ABSTRACT
Although segmented negative-sense RNA viruses (SNSRVs) have been frequently discovered in various fungi, most SNSRVs reported only the large segments. In this study, we investigated the diversity of the mycoviruses in the phytopathogenic fungus Fusarium asiaticum using the metatranscriptomic technique. We identified 17 fungal single-stranded RNA (ssRNA) viruses including nine viruses within Mitoviridae, one each in Narnaviridae, Botourmiaviridae, Hypoviridae, Fusariviridae, and Narliviridae, two in Mymonaviridae, and one trisegmented virus temporarily named Fusarium asiaticum mycobunyavirus 1 (FaMBV1). The FaMBV1 genome comprises three RNA segments, large (L), medium (M), and small (S) with 6,468, 2,639, and 1,420 nucleotides, respectively. These L, M, and S segments putatively encode the L protein, glycoprotein, and nucleocapsid, respectively. Phylogenetic analysis based on the L protein showed that FaMBV1 is phylogenetically clustered with Alternaria tenuissima negative-stranded RNA virus 2 (AtNSRV2) and Sclerotinia sclerotiorum negative-stranded RNA virus 5 (SsNSRV5) but distantly related to the members of the family Phenuiviridae. FaMBV1 could be vertically transmitted by asexual spores with lower efficiency (16.7%, 2/42). Comparison between FaMBV1-free and -infected fungal strains revealed that FaMBV1 has little effect on hyphal growth, pathogenicity, and conidium production, and its M segment is dispensable for viral replication and lost during subculture and asexual conidiation. The M and S segments of AtNSRV2 and SsNSRV5 were found using bioinformatics methods, indicating that the two fungal NSRVs harbor trisegmented genomes. Our results provide a new example of the existence and evolution of the segmented negative-sense RNA viruses in fungi.
IMPORTANCE Fungal segmented negative-sense RNA viruses (SNSRVs) have been frequently found. Only the large segment encoding RNA-dependent RNA polymerase (RdRp) has been reported in most fungal SNSRVs, except for a few fungal SNSRVs reported to encode nucleocapsids, nonstructural proteins, or movement proteins. Virome analysis of the Fusarium spp. that cause Fusarium head blight discovered a novel virus, Fusarium asiaticum mycobunyavirus 1 (FaMBV1), representing a novel lineage of the family Phenuiviridae. FaMBV1 harbors a trisegmented genome that putatively encodes RdRp, glycoproteins, and nucleocapsids. The putative glycoprotein was first described in fungal SNSRVs and shared homology with glycoprotein of animal phenuivirus but was dispensable for its replication in F. asiaticum. Two other trisegmented fungal SNSRVs that also encode glycoproteins were discovered, implying that three-segment bunyavirus infections may be common in fungi. These findings provide new insights into the ecology and evolution of SNSRVs, particularly those infecting fungi.
KEYWORDS: mycovirus, trisegmented bunyavirus, glycoprotein, Phenuiviridae, Fusarium asiaticum
INTRODUCTION
Fusarium head blight (FHB) is an economically devastating disease affecting cereal crops worldwide and has become more severe and frequent in the middle and lower regions of the Yangtze River in China (1, 2). FHB is induced by Fusarium graminearum species complexes that are mainly composed of Fusarium asiaticum and Fusarium graminearum in China (1). Although FHB can result in poor seed quality and low yields, the public is most concerned about FHB-infected crop contamination with mycotoxins, including deoxynivalenol and zearalenone, which pose severe threats to human and animal health and food safety (2). The strategy of virocontrol of FHB with hypovirulence-associated mycoviruses is a potential alternative to green management for sustainable and long-term disease control. Diverse mycoviruses have been discovered in over 22 Fusarium spp. (3–13), including a tripartite single-stranded DNA (ssDNA) mycovirus, Fusarium graminearum gemytripvirus 1 (12), and two capsidless multisegmented positive single-stranded RNA (+ssRNA) viruses, hadakavirus 1 strain 7n and hadakavirus 1 strain 1NL (5, 11). Only a few mycoviruses confer hypovirulence in Fusarium, including Fusarium graminearum virus 1, Fusarium graminearum virus China 9, Fusarium graminearum hypovirus 2, Fusarium oxysporum f. sp. dianthi mycovirus 1, Fusarium pseudograminearum megabirnavirus 1, Fusarium oxysporum ourmia-like virus 1, and Fusarium oxysporum alternavirus 1 (10, 14–16). Except for the reported Fusarium asiaticum victorivirus 1 (17), mycoviruses infecting F. asiaticum were rarely characterized.
Negative-sense RNA viruses (NSRVs) are widely distributed in vertebrates, invertebrates, and plants. Moreover, they are divided into two subphyla, Haploviricotina and Polyploviricotina, according to the reconstruction of the evolution of the RNA virus (18, 19). In fungi, most reported NSRVs, including the well-characterized Sclerotinia sclerotiorum negative-sense RNA virus 1 (20, 21), belong to the subphylum Haploviricotina, most of which are monosegmented. Another clade of NSRVs is the subphylum Polyploviricotina, most of which accommodates segmented negative-sense RNA viruses (SNSRVs), including bunyaviruses (19). Bunyaviruses are a diverse group of pathogens that cause diseases in plants and animals (22, 23). Now, the order Bunyavirales has expanded to comprise 14 families and 60 genera based on the recent International Committee on Taxonomy of Viruses (ICTV) release (24, 25). In contrast to nonsegmented negative-sense RNA viruses, bunyavirus diversity in fungi has been rarely explored, until recently with rapidly increasing viromic studies. Depending on the family and genus, viruses in Bunyavirales contain genome segments ranging from one to 10 for viruses in Tulasviridae and Fimoviridae (23, 26). The classic bunyaviruses have three negative-sense RNA segments, large (L), medium (M), and small (S), based on the segment length, which encode RdRp (L protein), enveloped glycoprotein (G), and nucleocapsid protein (N) (22). At least 36 fungal SNSRVs related to viruses in Bunyavirales have been reported, although most encode only RdRp (26–39). Similar results were reported in oomycetes, where all oomycete bunyaviruses were known to encode only RdRp (40–45). Although capsidless viruses in fungi are not rare, the genome of fungal bunyaviruses could be coated by nucleocapsid because six viruses at least encode nucleocapsid and other nonstructural proteins (26, 30, 31, 35–37). Lentinula edodes negative-strand RNA virus 2 and Entoleuca phenui-like virus 1 contain two segments encoding the RdRp, the nucleocapsid and putative movement protein (30, 31). Penicillium roseopurpureum negative-sense RNA virus 1 contains three segments encoding RdRp, nucleocapsids, and nonstructural proteins (37). The recently reported Botrytis cinerea bocivirus 1 has three segments encoding RdRp, movement protein-like protein, and nucleocapsid (35). Sutela et al. found a monosegmented bunyavirus in which the RdRp and nucleocapsid are located in the same segment (26). However, none of the known fungal bunyaviruses has been reported to encode a glycoprotein.
In the present study, the diversity of mycoviruses was investigated in phytopathogenic fungus F. asiaticum via metatranscriptomic techniques, and 16 complete or incomplete mycoviral genomes were reconstructed. A novel segmented negative-sense RNA virus, tentatively named Fusarium asiaticum mycobunyavirus 1 (FaMBV1), has been molecularly characterized. FaMBV1 has three negative-sense RNA segments coding for RdRp, glycoprotein, and nucleocapsid and shares lower similarity with phenuiviruses. Phylogenetic analysis showed that FaMBV1 might represent a new viral lineage within the Bunyavirales. FaMBV1 has lower efficiency of transmission by asexual spores, and the M segment seems dispensable for the replication of FaMBV1 in F. asiaticum. Moreover, we identified the M and S segments of Alternaria tenuissima negative-stranded RNA virus 2 (AtNSRV2) and Sclerotinia sclerotiorum negative-stranded RNA virus 5 (SsNSRV5) using a bioinformatics approach, which revealed hidden segments of multisegmented viruses. Studies related to fungal NSRVs will undoubtedly help refine hypotheses about the evolutionary origins of different fungal viruses. Moreover, defining the fungal negative-sense RNA virosphere is an interesting area for future research.
RESULTS
Metatranscriptomic screening of F. asiaticum isolates for virus infection.
To explore the mycovirus diversity and viral candidates for biocontrol agents of FHB disease, 397 wheat spikes with typical symptoms of FHB disease but lower severity were collected from three districts in the Yangtze Valley Plain, and 350 strains were isolated. Most purified strains were identified as F. asiaticum using species-specific primers Fg16F/R, as previously described (46, 47). Total RNA was extracted from 350 strains of F. asiaticum, mixed with an equal quantity, and subjected to RNA sequencing. Seventeen mycoviruses were discovered by de novo transcript assembly and annotation and verified using virus-specific primers (Table 1). Based on the replication-associated sequence analysis, 12 mycoviruses were positioned in Lenarviricota, eight mitoviruses (Fusarium asiaticum mitovirus 1 to 8) of which were first discovered in Fusarium spp. Fusarium poae mitovirus 1 that was previously reported to infect Fusarium poae was found to infect F. asiaticum (28). One narnavirus, Fusarium asiaticum narnavirus 1, had two RNA segments and was clustered with a group of previously reported bisegmented narnaviruses or ORFans (open reading frames [ORFs] with no detectable homology to other ORFs in a database) (33, 40). Two new botourmiaviruses, Fusarium asiaticum ourmiavirus 1 (FaOV1) and FaOV2, were first reported in Fusarium spp. FaOV2 is highly similar to Erysiphe necator-associated narnavirus 51 (Table 1), which forms a new clade “Narliviridae”, including plant ourmiaviruses and some narna-like viruses (Fig. 1), as recently proposed (48). Fusarium graminearum hypovirus 2 (FgHV2) was identified in F. asiaticum with 95% amino acid (aa) identity to FgHV2/JS16, as previously reported by Li et al. (49). Fusarium asiaticum fusarivirus 1 was highly similar to Fusarium graminearum double-stranded RNA (dsRNA) mycovirus 1 (50). To date, only three NSRVs infecting Fusarium spp. have been reported, including Fusarium poae negative-stranded RNA virus 1 (accession no. LC150618) (28), Fusarium poae negative-stranded RNA virus 2 (accession no. LC150619) (25), and Fusarium graminearum negative-stranded RNA virus 1 (accession no. MF276904) (51). In the present study, we identified three other NSRVs, including two mymonaviruses and one phenuivirus. The two mymonaviruses, Fusarium asiaticum negative-stranded RNA virus 1 (FaNSRV1) and Fusarium graminearum negative-stranded RNA virus 1, shared 82.69% and 98.51% identities, respectively, with soybean leaf-associated negative-stranded RNA virus 1 (accession no. KT598225) (Table 1). The putative phenuivirus with a trisegmented RNA genome was characterized, and its features are described in detail below.
TABLE 1.
Summary of the viruses detected in F. asiaticum isolates
| Tentative virus name | Genome | Length (nt) | Depth of coverage (fold) | Accession no. | Main gene product | Best hit in blastp | Hit accession no. | Query coverage (%) | E value | Identity (%) |
|---|---|---|---|---|---|---|---|---|---|---|
| Fusarium asiaticum mitovirus 1 | RNA | 2,629 | 2,989 | MZ969051 | RdRp | Plasmopara viticola lesion-associated mitovirus 13 | QIR30236.1 | 100 | 0 | 93.94 |
| Fusarium asiaticum mitovirus 2 | RNA | 2,636 | 431 | MZ969052 | RdRp | Soybean leaf-associated mitovirus 5 | ALM62240.1 | 97 | 0 | 93.36 |
| Fusarium asiaticum mitovirus 3 | RNA | 2,516 | 66 | MZ969053 | RdRp | Erysiphe necator-associated mitovirus 6 | QHD64817.1 | 99 | 0 | 74.08 |
| Fusarium asiaticum mitovirus 4 | RNA | 2,251 | 37 | MZ969054 | RdRp | Erysiphe necator-associated mitovirus 22 | QKI79970.1 | 99 | 0 | 89.1 |
| Fusarium asiaticum mitovirus 5 | RNA | 2,239 | 54 | MZ969055 | RdRp | Erysiphe necator-associated mitovirus 22 | QKI79970.1 | 99 | 0 | 90.87 |
| Fusarium asiaticum mitovirus 6 | RNA | 2,251 | 56 | MZ969056 | RdRp | Erysiphe necator-associated mitovirus 22 | QKI79970.1 | 99 | 0 | 89.54 |
| Fusarium asiaticum mitovirus 7 | RNA | 2,992 | 112 | MZ969057 | RdRp | Ophiostoma mitovirus 7 | AGT55877.1 | 98 | 6.00E−148 | 37.35 |
| Fusarium asiaticum mitovirus 8 | RNA | 3,038 | 2,334 | MZ969058 | RdRp | Grapevine-associated mitovirus 13 | QXN75366.1 | 97 | 3.00E−152 | 39.41 |
| Fusarium poae mitovirus 1 | RNA | 2,395 | 140 | MZ969059 | RdRp | Fusarium poae mitovirus 1 | YP_009272898.1 | 100 | 0 | 95.59 |
| Fusarium asiaticum narnavirus 1 | RNA1 | 3,051 | 819 | MZ969060 | RdRp | Plasmopara viticola lesion-associated orfanplasmovirus 5 | QNQ74067.1 | 100 | 0 | 60.47 |
| RNA2 | 2,478 | 5,154 | MZ969061 | Hypothetical protein | Insect narna-like virus 1 | QXP45024.1 | 14 | 2.00E−05 | 29.57 | |
| Fusarium asiaticum ourmiavirus 1 | RNA | 2,528 | 366 | MZ969062 | RdRp | Plasmopara viticola lesion-associated ourmia-like virus 4 | QGY72534.1 | 88 | 7.00E−153 | 45.37 |
| Fusarium asiaticum ourmiavirus 2 | RNA | 2,576 | 32 | MZ969063 | RdRp | Erysiphe necator-associated narnavirus 51 | QJT93783.1 | 99 | 0 | 97.28 |
| Fusarium graminearum hypovirus 2 | RNA | 12,809 | 164 | MZ969064 | Polyprotein | Fusarium graminearum hypovirus 2 | YP_009130646.1 | 100 | 0 | 95.92 |
| Fusarium asiaticum fusarivirus 1 | RNA | 6,612 | 318 | MZ969065 | RdRp | ORF1 (Fusarium graminearum dsRNA mycovirus 1) | QKX95871.1 | 100 | 0 | 98.65 |
| Fusarium asiaticum negative-stranded RNA virus 1 | RNA | 9,055 | 208 | MZ969066 | RdRp | Soybean leaf-associated negative-stranded RNA virus 1 | ALM62220.1 | 100 | 0 | 82.69 |
| Fusarium graminearum negative-stranded RNA virus 1 | RNA | 9,062 | 64 | MZ969067 | RdRp | Soybean leaf-associated negative-stranded RNA virus 1 | ALM62220.1 | 100 | 0 | 98.51 |
| Fusarium asiaticum mycobunyavirus 1 | L | 6,450 | 21 | MZ969068 | RdRp | Alternaria tenuissima negative-stranded RNA virus 2 | QDB75016.1 | 88 | 0 | 33.65 |
| M | 2,631 | 367 | MZ969069 | Glycoprotein | No hits | |||||
| S | 1,409 | 22 | MZ969070 | Nucleocapsid | Icoaraci virus | ABO65056.1 | 36 | 4.00E−10 | 31.73 |
FIG 1.
