency of hybridization, and variability in animal sensitivity to an infectious agent. We developed a microarray modification of the STM technique allowing the generation of statistically valid output data. The approach allowed the simultaneous identification of several candidate virulence-related genes in Y. pseudotuberculosis. Annotation of the identified genes was assisted by availability of the complete genome sequence of the closely related species Y. pestis. Whereas the genes present in both species were found to be almost identical, five Y. pseudotuberculosis genes identified through STM did not have counterparts in the Y. pestis genome and may contribute to the different tropisms in these closely related infectious agents.
Screening of 603 transposon derivatives resulted in the identification of 31 potentially attenuated mutant genes, 26 of which had similarities in the Y. pestis genome. The disrupted genes included those involved in the transport of iron and other molecules (13%), signal transduction and transcription regulation (13%), as well as the genes with no similarity or with similarity to genes with unknown functions (27%). A third of the genes were related to LPS biosynthesis, similar to that found in the STM study of Y. enterocolitica (13). Unexpectedly, however, and in contrast to the previously mentioned publication, no genes related to a type III secretion or virulence plasmid were found. For example, despite the importance of YopK of Y. pseudotuberculosis for colonization of both spleen and liver after oral and i.v. infection (23), none of our mutants contained inserts into this gene. One explanation for this unexpected finding would be an insufficient number of mutants analyzed. However, no mutations affecting YopK were found after screening as many as 2,000 mutants of Y. enterocolitica (13). As the efficiency of our approach has been validated by the detection of a number of virulence-related genes, including those involved in LPS biosynthesis, the absence of virulence plasmid-related genes among the hits found in this model of infection was surprising. The most likely explanation for not identifying STM plasmid virulence-related genes is that the output pool of STM mutants was selected on CRMOX plates to ensure that mutants identified retained the virulence plasmid.
In this study, we have identified a novel Yersinia phospholipase, PldA, as a virulence determinant of Y. pseudotuberculosis. The wide distribution of this enzyme among the members of the Enterobacteriaceae (5, 41) suggests that this enzyme may play an important role in these organisms. In E. coli, pldA mutants have been shown to have a normal phenotype in vitro (5, 15), but pldA mutants of any species have not previously been tested in animal models of disease. Our finding that PldA is a virulence determinant of Y. pseudotuberculosis suggests that this enzyme should be evaluated as a virulence determinant in these other pathogens.
The role of the Y. pseudotuberculosis PldA in disease is not clear. Bacterial phospholipases C are considered important virulence factors (reviewed in reference 51), but bacterial phospholipases A have been less well studied. Mammalian cytosolic phospholipases A play a key role in the mediation of inflammatory responses by releasing arachidonic acid from membrane phospholipids (44). The released arachidonic acid is able to serve as a substrate for the generation of a variety of inflammatory mediators, such as prostaglandins, leukotrienes, and platelet-activating factor (44). It is thought that bacterial phospholipases C mimic the effects of the mammalian phospholipases A on host cells, resulting in profound inflammatory responses (51). It seems equally likely that bacterial phospholipases A could elicit these effects on host cells. The bacterial phospholipases A might also contribute to the invasion of host cells.
It has recently been shown that PldA of Helicobacter pylori is essential for colonization of gastric mucosa (16). It may also be significant that Pace et al. (33) showed that a rapid increase in the levels of free intracellular calcium ions and activation of phospholipase A2 and 5-lipoxygenase in cultured epithelial cells were required for invasion by Salmonella enterica serovar Typhimurium. These enzymes appeared to be associated with leukotriene production by these cells. The addition of the leukotriene D4 to Henle-407 cells caused both an increase in calcium ions and the internalization of an invasion-defective mutant of S. enterica. It is proposed that the phospholipases A involved in this process are of eukaryotic origin, but it seems possible that surface-bound bacterial enzymes could also contribute to this process.
