Abstract
Background:
Recurrent miscarriage (RM) affects 1%–3% of pregnancies. However, in almost 50% of cases, the cause is unknown. Increasing evidence have shown that benzo(a)pyrene [B(a)P], a representative of polycyclic aromatic hydrocarbons (PAHs), is correlated with miscarriage. However, the underlying mechanisms of B(a)P/benzo(a)pyrene-7,8-dihydrodiol-9,10-epoxide (BPDE)-induced trophoblast cell dysfunctions and miscarriage remain largely unknown.
Objective:
The objective was to discover the role(s) of a novel lncRNA, lnc-HZ09, in the regulation of BPDE-inhibited migration and invasion of trophoblast cells and the occurrence of miscarriage.
Method:
Human trophoblast cells were treated with 0, 0.25, 0.5, BPDE with or without corresponding lnc-HZ09 silencing or overexpression. Using these cells, we evaluated cell migration and invasion, the mRNA and protein levels of members of the PLD1/RAC1/CDC42 pathway, the regulatory roles of lnc-HZ09 in PLD1 transcription and mRNA stability, and lnc-HZ09 transcription and stability. Human villous tissues were collected from RM () group and their matched healthy control (HC, ) group. We evaluated the levels of BPDE-DNA adducts, lnc-HZ09, and the mRNA and protein expression of members of the PLD1/RAC1/CDC42 pathway, and correlated their relative expression levels. We further constructed 0, 0.05 or B(a)P-induced mouse miscarriage model (each ), in which the mRNA and protein expression of members of the Pld1/Rac1/Cdc42 pathway were measured.
Results:
We identified a novel lnc-HZ09. Human trophoblast cells treated with lnc-HZ09 exhibited less cell migration and invasion. In addition, the levels of this lncRNA were higher in villous tissues from women with recurrent miscarriage than those from healthy individuals. SP1-mediated PLD1 mRNA levels were lower, and HuR-mediated PLD1 mRNA stability was less in trophoblast cells overexpressing lnc-HZ09. However, trophoblast cells treated with MSX1 had higher levels of lnc-HZ09, and METTL3-mediated m6A methylation on lnc-HZ09 resulted in greater lnc-HZ09 RNA stability. In BPDE-treated human trophoblast cells and in RM villous tissues, MSX1-mediated lnc-HZ09 transcription and METTL3-mediated lnc-HZ09 stability were both greater. In our mouse miscarriage model, B(a)P-treated mice had lower mRNA and protein levels of members of the Pld1/Rac1/Cdc42 pathway.
Discussion:
These results suggest that in human trophoblast cells, BPDE exposure up-regulated lnc-HZ09 level, suppressed PLD1/RAC1/CDC42 pathway, and inhibited migration and invasion, providing new insights in understanding the causes and mechanisms of unexplained miscarriage. https://doi.org/10.1289/EHP10477
Introduction
Miscarriage, one of the most common adverse pregnancy outcomes, is defined as a termination of pregnancy due to fetus exclusion prior to 22 wk of gestation or of fetal weight.1 Reportedly, there is an 11%–22% cumulative risk of miscarriages in 5–20 wk of gestation, and the risk of miscarriage is higher in early gestation () relative to that in the later period.2,3 Recurrent miscarriage (RM) refers to two or more consecutive miscarriages with the same spouse, affecting 1%–3% of pregnancies.4,5 Many studies have indicated that various risk factors are associated with miscarriage in the first trimester, such as chromosomal abnormalities, antiphospholipid syndrome, congenital structural abnormalities of the uterus, type I diabetes, and thyroid dysfunction.1,6,7 However, in approximately 50% of RM patients, the clinical causes of miscarriage are completely unknown, and those are collectively classified as unexplained recurrent miscarriage.8,9 Therefore, it is particularly urgent to explore the unknown causes to precisely prevent and treat miscarriage.
Polycyclic aromatic hydrocarbons (PAHs) are a typical class of persistent organic pollutants, produced from incomplete pyrolysis and/or combustion of domestic or industrial coal, cigarettes, fossil fuel, wood, and food items.10 PAHs persist in the environment and biota for long periods and exert toxic effects on organisms largely through inhalation and diet.11 Increasing studies have reported that many PAHs are associated with skin, lung, and bladder cancers.10 Recent studies also suggest that PAHs act as endocrine disrupters and have adverse effects on reproductive health.12 B(a)P, a prototypical representative of PAHs, is metabolized to generate the ultimate metabolite, benzo(a)pyrene-7,8-dihydrodiol-9,10-epoxide (BPDE). In a small study of 29 women undergoing in vitro fertilization, higher levels of B(a)P were measured in the follicular fluid of women who self-reported exposure to mainstream smoke than in those who self-reported no exposure to cigarette smoke ( vs. ).13 More B(a)P has also been found out in follicular fluid of nonpregnant women than those pregnant women in Lithuania during the period 1991–1995 ( vs. ).14 A case–control study of miscarriage suggests that higher level of BPDE-DNA adducts in maternal blood is associated with a higher miscarriage risk.15 In mice, exposure to B(a)P in early pregnancy impaired murine uterine receptivity,16 suppressed embryo implantation,16 and endometrial stromal cell decidualization.17 Therefore, it is important to explore how B(a)P or BPDE affects miscarriage and specific pathways involved.
As one of the most important components in the placenta, human extravillous trophoblast (EVT) invades into the pregnant uterus to establish a maternal–fetal interface.18 Their normal proliferation, migration and invasion are essential for a successful pregnancy.19 Dysfunctions of human trophoblast cells may lead to impaired uterine spiral artery rebuilding and trophoblast-related adverse pregnancy outcomes, such as miscarriage.20 In our recent work, we found that human trophoblast cell lines HTR-8/SVneo21 and Swan 7120,22 cells treated with BPDE had higher levels of apoptosis and exhibited less proliferation, invasion, and migration. Human villous explants treated with BPDE also exhibited less migration and invasion.21 However, the underlying mechanisms of these results are still largely unknown.
The invasion and migration of mammalian cells are regulated by many signaling pathways. Phospholipase D hydrolyze 1 (PLD1) is involved in the regulation of various cellular biological processes, such as cell growth, proliferation, migration, and intracellular protein transportation.22 In addition, RAC1 (Ras related C3 botulinum toxin substrate 1) and CDC42 (cell division control protein 42) also regulate the dynamics of cell motility and proliferation.23 Moreover, it was reported that PLD1 could interact with RAC1 and CDC42 in RBL-2H3 cells.24 Therefore, whether the PLD1/RAC1/CDC42 pathway might regulate migration and invasion of human trophoblast cells and villous tissues should be further explored.
LncRNAs are noncoding RNA molecules with more than 200 ribonucleotides in length, which have exhibited important regulatory functions at both transcriptional and posttranscriptional levels.25 The expression of lncRNA is developmentally regulated, and lncRNAs are also tissue-specific and/or cell-type–specific. Studies suggest that trophoblast cell functions may be influenced by certain lncRNAs, such as lncRNA EPB41L4A-AS1, which induces metabolic reprogramming in trophoblast cells and placenta tissue26; lnc-SLC4A1-1, which alters trophoblast functions via activation of immune responses and regulation of the axis27; and lncRNA MEG8, which is involved in the regulation of early trophoblast cell function.27 Recently, we have performed transcriptome sequencing of unexplained RM and HC villous tissues and BPDE-treated trophoblast Swan 71 cells.28 Based on these sequencing data, we identified a number of novel lncRNAs of interest, including lnc-HZ01, which was found to regulate BPDE-suppressed trophoblast cell proliferation and miscarriage by forming a lnc-HZ01/MXD1 feedback loop29; lnc-HZ03, which up-regulated p53/SAT1 pathway to promote trophoblast apoptosis and affected miscarriage28; lnc-HZ04, which served as a ceRNA for miR-hz04 and up-regulated the pathway30; and lnc-HZ08, which regulated BPDE-induced trophoblast cell dysfunctions and was associated with miscarriage.31 These works illustrate that these lncRNAs are closely associated with the occurrence of miscarriage. However, a large number of novel lncRNAs have yet to be identified. How these novel lncRNAs regulate BPDE-suppressed migration and invasion of human trophoblast cells and regulate miscarriage is still largely unclear.
Therefore, in this study, we aimed to explore the signaling pathway by which BPDE inhibited migration and invasion of trophoblast cells and to identify the regulatory roles of a novel lncRNA in both BPDE-treated trophoblast cells and RM villous tissues. For these aims, we conducted in vitro cellular experiments, human tissue experiments, and in vivo mouse model experiments, and we expected to discover the mechanism of BPDE-inhibited invasion and migration of human trophoblast cells in the occurrence of miscarriage, as well as the regulatory roles of a novel lncRNA. Hopefully, this work might provide novel insights in understanding the causes and mechanism of unexplained miscarriage.
Materials and Methods
Chemicals
Anhydrous DMSO, corn oil, benzo(a)pyrene [B(a)P, purity 99%] and actinomycin D were from Sigma-Aldrich. Benzo(a)pyren-7, 8-dihydrodiol-9, 10-epoxide (BPDE, 99.9%) was from MRIGlobal (MRIGlobal). BPDE was dissolved in DMSO at and stored at . B(a)P was dissolved in corn oil before use. All the vehicle control or cultures included an equal amount of DMSO (0.03%, v/v).
In Vitro Molecular and Cellular Biology Assays
Cell culture.
Swan 71 cells, first-trimester human trophoblast cells that have been immortalized by human telomerase, were constructed by the Gil Mor group at Yale University32 and were obtained from this group as a gift. HTR-8/SVneo cells, human first-trimester trophoblast cells that have been immortalized by SVneo virus, were purchased from Hunan Fenghui Biotechnology Co., Ltd. (CL0164). Swan 71 cells were cultured in DMEM/F12 medium (GIBCO, Invitrogen), and HTR-8/SVneo cells were cultured in RPMI 1640 medium (GIBCO), both of which were supplemented with 10% FBS (GIBCO), in a humidified atmosphere containing 5% at 37°C.