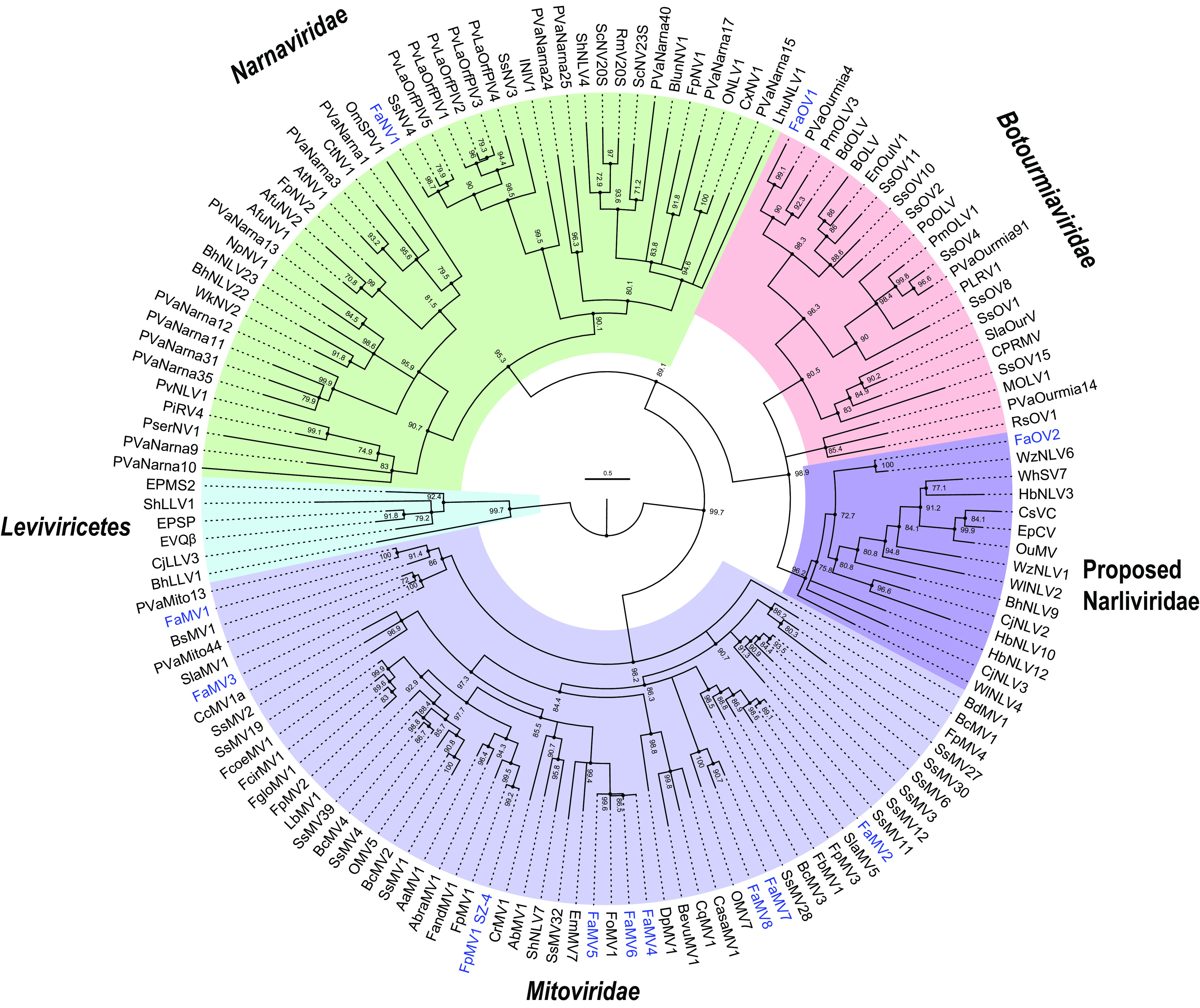
Phylogenetic analysis of viruses in phylum Lenarviricota detected in F. asiaticum isolates. Viruses identified in this study are in blue. Numbers at the branch nodes are ultrafast bootstrap support (percent) of 1,000 replicates. The full names and accession numbers of the viruses are listed in Table S1.
Discovery of a mycovirus infecting F. asiaticum related to phenuiviruses.
Two assembled virus contigs showed low similarities to the RdRp and nucleocapsid protein of viruses in Phenuiviridae (Table 1). We subsequently detected these two contigs in the strain XG-7 by reverse transcription-PCR (RT-PCR) with contig-specific primers. The full sequence was obtained using the RACE (rapid amplification of cDNA ends) method. The 5′ and 3′ termini of the virus segments were inverted complementary to each other, allowing the formation of a panhandle structure. Two inverted complementary sequences of 5′-ACACAA(G)AGAC-3′ at the 5′ terminus and 5′-CUUUGUGU-3′ at the 3′ terminus were found (Fig. 2B), which were identical to those of phleboviruses (Fig. 2C). To search for the putative viral segment coding for glycoproteins, we utilized the features of high-consensus terminal sequences in phleboviruses and the consensus terminal nucleotide sequences obtained in FaMBV1’s L and S segments (52) and designed a consensus primer (M-TF/TR, ctcagccctACACAAAGACC) to amplify an additional genomic segment as used by Zheng et al. (53). Two main bands were amplified and cloned for sequencing. The smaller band was the same as the contig coding for the nucleocapsid protein. The large band showed no hits using blastp against the NR database, and we found it in the assembled contigs with high coverage from the library. Furthermore, the M segment was not amplified from other F. asiaticum strain genomes or cDNA, which confirmed that the M segment existed only in XG-7. Because the terminus of the M segment shared consensus with the L and S segments, we hypothesized that the phenui-like virus, named FaMBV1, had three segments termed L, M, and S segments (Fig. 2A). The complete sequence of the L segment comprises 6,468 nucleotides (nt) with 38.7% GC content and has a single large open reading frame (ORF) in the antisense strand (viral cRNA). This large ORF is predicted to encode a 2,094-aa protein that shares 33% identity with replicases of AtNSRV2 (E value, 0; query coverage, 94%) by blastp search (Table 1). Two conserved domains, the DUF3370 domain and Bunya_RdRp domain (pfam04196), were found in the large ORF using the NCBI conserved domain search. Multiple sequence alignments with phenuiviruses revealed four conserved motifs (H…PD…D/E…K) at the N terminus of the L protein (Fig. 3), which is a typical feature of the endonuclease domain of RdRp in bunyavirus (54, 55). The predicted endonuclease domain implies that FaMBV1 may employ a cap-snatching process for viral mRNA transcription (56). Following the endonuclease domain, eight conserved motifs (GFAHBCDE) with high conservation with phleboviruses and tenuiviruses were found (Fig. 2A and Fig. 3). Furthermore, these were considered to be the RdRp active motifs (54, 57). Thus, the L protein is responsible for RdRp function in FaMBV1.
FIG 2.
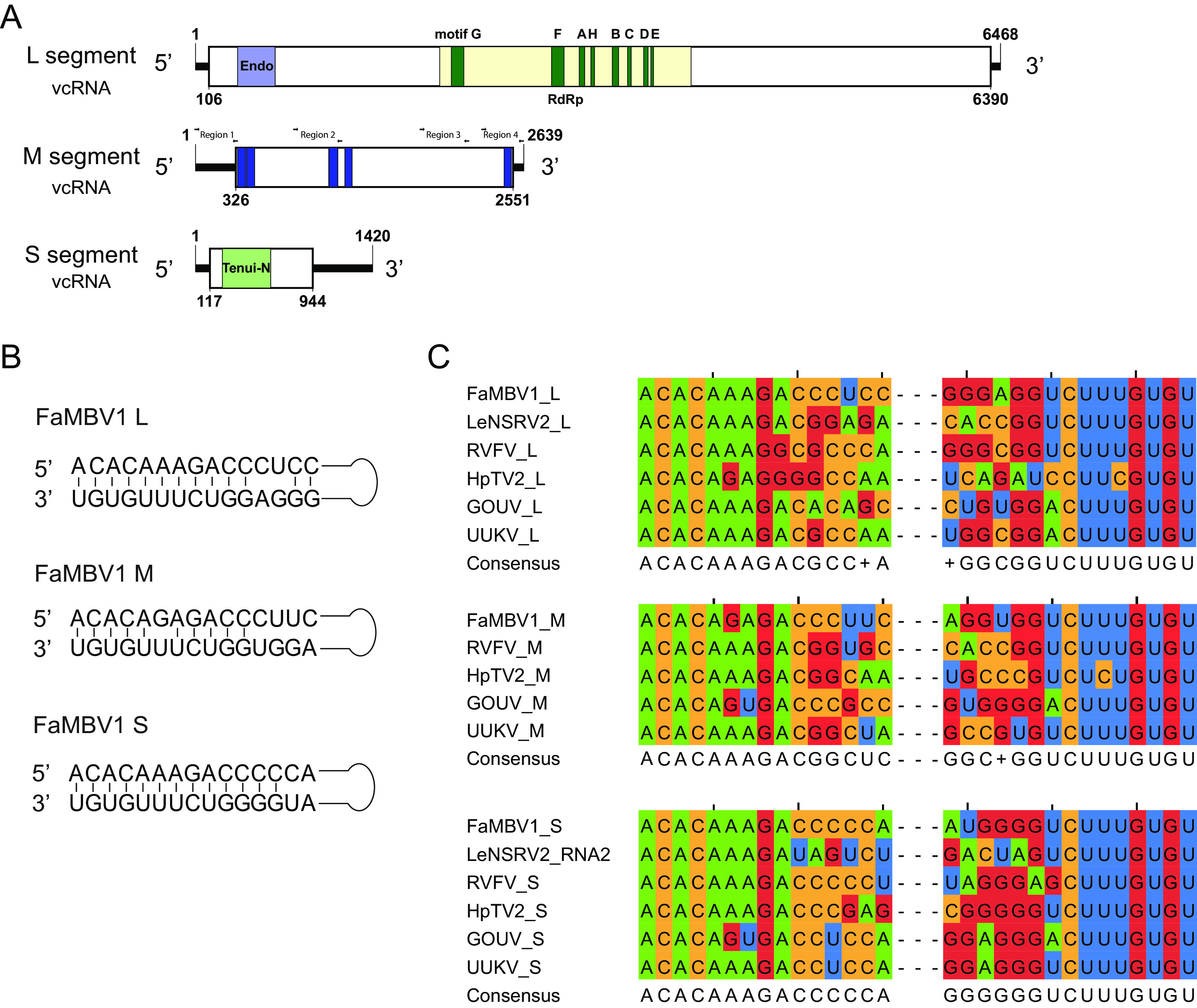
Schematic diagram of the FaMBV1 genome and features. (A) Schematic diagram of the FaMBV1 genome. vRNA, viral RNA strand; vcRNA, viral RNA complementary strand. Open reading frames are represented by rectangles. Conserved domains are presented in an acid green box. Four different regions with primers used for M segment detection by PCR are shown. (B) Panhandle structures are formed by the 5′ and 3′ termini of FaMBV1 L, M, and S segments. (C) Comparison of FaMBV1 L, M, and S segment terminal nucleotide sequences with other bunyaviruses; consensus sequences are presented. LeNSRV2, Lentinula edodes negative-stranded RNA virus 2; RVFV, Rift Valley fever virus; HpTV2, Huángpí tick virus 2; GOUV, Gouléako virus; UUKV, Uukuniemi virus.
FIG 3.
