SUMMARY
A collective cell motility event that occurs during Drosophila eye development, ommatidial rotation (OR), serves as a paradigm for signaling-pathway-regulated directed movement of cell clusters. OR is instructed by the EGFR and Notch pathways and Frizzled/planar cell polarity (Fz/PCP) signaling, all of which are associated with photoreceptor R3 and R4 specification. Here, we show that Abl kinase negatively regulates OR through its activity in the R3/R4 pair. Abl is localized to apical junctional regions in R4, but not in R3, during OR, and this apical localization requires Notch signaling. We demonstrate that Abl and Notch interact genetically during OR, and Abl co-immunoprecipitates in complexes with Notch in eye discs. Perturbations of Abl interfere with adherens junctional organization of ommatidial preclusters, which mediate the OR process. Together, our data suggest that Abl kinase acts directly downstream of Notch in R4 to fine-tune OR via its effect on adherens junctions.
In brief
Koca et al. show that dAbl negatively regulates ommatidial rotation (OR) within the photoreceptors R3 and R4 downstream of Notch signaling activation in R4. Abl and Notch interact genetically and physically to instruct the OR process by controlling the adherens junctional organization.
Graphical Abstract
INTRODUCTION
Cells often possess directional features that play essential roles during development, function, and homeostasis of organs and tissues. Cellular polarity across the plane of tissues, referred to as planar cell polarity (PCP), provides cells with positional information thereby allowing them to orient with respect to the body and tissue axes.1–4 PCP polarization and cellular orientation are also key for directed cellular movement within tissues.1,5,6 PCP has been best studied in Drosophila and its establishment is mediated by a specific set of evolutionarily conserved “core PCP” proteins, which include the transmembrane proteins Frizzled (Fz), Van Gogh (Vang; Vangl in vertebrates, also known as Stbm in Drosophila), and Flamingo (Fmi; Celsr in vertebrates) and the cytoplasmic factors Dishevelled (Dsh; Dvl in vertebrates), Diego (Dgo; Diversin/Inversin in vertebrates), and Prickle (Pk).1,2,7 During PCP establishment, interactions among these core factors lead to the formation of asymmetrically localized complexes of Fz-Dsh-Dgo and Vang-Pk on opposing sides of cells, which are stabilized via intercellular homophillic adhesion of Fmi between neighboring cells across apical junctional membranes. These complexes form separate signaling units, interacting with their set of effector proteins and thus initiate distinct tissue and cell type-specific responses.1,4,7 PCP-induced downstream effector cascades can range from the (re)organization of cytoskeletal elements and the remodeling of cell adhesion complexes, to transcriptional regulation and associated cell fate changes.1,4,5,8,9 In Drosophila, eye development is particularly well suited to study several aspects of PCP signaling, as it entails PCP-dependent cell fate differentiation and cell motility processes.8,10,11
The Drosophila eye consists of ~800 highly regularly arranged ommatidia, each of which is composed of 8 photoreceptor (R cell) neurons (R1–R8), arranged into a defined invariant trapezoidal pattern, and 12 accessory (cone, pigment, and bristle) cells.12,13 During larval stages, the eye develops from an epithelial imaginal disc, which is initially composed of identical pluripotent precursor cells. As a wave of cell proliferation and differentiation (referred to as morphogenetic furrow [MF]) travels across the disc from posterior to anterior, regularly spaced preclusters of differentiating cells start to form in its wake that will subsequently mature into ommatidial clusters (and ommatidia in the adult).12–15 At the 5-cell precluster stage, differential specification of the R3/R4 cell pair requires asymmetric Fz/PCP signaling followed by directional Notch pathway activation within the pair and EGFR signaling, breaking the initial symmetry of the precluster.16–20 Differential specification of R3and R4 fatesalso generates the directional cues that instruct the subsequent rotation of the precluster toward the dorsal-ventral (D/V) midline, often referred to as the equator, in a process called ommatidial rotation (OR).8,10,11,21 During OR, as new cells join the precluster and differentiate, the precluster collectively undergoes a 90° rotation in opposing directions in the dorsal and ventral halves of the eye. This establishes the mirror-symmetric pattern most apparent in adult ommatidia across the D/V midline8 (see also Figures 1A–1C).
Figure 1. dAbl LOF clones reveal OR defects in the eye.
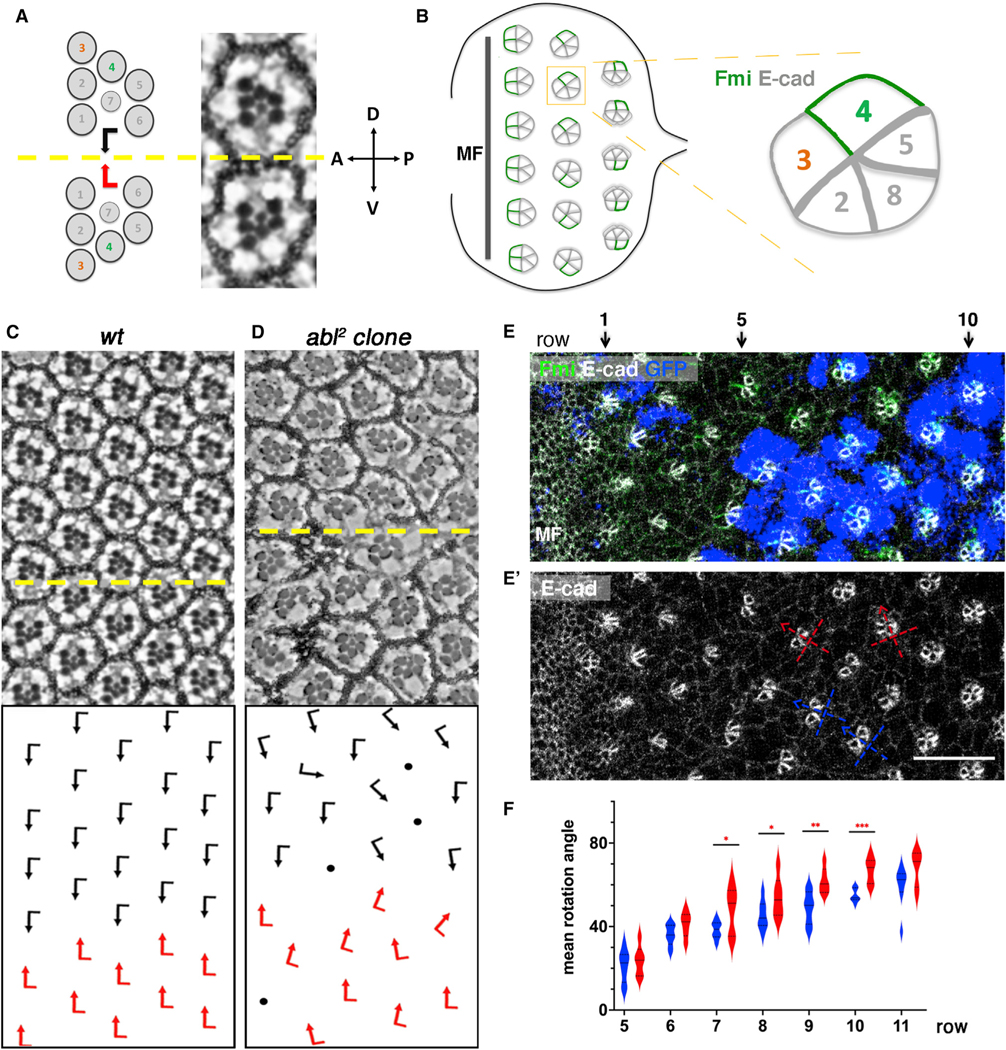
(A) Schematic and section view of the two distinct chiral forms of adult wild-type (WT) ommatidia, displaying mirror image symmetry across the equator (yellow line).
(B) Schematic of third-instar eye imaginal disc. As furrow (MF) moves across eye disc from posterior to anterior, ommatidial preclusters are forming in its wake, with R8 precursors inducing the sequential recruitment of R2/R5 and R3/R4 precursors pairs, resulting in the 5-cell precluster. After the symmetry of 5-cell preclusters breaks because of differential R3/R4 specification, they rotate toward the dorsoventral midline until they complete a 90° rotation and are aligned perpendicular to the equator. Fmi (green), initially detected in junctions of both R3/R4 precursors, becomes enriched to R4 junctional surfaces as the precursors mature. DE-cadherin (gray) is upregulated in R2/R5 and R8 cells (indicated by thicker junctional gray lines). Anterior is to the left and dorsal up in all panels.
(C) Adult eye section of WT with orientation schematics in lower panel (arrows as in A).
(D) abl2 mosaic adult eye with orientation schematics in lower panel; mutant tissue is marked by absence of pigment.
(E) abl2 mosaic third-instar larval eye imaginal disc stained for Fmi (green), DE-cad (gray), and GFP (blue) with MF on left. abl2 mutant clones are marked by the absence of blue/GFP.
(E′) E-cad monochrome staining to reveal cluster orientation angles, with the R2/R5 plane depicting the angle (see also B for schematic), of WT and mutant clusters; two examples each are marked by blue (WT) and red (mutant) lines, respectively.
(F) Quantification (violin plots) of average rotation angles observed in preclusters in rows 5–11 for WT (blue, 8 biological replicates) and abl2 mutant/mosaic (red, 6 biological replicates). Orientation angles were averaged per row per eye and plotted. Statistical analyses between the average values were performed for each row between the two genotypes. Note significant over-rotation in abl2 mutant clusters from row 7 onward.
Asterisks denote significance by t test (*p < 0.05, **p < 0.005, and ***p < 0.0005). Scale bar, 10 μm (E and E′).
In core PCP mutants, differential R3/R4 specification fails or becomes randomized and ommatidia are often misoriented,22–25 suggesting that PCP signaling not only dictates the R3/R4 cell fate specification,18,19 but also the direction and degree of OR. To date, several OR-specific regulators have been discovered on the basis of the ommatidial misorientation phenotypes associated with their mutants.26–34 For example, Fz/PCP signaling feeds into cadherin-based cell adhesion machinery through downstream effectors to regulate the OR process.33 Furthermore, cytoskeletal reorganization of ommatidial cells is coordinated with adhesion remodeling to drive the OR process downstream of several signaling pathways, including Fz/PCP, EGFR, and Notch signaling.26,29,31,34,35 Despite this knowledge, mechanistic insights into the OR process remain largely elusive.
Abelson (Abl) kinases are a family of non-receptor tyrosine kinases that govern a multitude of cellular processes in metazoans, including proliferation, differentiation, survival, and migration.36–38 Unlike master switch kinases, Abl kinases are transitionally activated, and subcellularly enriched, depending on the function they are involved in and therefore regulate distinct cellular processes.36–38 In the absence of activating signals, the SH2 and SH3 domains of Abl interact with each other and lock the protein in a kinase-inactive state.39,40 Although a direct mechanism for Abl kinase activation has not been identified, it is likely that upon stimulatory signals, the SH2 and SH3 domains interact locally with secondary molecules, unlocking the inhibitory conformation and locally enabling kinase activity. Abl kinases can be activated downstream of various signals, including growth factors, cellextracellular matrix (ECM) interactions, or adhesion receptors, highlighting the versatile nature of cellular Abl signaling.36–38 Drosophila Abl (dAbl) is primarily cytoplasmic and has been linked to the regulation of cytoskeletal and adhesion processes in several tissues. During early embryogenesis, for example, dAbl is required to localize actin polymerization and actomyosin activity to the apical domain by restricting Enabled (Ena) activity.41–45 In axon guidance, dAbl feeds into multiple signaling branches, including Ena and Rac GTPases, to regulate the balance of linear versus branched actin networks.46,47 Germband elongation requires dAbl to locally control the mobility of adhesion complexes through phosphorylation of Arm/β-catenin thuspromoting convergent extension movements.48 In addition, during photoreceptor morphogenesis dAbl has been shown to be essential for the maintenance of the apicobasal integrity in photoreceptors and associated proper organization of adherens junctions.49 Interestingly, dAbl was also shown to function within the core PCP pathway, through Dsh phosphorylation, to promote Fz/Dsh-PCP signaling within R3/R4 pairs.50 The involvement of Abl kinases in cell adhesion and cytoskeletal remodeling has been documented in various vertebrate contexts, suggesting that many of its functions are conserved across species.36–38
Here we show that during Drosophila eye patterning dAbl kinase negatively regulates OR downstream of the Notch receptor. Loss-of-function (LOF) and gain-of-function (GOF) genotypes of dAbl consistently cause opposite effects on the OR process. dAbl becomes apically localized in photoreceptors R8, R2/R5 and importantly R4, but not in R3, during OR. Apical junctional localization in R4 requires Notch signaling. Functionally, dAbl and Notch interact genetically during OR, and dAbl co-exists in complexes with Notch in developing eye discs. Our data collectively suggest that dAbl functions directly downstream of the Notch receptor in R4 by acting on the cadherin/β-catenin complexes and cell adhesion, via β-catenin phosphorylation. It thus serves a “brake function” during the OR process, fine-tuning OR cell motility for convergence to a 90° angle.