Cell transfection.
Empty vector pcDNA3.1 (Catalog no. V790-20) was purchased from Thermo Fisher Scientific Company. cDNAs that were used for construction of overexpression plasmid of lnc-HZ09 (pcDNA3.1-HZ09), METTL3 mRNA (pcDNA3.1-METTL3), MSX1 mRNA (pcDNA3.1-MSX1), SP1 mRNA (pcDNA3.1-SP1), or HuR mRNA (pcDNA3.1-HuR) were synthesized and constructed into pcDNA3.1 vector by Addgene (Table S1). The corresponding RNA sequences were obtained from National Center for Biotechnology Information (NCBI) database (Gene Bank, Homo sapiens, GRCh38.p14; sequences in Table S1). Empty vector pcDNA3.1 was used as a negative control. Si-HZ09, si-METTL3, si-MSX1, si-SP1, si-HuR, and si-NC (negative control) were customized by Thermo Fisher (sequences in Table S2). Swan 71 and HTR-8/SVneo cells ( cells/well) were seeded in 6-well plates and cultured to 80% confluence. Trophoblast cells were transfected with plasmids or siRNAs in Lipofectamine 3,000 (Invitrogen) medium for 24 h according to the manufacturer’s protocols. For all assays, cell quantification was performed using TC20 Automated Cell Counter (Bio-Rad Laboratories).
Cell migration and invasion.
Trophoblastic cells ( cells/well) in six-well plates were transfected with si-NC, si-HZ09, si-PLD1, si-HuR, or si-METTL3, or transfected with pcDNA3.1, pcDNA3.1-HZ09, pcDNA3.1-PLD1, or pcDNA3.1-METTL3 using Lipofectamine 3,000 media for 24 h. Media was removed, cells were detached using trypsin, and the surviving cells were resuspended into DMEM/F12 or RPMI 1640 medium. For migration assays, cells/well were seeded in 24-well transwell chambers (Corning) and cultured for 24 h. For invasion assays, aliquots of Matrigel (BD Biosciences) diluted in DMEM/F12 medium (dilution ratio 1:8) were coated on 24-well transwell chambers and were solidified at 37°C for 1 h. Cells ( cells/well) were plated on the top of Matrigel matrix and cultured for 24 h. The bottom chamber contained medium containing 10% FBS as chemoattractant of human trophoblast cells. After 24 h, the whole chambers were fixed with 4% paraformaldehyde for 20 min, stained with crystal violet for 15 min, and then washed thrice with phosphate-buffered saline (PBS). For visualization, the cells on the bottom surface of membrane were photographed by Axio Observer 3 (Zeiss) at magnification and counted in five random fields.
High-throughput mRNA sequencing and data processing.
Swan 71 cells ( cells) in a dish overexpressing lnc-HZ09 by transfecting with pcDNA3.1-HZ09 and equal number of control cells by transfecting with pcDNA3.1 were also used for mRNA sequencing. High-throughput mRNA sequencing was performed on HiSeq 2000 sequencing platform (BGI-Shenzhen) according to BGI commercial standard process (https://www.bgi.com/).28,29,31 Briefly, total RNAs were extracted by Trizol reagent (Thermo Fisher Scientific). The process included the removal of rRNA, synthesis of double-stranded cDNA, end repair, degradation of one strand, and enrichment of the other strand by polymerase chain reaction (PCR). The library quality was confirmed by sequencing. The differentially expressed mRNAs with differences and were generated from read counts using the online bioinformatic platform Dr. Tom provided by BGI (biosys.bgi.com). Differentially mRNAs were searched in the NCBI database (Gene Bank, Homo sapiens, GRCh38.p14) to determine their genome loci. These differentially expressed mRNAs were used for gene ontology (GO) analysis (http://geneontology.org/) to generate GO plots.33,34
5′ and 3′ rapid amplification of cDNA ends (RACE) assays.
Total RNAs were isolated with Trizol reagent. RACE-ready first-strand cDNA were performed with total RNAs using the SMARTer RACE 5′/3′ Kit (Takara Bio) according to the manufacturer’s instructions. The 5′- and 3′-RACE PCR reactions (including PCR Master Mix, SeqAmp DNA Polymerase, 5′- or 3′-RACE-ready cDNA, 5′ or 3′ GSP, UPM, SeqAmp Buffer, and four dNTPs) were performed on LightCycler480 II (Roche) following the manufacturer’s instructions (Takara Bio). The PCR programs are shown in Table S3. RACE PCR products were separated on a 1% agarose gel and extracted with the NuceloSpin Gel and PCR Clean-Up Kit (Takara Bio). Then, the DNA products were subcloned into pGH-T vectors (GV0108-C, Shanghai Generay Biotech Co., Ltd.) using In-Fusion Snap Assembly Master Mix (Takara Bio) for 15 min at 50°C and then sequenced bidirectionally using gene specific primers (Sagene) (Table S4). RNA integrity was determined with a Bioanalyzer 2100 using RNA 6000 nanochips (Agilent). DNA quantity was assessed using a NanoDrop One C spectrophotometer (Thermo Fisher Scientific). The full-length nucleotide sequence of lnc-HZ09 is shown in Table S5.
Total RNA extraction and quantitative real-time polymerase chain reaction (RT-qPCR).
RNA was extracted from Swan 71 cells, HTR-8/SVneo cells, human villous tissues, or mouse placental tissues using Trizol (Invitrogen) containing DNase I (Life Technologies), according to the manufacturer’s protocols. RNA quality and quantity were assessed using a NanoDrop 2000 UV spectrophotometer (Thermo Fisher Scientific). RT-qPCR analysis was performed as described previously.28,29,31 Briefly, the isolated RNAs () were converted into cDNAs using the first-strand cDNA synthesis kit (Invitrogen). Then, cDNAs were further amplified using a SYBR qPCR system (including Maxima SYBR Green qPCR Mix, which includes Maxima Hot Start Taq DNA polymerase, SYBR Green I, and dNTPs) following the manufacturer’s instructions (Thermo Fisher Scientific) (conditions in Table S6). The amplification results were automatically analyzed using method with iQ5 real-time detection system (Bio-Rad Laboratories). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA was used as normalization internal standard for lncRNAs and mRNAs. The primer (Sangon Biotech) sequences are listed in Table S7.
Western blot analysis.
Western blot analysis was performed as described previously.28,29,31 Briefly, proteins were extracted using RIPA lysis buffer (Thermo Fisher Scientific) and quantified using Pierce BCA Protein Assay Kit (Pierce). Proteins () were separated using 6%–12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and were then transferred to equilibrated polyvinylidene fluoride (PVDF) membrane (Amersham Biosciences). The membranes were blocked with 5% bovine serum albumin (BSA; Sigma-Aldrich) at 25°C for 1 h. Then, the membranes were incubated with primary antibody at 4°C overnight. The primary antibodies included anti-PLD1 (dilution 1:100; sc-28,314, Santa Cruz Biotechnology), anti-RAC1 (dilution 1:1,000; ab155938, Abcam), anti-CDC42 (dilution 1:1,000; ab1429, Abcam), anti-SP1 (dilution 1:1,000; ab255289, Abcam), anti-HuR (dilution 1:1,000; ab200342, Abcam), anti-MSX1 (dilution 1:600; LS-C30725, LifeSpan Biosciences), anti-METTL3 (dilution 1:1,000; ab195352, Abcam), anti-GAPDH (dilution 1:1,000; ab8245, Abcam) and (dilution 1:1,000; ab78078, Abcam). After washing thrice with Tris-buffered saline containing Tween-20 (TBST) for 10 min in each time, the membrane was incubated with secondary antibody in blocking solution at 25°C for 1 h. The secondary antibodies included goat anti-rabbit immunoglobulin G (IgG) (dilution 1:1,000; ab207995, Abcam) and goat antimouse IgG (dilution 1:1,000; ab207996, Abcam). The relative density of each protein band was analyzed by Image J with or GAPDH as the internal standard.
Fluorescence in situ hybridization (FISH).
Lnc-HZ09 in Swan 71 cells was detected by Cy3-labeled lnc-HZ09 probe (5′-CACGAGC-Cy3-TGCCCACGGTCT-Cy3-TCCTTT-3′) (Empire Genomics) according to FISH Kit procedure (RIBOBIO), as described previously.31 Briefly, Swan 71 cells ( cells/well) were seeded in 35-mm confocal dishes (Cat. No. 80100; NEST) for 24 h; then they were washed with PBS and fixed by 4% paraformaldehyde (Sigma-Aldrich) for 15 min and treated with 1% Triton X-100 (Sigma-Aldrich) for 30 min, following incubation with FISH probe of lnc-HZ09 that was pretreated at 73°C for 5 min after washing with PBS thrice. Afterward, these fixed cells were further stained with DAPI for 20 min. After washing with PBS thrice, the emission fluorescence was recorded at for DAPI and 570 nm for FISH probe on a confocal fluorescence microscope (; Nikon). The relative levels of lnc-HZ09 in the cytoplasm and nucleus were quantified using 20 random cells with software Image J ().
mRNA stability assays.
Swan 71 or HTR-8/SVneo cells ( cells/well) with overexpression or knockdown of lnc-HZ09, HuR, or METTL3 were seeded in a 6-well plate for 12 h. Then, the cells were treated with actinomycin D (Sigma-Aldrich) to block mRNA transcription. After 0, 1, 2, 3, 4, or 5 h, RNAs were extracted from cells, and the lnc-HZ09 or PLD1 mRNA were analyzed by RT-qPCR assays. GAPDH mRNA was used as the normalization internal standard.
Chromatin immunoprecipitation (ChIP) assays.
