ABSTRACT
Clostridium thermocellum is a cellulolytic thermophile that is considered for the consolidated bioprocessing of lignocellulose to ethanol. Improvements in ethanol yield are required for industrial implementation, but the incompletely understood causes of amino acid secretion impede progress. In this study, amino acid secretion was investigated via gene deletions in ammonium-regulated, nicotinamide adenine dinucleotide phosphate (NADPH)-supplying and NADPH-consuming pathways as well as via physiological characterization in cellobiose-limited or ammonium-limited chemostats. First, the contribution of the NADPH-supplying malate shunt was studied with strains using either the NADPH-yielding malate shunt (Δppdk) or a redox-independent conversion of PEP to pyruvate (Δppdk ΔmalE::Peno-pyk). In the latter, branched-chain amino acids, especially valine, were significantly reduced, whereas the ethanol yield increased from 46 to 60%, suggesting that the secretion of these amino acids balances the NADPH surplus from the malate shunt. The unchanged amino acid secretion in Δppdk falsified a previous hypothesis on an ammonium-regulated PEP-to-pyruvate flux redistribution. The possible involvement of another NADPH-supplier, namely, NADH-dependent reduced ferredoxin:NADP+ oxidoreductase (nfnAB), was also excluded. Finally, the deletion of glutamate synthase (gogat) in ammonium assimilation resulted in the upregulation of NADPH-linked glutamate dehydrogenase activity and decreased amino acid yields. Since gogat in C. thermocellum is putatively annotated as ferredoxin-linked, a claim which is supported by the product redistribution observed in this study, this deletion likely replaced ferredoxin with NADPH in ammonium assimilation. Overall, these findings indicate that a need to reoxidize NADPH is driving the observed amino acid secretion, likely at the expense of the NADH needed for ethanol formation. This suggests that metabolic engineering strategies that simplify the redox metabolism and ammonium assimilation can contribute to increased ethanol yields.
IMPORTANCE Improving the ethanol yield of C. thermocellum is important for the industrial implementation of this microorganism in consolidated bioprocessing. A central role of NADPH in driving amino acid byproduct formation was demonstrated by eliminating the NADPH-supplying malate shunt and separately by changing the cofactor specificity in ammonium assimilation. With amino acid secretion diverting carbon and electrons away from ethanol, these insights are important for further metabolic engineering to reach industrial requirements on ethanol yield. This study also provides chemostat data that are relevant for training genome-scale metabolic models and for improving the validity of their predictions, especially considering the reduced degree-of-freedom in the redox metabolism of the strains generated here. In addition, this study advances the fundamental understanding on the mechanisms underlying amino acid secretion in cellulolytic Clostridia as well as on the regulation and cofactor specificity in ammonium assimilation. Together, these efforts aid in the development of C. thermocellum for the sustainable consolidated bioprocessing of lignocellulose to ethanol with minimal pretreatment.
KEYWORDS: Acetivibrio thermocellus, Clostridium thermocellum, amino acids, ammonium assimilation, chemostat cultures, glutamate synthase, malate shunt, redox-cofactors
INTRODUCTION
Clostridium thermocellum is a promising candidate for cost-competitive cellulosic ethanol production through consolidated bioprocessing due to its native ability to efficiently solubilize lignocellulose (1). This anaerobic thermophile (also called Ruminiclostridium thermocellum, Hungateiclostridium thermocellum, and Acetivibrio thermocellus [2]) outperforms other cellulosic microorganisms as well as fungal cellulases in lignocellulose solubilization (3). Recently, this microorganism has been considered for the hybrid biological/catalytic conversion of cellulosic biomass to fuels, in which the ethanol produced by C. thermocellum is chemically upgraded to larger fuel molecules that are compatible with heavy-duty, difficult-to-electrify transportation modes (4). However, the industrial implementation of C. thermocellum would require improvements of the hitherto achieved ethanol titer (30 g L−1 [5]) and yield (75% of the theoretical maximum [6]) to >40 g L−1 at >90% of the theoretical maximum yield (4). An increased understanding of the complex and flexible central metabolism of C. thermocellum would directly benefit the design-build-test-learn cycles that are aiming to reach these targets.
The secretion of amino acids as byproducts is an atypical phenomenon that is observed in both wild-type and engineered C. thermocellum strains, and it diverts sugar away from ethanol formation. Amino acid secretion occurs at both low (5 g L−1) and high (93 g L−1) loadings of cellulose in batch cultures, in which up to 8.7% of the total consumed carbon is secreted as amino acids, mainly valine (up to 6.0% of the total carbon) and alanine (up to 1.6% of the total carbon) (7–9). In strains engineered and evolved for high ethanol titers (22 g L−1) and yields (75% of the theoretical maximum) on cellulose, amino acid secretion increases to 10% of the consumed carbon, becoming the second most abundant organic product, after ethanol (6). Redirecting this carbon and the associated available electrons from amino acids to ethanol will, especially in view of the low profit margins for the production of commodity chemicals and fuels, have a large impact on the economic viability of C. thermocellum-based ethanol production.
When investigating hypotheses for the observed amino-acid secretion by C. thermocellum, it is important to consider how conditions, substrate availability, and metabolic fluxes deviate between laboratory or industrial applications and its natural environment (10, 11). Additionally, it is important to consider the metabolic impact of free-energy conserving mechanisms in its close-to-equilibrium glycolysis (12–14). In one hypothesis, the high nitrogen content in laboratory batch media is thought to result in the accumulation of amino acids (8). Based on this, it was expected that amino acid secretion would be lower in nitrogen-limited conditions. Surprisingly, a study by Holwerda et al. (15) showed the opposite effect in that nitrogen-limited chemostats resulted in a 6-fold higher production rate of secreted amino acids than did carbon-limited chemostats. Specifically, valine was the most abundant, displaying a 20-fold increase, and this was followed by an 8-fold increase in isoleucine and a coinciding 50-fold higher pyruvate secretion. Cell lysis was excluded as a major mechanism, based on calculations on the amino acid distribution in the protein fraction of the biomass as well as on the observed concentrations and patterns in the extracellular amino acids (15). The observation of increased amino acid formation under nitrogen limitation at otherwise identical growth rates in chemostats also makes it unlikely that the increased reversibility of the tRNA charging in protein synthesis, which is caused by C. thermocellum lacking a cytosolic pyrophosphatase, is a major contributor to amino acid secretion (15). A third hypothesis by van der Veen et al. (7) proposes that amino acid secretion in C. thermocellum is related to redox-cofactor balancing. Investigating this hypothesis requires an understanding of sources and sinks, especially those of NADPH, and of the regulation of the involved pathways by, among other regulators, ammonium. Underlying the hypothesis of van der Veen et al. (7) is the reoxidation of NADPH in amino acid biosynthesis. Based on a current omics-based genome-scale metabolic model (GEM) of C. thermocellum (16) and on extrapolation from model microorganisms, the biosynthetic pathways of amino acids from their central building block are expected to predominantly use NADPH (Table 1). For instance, the production of valine from two pyruvate requires two NADPH (Table 1). When considering this possible role of amino acid secretion as an NADPH sink, it is important to note that in C. thermocellum, NADPH can additionally be reoxidized through hydrogen formation (Fig. 1C) (17, 18). However, in pure culture laboratory or industrial applications, hydrogen formation has severe thermodynamic limitations, whereas in a natural environment, this is often dealt with through an interspecies hydrogen transfer to methanogens (11, 19, 20).
TABLE 1.
Precursors and stoichiometric coefficients for cofactors used in amino acid synthesis from precursors in C. thermocellum, based on the genome-scale metabolic model iCBI655 (16)a
| Amino acid | Precursor | NH4+ | NADPH | NADH | Fdreduced | ATP |
|---|---|---|---|---|---|---|
| Alanine | 1 pyruvate | −1 | −1 (0) | 0 (0) | 0 (−1) | 0 (0) |
| Arginine | 1 2-ketoglutarate, 1 oxaloacetate | −4 | −4 (−1) | 0 (0) | 0 (−3) | −5 (−8) |
| Asparagine | 1 oxaloacetate | −2 | −1 (0) | 0 (0) | 0 (−1) | −2 (−3) |
| Aspartate | 1 oxaloacetate | −1 | −1 (0) | 0 (0) | 0 (−1) | 0 (−1) |
| Cysteine | 1 3-phosphoglycerate, 1 AcCoA | −1 | −5 (−4) | 1 (1) | 0 (−1) | −2 (−3) |
| Glutamine | 1 2-ketoglutarate | −2 | −1 (0) | 0 (0) | 0 (−1) | −1 (−2) |
| Glycine | 1 3-phosphoglycerate | −1 | 0 (1) | 1 (1) | 0 (−1) | 1 (0) |
| Glutamate | 1 2-ketoglutarate | −1 | −1 (0) | 0 (0) | 0 (−1) | 0 (−1) |
| Histidine | 1 ribose 5-phosphate | 2 | 0 (0) | 1 (1) | 0 (0) | 4 (4) |
| Isoleucine | 2 pyruvate, 1 AcCoA | −1 | −2 (−1) | 1 (1) | 0 (−1) | 0 (−1) |
| Leucine | 2 pyruvate, 1 AcCoA | −1 | −2 (−1) | 1 (1) | 0 (−1) | 0 (−1) |
| Lysine | 1 oxaloacetate, 1 pyruvate | −2 | −3 (−1) | −1 (−1) | 0 (−2) | 0 (−3) |
| Methionine | 1 oxaloacetate, 1 AcCoA | −1 | −8 (−7) | −1 (−1) | 0 (−1) | −4 (−5) |
| Phenylalanine | 2 PEP, 1 erythrose 4-phosphate | −1 | −2 (−1) | 0 (0) | 0 (−1) | −1 (−2) |
| Proline | 1 2-ketoglutarate | −1 | −2 (−1) | −1 (−1) | 0 (−1) | −1 (−2) |
| Serine | 1 3-phosphoglycerate | −1 | −1 (0) | 1 (1) | 0 (−1) | 0 (−1) |
| Threonine | 1 oxaloacetate | −1 | −2 (−1) | −1 (−1) | 0 (−1) | −2 (−3) |
| Tryptophan | 2 PEP, 1 erythrose 4-phosphate, 1 3-phosphoglycerate, 1 ribose 5-phosphate |
−2 | −2 (−1) | 1 (1) | 0 (−1) | −3 (−4) |
| Tyrosine | 2 PEP, 1 erythrose 4-phosphate | −1 | −2 (−1) | 1 (1) | 0 (−1) | −1 (−2) |
| Valine | 2 pyruvate | −1 | −2 (−1) | 0 (0) | 0 (−1) | 0 (−1) |
The numbers in parentheses represent cofactor usage if glutamate production occurred via a ferredoxin-dependent and ATP-dependent glutamine synthetase (GS)-glutamate synthase (GOGAT) cycle instead of a NADPH-dependent glutamate dehydrogenase (GDH).
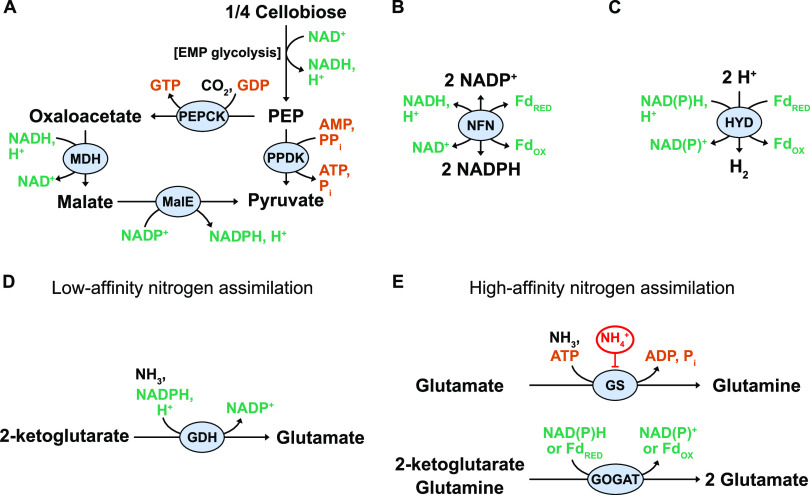
FIG 1.
Cofactor usage of C. thermocellum in (A) the PEP-to-pyruvate conversion, either via the redox-independent PPDK or via the NADPH-producing malate shunt, (B) the NADH-dependent NFN reaction, (C) a summarized HYD reaction, (D) the low-affinity GDH reaction, and (E) the high-affinity GS-GOGAT cycle. EMP, Embden-Meyerhof-Parnas; PEP, phosphoenolpyruvate; PPDK, pyruvate phosphate dikinase; PEPCK, PEP carboxykinase; MDH, malate dehydrogenase; MalE, malic enzyme; NFN, NADH-dependent reduced ferredoxin:NADP+ oxidoreductase; HYD, hydrogenase; GDH, glutamate dehydrogenase; GS, glutamine synthetase; GOGAT, glutamate synthase.