RESULTS
Abl kinase regulates OR
Our past studies suggested that dAbl could have a role in the OR process (see Figures 1A and 1B for schematic of retinal development features specific to OR), as overexpression of dAbl caused OR defects, but this potential dAbl contribution to Drosophila eye patterning and morphogenesis remained unexplored.50 To investigate how dAbl contributes to OR during retinal patterning, we analyzed the phenotypes of LOF clones of dAbl in mosaic eye tissue. Although clones of the abl2 allele can be recovered in adult eyes, theydisplay pleiotropic developmental and morphogenetic defects (Figures 1C and 1D). Besides previously reported phenotypes of loss and malformation of photoreceptors49,50 and loss of chirality,50 these clones also display misoriented ommatidia, consistent with the hypothesis that dAbl also regulate the OR process (Figures 1C and 1D). Adult dAbl mutant clones are small with mutant ommatidia and photoreceptors being often misshapen due to defects incellular specification and morphogenesis/maturation (as documented by Xiong and Rebay49). Therefore, to test the hypothesis that OR defects observed in LOF clones are a primary phenotype of dAbl mutant clusters, we examined abl2 clonal tissue in eye imaginal discs at the time of OR. dAbl mutant or mosaic ommatidial clusters frequently displayed an over-rotation phenotype in rows 5–11 compared with wild-type (WT) clusters (Figures 1E and 1F). In these early dAbl deficient ommatidial (pre)clusters, the organization of the 5-cell precluster was largely normal (Figures 1E and 1E′), suggesting that Abl has an OR-specific function.
To get further insight into the role of dAbl in OR, we analyzed the effects of Abl GOF posterior to the MF by using the sevGal4 driver (which drives expression broadly in R cells and cone cells behind the MF; see Figure S1 for expression pattern). In sevGal4, UAS-dAbl adult eyes (sev>Abl), most aspects of eye development were normal, including a correct chiral ommatidial arrangement, with the exception of many ommatidia being misoriented. Notably, ommatidial under-rotation was most frequently observed (Figures 2A and 2B′). To confirm that these OR defects observed upon sev>Abl expression arise as primary defects, we analyzed the respective larval eye discs during ommatidial patterning. Consistently, dAbl overexpressing ommatidia showed a slower rate of rotation, compared with WT (Figures 2C–2E). These data, together with the LOF clonal phenotypes displaying over-rotation features, suggest that Abl regulates the rate of rotation and that it has a specific inhibitory or “braking” function in OR.
Figure 2. Abl overexpression in eye discs posterior to MF slows down OR.
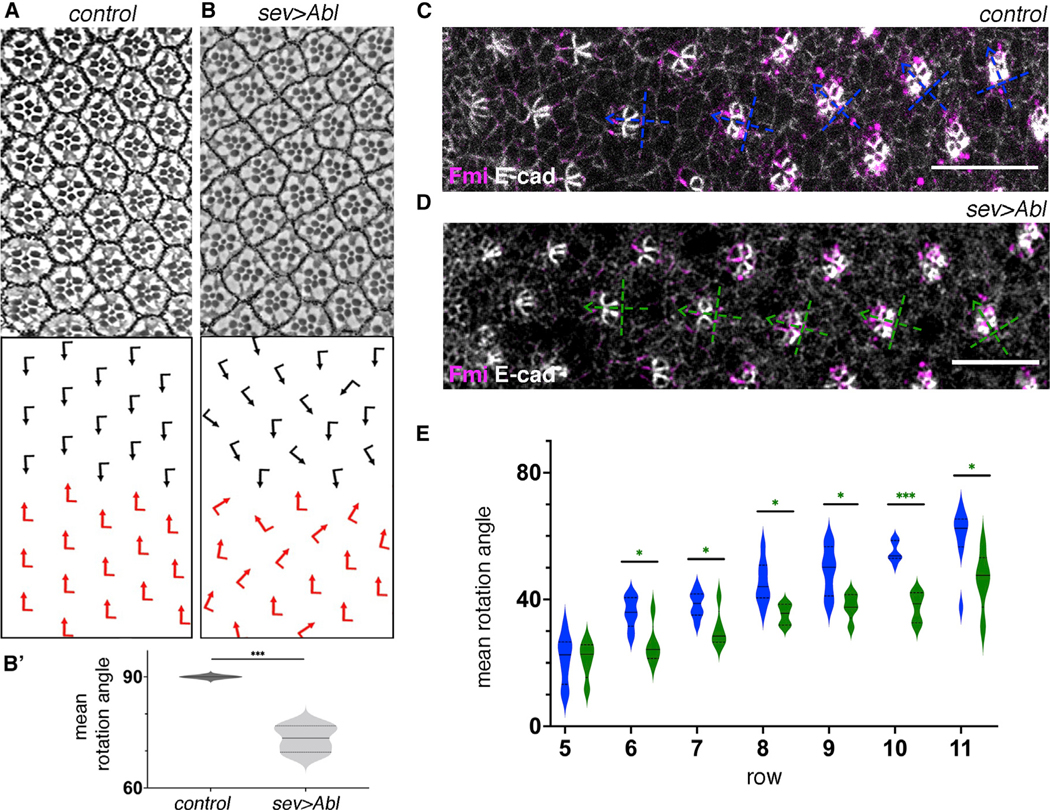
(A and B) Adult eye sections with ommatidial orientation schematics (arrows as in Figures 1A and 1C) of the indicated genotypes: (A) WT and (B) sev>Abl.
(B′) Quantification (violin plots) of adult average ommatidial orientation angles for the genotypes indicated (4 biological replicates each). Note marked under rotation in sev-Abl eyes compared with WT. Asterisks denote signifinace by t test (***p < 0.0005).
(C and D) Third-instar larval eye imaginal discs stained for Fmi (magenta) and E-cad (gray) in WT (C) and sev>Abl (D) backgrounds. Blue (WT) and green (sev-Abl) dashed cross arrows indicate orientation of ommatidial (pre)clusters.
(E) Quantification (violin plots) of average rotation angles observed in preclusters in rows 5–11, plotted for WT (blue, 8 biological replicates) and sev>Abl (green, 5 biological replicates). Statistical analyses were performed for each row between WT (blue) and sev-Abl (Abl overexpression, green) genotypes.
Asterisks denote significance by t test (*p < 0.05 and ***p < 0.0005). Note marked under-rotation trend of clusters in multiple rows of sev>Abl genotype compared with WT. Scale bars, 10 μm. See also Figure S1.
Abl kinase is apically localized and required in R4 to slow down OR
To gain insight into how dAbl may regulate rotation, we next analyzed the expression and localization pattern of dAbl in eye imaginal discs. dAbl expression was detected prominently in photoreceptors starting from a few rows posterior to the MF and persisting throughout ommatidial development. Notably, dAbl became localized apically at the level of the adherens junctions in R8, R2/R5, and R4 during later stages of OR (from rows 8 and 9 onward; Figures 3A–3C′), a localization maintained as the clusters further matured. Importantly, we did not observe apical localization of dAbl in R3 at any stage posterior to the MF. Thus, dAbl displayed a striking localization difference in apical junctional areas between the two cells of the R3/R4 pair (summarized in Figure 3B‴). Unlike this apical pattern, Abl localization and intensity did not show a notable difference between R3 and R4 more basally (Figure S2), suggesting that Abl is specifically localized to the apical junctional domain of R4 during OR.
Figure 3. Abl is apically localized in all photoreceptors of the 5-cell precluster except for R3.
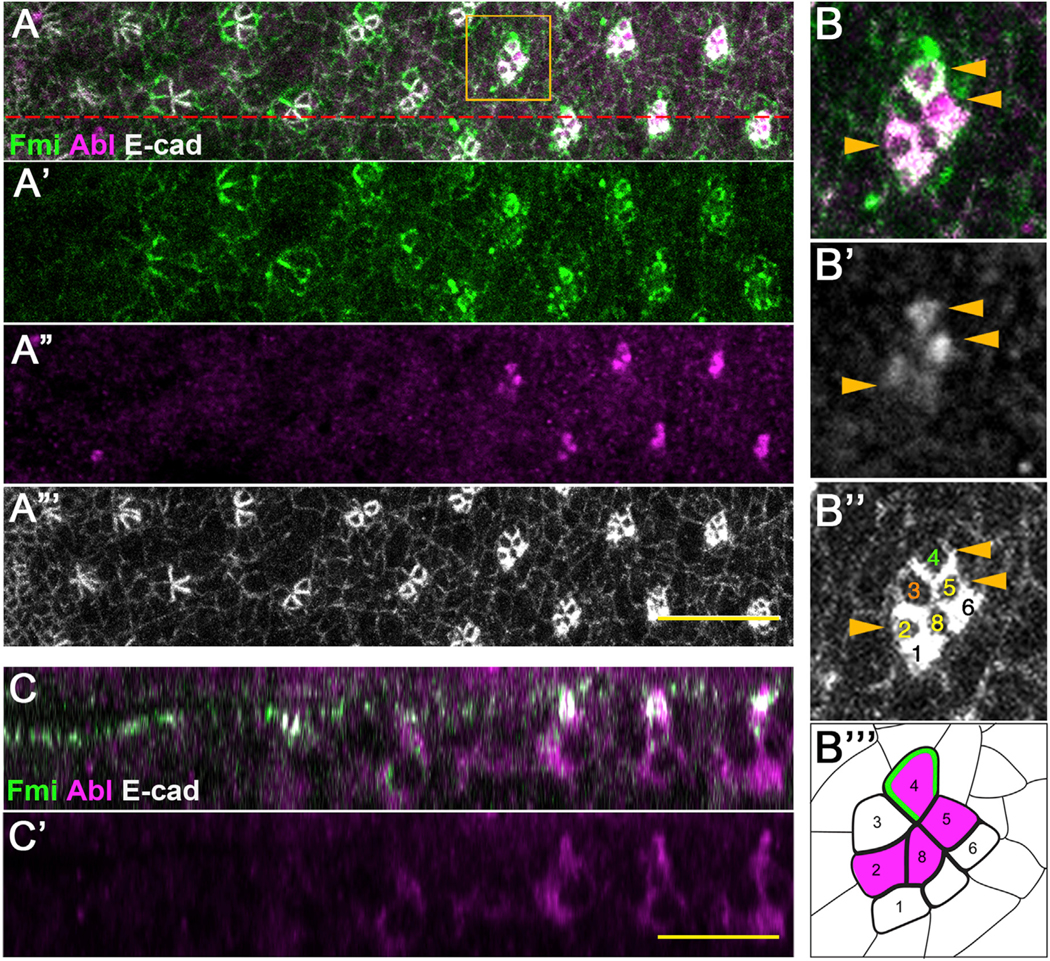
(A–A′′′) Control third-instar larval eye imaginal disc stained for Fmi (green, also A′), Abl (magenta, also A′′), and E-cad (gray, also A′′′).
(B–B′′′) Enlarged view of an individual cluster (boxed in A), with merge (B), and monochrome of Abl (B′) or E-cad (B′′, to highlight individual R cells, as numbered). Cells with high Abl levels in apical junctional region are marked with yellow arrowheads. (B′′′) Schematic of cluster with numbered R cells and Abl (magenta) as detected. Note that Abl is present in apical junctional regions in R8, R2/R5, and R4, but not in R3.
(C and C′) xz-view staining of the same eye disc, at the section marked by the red dashed line in A. Note the localization of Abl at the level of apical E-cad and Fmi. Scale bars, 10 μm. See also Figure S2.