The PldA that we have identified is not similar to the YplA phospholipase A of Y. enterocolitica that is essential for virulence in Y. enterocolitica (43), as they have no sequence similarity. Also, whereas YplA is an extracellular enzyme, PldA-related proteins are outer membrane proteins (3). The pldA and yplA genes are found in both the Y. pseudotuberculosis and Y. pestis genomes. Our finding that inactivation of the pldA gene in Y. pseudotuberculosis resulted in significant reduction but not complete elimination of phospholipase activity suggests that the residual phospholipase A activity in the mutant may be attributable to YplA. Our findings also indicate that the pldA gene encodes a phospholipase which is active towards both phosphatidylcholine and sphingomyelin. These phospholipids are the major phospholipid components of the outer leaflet of eukaryotic cell membranes. Since the liposome assay presented phospholipids to the enzyme which were packed in a form similar to that found in cell membranes, it is possible that the PldA enzyme we have isolated would be active towards host cell membranes. A similar reduction in activity towards phosphatidylcholine and sphingomyelin liposomes was found in a whole-cell extract from the pldA mutant. This is consistent with a single enzyme's catalyzing both reactions. Our finding that residual phospholipase activity was detected in cell extracts from the pldA mutant is also consistent with the presence of a second phospholipase in Y. pseudotuberculosis. However, the fact that the pldA mutant showed a 200-fold reduction in the expected MLD indicates that the two phospholipase A activities are not simply a result of redundancy and do not complement each other.
A number of Y. pseudotuberculosis genes identified in this study are related to LPS, and more specifically, to O-antigen biosynthesis (e.g. manC, fcl, wbyJ, and ddhB). A significant proportion of virulence-related genes identified by STM of Y. enterocolitica (13) and V. cholerae (9) were also found to be LPS related. One of the functions of the O-antigen is thought to be to enhance survival of enteric pathogens in the gastrointestinal tract (38). In support of this suggestion, an O-antigen mutant of Y. enterocolitica has been shown to be 50-fold less virulent than the wild-type strain when given orally to mice (2). Also it has been shown that a rough mutant of Salmonella enterica var. choleraesuis was attenuated by the oral but not by the i.v. route of challenge.
In Y. pestis, which is transmitted to humans by flea vectors or by the inhalation route, many of the O-antigen biosynthesis genes (including fcl and ddhB) are inactivated, resulting in a rough phenotype (36, 50). These mutations have been suggested to be a consequence of the change in life style from the ancestral Y. pseudotuberculosis strain (50). Our studies were carried out with Y. pseudotuberculosis administered by the i.v. route and clearly indicate that the O-antigen is important for infection by this route, probably because of the protection afforded against complement-mediated lysis (38). Therefore, it seems likely that the loss of the O-antigen by Y. pestis is not simply a consequence of the change in life style from an enteric to a blood-borne pathogen. Since Y. pestis is clearly able to cause infection when given by the i.v. route, it seems likely that other mechanisms have evolved to allow survival in the host.
The identification of 31 potential attenuating strains among 603 tagged transposon mutants is higher than that reported in the majority of STM studies. The higher hit rate achieved in this study may result from the modifications to the standard STM procedure. These include the use of double-tagged minitransposons and quantitative hybridization analysis using Affymetrix oligonucleotide microarray technology. The use of selected double tags further increases the reliability of the procedure. This set of optimized 96 double-tagged transposons cloned on a suicide vector can be used in construction of mutants of other microorganisms. Furthermore, the technique offers automaton and quantitative analysis of the data. In addition to studying tagged microbes in animal models, the increased reliability and sensitivity of quantifying tagged microbes described here should facilitate the application of STM to the population dynamics of microbes in other complex environments, such as biofilms, mixed cultures, and food.
ACKNOWLEDGMENTS
This work was supported by the Wellcome Trust and DERA, United Kingdom. E.W. was supported by the John Wasmuth fellowship in genomic analysis (HG00185-02).
We are grateful to staff at the Sanger Center for sequencing the Y. pestis genome and releasing the data and Beowulf Genomics for funding the genome sequencing project. We are grateful to B. L. Wanner for strain E. coli 19851. We thank Lynne Batty, Debbie Rogers, and Mark Richards for technical assistance.