ChIP assays were performed with EZ-Magna ChIP Chromatin Immunoprecipitation Kit (Millipore) based on the manufacturer’s protocols.29,31 Briefly, Swan 71 or HTR-8/SVneo cells ( cells) were transfected with pcDNA3.1-HZ09 or si-HZ09 or were treated with BPDE. Then, the cells were cross-linked with 1% formaldehyde at 37°C for 15 min and were quenched in glycine for 5 min. DNA fragments ranged from 300 to 600 bp were generated after sonication using a Bioruptor (Diagenode SA). Subsequently, the antibody against MSX1 (dilution 1:200; sc-517256, Santa Cruz Biotechnology) or SP1 (dilution 1:200; ab227383, Abcam) was used for immunoprecipitation at 4°C overnight on an inverse rotator, with equal weight of IgG (dilution 1:200; ab172730, Abcam) as negative control. The promoter regions of lnc-HZ09 or PLD1 were immunoprecipitated, extracted, and amplified () by qPCR (programs in Table S6). The specific qPCR primers are listed in Table S8.
RNA immunoprecipitation (RIP) assays.
RIP assays were performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) based on the manufacturer’s protocols.29 Briefly, trophoblastic cells ( cells) were lysed in lysis buffer containing RNase inhibitor and protease inhibitor. Cell lysates were incubated with magnetic beads attached with human HuR antibody (ab200342; Abcam) or mouse IgG (5873S; Cell Signaling Technology) as negative control. The immunoprecipitated RNAs () were extracted by Trizol and analyzed by RT-qPCR (programs in Table S6) using primers (Sangon Biotech) listed in Table S8.
Methylated RNA immunoprecipitation (MeRIP).
(m6A RNA methylation) modification on lnc-HZ09 was determined by MeRIP-qPCR assays, as described previously.29 Briefly, the purified and fragmented RNAs were incubated with Pierce Protein A/G Magnetic Beads (Thermo Fisher Scientific) coupled with anti-m6A antibody (ab208577; Abcam) or rabbit IgG as negative control for 2 h at 4°C with rotation. After washes, m6A-modified RNAs were eluted and detected by RT-qPCR (programs in Table S6) using the primers listed in Table S8. One tenth of the amount of total RNAs was used as input. The corresponding m6A modification level in each sample was calculated by normalizing against the level in input.
In vitro RNA pull-down assays.
In vitro RNA pull-down assays were performed as described previously.31 Briefly, lnc-HZ09, lnc-HZ09-AS (antisense sequence), PLD1 mRNA or PLD1 mRNA-AS was respectively in vitro transcribed from pGEM-T-HZ09, pGEM-T-HZ09-AS, pGEM-T-PLD1, or pGEM-T-PLD1-AS, all of which were customized and synthesized using pGEM-T empty vector (A3600; Promega), using TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher Scientific) and were purified with GeneJET RNA Purification Kit (Thermo Fisher Scientific). Then, each of the RNAs was labeled with biotin using Biotin RNA labeling mix (Roche) and was mixed with lysate proteins in protein-RNA binding buffer for 1 h at 4°C. Lnc-HZ09-AS or PLD1 mRNA-AS was used as negative control (sequences in Table S9). Subsequently, the RNA-protein complexes were pulled down by streptavidin attached on magnetic beads using Pierce Magnetic RNA-Protein Pull-Down Kit (Thermo Fisher Scientific), and the proteins were analyzed by Western blotting.
Human Tissue Assays
Tissues collection and statement.
Villous tissues were collected from 15 patients with unexplained recurrent miscarriage (RM group) and 15 women who had elective abortion as healthy control (HC group) ages 25–30 y old and were treated at the West China Fourth Hospital (Chengdu, China) from December 2018 to December 2019, as described previously.28,29,31 All RM patients had consecutive unexplained miscarriages. Any women with one of the following features was excluded35: a) eclampsia or preeclampsia; b) viral infectious disease (e.g., AIDS, syphilis, tuberculosis, gonorrhea); c) polycystic ovarian syndrome; d) uterine abnormalities or cervical incompetence; e) luteal phase defects, autoimmune abnormality, hyperandrogenemia, hyperprolactinemia, or antiphospholipid antibody syndrome; f) abnormal karyotype of the parents or abortus; g) the symptoms of endocrine or metabolic diseases (e.g., hyperthyroidism, diabetes, and hypothyroidism); and h) tuberculosis, HIV, HBV, HCV, or with positive results from release tests. All of the HC group lacked any of the eight features as described above and had previous pregnancies. A piece of villous tissue with dimensions of approximate was manually dissected from the fetal side of the placenta and cleared of maternal decidua from these two groups at 6–10 wk of gestation. These samples were serially washed and immediately frozen in liquid nitrogen. Villous tissues were homogenized using Silica beads (107735; Merck) via shaking for a 1-min burst using a TissueLyser LT instrument (Qiagen). Approximately villous tissues were homogenized in Trizol reagent (Invitrogen) for total RNA extraction and in RIPA lysis buffer (Thermo Fisher Scientific) containing a protease inhibitor cocktail for total protein separation. The experiment protocols have been authorized by the Medical Ethics Committee of the West China Fourth Hospital, Sichuan University. Written informed consent was signed before the study.
Determination of BPDE-DNA adduct levels in villous tissues.
Genomic DNA was isolated from human villous tissues using Tissue/Cell Genomic DNA extraction and Purification Kit (K1442-100; BioVision). The levels of BPDE-DNA adducts were assessed using BPDE-DNA adduct ELISA kit (STA-357; Cell Biolabs). Briefly, DNA samples () were sonicated into fragments with 200–1,000 bp as determined by 1% agarose gel and then incubated with anti-BPDE antibody (dilution 1:1,000; 235601, Cell Biolabs) in 96-well plates for 2 h at room temperature. After washing with wash buffer (310806; Cell Biolabs), the secondary antibody (10902; Cell Biolabs) was added to each well for 1 h, 3,3′,5,5′-tetramethylbenzidine buffer was added, and then the mixture was incubated for 20 min at room temperature. After termination of reaction, the relative levels of BPDE-DNA adducts were determined by measuring the absorbance at using a microplate reader (VL0L00D0; Thermo Fisher Scientific), with the Reduced DNA Standard (235602; Cell Biolabs) as absorbance blank. The amount of BPDE-DNA adducts was quantified using a BPDE-DNA standard curve. The results were expressed as nanograms of BPDE-DNA adducts per microgram of DNA.
In Vivo Mouse Model Assays
A B(a)P-induced miscarriage mouse model was constructed as described previously.28,29,31 Briefly, C57BL/6 mice were from the Charles River Company and were housed in the local laboratory animal center. Mice (6–8 wk old) were housed under standard environmental conditions (12 h light/dark cycle, 22°C). The mice had a 1-wk acclimation, during which time they received standard chow and tap water ad libitum. Female mice were mated with male mice overnight, and the appearance of vaginal plug was considered as the first day of pregnancy (D1), which was further validated by monitoring the increase in weight. Three groups of pregnant mice (each ) were daily given B(a)P (0, 0.05, or , dissolved in corn oil) by oral gavage from D1 to D13, with an equal volume of corn oil as vehicle control. All mice were weighed daily, and they were euthanized by injection with Nembutal () on D14 to collect uteri. A random placenta was collected from every mouse in each group. The placental tissues were manually dissected and then snap-frozen in liquid nitrogen and stored at for subsequent RT-qPCR and Western blotting analysis. The protocol was approved by the Medical Ethics Committee of West China Medical Center.
Software Prediction and Data Analysis
Coding Potential Assessment Tool [CPAT (https://wlcb.oit.uci.edu/cpat/)]36 was used to analyze the protein-coding potential of lncRNAs. The open reading frame (ORF) was predicted with the NCBI Open Reading Frame Finder [ORF finder (https://www.ncbi.nlm.nih.gov/orffinder/)]. Conserved Domain Database [CDD (https://www.ncbi.nlm.nih.gov/cdd/)] and Pfam 34.0 (http://pfam.xfam.org/)37 were used to analyze the conserved domains in lncRNAs. PROMO software [version 3.0.2 (http://alggen.lsi.upc.es/cgi-bin/promo_v3/promo/promoinit.cgi?dirDB=TF_8.3)]38,39 was used to predict the transcription factors. The proteins that might be regulated by PLD1 were analyzed using STRING [version 11.5 (https://cn.string-db.org/)].40 Last, the m6A methylation modification site on RNA was identified with SRAMP software [version 2.15 (http://www.cuilab.cn/sramp/)].41
All experiments were replicated thrice independently, and the data were presented as (SD; ). Statistical analysis was operated using SPSS software (version 24.0; SPSS Inc.). Independent-samples Student’s -test was used between two groups; and one-way analysis of variance was used for more groups with Dunnett’s or LSD post hoc test. The correlation analysis of the relative expression levels was performed using Pearson analysis. The dotted box was used to visually distinguish the data points of RM and HC groups. All graphs were made with GraphPad Prism (version 8.0; GraphPad Inc.). Differences were considered significant when *, **, or ***.
Results
Characterization of a Novel lnc-HZ09 Highly Expressed in RM Villous Tissues and in BPDE-Exposed Human Trophoblast Cells
In our previous work, we identified 22 novel lncRNAs that were significantly highly expressed in human Swan 71 cells after BPDE treatment and 10 novel lncRNAs that were significantly highly expressed in RM tissues relative to HC tissues by high-throughput transcriptome sequencing.28 In these sequencing data, a novel lncRNA, lnc-32238, was one of the most considerably highly expressed lncRNAs in BPDE-exposed trophoblast cells (Figure S1A) and in RM tissues relative to HC tissues (Figure S1B), implying that this lncRNA might regulate the dysfunctions of BPDE-exposed trophoblast cells and the occurrence of miscarriage in an unidentified regulatory approach. In the current work, we focused on this lncRNA. Lnc-32238 was further confirmed to be significantly highly expressed in BPDE-exposed Swan 71 and HTR-8/SVneo cells by RT-qPCR analysis (Figure 1A,B). This lncRNA was identified as a sense transcript with 365 nucleotides (nt) in length by rapid amplification of cDNA ends (RACE) assays (Figure S1C; Tables S4 and S5) and resided at chromosome 16 (chr 16: 3,220,699 - 3,221,017). Then, this lnc-32238 was termed as lnc-HZ09, and its sequence was submitted to NCBI with accession No. MW675687. The protein-coding potential of lnc-HZ09 was analyzed to be very weak using NCBI ORF finder and CPAT (coding probability ).36 Additionally, lnc-HZ09 was found to have no conserved domains using CDD and Pfam.42 The data indicated that lnc-HZ09 might not encode a protein (Table. S10). Lnc-HZ09 was distributed to both the cytoplasm and nucleus of Swan 71 cells, as detected by FISH assays (Figure 1C).