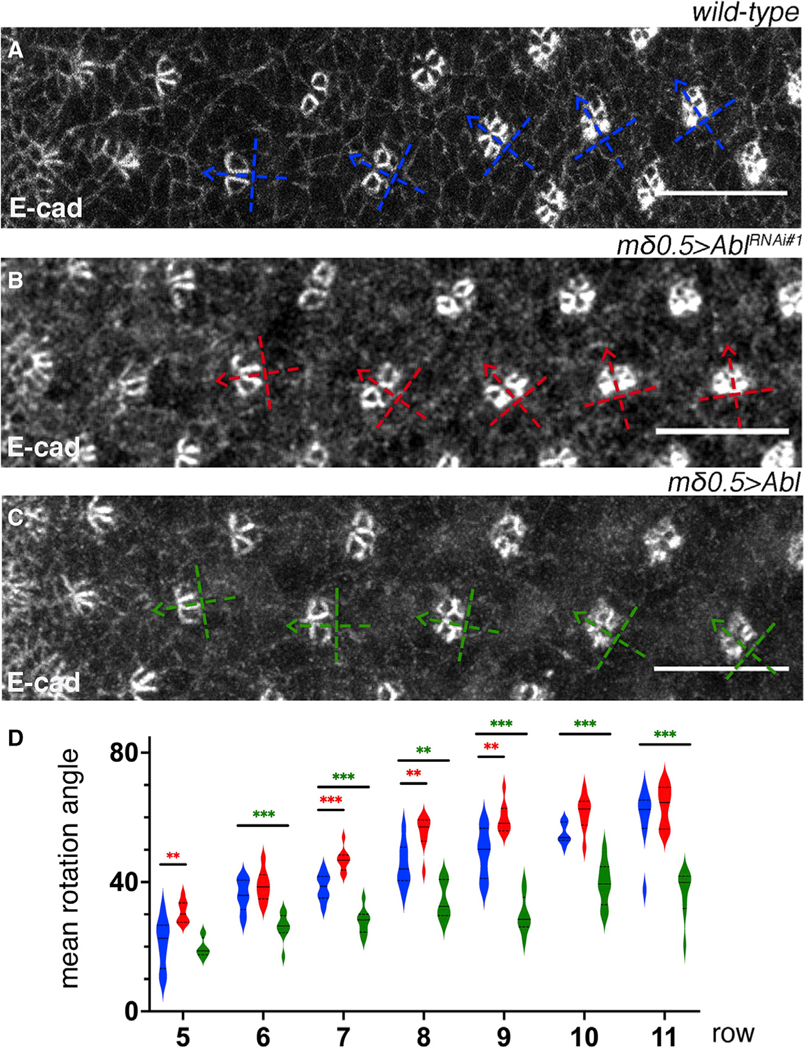
Differential specification of R3/R4 by Fz/PCP and Notch signaling and associated signaling events are critical for the correct direction and execution of OR.10,11,21 To better understand the requirement of dAbl for OR within the R3/R4 pair, with its differential apical localization pattern between the R3 and R4 cells, we perturbed dAbl function and localization specifically in the R3/R4 cells. To this end, we employed the mδ0.5-Gal4 driver, which is initially active in both R3/R4 precursors and later upregulated in R4 as a result of Notch-mediated R4 specification.19,35 mδ0.5-Gal4 mediated knockdown (KD) of dAbl within R3/R4 cells led to an aberrant rotation pattern of ommatidial clusters (compared with WT), with clusters generally rotating faster (Figures 4A, 4B, 4D, and S3). Conversely, dAbl overexpression in R3/R4 with mδ0.5-Gal4 caused significant under-rotation starting from early stages of OR (Figures 4A, 4C, and 4D; also Figure S3). In these backgrounds, R3/R4 cell fate specification was not affected, as R4-specific Fmi upregulation23 and associated apical localization remained intact (Figure S3). Taken together, these data suggest that differential dAbl localization, with dAbl localized apically only in R4, is required to fine-tune the slowing rate of rotation at later stages of OR.
Figure 4. Abl signaling within R3/R4 pair negatively regulates OR.
(A–C) Third-instar larval eye imaginal discs stained for E-cad (gray) in WT (A), mδ0.5>AblRNAi#1 (B), and mδ0.5>Abl (C). Blue, red, and green dashed cross arrows, respectively, indicate the orientation of ommatidial clusters for each genotype.
(D) Quantification (violin plots) of average rotation angles observed in rows 5–11, plotted for WT (blue), mδ0.5>AblRNAi#1 (red), and mδ0.5>Abl (green, Abl overexpression/OE in R3/R4) (8 biological replicates each). Statistical analyses were performed for each row between WT (blue) and AblRNAi (red) or Abl OE (green) genotypes.
Asterisks denote significance by one-way ANOVA (*p < 0.05, **p < 0.005, and ***p < 0.0005). Note over- and under-rotation observed in AblRNAi (red) and Abl OE (green) genotypes, respectively, relative to WT. Scale bars, 10 μm. See also Figure S3.
Notch activation is required for apical R4 localization of dAbl
The stark difference in apical localization of dAbl between the R3 and R4 cells (Figure 3) raised the possibility that either Fz/PCP or Notch signaling, both required to specify R4, instruct apical dAbl localization. This possibility was investigated in the R3/R4 cell pair both in core PCP mutant eye clones and upon Notch signaling interference. Firstly, we did not observe any difference in apical dAbl localization pattern of R3/R4 pairs in pk−/−, stbm−/− double-mutant clones, compared with WT (Figures 5A and 5A′). Among the core Fz/PCP signaling genes, both pk and stbm/Vang are functionally required in R4,22,51 therefore these data suggest that Fz/PCP signaling is not directly required for apical dAbl localization in R4, and its associated differential localization pattern between cells of the R3/R4 pair. Nevertheless, dAbl appears to interact with core PCP factors to regulate OR. For example, although overexpression of Fmi in R3/R4 pairs with the mδ0.5-Gal4 driver leads to the randomization of chirality, it only occasionally leads to misorientation in the adults; however, when Fmi and dAbl were co-overexpressed (mδ0.5>Fmi, >Abl), the misorientation phenotype was exacerbated in comparison to the individual backgrounds (mδ0.5>Abl and mδ0.5>Fmi; Figures S4A–S4D, see figure legend for more details).
Figure 5. Notch signaling, but not core PCP factors, is required for apical Abl localization in R4.
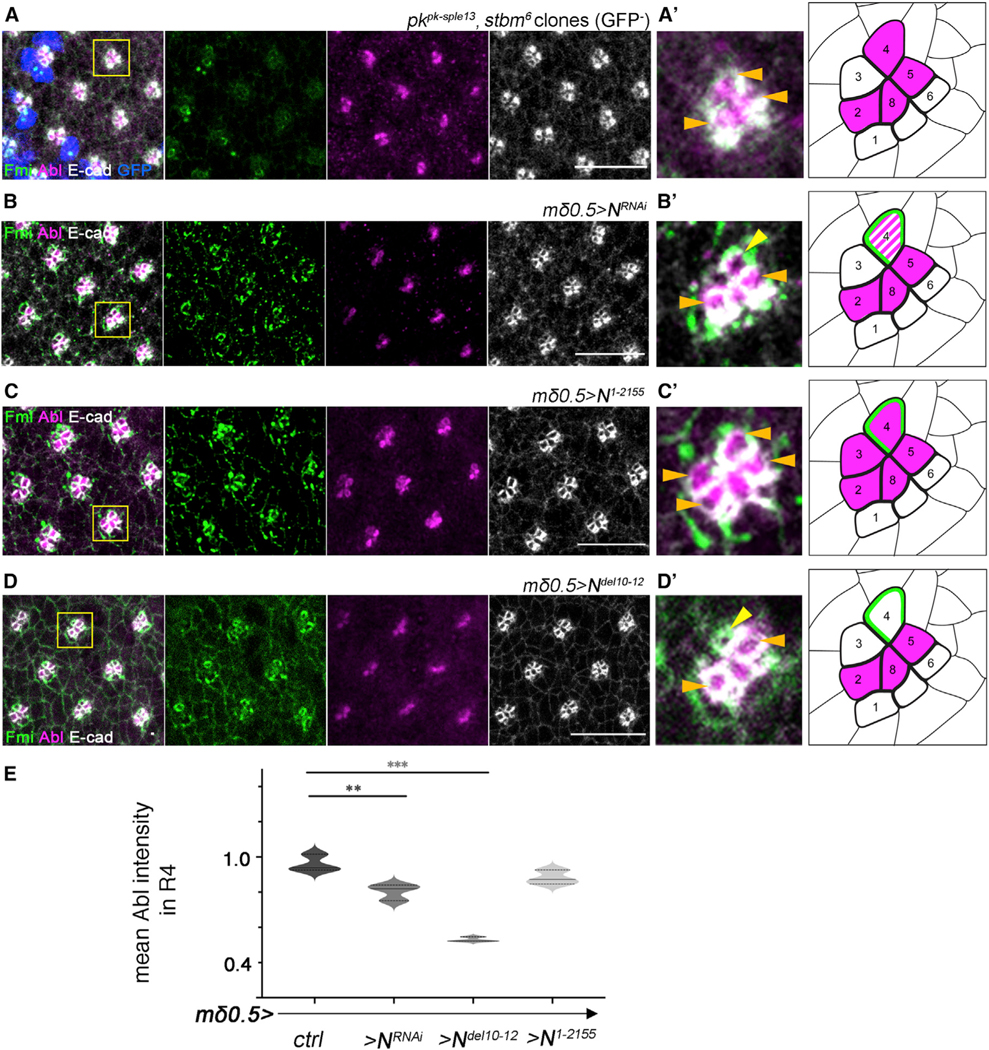
(A–D) Third-instar larval eye imaginal discs of the designated genotypes stained for Fmi (green), Abl (magenta), and E-cad (white): (A) pkpk-sple13stbm6 double-mutant mosaics, mutant clones are marked by the absence of GFP (blue), (B) mδ0.5>NRNAi (partial Notch LOF) (C) mδ0.5>N1−2155 (Notch GOF), and (D) mδ0.5>Ndel10−12 (Notch signaling defective).
(A′–D′) Enlarged view of individual clusters for each genotype (as marked by yellow boxes in A–D) are depicted in left panels, right panels show the respective schematics (same schematic drawing as in Figure 3B′′′). Orange horizontal arrows mark cells with apical Abl, and yellow arrows mark R4 cells with Abl staining loss. Note that apical junctional localization is not affected in pk, Vang double-mutant clusters but is reduced or lost in R4 in genotypes impairing Notch signaling (mδ0.5>NRNAi and mδ0.5>Ndel10−12 backgrounds, B′ and D′, respectively); in contrast Notch GOF in R3 recruits Abl to junctional apical regions in R3 as well (C′).
(E) Quantification (violin plots) of average apical Abl intensity in R4 (normalized to that in R2), plotted for WT, mδ0.5>NRNAi, mδ0.5>Ndel10−12, and mδ0.5>N1−2155 (3 biological replicates each).
Asterisks denote significance by one-way ANOVA (*p < 0.05 and ***p < 0.0005). Note reduction of apical Abl intensity in R4 in the mδ0.5>NRNAi and mδ0.5>Ndel10−12 backgrounds, with reduced or absent Notch signaling, compared with WT. Scale bars, 10μm. See also Figure S4.
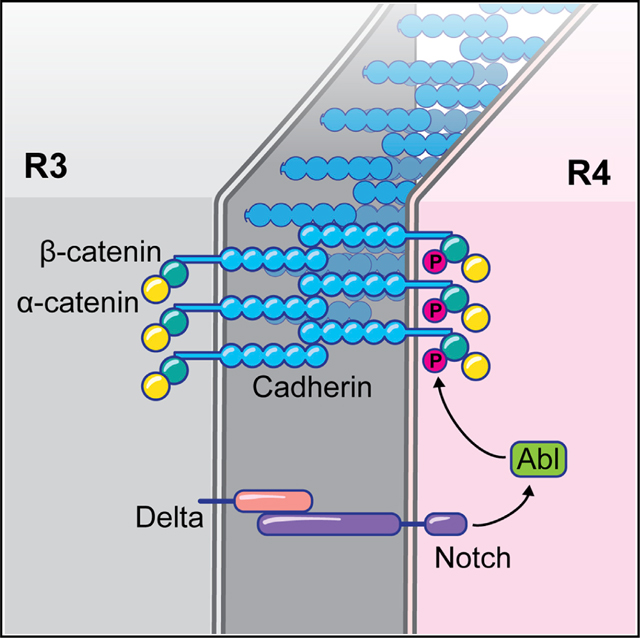
Notch is specifically and asymmetrically activated in R4, as a result of directional Fz/PCP signaling between R3/R4, and thus we next asked whether Notch signaling was required for apical dAbl localization in R4. Apical junctional localization of dAbl was largely lost in R4 upon Notch knockdown (via mδ0.5-Gal4) (Figures 5B, 5B′, and 5E), while the apical, junctional R4 marker Fmi remained intact. Furthermore, expression of a stable, truncated isoform of Notch (which behaves like an activated isoform; Figures S4G and S4G′) in the R3/R4 pair (mδ0.5-Gal4, UAS-N1−2155) caused apical dAbl localization in both cells of the pair (Figures 5C and 5C′). These data are consistent with the hypothesis that (activated) Notch is sufficient for apical dAbl localization, which in WT is restricted to R4. Hence, we hypothesize that apical R4-specific accumulation of dAbl requires Notch activation.