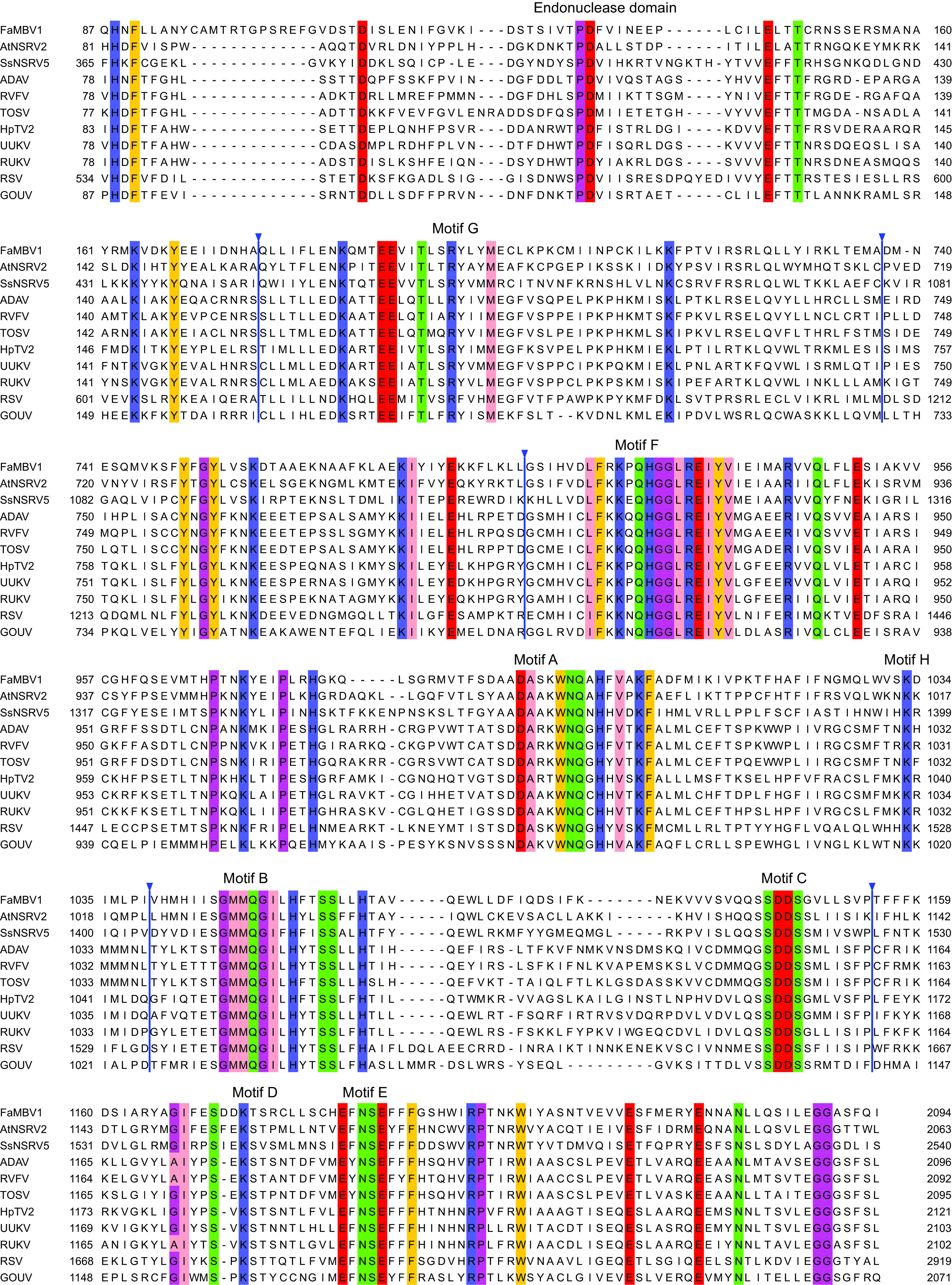
Alignment of L protein of FaMBV1 and phenuivirus revealed endonuclease and RNA-dependent RNA polymerase domains. The alignment was generated using MAFFT as described in Materials and Methods. Ambiguously aligned and gapped regions are hidden and marked with an inverted triangle. Conserved amino acids with 50% conservation are colored in Zappo color mode in Jalview. ADAV, Adana virus (AJK91618.1); RVFV, Rift Valley fever virus (ABD51507.1); TOSV, Toscana virus (CAA48478.1); HpTV2, Huángpí tick virus 2 (AJG39238.1); UUKV, Uukuniemi virus (BAA01590.1); RUKV, Rukutama virus (AII79372.1); RSV, rice stripe tenuivirus (BAA06677.1); GOUV, Gouléako virus (AEJ38175.1).
To link the new FaMBV1 to the known diversity of bunyaviruses, the core RdRp protein sequences of FaMBV1 and the representative members of Bunyavirales were selected to construct a phylogenetic tree. Phylogenetic analysis suggested that FaMBV1 is clustered with two other fungal NSRVs, AtNSRV2 and SsNSRV5, and forms a fungus-related bunyavirus group. Furthermore, this bunyavirus group is related to the Phenuiviridae groups but formed an independent phylogenetic branch (Fig. 4), suggesting that a new Mycobunyavirus genus in the family Phenuiviridae should be proposed to accommodate these mycoviruses.
FIG 4.
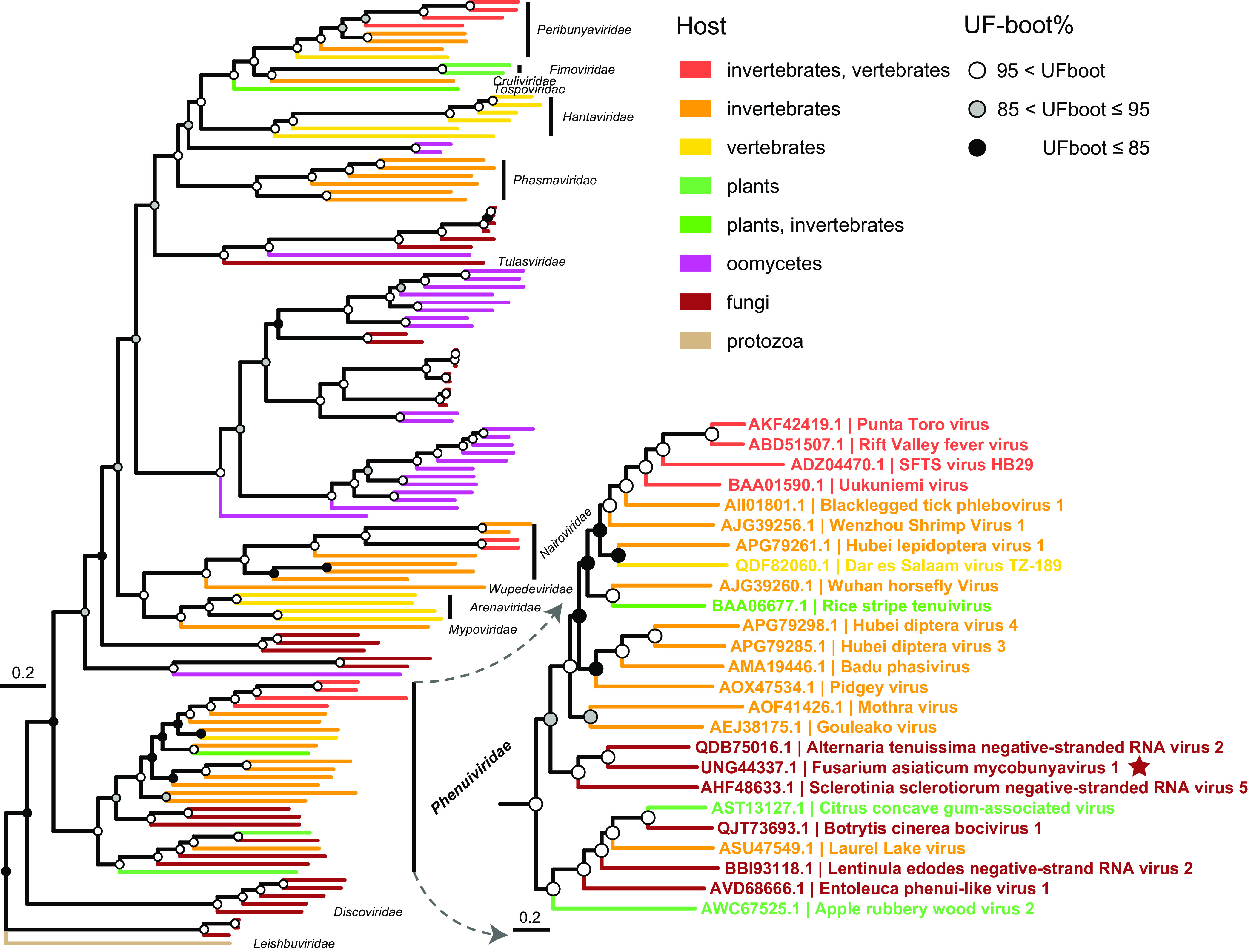
Phylogenetic position of FaMBV1 within the order Bunyavirales. L segments encoding the L protein were aligned and trimmed as described in Materials and Methods. Maximum-likelihood tree and the root were inferred and evaluated using IQ-TREE with the NONREV+FO substitution model and –root-test (95, 100). FaMBV1 is marked with a red star. The names of mycoviruses are marked orange and magenta. Circles at the branch nodes are ultrafast bootstrap support (percent) value. The tree was visualized by ggtree (101, 102), and the enlarged tree shows the Phenuiviridae.
The FaMBV1 M segment encoded a 741-amino-acid protein with no hits found using blastp. However, the glycoproteins of the rice stripe virus and Rift Valley fever virus shared homology with the M protein using HHpred tools (Fig. 5A). Unlike the glycoprotein in Phenuiviridae, no signal peptide was predicted using SignalP-5.0 in the M protein amino terminus. The M protein was predicted to contain four transmembrane helices by the TMHMM-2.0 server (Fig. 5B). The arrangement of the first two transmembrane helices located at the N terminus was similar to that of the large signal peptide in the Lassa virus, which was cleaved by signal peptidases after the second transmembrane helix (58). A hydrophobic region was predicted after the third transmembrane helix, which may function as a signal peptide (59). Host peptidase may cleave this protein into two parts, like the glycoproteins Gn and Gc (59). Four potential N-glycosylation sites were predicted in the M protein using NetNGlyc 1.0 (Fig. 5C). These results revealed that the M segment-encoded proteins might be glycoproteins related to bunyaviruses.
FIG 5.
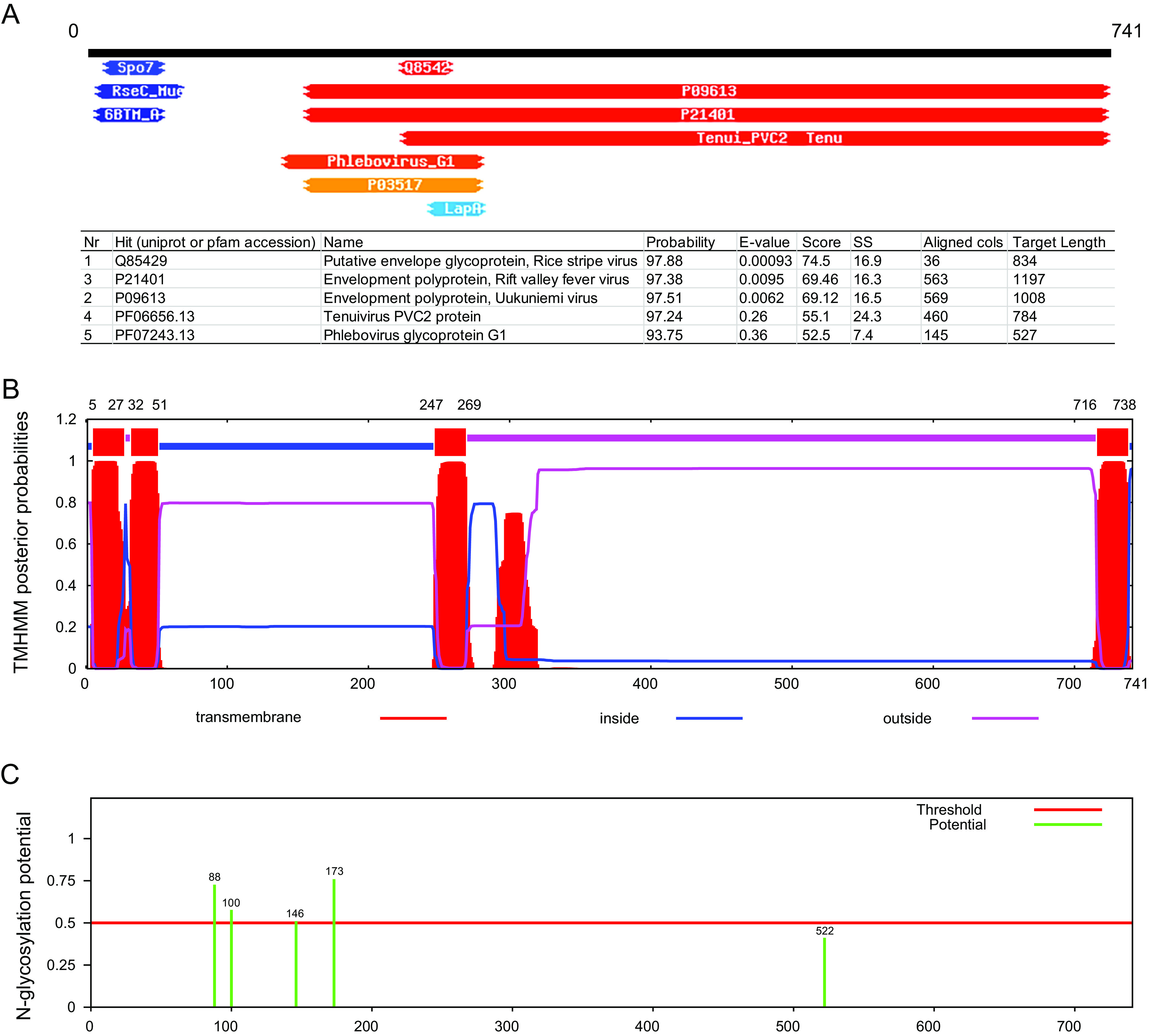
FaMBV1 M protein is a putative glycoprotein. (A) Homology detection and structure prediction of FaMBV1 M protein using HHpred (90). The top five hits are listed in the table. (B) TMHMM detection of the transmembrane helix in FaMBV1 M protein. The position of the transmembrane helix is on top. (C) FaMBV1 M protein N-glycosylation site prediction using NetNGlyc 1.0. The putative N-glycosylation sites are labeled in green.