Figure 1.
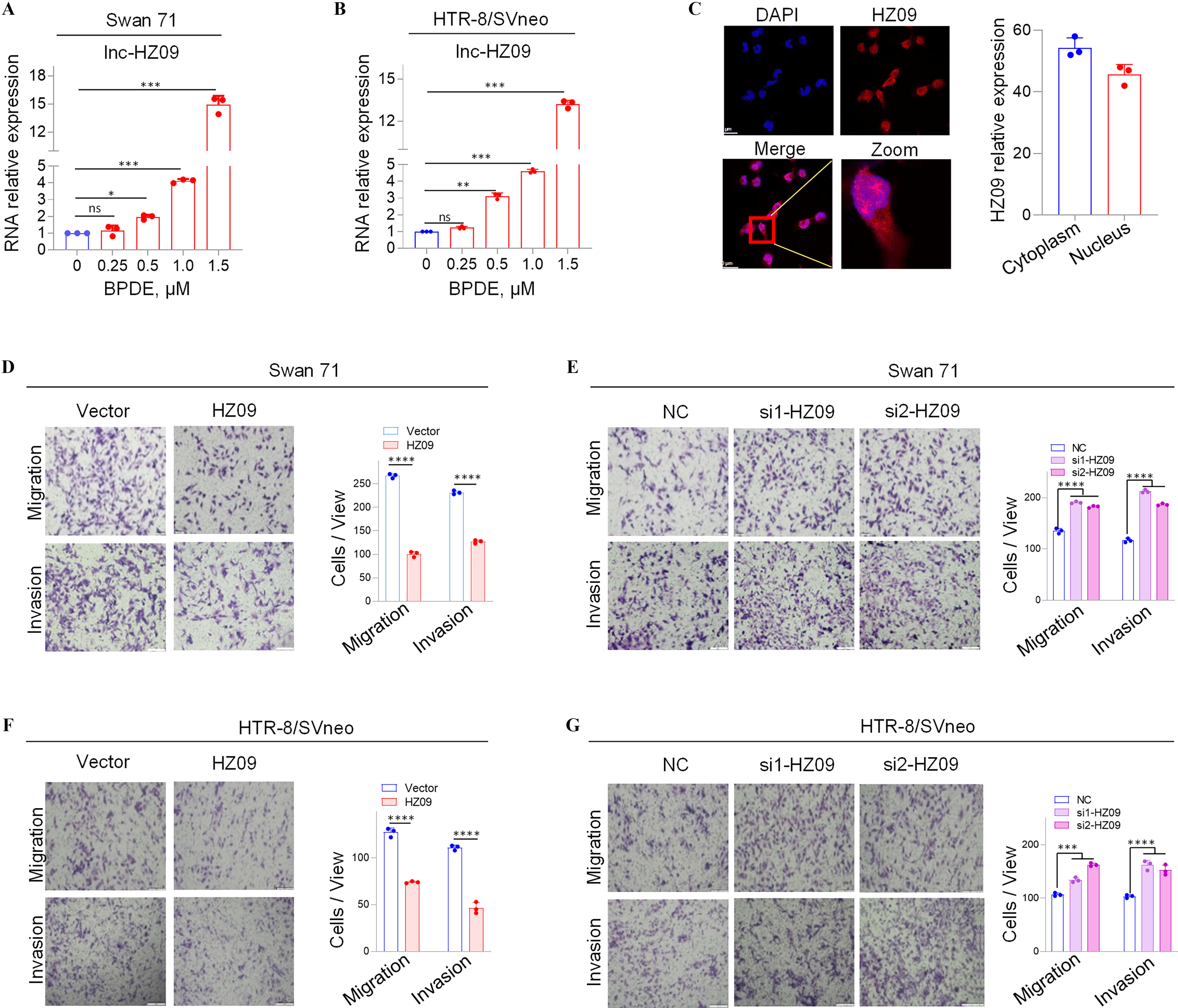
Expression levels of a novel lnc-HZ09 and the migration and invasion of human trophoblast cells with overexpression or knockdown of lnc-HZ09. (A–B) RT-qPCR analysis (each ) of lnc-HZ09 expression levels in BPDE-treated Swan 71 (A) or HTR-8/SVneo (B) cells. (C) FISH analysis (each ) of the distribution of lnc-HZ09 (red) in the nucleus and cytoplasm of Swan 71 cells (scale bar, ) and the relative levels of lnc-HZ09 foci. (D–G) Transwell assay analysis of the migration and invasion of Swan 71 (D–E) or HTR-8/SVneo (F–G) cells with overexpression (D and F) or knockdown (E and G) of lnc-HZ09 (scale bar, ). The number of cells per view was quantified. The summary data of these bar charts were shown in Excel Table S1. The RNA level in the untreated cells was set as “1” in RT-qPCR assays. C–G show the representative data from three independent experiments. Data in (A–G) show of three independent experiments. Two-tailed Student’s -test for (D,F); one-way ANOVA analysis for (A,B,E,G); *, **, and ***. Note: ANOVA, analysis of variance; BPDE, benzo(a)pyrene-7,8-dihydrodiol-9,10-epoxide; FISH, fluorescence in situ hybridization; HZ09, overexpression of lnc-HZ09; NC, negative control of siRNA; ns, nonsignificance; RT-qPCR, quantitative reverse transcription polymerase chain reaction; SD, standard deviation; si-HZ09, knockdown of lnc-HZ09; Vector, empty vector of pcDNA3.1.
Measurement of Migration Ad Invasion of Human Trophoblast Cells with Overexpression or Silence of lnc-HZ09
Because lnc-HZ09 was highly expressed in BPDE-treated trophoblast cells, its roles in the regulation of trophoblast cell functions were explored. To identify this, lnc-HZ09 was overexpressed in human trophoblast Swan 71 cells by transfecting with pcDNA3.1-HZ09 (validation shown in Figure S1D); and these cells and the corresponding control cells were used for mRNA sequencing. In sequencing data, we found that 1,055 mRNAs were down-regulated, and 710 mRNAs were up-regulated with differences and -values with overexpression of lnc-HZ09 (sequencing data in Excel Table S2). Subsequently, GO biological process analysis showed that cell migration might be significantly regulated by lnc-HZ09 overexpression (Figure S1E). To experimentally validate this, lnc-HZ09 was overexpressed by transfecting with pcDNA3.1-HZ09 or silenced by transfecting with its two distinct siRNAs (si1-HZ09 or si2-HZ09) in both Swan 71 and HTR-8/SVneo cells, and their efficiencies were validated by RT-qPCR analysis (Figure S1D, F–H). Migration and invasion of the respective cells were assessed. We found that lnc-HZ09-overexpressing cells demonstrated significantly less migration and invasion in comparison with cells transfected with empty vector. In contrast, lnc-HZ09-silenced cells demonstrated significantly greater migration and invasion (Figure 1D–G).
The Regulation of lnc-HZ09 on Trophoblast Cell Migration and Invasion through PLD1/RAC1/CDC42 Pathway
Next, the potential signaling pathway that might be regulated by lnc-HZ09 was further studied. First, the key molecules that were differentially expressed were discovered. In RNA sequencing data of BPDE-treated Swan 71 cells, human RM vs HC villous tissues, and lnc-HZ09-overexpressed cells, we identified nine down-regulated mRNAs with expression level difference between experimental and control (e.g., BPDE-treated vs. untreated Swan 71 cells, RM vs. HC villous tissues, and lnc-HZ09-overexpressed vs. control Swan 71 cells) with -values in the intersection of these three sets of sequencing data (Figure 2A). RT-qPCR analysis further confirmed that they were all up-regulated in Swan 71 cells with lnc-HZ09 knockdown via siRNA (Figure S2A). Among them, PLD1 was one of the most considerably altered mRNAs (Figure S2A). String analysis24 showed that PLD1 might interact with RAC1 and CDC42 (Figure 2B), implying that this PLD1/RAC1/CDC42 pathway might regulate migration and invasion of trophoblast cells and might also be regulated by lnc-HZ09 in trophoblast cells.
Figure 2.