To further elucidate how Notch mediates apical dAbl localization and whether it requires ligand-mediated activation and Notch pathway signaling down to its transcriptional activity, we have analyzed several truncated versions of Notch (expressed under mδ0.5-Gal4) and investigated if and how these could affect apical recruitment of dAbl in R4. Strikingly, expression of a deletion isoform of Notch that cannot bind to its ligand Delta, termed Ndel10−12 (lacking EGF repeats 10–12 in its extracellular ligand binding domain, which are required for its interaction with Delta52,53), displayed a marked loss of apical dAbl localization in R4 (Figures 5D and 5E; while localization of the R4 apical marker Fmi appeared normal, as control). Notably, in this background, the preceding Notch signaling-mediated R4 fate specification was unaffected and the R4 fate-associated Notch transcriptional reporter, mδ0.5-lacZ, was active in R4 cells of rotating preclusters (Figures S4E and S4F), suggesting that Dl-Notch binding is required for apical dAbl localization, via a transcription-independent mechanism. As adult ommatidia of this background, mδ0.5>Ndel10−12, displayed consistently misorientations (Figure S4H), we tested whether this phenotype could be rescued by Abl co-overexpression in R3/R4 pairs. However, rotation defects in such eyes (mδ0.5>Ndel10−12 >Abl) were not significantly rescued (Figure S4H). As Notch signaling has multiple roles in eye development (see introduction for references), it is likely that its OR function cannot be rescued by simply increasing dAbl levels in the absence of normal Notch signaling.
dAbl associates with a junctional Notch complex
We next tested whether dAbl can physically associate with Notch in vivo. Strikingly, Notch co-immunoprecipitated with dAbl in extracts from third-instar larval eye discs (Figures 6A and 6B; also see Figures S5A–S5D). Consistently, Notch also co-immunoprecipitated with the adherens junction component Arm/β-catenin (a known Abl phosphorylation target48 in eye disc tissue, suggesting that Notch interacts with adherens junction components. Nevertheless, Notch and dAbl did not interact directly in a GST pull-down assay (Figure 6C), indicating that Notch and dAbl co-exist in membrane-associated junctional complexes during eye development, but that additional factors are required to link the two proteins. To further assess whether Notch can generally instruct localization of dAbl to junctional membrane regions, we turned to a salivary glands assay, where cells are large in size and cell compartments are relatively easy to analyze. In WT salivary glands, dAbl is detected in the cytoplasm and at junctional membranes, where it co-localizes with E-cad and Notch (Figure 6G). Importantly, junctional dAbl localization was increased in salivary gland cells upon Notch overexpression (Figures 6H–6J, S5E, and S5F), while total dAbl levels were unchanged in this background (Figures S5E and S5F), suggesting that Notch can generally potentiate dAbl localization to adherens junctions. Taken together, these results argue that the Notch receptor promotes recruitment of dAbl to apical junctional complexes.
Figure 6. Abl and Notch physically and genetically interact to regulate OR.
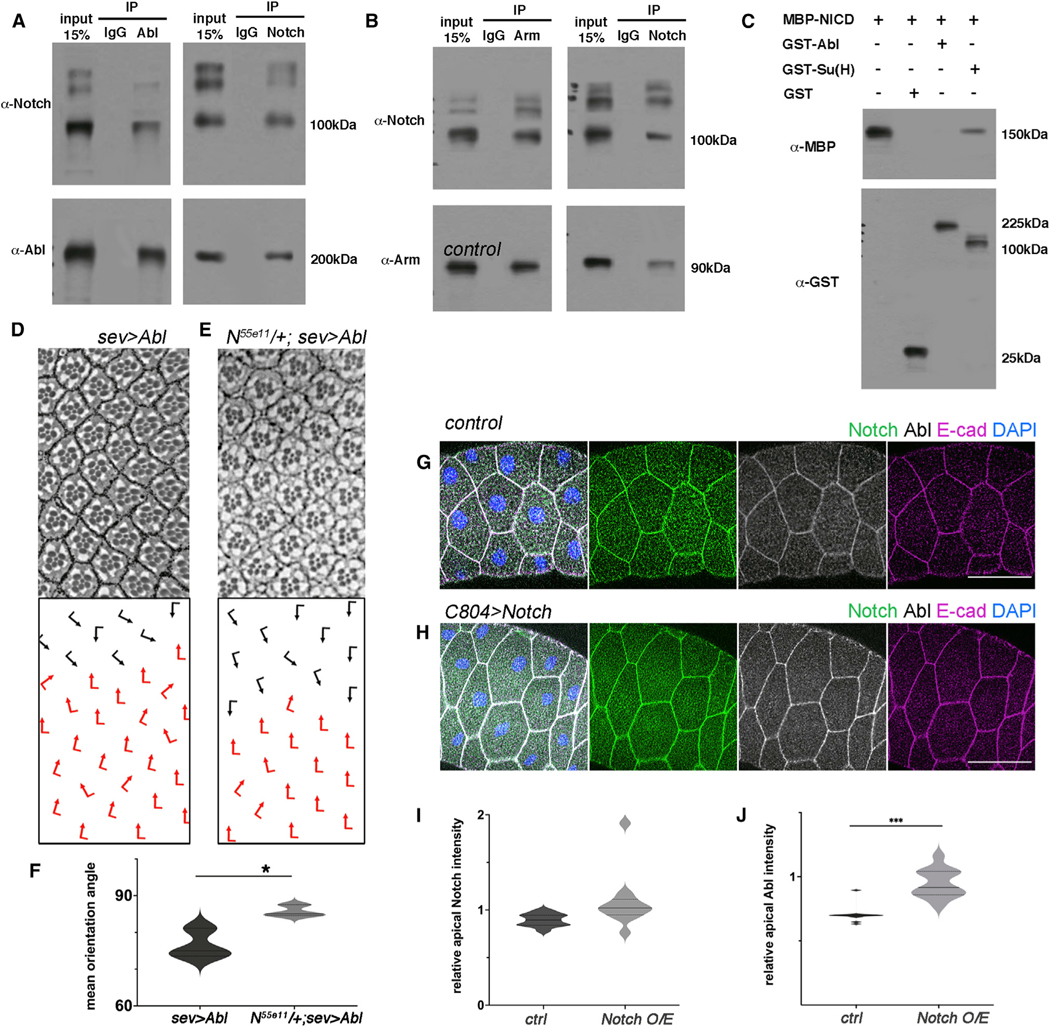
(A) WT third-instar larval eye disc lysates immunoprecipitated by α-IgG, α-Abl or α-Notch, blotted for Abl and Notch. Note that Abl and Notch immunoprecipitated each other.
(B) WT third-instar larval eye disc lysates immunoprecipitated by α-IgG, α-Arm or α-Notch, blotted for Arm and Notch, showing that Arm and Notch immunoprecipitated each other.
(C) MBP-NICD was pulled down by GST-Su(H) but not GST-Abl or GST.
(D and E) Adult eye sections of the genotypes indicated: (D) sev>Abl and (E) N55e11/+; sev>Abl, with ommatidial orientation schematics in lower panels.
(F) Quantification (violin plots) of average ommatidial orientation angles of sev>Abl and N55e11/+; sev>Abl genotypes (3 biological replicates each). Asterisk shows significance by t test (*p < 0.05).
(G and H) WT (G) and Notch overexpressing (H) salivary gland cells stained for Notch (green), Abl (white), E-cad (magenta), and DAPI (nuclei, blue). Note enrichment of Abl at apical junctions upon Notch overexpression. Scale bars, 50 μm.
(I and J) Quantification (violin plots) of average junctional Abl and Notch intensities (relative to junctional E-cad) (as detected in samples equivalent to those shown in (G) and (H), 12 biological replicates each). Note increase in junctional Abl levels upon Notch overexpression (J). ***p < 0.0005 by t test.
See also Figure S5.
To confirm that the association of Notch and dAbl is critical for the function of dAbl during OR, we tested for genetic interactions between Notch and dAbl specifically in the OR context. To this end, we used the OR-specific dAbl GOF genotype, sev>Abl, which displays ommatidial under-rotation (see above, Figure 2). Removing a gene copy of Notch (and thus reducing Notch protein levels) caused a marked suppression of the sev>Abl misrotation phenotype in adult eyes (Figures 6D–6F), which is consistent with the notion that dAbl acts downstream of the Notch receptor and requires Notch for its localization and/or function to regulate the OR process.
dAbl interacts with junctional components during OR
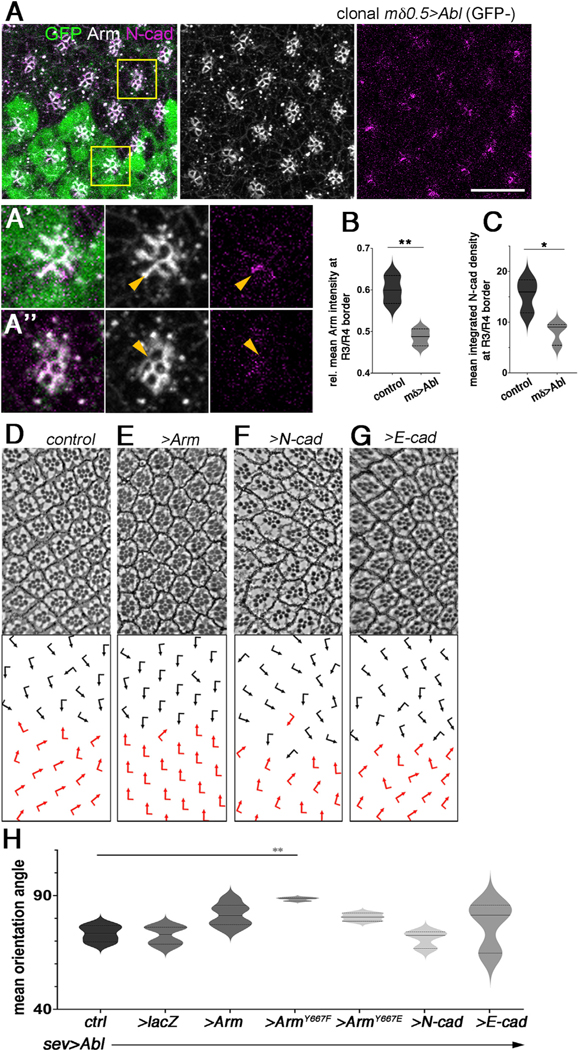
Abl kinases mediate cytoskeletal and junctional dynamics in various contexts.36–38 Adherens junction components have been shown to be critical for the OR process.32,33 As the Abl GOF genotypes (e.g., sev>Abl or mδ0.5>Abl, see above) caused (largely) clean OR defects, and dAbl is localized to adherens junction regions of R4, we asked whether dAbl affects junctional components in the OR context. Consistent with the notion that adherens junction composition and regulation could cause OR defects, we observed ommatidial misorientations, when E-cad or N-cad were knocked down with the same Gal4 drivers as we used for the Abl experiments (Figure S6). To test the above hypothesis and examine dAbl effects on junctional components, we used a clonal GOF strategy in mosaic third-instar larval eye discs. During OR, N-cadherin (N-cad) becomes enriched at the junctional border between the R3/R4 cells.32 Strikingly, clonal overexpression of dAbl in R3/R4 cells led to a reduction in N-cad, and associated Arm/β-catenin levels, at the junctional R3/R4 borders (Figures 7A–7C), suggesting that dAbl regulates the R3/R4 adherens junction organization (a consistent effect with N-cad is also observed in abl2 LOF eye disc clones; Figure S6C). In contrast, comparing the apical F-actin pattern of R3/R4 cells between dAbl GOF (mδ0.5>Abl clones) and control wild-type eye disc regions, we did not detect apparent differences, with F-actin being densely localized as an apical junctional ring within each R cell in both genetic backgrounds (Figure S6D).
Figure 7. Abl affects the levels of junctional components during the OR process.
(A) mδ0.5>Abl mosaic eye disc stained for GFP (green), Arm (gray), and N-cad (magenta). mδ0.5>Abl tissue is marked by the absence of GFP.
(A′ and A′′) Enlarged view of a WT (A′) and a mδ0.5>Abl cluster (A′′). Orange arrows mark the R3/R4 junction; note the reduction in N-cad and Arm signal at R3/R4 border in A′′. Scale bar, 10 μm.
(B and C) Quantification of average Arm (B) and N-cad intensity (C) at the R3/R4 borders, plotted for WT (gray) and mδ0.5>Abl (light gray) (3 biological replicates each). Asterisks denote significance by t test (*p < 0.005 and **p < 0.005).
(D–G) Adult eye sections with ommatidial orientation schematics in lower panels for the genotypes indicated: (D) sev>Abl; (E) sev>Abl, >Arm; (F) sev>Abl, >N-cad; and (G) sev>Abl, >E-cad.
(H) Violin plots showing quantification of average ommatidial orientation angles for the genotypes indicated (3–6 biological replicates each). Asterisks denote significance by one-way ANOVA (**p < 0.005), demonstrating significant rescue of the sev>Abl under-rotation phenotype by co-expression of ArmY667F. Other genotypes are not significantly different in phenotype, but note that Arm and ArmY667E show a rescue trend.
See also Figure S6.