REFERENCES
- 1.Achtman M, Zurth K, Morelli G, Torrea G, Guiyoule A, Carniel E. Yersinia pestis, the cause of plague, is a recently emerged clone of Yersinia pseudotuberculosis. Proc Natl Acad Sci USA. 1999;96:14043–14048. doi: 10.1073/pnas.96.24.14043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.al-Hendy A, Toivanen P, Skurnik M. Lipopolysaccharide O side chain of Yersinia enterocolitica O:3 is an essential virulence factor in an orally infected murine model. Infect Immun. 1992;60:870–875. doi: 10.1128/iai.60.3.870-875.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Brok R G, Belandia I U, Dekker N, Tommassen J, Verheij H M. Escherichia coli outer membrane phospholipase A: role of two serines in enzymatic activity. Biochemistry. 1996;35:7787–7793. doi: 10.1021/bi952970i. [DOI] [PubMed] [Google Scholar]
- 4.Brok R G, Boots A P, Dekker N, Verheij H M, Tommassen J. Sequence comparison of outer membrane phospholipases A: implications for structure and for the catalytic mechanism. Res Microbiol. 1998;149:703–710. doi: 10.1016/s0923-2508(99)80017-5. [DOI] [PubMed] [Google Scholar]
- 5.Brok R G, Brinkman E, van Boxtel R, Bekkers A C, Verheij H M, Tommassen J. Molecular characterization of enterobacterial pldA genes encoding outer membrane phospholipase A. J Bacteriol. 1994;176:861–870. doi: 10.1128/jb.176.3.861-870.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brubaker R R. Factors promoting acute and chronic diseases caused by yersiniae. Clin Microbiol Rev. 1991;4:309–324. doi: 10.1128/cmr.4.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Camacho L R, Ensergueix D, Perez E, Giequel B, Guilhot C. Identification of a virulence gene cluster of Mycobacterium tuberculosis by signature-tagged transposon mutagenesis. Mol Microbiol. 1999;34:257–267. doi: 10.1046/j.1365-2958.1999.01593.x. [DOI] [PubMed] [Google Scholar]
- 8.Cheng L W, Schneewind O. Type III machines of Gram-negative bacteria: delivering the goods. Trends Microbiol. 2000;8:214–220. doi: 10.1016/s0966-842x(99)01665-0. [DOI] [PubMed] [Google Scholar]
- 9.Chiang S L, Mekalanos J J. Use of signature-tagged transposon mutagenesis to identify Vibrio cholerae genes critical for colonization. Mol Microbiol. 1998;27:797–805. doi: 10.1046/j.1365-2958.1998.00726.x. [DOI] [PubMed] [Google Scholar]
- 10.Claus H, Frosch M, Vogel U. Identification of a hotspot for transformation of Neisseria meningitidis by shuttle mutagenesis using signature-tagged transposons. Mol Gen Genet. 1998;259:363–371. doi: 10.1007/s004380050823. [DOI] [PubMed] [Google Scholar]
- 11.Coulter S N, Schwan W R, Ng E Y, Langhorne M H, Ritchie H D, Westbrock-Wadman S, Hufnagle W O, Folger K R, Bayer A S, Stover C K. Staphylococcus aureus genetic loci impacting growth and survival in multiple infection environments. Mol Microbiol. 1998;30:393–404. doi: 10.1046/j.1365-2958.1998.01075.x. [DOI] [PubMed] [Google Scholar]
- 12.Cox J S, Chen B, McNeil M, Jacobs W R. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature. 1999;402:79–83. doi: 10.1038/47042. [DOI] [PubMed] [Google Scholar]
- 13.Darwin A J, Miller V L. Identification of Yersinia enterocolitica genes affecting survival in an animal host using signature-tagged transposon mutagenesis. Mol Microbiol. 1999;32:51–62. doi: 10.1046/j.1365-2958.1999.01324.x. [DOI] [PubMed] [Google Scholar]