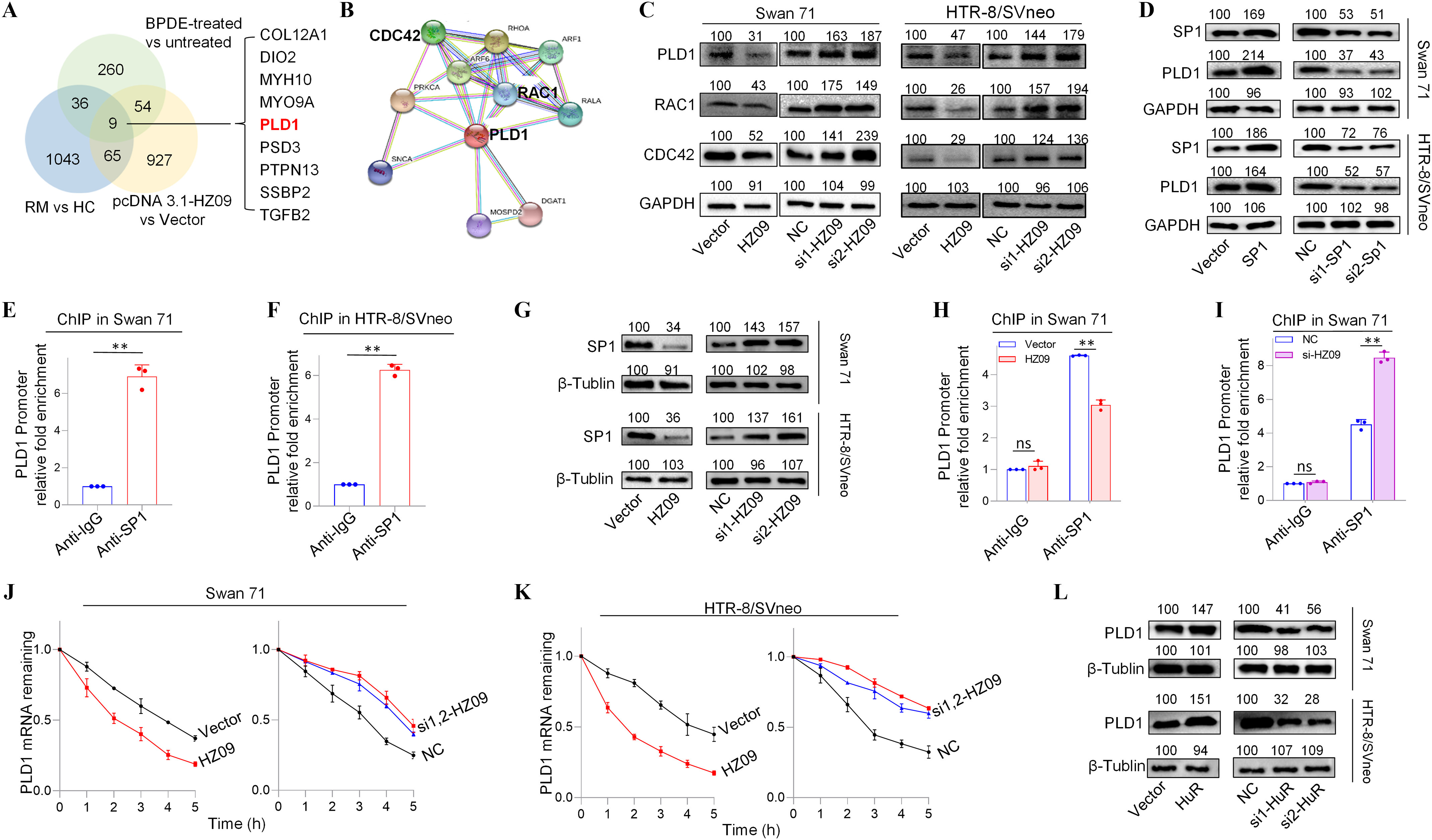
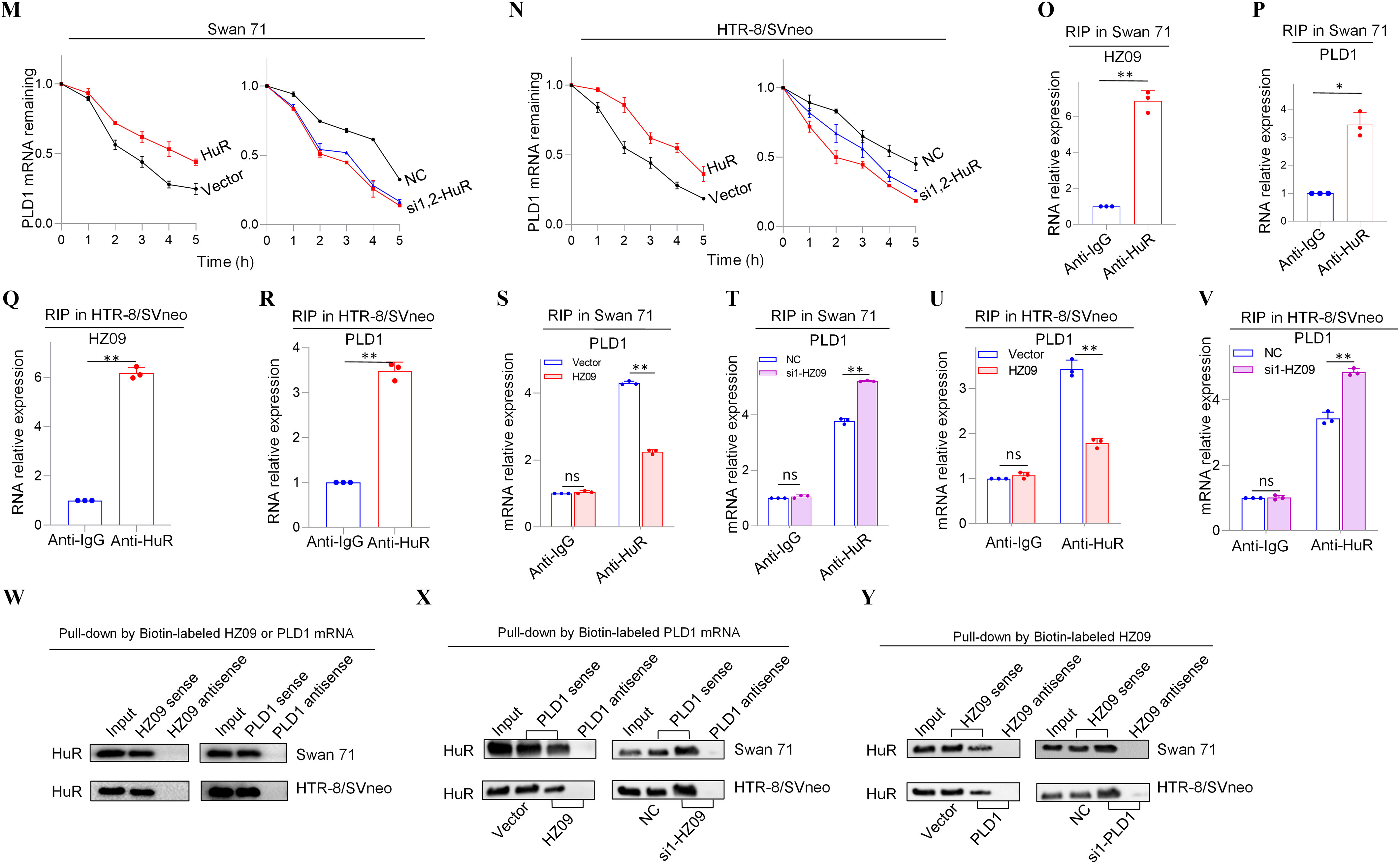
Expression levels of members of PLD1/RAC1/CDC42 pathway regulated by lnc-HZ09 in human trophoblast cells. (A) The significantly down-regulated mRNAs in the intersection of mRNA sequencing data of BPDE-treated vs. untreated Swan 71 cells, lnc-HZ09-overexpressed vs control Swan 71 cells, and RM vs HC villous tissues. (B) String analysis of PLD1, RAC1 and CDC42. (C) Representative western blot analysis of the protein levels of PLD1, RAC1 and CDC42 in Swan 71 or HTR-8/SVneo cells with overexpression or knockdown of lnc-HZ09, with GAPDH as internal standard. The relative intensity of each band was quantified and the of three replicates was shown in Figure S3A. (D) Representative western blot analysis of the protein levels of SP1 and PLD1 in Swan 71 or HTR-8/SVneo cells with overexpression or knockdown of SP1, with GAPDH as internal standard. The relative intensity of each band was quantified and the of three replicates was shown in Figure S3G. (E–F) SP1 ChIP assay analysis (each ) of the relative enrichment of SP1 in the promoter region of PLD1 gene in Swan 71 (E) or HTR-8/SVneo (F) cells. (G) Representative western blot analysis of the protein levels of SP1 in Swan 71 or HTR-8/SVneo cells with overexpression or knockdown of lnc-HZ09, with as internal standard. The relative intensity of each band was quantified and the of three replicates was shown in Figure S3H. (H–I) SP1 ChIP assay analysis (each ) of the relative enrichment of SP1 in the promoter region of PLD1 gene in Swan 71 cells with overexpression (H) or knockdown (I) of lnc-HZ09. (J–K) The mRNA stability of PLD1 (each ) in Swan 71 (J) or HTR-8/SVneo (K) cells with overexpression or knockdown of lnc-HZ09. (L) Representative western blot analysis of the protein levels of PLD1 in Swan 71 or HTR-8/SVneo cells with overexpression or knockdown of HuR, with as internal standard. The relative intensity of each band was quantified and the of three replicates was shown in Figure S5E. (M–N) The mRNA stability of PLD1 (each ) in Swan 71 (M) or HTR-8/SVneo (N) cells with overexpression or knockdown of HuR. (O–R) RIP assay analysis (each ) of the relative levels of lnc-HZ09 (O and Q) or PLD1 mRNA (P and R) that was pulled down by HuR protein in Swan 71 (O and P) or HTR-8/SVneo (Q and R) cells. (S–V) RIP assay analysis (each ) of the relative levels of PLD1 mRNA that was pulled down by HuR protein in Swan 71 (S and T) or HTR-8/SVneo (U and V) cells with overexpression (S and U) or knockdown (T and V) of lnc-HZ09. (W) Representative western blot analysis of HuR that was pulled down by biotin-labeled lnc-HZ09 or PLD1 mRNA in Swan 71 or HTR-8/SVneo cells in pull-down assays. (X) Representative western blot analysis of HuR that was pulled down by biotin-labeled PLD1 mRNA in Swan 71 or HTR-8/SVneo cells with overexpression or knockdown of lnc-HZ09 in pull-down assays. (Y) Representative western blot analysis of HuR that was pulled down by biotin-labeled lnc-HZ09 in Swan 71 or HTR-8/SVneo cells with overexpression or knockdown of PLD1 in pull-down assays. The summary data of these bar charts and diagrams were shown in Excel Table S1. The expression level in NC or Vector group was set as “1” in all of mRNA stability assays; the levels of DNA or RNA pulled down by IgG were set as “1” in all of ChIP and RIP assays, respectively; and the band intensity in NC or Vector group was set as ‘100’ in all of western blot assays. C,D,G,L,W–Y show the representative data from three independent experiments. Data in (E–F, H–K, M–V) show of three independent experiments. Two-tailed Student’s -test for (E–F,H–I,O–V); *, **. Note: BPDE, benzo(a)pyrene-7,8-dihydrodiol-9,10-epoxide; ChIP, chromatin immunoprecipitation; GADPH, glyceraldehyde-3-phosphate dehydrogenase; HC, healthy control; HuR, overexpression of HuR; HZ09, overexpression of lnc-HZ09; NC, negative control of siRNA; ns, nonsignificance; PLD1, phospholipase D hydrolyze 1; RIP, RNA immunoprecipitation; RM, recurrent miscarriage; SD, standard deviation; si-HuR, knockdown of HuR; si-SP1, knockdown of SP1; SP1, overexpression of SP1; Vector, empty vector of pcDNA3.1; si-HZ09, knockdown of lnc-HZ09.