To mechanistically address the effect of dAbl on adherens junctions during OR, we used again the rotation specific dAbl GOF scenario (sev>Abl). sev>Abl eyes displayed a strong trend for suppression of under-rotation features when dAbl was co-over-expressed with the junctional component Arm/β-catenin (sev>Abl, >Arm) (Figures 7D and 7E). Note that sev>Abl-associated under-rotation phenotype (with many clusters rotating significantly less than 90°; quantified in Figure 7H) was ameliorated with average rotation closer to 90° in the sev>Abl, >Arm background (Figures 7D, 7E and 7H). These data were consistent with the notion that dAbl regulates OR via effects on adherens junctions. As co-overexpression of Drosophila E-cad or N-cad did not suppress sev>Abl phenotype (Figures 7D–7H), it appeared that Arm/β-catenin might be a more direct target of Abl, which is consistent with dAbl phosphorylating Arm.48 We thus investigated the potential involvement of the previously identified Abl target site on Arm (ArmY667)48 during the rotation process. Weasked whether co-overexpression of a non-phosphorylatable isoform (ArmY667F), or a potentially phosphomimetic version (ArmY667E) of Arm for this site altered the sev>Abl phenotype. Strikingly, the non-phosphorylatable ArmY667F isoform, but not the phosphomimetic ArmY667E form, showed an almost complete rescue of the sev>Abl phenotype, rescuing it largely to WT (Figure 7H), indicating that Abl phosphorylation of Arm at Y667 is important for the Abl effect, with Abl GOF (as in sev>Abl) being counteracted by a phosphorylation resistant Arm isoform (see discussion).
DISCUSSION
We demonstrate that dAbl regulates cell motility during OR. Although loss of Abl function interferes with multiple aspects of photoreceptor development and morphogenesis, overexpression of dAbl in developing ommatidial clusters in eye discs affects specifically OR, suggesting that dAbl has a defined function in rotation. During OR, dAbl appears to have an inhibitory role, as ommatidial clusters with increased dAbl levels under-rotate, whereas dAbl mutant ommatidia tend to rotate faster.
The localization pattern of dAbl posterior to the MF provides further insight about its role in OR. dAbl becomes apically localized in photoreceptors R8, R2/R5, and R4, following a steady phase of rotation, at the time when clusters slow down and refine their motility until the completion of the 90° angle. Prominent Abl localization within the apical plane of specific photoreceptors suggests that Abl is likely to have a local function in the apical junctional domain. Under-rotation features observed upon dAbl overexpression are consistent with the notion that dAbl becomes apically localized in specific R cells, toward the later stages of OR, to slow down the process. Interestingly, there is a differential localization of dAbl between R3 and R4 in the apical junctional domain. Considering the role of the R3/R4 pair and associated signaling pathways in OR, it is tempting to speculate that this differential dAbl localization is comparable to the requirement of the Nmo kinase within R3/R4, with Nmo providing a directional impulse to rotation in R433 and dAbl regulating its slowing down. Our data argue that dAbl activity within R3/R4 pairs is indeed important for fine-tuning rotation. Knockdown and overexpression of dAbl in R3/R4 pairs lead to over-rotation and under-rotation, respectively, during the active rotation process in eye discs, suggesting that Abl activity negatively regulates rotation. Specifically, knockdown of Abl in R3/R4 leads to over-rotation of ommatidia, which, taken together with the WT localization of Abl being restricted to the R4 apical junctional domain, suggests that Abl is required in R4 within the apical region to slow down rotation (Figures 4 and S3). In the case of under-rotation caused by mδ0.5>Abl overexpression, we detect apical dAbl in both cells of the R3/R4 pair and, importantly, temporally earlier in this background compared with WT, suggesting that early dAbl expression in both cells causes an under-rotation phenotype by interfering with rotation (Figures 4 and S3). Taken together, these observations are consistent with the hypothesis that the timing and specificity of apical localization of dAbl in R4 is critical for its normal function in OR.
Regulation of OR by Abl appears not to require its potential effect on core PCP signaling molecules.50 Notably, Abl overexpression does not appear to affect ommatidial chirality and the localization of PCP factors, as Fmi expression and localization remain intact. Furthermore, Abl overexpression causes a specific and severe under-rotation defect, unlikely resulting from deregulation of core PCP factors, which are commonly associated with random ommatidial chirality and rotation. It is most likely that Abl overexpression, under sev- or mδ0.5-Gal4 drivers, is temporally too late to interfere with Fz/PCP signaling-mediated R3/R4 cell fate decisions, and thus specifically affects OR.
Notch recruits dAbl to apical junctional membrane complexes in R4
Fz/PCP signaling appears dispensable for the R4-specific apical dAbl localization, as the pattern is maintained in core PCP mutant ommatidia. Yet dAbl does synergize with Fmi, when co-overexpressed in the R3/R4 pair, in a rotation specific manner. This OR-associated functional interaction of Abl with membrane-associated core PCP factors, along with the localization pattern of Abl in the apical domain further suggests that dAbl activity is important in R4 in the apical junctional domain. Our results identify Notch and Notch signaling in R4 as critical for apical dAbl localization. Notch over-activation within the R3/R4 pair (via expression of stable isoforms of the receptor) induces apical dAbl localization in both cells of the pair. In contrast, expression in R3/R4 pairs of a version of Notch deficient in Delta binding, the key Notch ligand in the eye, (Ndel10−12), and thus interference with ligand induced Notch activation, leads to a loss of apical dAbl in R4. Similarly, reduction of Notch levels in R3/R4 cells (via RNAi-mediated knockdown) also causes a marked decrease in apical dAbl levels in R4. As Notch-dependent transcription is still active in these backgrounds, the combination of these results suggests that Notch-mediated dAbl apical localization is rather direct, and not via a secondary mechanism through transcriptional regulation. This conclusion is corroborated by the co-immunoprecipitation experiments.
Several experimental lines support the hypothesis that the Notch receptor physically recruits dAbl to the membrane. In salivary glands, Notch overexpression augments junctional dAbl localization, leaving total dAbl levels unaffected. dAbl co-immunoprecipitates with Notch in third-instar larval eye disc extracts, supporting a membrane-associated Notch-Abl interaction in vivo, independent of nuclear Notch signaling activity. The sev>Abl GOF rotation phenotype is markedly suppressed upon removal of one copy of Notch, further supporting the idea that a functional N-Abl signaling module in the apical domain of R4 regulates OR.
dAbl localization appears to be within the apical region and not restricted to the apical membrane ring. There may be multiple reasons for this. As the Notch receptor is cleaved upon ligand binding and its intracellular domain is released to the cytoplasm, distribution of Abl molecules in the apical region may be broader than restricted to the transmembrane fraction of Notch. Abl-Notch interactions likely last after Notch cleavage, considering efficient Abl co-immunoprecipitation with the Notch ICD (Figure 6A). Abl can also interact with actomyosin cytoskeletal elements, which are apically enriched in R cells.49 As the apical diameter of R cells in this region is less than 2 μm, the imaging resolution does not separate the membrane Abl signal from the juxta-membrane cytoplasmic signal. Notably, in Notch overexpression contexts, Abl signal is often detected as a ring at the apical membrane (Figure 5C), likely attributable to the presence of more uncleaved membrane-associated Notch. Furthermore, we can detect and quantify Abl at junctions in salivary glands, and thus document the increased levels of membrane-associated Abl upon higher Notch levels. All these data are consistent with the notion that Abl is specifically recruited to the apical junctional membrane domain by Notch.
Abl kinase and non-canonical Notch signaling
In Drosophila, dAbl has been suggested to act downstream of Notch during axonal pathfinding in embryos. Compelling evidence suggests that a non-canonical Notch signaling branch, which does not entail nuclear Notch activity, instructs axonal pathfinding52 and axon-guidance-specific genetic interactions between dAbl and Notch argue that a non-canonical Notch signaling pathway via dAbl may be at work in this context.54 Our results are in accordance with these observations and provide further evidence for a non-canonical Notch-Abl signaling module during morphogenesis. Recently, a non-canonical Notch pathway has been reported in the regulation of adherens junction organization during human vascular barrier formation,55 with the transmembrane domain of Notch forming complexes with the tyrosine phosphatase LAR, vascular endothelial cadherin, and Rac1GEF Trio to confer barrier function in human engineered microvessels. The Notch transmembrane domain requires the cleavage of the Notch extracellular and intracellular domains in this context.55 Our data during OR indicate that apical dAbl recruitment in R4 similarly requires Notch activation by Delta. Whether the transmembrane domain of Notch is an essential component of dAbl recruitment and/or regulation remains to be confirmed. There is a growing body of evidence that Notch uses alternative downstream signaling events to regulate cellular morphogenesis and organization, besides canonical transcriptional target gene regulation.52,54–56
Abl kinase regulation of adherens junctions
Abl appears to affect junctional N-cad and Arm levels in the R3/R4 pair. N-cad mutants show OR defects.32 Although the mechanism of N-cad involvement remains unclear, N-cad and/or Arm at the R3/R4 boundary could mediate the communication between these cells to determine relative force generation or other directional behavior to give the rotation direction or impulse/force. Such mechanisms have been suggested in border cell migration through E-cad.57 N-cad mutant ommatidia appear to over-rotate32 unlike Abl-overexpressing ommatidia (in which N-cad is downregulated at the R3/R4 border). Although this seems like a discrepancy, Abl overexpression by mδ0.5-Gal4 (unlike N-cad mutations) is spatially and temporally restricted to R3/R4s, possibly accounting for the differences observed in these backgrounds. Furthermore, Abl likely affects OR via regulating several downstream effectors, including cytoskeletal regulators, in parallel to N-cad and thus has a more complex impact on OR than N-cad alone.
The observation that the non-phosphorylatable isoform of Arm/β-catenin, ArmY667F, rescues the Abl GOF defects, supports the idea that Arm is a key and direct target of dAbl in the OR context. dAbl is involved in the regulation of multi-cellular reorganization in the context of Drosophila germband elongation through the phosphorylation of Arm/β-catenin on tyrosine 667 (Y667), by which it controls adherens junction turnover to promote convergent extension cell movements.48 Our data argue that dAbl may similarly be involved in regulating Arm/β-catenin dynamics through the same residue during the OR process. The under-rotation phenotype associated with the dAbl GOF (sev>Abl) showed a trend toward rescue by co-(over)expression of Arm-WT and ArmY667E, which is likely due to the fact that exogenously overexpressed Arm isoforms compete with endogenous Arm for dAbl binding.Further experiments will be needed to test these hypotheses.
Abl function in OR relative to other regulators of the process
The requirement of Abl in R4 for accurate rotation suggests that it acts antagonistically to Nemo which is enriched at junctions in R4 early via core PCP factors and its function is to promote rotation.33 There isa temporal sequence of apicalplane enrichment of factors in R4 with Nemo first to initiate rotation,33 and Abl a few hours later to slow it down (this work). It was originally proposed that OR is a two-step process, with an initial fast rotation to 45° and a subsequent slower step to achieve the full 90°. However, this idea goes back to the identification of the original allele of nemo, which is a hypomorph, and only affected the rotation process partially.26–34 Recent live imaging studies documenting OR dynamics have established that rotation is continuous with comparable speed throughout.58 Similarly, there is growing evidence that for rotation to occur correctly, adherens junctions need to be dynamically regulated at the interface between all photoreceptors and the non-rotating inter-ommatidial cells, and possibly between individual inter-ommatidial cells.58 It is thus very likely that Abl overexpression with mδ0.5 and sev drivers interferes with rotation by affecting adherens junction regulation and dynamics in all or multiple R cells, like Nemo.33,58
Localization of Abl within the apical plane of R4, as well as R2/R5, is detected at late stages of rotation (from rows 7 and 8 onward), when rotation needs to be slowed down and stopped at 90, indicating that Abl has a role at the late phases of the process, to terminate rotation. There are additional cues that appear to signal within ommatidia to stop rotation. For example, EGFR signaling via Argos (the original allele of argos being “roulette/rlt”) certainly feeds into slowing down rotation, as without the inhibitory EGFR ligand, argosrlt mutant clusters rotate beyond 90 (as the name “roulette” indicates). Similarly, Scabrous (Sca), a secreted fibrinogen-like factor,59,60 has been suggested to regulate the properties of the extracellular matrix to create a barrier to rotation.28 Although the mechanism of Sca function remains unknown, a direct involvement of the ECM in rotation has been reported with a specific link of Integrin signaling and ECM in the OR process.61 A model is thus emerging that suggests the degree of rotation depends on an interplay between multiple signaling pathways, including Notch-Abl signaling, and their regulatory input to cell adhesion and cytoskeletal elements.