The S segment of FaMBV1 was 1,420 nt long and had a GC content of 38.7%. A single ORF was predicted in the antisense segment and encoded a 275-aa nucleocapsid protein containing a Tenui_N domain (pfam05733). Using blastp comparison, it was shown that the nucleocapsid protein is similar, with a lower identity of 29.17% and 28.12%, to the Cocle virus (E value, 3e−04; query coverage, 34%) and Punta Toro virus (E value, 0.001; query coverage, 34%), respectively, belonging to Phenuiviridae.
Transmission of FaMBV1 and its roles in the biological features of F. asiaticum.
To study the biological effect of FaMBV1 on F. asiaticum, the virome of the strain XG-7 was analyzed by transcriptome sequencing (RNA-seq). Combined with RT-PCR results, XG-7 is infected by FaMBV1 alone. To eliminate FaMBV1 from strain XG-7, 42 single conidial isolates were obtained. FaMBV1 was detected via RT-PCR using FaMBV1-specific primers, and the results suggested that two conidium isolates harbored FaMBV1. Furthermore, the M segment was lost in these two conidial progenies. These results indicated that FaMBV1 has low vertical transmission efficiency (2/42) in asexual spores. To explore the effect of FaMBV1 on F. asiaticum, the two conidial progeny strains, XG-7-13 and XG-7-15, were selected for further study. Compared with the strain XG-7, the strain XG-7-13 was infected with the M segment-free isolate of FaMBV1 (FaMBV1M-free), and strain XG-7-15 was FaMBV1 free, according to the RT-PCR results (Fig. 6A). FaMBV1- and FaMBV1M-free-infected strains showed normal colony morphology (Fig. 6B). FaMBV1-infected strain XG-7 showed slightly slower growth than the virus-free strain XG-7-15. The FaMBV1M-free-infected strain XG-7-13 grew slightly faster but was not statistically different from the XG-7 strain (Fig. 6B). As FaMBV1 was mostly lost in most conidial progeny, mycelium plug inoculation experiments on corn silk and wheat head showed that FaMBV1 did not affect the pathogenicity of the F. asiaticum strain (Fig. 6C and D). On the further assessment of the impact of FaMBV1 on F. asiaticum strains, we observed no difference in the sporulation, the number of conidial septa, and the length of conidia of the strains in the mung bean soup medium (Fig. 6E to H). We found that FaMBV1 showed cryptic infection in F. asiaticum and was easily lost in the conidial progeny.
FIG 6.
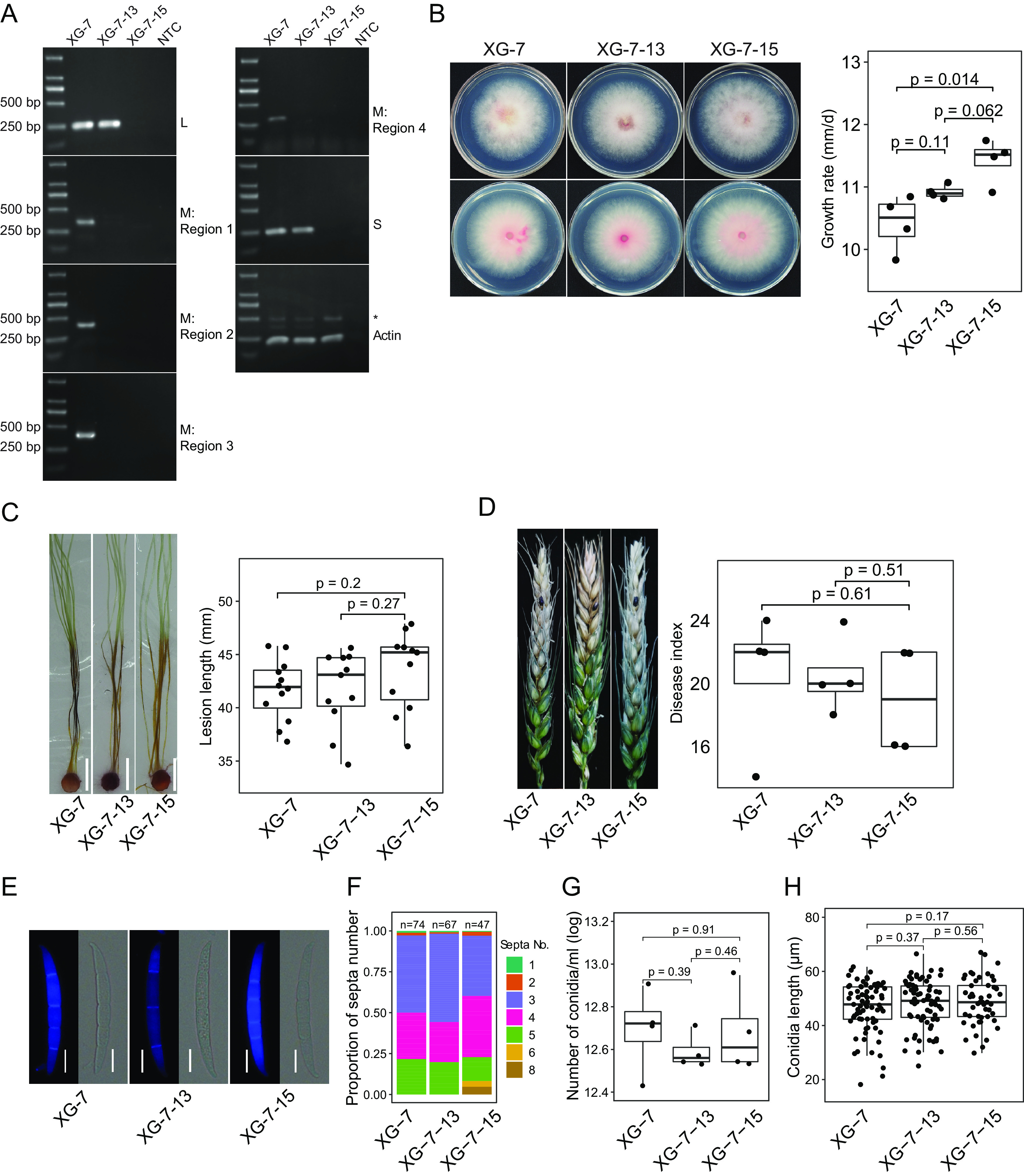
Vertical transmission of FaMBV1 and its effects on biological features of F. asiaticum. (A) RT-PCR detection of FaMBV1 L, M, and S segments in asexual spore progeny with virus-specific primers. Four different regions, as shown in Fig. 2A, were included for the detection of the M segment in XG-7-13 to exclude negative RT-PCR results from small deletions. The actin gene was used as a control. NTC, no-template control. A larger band (*) above the actin band was amplified from the genomic DNA. (B) (Left) Colony morphology after 3 days of incubation at 28°C. XG-7-13 was infected with L and S segments of FaMBV1. XG-7-15 was free of FaMBV1. The front and reverse sides of the colony are presented. (Right) Boxplot showing the growth rates of XG-7, XG-7-13, and XG-7-15. Student’s t test was used for the statistical test. (C) (Left) Corn silks inoculated with the mycelium plugs of strains XG-7, XG-7-13, and XG-7-15 were examined after 5 days. White bar, 1 cm. (Right) Lesion length of XG-7, XG-7-13, and XG-7-15 inoculated on the corn silk. P values were derived from the Student t test. (D) (Left) Representative picture of wheat heads of cultivar XiaoYan 22 inoculated with mycelium plugs. (Right) Boxplot showing disease index measured at 14 days post-inoculation (dpi). P values were derived from the Student t test. (E) Representative conidia of the strains stained using calcofluor white for septa. Bars, 10 μm. (F) The proportion of the septum number is shown in the bar plot, and total conidial number is indicated on the top of the bar. (G) Conidial production in mung bean soup medium. The conidial numbers were transformed with logarithms for the Student t test. (H) Boxplot shows the conidium length. Significance analysis was performed using the Student t test.
Missed bunyavirus segments discovered using newly found viral proteins as bait.
FaMBV1 was found to be distantly related to two mycobunyaviruses, AtNSRV2 and SsNSRV5. We hypothesized that the two viruses would also have trisegmented genomes. To search for hidden virus segments, we analyzed the library F2 (SRR8303986), which contained the AtNSRV2 genome sequence, and the library Fungal_2011 (SRR3482281), which contained the SsNSRV5 genome sequence. For SsNSRV5, an assembled contig, named the SsNSRV5 M segment, is 2,770 nt long with a putative ORF coding for 850 amino acids. A signal peptide was predicted at the N terminus of the SsNSRV5 M segment using SignalP 6.0 (60). Moreover, two transmembrane helices spanning residues 292–314 and 823–846 were predicted using TOPCONS (Fig. 7A). For AtNSRV2, a discovered contig, named the AtNSRV2 M segment, is 2,978 nt long with a putative ORF coding for 868 amino acids. No signal peptide was predicted at the N terminus of the AtNSRV2 M segment. However, two consecutive transmembrane helices lying at residues 70–90 and 96–115 were predicted to be similar to the large signal peptide of FaMBV1 glycoprotein. Four additional transmembrane domains were predicted, in addition to the large signal peptide of the AtNSRV2 M segment (Fig. 7A). HHpred results indicated that the SsNSRV5 M and AtNSRV2 M segments shared similarities with the glycoproteins of phenuiviruses. Based on the domain organization of phlebovirus and multiple sequence alignment, three Pfam domains, Phlebovirus_G1, Phlebovirus_G2, and Phlebo_G2_C, are outlined (Fig. 7A and B). For the nucleocapsid, two contigs were found, which were arbitrarily named the SsNSRV5 S segment and AtNSRV2 S segment. The SsNSRV5 S segment is 1,214 nt long with a putative 282-aa ORF coding for the nucleocapsid protein. The AtNSRV2 S segment is 1,214 nt long with a putative 285-aa ORF coding for nucleocapsid protein. These nucleocapsids share a small but conserved site with the Phenuiviridae nucleocapsid (Fig. 8A). Since capsid proteins are usually more similar in three-dimensional (3D) structures than in primary sequences, the FaMBV1 nucleocapsid structure was predicted using the I-TASSER server (61–63). The best model predicted using I-TASSER was found to have a C score of −0.50, the estimated TM score of 0.65 ± 0.13, and a root mean square deviation (RMSD) of 7.1 ± 4.2 Å (Fig. 8B). This model was predicted to bind nucleic acids using consensus approach (COACH), indicating its viral RNA strand-binding property. FaMBV1 nucleocapsid shares a similar structure to the known phlebovirus Rift Valley fever virus nucleocapsid protein (PDB 4h5o), with an N-terminal arm involved in multimerization and an RNA binding slot (Fig. 8B). Tyr37 and Phe40 were involved in base stacking with RNA, and Asp72 and Arg118 involved in stabilizing the phosphate backbone were the most conserved (Fig. 8A and B). The three nucleocapsid proteins of mycobunyavirus clustered together with the nucleocapsid protein of apple rubbery wood virus 2 (Fig. 8C). Notably, SsNSRV5 nucleocapsid showed a low bootstrap rate, and more related viruses are needed to clarify the evolutionary relationship. We identified the M and S segments of AtNSRV2 and SsNSRV5 based on FaMBV1.
FIG 7.