Subsequently, the functions of this pathway were explored in human trophoblast cells. Cells (both Swan 71 and HTR-8/SVneo) with PLD1 overexpression had higher protein levels of RAC1 and CDC42, whereas cells with PLD1 knockdown had lower protein levels of RAC1 and CDC42 (Figure S2B–E). Furthermore, Swan 71 or HTR-8/SVneo cells withPLD1 overexpression had less migration and invasion, whereas cells with PLD1 knockdown had greater migration and invasion (Figure S2F–I). Subsequently, the role of lnc-HZ09 in regulation of this pathway was further explored. The mRNA and protein expression levels of PLD1, RAC1 and CDC42 were all lower in lnc-HZ09-overexpressed Swan 71 or HTR-8/SVneo cells and were all higher in lnc-HZ09-silenced cells compared to relevant controls (Figure 2C; S3A–B).
The Effects of lnc-HZ09 on SP1-Mediated PLD1 mRNA Transcription in Human Trophoblast Cells
We next explored how lnc-HZ09 regulated PLD1 expression. First, how lnc-HZ09 affected PLD1 mRNA transcription was studied. It has been reported that SP1 acted as a transcription factor to promote PLD1 mRNA transcription in hepatocytes.43 As analyzed by PROMO software, SP1 might recognize the promoter sequence of PLD1 (Figure S4). Experimentally, Swan 71 and HTR-8/SVneo cells with overexpression of SP1 had higher, whereas cells with SP1 knockdown had lower mRNA and protein expression levels of PLD1 (Figure 2D, S3C–G). SP1 ChIP assays further confirmed that SP1 could bind with the promoter region of PLD1 (Figure 2E–F), indicating that SP1 may act as a transcription factor to facilitate PLD1 transcription in human trophoblast cells. Moreover, cells with overexpression of lnc-HZ09 had lower, whereas cells with lnc-HZ09 knockdown had higher SP1 mRNA and protein levels (Figure 2G; Figure S3H). SP1 ChIP assays further showed that cells with lnc-HZ09 overexpression had greater, whereas cells with knockdown of lnc-HZ09 had lower occupancy of SP1 on the promoter region of PLD1 (Figure 2H,I; Figure S3J,K). Collectively, these results supported that lnc-HZ09 might suppress SP1-mediated PLD1 mRNA transcription.
The Effects of lnc-HZ09 on PLD1 mRNA Stability in Human Trophoblast Cells
Subsequently, we further investigated whether lnc-HZ09 might affect PLD1 mRNA stability. For both Swan 71 and HTR-8/SVneo cells, lnc-HZ09-overexpressed cells had lower, whereas lnc-HZ09-silenced cells had higher PLD1 mRNA stability (Figure 2J,K). As control, alteration of lnc-HZ09 did not affect the mRNA stability of GAPDH in both cells (Figure S5A,B).
It has been reported that HuR is an RNA binding protein, which could bind RNAs containing AUUU specific sequence and enhance their RNA stability.44 Here, we explored whether HuR protein could promote PLD1 mRNA stability in human trophoblast cells. We found that in both Swan 71 and HTR-8/SVneo cells, those with overexpression of HuR had higher, whereas those with HuR knockdown had lower PLD1 mRNA and protein levels (Figure 2L; S5C–F). Similarly, HuR-overexpressed cells had higher mRNA stability and those with HuR knockdown had lower mRNA stability (Figure 2M,N). As control, alteration of HuR did not affect GAPDH mRNA stability in both cells (Figure S5G,H). Furthermore, cells with overexpression of HuR exhibited greater, whereas cells with HuR knockdown exhibited less migration and invasion (Figure S5I–L). Notably, alteration of lnc-HZ09 did not affect the protein level of HuR in both trophoblast cells (Figure S5M,N).
Because HuR is an RNA binding protein, whether HuR could competitively bind with lnc-HZ09 or PLD1 mRNA was determined in human trophoblast cells. RIP assays showed that both PLD1 mRNA and lnc-HZ09 could be pulled down by HuR protein in both Swan 71 and HTR-8/SVneo cells (Figure 2O–R). Furthermore, cells with lnc-HZ09 overexpression had lower levels of PLD1 mRNA that were pulled down by HuR protein in both cells, whereas those with lnc-HZ09 knockdown had higher levels of PLD1 mRNA that were pulled down by HuR protein in both cells (Figure 2S–V). RNA pull-down assays further confirmed that HuR protein could be pulled down by biotin-labeled lnc-HZ09 or PLD1 mRNA but not by their antisense RNAs (Figure 2W). Moreover, cells with overexpression of lnc-HZ09 had lower levels of HuR protein that were pulled down by biotin-labeled PLD1 mRNA, whereas cells with knockdown of lnc-HZ09 had higher levels of HuR protein that were pulled down by biotin-labeled PLD1 mRNA (Figure 2X). Similarly, cells with PLD1 overexpression had lower levels of HuR protein pulled down by biotin-labeled lnc-HZ09, whereas cells with knockdown of PLD1 had higher levels of HuR protein pulled down by biotin-labeled lnc-HZ09 (Figure 2Y).
Regulatory Roles of MSX1 in lnc-HZ09 Transcription in Human Trophoblast Cells
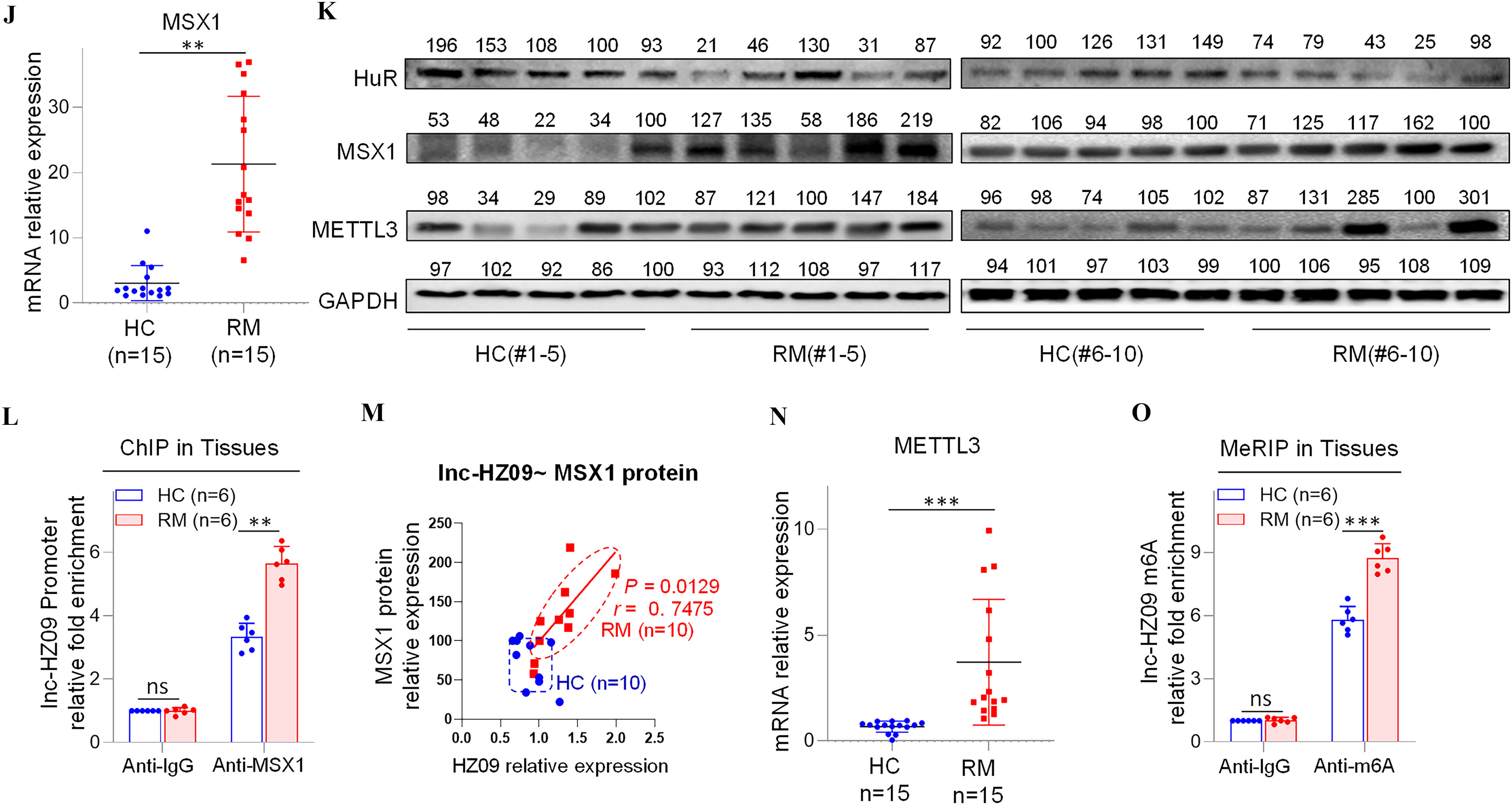
We next explored what regulated lnc-HZ09 expression level in human trophoblast cells. First, the transcription of lnc-HZ09 was studied. It has been reported that MSX1 was a transcription factor that promoted mRNA transcription of fibroblast growth factor 9 in murine myoblast C2C12 cells.45 As identified by PROMO software, MSX1 might recognize the promoter region of lnc-HZ09 (Figure S6). Experimentally, in both Swan 71 and HTR-8/SVneo cells, cells with overexpression of MSX1 had higher expression level of lnc-HZ09, whereas those with knockdown of MSX1 had lower expression level of lnc-HZ09 (Figure 3A–B; Figure S7A). Moreover, MSX1 ChIP assays showed that MSX1 could bind to the promoter region of lnc-HZ09 (Figure 3C,D), indicating that MSX1 may act as a transcription factor to promote lnc-HZ09 transcription.