Multiple Notch regulated inputs into the OR process
Notch signaling in R3/R4 pairs is critical to coordinate OR via its feeding into the transcriptional regulation of argos,35 with Notch signaling directly promoting the transcription of argos, the inhibitory ligand to EGFR, required to fine-tune EGFR signaling activity during OR.31 Here, we show that Notch signaling regulates OR via apical junctional recruitment of dAbl in R4, linking Notch activity to non-canonical, Abl-mediated Notch signaling and associated local cellular processes, with Abl modulating cadherin/β-catenin-based junctional complexes. Involvement of Notch signaling in cellular morphogenesis has been suggested in various contexts, including Drosophila oogenesis and neuronal pathfinding, zebra-fish sensory organ development and human vascular barrier formation among others.52,54–56,62–65 Besides the reported Notch signaling-mediated transcriptional inputs into adhesion and cytoskeletal dynamics,63,65,66 a direct link from the Notch receptor to cell adhesion has been revealed.55 Our work also suggests a direct input from Notch signaling to cell adhesion dynamics.
Many regulators of OR show conservation across developmental processes in vertebrates. The role of Notch signaling in OR suggests a potential involvement for Notch in PCP-mediated morphogenetic events in vertebrates, which has not been reported thus far. Similarly, Abl kinase may have a role in such processes in its interaction with PCP and Notch signaling pathways. Strikingly, the mouse abl−/− arg−/− double mutants exhibit defects in neurulation and delays in neural tube closure,67 a process generally requiring PCP-regulated features.1,5,6,68
Limitations of the study
The work described here provides insight into Notch-Abl signaling in a tissue remodeling, cell motility process. Although all data are consistent with the proposed model (see above), this model is generated by inference from analyses of static fixed tissue samples, genetics, and biochemical studies. As it involves a cell motility process, it would be desirable to analyze the respective mutant genotypes via live imaging in vivo, including studies applying FRAP and other technologies. This would allow a more complete understanding of how Abl affects junctional dynamics during OR. Future studies will be needed to provide insight into the mechanistic details of how Notch and Abl cooperate in regulating junctional complexes and their dynamics during OR and other morphogenetic developmental and disease processes.
STAR★METHODS
RESOURCE AVAILABILITY
Lead contact
Further information and requests for resources and reagents should be directed to and will be fulfilled by the lead contact, Marek Mlodzik (marek.mlodzik@mssm.edu).
Materials availability
This study did not generate new, unique reagents.
Data and code availability
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Drosophila melanogaster
All genotypes were selected from males unless otherwise stated.
Mδ0.5>NRNAi BL31383 (mδ0.5-Gal4/+; UAS-NRNAi BL31383/+) were obtained at 25°C.
mδ0.5>N1−2155 (mδ0.5-Gal4/+; UAS-N1−2155/+) were obtained at 25°C.
mδ0.5>Ndel10−12 (mδ0.5-Gal4/UAS-Ndel10−12) eye discs were obtained at 29°C.
mδ0.5>Ndel10−12 (mδ0.5-Gal4 UAS-Ndel10−12/+) adult eyes were obtained at 25°C.
mδ0.5>Ndel10−12 (mδ0.5-Gal4 UAS-Ndel10−12/+;UAS-Abl/+) adult eyes were obtained at 25°C.
mδ0.5>AblRNAi#2 BL35327 (mδ0.5-Gal4/+; UAS-AblRNAi#2 BL35327/+) were obtained at 29°C.
mδ0.5>AblRNAi#1 VDRC110186 (mδ0.5-Gal4/+; UAS-AblRNAi#1 VDRC110186/+) were obtained at 29°C.
mδ0.5>Abl (mδ0.5-Gal4, UAS-Abl/+) were obtained at 29°C.
sev>Abl (sev-Gal4/+; UAS-Abl/+) were obtained at 18°C.
Control eye disc stainings were done in mδ0.5-Gal4 FRT40/+ backgrounds.
Genetic interactions were tested at 18°C between sev-Gal4/+; UAS-Abl/+ and the heterozygosity of the respective transgenic flies.
For N55e11/+ interaction, females were analyzed.
abl2 mutant clones were generated with the following genotype:
eyFLP/+; abl2 FRT80/ubiGFP FRT80
pkpk-sple13stbm6 double mutants were generated with the following genotype:
eyFLP/+; pkpk-sple13stbm6 FRT42/ubiGFP FRT42
mδ0.5>UAS-Abl clones were obtained at 29°C by employing FLP/FRT mediated mitotic recombination with the following genotypes:
eyFLP/+; mδ0.5-Gal4 FRT40/ubiGFP FRT40; UAS-Abl/+
METHOD DETAILS
Immunohistochemistry and histology
Adult eye sectioning was performed as described75: Briefly, the eyes were transferred into 2% osmium tetroxide solution in phosphate buffer (NaH2PO4, 0.5M) and incubated on ice for up to an hour followed by incubation on ice in 4% osmium tetroxide solution for 1–6 hours. The eyes were then dehydrated by incubating for 10 minutes in 30%, 50%, 70%, 90%, 100% ethanol solutions consecutively. They were washed in propylene oxide at room temperature two times for 10 minutes and incubated in 1:1 mix of propylene oxide and Durcupan resin overnight. Next day, the solution was replaced with pure resin, the eyes were incubated for 2–4 hours at room temperature and embedded in molds. The molds were baked at 70°C overnight and sectioning was performed.
Third larval instar eye discs were dissected in ice-cold PBS and fixed in PBT (PBS+0.1% Triton-X)-4% formaldehyde for 12 minutes at room temperature. For immunohistochemistry, following primary antibodies were used: rabbit anti-Abl (1:125, Mlodzik Lab47), rat anti-DE-cad (1:20, DSHB), mouse anti-Fmi (1:10, DSHB), rat anti-DN-cad (1:20, DSHB), mouse anti-Arm (1:20, DSHB), chicken anti-GFP (1:1000, Aves Labs) and rhodamine phalloidin (1:1000, Invitrogen). Secondary antibodies were obtained from Jackson Laboratories. Images were acquired by using Leica SP5 DMI microscope.
Biochemical interaction assays
For in vitro GST pull downs, the recombinant proteins GST-Notch ICD, GST-Su(H) and MBP-Abl were expressed in bacteria and purified. For the amplification of Notch ICD, Abl and Su(H) fragments, the following primers were used:
Notch forward primer: 5′ CATGCCCGGGACAAAGAAAGCGGGCACATG
Notch reverse primer: 5′ CTAGGCGGCCGCTCAAATGTAGATGGCCTC
Abl forward primer: 5′ CATGTCTAGAGGGGCTCAGCAGGGCAAGG
Abl reverse primer: 5′ CATGGTCGACTTACCTGTTAAGCGCATTGG
Su(H) forward primer: 5′ ATGAGAATTCATGTGTGATTAGTCGTGAATC
Su(H) reverse primer: 5′ ATGAGTCGACTCATTTAGATCTTTGGAAATTCAT
Amplified fragments were cloned into pGEX-4T1 vector for GST tagging and pMAL-c2X vector for MBD tagging. Bacteria lysates were prepared as described.76 Glutathion Sepharose 4B beads (GE Healthcare) with GST or with GST-fusion proteins were incubated with lysates containing MBP-Abl O/N at 4°C. After that, the samples were washed 3 times with pull-down buffer (20 mM Tris pH 7.6, 150 mM NaCl, 0.5 mM EDTA, 10% glycerol, 0.1% Triton X 100, 1mM DTT, and protease inhibitor cocktail). Pull-down samples were resuspended in SDS sample buffer, boiled for 5 min and the proteins were separated by SDS PAGE. For Western blot, the nitrocellulose membrane was incubated with mouse-anti-GST and rabbit-anti-MBP antibody. Proteins were detected by enhanced chemiluminescence (Millipore).
For in vivo co-immunoprecipitations eye imaginal discs were dissected from third instar larvae. Lysates from 30 wt eye imaginal discs (1 mg of total protein) was precleared by incubating with protein A-sepharose beads (Thermo Scientific) for 1h at 4°C followed by centrifugation. A-sepharose beads were immunoprecipitated with anti-Abl47 or anti-Notch antibody (https://dshb.biology.uiowa.edu/C17-9C6) and the lysates at 4C O/N. Immunoprecipitates were captured by protein A-sepharose at 4°C in IP buffer (20 mM 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) pH 7.5, 100 mM NaCl, 0.05% Triton X100, 1 mM EDTA, 5 mM DTT, 10 mM NaVO4, 10% glycerol and protease inhibitor cocktail). After several washes with IP buffer, immunoprecipitates were resuspended in SDS sample buffer; beads were boiled for 5 min, and proteins were resolved on SDS-PAGE. For western blotting, proteins were transferred onto nitrocellulose membrane, blocked in 5% skim milk (Labscientific) and incubated with primary rabbit anti-Abl or mouse anti-NotchICD, and secondary anti-rabbit or anti-mouse antibody. Protein was detected by enhanced chemiluminescence (Millipore).
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantitative analysis of adult eye sections
The orientation of each ommatidium was marked based on the trapezoidal organization of the R-cells (see Figure 1A). A linear equator has been drawn along the boundary where two chiral forms meet. Clockwise and counter-clockwise angles from the equator to each ommatidia were measured for the black and red chiral forms respectively (see Figures 1A and 1C). Measurements were done by using ImageJ (National Institute of Health). The absolute values of measured angles were averaged and plotted for each animal (n>=3) using PRISM. For statistical analyses, a two-tailed t-test or one-way ANOVA was performed on averaged angles from each genotype and compared among the different genotypes.
Quantitative analysis of eye discs
The orientation of each ommatidium was marked perpendicular to the plane of R2/R5 cells (See Figures 1B, 2C and 2D). A linear equator was drawn perpendicular to the MF at the dorsoventral midline. Clockwise and counter-clockwise angles from the equator to each ommatidia were measured for the dorsal and ventral halves respectively. To avoid a potential bias due to the developmental delay in rotation from equator to the poles, the measurements were limited to the first 8 ommatidia from the equator for each row. Measurements were done by using ImageJ (National Institute of Health). The absolute values of measured angles from each row were averaged and plotted for each animal (n>=5) using PRISM. For statistical analyses, a two-tailed t-test or one-way ANOVA was performed on averaged angles for each row and compared between the respective genotypes.
For Abl quantifications in control and Notch-perturbed backgrounds, apical confocal stacks were maximum projected and individual cell intensities were measured in R4 and R2 cells between rows 9–11 by using ImageJ. Intensities in R4 cells were normalized to their R2 neighbors within each ommatidia. Measurements were averaged for each animal (n>=3) and plotted in PRISM. For statistical analyses, one-way ANOVA was performed on averaged intensities to compare between the respective genotypes.
For N-cad quantifications in mosaic eye discs, apical confocal stacks were maximum projected and integrated N-cad intensities were measured in ImageJ. Within GFP+ and GFP− tissue, N-cad integrated intensity within the R3/R4 boundary was measured for each ommatidial cluster between rows 5–11. Measurements were averaged for each animal (n>=3) and plotted in PRISM. For statistical analyses, t-test was performed on averaged integrated densities to compare genotypes.
For Arm/β-catenin quantifications in mδ0.5>Abl mosaic eye discs, apical confocal stacks were maximum projected and Arm intensities between R3/R4 borders were measured in ImageJ. Within GFP+ and GFP− tissue, Arm intensity at multiple points along the R3/R4 boundary was measured for each ommatidia between rows 5–11 and the mean intensity for each ommatidia was measured in ImageJ. Measurements were averaged for each fly (n>=3) and plotted in PRISM. For statistical analyses, t-test was performed on averaged mean intensities to compare genotypes.
Quantitative analysis of salivary glands
Salivary gland staining images were processed using ImageJ. For apical Notch and Abl quantifications, apical confocal stacks were maximum projected, apical junctional intensities were measured at multiple points and normalized to the corresponding DE-cad intensities followed by averaging for each salivary gland. Mean measurements were plotted for 12 salivary glands. For statistical analyses, a two tailed t-test was performed on normalized mean intensity measurements to compare genotypes.