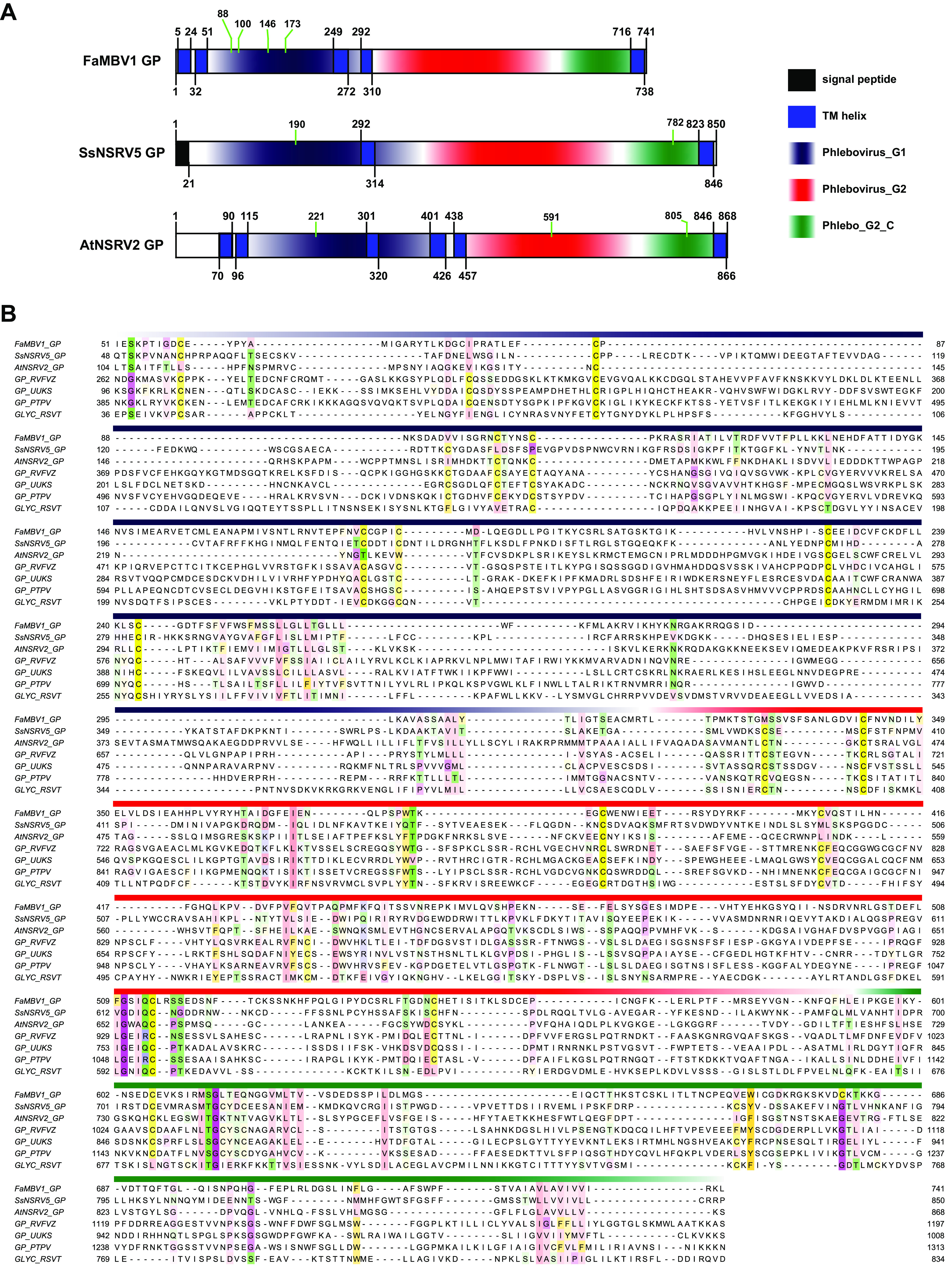
The features of FaMBV1, AtNSRV2, and SsNSRV5 glycoproteins (GP). (A) Glycoprotein domain structures of FaMBV1, AtNSRV2, and SsNSRV5. Transmembrane domains were predicted using TOPCONS servers according to PolyPhobious results (89). The putative N-glycosylation site is indicated by a green line. (B) Alignment of the glycoprotein with domain is presented with the same color bar as in panel A. Conservation sites above 30% are highlighted in Zappo color mode in Jalview. GP_RVFVZ, glycoprotein of Rift Valley fever virus (strain ZH-548 M12); GP_UUKS, glycoprotein of Uukuniemi virus (strain S23); GP_PTPV, glycoprotein of Punta Toro phlebovirus; GLYC_RSVT, glycoprotein of rice stripe virus (isolate T).
FIG 8.
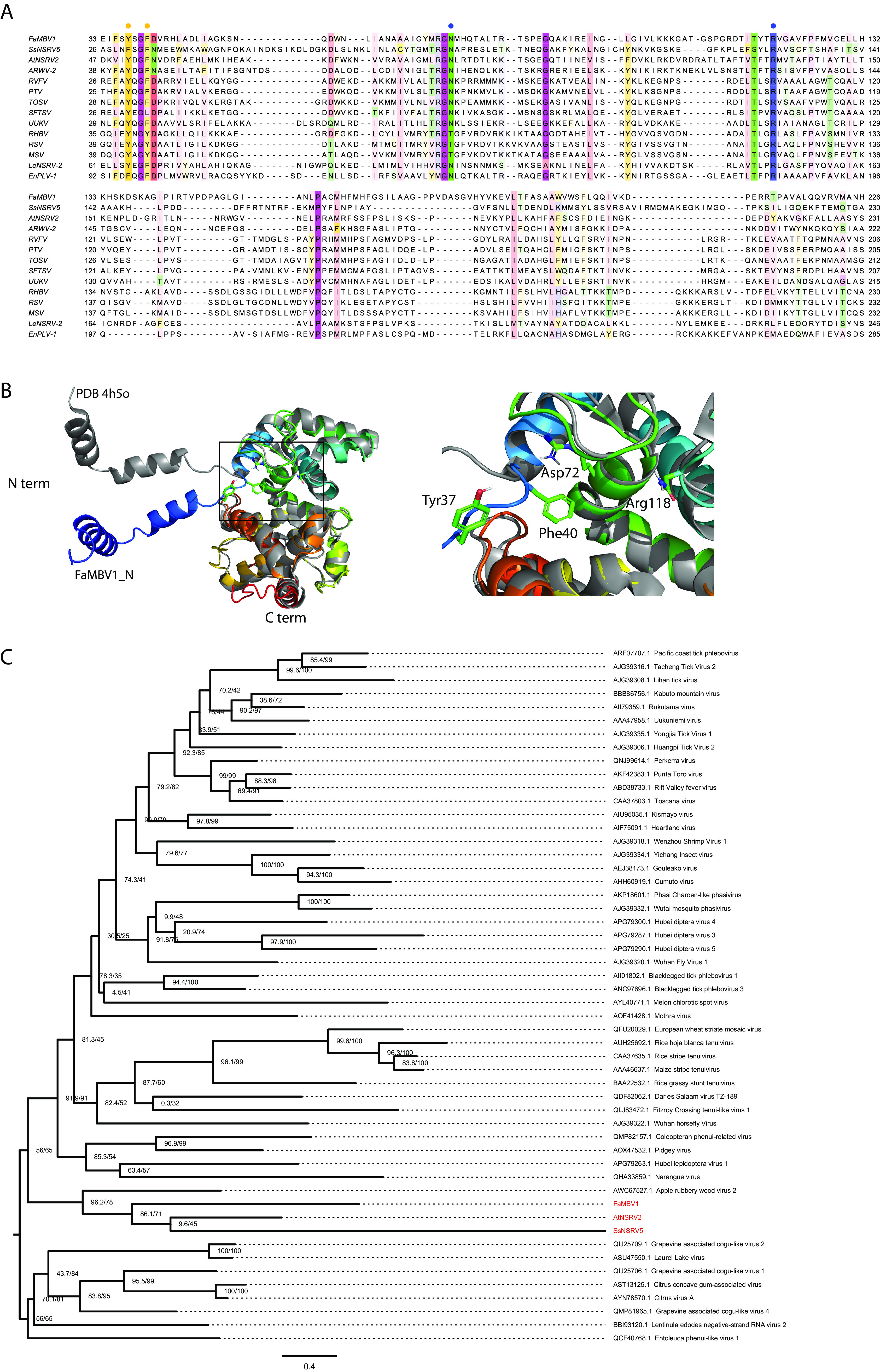
The alignment and phylogenetic analysis of FaMBV1, AtNSRV2, and SsNSRV5 nucleocapsids (N). (A) Alignment of the nucleocapsids of FaMBV1, AtNSRV2, and SsNSRV5 with the related phenuiviruses. The orange circles indicate the sites involved in base stacking. The blue circles indicate the sites involved in the stabilization of the phosphate backbone. (B) The left panel shows the 3D structure alignment of FaMBV1 and Rift Valley fever virus nucleocapsids using PyMOL. The right panel displays the conserved sites noted in (A), which are shown as sticks in a closeup view. N term, N terminus; C term, C terminus. (C) Phylogenetic tree of Phenuiviridae nucleocapsids. S segments encoding nucleocapsids were aligned and trimmed as described in Materials and Methods. The maximum-likelihood tree was inferred using IQ-TREE (95). The model Q.pfam+F+R7 was chosen according to the Bayesian information criterion. FaMBV1, AtNSRV2, and SsNSRV5 are highlighted in red. Numbers at the branch nodes are SH-aLRT support (percent)/ultrafast bootstrap support (percent) value.
DISCUSSION
In this study, we identified the virome of 350 F. asiaticum strains isolated from wheat spikes with typical wheat scab symptoms. Seventeen viruses have been identified, belonging to the families Mitoviridae, Narnaviridae, Botourmiaviridae, the proposed Narliviridae, Hypoviridae, Fusariviridae, Mymonaviridae, and Phenuiviridae. FaMBV1 is a new trisegmented negative-strand RNA virus, putatively belonging to a new genus in Phenuiviridae. The M segment coding for glycoprotein seems not to be required for FaMBV1 replication in F. asiaticum. In addition, the M and S segments of AtNSRV2 and SsNSRV5 were discovered, implying the existence of glycoprotein-encoding trisegmented NSRVs in fungi.
Through our metaviromic study of F. asiaticum, we identified 17 viruses from 350 strains. At least nine virus lineages were first discovered in Fusarium spp. Compared with other metaviromic studies of pathogenic fungi, our study revealed that the virome of F. asiaticum shows low diversity (33–35, 39, 64, 65). One possible reason was that F. asiaticum and F. graminearum may harbor low virus diversity, with about 2% virus incidence in F. graminearum, lower than the 14% to 20% in Rosellinia necatrix (65, 66). Subjectively selected nonnormal strains or dsRNA-infecting strains for sequencing may contribute to uncovering more mycoviruses. Interestingly, in some Fusarium genera, a single strain of F. poae (MAFF 240374) and Fusarium mangiferae isolate SP1 harbor 16 and 11 viruses, respectively (13, 28). The reasons for these differences may be due to different lifestyles and ecological niches. For example, as important pathogens of wheat and other cereal crops in the agroecological system, F. asiaticum and F. graminearum occupy a unique niche and are more aggressive than F. poae (67). Perhaps the spore-producing aggressive lifestyle made the virus content very low, as exemplified by FaMBV1 erased through asexual conidiation.
The NSRV was described in fungi 8 years ago (20). Moreover, SNSRVs were recently discovered using metagenomics analysis (30, 31, 35). Here, we reported a novel SNSRV, FaMBV1. Furthermore, each segment of it was found to have a single ORF encoding the L protein, putative glycoprotein, and nucleocapsid (Fig. 2A). Phylogenetic analysis of the RdRp domain of the L protein and nucleocapsid proteins demonstrated that FaBMV1 clustered with AtNSRV2 and SsNSRV5 within the family Phenuiviridae, which may represent a new genus.
We found that the FaMBV1 M segment was lost during regular subculture or conidial progeny, implying that glycoproteins are dispensable for FaMBV1 replication in F. asiaticum. Analogous observations have been made for envelope-deficient mutants of tomato spotted wilt virus generated from consecutive mechanical passages in plants (68, 69). Generally, the virion-forming glycoproteins of bunyaviruses are involved in viral entry and are dispensable for replication (70–72). It is conceivable that fungal bunyaviruses would lose genes coding for glycoproteins to adapt to intracellular life. For example, some plant viruses in the Coguvirus genus and some mycoviruses, including Entoleuca phenui-like virus 1, Botrytis cinerea bocivirus 1, and Lentinula edodes negative-stranded RNA virus 2, do not have genome segments coding for glycoproteins (30, 31, 35, 73, 74). However, there may be some transmission vectors for mycoviruses; for instance, Lycoriella ingenua can transmit Sclerotinia sclerotiorum hypovirulence-associated DNA virus 1 (75). Considering the unprecedented NSRV diversity in invertebrates (76, 77) and the interaction between Fusarium spp. and herbivorous arthropods in cereals (78), FaMBV1, which possesses the ability to encode a glycoprotein, is likely to be transmitted by invertebrate vectors. The virome of various invertebrates and vertebrates in wheat fields needs to be investigated to prove this conjecture in future research.
Bunyaviruses often attract attention for causing overt diseases. Although some bunyavirus-related mycoviruses have been reported, little is known about their effect on the host. In this study, we found that FaMBV1 did not affect F. asiaticum by comparing the biological features of XG-7 and the FaMBV1-free strain XG-7-15. This is not surprising, since only a few attenuated viruses have been found in the pathogenic fungi of FHB. FaMBV1 was mostly lost in the conidial progeny and subculture, implying its low adaptation to F. asiaticum. Moreover, further studies are required to find out whether any aggressive fungal bunyaviruses can be used for biocontrol.
Metagenomic sequencing has dramatically facilitated the discovery of viruses (79, 80). However, due to the high mutation rate and abundant diversity of RNA viruses, general annotation tools such as BLAST are not sufficient to detect some “dark” virus sequences and ORFans. Some attention has been recently paid to ORFans, which were often missed during regular virus mining. Moreover, these ORFans would be new viruses or other fragments of multisegment viruses (81, 82). Because AtNSRV2 and SsNSRV5 clustered with FaMBV1, we reanalyzed the published data and identified the M and S segments of the two viruses. The M and S segments showed little similarity to known viruses. Moreover, the glycoproteins showed domain organization and transmembrane structure similar to those of phleboviruses, implying that they may share similar functions in virion formation and virus replication. Considering that most phenuiviruses with glycoproteins are transmitted by arthropods, AtNSRV2, SsNSRV5, and FaMBV1 could hypothetically be similarly transmitted. The nucleocapsids of AtNSRV2, SsNSRV5, and FaMBV1 also showed low similarity to those of the known viruses. Furthermore, the FaMBV1 nucleocapsid shares a conserved 3D structure with the phlebovirus nucleocapsid protein. RNA-binding slots and ribonucleoprotein formation might be conserved among fungal and other animal phenuiviruses. Many fungal bunya-like viruses have been discovered, but only a few are multisegmented and no glycoprotein-encoding segment has been reported (26–28, 33–35, 39, 40). In this study, we characterized three glycoproteins from three fungal bunyaviruses, putatively being a new genus in Phenuiviridae. Future studies are needed to identify other segments and transmission vectors of mycobunyaviruses, which might provide new insights into fungal SNSRVs’ evolution and ecology. More fungal NSRVs will undoubtedly help to refine hypotheses about the evolutionary origins of different fungal viruses. Defining the fungal negative-sense RNA virosphere would be an interesting area for future research.