- 14.de Lorenzo V, Herrero M, Jakubzik U, Timmis K N. Mini-Tn5 transposon derivatives for insertion mutagenesis, promoter probing, and chromosomal insertion of cloned DNA in gram-negative eubacteria. J Bacteriol. 1990;172:6568–6572. doi: 10.1128/jb.172.11.6568-6572.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Doi O, Nojima S. Nature of Escherichia coli mutants deficient in detergent-resistant and/or detergent-sensitive phospholipase A. J Biochem (Tokyo) 1976;80:1247–1258. doi: 10.1093/oxfordjournals.jbchem.a131396. [DOI] [PubMed] [Google Scholar]
- 16.Dorrell N, Martino M C, Stabler R A, Ward S J, Zhang Z W, McColm A A, Farthing M J, Wren B W. Characterization of Helicobacter pylori PldA, a phospholipase with a role in colonization of the gastric mucosa. Gastroenterology. 1999;117:1098–1104. doi: 10.1016/s0016-5085(99)70394-x. [DOI] [PubMed] [Google Scholar]
- 17.Edelstein P H, Edelstein A C, Higa F, Falkow S. Discovery of virulence genes of Legionella pneumophila by using signature tagged mutagenesis in guinea pig pneumonia model. Proc Natl Acad Sci USA. 1999;96:8190–8195. doi: 10.1073/pnas.96.14.8190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fukushima H, Sato T, Nagasako R, Takeda I. Acute mesenteric lymphadenitis due to Yersinia pseudotuberculosis lacking a virulence plasmid. J Clin Microbiol. 1991;29:1271–1275. doi: 10.1128/jcm.29.6.1271-1275.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grinter N J. A broad-host-range cloning vector transposable to various replicons. Gene. 1983;21:133–143. doi: 10.1016/0378-1119(83)90155-5. [DOI] [PubMed] [Google Scholar]
- 20.Hensel M, Shea J E, Gleeson C, Jones M D, Dalton E, Holden D W. Simultaneous identification of bacterial virulence genes by negative selection. Science. 1995;269:400–403. doi: 10.1126/science.7618105. [DOI] [PubMed] [Google Scholar]
- 21.Herrero M, de Lorenzo V, Timmis K N. Transposon vectors containing non-antibiotic resistance selection markers for cloning and stable chromosomal insertion of foreign genes in gram-negative bacteria. J Bacteriol. 1990;172:6557–6567. doi: 10.1128/jb.172.11.6557-6567.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Holden D W, Hensel M. Signature tagged mutagenesis. Methods Microbiol. 1998;27:359–369. [Google Scholar]
- 23.Holmstrom A, Rosqvist R, Wolf-Watz H, Forsberg A. YopK, a novel virulence determinant of Yersinia pseudotuberculosis. Contrib Microbiol Immunol. 1995;13:239–243. [PubMed] [Google Scholar]
- 24.Hong P C, Tsolis R M, Ficht T A. Identification of genes required for chronic persistence of Brucella abortus in mice. Infect Immun. 2000;68:4102–7. doi: 10.1128/iai.68.7.4102-4107.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jones A L, Knoll K M, Rubens C E. Identification of Streptococcus agalactiae virulence genes in the neonatal rat sepsis model using signature-tagged mutagenesis. Mol Microbiol. 2000;37:1444–1455. doi: 10.1046/j.1365-2958.2000.02099.x. [DOI] [PubMed] [Google Scholar]
- 26.Karlyshev A V, Pallen M J, Wren B W. Single-primer PCR procedure for rapid identification of transposon insertion sites. BioTechniques. 2000;28:1078–1082. doi: 10.2144/00286bm05. [DOI] [PubMed] [Google Scholar]