Figure 3.
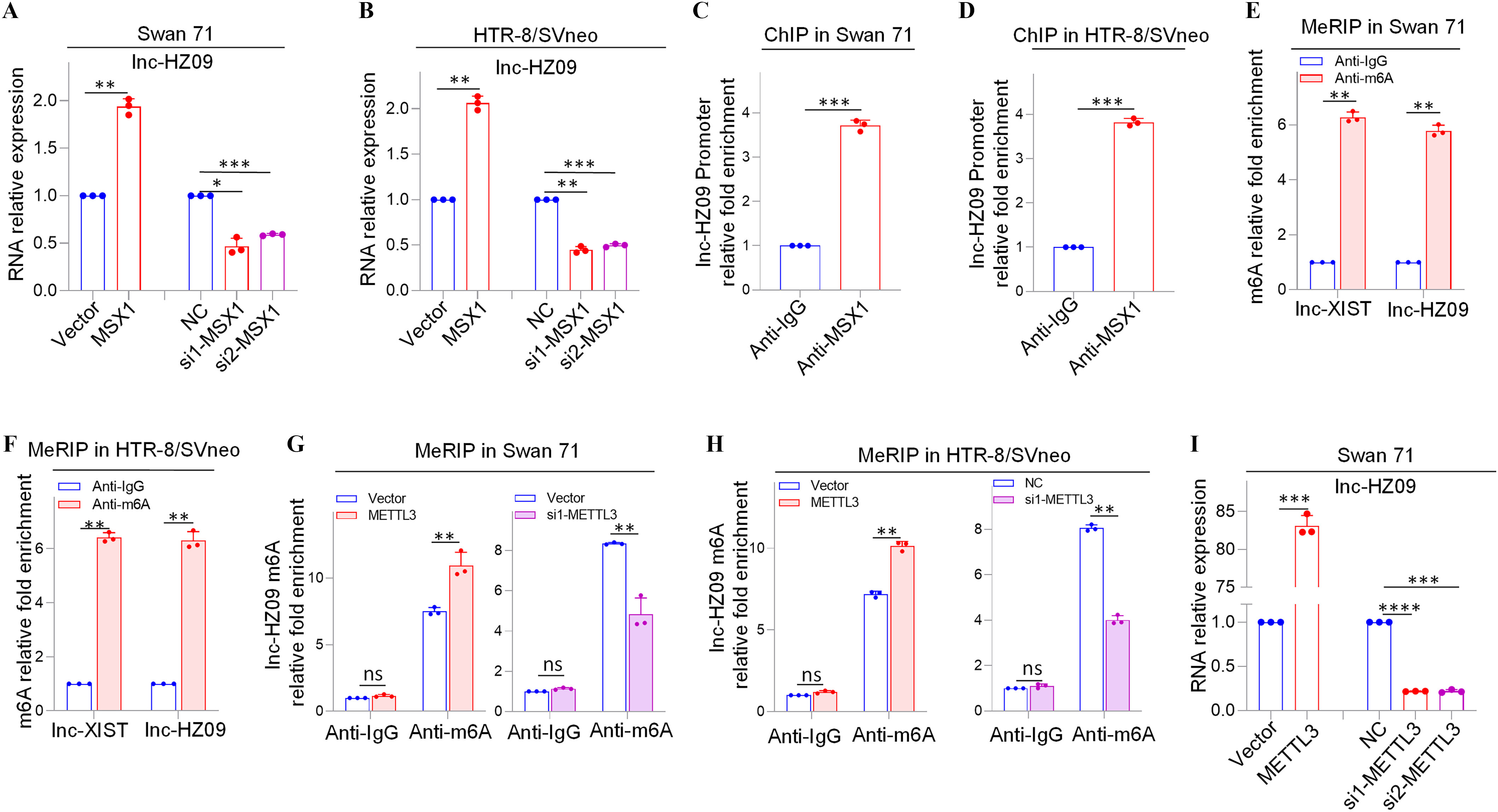
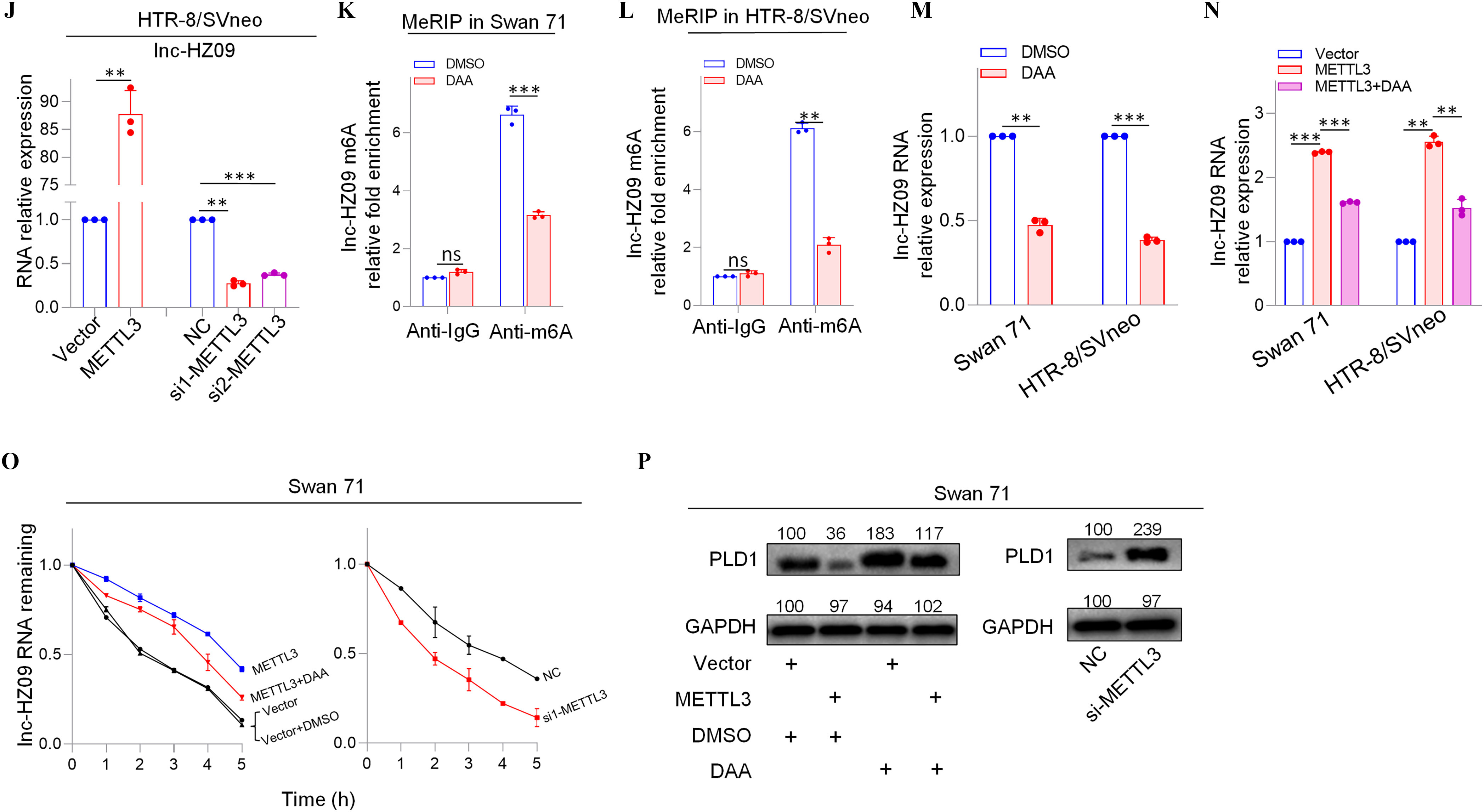
Expression levels of lnc-HZ09 regulated by MSX1 and m6A RNA methylation in human trophoblast cells. (A–B) RT-qPCR analysis (each ) of the levels of lnc-HZ09 in Swan 71 (A) or HTR-8/SVneo (B) cells with overexpression or knockdown of MSX1. (C–D) MSX1 ChIP assay analysis (each ) of the relative enrichment of MSX1 in the promoter region of lnc-HZ09 in Swan 71 (C) or HTR-8/SVneo (D) cells. (E–F) MeRIP assay analysis (each ) of the levels of m6A RNA methylation on lnc-XIST or lnc-HZ09 in Swan 71 (E) or HTR-8/SVneo (F) cells. (G–H) MeRIP assay analysis (each ) of the levels of m6A RNA methylation on lnc-HZ09 in Swan 71 (G) or HTR-8/SVneo (H) cells with overexpression or knockdown of METTL3. (I–J) RT-qPCR analysis (each ) of the levels of lnc-HZ09 in Swan 71 (I) or HTR-8/SVneo (J) cells with overexpression or knockdown of METTL3. (K–L) MeRIP assay analysis (each ) of the levels of m6A RNA methylation on lnc-HZ09 in Swan 71 (K) or HTR-8/SVneo (L) cells with DAA treatment. (M) RT-qPCR analysis (each ) of the levels of lnc-HZ09 in Swan 71 or HTR-8/SVneo cells with DAA treatment. (N) RT-qPCR analysis (each ) of the levels of lnc-HZ09 in Swan 71 or HTR-8/SVneo cells with overexpression of METTL3 or overexpression of METTL3 together with DAA treatment. (O) The RNA stability (each ) of lnc-HZ09 in Swan 71 cells with overexpression or knockdown of METTL3, or with overexpression of METTL3 together with DAA treatment. (P) Representative western blot analysis of PLD1 protein levels in Swan 71 cells with overexpression or knockdown of METTL3, or with overexpression of METTL3 together with DAA treatment, with GAPDH as internal standard. The relative intensity of each band was quantified and their of three replicates was shown in Figure S7F. The summary data of these bar charts and diagrams were shown in Excel Table S1. The expression level in NC or Vector group was set as “1” in all of RT-qPCR and mRNA stability assays; the levels of DNA or RNA pulled down by IgG were set as “1” in all ChIP and MeRIP assays; and the band intensity in NC or Vector group was set as “100” in all western blot assays. (P) shows the representative data from three independent experiments. Data in (A–O) show of three independent experiments. Two-tailed Student’s -test for (A–M); one-way ANOVA analysis for (A,B,I,J,N). *, **, and ***. Note: ANOVA, analysis of variance; ChIP, chromatin immunoprecipitation; DAA, 3-deazaadenosine; GADPH, glyceraldehyde-3-phosphate dehydrogenase; IgG, immunoglobulin G; MeRIP, methylated RNA immunoprecipitation; METTL3, overexpression of METTL3; MSX1, overexpression of MSX1; NC, negative control of siRNA; ns, nonsignificance; PLD1, phospholipase D hydrolyze 1; RT-qPCR, quantitative reverse transcription polymerase chain reaction; SD, standard deviation; si-METTL3, knockdown of METTL3; si-MSX1, knockdown of MSX1; Vector, empty vector of pcDNA3.1.