MATERIALS AND METHODS
Fungal strain isolation and culture conditions.
Wheat spikes with typical wheat scab symptoms were collected from fields in Hubei and Nanjing provinces, the two major wheat-producing regions in the Yangtze Plain of China (see Table S3 in the supplemental material). The candidate Fusarium strains were isolated from FHB-infected wheat via conidial isolation on potato dextrose agar (PDA) at 25 to 28°C and routinely subcultured for purification. After the strains were stable in colony morphology on PDA, they were stored on PDA slants at 4°C.
Nucleic acid extraction, sequencing, and de novo metatranscriptome assembly.
All the fungal strains were cultured on PDA laid with cellophane for 3 to 4 days. The mycelium was then harvested for nucleic acid extraction. Genomic DNA samples of all the purified strains were extracted using the cetyltrimethylammonium bromide (CTAB) method (83) and used for molecular identification with specific primers Fg16F/R (84).
Total RNA samples from all the Fusarium strains were obtained using the RNAiso Plus kit (TaKaRa Biomedical Technology, Beijing, China) and treated with DNase I to remove the DNA contamination. The total RNA samples were quantified using NanoDrop 2000, and the integrity was checked using Agilent Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA). The RNA-seq library was prepared after the rRNA depletion, and metatranscriptomic sequencing was performed using Illumina HiSeq X (Shanghai Biotechnology Corporation, Shanghai, China). To obtain clean sequences, the raw reads from deep sequencing were processed to remove the reads containing low-quality bases and eliminate adaptor contamination using Trimmomatic V0.32 (85). The read quality was checked using FastQC (v0.11.7) (https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). The clean reads were then de novo assembled into contigs using the SPAdes genome assembler in rnaSPAdes mode (v3.13.1) (86). The assembled contigs were translated using TransDecoder-v5.5.0 and annotated using DIAMOND blastp (v0.9.10) against the NCBI nonredundant (NCBI-nr) database (87). The putative viral contigs were manually filtered.
Searching additional virus segments for bunyavirus.
To search for other virus segments of a bunyavirus, SRA data were retrieved and FASTQ data were extracted using the NCBI SRA toolkit with accession no. SRR8303986 for AtNSRV2 and SRR3482281 for SsNSRV5 according to the published data of Nerva et al. and Marzano et al. (27, 39). The read assembly and contig annotation were performed as previously described. To search for glycoproteins, dark contigs with amino acid lengths between 600 and 900 were selected for transmembrane analysis by TMHMM (88) and TOPCONS (89). Furthermore, the transmembrane protein was sent for HHpred analysis to search for homologs with low similarity (90). To search for nucleocapsids, multiple sequence alignment of the nucleocapsid protein of FaMBV1 and related viruses was used for profile construction using hmmbuild. Subsequently, hmmsearch was used to search the customized nucleocapsid profile against the assembled contigs (http://hmmer.org/). The candidate contig was further confirmed using HHpred (90). TBtools were used for routine FASTA sequence manipulation (91).
Identification of the virus sequences derived from metatranscriptome data using RT-PCR.
Total RNA was isolated from the hyphae of individual strains and then transcribed to cDNA using reverse transcriptase (RTase) from Moloney murine leukemia virus (M-MLV) (RNase H−) according to the manufacturer’s instructions (TaKaRa Biomedical Technology, Beijing, China). Briefly, the reaction was performed in a final volume of 10 μL containing 2 μL of 5× M-MLV buffer, 0.5 μL deoxynucleoside triphosphate (dNTP) mixture (10 mM), 1 μL of RNA (100 ng to 1 μg), 0.25 μL RNase inhibitor (40 U/μL), 0.25 μL RTase from M-MLV (200 U/μL), and 6 μL of RNase-free water. The mixture was reacted with one cycle at 30°C for 10 min, one cycle at 42°C for 60 min, and one final cycle at 70°C for 15 min. Specific primers (Table S2) were designed to detect individual putative mycoviruses based on the viral contig sequences.
Determination of the terminal sequences of FaMBV1.
To obtain the 5′- and 3′-terminal sequences of a bunyavirus, the SMARTer RACE 5′/3′ kit (Clontech Laboratories, Inc.) was used according to the manufacturer’s protocol. The nested mycovirus-specific primers (Table S2) close to the end of the known sequence were designed and used for the amplification of the terminal sequence using universal primer mix provided in the kit. Briefly, to obtain 5′-terminal sequences, an additional RNA template was generated by the joint action of the SMARTer II A oligonucleotide and reverse transcriptase. Then cDNA was generated at 42°C for 90 min. The resulting cDNA was used as a template for nested PCR using universal primer mix and mycovirus-specific reverse primers (Table S2). To obtain 3′-terminal sequences, a polyadenylated tail was added to the viral RNA strand 3′ terminus using poly(A) polymerase (TaKaRa Biomedical Technology, Beijing, China). Furthermore, the first-strand 3′ cDNA was synthesized after incubation at 42°C for 90 min in an air incubator according to the manufacturer’s instructions. The cDNA was then amplified using mycovirus-specific reverse primers and the universal primer mix provided in the kit. The PCR products of the 3′/5′ terminus were separated on agarose gels for purification, ligated into the pMD18-T cloning vector, and subsequently sequenced. Alternatively, a terminally conserved primer was used to amplify the full-length cDNA, subsequently sequenced as previously described.
For a search for additional segments of FaMBV1, AC primer (ACACAAAGACC) was used for reverse transcription. Subsequently, primer M-TF/TR was used for PCR. The PCR products were sequenced as previously described. A classic RACE was employed to confirm the exact sequences of the M segment of FaMBV1, as reported previously (92).
Multiple sequence alignment and phylogenetic analysis.
The amino acid sequences of FaMBV1 and bunyaviruses retrieved from the NCBI virus database were aligned using MAFFT (version 7.243) with the E-INS-i option (93). The resulting alignment was visualized using Jalview software. For phylogenetic analysis, the aligned sequences were trimmed using trimAl with the -automated1 option (94). The trimmed multiple sequence alignment was used to infer a maximum-likelihood tree using IQ-TREE (95). The best-fit model was automatically selected using ModelFinder and chosen according to the Bayesian information criterion (96). After the tree search, an SH-aLRT test with 1,000 replicates and ultrafast bootstrapping with 10,000 samples were performed (95, 97). Finally, the tree was displayed using FigTree v1.44 (https://github.com/rambaut/figtree/releases).
Colony morphology observation, growth rate determination, and conidiation assay.
For vegetative growth, a mycelial plug (6 mm in diameter) of the individual strain was inoculated onto the center of a petri dish (9 cm in diameter) containing PDA. The plates were incubated at 25°C in complete darkness for fungal colony morphology observation. The mycelial growth was assessed every 24 h by measuring the diameter of the colony in two right-angled directions.
For the conidium production and morphology assay, the individual strain was cultured in mung bean soup medium for 7 days. Furthermore, the conidia were collected. The conidia were observed under a Nikon Eclipse 80i microscope. Moreover, the concentration of the conidia was measured using a hemocytometer. The septa were stained using calcofluor white stain (Sigma-Aldrich 18909) and examined under UV light.
A virulence assay was conducted on corn silk as described previously, with minor modifications (98). Fresh corn silk was bought and cut from the root to a length of 8 to 9 cm. The mycelium plugs were inoculated into the corn whisker incision. The corn silks were placed on a tray with wetted filter paper overlaid with plastic wrap and then incubated at 25 to 28°C under dark conditions for 7 days.
For the virulence assay on the wheat head, mycelium plugs were placed into the wheat spikelets at the poplar flowering stage. After spraying the wheat ear with sterile water, the wheat ear was covered with a sealable plastic food storage bag, and the bag was removed after 36 h. The disease index was reported as the number of symptomatic spikelets on both sides of the wheat ear at 14 days post-inoculation (99).
Statistical analyses were performed with the data from at least three independent biological replicates using the Student t test.
Data availability.
The detailed information on the research is available under the BioProject accession number PRJNA841045. Virus genomic sequences are under the accession numbers MZ969051 to MZ969070 in the GenBank database. Additional nucleotide sequence data reported for SsNSRV5 and AtNSRV2 are available in the Third Party Annotation (TPA) Section of the DDBJ/ENA/GenBank databases under the accession numbers TPA BK061361 to BK061364.
ACKNOWLEDGMENTS
This work was financially supported by the Fundamental Research Funds for the Central Universities (2021ZKPY005) and the Natural Science Foundation of China (32072475).
Footnotes
Supplemental material is available online only.
Contributor Information
Jiatao Xie, Email: jiataoxie@mail.hzau.edu.cn.
Colin R. Parrish, Cornell University
REFERENCES
- 1.van der Lee T, Zhang H, van Diepeningen A, Waalwijk C. 2015. Biogeography of Fusarium graminearum species complex and chemotypes: a review. Food Addit Contam Part A Chem Anal Control Expo Risk Assess 32:453–460. 10.1080/19440049.2014.984244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chen Y, Kistler HC, Ma Z. 2019. Fusarium graminearum trichothecene mycotoxins: biosynthesis, regulation, and management. Annu Rev Phytopathol 57:15–39. 10.1146/annurev-phyto-082718-100318. [DOI] [PubMed] [Google Scholar]
- 3.Mizutani Y, Uesaka K, Ota A, Calassanzio M, Ratti C, Suzuki T, Fujimori F, Chiba S. 2021. De novo sequencing of novel mycoviruses from Fusarium sambucinum: an attempt on direct RNA sequencing of viral dsRNAs. Front Microbiol 12:641484. 10.3389/fmicb.2021.641484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mahillon M, Decroës A, Caulier S, Tiendrebeogo A, Legrève A, Bragard C. 2021. Genomic and biological characterization of a novel partitivirus infecting Fusarium equiseti. Virus Res 297:198386. 10.1016/j.virusres.2021.198386. [DOI] [PubMed] [Google Scholar]
- 5.Khan HA, Sato Y, Kondo H, Jamal A, Bhatti MF, Suzuki N. 2021. A second capsidless hadakavirus strain with 10 positive-sense single-stranded RNA genomic segments from Fusarium nygamai. Arch Virol 166:2711–2722. 10.1007/s00705-021-05176-x. [DOI] [PubMed] [Google Scholar]
- 6.Yao Z, Zou C, Peng N, Zhu Y, Bao Y, Zhou Q, Wu Q, Chen B, Zhang M. 2020. Virome identification and characterization of Fusarium sacchari and F. andiyazi: causative agents of pokkah boeng disease in sugarcane. Front Microbiol 11:240. 10.3389/fmicb.2020.00240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mahillon M, Romay G, Liénard C, Legrève A, Bragard C. 2020. Description of a novel mycovirus in the phytopathogen Fusarium culmorum and a related EVE in the yeast Lipomyces starkeyi. Viruses 12:523. 10.3390/v12050523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jacquat AG, Theumer MG, Cañizares MC, Debat HJ, Iglesias J, García Pedrajas MD, Dambolena JS. 2020. A survey of mycoviral infection in Fusarium spp. isolated from maize and sorghum in Argentina identifies the first mycovirus from Fusarium verticillioides. Viruses 12:1161. 10.3390/v12101161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mahillon M, Decroës A, Liénard C, Bragard C, Legrève A. 2019. Full genome sequence of a new polymycovirus infecting Fusarium redolens. Arch Virol 164:2215–2219. 10.1007/s00705-019-04301-1. [DOI] [PubMed] [Google Scholar]