In contrast to many model heterotrophic microorganisms, in which the pentose phosphate pathway provides most of the NADPH, C. thermocellum predominantly uses the malate shunt to supply NADPH by converting phosphoenolpyruvate (PEP) to pyruvate through PEP carboxykinase (PEPCK), NADH-dependent malate dehydrogenase (MDH), and NADP+-dependent malic enzyme (encoded by malE, Clo1313_1879) (Fig. 1A) (13, 14, 21, 22). An alternative to the malate shunt, namely, PEP-to-pyruvate conversion, can occur through the alternative non-NADPH-producing (redox-independent) pyruvate phosphate dikinase (encoded by ppdk, Clo1313_0949) (Fig. 1A). By changing the flux distribution through the malate shunt with flux through Ppdk, more or less NADPH can be formed. Interestingly, both MalE and Ppdk are activated by ammonium, but these activations occur with different activation constants (Ka). MalE shows a Ka for ammonium of 0.7 to 0.8 mM (14, 23), whereas Ppdk requires a higher ammonium concentration, with an estimated Ka of 3.8 mM (15). Hence, it has been suggested that at lower intracellular ammonium levels, as is likely under nitrogen limitation, a flux redistribution between Ppdk and MalE that favors NADPH formation may occur (15). If this flux distribution results in an oversupply of NADPH, the increased production of amino acids, such as valine, could act as a NADPH sink. C. thermocellum also contains an NADH-dependent reduced ferredoxin:NADP+ oxidoreductase (encoded by nfnAB, Clo1313_1848-1849) (Fig. 1B) that might contribute to the formation of NADPH by transferring electrons from NADH and reduced ferredoxin to NADP+ or, alternatively, reoxidize excess NADPH by producing reduced-ferredoxin and NADH (17).
In addition to the regulatory role on Ppdk and MalE, (intracellular) ammonium levels commonly regulate the activities of the ammonium-assimilation pathways through either glutamate dehydrogenase (GDH) or the higher-affinity glutamine synthetase (GS)-glutamate synthase (GOGAT) (Fig. 1D and E) (24, 25). C. thermocellum has genes encoding one NADPH-dependent GDH (Clo1313_1847), four GS (three Type III GS, glnN, Clo1313_1357, 2038, 2303; and one Type Iα GS, glnA, Clo1313_2031), and a GOGAT cluster (gogat, Clo1313_2032-2036) (26, 27). The enzyme activities for GDH and GS were confirmed in vitro (15, 28). In line with expectations, GS activity in C. thermocellum increases under nitrogen-depleted conditions (28). Despite several attempts, GOGAT activity has hitherto not been measured in C. thermocellum (28, 29). Failure to measure NADH- or NADPH-dependent activity is in line with the current KEGG genome annotation of a ferredoxin binding-subunit (Clo1313_2035) in the gogat operon of C. thermocellum (26, 28, 29). This would mean that a switch from the NADPH-dependent GDH (Fig. 1D) to a higher-affinity ferredoxin-linked GS-GOGAT system (Fig. 1E), would decrease the use of NADPH reoxidized in amino acid biosynthesis (Table 1). In the example of valine, the decrease from two to one NADPH (Table 1) would mean that the regeneration of the same amount of NADPH would require a doubling of the valine secretion flux, possibly contributing to the observed increased amino acid secretion under nitrogen limitation.
The aim of this study was to investigate the role of the NADPH-supplying and NADPH-consuming pathways in C. thermocellum on amino acid secretion. The contribution of the NADPH-supplying malate shunt was investigated in strains relying fully on either the NADPH-yielding malate shunt (in a Δppdk strain) or the redox-independent conversion of PEP-to-pyruvate (in a Δppdk ΔmalE::Peno-pyk strain). To investigate another potential NADPH source, a ΔnfnAB strain was tested under identical conditions. Finally, the role of the GDH/GS-GOGAT node was investigated by testing a Δgogat strain. All strains were characterized and compared to the reference strain in cellobiose-limited or ammonium-limited chemostat cultures through the determination of the substrate, biomass, and extracellular metabolite yields at a fixed dilution rate of 0.1 h−1. Enzyme activity assays were performed to confirm the targeted gene deletions as well as to investigate the regulation due to the gene deletions or nutrient limitation.
RESULTS
The elimination of the malate shunt decreased amino acid secretion.
The contribution of NADPH formation at the PEP-to-pyruvate node to amino acid secretion was investigated by constructing strains that fully relied on either the NADPH-forming malate shunt or on the alternative redox-independent conversion by Pyk. The removal of ppdk resulted in a strain fully relying on the NADPH-forming malate shunt (Δppdk, named AVM003 [30]). Hitherto, the construction of a strain relying solely on Ppdk for PEP-to-pyruvate conversion has not been successful (13). To obtain a strain fully relying on the redox-independent conversion of PEP-to-pyruvate, the native malE gene was swapped with a heterologous pyruvate kinase (pyk) from Thermoanaerobacterium saccharolyticum (13, 31) in the Δppdk strain, resulting in strain AVM064 (Δppdk ΔmalE::Peno-pyk). Using the Δppdk background for AVM064 simultaneously avoids competition/interference between Ppdk and Pyk as well as any influence of ammonium activation on Ppdk. The removal of Ppdk and MalE in the respective strains was confirmed via enzyme activity assays, and a high Pyk activity was measured in AVM064 (Table 2). The reference strain showed little background Pyk activity that can be attributed to the combined reaction of PEPCK (13) and MDH (Table 2; Fig. 1) (32).
TABLE 2.
Activities of Ppdk, MalE, and heterologously expressed Pyk (T. saccharolyticum) in batch culturesa,b
| Strain | Enzyme activity (μmol mgprotein−1 min−1) |
||
|---|---|---|---|
| Ppdk | MalE | Pyk | |
| Reference strain (DSM 1313) | 0.28 ± 0.02 | 5.8 ± 0.3 | 0.08 ± 0.03 |
| AVM003 (Δppdk) | <0.05 | 12.5 ± 1.3 | 0.06 ± 0.03 |
| AVM064 (Δppdk ΔmalE::Peno-pyk) | <0.05 | <0.05 | 38.4 ± 2.2 |
The detection limit was 0.05 μmol mgprotein−1 min−1.
Data are shown as the average ± standard deviation for four technical replicates of biological duplicates.
The hypothesis that changes in the flux distribution at the PEP-to-pyruvate node that are caused by various intracellular ammonium concentrations influence amino acid secretion was investigated using the engineered and wild-type strains in chemostat cultures that were limited in either ammonium (N-source) or cellobiose (C-source). If the previously observed increased amino acid secretion by the wild-type under nitrogen limitation (15) is indeed caused by higher activity and a larger flux distribution through the malate shunt, compared to Ppdk, thereby resulting in an NADPH oversupply, amino acid secretion should be high and independent of the nutrient limitation in the strain solely relying on the malate shunt, and amino acid secretion should be constitutively low in the strain solely relying on pyruvate kinase. The chemostats were designed with a feed containing 5 g L−1 cellobiose and either 0.09 or 0.7 g L−1 ammonium to achieve ammonium-limitation or cellobiose-limitation, respectively, based on the rigorous study by Holwerda et al. (15) that evaluated different C/N ratios. The targeted limitation was confirmed via residual cellobiose and ammonium, respectively, being below the detection limit when the respective substrate was limited and in excess when not limited (Table 3). As commonly seen for various microorganisms when growth is not limited by the energy source (31, 33–35), the ammonium-limited chemostats showed higher cellobiose uptake rates and slightly lower biomass yields on cellobiose (Table 3).
TABLE 3.
Physiological parameters in chemostats with either cellobiose or ammonium limitation, at a dilution rate of 0.10 h−1a,b
| Parameter | Cellobiose limitation |
Ammonium limitation |
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Reference strain (LL345) | AVM064 (Δppdk ΔmalE::peno-pyk) | AVM003 (Δppdk) | LL1084 (ΔnfnAB) | AG1715 (Δgogat) | Reference strain (LL345) | AVM064 (Δppdk ΔmalE::peno-pyk) | AVM003 (Δppdk) | LL1084 (ΔnfnAB) | AG1715 (Δgogat) | |
| Cellobiose in feed (g L−1) | 4.70 ± 0.03 | 4.98 ± 0.03 | 4.76 ± 0.08 | 4.87 ± 0.02 | 4.80 ± 0.04 | 4.69 ± 0.04 | 4.99 ± 0.00 | 4.63 ± 0.04 | 4.85 ± 0.02 | 4.77 ± 0.06 |
| Residual cellobiose (g L−1) | <0.01 | <0.01 | <0.01 | <0.01 | <0.01 | 1.83 ± 0.03 | 1.10 ± 0.04 | 1.66 ± 0.11 | 1.57 ± 0.10 | 2.23 ± 0.30 |
| Ammonium in feed (g L−1) | 0.69 ± 0.00 | 0.71 ± 0.02 | 0.67 ± 0.02 | 0.70 ± 0.01 | 0.70 ± 0.01 | 0.087 ± 0.003 | 0.089 ± 0.001 | 0.087 ± 0.001 | 0.089 ± 0.001 | 0.089 ± 0.001 |
| Residual ammonium (g L−1) | 0.51 ± 0.01 | 0.53 ± 0.01 | 0.50 ± 0.01 | 0.45 ± 0.01 | 0.45 ± 0.01 | <0.002 | <0.002 | <0.002 | <0.002 | <0.002 |
| qCellobiose (mmol gbiomass−1 h−1) | −2.19 ± 0.03 | −2.36 ± 0.08 | −2.07 ± 0.09 | −2.20 ± 0.02 | −2.15 ± 0.02 | −2.49 ± 0.02 | −2.54 ± 0.10 | −2.40 ± 0.10 | −2.45 ± 0.12 | −2.11 ± 0.06 |
| qAmmonium (mmol gbiomass−1 h−1) | −1.60 ± 0.04 | −1.62 ± 0.07 | −1.56 ± 0.04 | −2.14 ± 0.12 | −2.07 ± 0.06 | −1.43 ± 0.05 | −1.10 ± 0.05 | −1.33 ± 0.10 | −1.26 ± 0.03 | −1.41 ± 0.11 |
| Biomass yield (g gcellobiose−1) | 0.13 ± 0.00 | 0.12 ± 0.01 | 0.15 ± 0.01 | 0.13 ± 0.00 | 0.14 ± 0.00 | 0.12 ± 0.00 | 0.11 ± 0.00 | 0.13 ± 0.01 | 0.12 ± 0.01 | 0.14 ± 0.00 |
| Glycogen content (%, g gbiomass−1) | 1.1 ± 0.6 | NDg | ND | ND | ND | 31.5 ± 0.6 | ND | ND | ND | ND |
| Ethanol yield (mol molcellobiose−1) | 0.97 ± 0.06 | 1.55 ± 0.01 | 0.96 ± 0.03 | 0.96 ± 0.02 | 0.84 ± 0.07 | 1.07 ± 0.03 | 1.55 ± 0.05 | 0.84 ± 0.05 | 1.00 ± 0.02 | 0.77 ± 0.03 |
| Acetate yield (mol molcellobiose−1) | 1.38 ± 0.01 | 0.65 ± 0.02 | 1.46 ± 0.04 | 1.38 ± 0.02 | 1.38 ± 0.01 | 0.94 ± 0.01 | 0.61 ± 0.01 | 1.17 ± 0.12 | 1.01 ± 0.01 | 0.84 ± 0.07 |
| Formate yield (mol molcellobiose−1) | 0.11 ± 0.04 | 0.22 ± 0.02 | 0.10 ± 0.01 | 0.31 ± 0.01 | 0.07 ± 0.01 | 0.15 ± 0.01 | 0.32 ± 0.03 | 0.18 ± 0.05 | 0.13 ± 0.03 | 0.30 ± 0.12 |
| Lactate yield (mol molcellobiose−1) | 0.01 ± 0.00 | 0.02 ± 0.00 | 0.02 ± 0.00 | 0.01 ± 0.00 | 0.02 ± 0.00 | 0.07 ± 0.01 | 0.04 ± 0.02 | 0.06 ± 0.02 | 0.06 ± 0.00 | 0.04 ± 0.02 |
| Pyruvate yield (mol molcellobiose−1) | <0.01 | <0.01 | 0.01 ± 0.01 | <0.01 | 0.01 ± 0.00 | 0.09 ± 0.00 | 0.01 ± 0.00 | 0.02 ± 0.00 | 0.03 ± 0.00 | 0.02 ± 0.01 |
| Malate yield (mol molcellobiose−1) | <0.01 | 0.11 ± 0.02 | <0.01 | <0.01 | <0.01 | <0.01 | 0.03 ± 0.00 | <0.01 | <0.01 | <0.01 |
| Total amino acids yield (mol molcellobiose−1)c | 0.026 ± 0.001 | 0.031 ± 0.002 | 0.028 ± 0.002 | 0.030 ± 0.001 | 0.022 ± 0.001 | 0.116 ± 0.002 | 0.044 ± 0.006 | 0.107 ± 0.026 | 0.115 ± 0.009 | 0.038 ± 0.010 |
| CO2 yield (mol molcellobiose−1)d | 2.38 ± 0.02 | 2.00 ± 0.04 | 2.48 ± 0.06 | 2.18 ± 0.03 | 2.29 ± 0.05 | 2.07 ± 0.03 | 1.96 ± 0.01 | 2.06 ± 0.08 | 2.11 ± 0.02 | 1.48 ± 0.12 |
| H2 yield (mol molcellobiose−1) | 4.85 ± 0.06 | ND | ND | ND | 4.73 ± 0.05 | 2.44 ± 0.09 | ND | ND | ND | 4.26 ± 0.46 |
| Ex. protein yield (mol molcellobiose−1)e | 0.09 ± 0.00 | 0.06 ± 0.00 | 0.08 ± 0.00 | 0.10 ± 0.00 | 0.08 ± 0.00 | 0.05 ± 0.00 | 0.04 ± 0.00 | 0.05 ± 0.00 | 0.04 ± 0.00 | 0.04 ± 0.01 |
| Carbon recovery (%)f | 81 ± 1 | 77 ± 0 | 84 ± 2 | 81 ± 1 | 77 ± 1 | 77 ± 1 | 75 ± 1 | 77 ± 3 | 76 ± 1 | 64 ± 1 |
| Nitrogen recovery (%)f | 71 ± 2 | 66 ± 2 | 70 ± 2 | 54 ± 3 | 53 ± 2 | 87 ± 2 | 95 ± 3 | 96 ± 2 | 97 ± 1 | 72 ± 6 |
| Degree-of-reduction recovery (%)f | 70 ± 1 | 73 ± 0 | 72 ± 2 | 70 ± 1 | 66 ± 1 | 69 ± 1 | 71 ± 1 | 68 ± 2 | 68 ± 1 | 59 ± 1 |
| Degree-of-reduction recovery with H2 (%)f | 90 ± 1 | ND | ND | ND | 86 ± 2 | 79 ± 1 | ND | ND | ND | 76 ± 1 |
Data are shown as the average ± standard deviation for three biological replicates.