Western blot images were processed using ImageJ. For relative protein expression quantifications of Notch and Abl, scans of developed films were converted to 8-bit images to perform uncalibrated optical density. Each band was individually selected and circumscribed with the rectangular ROI selection, followed by quantification of peak area of obtained histograms. Data were acquired with arbitrary area values. The intensity of protein bands was normalized to expression of Tubulin. For statistical analyses, a two tailed t-test was performed on normalized intensity measurements.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
|
| ||
| Antibodies | ||
|
| ||
| Mouse anti-Arm | DSHB | N2 7A1;RRID: AB_528089 |
| Rat anti-DE cad | DSHB | DCAD2;RRID: AB_528120 |
| Rabbit anti-MBP | Sigma-Aldrich | Cat# SAB2104171; RRID: AB_10668141 |
| Mouse anti-GST | Invitrogen | Cat# MA4-004-HRP; RRID: AB_2537634 |
| Chicken anti-GFP | Aves Labs | Cat# GFP-1020; RRID: AB_10000240 |
| Mouse anti-γ-tubulin | Sigma-Aldrich | Cat# T6557; RRID: AB_477584 |
| Mouse anti-GST | Invitrogen | Cat# MA4-004-HRP; RRID: AB_2537634 |
| Rat anti-DNcad | DSHB | MNCD2;RRID: AB_528119 |
| Rabbit anti-Abl | Mlodzik Lab | Song et al.47 |
| Mouse anti-Fmi | DSHB | Flamingo #74;RRID: AB_2619583 |
| Mouse anti-NICD | DSHB | C17.9C6;RRID: AB_528410 |
| Anti Mouse Alexa Fluor 647 | Jackson ImmunoResearch | Cat# 715-605-151;RRID: AB_2340863 |
| Anti Chicken FITC | Jackson ImmunoResearch | Cat# 703-095-155; RRID: AB_2340356 |
| Anti Rat TRITC | Jackson ImmunoResearch | Cat# 712-025-153; RRID: AB_2340636 |
| Anti Rabbit Alexa Fluor 568 | Invitrogen | Cat# A11036;RRID: AB_10563566 |
| Anti Rat Alexa Fluor 647 | Jackson ImmunoResearch | Cat# 712-605-150; RRID: AB_2340693 |
| Anti Mouse Alexa Fluor 488 | Invitrogen | Cat# A21202;RRID: AB_141607 |
| Anti Rabbit HRP | Jackson ImmunoResearch | Cat# 711-035-152; RRID: AB_10015282 |
| Anti Rat HRP | Jackson ImmunoResearch | Cat# 712-035-153; RRID: AB_2340639 |
| Anti Mouse HRP | Jackson ImmunoResearch | Cat# 715-035-150; RRID: AB_2340770 |
|
| ||
| Experimental models: Organisms/strains | ||
|
| ||
| D.melanogaster :UAS-Abl | Ed Giniger | Le Gall et al.52 |
| D.melanogaster :UAS-Arm | Jennifer Zallen | Tamada et al.48 |
| D.melanogaster :UAS-ArmY667F | Jennifer Zallen | Tamada et al.48 |
| D.melanogaster :UAS-ArmY667E | Jennifer Zallen | Tamada et al.48 |
| D.melanogaster :UAS-Notch RNAi | BDCS | BDSC: 31383;FlyBase: FBti0130800 |
| D.melanogaster :UAS-Abl RNAi#1 | VDRC | VDRC: 110186;FlyBase: FBst0481767 |
| D.melanogaster :UAS-Abl RNAi#2 | BDSC | BDSC: 35327;FlyBase: FBti0144320 |
| D.melanogaster :UAS-DECad RNAi | BDSC | BDSC: 32904;FlyBase: FBti0140407 |
| D.melanogaster :UAS-DECad RNAi | BDSC | BDSC: 27689;FlyBase: FBti0128871 |
| D.melanogaster :UAS-DNCad RNAi | VDRC | VDRC: 1092;FlyBase: FBst0450184 |
| D.melanogaster :UAS-DNCad RNAi | BDSC | BDSC: 41982;FlyBase: FBti0150007 |
| D.melanogaster :UAS-DE-Cad | Tadashi Uemura | Lee et al.69 |
| D.melanogaster :UAS-DN- Cad | Tadashi Uemura | Oda et al.70 |
| D.melanogaster :UAS-lacZ | BDSC | BDSC: 1776;FlyBase: FBtp0000355 |
| D.melanogaster : mδ0.5-Gal4 FRT40/SM3:TM6b | Mlodzik Lab | Mirkovic et al.33 |
| D.melanogaster : sev-Gal4/TM3 | Mlodzik Lab | Boutros et al.71 |
| D.melanogaster : pkpk-sple13stbm6 FRT42/CyO | Mlodzik Lab | Jenny et al.72 |
| D.melanogaster : abl2 FRT80/TM6b | BDSC | BDSC: 8565;FlyBase: FBst0008565 |
| D.melanogaster : eyFLP; ubiGFP FRT80 | Mlodzik Lab | Mirkovic et al.33 |
| D.melanogaster : eyFLP; ubiGFP FRT42 | Mlodzik Lab | Jenny et al.72 |
| D.melanogaster : eyFLP; ubiGFP FRT40 | Mlodzik Lab | Jenny et al.72 |
| D.melanogaster : UAS-Fmi | Tadashi Uemura | Usui et al.73 |
| D.melanogaster : N 55e11 /FM6b | BDSC | BDSC: 28813;FlyBase: FBst0028813 |
| D.melanogaster : C805-Gal4 | BDSC | BDSC:6978;FlyBase: FBaI0141532 |
|
| ||
| Oligonucleotides | ||
|
| ||
| Notch forward primer: 5′ CATGCCCGGGACAAA GAAAGCGGGCACATG |
Eurofins | N/A |
| Notch reverse primer: 5′ CTAGGCGGCCGCTC AAATGTAGATGGCCTC |
Eurofins | N/A |
| Abl forward primer: 5′ CATGTCTAGAGGGG CTCAGCAGGGCAAGG |
Eurofins | N/A |
| Abl reverse primer: 5′ CATGGTCGACTTACC TGTTAAGCGCATTGG |
Eurofins | N/A |
| Su(H) forward primer: 5′ ATGAGAATTCATGTG TGATTAGTCGTGAATC |
Eurofins | N/A |
| Su(H) reverse primer: 5′ ATGAGTCGACTCATT TAGATCTTTGGAAATTCAT |
Eurofins | N/A |
|
| ||
| Recombinant DNA | ||
|
| ||
| pGEX-4T-1 | Milipore Sigma | Cat# GE28-9545-49 |
| pMAL-c2X | Walker et al.74 | Addgene;Cat# 75286 |
Highlights.
dAbl negatively regulates ommatidial rotation through β-catenin at adherens junctions
dAbl localizes apically in R4, but not R3, during ommatidial rotation
Apical localization and activity of dAbl requires Notch signaling
dAbl interacts genetically and physically with Notch during ommatidial rotation
ACKNOWLEDGMENTS
We thank the Bloomington Drosophila Research Center and Jennifer Zallen for fly strains and reagents. We are grateful to all Mlodzik lab members, past and present, for helpful input and discussions and Robert Krauss, Cathie Pfleger, Timothy Blenkinsop, and Jennifer Zallen for helpful comments and suggestions on the manuscript. Confocal laser scanning microscopy was performed at the ISMMS-Microscopy Core Facility supported by the Tisch Cancer Institute grant P30 CA196521 from the National Cancer Institute (NCI). This work was supported by NIH/National Eye Institute (NEI) grant R01 EY13256 to M.M.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental information can be found online at https://doi.org/10.1016/j.celrep.2022.111788.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- 1.Humphries AC, and Mlodzik M. (2018). From instruction to output: Wnt/PCP signaling in development and cancer. Curr. Opin. Cell Biol 51, 110–116. 10.1016/j.ceb.2017.12.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goodrich LV, and Strutt D. (2011). Principles of planar polarity in animaldevelopment. Development 138, 1877–1892. 10.1242/dev.054080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Peng Y, and Axelrod JD (2012). Asymmetric protein localization inplanar cell polarity: mechanisms, puzzles, and challenges. Curr. Top. Dev. Biol 101, 33–53. 10.1016/B978-0-12-394592-1.00002-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Adler PN (2012). The frizzled/stan pathway and planar cell polarity in theDrosophila wing. Curr. Top. Dev. Biol 101, 1–31. 10.1016/B978-0-12-394592-1.00001-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Davey CF, and Moens CB (2017). Planar cell polarity in moving cells: think globally, act locally. Development 144, 187–200. 10.1242/dev.122804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Butler MT, and Wallingford JB (2017). Planar cell polarity in development and disease. Nat. Rev. Mol. Cell Biol 18, 375–388. 10.1038/nrm.2017.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu J, and Mlodzik M. (2009). A quest for the mechanism regulatingglobal planar cell polarity of tissues. Trends Cell Biol. 19, 295–305. 10.1016/j.tcb.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jenny A. (2010). Planar cell polarity signaling in the Drosophila eye. Curr.Top. Dev. Biol 93, 189–227. 10.1016/B978-0-12-385044-7.00007-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Devenport D. (2016). Tissue morphodynamics: translating planar polaritycues into polarized cell behaviors. Semin. Cell Dev. Biol 55, 99–110. 10.1016/j.semcdb.2016.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mlodzik M. (1999). Planar polarity in the Drosophila eye: a multifacetedview of signaling specificity and cross-talk. EMBO J. 18, 6873–6879. 10.1093/emboj/18.24.6873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Strutt H, and Strutt D. (1999). Polarity determination in the Drosophilaeye. Curr. Opin. Genet. Dev 9, 442–446. 10.1016/S0959-437X(99)80067-7. [DOI] [PubMed] [Google Scholar]
- 12.Tomlinson A, and Ready DF (1987). Neuronal differentiation in Drosophila ommatidium. Dev. Biol 120, 366–376. 10.1016/0012-1606(87)90239-9. [DOI] [PubMed] [Google Scholar]
- 13.Wolff T, and Ready DF (1991). The beginning of pattern formation in the Drosophila compound eye: the morphogenetic furrow and the second mitotic wave. Development 113, 841–850. [DOI] [PubMed] [Google Scholar]
- 14.Cagan RL, and Ready DF (1989). The emergence of order in theDrosophila pupal retina. Dev. Biol 136, 346–362. 10.1016/0012-1606(89)90261-3. [DOI] [PubMed] [Google Scholar]
- 15.Roignant JY, and Treisman JE (2009). Pattern formation in theDrosophila eye disc. Int. J. Dev. Biol 53, 795–804. 10.1387/ijdb.072483jr. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber U, Pataki C, Mihaly J, and Mlodzik M. (2008). Combinatorial signaling by the Frizzled/PCP and Egfr pathways during planar cell polarity establishment in the Drosophila eye. Dev. Biol 316, 110–123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weber U, Paricio N, and Mlodzik M. (2000). Jun mediates Frizzled-induced R3/R4 cell fate distinction and planar polarity determination in the Drosophila eye. Development 127, 3619–3629. [DOI] [PubMed] [Google Scholar]
- 18.Fanto M, and Mlodzik M. (1999). Asymmetric Notch activation specifies photoreceptors R3 and R4 and planar polarity in the Drosophila eye. Nature 397, 523–526. 10.1038/17389. [DOI] [PubMed] [Google Scholar]
- 19.Cooper MT, and Bray SJ (1999). Frizzled regulation of Notch signalling polarizes cell fate in the Drosophila eye. Nature 397, 526–530. 10.1038/17395. [DOI] [PubMed] [Google Scholar]
- 20.Tomlinson A, and Struhl G. (1999). Decoding vectorial information from a gradient: sequential roles of the receptors Frizzled and Notch in establishing planar polarity in the Drosophila eye. Development 126, 5725–5738. [DOI] [PubMed] [Google Scholar]
- 21.Blair SS (1999). Eye development: Notch lends a handedness. Curr. Biol 9, R356–R360. 10.1016/s0960-9822(99)80226-7. [DOI] [PubMed] [Google Scholar]
- 22.Wolff T, and Rubin GM (1998). Strabismus, a novel gene that regulates tissue polarity and cell fate decisions in Drosophila. Development 125, 1149–1159. [DOI] [PubMed] [Google Scholar]
- 23.Das G, Reynolds-Kenneally J, and Mlodzik M. (2002). The atypical cadherin Flamingo links Frizzled and Notch signaling in planar polarity establishment in the Drosophila eye. Dev. Cell 2, 655–666. [DOI] [PubMed] [Google Scholar]
- 24.Wu J, Klein TJ, and Mlodzik M. (2004). Subcellular localization of frizzled receptors, mediated by their cytoplasmic tails, regulates signaling pathway specificity. PLoS Biol. 2, E158. 10.1371/journal.pbio.0020158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng L, Zhang J, and Carthew RW (1995). Frizzled regulates mirror-symmetric pattern formation in the Drosophila eye. Development 121, 3045–3055. [DOI] [PubMed] [Google Scholar]
- 26.Brown KE, and Freeman M. (2003). Egfr signalling defines a protectivefunction for ommatidial orientation in the Drosophila eye. Development 130, 5401–5412. 10.1242/dev.00773. [DOI] [PubMed] [Google Scholar]
- 27.Choi KW, and Benzer S. (1994). Rotation of photoreceptor clusters inthe developing Drosophila eye requires the nemo gene. Cell 78,125–136. 10.1016/0092-8674(94)90579-7. [DOI] [PubMed] [Google Scholar]
- 28.Chou YH, and Chien CT (2002). Scabrous controls ommatidial rotation in the Drosophila compound eye. Dev. Cell 3, 839–850. [DOI] [PubMed] [Google Scholar]
- 29.Fiehler RW, and Wolff T. (2007). Drosophila Myosin II, Zipper, is essential for ommatidial rotation. Dev. Biol 310, 348–362. 10.1016/j.ydbio.2007.08.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fiehler RW, and Wolff T. (2008). Nemo is required in a subset of photoreceptors to regulate the speed of ommatidial rotation. Dev. Biol 313, 533–544. 10.1016/j.ydbio.2007.10.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gaengel K, and Mlodzik M. (2003). Egfr signaling regulates ommatidialrotation and cell motility in the Drosophila eye via MAPK/Pnt signaling and the Ras effector Canoe/AF6. Development 130, 5413–5423. 10.1242/dev.00759. [DOI] [PubMed] [Google Scholar]