- 10.Li P, Bhattacharjee P, Wang S, Zhang L, Ahmed I, Guo L. 2019. Mycoviruses in Fusarium species: an update. Front Cell Infect Microbiol 9:257. 10.3389/fcimb.2019.00257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sato Y, Shamsi W, Jamal A, Bhatti MF, Kondo H, Suzuki N. 2020. Hadaka virus 1: a capsidless eleven-segmented positive-sense single-stranded RNA virus from a phytopathogenic fungus, Fusarium oxysporum. mBio 11:e00450-20. 10.1128/mBio.00450-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li P, Wang S, Zhang L, Qiu D, Zhou X, Guo L. 2020. A tripartite ssDNA mycovirus from a plant pathogenic fungus is infectious as cloned DNA and purified virions. Sci Adv 6:eaay9634. 10.1126/sciadv.aay9634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khan HA, Shamsi W, Jamal A, Javaied M, Sadiq M, Fatma T, Ahmed A, Arshad M, Waseem M, Babar S, Dogar MM, Virk N, Janjua HA, Kondo H, Suzuki N, Bhatti MF. 2021. Assessment of mycoviral diversity in Pakistani fungal isolates revealed infection by 11 novel viruses of a single strain of Fusarium mangiferae isolate SP1. J Gen Virol 102. 10.1099/jgv.0.001690. [DOI] [PubMed] [Google Scholar]
- 14.Zhao Y, Zhang Y, Wan X, She Y, Li M, Xi H, Xie J, Wen C. 2020. A novel ourmia-like mycovirus confers hypovirulence-associated traits on Fusarium oxysporum. Front Microbiol 11:569869. 10.3389/fmicb.2020.569869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xie Y, Wang Z, Li K, Liu D, Jia Y, Gao F, Dai J, Zhang S, Zhang X, Li H. 2022. A megabirnavirus alleviates the pathogenicity of Fusarium pseudograminearum to wheat. Phytopathology 112:1175–1184. 10.1094/PHYTO-03-21-0126-R. [DOI] [PubMed] [Google Scholar]
- 16.Wen C, Wan X, Zhang Y, Du H, Wei C, Zhong R, Zhang H, Shi Y, Xie J, Fu Y, Zhao Y. 2021. Molecular characterization of the first alternavirus identified in Fusarium oxysporum. Viruses 13:2026. 10.3390/v13102026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li W, Xia Y, Zhang H, Zhang X, Chen H. 2019. A victorivirus from Fusarium asiaticum, the pathogen of Fusarium head blight in China. Arch Virol 164:313–316. 10.1007/s00705-018-4038-9. [DOI] [PubMed] [Google Scholar]
- 18.Wolf YI, Kazlauskas D, Iranzo J, Lucía-Sanz A, Kuhn JH, Krupovic M, Dolja VV, Koonin EV. 2018. Origins and evolution of the global RNA Virome. mBio 9:e02329-18. 10.1128/mBio.02329-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koonin EV, Dolja VV, Krupovic M, Varsani A, Wolf YI, Yutin N, Zerbini FM, Kuhn JH. 2020. Global organization and proposed megataxonomy of the virus world. Microbiol Mol Biol Rev 84:e00061-19. 10.1128/MMBR.00061-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Liu L, Xie J, Cheng J, Fu Y, Li G, Yi X, Jiang D. 2014. Fungal negative-stranded RNA virus that is related to bornaviruses and nyaviruses. Proc Natl Acad Sci USA 111:12205–12210. 10.1073/pnas.1401786111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gao Z, Wu J, Jiang D, Xie J, Cheng J, Lin Y. 2020. ORF Ι of Mycovirus SsNSRV-1 is associated with debilitating symptoms of Sclerotinia sclerotiorum. Viruses 12:456. 10.3390/v12040456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Elliott RM, Schmaljohn CS. 2013. Bunyaviridae, p 1244–1282. In Knipe DM, Howley PM, Cohen JI, Griffin DE, Lamb RA, Martin MA, Racaniello VR, Roizman B (ed), Fields virology, 6th ed. Lippincott Williams & Wilkins, Philadelphia, PA. [Google Scholar]
- 23.Kormelink R, Verchot J, Tao X, Desbiez C. 2021. The Bunyavirales: the plant-infecting counterparts. Viruses 13:842. 10.3390/v13050842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kuhn JH, Adkins S, Agwanda BR, Al Kubrusli R, Alkhovsky SV, Amarasinghe GK, Avšič-Županc T, Ayllón MA, Bahl J, Balkema-Buschmann A, Ballinger MJ, Basler CF, Bavari S, Beer M, Bejerman N, Bennett AJ, Bente DA, Bergeron É, Bird BH, Blair CD, Blasdell KR, Blystad D-R, Bojko J, Borth WB, Bradfute S, Breyta R, Briese T, Brown PA, Brown JK, Buchholz UJ, Buchmeier MJ, Bukreyev A, Burt F, Büttner C, Calisher CH, Cao M, Casas I, Chandran K, Charrel RN, Cheng Q, Chiaki Y, Chiapello M, Choi I-R, Ciuffo M, Clegg JCS, Crozier I, Dal Bó E, de la Torre JC, de Lamballerie X, de Swart RL, et al. 2021. Correction to: 2021 taxonomic update of phylum Negarnaviricota (Riboviria: Orthornavirae), including the large orders Bunyavirales and Mononegavirales. Arch Virol 166:3567–3579. 10.1007/s00705-021-05266-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Walker PJ, Siddell SG, Lefkowitz EJ, Mushegian AR, Adriaenssens EM, Alfenas-Zerbini P, Dempsey DM, Dutilh BE, García ML, Curtis Hendrickson R, Junglen S, Krupovic M, Kuhn JH, Lambert AJ, Łobocka M, Oksanen HM, Orton RJ, Robertson DL, Rubino L, Sabanadzovic S, Simmonds P, Smith DB, Suzuki N, Van Doorslaer K, Vandamme A-M, Varsani A, Zerbini FM. 2022. Recent changes to virus taxonomy ratified by the International Committee on Taxonomy of Viruses (2022). Arch Virol 167:2429–2440. 10.1007/s00705-022-05516-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sutela S, Forgia M, Vainio EJ, Chiapello M, Daghino S, Vallino M, Martino E, Girlanda M, Perotto S, Turina M. 2020. The virome from a collection of endomycorrhizal fungi reveals new viral taxa with unprecedented genome organization. Virus Evol 6:veaa076. 10.1093/ve/veaa076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marzano S-YL, Nelson BD, Ajayi-Oyetunde O, Bradley CA, Hughes TJ, Hartman GL, Eastburn DM, Domier LL. 2016. Identification of diverse mycoviruses through metatranscriptomics characterization of the viromes of five major fungal plant pathogens. J Virol 90:6846–6863. 10.1128/JVI.00357-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Osaki H, Sasaki A, Nomiyama K, Tomioka K. 2016. Multiple virus infection in a single strain of Fusarium poae shown by deep sequencing. Virus Genes 52:835–847. 10.1007/s11262-016-1379-x. [DOI] [PubMed] [Google Scholar]
- 29.Donaire L, Pagán I, Ayllón MA. 2016. Characterization of Botrytis cinerea negative-stranded RNA virus 1, a new mycovirus related to plant viruses, and a reconstruction of host pattern evolution in negative-sense ssRNA viruses. Virology 499:212–218. 10.1016/j.virol.2016.09.017. [DOI] [PubMed] [Google Scholar]
- 30.Velasco L, Arjona-Girona I, Cretazzo E, López-Herrera C. 2019. Viromes in Xylariaceae fungi infecting avocado in Spain. Virology 532:11–21. 10.1016/j.virol.2019.03.021. [DOI] [PubMed] [Google Scholar]
- 31.Lin Y-H, Fujita M, Chiba S, Hyodo K, Andika IB, Suzuki N, Kondo H. 2019. Two novel fungal negative-strand RNA viruses related to mymonaviruses and phenuiviruses in the shiitake mushroom (Lentinula edodes). Virology 533:125–136. 10.1016/j.virol.2019.05.008. [DOI] [PubMed] [Google Scholar]
- 32.Picarelli MASC, Forgia M, Rivas EB, Nerva L, Chiapello M, Turina M, Colariccio A. 2019. Extreme diversity of mycoviruses present in isolates of Rhizoctonia solani AG2-2 LP from Zoysia japonica from Brazil. Front Cell Infect Microbiol 9:244. 10.3389/fcimb.2019.00244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jia J, Fu Y, Jiang D, Mu F, Cheng J, Lin Y, Li B, Marzano S-YL, Xie J. 2021. Interannual dynamics, diversity and evolution of the virome in Sclerotinia sclerotiorum from a single crop field. Virus Evol 7:veab032. 10.1093/ve/veab032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang J, Ni Y, Liu X, Zhao H, Xiao Y, Xiao X, Li S, Liu H. 2020. Divergent RNA viruses in Macrophomina phaseolina exhibit potential as virocontrol agents. Virus Evol 7:veaa095. 10.1093/ve/veaa095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ruiz-Padilla A, Rodríguez-Romero J, Gómez-Cid I, Pacifico D, Ayllón MA. 2021. Novel Mycoviruses Discovered in the Mycovirome of a Necrotrophic Fungus. mBio 12:e03705-20. 10.1128/mBio.03705-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Li Y, Zhou M, Yang Y, Liu Q, Zhang Z, Han C, Wang Y. 2021. Characterization of the mycovirome from the plant-pathogenic fungus Cercospora beticola. Viruses 13:1915. 10.3390/v13101915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nerva L, Forgia M, Ciuffo M, Chitarra W, Chiapello M, Vallino M, Varese GC, Turina M. 2019. The mycovirome of a fungal collection from the sea cucumber Holothuria polii. Virus Res 273:197737. 10.1016/j.virusres.2019.197737. [DOI] [PubMed] [Google Scholar]
- 38.Chen F, Pu Z, Ni H, Wang Y, Yan B. 2021. Multiple mycoviruses identified in Pestalotiopsis spp. from Chinese bayberry. Virol J 18:43. 10.1186/s12985-021-01513-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nerva L, Turina M, Zanzotto A, Gardiman M, Gaiotti F, Gambino G, Chitarra W. 2019. Isolation, molecular characterization and virome analysis of culturable wood fungal endophytes in esca symptomatic and asymptomatic grapevine plants. Environ Microbiol 21:2886–2904. 10.1111/1462-2920.14651. [DOI] [PubMed] [Google Scholar]
- 40.Chiapello M, Rodríguez-Romero J, Ayllón MA, Turina M. 2020. Analysis of the virome associated to grapevine downy mildew lesions reveals new mycovirus lineages. Virus Evol 6:veaa058. 10.1093/ve/veaa058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sasai S, Tamura K, Tojo M, Herrero M-L, Hoshino T, Ohki ST, Mochizuki T. 2018. A novel non-segmented double-stranded RNA virus from an Arctic isolate of Pythium polare. Virology 522:234–243. 10.1016/j.virol.2018.07.012. [DOI] [PubMed] [Google Scholar]
- 42.Botella L, Janoušek J, Maia C, Jung MH, Raco M, Jung T. 2020. Marine oomycetes of the genus Halophytophthora harbor viruses related to bunyaviruses. Front Microbiol 11:1467. 10.3389/fmicb.2020.01467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Botella L, Jung T. 2021. Multiple viral infections detected in Phytophthora condilina by total and small RNA sequencing. Viruses 13:620. 10.3390/v13040620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Poimala A, Parikka P, Hantula J, Vainio EJ. 2021. Viral diversity in Phytophthora cactorum population infecting strawberry. Environ Microbiol 23:5200–5221. 10.1111/1462-2920.15519. [DOI] [PubMed] [Google Scholar]
- 45.Raco M, Vainio EJ, Sutela S, Eichmeier A, Hakalová E, Jung T, Botella L. 2022. High diversity of novel viruses in the tree pathogen Phytophthora castaneae revealed by high-throughput sequencing of total and small RNA. Front Microbiol 13:911474. 10.3389/fmicb.2022.911474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qu B, Li HP, Zhang JB, Xu YB, Huang T, Wu AB, Zhao CS, Carter J, Nicholson P, Liao YC. 2008. Geographic distribution and genetic diversity of Fusarium graminearum and F. asiaticum on wheat spikes throughout China. Plant Pathol 57:15–24. [Google Scholar]
- 47.Zhang X, Ma H-X, Zhou Y-J, Xing J-C, Chen J-H, Yu G-H, Sun X-B, Wang L. 2014. Identification and genetic division of Fusarium graminearum and Fusarium asiaticum by species-specific SCAR markers. J Phytopathol 162:81–88. 10.1111/jph.12155. [DOI] [Google Scholar]
- 48.Sadiq S, Chen Y-M, Zhang Y-Z, Holmes EC. 2022. Resolving deep evolutionary relationships within the RNA virus phylum Lenarviricota. Virus Evol 8:veac055. 10.1093/ve/veac055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Li P, Zhang H, Chen X, Qiu D, Guo L. 2015. Molecular characterization of a novel hypovirus from the plant pathogenic fungus Fusarium graminearum. Virology 481:151–160. 10.1016/j.virol.2015.02.047. [DOI] [PubMed] [Google Scholar]
- 50.Zhang L, Chen X, Bhattacharjee P, Shi Y, Guo L, Wang S. 2020. Molecular characterization of a novel strain of Fusarium graminearum virus 1 infecting Fusarium graminearum. Viruses 12:357. 10.3390/v12030357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Wang L, He H, Wang S, Chen X, Qiu D, Kondo H, Guo L. 2018. Evidence for a novel negative-stranded RNA mycovirus isolated from the plant pathogenic fungus Fusarium graminearum. Virology 518:232–240. 10.1016/j.virol.2018.03.008. [DOI] [PubMed] [Google Scholar]
- 52.Elliott RM, Brennan B. 2014. Emerging phleboviruses. Curr Opin Virol 5:50–57. 10.1016/j.coviro.2014.01.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zheng Y, Navarro B, Wang G, Wang Y, Yang Z, Xu W, Zhu C, Wang L, Serio FD, Hong N. 2017. Actinidia chlorotic ringspot-associated virus: a novel emaravirus infecting kiwifruit plants. Mol Plant Pathol 18:569–581. 10.1111/mpp.12421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Amroun A, Priet S, de Lamballerie X, Quérat G. 2017. Bunyaviridae RdRps: structure, motifs, and RNA synthesis machinery. Crit Rev Microbiol 43:753–778. 10.1080/1040841X.2017.1307805. [DOI] [PubMed] [Google Scholar]
- 55.Sun Y, Li J, Gao GF, Tien P, Liu W. 2018. Bunyavirales ribonucleoproteins: the viral replication and transcription machinery. Crit Rev Microbiol 44:522–540. 10.1080/1040841X.2018.1446901. [DOI] [PubMed] [Google Scholar]
- 56.Olschewski S, Cusack S, Rosenthal M. 2020. The cap-snatching mechanism of bunyaviruses. Trends Microbiol 28:293–303. 10.1016/j.tim.2019.12.006. [DOI] [PubMed] [Google Scholar]