- 27.Lashkari D A, Hunicke-Smith S P, Norgren R M, Davis R W, Brennan T. Proc. Natl Acad Sci USA. 1995;92:7912–7915. doi: 10.1073/pnas.92.17.7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lehoux D E, Levesque R C. An automated multiplex oligonucleotide synthesizer: development of high-throughput, low-cost DNA synthesis. Detection of genes essential in specific niches by signature-tagged mutagenesis. Curr Opin Biotechnol. 2000;11:434–439. doi: 10.1016/s0958-1669(00)00124-5. [DOI] [PubMed] [Google Scholar]
- 29.Lestrate P, Delrue R M, Danese I, Didembourg C, Taminiau B, Mertens P, De Bolle X, Tibor A, Tang C M, Letesson J J. Identification and characterization of in vivo attenuated mutants of Brucella melitensis. Mol Microbiol. 2000;38:543–551. doi: 10.1046/j.1365-2958.2000.02150.x. [DOI] [PubMed] [Google Scholar]
- 30.Mei J M, Nourbakhsh F, Ford C W, Holden D W. Identification of Staphylococcus aureus virulence genes in a murine model of bacteraemia using signature-tagged mutagenesis. Mol Microbiol. 1997;26:399–407. doi: 10.1046/j.1365-2958.1997.5911966.x. [DOI] [PubMed] [Google Scholar]
- 31.Metcalf W W, Jiang J, Daniels L L, Kim S-K, Haldimann A, Wanner B L. Conditionally replicative and conjugative plasmids carrying lacZα for cloning, mutagenesis, and allele replacement in bacteria. Plasmid. 1996;35:1–13. doi: 10.1006/plas.1996.0001. [DOI] [PubMed] [Google Scholar]
- 32.Oyston P C, Russell P, Williamson E D, Titball R W. An aroA mutant of Yersinia pestis is attenuated in guinea-pigs, but virulent in mice. Microbiology. 1996;142:1847–1853. doi: 10.1099/13500872-142-7-1847. [DOI] [PubMed] [Google Scholar]
- 33.Pace J, Hayman M J, Galan J E. Signal transduction and invasion of epithelial cells by S. typhimurium. Cell. 1993;72:505–514. doi: 10.1016/0092-8674(93)90070-7. [DOI] [PubMed] [Google Scholar]
- 34.Perry R D. Signature-tagged mutagenesis and the hunt for virulence factors. Trends Microbiol. 1999;7:385–389. doi: 10.1016/s0966-842x(99)01582-6. [DOI] [PubMed] [Google Scholar]
- 35.Polissi A, Pontiggia A, Feger G, Altieri M, Mottl H, Ferrari L, Simon D. Large-scale identification of virulence genes from Streptococcus pneumoniae. Infect Immun. 1998;66:5620–5629. doi: 10.1128/iai.66.12.5620-5629.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Prior J, Hitchin P G, Williamson E D, Reason A J, Morris H R, Dell A, Wren B, Titball R W. Characterisation of the lipopolysaccharide of Yersinia pestis. Microb Pathog. 2001;30:48–57. doi: 10.1006/mpat.2000.0411. [DOI] [PubMed] [Google Scholar]
- 37.Reed L J, Muench H. A simple method for estimating fifty percent endpoints. Am J Hyg. 1938;27:493–497. [Google Scholar]
- 38.Reeves P. Role of O-antigen variation in the immune response. Trends Microbiol. 1995;3:381–386. doi: 10.1016/s0966-842x(00)88983-0. [DOI] [PubMed] [Google Scholar]
- 39.Riley G, Toma S. Detection of pathogenic Yersinia enterocolitica by using congo-red magnesium oxalate agar medium. J Clin Microbiol. 1989;27:213–214. doi: 10.1128/jcm.27.1.213-214.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rosquist R, Skurnik M, Wolf-Watz H. Increased virulence of Yersinia pseudotuberculosis by two independent mutations. Nature. 1988;334:522–525. doi: 10.1038/334522a0. [DOI] [PubMed] [Google Scholar]
- 41.Saxena M, Gupta J K, Vadehra D V. Phospholipase A production by isolates of Salmonella species. Indian J Med Res. 1990;91:177–181. [PubMed] [Google Scholar]
- 42.Schena M, Heller R A, Theriault T P, Konrad K, Lachenmeier E, Davis R W. Microarrays: biotechnology's discovery platform for functional genomics. Trends Biotechnol. 1998;16:301–306. doi: 10.1016/s0167-7799(98)01219-0. [DOI] [PubMed] [Google Scholar]