- 32.Mirkovic I, and Mlodzik M. (2006). Cooperative activities of drosophilaDE-cadherin and DN-cadherin regulate the cell motility process of ommatidial rotation. Development 133, 3283–3293. 10.1242/dev.02468. [DOI] [PubMed] [Google Scholar]
- 33.Mirkovic I, Gault WJ, Rahnama M, Jenny A, Gaengel K, Bessette D, Gottardi CJ, Verheyen EM, and Mlodzik M. (2011). Nemo kinase phosphorylates beta-catenin to promote ommatidial rotation and connects core PCP factors to E-cadherin-beta-catenin. Nat. Struct. Mol.Biol 18, 665–672. 10.1038/nsmb.2049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Winter CG, Wang B, Ballew A, Royou A, Karess R, Axelrod JD,and Luo L. (2001). Drosophila Rho-associated kinase (Drok) links Frizzled-mediated planar cell polarity signaling to the actin cytoskeleton.Cell 105, 81–91. 10.1016/s0092-8674(01)00298-7. [DOI] [PubMed] [Google Scholar]
- 35.Koca Y, Housden BE, Gault WJ, Bray SJ, and Mlodzik M. (2019). Notch signaling coordinates ommatidial rotation in the Drosophila eye via transcriptional regulation of the EGF-Receptor ligand Argos. Sci.Rep 9, 18628. 10.1038/s41598-019-55203-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bradley WD, and Koleske AJ (2009). Regulation of cell migration and morphogenesis by Abl-family kinases: emerging mechanisms and physiological contexts. J. Cell Sci 122, 3441–3454. 10.1242/jcs.039859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hernández SE, Krishnaswami M, Miller AL, and Koleske AJ (2004). How do Abl family kinases regulate cell shape and movement? Trends Cell Biol. 14, 36–44. 10.1016/j.tcb.2003.11.003. [DOI] [PubMed] [Google Scholar]
- 38.Wang JYJ (2014). The capable ABL: what is its biological function? Mol.Cell Biol 34, 1188–1197. 10.1128/MCB.01454-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hantschel O, Nagar B, Guettler S, Kretzschmar J, Dorey K, Kuriyan J, and Superti-Furga G. (2003). A myristoyl/phosphotyrosine switch regulates c-Abl. Cell 112, 845–857. 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 40.Nagar B, Hantschel O, Young MA, Scheffzek K, Veach D, Bornmann W, Clarkson B, Superti-Furga G, and Kuriyan J. (2003). Structural basis for the autoinhibition of c-Abl tyrosine kinase. Cell 112, 859–871. 10.1016/s0092-8674(03)00194-6. [DOI] [PubMed] [Google Scholar]
- 41.Baum B, and Perrimon N. (2001). Spatial control of the actin cytoskeleton in Drosophila epithelial cells. Nat. Cell Biol 3, 883–890. 10.1038/ncb1001-883. [DOI] [PubMed] [Google Scholar]
- 42.Fox DT, and Peifer M. (2007). Abelson kinase (Abl) and RhoGEF2 regulate actin organization during cell constriction in Drosophila. Development 134, 567–578. 10.1242/dev.02748. [DOI] [PubMed] [Google Scholar]
- 43.Grevengoed EE, Loureiro JJ, Jesse TL, and Peifer M. (2001). Abelson kinase regulates epithelial morphogenesis in Drosophila. J. Cell Biol 155, 1185–1198. 10.1083/jcb.200105102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Grevengoed EE, Fox DT, Gates J, and Peifer M. (2003). Balancingdifferent types of actin polymerization at distinct sites: roles for Abelson kinase and Enabled. J. Cell Biol 163, 1267–1279. 10.1083/jcb.200307026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jodoin JN, and Martin AC (2016). Abl suppresses cell extrusion andintercalation during epithelium folding. Mol. Biol. Cell 27, 2822–2832. 10.1091/mbc.E16-05-0336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kannan R, Song JK, Karpova T, Clarke A, Shivalkar M, Wang B,Kotlyanskaya L, Kuzina I, Gu Q, and Giniger E. (2017). The Abl pathway bifurcates to balance Enabled and Rac signaling in axon patterning in Drosophila. Development 144, 487–498. 10.1242/dev.143776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Song JK, Kannan R, Merdes G, Singh J, Mlodzik M, and Giniger E(2010). Disabled is a bona fide component of the Abl signaling network. Development 137, 3719–3727. 10.1242/dev.050948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Tamada M, Farrell DL, and Zallen JA (2012). Abl regulates planarpolarized junctional dynamics through beta-catenin tyrosine phosphorylation. Dev. Cell 22, 309–319. 10.1016/j.devcel.2011.12.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xiong W, and Rebay I. (2011). Abelson tyrosine kinase is required forDrosophila photoreceptor morphogenesis and retinal epithelial patterning. Dev. Dyn 240, 1745–1755. 10.1002/dvdy.22674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Singh J, Yanfeng WA, Grumolato L, Aaronson SA, and Mlodzik M(2010). Abelson family kinases regulate Frizzled planar cell polarity signaling via Dsh phosphorylation. Genes Dev. 24, 2157–2168. 10.1101/gad.1961010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Jenny A, Darken RS, Wilson PA, and Mlodzik M. (2003). Prickle and Strabismus form a functional complex to generate a correct axis during planar cell polarity signaling. EMBO J. 22, 4409–4420. 10.1093/emboj/cdg424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Le Gall M, De Mattei C, and Giniger E. (2008). Molecular separation oftwo signaling pathways for the receptor, Notch. Dev. Biol 313, 556–567. 10.1016/j.ydbio.2007.10.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Rebay I, Fleming RJ, Fehon RG, Cherbas L, Cherbas P, and Artavanis-Tsakonas S. (1991). Specific EGF repeats of Notch mediate interactions with Delta and Serrate: implications for Notch as a multifunctional receptor. Cell 67, 687–699. 10.1016/0092-8674(91)90064-6. [DOI] [PubMed] [Google Scholar]
- 54.Giniger E. (1998). A role for Abl in Notch signaling. Neuron 20, 667–681. 10.1016/s0896-6273(00)81007-7. [DOI] [PubMed] [Google Scholar]
- 55.Polacheck WJ, Kutys ML, Yang J, Eyckmans J, Wu Y, Vasavada H, Hirschi KK, and Chen CS (2017). A non-canonical Notch complex regulates adherens junctions and vascular barrier function. Nature 552, 258–262. 10.1038/nature24998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Song JK, and Giniger E. (2011). Noncanonical Notch function in motoraxon guidance is mediated by Rac GTPase and the GEF1 domain of Trio. Dev. Dyn 240, 324–332. 10.1002/dvdy.22525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Cai D, Chen SC, Prasad M, He L, Wang X, Choesmel-Cadamuro V, Sawyer JK, Danuser G, and Montell DJ (2014). Mechanical feedback through E-cadherin promotes direction sensing during collective cell migration. Cell 157, 1146–1159. 10.1016/j.cell.2014.03.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Founounou N, Farhadifar R, Collu GM, Weber U, Shelley MJ, and Mlodzik M. (2021). Tissue fluidity mediated by adherens junction dynamics promotes planar cell polarity-driven ommatidial rotation. Nat. Commun 12, 6974. 10.1038/s41467-021-27253-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Baker NE, Mlodzik M, and Rubin GM (1990). Spacing differentiation in the developing Drosophila eye: a fibrinogen-related lateral inhibitor encoded by scabrous. Science 250, 1370–1377. 10.1126/science.2175046. [DOI] [PubMed] [Google Scholar]
- 60.Mlodzik M, Baker NE, and Rubin GM (1990). Isolation and expressionof scabrous, a gene regulating neurogenesis in Drosophila. Genes Dev. 4, 1848–1861. 10.1101/gad.4.11.1848. [DOI] [PubMed] [Google Scholar]
- 61.Thuveson M, Gaengel K, Collu GM, Chin ML, Singh J, and Mlodzik M. (2019). Integrins are required for synchronous ommatidial rotation in the Drosophila eye linking planar cell polarity signalling to the extracellular matrix. Open Biol. 9, 190148. 10.1098/rsob.190148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dobens LL, and Raftery LA (2000). Integration of epithelial patterningand morphogenesis in Drosophila ovarian follicle cells. Dev. Dyn 218, 80–93. . [DOI] [PubMed] [Google Scholar]
- 63.Kozlovskaja-Gumbriene A, Yi R, Alexander R, Aman A, Jiskra R, Na-gelberg D, Knaut H, McClain M, and Piotrowski T. (2017). Proliferation-independent regulation of organ size by Fgf/Notch signaling. Elife 6, e21049. 10.7554/eLife.21049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Cheng D, Yan X, Qiu G, Zhang J, Wang H, Feng T, Tian Y, Xu H,Wang M, He W, et al. (2018). Contraction of basal filopodia controls periodic feather branching via Notch and FGF signaling. Nat. Commun 9, 1345. 10.1038/s41467-018-03801-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Pang J, Le L, Zhou Y, Tu R, Hou Q, Tsuchiya D, Thomas N, Wang Y, Yu Z, Alexander R, et al. (2021). NOTCH signaling controls ciliary body morphogenesis and secretion by directly regulating nectin protein expression. Cell Rep. 34, 108603. 10.1016/j.celrep.2020.108603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Grammont M. (2007). Adherens junction remodeling by the Notch pathway in Drosophila melanogaster oogenesis. J. Cell Biol 177, 139–150. 10.1083/jcb.200609079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Koleske AJ, Gifford AM, Scott ML, Nee M, Bronson RT, Miczek KA, and Baltimore D. (1998). Essential roles for the Abl and Arg tyrosine kinases in neurulation. Neuron 21, 1259–1272. 10.1016/s0896-6273(00)80646-7. [DOI] [PubMed] [Google Scholar]
- 68.Koca Y, Collu GM, and Mlodzik M. (2022). Wnt-frizzled planar cell polarity signaling in the regulation of cell motility. Curr. Top. Dev. Biol 150, 255–297. 10.1016/bs.ctdb.2022.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lee M, Lee S, Zadeh AD, and Kolodziej PA (2003). Distinct sites in E-cadherin regulate different steps in Drosophila tracheal tube fusion.Development 130, 5989–5999. 10.1242/dev.00806. [DOI] [PubMed] [Google Scholar]
- 70.Oda H, and Tsukita S. (1999). Nonchordate classic cadherins have astructurally and functionally unique domain that is absent from chordate classic cadherins. Dev. Biol 216, 406–422. 10.1006/dbio.1999.9494. [DOI] [PubMed] [Google Scholar]
- 71.Boutros M, Paricio N, Strutt DI, and Mlodzik M. (1998). Dishevelled activates JNK and discriminates between JNK pathways in planar polarity and wingless signaling. Cell 94, 109–118. 10.1016/s0092-8674(00)81226-x. [DOI] [PubMed] [Google Scholar]
- 72.Jenny A, Reynolds-Kenneally J, Das G, Burnett M, and Mlodzik M(2005). Diego and Prickle regulate Frizzled planar cell polarity signalling by competing for Dishevelled binding. Nat. Cell Biol 7, 691–697. 10.1038/ncb1271. [DOI] [PubMed] [Google Scholar]
- 73.Usui T, Shima Y, Shimada Y, Hirano S, Burgess RW, Schwarz TL,Takeichi M, and Uemura T. (1999). Flamingo, a seven-pass transmembrane cadherin, regulates planar cell polarity under the control of Frizzled. Cell 98, 585–595. 10.1016/s0092-8674(00)80046-x. [DOI] [PubMed] [Google Scholar]
- 74.Walker IH, Hsieh PC, and Riggs PD (2010). Mutations in maltosebinding protein that alter affinity and solubility properties. Appl. Microbiol. Biotechnol 88, 187–197. 10.1007/s00253-010-2696-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Jenny A. (2011). Preparation of adult Drosophila eyes for thin sectioning and microscopic analysis. J. Vis. Exp 10, 2959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Frangioni JV, and Neel BG (1993). Solubilization and purification ofenzymatically active glutathione S-transferase (pGEX) fusion proteins. Anal. Biochem 210, 179–187. 10.1006/abio.1993.1170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data reported in this paper will be shared by the lead contact upon request.
This paper does not report original code.
Any additional information required to reanalyze the data reported in this paper is available from the lead contact upon request.