Negative values represent consumption, and positive values represent production.
Excluding cysteine.
Calculated from the fermentation product yields and biomass yield (see Materials and Methods).
Based on the molecular weight of a generalized protein composition (see Table S1 and Materials and Methods).
Calculated as the ratio of the total element or degree-of-reduction found in the products to the total element or degree-of-reduction in the consumed substrate (Tables S3–S5).
ND, not determined.
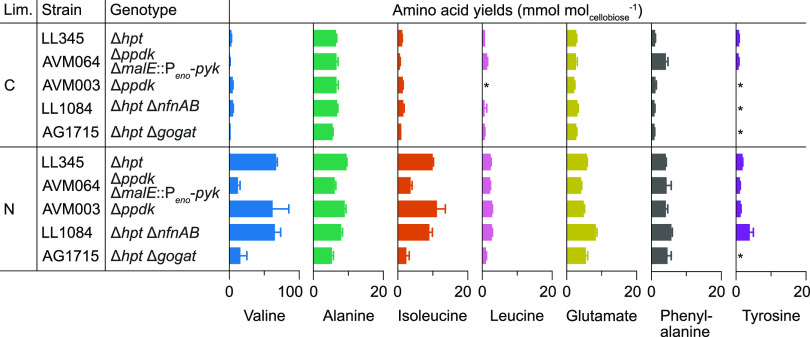
To create a baseline for the subsequent comparisons of the other strains under identical conditions, the reference strain LL345 was grown in cellobiose-limited or ammonium-limited chemostats (Table 3). The total amino acid yield increased 4.5-fold, from 0.026 ± 0.001 mol molcellobiose−1 under cellobiose limitation to 0.116 ± 0.002 mol molcellobiose−1 under ammonium limitation (Table 3). Not only did the yields of the pyruvate-derived amino acids alanine (1.5-fold), isoleucine (7.8-fold), leucine (3.1-fold), and valine (20.4-fold) increase, but the extracellular pyruvate (from <0.01 to 0.09 ± 0.00 mmol molcellobiose−1) and lactate yields (4.7-fold) also increased (Table 3; Fig. 2; Table S2). Other amino acid yields that increased were methionine (from undetected to 6.3 ± 0.5 μmol molcellobiose−1), phenylalanine (3.9-fold), glutamate (2.1-fold), and tyrosine (2.1-fold) (Fig. 2; Table S2). No significant changes were observed for asparagine, aspartate, glutamine, arginine, glycine, histidine, threonine, or tryptophan secretion, whereas the serine yield dropped by 26% (Table S2). In absolute numbers, valine, alanine, and glutamate each accounted for approximately 15 to 20% of the secreted amino acids under cellobiose limitation (Fig. 2). Under ammonium limitation, valine was by far the most abundant, at 58% of the secreted amino acids (Fig. 2). As a result, secreted amino acids accounted for 4.9% of the carbon and 5.7% of the degree-of-reduction of the consumed cellobiose under ammonium limitation, compared to only 0.9% and 0.9%, respectively, under cellobiose limitation (Table S3 and S4). Similarly, 20.3% of the total consumed nitrogen ended up in secreted amino acids under ammonium limitation, compared to 3.7% under cellobiose limitation (Table S5). Among the traditional fermentation products, the largest change was observed for the H2 yield, which dropped by 50% under ammonium limitation. Additionally, the intracellular glycogen content for the reference strain increased from 1.1 ± 0.6% (wt/wt) under cellobiose limitation to 31.5 ± 0.6% (wt/wt) under ammonium limitation. The results of this baseline characterization of the reference strain in cellobiose-limited/ammonium-limited chemostats are consistent with the previous results of Holwerda et al. (15).
FIG 2.
Amino acid yields in chemostats with either cellobiose (C) or ammonium (N) as the sole limiting nutrient at a dilution rate of 0.10 h−1. The remaining amino acids can be found in Table S6 and were either below the detection limit (*, <0.5 mmol molcellobiose−1) or only showed small changes. The error bars represent the standard deviation of three biological replicates.
Under identical conditions, the strain fully relying on Pyk for the conversion of PEP-to-pyruvate (AVM064; Δppdk ΔmalE::Peno-pyk), and thereby lacking the NADPH provided by the malate shunt, only showed a 1.4-fold increase (P < 0.05) in the total amino acid yield from cellobiose limitation to ammonium limitation, compared to the 4.5-fold increase observed for the reference strain (Table 3). Although valine secretion increased 10-fold (P < 0.05) from cellobiose to ammonium limitation, the valine yield under ammonium limitation was still 81% lower than that of the reference strain (Fig. 2; Table S2). Increases in the glutamate (1.6-fold), isoleucine (5.6-fold), leucine (1.6-fold), and tyrosine (1.4-fold) yields (P < 0.05) under ammonium limitation were also lower than that of the reference strain by 28%, 63%, 8%, and 40%, respectively (P < 0.05) (Fig. 2). No significant differences between the two limitations for AVM064 were observed for alanine and lactate (Fig. 2), whereas pyruvate increased from <0.01 to 0.01 ± 0.00 mol molcellobiose−1 (P < 0.05), which was 87% less than that of the reference strain under ammonium limitation (Table 3). Overall, rerouting the PEP-to-pyruvate conversion through Pyk rather than through the malate shunt decreased the total carbon, degree-of-reduction, and nitrogen ending up in the secreted amino acids under nitrogen limitation to 1.9%, 2.1%, and 10.8%, respectively, compared to 4.9%, 5.7%, and 20.3% for the reference strain under the same conditions (Tables S3–S5). The malate shunt in AVM064 was interrupted through the removal of malic enzyme while maintaining malate dehydrogenase, resulting in the secretion of malate with a yield of 0.11 ± 0.02 mol molcellobiose−1 under cellobiose limitation and 0.03 ± 0.00 mol molcellobiose−1 under ammonium limitation (Table 3). In line with a switch from the malate shunt, which transfers electrons from NADH to NADP+, to the redox-independent PEP-to-pyruvate conversion in the Pyk-dependent strain, the ethanol yield increased by 46 to 60% to 1.55 mol molcellobiose−1 under either nutrient limitation, compared to the reference strain (Table 3).
The physiological impact of deleting ppdk, which thereby routed the entire PEP-to-pyruvate conversion through the NADPH-yielding malate shunt, did not result in any drastic changes in product yields, compared to the reference strain (Table 3). In the ammonium-limited chemostat cultures, the impact of ppdk deletion was slightly higher, with a 21% decrease in the ethanol yield, from 1.07 ± 0.03 mol molcellobiose−1 in the reference strain to 0.84 ± 0.05 mol molcellobiose−1 in the Δppdk strain, and a corresponding increase in the acetate yield (Table 3). The amino acid yields under both limitations were similar to those of the reference strain (Fig. 2; Table S2).
The lack of a physiological impact of the ppdk deletion in the cellobiose-limited chemostats (Table 3) suggests that the flux through Ppdk in the wild-type under this condition was likely insignificant. Even though NADPH supply through the malate shunt is clearly required for surplus amino acid secretion under ammonium-limited conditions, as is seen from the strongly decreased total amino acid yield observed for AVM064 (Δppdk ΔmalE::Peno-pyk) (Table 3), this suggests a more complex underlying mechanism than only the ammonium-dependent redistribution of the PEP-to-pyruvate conversion fluxes between Ppdk and MalE.
No impact of nfnAB deletion on amino acid secretion.
To investigate the role of the NADH-dependent reduced ferredoxin:NADP+ oxidoreductase (NfnAB) (Fig. 1B) in amino acid secretion, a ΔnfnAB strain (LL1084) (17) was characterized in cellobiose-limited or ammonium-limited chemostats. A comparison of this strain with the reference strain in cellobiose-limited chemostats showed the same biomass yield and cellobiose uptake rate under both conditions (Table 3). Under cellobiose limitation, there was a 2.7-fold increase (P < 0.05) in the formate yield, compared to that of the reference strain at identical acetate and ethanol yields (Table 3). This suggests a small shift from pyruvate ferredoxin oxidoreductase (PFOR), which reduces ferredoxin, to pyruvate formate-lyase (PFL) in the ΔnfnAB strain. Interestingly, under ammonium limitation, the formate yield was the same as that of the reference strain, suggesting a negligible flux through NfnAB in the reference strain under this condition or the full functional complementation by the other NADPH-supplying routes. In line with this, the deletion of nfnAB also did not result in a meaningful change in the total amino acid formation (Fig. 2). Similar to the cellobiose-limited chemostats, the ammonium limitation of the ΔnfnAB strain showed the same biomass yield and cellobiose uptake rate as did the reference strain (Table 3). Only a minor decrease of the pyruvate yield from 0.09 ± 0.00 mol molcellobiose−1 in the reference strain to 0.03 ± 0.00 mol molcellobiose−1 in the ΔnfnAB strain, as well as an accompanying increase of the acetate yield by the same magnitude (P < 0.05), were observed.
The deletion of gogat decreased amino acid secretion under nitrogen limitation.
To investigate whether GOGAT upregulation and the predicted accompanying shift from NADPH-dependent to partially ferredoxin-linked ammonium assimilation contribute to amino acid secretion under nitrogen limitation, a Δgogat strain (AG1715) was constructed and characterized in cellobiose-limited or ammonium-limited chemostats. Under cellobiose limitation, in which both strains showed high NADPH-linked GDH activities (25.0 ± 2.5 and 32.3 ± 1.3 μmol mgprotein−1 min−1) (Table 4) and in which GDH is expected to carry the majority of the ammonium-fixation flux, the Δgogat strain and the reference strain showed the same yields and rates for canonical fermentation products (Table 3). Under these conditions, a small decrease of 15% (P < 0.05) was observed in total amino acid secretion in the Δgogat strain, compared to the reference strain (Table 3).
TABLE 4.
Glutamate dehydrogenase activities in LL345 and AG1715 from steady-state cultures grown in cellobiose-limited or ammonium-limited chemostats, expressed in μmol mgprotein−1 min−1a
| GDH- | Cellobiose limitation |
Ammonium limitation |
||
|---|---|---|---|---|
| Reference strain (LL345) | AG1715 (Δgogat) | Reference strain (LL345) | AG1715 (Δgogat) | |
| NADPH | 25.0 ± 2.5 | 32.3 ± 1.3 | 0.39 ± 0.04 | 62.3 ± 10.3 |
| NADH | 0.16 ± 0.01 | 0.38 ± 0.03 | <0.05 | 0.36 ± 0.02 |
Data are shown as the average ± standard deviation for four technical replicates of biological duplicates.
In ammonium-limited chemostats, the GDH activity of the reference strain dramatically decreased 64-fold to 0.39 ± 0.04 μmol mgprotein−1 min−1 (Table 4), which is in line with a textbook switch to the higher-affinity GS-GOGAT system (activity not measured). With GDH being the sole ammonium-assimilating option in the Δgogat strain, GDH activity was upregulated 160-fold to 62.3 ± 10.3 μmol mgprotein−1 min−1, compared to the reference strain under ammonium limitation (Table 4). Interestingly, comparing the Δgogat strain between growth conditions also showed a 2-fold upregulation (P < 0.05) of GDH activity under ammonium limitation. This upregulation was likely needed to compensate for the (predicted) lower intracellular ammonium concentration and to maintain the same ammonium-assimilation flux that was needed at the identical growth rates of 0.10 h−1.