- 57.Ferron F, Weber F, de la Torre JC, Reguera J. 2017. Transcription and replication mechanisms of Bunyaviridae and Arenaviridae L proteins. Virus Res 234:118–134. 10.1016/j.virusres.2017.01.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Nunberg JH, York J. 2012. The curious case of arenavirus entry, and its inhibition. Viruses 4:83–101. 10.3390/v4010083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hulswit RJG, Paesen GC, Bowden TA, Shi X. 2021. Recent advances in bunyavirus glycoprotein research: precursor processing, receptor binding and structure. Viruses 13:353. 10.3390/v13020353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Teufel F, Almagro Armenteros JJ, Johansen AR, Gíslason MH, Pihl SI, Tsirigos KD, Winther O, Brunak S, von Heijne G, Nielsen H. 2022. SignalP 6.0 predicts all five types of signal peptides using protein language models. Nat Biotechnol 40:1023–1025. 10.1038/s41587-021-01156-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Yang J, Zhang Y. 2015. I-TASSER server: new development for protein structure and function predictions. Nucleic Acids Res 43:W174–W181. 10.1093/nar/gkv342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zhang C, Freddolino PL, Zhang Y. 2017. COFACTOR: improved protein function prediction by combining structure, sequence and protein-protein interaction information. Nucleic Acids Res 45:W291–W299. 10.1093/nar/gkx366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zheng W, Zhang C, Li Y, Pearce R, Bell EW, Zhang Y. 2021. Folding non-homologous proteins by coupling deep-learning contact maps with I-TASSER assembly simulations. Cell Rep Methods 1:100014. 10.1016/j.crmeth.2021.100014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Mu F, Xie J, Cheng S, You MP, Barbetti MJ, Jia J, Wang Q, Cheng J, Fu Y, Chen T, Jiang D. 2017. Virome characterization of a collection of Sclerotinia sclerotiorum from Australia. Front Microbiol 8:2540. 10.3389/fmicb.2017.02540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Arjona-Lopez JM, Telengech P, Jamal A, Hisano S, Kondo H, Yelin MD, Arjona-Girona I, Kanematsu S, Lopez-Herrera CJ, Suzuki N. 2018. Novel, diverse RNA viruses from Mediterranean isolates of the phytopathogenic fungus, Rosellinia necatrix: insights into evolutionary biology of fungal viruses. Environ Microbiol 20:1464–1483. 10.1111/1462-2920.14065. [DOI] [PubMed] [Google Scholar]
- 66.Chu Y-M, Jeon J-J, Yea S-J, Kim Y-H, Yun S-H, Lee Y-W, Kim K-H. 2002. Double-stranded RNA mycovirus from Fusarium graminearum. Appl Environ Microbiol 68:2529–2534. 10.1128/AEM.68.5.2529-2534.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Tan J, Ameye M, Landschoot S, De Zutter N, De Saeger S, De Boevre M, Abdallah MF, Van der Lee T, Waalwijk C, Audenaert K. 2020. At the scene of the crime: new insights into the role of weakly pathogenic members of the fusarium head blight disease complex. Mol Plant Pathol 21:1559–1572. 10.1111/mpp.12996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.de Oliveira Resende R, de Haan P, de Avila AC, Kitajima EW, Kormelink R, Goldbach R, Peters D. 1991. Generation of envelope and defective interfering RNA mutants of tomato spotted wilt virus by mechanical passage. J Gen Virol 72:2375–2383. 10.1099/0022-1317-72-10-2375. [DOI] [PubMed] [Google Scholar]
- 69.Nagata T, Inoue-Nagata AK, Prins M, Goldbach R, Peters D. 2000. Impeded thrips transmission of defective tomato spotted wilt virus isolates. Phytopathology 90:454–459. 10.1094/PHYTO.2000.90.5.454. [DOI] [PubMed] [Google Scholar]
- 70.Shi X, Elliott RM. 2007. Analysis of glycoproteins of viruses in the family Bunyaviridae, p 137–148. In Sugrue RJ (ed), Glycovirology protocols. Humana Press, Totowa, NJ. [DOI] [PubMed] [Google Scholar]
- 71.Feng M, Cheng R, Chen M, Guo R, Li L, Feng Z, Wu J, Xie L, Hong J, Zhang Z, Kormelink R, Tao X. 2020. Rescue of tomato spotted wilt virus entirely from complementary DNA clones. Proc Natl Acad Sci USA 117:1181–1190. 10.1073/pnas.1910787117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Spiegel M, Plegge T, Pöhlmann S. 2016. The role of phlebovirus glycoproteins in viral entry, assembly and release. Viruses 8:202. 10.3390/v8070202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Navarro B, Minutolo M, De Stradis A, Palmisano F, Alioto D, Di Serio F. 2018. The first phlebo-like virus infecting plants: a case study on the adaptation of negative-stranded RNA viruses to new hosts. Mol Plant Pathol 19:1075–1089. 10.1111/mpp.12587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Navarro B, Zicca S, Minutolo M, Saponari M, Alioto D, Di Serio F. 2018. A negative-stranded RNA virus infecting citrus trees: the second member of a new genus within the order Bunyavirales. Front Microbiol 9:2340. 10.3389/fmicb.2018.02340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Liu S, Xie J, Cheng J, Li B, Chen T, Fu Y, Li G, Wang M, Jin H, Wan H, Jiang D. 2016. Fungal DNA virus infects a mycophagous insect and utilizes it as a transmission vector. Proc Natl Acad Sci USA 113:12803–12808. 10.1073/pnas.1608013113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Li C-X, Shi M, Tian J-H, Lin X-D, Kang Y-J, Chen L-J, Qin X-C, Xu J, Holmes EC, Zhang Y-Z. 2015. Unprecedented genomic diversity of RNA viruses in arthropods reveals the ancestry of negative-sense RNA viruses. Elife 4:e05378. 10.7554/eLife.05378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Shi M, Lin X-D, Tian J-H, Chen L-J, Chen X, Li C-X, Qin X-C, Li J, Cao J-P, Eden J-S, Buchmann J, Wang W, Xu J, Holmes EC, Zhang Y-Z. 2016. Redefining the invertebrate RNA virosphere. Nature 540:539–543. 10.1038/nature20167. [DOI] [PubMed] [Google Scholar]
- 78.Drakulic J, Bruce TJA, Ray RV. 2017. Direct and host-mediated interactions between Fusarium pathogens and herbivorous arthropods in cereals. Plant Pathol 66:3–13. 10.1111/ppa.12546. [DOI] [Google Scholar]
- 79.Neri U, Wolf YI, Roux S, Camargo AP, Lee B, Kazlauskas D, Chen IM, Ivanova N, Allen LZ, Paez-Espino D, Bryant DA, Bhaya D, RNA Virus Discovery Consortium, Krupovic M, Dolja VV, Kyrpides NC, Koonin EV, Gophna U. 2022. Expansion of the global RNA virome reveals diverse clades of bacteriophages. bioRxiv. 10.1101/2022.02.15.480533. [DOI] [PubMed]
- 80.Zayed AA, Wainaina JM, Dominguez-Huerta G, Pelletier E, Guo J, Mohssen M, Tian F, Pratama AA, Bolduc B, Zablocki O, Cronin D, Solden L, Delage E, Alberti A, Aury J-M, Carradec Q, da Silva C, Labadie K, Poulain J, Ruscheweyh H-J, Salazar G, Shatoff E, Tara Oceans Coordinators, Bundschuh R, Fredrick K, Kubatko LS, Chaffron S, Culley AI, Sunagawa S, Kuhn JH, Wincker P, Sullivan MB, Acinas SG, Babin M, Bork P, Boss E, Bowler C, Cochrane G, de Vargas C, Gorsky G, Guidi L, Grimsley N, Hingamp P, Iudicone D, Jaillon O, Kandels S, Karp-Boss L, Karsenti E, Not F, Ogata H, Poulton N, Pesant S, Sardet C, Speich S, Stemmann L, Sullivan MB, Sungawa S, Wincker P. 2022. Cryptic and abundant marine viruses at the evolutionary origins of Earth’s RNA virome. Science 376:156–162. 10.1126/science.abm5847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Forgia M, Chiapello M, Daghino S, Pacifico D, Crucitti D, Oliva D, Ayllon M, Turina M, Turina M. 2022. Three new clades of putative viral RNA-dependent RNA polymerases with rare or unique catalytic triads discovered in libraries of ORFans from powdery mildews and the yeast of oenological interest Starmerella bacillaris. Virus Evol 8:veac038. 10.1093/ve/veac038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Obbard DJ, Shi M, Roberts KE, Longdon B, Dennis AB. 2020. A new lineage of segmented RNA viruses infecting animals. Virus Evol 6:vez061. 10.1093/ve/vez061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Allen GC, Flores-Vergara MA, Krasynanski S, Kumar S, Thompson WF. 2006. A modified protocol for rapid DNA isolation from plant tissues using cetyltrimethylammonium bromide. Nat Protoc 1:2320–2325. 10.1038/nprot.2006.384. [DOI] [PubMed] [Google Scholar]
- 84.Nicholson P, Simpson DR, Weston G, Rezanoor HN, Lees AK, Parry DW, Joyce D. 1998. Detection and quantification of Fusarium culmorum and Fusarium graminearumin cereals using PCR assays. Physiol Mol Plant Pathol 53:17–37. 10.1006/pmpp.1998.0170. [DOI] [Google Scholar]
- 85.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30:2114–2120. 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Bushmanova E, Antipov D, Lapidus A, Prjibelski AD. 2019. rnaSPAdes: a de novo transcriptome assembler and its application to RNA-Seq data. Gigascience 8:giz100. 10.1093/gigascience/giz100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Buchfink B, Xie C, Huson DH. 2015. Fast and sensitive protein alignment using DIAMOND. Nat Methods 12:59–60. 10.1038/nmeth.3176. [DOI] [PubMed] [Google Scholar]
- 88.Krogh A, Larsson B, von Heijne G, Sonnhammer ELL. 2001. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol 305:567–580. 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 89.Tsirigos KD, Peters C, Shu N, Käll L, Elofsson A. 2015. The TOPCONS web server for consensus prediction of membrane protein topology and signal peptides. Nucleic Acids Res 43:W401–W407. 10.1093/nar/gkv485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gabler F, Nam S-Z, Till S, Mirdita M, Steinegger M, Söding J, Lupas AN, Alva V. 2020. Protein sequence analysis using the MPI Bioinformatics toolkit. Curr Protoc Bioinformatics 72:e108. 10.1002/cpbi.108. [DOI] [PubMed] [Google Scholar]
- 91.Chen C, Chen H, Zhang Y, Thomas HR, Frank MH, He Y, Xia R. 2020. TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol Plant 13:1194–1202. 10.1016/j.molp.2020.06.009. [DOI] [PubMed] [Google Scholar]
- 92.Deflubé LR, Cressey TN, Hume AJ, Olejnik J, Haddock E, Feldmann F, Ebihara H, Fearns R, Mühlberger E. 2019. Ebolavirus polymerase uses an unconventional genome replication mechanism. Proc Natl Acad Sci USA 116:8535–8543. 10.1073/pnas.1815745116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Katoh K, Standley DM. 2016. A simple method to control over-alignment in the MAFFT multiple sequence alignment program. Bioinformatics 32:1933–1942. 10.1093/bioinformatics/btw108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinformatics 25:1972–1973. 10.1093/bioinformatics/btp348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Nguyen L-T, Schmidt HA, von Haeseler A, Minh BQ. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol 32:268–274. 10.1093/molbev/msu300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kalyaanamoorthy S, Minh BQ, Wong TKF, von Haeseler A, Jermiin LS. 2017. ModelFinder: fast model selection for accurate phylogenetic estimates. Nat Methods 14:587–589. 10.1038/nmeth.4285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hoang DT, Chernomor O, von Haeseler A, Minh BQ, Vinh LS. 2018. UFBoot2: improving the ultrafast bootstrap approximation. Mol Biol Evol 35:518–522. 10.1093/molbev/msx281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu N, Yun Y, Yin Y, Hahn M, Ma Z, Chen Y. 2019. Lipid droplet biogenesis regulated by the FgNem1/Spo7-FgPah1 phosphatase cascade plays critical roles in fungal development and virulence in Fusarium graminearum. New Phytol 223:412–429. 10.1111/nph.15748. [DOI] [PubMed] [Google Scholar]
- 99.Ding S, Mehrabi R, Koten C, Kang Z, Wei Y, Seong K, Kistler HC, Xu J-R. 2009. Transducin beta-like gene FTL1 is essential for pathogenesis in Fusarium graminearum. Eukaryot Cell 8:867–876. 10.1128/EC.00048-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Naser-Khdour S, Quang Minh B, Lanfear R. 2022. Assessing confidence in root placement on phylogenies: an empirical study using nonreversible models for mammals. Syst Biol 71:959–972. 10.1093/sysbio/syab067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Yu G, Lam TT-Y, Zhu H, Guan Y. 2018. Two methods for mapping and visualizing associated data on phylogeny using Ggtree. Mol Biol Evol 35:3041–3043. 10.1093/molbev/msy194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Yu G. 2020. Using ggtree to visualize data on tree-like structures. Curr Protoc Bioinformatics 69:e96. 10.1002/cpbi.96. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 to S3. Download jvi.01381-22-s0001.pdf, PDF file, 0.1 MB (182.7KB, pdf)
Data Availability Statement
The detailed information on the research is available under the BioProject accession number PRJNA841045. Virus genomic sequences are under the accession numbers MZ969051 to MZ969070 in the GenBank database. Additional nucleotide sequence data reported for SsNSRV5 and AtNSRV2 are available in the Third Party Annotation (TPA) Section of the DDBJ/ENA/GenBank databases under the accession numbers TPA BK061361 to BK061364.