- 43.Schmiel D H, Wagar E, Karamanou L, Weeks D, Miller V L. Phospholipase A of Yersinia enterocolitica contributes to pathogenesis in a mouse model. Infect Immun. 1998;66:3941–3951. doi: 10.1128/iai.66.8.3941-3951.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schoenberg M H, Mayer J M, Beger H G. Phospholipase A2: from basic research to clinical reality. Chirurg. 1997;68:1112–1118. doi: 10.1007/s001040050330. [DOI] [PubMed] [Google Scholar]
- 45.Schwan W R, Coulter S N, Ng E Y, Langhorne M H, Ritchie H D, Brody L L, Westbrock-Wadman S, Bayer A S, Folger K R, Stover C K. Identification and characterization of the PutP proline permease that contributes to in vivo survival of Staphylococcus aureus in animal models. Infect Immun. 1998;66:567–572. doi: 10.1128/iai.66.2.567-572.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Schweizer H P. A method for construction of bacterial hosts for lac-based cloning and expression vectors: alpha-complementation and regulated expression. Biotechniques. 1994;17:452–456. [PubMed] [Google Scholar]
- 47.Shea J E, Hensel M, Gleeson C, Holden D W. Identification of a virulence locus encoding a second type III secretion system in Salmonella typhimurium. Proc Natl Acad Sci USA. 1996;93:2593–2597. doi: 10.1073/pnas.93.6.2593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47a.Shoemaker D. Ph.D. thesis. Stanford, Calif: Stanford University; 1997. [Google Scholar]
- 48.Shoemaker D D, Lashkari D A, Morris D, Mittmann M, Davis R W. Quantitative phenotypic analysis of yeast deletion mutants using a highly parallel molecular bar-coding strategy. Nat Genet. 1996;14:450–456. doi: 10.1038/ng1296-450. [DOI] [PubMed] [Google Scholar]
- 49.Simonet M, Mazigh D, Berche P. Growth of Yersinia plesudotuberculosis in mouse spleen despite loss of a virulence plasmid of mol. wt. 47 × 106. J Med Microbiol. 1984;18:371–375. doi: 10.1099/00222615-18-3-371. [DOI] [PubMed] [Google Scholar]
- 50.Skurnik M, Peippo A, Ervela E. Characterization of the O-antigen gene clusters of Yersinia pseudotuberculosis and the cryptic O-antigen gene cluster of Yersinia pestis shows that the plague bacillus is most closely related to and has evolved from Y. pseudotuberculosis serotype O:1b. Mol Microbiol. 2000;37:316–330. doi: 10.1046/j.1365-2958.2000.01993.x. [DOI] [PubMed] [Google Scholar]
- 51.Titball R W. Bacterial phospholipases. Soc Appl Bacteriol Symp Ser. 1998;27:127S–137S. [PubMed] [Google Scholar]
- 52.Une T, Brubaker R R. In vivo comparison of avirulent Vwa− and Pgm− or Pstr phenotypes of yersiniae. Infect Immun. 1984;43:895–900. doi: 10.1128/iai.43.3.895-900.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Winzeler E A, Shoemaker D D, Astronomoff A, Liang H, et al. Functional characteristics of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 54.Wodicka L, Dong H, Mittmann M, Ho M H, Lockhart D J. Genome-wide expression monitoring in Saccharomyces cerevisiae. Nat Biotechnol. 1997;15:1359–1367. doi: 10.1038/nbt1297-1359. [DOI] [PubMed] [Google Scholar]
- 55.Zhao H, Li X, Johnson D E, Mobley H L T. Identification of protease and rpoN-associated genes of uropahogenic Proteus mirabilis by negative selection on a mouse model of ascending urinary tract infection. Microbiology. 1999;145:185–195. doi: 10.1099/13500872-145-1-185. [DOI] [PubMed] [Google Scholar]