In the ammonium-limited chemostats, the total amino acid secretion decreased by 67%, from 0.116 ± 0.002 mol molcellobiose−1 in the reference strain to 0.038 ± 0.010 mol molcellobiose−1 in the Δgogat strain (Table 3). Valine decreased the most and had a yield of 16.0 ± 9.4 mmol molcellobiose−1 under ammonium limitation, which was 76% lower than that of the reference strain (P < 0.05) (Fig. 2). The yields of other pyruvate-derived amino acids also significantly decreased, compared to the reference strain, with 44% lower alanine, 75% lower isoleucine, and 56% lower leucine under ammonium limitation (P < 0.05) (Fig. 2). All remaining amino acid yields, except that of phenylalanine, were significantly lower in the Δgogat strain (P < 0.05) (Fig. 2; Table S2). The relative abundance of valine dropped from 58% of the total secreted amino acids in the reference strain to 42% in the Δgogat strain (Table S2). Phenylalanine, glutamate, and alanine followed, corresponding to roughly 12%, 15%, and 14%, respectively. Overall, the deletion of gogat decreased the amounts of carbon, degree-of-reduction, and nitrogen consumed that were diverted to the amino acid byproducts under ammonium limitation from 4.9%, 5.7%, and 20.3% in the reference strain to 1.6%, 1.8%, and 5.9% in the Δgogat strain (Tables S3–S5).
Surprisingly, the deletion of gogat resulted in a large increase of the H2 yield under ammonium limitation. The reference strain had a H2 yield of 2.44 ± 0.09 mol molcellobiose−1, whereas the Δgogat strain showed a H2 yield of 4.26 ± 0.46 mol molcellobiose−1 (Table 3), which might reflect an increased need for ferredoxin reoxidation through the hydrogenases. Concomitantly, the ethanol yield decreased with 0.3 mol molcellobiose−1 (27%) (P < 0.05), whereas the formate yield doubled to 0.30 ± 0.12 mol molcellobiose−1 (P < 0.05) (Table 3). These changes in the redox metabolism would be in line with the increased stoichiometric need for NADPH in the ammonium assimilation if the NADPH that was formed through the malate shunt were to go at the expense of NADH. The biomass yield increased by 19%, compared to the reference strain, which can (at least, directionally) be explained by the ATP savings made in biosynthesis when shifting from ammonium assimilation via the ATP-consuming GOGAT-GS cycle to the ATP-neutral GDH reaction.
DISCUSSION
Using targeted gene deletions and cellobiose-limited/ammonium-limited chemostats as diagnostic tools, this study found several indications that amino acid secretion is driven by a cellular need to balance NADPH. This was illustrated by decreased amino acid yields upon the elimination of the NADPH-supplying malate shunt and separately, likely by the changing of the cofactor specificity of ammonium assimilation. Although the hypothesis of the differential regulation of the malate shunt and Ppdk by the ammonium concentration was falsified, the mechanisms underlying amino acid secretion clearly involve an oversupply of NADPH that, due to transhydrogenation in the malate shunt, comes at the expense of the NADH supply. The lower NADH supply and an apparent minimal contribution of NfnAB for NADPH-to-NADH conversion might limit the conversion of pyruvate to lactate and ethanol and instead favor the production of NADPH-linked, pyruvate-derived amino acids. Using amino acid formation to regenerate NADP+ has been observed in other species, as well. This mechanism has been proposed in the fungus Aspergillus nidulans (36) and in the alanine-producing archaeon Pyrococcus furiosus (37). Anaerobic rumen bacteria also commonly produce amino acids that regenerate NADP+ at a low ATP cost (38). Amino acid secretion is found among other Clostridia species as well (34, 39), which generally have 5 to 20-fold higher intracellular amino acid levels than do Gram-negative bacteria (40).
In addition to the NADPH originating from the malate shunt, specifically under ammonium limitation, a shift from GDH to GOGAT likely decreases the NADPH reoxidized per amino acid and thereby increases amino acid secretion in wild-type C. thermocellum under these laboratory conditions. Underlying that hypothesis is the proposed use of ferredoxin as the redox cofactor of GOGAT. This currently unconfirmed annotation (26) is supported by the observed changes in the redox metabolism of the reference strain and Δgogat strain under ammonium limitation (Table 3; Fig. 2). The drop in the H2 yield of the reference strain under ammonium limitation (Table 3) is in line with a switch from (mainly) NADPH-dependent GDH activity in cellobiose limitation to (mainly) ferredoxin-linked GS-GOGAT activity in ammonium assimilation, which decreases the availability of reduced ferredoxin for H2 formation. In addition to this switch, a 17% decrease in the production of reduced ferredoxin by pyruvate ferredoxin oxidoreductase (PFOR) (Table 3), calculated as , is likely also contributing to the drop in the H2 yield. The Δgogat strain would likely require other mechanisms to reoxidize ferredoxin, which, in C. thermocellum, can (in theory) occur through: (i) H2 production via a proton-pumping, energy-converting hydrogenase (ECH) or a bifurcating NAD(P)H-hydrogenase; (ii) a proton-pumping ferredoxin:NAD+ oxidoreductase (RNF) that increases the NADH pool; (iii) NfnAB, by consuming NADH and producing NADPH; or (iv) a cycle with reverse PFOR and forward PFL that assimilates CO2 and generates formate (17). The decreased ethanol yield (suggesting a lower NADH availability), increased formate yield, decreased amino acid yields, and increased H2 yield of the Δgogat strain under ammonium limitation suggest that the combination of hydrogenases and a 40% lower net flux through PFOR, calculated as , (Table S6) replace GOGAT as a ferredoxin sink (Table 3; Fig. 2).
Current stoichiometric metabolic models of C. thermocellum contain the “free” shuffling of electrons between NAD(P) and ferredoxin via the transhydrogenases NFN and RNF, which ultimately results in H2 instead of amino acids as an electron sink (16, 41). However, the metabolic flux and a thermodynamic analysis suggest a narrow window for thermodynamically favorable conversion via the (trans)hydrogenases (42). The lack of a significant effect on the fermentation product profile of ΔnfnAB in chemostat cultures in the present study and in the batch cultures in the study of Lo et al. (17) suggests either a minimal contribution or the full complementarity of NFN activity under the tested conditions in these strain backgrounds. In laboratory environments, H2 production via hydrogenases is likely further thermodynamically limited by H2 supersaturation due to poor liquid-to-gas transfer (43). Physiological predictions and the design of metabolic engineering strategies would benefit from including these thermodynamic limitations into their models.
The amino acid secretion observed under these laboratory conditions might be less common in the natural environments of cellulolytic Clostridia, as these niches not only are deficient in bioavailable nitrogen but also commonly contain H2-consuming methanogens (10, 11). In the presence of these methanogens, hydrogen partial pressures are maintained at low levels, which makes H2 production as an electron sink more favorable and increases the energy efficiency in the metabolism (10, 11). Indeed, cocultures with H2-consuming methanogens result in higher acetate-to-ethanol ratios (11), whereas increasing the (dissolved) H2 partial pressure results in a higher ethanol-to-acetate ratio, a decreased H2 yield, and reverse NAD(P)H-hydrogenase activity (44–46). Sparging laboratory cultures with N2, which is believed to make H2 production more favorable, shifted the fermentation profile, with increases in the ethanol and acetate titers and a decrease in the valine titer (42). On top of thermodynamic limitations, hydrogenases are complex organometallic proteins that require specific systems for posttranslational assembly (18, 47). Hence, their synthesis might consume more ATP than that of proteins involved in the valine and alanine pathways. Such kinetic limitations were observed with Clostridium cellulolyticum, in which chemostat cultures at various dilution rates showed that the regeneration of cofactors through hydrogen gas formation was too slow at high carbon fluxes and resulted in more lactate and polysaccharides (33, 48). Importantly, the diversion of electrons to either hydrogen and/or methane in mixed cultures goes at the expense of the ethanol yield on the substrate, necessitating a choice between lower theoretical ethanol yields or the continued engineering of C. thermocellum for improved product yields.
Aiming to get closer to the maximum theoretical ethanol yield would require further metabolic engineering to decrease the 10% of the consumed carbon that is currently diverted to amino acids in the C. thermocellum strain with the highest yield (6). One strategy to reduce amino acid secretion would be to simplify the redox metabolism by reducing the diversity in the redox cofactors. Similar to Saccharomyces cerevisiae, which relies solely on NADH for glycolysis and ethanol production, this would likely reduce the NADPH-driven amino acid secretion observed in C. thermocellum. As a first step, this study has shown that deleting the NADPH-supplying MalE (Δppdk ΔmalE::Peno-pyk) increased the ethanol and malate yields, whereas the amino acid yields in chemostat cultures dropped, reflecting a higher NADH availability and a lower NADPH availability. This is in line with the results of a study by Olson et al. (32), in which a similarly constructed strain (Δppdk ΔmalE-mdh Δldh::Peno-pyk) in batch cultures showed a 3-fold increase in ethanol production, whereas valine production dropped 3-fold. However, engineering of the steps required to convert pyruvate to ethanol in a scenario with a simplified redox metabolism, through either (i) PFOR, AdhE, and RNF or (ii) through a pyruvate decarboxylase (PDC: pyruvate acetaldehyde + CO2) and NADH-dependent alcohol dehydrogenase, has had limited success in achieving a sufficiently high ethanol yield and titer and requires more research (49–51). Alternatively, the ethanol production pathway could be changed to reoxidize the NADPH that is otherwise diverted to amino acid secretion. For instance, the laboratory evolution of a Δpta Δldh strain resulted in a point mutation in the bifunctional alcohol/aldehyde dehydrogenase (adhE) that allowed for the use of NADPH on top of NADH as a cofactor, thereby producing more ethanol and less amino acids (5, 7, 15). Combining the above interventions with a deletion in glutamate synthase and/or gogat might further reduce amino acid secretion. In line with that, Rydzak et al. (27) showed that the deletion of the GS gene glnA increased the ethanol yield and reduced the secretion of amino acids significantly. Overall, these findings can guide engineering to decrease the amino acid secretion and improve the ethanol yield of C. thermocellum for the sustainable, consolidated bioprocessing of lignocellulosic biomass.
MATERIALS AND METHODS
Strains and maintenance.
The C. thermocellum DSM 1313 wild-type (WT) strain was purchased from the DSMZ microorganism collection (www.dsmz.de). All strains used or constructed in this study are listed in Table 5. Freezer stocks were prepared by growing strains in the complex medium CTFUD (described by Olson and Lynd [52]) at 55°C to an optical density (OD) of 0.6 to 2.0. After the addition of glycerol to 25% (vol/vol), 1 mL aliquots were stocked in cryo-vials (VWR, Stockholm, Sweden) at −80°C. Strain construction and stocking were performed in an anaerobic vinyl chamber from Coy Laboratory Products (TG Instruments, Helsingborg, Sweden) with an atmosphere of 5% H2, 10% CO2, and 85% N2 (Strandmöllen AB, Ljungby, Sweden).
TABLE 5.
Strains used in this study
| Strain name | Parent strain | Genotype | Reference |
|---|---|---|---|
| Wild-type (WT) | WT | DSM 1313 | DSMZa |
| LL345 or M1354 | WT | DSM 1313 Δhpt | (63) |
| AVM003 | WT | DSM 1313 Δppdk | (30) |
| AVM064 | AVM003 | DSM 1313 Δppdk ΔmalE::Peno-pykb | This study |
| LL1084 | LL345 | DSM 1313 Δhpt ΔnfnAB | (17) |
| AG1715 | LL345 | DSM 1313 Δhpt Δgogat (Clo1313_2032-2036) | This study |
DSMZ, German Collection of Microorganisms and Cell Cultures GmbH, Braunschweig, Germany (www.dsmz.de).
The pyk gene (tsac_1363) is from the Thermoanaerobacterium saccharolyticum JW/SL-YS 485 genome.
Plasmid and strain construction.
The plasmids and primers used in this study are listed in Tables 5 and 6. Plasmid construction and propagation were performed in Escherichia coli Top10 (dam+ dcm+) (Invitrogen, Carlsbad, CA) and BL21 (dam+ dcm−) (New England Biolabs, Ipswich, MA) aerobically in LB medium supplemented with 12 to 25 μg mL−1 chloramphenicol. The purification of the plasmid DNA, genomic DNA, and PCR products were performed with commercially available kits from GeneJET (Thermo Fisher Scientific) and QIAprep (Qiagen, Germantown, MD). Primers were purchased from Invitrogen (Thermo Fischer Scientific). The plasmids were designed for markerless gene deletion according to the methods of Olson and Lynd (52), using pDGO145 as a backbone (53). Phusion High-Fidelity DNA polymerase (Thermo Scientific) was used to amplify fragments from C. thermocellum genomic DNA that corresponded to 500 to 1,000 bp upstream (5′-flank), downstream (3′-flank), and internal (int) regions of the coding sequences of interest (malE, Clo1313_1879; gogat, Clo1313_2032-2036) as well as to the promoter for the enolase gene (178 bp upstream of Clo1313_2090 on the reverse strand) (54), using the primers listed in Table 7. The pyk gene (Tsac_1363) was amplified from the genomic DNA of T. saccharolyticum JW/SL-YS 485 (DSM 8691), using primers 0553 and 0564. The fragments were assembled into the backbone according to the Gibson protocol (55), generating plasmids pJY19 (simultaneous deletion of malE and insertion of Peno-pyk) and pNJ22::GOGAT_del (Δgogat). For the simultaneous deletion and insertion, the Peno-pyk gene was placed between the 5′-flank and the 3′-flank (56). The plasmids were verified via diagnostic PCR and sequencing. Finally, the propagation of plasmids in E. coli BL21 (dam+ dcm−) ensured correct methylation before transformation into C. thermocellum (57). The transformation and selection for markerless gene editing in C. thermocellum were performed according to the methods of Olson and Lynd in CTFUD medium (52). malE was deleted while simultaneously inserting Peno-pyk at the malE locus in strain AVM003 (Δppdk), thereby generating AVM064 (Δppdk ΔmalE::Peno-pyk). The gogat gene cluster was deleted in LL345 (Δhpt) using pNJ22::GOGAT_del, thereby generating strain LL1715 (Δhpt Δgogat). The sequencing of the target loci and the 16S rRNA locus, using the primers listed in Table 7, was performed to confirm the correctness of the gene edits and the culture purity.
TABLE 6.
Plasmids used in this study
TABLE 7.
Primers used in this study
| ID | Description | Sequence (5′ to 3′)a |
|---|---|---|
| 0001 | 16S rRNA fragment to confirm culture purity | ACGGCTACCTTGTTACGACTT |
| 0002 | 16S rRNA fragment to confirm culture purity | ACGGCTACCTTGTTACGACTT |
| 0224 | pDGO145 backbone | GATATCGCCTCGTGATACGC |
| 0225 | pDGO145 backbone | CAGCTGCTAATAGTAGTGAAAAAATCAG |
| 0226 | PgapDH-cat-hpt selection cassette from pDGO145 | CTGAACTACTGGGCCAGGTATG |
| 0227 | PgapDH-cat-hpt selection cassette from pDGO145 | ATCGTGGGAATAGGCATGG |
| 0557 | Internal region of malE with an overhang matching the pDGO145 backbone | gcggcattatccctgattttttcactactattagcagctgGTTTATCTGGTTCGGGAAGTCG |
| 0558 | Internal region of malE with an overhang matching the PgapDH-cat-hpt cassette | cctccccatgctttaatacatacctggcccagtagttcagCGAAGATATATCCGCTCCGAGATG |
| 0559 | 5′-flank to malE with an overhang matching the PgapDH-cat-hpt cassette | tccgggaaaacaaatcaaccTTGATTCTACCTCCAAATTA |
| 0560 | 5′-flank to malE with an overhang matching the pDGO145 backbone | acattaacctataaaaataggcgtatcacgaggcgatatcCTGTGGGAAAAGTTATAGGT |
| 0561 | 3′-flank to malE with an overhang matching the PgapDH-cat-hpt cassette | cctcgggcaaaaaaatcttttccatgcctattcccacgatAAGCCCAATATAATTACTGTCGC |
| 0562 | 3′-flank to malE with an overhang matching the pyk gene from T. saccharolyticum | gtatagtgaatataaaatagCCGGAGTTGCAAGAATATAAAAC |
| 0563 | pyk gene from T. saccharolyticum with an overhang matching the 3′-flank of malE | ttatattcttgcaactccggCTATTTTATATTCACTATACCTTTGTAGACCA |
| 0552 | pyk gene from T. saccharolyticum with an overhang matching the Peno promoter | atatgaagggagaatggagaATGCGTAGAACTAAGATAATATGCACG |
| 0553 | Peno promoter with an overhang matching pyk | attatcttagttctacgcatTCTCCATTCTCCCTTCATATAGC |
| 0564 | Peno promoter with an overhang matching the 5′-flank of malE | taatttggaggtagaatcaaGGTTGATTTGTTTTCCCGGAA |
| 1001 | 5′-flank to gogat with an overhang matching the pDGO145 backbone | attttgtttcccataggcgcgccgatTCCCCCCTCATCATGGGGAATATATGTGGG |
| 1002 | 5′-flank to gogat with an overhang matching its 3′-flank | tattttgcagaatacatgaggcggccgcTAGCCGAGAATATGGCCAAAGGAGGT |
| 1003 | 3′-flank to gogat with an overhang matching its 5′-flank | tagcggccgcCTCATGTATTCTGCAAAATACATTTTTT |
| 1004 | 3′-flank to gogat with an overhang matching the PgapDH-cat-hpt cassette | attaattttttaaATACTTTCGCTTCTCCTCTATGAGCT |
| 1005 | PgapDH-cat-hpt cassette with an overhang matching the 3′-flank of gogat | agaggagaagcgaaagtatTTAAAAAATTAATTATTTTTTATCTAAACTATTGAA |
| 1006 | PgapDH-cat-hpt cassette with an overhang matching the int region of gogat | tccggaggatgtcggagagttTTATGAATACATTTCAGGTTTCAAAACGCC |
| 1007 | int region of gogat with an overhang matching the PgapDH-cat-hpt | aatgtattcataaAACTCTCCGACATCCTCCGGAAAACC |
| 1008 | int region of gogat with an overhang matching the pDGO145 backbone | tatacactccgctagcgcggatccgattTGAAAGAAGGTCAAATAAGGATACCGTCGGG |
| 0033 | Sequencing of hpt region in pJY19; confirmation of PgapDH-cat-hpt selection cassette removal | GCTATCTTTACAGGTACATCATTCTGTTTGTG |
| 0034 | Sequencing of hpt region in pJY19; confirmation of PgapDH-cat-hpt selection cassette removal | TTTCATCAAAGTCCAATCCATAACCC |
| 0050 | Sequencing of int region in pJY19 | CCAGACGAAAAAGTTTTGAC |
| 0051 | Sequencing of int region in pJY19 | TATGTCACGCTTACATTCAC |
| 0284 | Sequencing of int to hpt region in pJY19 | GTTAGAGCGGCATTATCCCT |
| 0555 | Sequencing of backbone to 5′-flank in pJY19 | GAGCGGATACATATTTGAATG |
| 0556 | Sequencing of 5′-flank to Peno-pyk region in pJY19 | AGTTGGTATGAATTTATTCC |
| 0565 | Sequencing of 5′-flank to backbone in pJY19 | TCTATTCCATACATGCCG |
| 0566 | Sequencing of 3′-flank to PgapDH region in pJY19 | GCATATAAGATCCGCTTTC |
| 0567 | Sequencing of 3′-flank to pyk region in pJY19 and AVM064 | GAAGCCGTAAGTGTGGATG |
| 0568 | Sequencing of 5′-flank to pyk in AVM064 | ATCCATCGTAAATTCCGAAG |
| 0281 | Sequencing of PgapDH to 3′-flank region in pJY19 | AAGAAAACAGACGCGCCC |
| 0283 | Sequencing of pyk to 3′-flank region in pJY19 and AVM064 | ATCATCGTTGCTCAAAAAACTG |
| 0159 | Sequencing of tdk region in pJY19 | GGTACGAATGTATAAGATGGTGC |
| 0267 | Sequencing of tdk region in pJY19 | TTCTCCTGCCACATGAAG |
| 0286 | Sequencing of tdk region in pJY19 | GCATGTTGTCGCCGTTATGA |
| 0232 | Sequencing of pyk region in pJY19 | ATGCGTAGAACTAAGATAATATGCACG |
| 0275 | Sequencing of pyk region in pJY19 and AVM064 | GAGATATAGGCGCAACTGCC |
| 0276 | Sequencing of pyk region in pJY19 and AVM064 | GCGACAGGTATTCCTGCCGA |
| 0277 | Sequencing of pyk region in pJY19 and AVM064; confirmation of Peno-pyk insertion in AVM064 | GGAATCGACATGATTGCAGCG |
| 0278 | Sequencing of pyk region in pJY19 and AVM064 | TTTCGACACCCAAATCGCCG |
| 0279 | Sequencing of pyk to Peno-5′-flank in pJY19 and AVM064 | CAGGCCCTTTTGTATCAAGC |
| 1009 | Sequencing of gogat region in pNJ22::GOGAT_del | ACGCACGAAAAGCCCTCTAG |
| 1010 | Sequencing of gogat region in pNJ22::GOGAT_del | AGGCGAGTCGGATAAATTTC |
| 1011 | Sequencing of gogat region in pNJ22::GOGAT_del | AATAAAAACGGTGAGTTTCC |
| 1012 | Sequencing of gogat region in pNJ22::GOGAT_del | GACGGAGAGTTAGGTTATTGGG |
| 1013 | Sequencing of gogat region in pNJ22::GOGAT_del | ACAGAAGAAGAGTTGAAGGAAAAG |
| 1014 | Sequencing of gogat region in pNJ22::GOGAT_del | CATCCTCCGGAAAACCTACC |
| 0285 | Confirmation of Pcbp-tdk selection marker removal | ACGTTATATTGCTTGCCGGG |
| 0289 | Confirmation of Pcbp-tdk selection marker removal | AAGACTCCTTTGCTCCAACC |
| 0569 | Confirmation of malE deletion, Peno-pyk insertion | CTTGTAGTATCCAATCCTGTTGACA |
| 0472 | Confirmation of malE deletion, Peno-pyk insertion | GTTGTAATGGTAAGCTGTTGCG |
| 0544 | Confirmation of malE deletion | GAGACATAGGACCTGAAGCC |
| 0570 | Confirmation of malE deletion | GCTCTTACATCAAGCGCACC |
| 0238 | Confirmation of hpt deletion | CACTTTCTTGTTGGCTCTGGCAGC |
| 0239 | Confirmation of hpt deletion | CCGGAGATGAGGCTTTTGTTGAGAAC |
| 0003 | Confirmation of nfnAB deletion | TCATCCACCCACGGTACT |
| 0004 | Confirmation of nfnAB deletion | GGGGGAAATGTATAAGAGGGGA |
| 0681 | Confirmation of gogat deletion | AGAACATATTAAAACCCCGGCA |
| 0682 | Confirmation of gogat deletion | TGGGTAAAATGGGTTGCTCC |
| 1015 | Confirmation of gogat deletion | CTATCTTTTTGTTTATCTTCATTACACATC |
| 1016 | Confirmation of gogat deletion | GATTTGCAGGTTATGGAATCTATC |
| 1017 | Confirmation of gogat deletion | CAGACAATTTTCTATGACAATATCTTTATC |
| 1018 | Confirmation of gogat deletion | TACAGGTTTGATAAAAGATATAAACGG |
| 1019 | Confirmation of gogat deletion | TCTATTTCATGTTGCCCGGG |
| 1020 | Confirmation of gogat deletion | AACCATACTTCCGAATACATTGGG |
The annealing sequences are represented by uppercase letters. The overhang sequences used for the Gibson assembly are represented by lowercase letters.
Media and culture conditions.
Serum bottle cultivations were carried out in 125-mL Wheaton serum bottles (DWK Life Sciences, Millville, NJ, USA) that were closed with blue butyl rubber stoppers (number CLS-4209-14, Chemglass Life Sciences, NJ, US) and contained 50 mL of a defined low-carbon (LC) medium (56, 58). Per L, the medium contained 5 g D-(+)-cellobiose, 0.5 g urea, 5 g MOPS, 2 g KH2PO4, 3 g K2HPO4, 0.1 g Na2SO4, 0.2 g MgCl2·6H2O, 0.05 g CaCl2·2H2O, 0.0035 g FeSO4·7H2O, 0.025 g FeCl2·4H2O, 0.1 g l-cysteine-HCl-monohydrate, vitamins (0.02 g pyrodoxamine dihydrochloride, 0.004 g p-aminobenzoic acid, 0.002 g d-biotin, and 0.002 g vitamin B12), and trace elements (1.25 mg MnCl2·4H2O, 0.5 mg ZnCl2, 0.125 mg CoCl2·6H2O, 0.125 mg NiCl2·6H2O, 0.125 mg CuSO4·5H2O, 0.125 mg H3BO3, and 0.125 mg Na2MoO4·2H2O). The suppliers of the chemicals are listed in Yayo et al. (56). The final medium for the closed-batch serum bottles was prepared from sterile anaerobic stock solutions as described by Kuil et al. (30) and purged with 20% CO2 and 80% N2 (Strandmöllen AB) prior to inoculation by alternating between vacuum and gas for five cycles.
For chemostats, the LC medium described above was used, except with 1 g L−1 cysteine, no MOPS, and 5 times higher trace element concentrations, as described by Yayo et al. (56), and the nitrogen source was changed from urea to ammonium to avoid the cyclic CO2 production observed in preliminary carbon-limited chemostats with urea (at a dilution rate of 0.1 h−1). With urea in the feed, a spike in the off-gas CO2 concentration every 30 h was found to be coupled to the alternating breakdown and accumulation of urea and ammonium (data not shown). Instead, the chemostat feed contained 0.25 g L−1 or 2 g L−1 ammonium chloride (equal to 0.07 or 0.9 g L−1 ammonium) (Sigma) for the ammonium-limited or cellobiose-limited cultivations, respectively (15). The final medium (10 L) was prepared in 10 L Duran flasks (Saveen and Werner AB, Malmö, Sweden) from stock solutions as described by Yayo et al. (56). The feed vessels were covered in aluminum foil and were continuously stirred and sparged with filtered N2 gas (99.999%) (Nippon Gases, Köping, SE, or Air Liquide Gas AB, Malmö, Sweden) at room temperature.
The inocula for serum bottle cultivations and chemostats were prepared by two transfers from the freezer stock in serum bottles. First, an overnight culture was inoculated into 50 mL of fresh medium. At an OD of 0.5 to 1.0, this preculture was transferred to the main cultivation (serum bottle or bioreactor) with an inoculation volume of 5%. All of the serum bottles were incubated in a Jeio Tech ISS-4075R incubator (Milmedtek AB, Karlskrona, Sweden) at 55°C and 180 rpm.
Chemostats.
Chemostats were performed in a multiparallel stainless-steel bioreactor system (GRETA from Belach AB, Stockholm, Sweden) with up to 6 unpressurized bioreactors running simultaneously at 55°C with 400 rpm agitation. The working volume was maintained at 0.8 L via feedback regulation on the effluent pump via a level sensor. The headspace of 0.45 L was continuously purged with filtered N2 gas (N5.0, Nippon Gases or Air Liquide Gas) at 0.2 L min−1, and the outgas was passed through a condenser. In order to minimize O2 permeability, Viton O-rings, Viton septa, and black Tygon tubing (A-60-G) were used (Sigma-Aldrich). The pH was measured with a SteamLine pH electrode (SL80-225pH from VWR) and was controlled at 7.0 via the addition of filter-sterilized 4.0 M KOH (0.2 μm PES filters) (VWR). The base vessel and 10 L feed vessels were connected via sterile 0.6 mm needles through septa on the head-plate (one feed vessel per reactor). A feed rate of 0.08 L per min, corresponding to a dilution rate of 0.10 h−1, was set and resulted in minimal intervals between medium droplets, thereby avoiding feast-famine cycles. The flow rate was checked daily with in line glass serological pipettes (25 mL) that were connected to the feed tube via a T-connection. Effluent was collected from a steam-sterilized sample valve in the bottom of the bioreactor into sterilized plastic Nalgene bottles (VWR) equipped with gas exhaust filters. The septa were cleaned with 70% isopropanol before and after the inoculation, sampling, and addition of feed and base tubes.
A batch cultivation preceded the chemostat phase in the bioreactors and was monitored via base titration or offline optical density measurements (see below). The feed pump, effluent pump, and level sensor were started at the end of the batch or shortly thereafter.
Steady-state was defined as a change of less than 5% in the biomass concentration over 3 residence times and after at least 4 residence times from the feed start. Pre-steady-state samples (35 mL) were taken in order to measure the OD and cell dry weight (CDW, see below) and thereby determine whether steady-state had been reached. After this, steady-state samples (described below) were taken (9 to 12 residence times after the feed start).
To investigate chemostat homogeneity, the OD and CDW of both the reactor and the effluent were compared. No significant differences (>5%) were observed for any of the characterized strains or conditions.
The culture purity of the steady-state cultures was assessed via the microscopy and sequencing of the 16S rRNA locus, using primers 0001 and 0002 (Table 7). Cross-contamination was excluded via the targeted PCR amplification of the strain-specific genotypes.
H2 analysis.
The headspace H2 mole fraction was measured via offline mass spectrometry. Samples were collected for each bioreactor in serum bottles that were closed with thick butyl rubber stoppers (number CLS-4209-14, Chemglass Life Sciences) to prevent gas diffusion (only for strains LL345 and LL1715). Gas was flowing through the bottles continuously from the start of the cultivations. These bottles were connected to the tops of the condensers via septa, sterile 0.6 mm needles, 0.2 μm syringe filters (PES), and Norprene tubing (3 mm ID). The bottles were immediately detached prior to the liquid sampling, and they were stored at room temperature until analysis on the same or next day.
The mass spectrometer consisted of an Extrel RGA mounted on an ultrahigh vacuum (<10−7 Pa) chamber. Sample gases were injected through a blue butyl rubber stopper (number CLS-4209-14, Chemglass Life Sciences) into a small, high-pressure region (pumped between samples), from where the gas was let into the UHV region with a precision leak valve. The pressure in the UHV chamber was monitored and controlled during the measurements, typically at 4.5 ± 0.8 · 10−5 Pa. 20 mass spectra were averaged to reduce the effects of any possible changes in pressure during each measurement. The mass spectra for the background subtraction were measured prior to each sample.
Ions were detected using a Faraday cup. The different ionization efficiencies, the fragmentation of the different species, and the m/z-dependent transmission efficiency of the quadrupole were accounted for via calibration with gas mixtures of known composition (0.2%, 0.5%, and 2% H2 in 20% O2 and balance N2; 1% H2 in 99% N2 [Skandinaviska Gasprodukter AB, Södertälje, Sweden]; 5% H2 in 10% CO2 and 85% N2 [Strandmöllen, Ljungby, Sweden]). This was done by determining calibration values for each of the parent ions that yielded the correct, known composition. This simple method was only valid for the species in the calibration mixtures but sufficed, as other species (e.g., ethanol) were not observed in the mass spectra of the samples.
Extracellular metabolite analysis.
A culture sample of approximately 100 mL was quickly withdrawn from the reactor and stored on ice. Samples from the feed vessels were collected at the beginning and end of the cultivation. Aliquots of 1 mL were centrifuged in a tabletop centrifuge (Eppendorf 5424, Thermo Fisher Scientific, Stockholm, Sweden) at 20,238 × g for 2 min. The supernatant was transferred to a nylon, low protein-binding, 0.22 μm Corning Costar Spin-X centrifuge tube filter (Sigma-Aldrich) and centrifuged at 20,238 × g for 2 min. Part of the filtrate was stored at 4°C until the analysis of the sugars and canonical fermentation products on an HPLC system, whereas the rest of the filtrate was stored at −20°C until the analysis of the ammonium, proteins, and amino acids (described below).
The supernatant samples stored at 4°C were analyzed within 1 week for cellobiose, glucose, ethanol, acetate, formate, lactate, pyruvate, and malate on a Waters Alliance 2695 HPLC system (Waters Sverige AB, Solna, Sweden) equipped with a Bio-Rad Aminex HPX-87H column (Bio-Rad, Solna, Sweden) that was operated at 60°C and with a 0.6 mL min−1 flow rate. The formate, pyruvate, and malate were separated using a 75 mM H2SO4 mobile phase, whereas the remaining components were separated using a 5 mM H2SO4 mobile phase. Cellobiose, glucose, and ethanol were detected using a Waters 2414 refractive-index detector, and the acids were detected using a Waters 2996 photodiode-array detector at 210 nm.
The amino acids were analyzed via derivatization, using a Waters AccQ-Tag Ultra commercial kit (cat. no. WAT052880) (Waters Sverige AB, Solna, Sweden). First, 2-aminobutyric acid (AABA) (Sigma-Aldrich) was added as an internal standard. Then, supernatant protein was removed via acidification with 5 g L−1 trichloroacetic acid (Supelco from Sigma-Aldrich), incubation for 20 min at room temperature, and centrifugation at 20,238 × g for 10 min in a tabletop centrifuge (Eppendorf 5424, Thermo Fisher Scientific, Stockholm, Sweden). The supernatant was derivatized according to the AccQ-Tag commercial kit protocol (Waters “UPLC Amino Acid Analysis Solution System Guide,” 71500129702, revision B). Separation was performed in a Waters ACQUITY Ultra-Performance Liquid Chromatography (UPLC) system with a reverse phase-UPLC AccQ-Tag Ultra column (cat.no. WAT052885) at 55°C with a flow rate of 0.70 mL min−1 and a run time of 10 min. The mobile phase consisted of eluent A, prepared by diluting Waters AccQ-Tag Ultra Eluent A concentrate (cat. no. 186003838) 20 times in ultrapure water (Purelab Chorus from AB Ninolab [Stockholm, Sweden]), and eluent B, as provided by Waters (cat. no. 186003839). The following elution profile was used: initial 99.9% A, 0.1% B; 0.54 min 99.9% A, 0.1% B, curve 6; 3.80 min, 96.0% A, 4.0% B, curve 6; 4.20 min, 96.0% A, 4.0% D, curve 6; 5.74 min, 90.9% A, 9.1% B, curve 7; 7.74 min, 78.8% A, 21.2% B, curve 6; 8.04 min 40.4% A, 59.6% B, curve 6; 8.05 min, 10.0% A, 90.0% B, curve 6; 8.64 min, 10.0% A, 90.0% B, curve 6; 8.73 min, 99.9% A, 0.1% B, curve 6; and 9.5 min, 99.9% A, 0.1% B, curve 6. Tryptophan, cysteine, and tyrosine were quantified using an ACQUITY TUV detector at 250 nm. Alanine, arginine, asparagine, aspartate, glutamine, glycine, glutamate, histidine, isoleucine, leucine, methionine, phenylalanine, serine, threonine, and valine were detected using a ACQUITY FLR detector with excitation at 266 nm and emission at 473 nm. The UPLC was set-up according to the AccQ-Tag commercial kit protocol. As standards, a Waters amino acid standard solution with 17 amino acids (cat. no. WAT088122) was mixed with glutamine (Irvine Scientific, Tilburg, The Netherlands), asparagine, and tryptophan (Sigma-Aldrich). A coelution of arginine and glutamine was observed.
Extracellular ammonium in the culture supernatants and medium samples was measured spectrophotometrically, using a commercial MegaZyme Ammonium Assay Kit (cat. no. KURAMR, Megazyme, Bray, Ireland) and a standard curve with ammonium chloride (Sigma-Aldrich).
Extracellular protein analysis.
The extracellular supernatant protein was quantified using a Bradford assay (59) that was modified for the detection of lower protein concentrations (1 to 10 μg mL−1). The protocol described in a Sigma-Aldrich technical bulletin (cat. no. B6916) was used with a 96-well microplate (flat-bottom, clear, PS) (Sigma-Aldrich) and with bovine serum albumin (Sigma-Aldrich) as the standard. A conversion to molar carbon, nitrogen, and degree-of-reduction was done using an estimated general amino acid composition for C. thermocellum, which was derived by counting the occurrences of each amino acid in all of the open-reading frames of the genome and correcting for polymerization (loss of H2O) (Table S1). These elemental concentrations were used to calculate the respective recovery on the substrate (described below) (Tables S3–S5).
CDW and OD measurements.
The cell dry weight was quantified in technical triplicates by first transferring 10 mL of culture into dried, preweighed, conical, glass centrifuge tubes and centrifuging at 2,250 × g in a tabletop centrifuge (Z206 A, Hermle Labortechnik GmbH, Wehingen, Germany) for 20 min. After washing the pellet in an equal volume of ultrapure water (Purelab Chorus), another round of centrifugation was performed. The washed pellet was dried overnight in a VENTI-Line forced-convection oven (VWR) set at 105°C. Upon cooling for 1 h in a desiccator, the dried glass tube was weighed. The CDW was calculated by dividing the difference in tube weights with the sample volume. The optical density was measured in technical triplicates in 1 mL polystyrene cuvettes in a V-1200 spectrophotometer (VWR) at 600 nm. Deionized water was used as a blank and for the dilutions.
Glycogen determination.
The sampling and assay of the glycogen content was performed as previously described (30). Briefly, 1 mL of sample was mixed with 5 mL of ice-cold methanol (–80°C) and centrifuged (10,000 × g, 10 min, −10°C). The pellet was washed with 5 mL of methanol (–80°C) and centrifuged again. After removing the supernatant, the pellet was stored at −80°C until analysis. Glycogen was hydrolyzed to glucose and was analyzed on a HPLC system (as described above).
Calculation of yields and rates.
The quantification of the extracellular metabolites and the biomass concentration in the chemostats allowed for the calculation of the yields on the substrate (Yi/S) and the biomass-specific rates (q) at steady-state. A general mass balance for a metabolite in the liquid phase is described by Equation 1.
| (1) |
Here, denotes concentrations (mmol L−1) for metabolite or biomass , is the liquid volume (L), is the flow rate (L h−1), is the biomass-specific conversion rate (mmol gx−1 h−1), and is the transfer rate between the gas and liquid phases for volatile compounds (mmol gx−1 h−1). Both and are defined as positive numbers, and the sign in front of each term depends on the production/transfer into liquid (+) or consumption/transfer out of liquid (−). Assuming steady-state , ideal mixing (), a constant volume, , and the definition of the dilution rate as (h−1), an expression for can be derived.
| (2) |
With a condenser to minimize evaporation, the transfer rate, , was assumed to be zero for all of the aqueous metabolites, except ethanol. For ethanol, a first-order transfer rate coefficient was determined to be = 0.018 ± 0.01 h−1 (n = 3) in the specified bioreactor system at 55°C and 400 rpm with 0.2 L min−1 overhead purging with N2 gas. This determination was made by measuring the dissipating ethanol concentrations in the liquid phase over time (data not shown). Therefore, the transfer term in Equation 2 was , giving the following expression for the ethanol production rate (where ).
| (3) |
Based on the biomass-specific conversion rates, the yield of product on the consumed substrate cellobiose was calculated as follows.
| (4) |
In order to calculate the molar yield of biomass on cellobiose, a molecular weight of 24.66 g Cmol−1 and a degree-of-reduction of 4.30 were used, based on the biomass composition of CH1.71O0.43N0.20S0.01 with 4.32% ash content from Hogsett (60) and the following elemental degree-of-reductions: C = 4, H = 1, O = −2, N = −3, S = 6.
The estimated rate of CO2 production was calculated based on the current biochemical knowledge regarding the C. thermocellum metabolism and the genome-scale metabolic model iCBI655 (16). In addition, in the conversion of cellobiose into biomass, CO2 is produced as a byproduct to balance the carbon, as biomass is slightly more reduced per carbon than is cellobiose. Based on the carbon and degree-of-reduction balancing of that biosynthesis reaction, it was estimated that 3.04 mmol CO2 per g biomass is formed. With this, the total CO2 production rate was calculated based on measured conversion rates, as shown in Equation 5 below.
| (5) |
The H2 production rate was derived by combining the mass balances for the gas and liquid phases. First, the general mass balance in the gas phase, as shown in Equation 6, can be simplified for H2, as the inlet gas only consists of N2.
| (6) |
At steady-state, Equation 7 is valid.
| (7) |
Here, denotes the mole fraction of the gas, is the total gas amount (mol), and is the gas flow in or out of the bioreactor (mol h−1). The H2 production rate was derived by combining Equation 1 for H2 (simplified by neglecting the inflow and outflow of H2 in the liquid due to low solubility) with Equation 7.
| (8) |
Here, (mol h−1) could be estimated from a nitrogen balance on the inlet (pure N2) and outlet (N2, CO2, and H2) gas streams (N2 is inert) and the use of the ideal gas law.
Calculation of the carbon, nitrogen, and degree-of-reduction recoveries on the consumed substrate.
The recovery on the substrate was defined as the ratio between the products and the substrates, in regard to their carbon, nitrogen, or degree-of-reduction contents. The substrate for the carbon and degree-of-reduction calculations was cellobiose, and the substrate for the nitrogen calculations was ammonium. In the degree-of-reduction balancing, the following definitions were made for the elements: C = 4, H = 1, O = −2, N = −3, S = 6. The molar concentrations of biomass and protein were estimated based on the molecular weights specified above.
Enzyme activity assays.
The cell-free extracts were prepared as described previously (56). In the case of steady-state cultures, 50 mL of sample was harvested. Enzyme activities were assayed aerobically at 55°C in a Cary 50 UV-visible spectrophotometer with a single-cell Peltier element from Varian AB (Solna, Sweden). The conversion of NAD(P)+ to NAD(P)H was followed at 340 nm in quartz cuvettes (VWR) with a path length of 1 cm and a reaction volume of 1 mL. An extinction coefficient of 6.22 AU mmol−1 L cm−1 was used for NAD(P)H. The proportionality between the activity and the amount of cell extract was confirmed by assaying two different concentrations of cell extract in technical duplicates. Each activity is reported for biological duplicates. An assay was carried out by first incubating all reaction mixture components, except for the coupling enzymes and the cell extract, at 55°C for 5 min. Then, the coupling enzymes (if used) and the cell extract were added and equilibrated for an additional 4 min, with the last minute serving as a background slope. The reaction was initiated by adding the indicated metabolites. The slope for the first 30 s was used to calculate the activity.
The glutamate dehydrogenase activity was measured based on the oxidation rate of NAD(P)H as 2-ketoglutarate was converted to glutamate (15, 61). The assay mixture contained 50 mM Tris-HCl (pH 8.0 at 55°C), 5 mM DTT, 50 mM NH4Cl, 5 mM 2-ketoglutarate (pH 8.0 at 25°C), and 0.3 mM NADPH or NADH. The reaction was started via the addition of 2-ketoglutarate.
The malic enzyme activity was measured by following the reduction of NADP+ as malate was converted to pyruvate (32). The assay mixture contained 50 mM Tris-HCl (pH 7.5 at 55°C), 5 mM DTT, 2 mM NADP+, 20 mM NH4Cl, 2.5 mM malate, and 50 or 100 μL cell extract. The reaction was started via the addition of malate.
The pyruvate phosphate dikinase activity was assayed by coupling the formation of pyruvate from PEP to the formation of lactate from pyruvate via added lactate dehydrogenase, resulting in the oxidation of NADH (32). The assay mixture contained 50 mM Tris-HCl (pH 7.5 at 55°C), 5 mM DTT, 0.3 mM NADH, 5 mM MgCl2·6H2O, 20 mM NH4Cl, 2 mM PEP, 2 mM AMP, 13 U mL−1 l-lactate dehydrogenase (from bovine heart, Sigma L2625), 1 mM K4PPi, and 50 or 100 μL cell extract. The reaction was started via the addition of PPi.
The pyruvate kinase activity was measured by coupling the formation of pyruvate from PEP to the formation of lactate from pyruvate via added lactate dehydrogenase, resulting in the oxidation of NADH (32). The assay mixture contained 50 mM Tris-HCl (pH 7.5 at 55°C), 5 mM DTT, 0.3 mM NADH, 12 mM MgCl2·6H2O, 10 mM KCl, 10 mM ADP, 0.1 mM 3-phosphoglyceric acid, 5 mM PEP, 13 U mL−1 l-lactate dehydrogenase (from bovine heart, Sigma L2625), and 50 or 100 μL cell extract. The reaction was initiated via the addition of PEP.
The lactate dehydrogenase (LDH) activity was assayed by measuring the oxidation of NADH as pyruvate was converted to lactate (62). The mixture contained 10 mM pyruvate, 1 mM fructose 1,6-bisphosphate, 0.22 mM NADH, 200 mM Tris-HCl (pH 7.3 at 55°C), and 50 or 100 mL of cell extract. The reaction was initiated via the addition of pyruvate. LDH activity was used routinely as a quality control on all cell extracts (Table S8).
Data analysis.
Student’s t tests were used for the unpaired comparisons between values in this study.
Data availability.
The data on the measured concentrations from the chemostats that were used in the calculations of the yields and rates are presented in Table S7. The GenBank accession numbers for the plasmids generated in this study are presented in Table 6.
ACKNOWLEDGMENTS
Funding for J.Y., T.K., A.K., and A.J.A.M. was provided by the Formas grant 2017-00973 and by the Novo Nordisk Foundation grant NNF20OC0064164. D.J.H. was funded by the Swedish Foundation for Strategic Research (SSF) (ITM17-0236). T.R. and A.M.G. were supported by the Center for Bioenergy Innovation, U.S. Department of Energy (DOE) Bioenergy Research Center, supported by the Office of Biological and Environmental Research in the DOE Office of Science. Oak Ridge National Laboratory is managed by UT-Battelle, LLC, for the U.S. DOE under contract DE-AC05-00OR22725.
We thank Gustav Sjöberg, Jeroen G. Koendjbiharie, and Chonticha Phongsawat for their insightful comments on the data analysis as well as for their help in setting up and sampling the chemostats. We are also thankful to Amparo Jiménez Quero for the experimental help with the metabolite analysis.
Footnotes
Supplemental material is available online only.
Contributor Information
Antonius J. A. van Maris, Email: tonvm@kth.se.
Haruyuki Atomi, Kyoto Daigaku.
REFERENCES
- 1.Lynd LR, Liang X, Biddy MJ, Allee A, Cai H, Foust T, Himmel ME, Laser MS, Wang M, Wyman CE. 2017. Cellulosic ethanol: status and innovation. Curr Opin Biotechnol 45:202–211. 10.1016/j.copbio.2017.03.008. [DOI] [PubMed] [Google Scholar]
- 2.Tindall BJ. 2019. The names Hungateiclostridium Zhang et al. 2018, Hungateiclostridium thermocellum (Viljoen et al. 1926) Zhang et al. 2018, Hungateiclostridium cellulolyticum (Patel et al. 1980) Zhang et al. 2018, Hungateiclostridium aldrichii (Yang et al. 1990) Zhang et al. 2018, Hungateiclostridium alkalicellulosi (Zhilina et al. 2006) Zhang et al. 2018, Hungateiclostridium clariflavum (Shiratori et al. 2009) Zhang et al. 2018, Hungateiclostridium straminisolvens (Kato et al. 2004) Zhang et al. 2018 and Hungateiclostridium saccincola (Koeck et al. 2016) Zhang et al. 2018 contravene Rule 51b of the International Code of Nomenclature of Prokaryotes and require replacement names in the genus Acetivibrio Patel et al. 1980. Int J Syst Evol Microbiol 69:3927–3932. 10.1099/ijsem.0.003685. [DOI] [PubMed] [Google Scholar]
- 3.Paye JMD, Guseva A, Hammer SK, Gjersing E, Davis MF, Davison BH, Olstad J, Donohoe BS, Nguyen TY, Wyman CE, Pattathil S, Hahn MG, Lynd LR. 2016. Biological lignocellulose solubilization: comparative evaluation of biocatalysts and enhancement via cotreatment. Biotechnol Biofuels 9:8. 10.1186/s13068-015-0412-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lynd LR, Beckham GT, Guss AM, Jayakody LN, Karp EM, Maranas C, McCormick RL, Amador-Noguez D, Bomble YJ, Davison BH, Foster C, Himmel ME, Holwerda EK, Laser MS, Ng CY, Olson DG, Román-Leshkov Y, Trinh CT, Tuskan GA, Upadhayay V, Vardon DR, Wang L, Wyman CE. 2022. Toward low-cost biological and hybrid biological/catalytic conversion of cellulosic biomass to fuels. Energy Environ Sci 15:938–990. 10.1039/D1EE02540F. [DOI] [Google Scholar]
- 5.Holwerda EK, Olson DG, Ruppertsberger NM, Stevenson DM, Murphy SJL, Maloney MI, Lanahan AA, Amador-Noguez D, Lynd LR. 2020. Metabolic and evolutionary responses of Clostridium thermocellum to genetic interventions aimed at improving ethanol production. Biotechnol Biofuels 13:40. 10.1186/s13068-020-01680-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tian L, Papanek B, Olson DG, Rydzak T, Holwerda EK, Zheng T, Zhou J, Maloney M, Jiang N, Giannone RJ, Hettich RL, Guss AM, Lynd LR. 2016. Simultaneous achievement of high ethanol yield and titer in Clostridium thermocellum. Biotechnol Biofuels 9:116. 10.1186/s13068-016-0528-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Veen D, Lo J, Brown SD, Johnson CM, Tschaplinski TJ, Martin M, Engle NL, van den Berg RA, Argyros AD, Caiazza NC, Guss AM, Lynd LR. 2013. Characterization of Clostridium thermocellum strains with disrupted fermentation end-product pathways. J Ind Microbiol Biotechnol 40:725–734. 10.1007/s10295-013-1275-5. [DOI] [PubMed] [Google Scholar]
- 8.Holwerda EK, Thorne PG, Olson DG, Amador-Noguez D, Engle NL, Tschaplinski TJ, van Dijken JP, Lynd LR. 2014. The exometabolome of Clostridium thermocellum reveals overflow metabolism at high cellulose loading. Biotechnol Biofuels 7:144. 10.1186/s13068-014-0155-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ellis LD, Holwerda EK, Hogsett D, Rogers S, Shao X, Tschaplinski T, Thorne P, Lynd LR. 2012. Closing the carbon balance for fermentation by Clostridium thermocellum (ATCC 27405). Bioresour Technol 103:293–299. 10.1016/j.biortech.2011.09.128. [DOI] [PubMed] [Google Scholar]
- 10.Leschine SB. 1995. Cellulose degradation in anaerobic environments. Annu Rev Microbiol 49:399–426. 10.1146/annurev.mi.49.100195.002151. [DOI] [PubMed] [Google Scholar]
- 11.Weimer PJ, Zeikus JG. 1977. Fermentation of cellulose and cellobiose by Clostridium thermocellum in the absence of Methanobacterium thermoautotrophicum. Appl Environ Microbiol 33:289–297. 10.1128/aem.33.2.289-297.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jacobson TB, Korosh TK, Stevenson DM, Foster C, Maranas C, Olson DG, Lynd LR, Amador-Noguez D. 2020. In vivo thermodynamic analysis of glycolysis in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum using 13C and 2H tracers. mSystems 5:e00736-19. 10.1128/mSystems.00736-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou J, Olson DG, Argyros DA, Deng Y, van Gulik WM, van Dijken JP, Lynd LR. 2013. Atypical glycolysis in Clostridium thermocellum. Appl Environ Microbiol 79:3000–3008. 10.1128/AEM.04037-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Taillefer M, Rydzak T, Levin DB, Oresnik IJ, Sparling R. 2015. Reassessment of the transhydrogenase/malate shunt pathway in Clostridium thermocellum ATCC 27405 through kinetic characterization of malic enzyme and malate dehydrogenase. Appl Environ Microbiol 81:2423–2432. 10.1128/AEM.03360-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Holwerda EK, Zhou J, Hon S, Stevenson DM, Amador-Noguez D, Lynd LR, van Dijken JP. 2020. Metabolic fluxes of nitrogen and pyrophosphate in chemostat cultures of Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. Appl Environ Microbiol 86:e01795-20. 10.1128/AEM.01795-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Garcia S, Thompson RA, Giannone RJ, Dash S, Maranas CD, Trinh CT. 2020. Development of a genome-scale metabolic model of Clostridium thermocellum and its applications for integration of multi-omics datasets and computational strain design. Front Bioeng Biotechnol 8:772. 10.3389/fbioe.2020.00772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lo J, Olson DG, Murphy SJ-L, Tian L, Hon S, Lanahan A, Guss AM, Lynd LR. 2017. Engineering electron metabolism to increase ethanol production in Clostridium thermocellum. Metab Eng 39:71–79. 10.1016/j.ymben.2016.10.018. [DOI] [PubMed] [Google Scholar]
- 18.Biswas R, Zheng T, Olson DG, Lynd LR, Guss AM. 2015. Elimination of hydrogenase active site assembly blocks H2 production and increases ethanol yield in Clostridium thermocellum. Biotechnol Biofuels 8:20. 10.1186/s13068-015-0204-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kouzuma A, Kato S, Watanabe K. 2015. Microbial interspecies interactions: recent findings in syntrophic consortia. Front Microbiol 6:477. 10.3389/fmicb.2015.00477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jackson BE, McInerney MJ. 2002. Anaerobic microbial metabolism can proceed close to thermodynamic limits. Nature 415:454–456. 10.1038/415454a. [DOI] [PubMed] [Google Scholar]
- 21.Lamed R, Zeikus JG. 1980. Ethanol production by thermophilic bacteria: relationship between fermentation product yields of and catabolic enzyme activities in Clostridium thermocellum and Thermoanaerobium brockii. J Bacteriol 144:569–578. 10.1128/jb.144.2.569-578.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rydzak T, McQueen PD, Krokhin OV, Spicer V, Ezzati P, Dwivedi RC, Shamshurin D, Levin DB, Wilkins JA, Sparling R. 2012. Proteomic analysis of Clostridium thermocellum core metabolism: relative protein expression profiles and growth phase-dependent changes in protein expression. BMC Microbiol 12:214. 10.1186/1471-2180-12-214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lamed R, Zeikus JG. 1981. Thermostable, ammonium-activated malic enzyme of Clostridium thermocellum. Biochim Biophys Acta 660:251–255. 10.1016/0005-2744(81)90167-4. [DOI] [PubMed] [Google Scholar]
- 24.Merrick MJ, Edwards RA. 1995. Nitrogen control in bacteria. Microbiol Rev 59:604–622. 10.1128/mr.59.4.604-622.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van Heeswijk WC, Westerhoff HV, Boogerd FC. 2013. Nitrogen assimilation in Escherichia coli: putting molecular data into a systems perspective. Microbiol Mol Biol Rev 77:628–695. 10.1128/MMBR.00025-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kanehisa M, Goto S. 2000. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28:27–30. 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rydzak T, Garcia D, Stevenson DM, Sladek M, Klingeman DM, Holwerda EK, Amador-Noguez D, Brown SD, Guss AM. 2017. Deletion of Type I glutamine synthetase deregulates nitrogen metabolism and increases ethanol production in Clostridium thermocellum. Metab Eng 41:182–191. 10.1016/j.ymben.2017.04.002. [DOI] [PubMed] [Google Scholar]
- 28.Bogdahn M, Kleiner D. 1986. Inorganic nitrogen metabolism in two cellulose-degrading clostridia. Arch Microbiol 145:159–161. 10.1007/BF00446774. [DOI] [Google Scholar]
- 29.Zhou J. 2016. Comparative analysis of central metabolism in three thermophilic saccharolytic bacteria. Thayer School of Engineering, Dartmouth College, Hanover, New Hampshire. [Google Scholar]
- 30.Kuil T, Hon S, Yayo J, Foster C, Ravagnan G, Maranas CD, Lynd LR, Olson DG, van Maris AJA. 2022. Functional analysis of H+-pumping membrane-bound pyrophosphatase, ADP-glucose synthase, and pyruvate phosphate dikinase as pyrophosphate sources in Clostridium thermocellum. Appl Environ Microbiol 88:e01857-21. 10.1128/aem.01857-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Larsson C, von Stockar U, Marison I, Gustafsson L. 1993. Growth and metabolism of Saccharomyces cerevisiae in chemostat cultures under carbon-, nitrogen-, or carbon- and nitrogen-limiting conditions. J Bacteriol 175:4809–4816. 10.1128/jb.175.15.4809-4816.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Olson DG, Hörl M, Fuhrer T, Cui J, Zhou J, Maloney MI, Amador-Noguez D, Tian L, Sauer U, Lynd LR. 2017. Glycolysis without pyruvate kinase in Clostridium thermocellum. Metab Eng 39:169–180. 10.1016/j.ymben.2016.11.011. [DOI] [PubMed] [Google Scholar]
- 33.Desvaux M, Guedon E, Petitdemange H. 2001. Carbon flux distribution and kinetics of cellulose fermentation in steady-state continuous cultures of Clostridium cellulolyticum on a chemically defined medium. J Bacteriol 183:119–130. 10.1128/JB.183.1.119-130.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Desvaux M, Petitdemange H. 2001. Flux analysis of the metabolism of Clostridium cellulolyticum grown in cellulose-fed continuous culture on a chemically defined medium under ammonium-limited conditions. Appl Environ Microbiol 67:3846–3851. 10.1128/AEM.67.9.3846-3851.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Senior PJ. 1975. Regulation of nitrogen metabolism in Escherichia coli and Klebsiella aerogenes: studies with the continuous-culture technique. J Bacteriol 123:407–418. 10.1128/jb.123.2.407-418.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Shimizu M, Fujii T, Masuo S, Takaya N. 2010. Mechanism of De Novo branched-chain amino acid synthesis as an alternative electron sink in hypoxic Aspergillus nidulans cells. Appl Environ Microbiol 76:1507–1515. 10.1128/AEM.02135-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ward DE, Kengen SWM, van der Oost J, de Vos WM. 2000. Purification and characterization of the alanine aminotransferase from the hyperthermophilic archaeon Pyrococcus furiosus and its role in alanine production. J Bacteriol 182:2559–2566. 10.1128/JB.182.9.2559-2566.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stevenson IL. 1978. The production of extracellular amino acids by rumen bacteria. Can J Microbiol 24:1236–1241. 10.1139/m78-198. [DOI] [PubMed] [Google Scholar]
- 39.Szech U, Braun M, Kleiner D. 1989. Uptake and excretion of amino acids by saccharolytic clostridia. FEMS Microbiol Lett 58:11–14. 10.1111/j.1574-6968.1989.tb03009.x. [DOI] [Google Scholar]
- 40.Tempest DW, Meers JL, Brown CM. 1970. Influence of environment on the content and composition of microbial free amino acid pools. J Gen Microbiol 64:171–185. 10.1099/00221287-64-2-171. [DOI] [PubMed] [Google Scholar]
- 41.Thompson RA, Dahal S, Garcia S, Nookaew I, Trinh CT. 2016. Exploring complex cellular phenotypes and model-guided strain design with a novel genome-scale metabolic model of Clostridium thermocellum DSM 1313 implementing an adjustable cellulosome. Biotechnol Biofuels 9:194. 10.1186/s13068-016-0607-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Thompson RA, Trinh CT. 2017. Overflow metabolism and growth cessation in Clostridium thermocellum DSM1313 during high cellulose loading fermentations: overflow metabolism and growth cessation in C. thermocellum. Biotechnol Bioeng 114:2592–2604. 10.1002/bit.26374. [DOI] [PubMed] [Google Scholar]
- 43.Zhang F, Zhang Y, Chen M, Zeng RJ. 2012. Hydrogen supersaturation in thermophilic mixed culture fermentation. Int J Hydrog Energy 37:17809–17816. 10.1016/j.ijhydene.2012.09.019. [DOI] [Google Scholar]
- 44.Rydzak T, Levin DB, Cicek N, Sparling R. 2011. End-product induced metabolic shifts in Clostridium thermocellum ATCC 27405. Appl Microbiol Biotechnol 92:199–209. 10.1007/s00253-011-3511-0. [DOI] [PubMed] [Google Scholar]
- 45.Lamed RJ, Lobos JH, Su TM. 1988. Effects of stirring and hydrogen on fermentation products of Clostridium thermocellum. Appl Environ Microbiol 54:1216–1221. 10.1128/aem.54.5.1216-1221.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bothun GD, Knutson BL, Berberich JA, Strobel HJ, Nokes SE. 2004. Metabolic selectivity and growth of Clostridium thermocellum in continuous culture under elevated hydrostatic pressure. Appl Microbiol Biotechnol 65:149–157. 10.1007/s00253-004-1554-1. [DOI] [PubMed] [Google Scholar]
- 47.Mulder DW, Shepard EM, Meuser JE, Joshi N, King PW, Posewitz MC, Broderick JB, Peters JW. 2011. Insights into [FeFe]-hydrogenase structure, mechanism, and maturation. Structure 19:1038–1052. 10.1016/j.str.2011.06.008. [DOI] [PubMed] [Google Scholar]
- 48.Guedon E, Payot S, Desvaux M, Petitdemange H. 1999. Carbon and electron flow in Clostridium cellulolyticum grown in chemostat culture on synthetic medium. J Bacteriol 181:3262–3269. 10.1128/JB.181.10.3262-3269.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tian L, Perot SJ, Hon S, Zhou J, Liang X, Bouvier JT, Guss AM, Olson DG, Lynd LR. 2017. Enhanced ethanol formation by Clostridium thermocellum via pyruvate decarboxylase. Microb Cell Fact 16:171. 10.1186/s12934-017-0783-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kannuchamy S, Mukund N, Saleena LM. 2016. Genetic engineering of Clostridium thermocellum DSM1313 for enhanced ethanol production. BMC Biotechnol 16 Suppl 1:34. 10.1186/s12896-016-0260-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Mazzoli R, Olson DG. 2020. Clostridium thermocellum: a microbial platform for high-value chemical production from lignocellulose, p 111–161. In Advances in Applied Microbiology. Elsevier. [DOI] [PubMed] [Google Scholar]
- 52.Olson DG, Lynd LR. 2012. Transformation of Clostridium thermocellum by electroporation, p 317–330. In Methods in Enzymology. Elsevier. [DOI] [PubMed] [Google Scholar]
- 53.Hon S, Olson DG, Holwerda EK, Lanahan AA, Murphy SJL, Maloney MI, Zheng T, Papanek B, Guss AM, Lynd LR. 2017. The ethanol pathway from Thermoanaerobacterium saccharolyticum improves ethanol production in Clostridium thermocellum. Metab Eng 42:175–184. 10.1016/j.ymben.2017.06.011. [DOI] [PubMed] [Google Scholar]
- 54.Olson DG, Maloney M, Lanahan AA, Hon S, Hauser LJ, Lynd LR. 2015. Identifying promoters for gene expression in Clostridium thermocellum. Metab Eng Commun 2:23–29. 10.1016/j.meteno.2015.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gibson DG. 2011. Enzymatic assembly of overlapping DNA fragments, p 349–361. In Methods in Enzymology. Elsevier. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yayo J, Kuil T, Olson DG, Lynd LR, Holwerda EK, van Maris AJA. 2021. Laboratory evolution and reverse engineering of Clostridium thermocellum for growth on glucose and fructose. Appl Environ Microbiol 87:e03017-20. 10.1128/AEM.03017-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guss AM, Olson DG, Caiazza NC, Lynd LR. 2012. Dcm methylation is detrimental to plasmid transformation in Clostridium thermocellum. Biotechnol Biofuels 5:30. 10.1186/1754-6834-5-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Holwerda EK, Hirst KD, Lynd LR. 2012. A defined growth medium with very low background carbon for culturing Clostridium thermocellum. J Ind Microbiol Biotechnol 39:943–947. 10.1007/s10295-012-1091-3. [DOI] [PubMed] [Google Scholar]
- 59.Bradford MM. 1976. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem 72:248–254. 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 60.Hogsett DAL. 1995. Cellulose hydrolysis and fermentation by Clostridium thermocellum for the production of ethanol. PhD thesis. Thayer School of Engineering, Dartmouth College, Hanover, New Hampshire. [Google Scholar]
- 61.Meers JL, Tempest DW, Brown CM. 1970. “Glutamine(amide): 2-oxoglutarate amino transferase oxido-reductase (NADP)”, an enzyme involved in the synthesis of glutamate by some bacteria. J Gen Microbiol 64:187–194. 10.1099/00221287-64-2-187. [DOI] [PubMed] [Google Scholar]
- 62.Lo J, Zheng T, Hon S, Olson DG, Lynd LR. 2015. The bifunctional alcohol and aldehyde dehydrogenase gene, adhE, is necessary for ethanol production in Clostridium thermocellum and Thermoanaerobacterium saccharolyticum. J Bacteriol 197:1386–1393. 10.1128/JB.02450-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Argyros DA, Tripathi SA, Barrett TF, Rogers SR, Feinberg LF, Olson DG, Foden JM, Miller BB, Lynd LR, Hogsett DA, Caiazza NC. 2011. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol 77:8288–8294. 10.1128/AEM.00646-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S8. Download aem.01753-22-s0001.pdf, PDF file, 0.4 MB (438.8KB, pdf)
Data Availability Statement
The data on the measured concentrations from the chemostats that were used in the calculations of the yields and rates are presented in Table S7. The GenBank accession numbers for the plasmids generated in this study are presented in Table 6.