The Effects of METTL3-Mediated m6A RNA Methylation on lnc-HZ09 Stability in Human Trophoblast Cells
Subsequently, lnc-HZ09 RNA stability was investigated. It has been shown that m6A modification on lncRNAs may regulate the stability of lncRNAs.46 M6A modification site (5′-GGACU-3′41) was identified in lnc-HZ09 sequence using SRAMP software (Table S11). Subsequently, MeRIP assays confirmed the presence of m6A RNA modification on lnc-HZ09 (Figure 3E,F). LncRNA XIST containing m6A RNA modification47 was used as a positive control.
METTL3 is an important RNA methyltransferase to produce m6A RNA methylation on RNAs.48 Herein, in both Swan 71 and HTR-8/SVneo cells, those with overexpression of METTL3 had higher levels of m6A RNA modification on lnc-HZ09, whereas cells with knockdown of METTL3 had lower levels of m6A RNA modification on lnc-HZ09 (Figure 3G,H; Figure S7B) and expression level of lnc-HZ09 (Figure 3I,J), as determined by MeRIP assays and RT-qPCR analysis, respectively. The addition of 3-deazaadenosine (DAA), an inhibitor of m6A RNA methylation,49 resulted in lower m6A modification level on lnc-HZ09 (Figure 3K,L) and lower lnc-HZ09 expression level (Figure 3M). Furthermore, the up-regulation of lnc-HZ09 by overexpressing METTL3 was diminished by the addition of DAA (Figure 3N). Moreover, RNA stability assays also showed that, in Swan 71 and HTR-8/SVneo cells, cells overexpressing METTL3 had greater lnc-HZ09 stability, whereas cells with knockdown of METTL3 had lower lnc-HZ09 stability (Figure 3O; S7C). As control, alteration of METTL3 did not affect GAPDH mRNA stability in both cells (Figure S7D,E). Furthermore, the enhancement of lnc-HZ09 stability by overexpressing METTL3 was diminished by treating trophoblast cells with DAA (Figure 3O; Figure S7C).
Subsequently, the effects of METTL3 on PLD1 expression levels and trophoblast cell functions were also explored. In Swan 71 and HTR-8/SVneo cells, METTL3-overexpressed cells had lower PLD1 protein levels, whereas cells with knockdown of METTL3 or treated with DAA had higher PLD1 protein levels (Figure 3P; Figure S7F–H). Moreover, cells with overexpression of METTL3 had less migration and invasion, an effect that was mitigated by treating cells with DAA (Figure S7I). In contrast, cells with knockdown of METTL3 had greater migration and invasion of (Figure S7J).
The Effects of lnc-HZ09 on Migration and Invasion of BPDE-Exposed Human Trophoblast Cells through PLD1/RAC1/CDC42 Pathway
To discover the underlying mechanism of BPDE effects on migration and invasion of human trophoblast cells, the regulation of lnc-HZ09 on the PLD1/RAC1/CDC42 pathway was investigated in BPDE-exposed trophoblast cells. First, we found that the expression levels of mRNAs and proteins of members in this PLD1/RAC1/CDC42 pathway were all lower (Figure 4A; Figure S8A–D), whereas the levels of lnc-HZ09 were higher (Figure 1A–B), with increasing BPDE concentrations in BPDE-treated Swan 71 or HTR-8 cells. Of BPDE-treated Swan 71 or HTR-8/SVneo cells, those overexpressing lnc-HZ09 had less migration and invasion, whereas cells with knockdown of lnc-HZ09 had greater migration and invasion (Figure 4B–E; Figure S8E–F). In the PLD1/RAC1/CDC42 pathway, the levels of mRNAs and proteins were all lower with lnc-HZ09 overexpression and were all higher with lnc-HZ09 knockdown in BPDE-treated both trophoblast cells (Figure 4F; Figure S8G–N).
Figure 4.
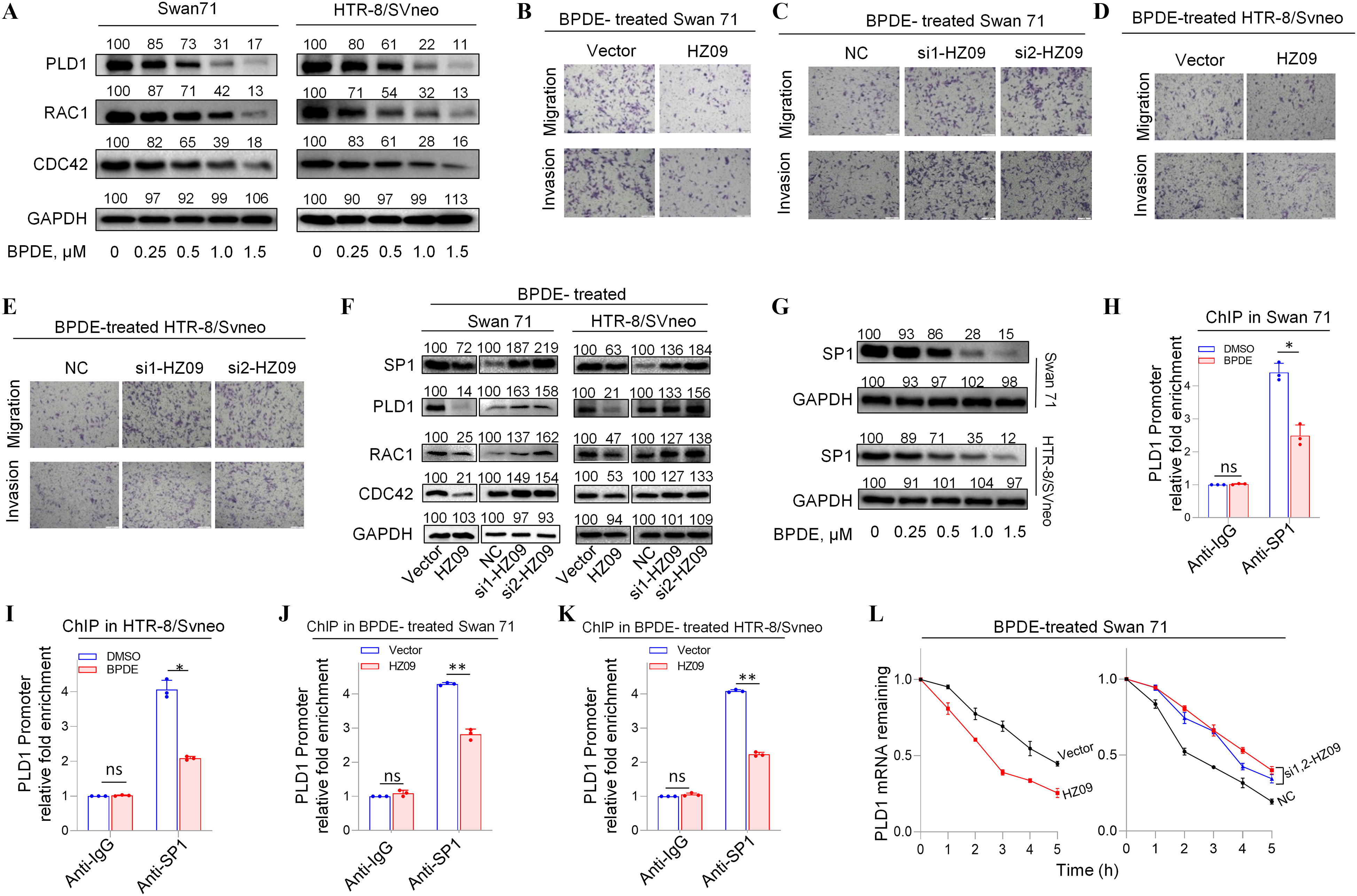
PLD1/RAC1/CDC42 pathway regulated by lnc-HZ09 in BPDE-exposed human trophoblast cells. (A) Representative western blot analysis of the protein levels of PLD1, RAC1 and CDC42 in BPDE-treated Swan 71 or HTR-8/SVneo cells, with GAPDH as internal standard. The relative intensity of each band was quantified and their of three replicates was shown in Figure S8A,B. (B–E) Representative transwell assay analysis of the migration and invasion of BPDE-treated Swan 71 (B and C) or HTR-8/SVneo (D and E) cells with overexpression or knockdown of lnc-HZ09 (scale bar, ). (F) Representative western blot analysis of the protein levels of SP1, PLD1, RAC1 and CDC42 in BPDE-treated Swan 71 or HTR-8/SVneo cells with overexpression or knockdown of lnc-HZ09, with GAPDH as internal standard. The relative intensity of each band was quantified and their of three replicates was shown in Figure S8G–J. (G) Representative western blot analysis of the protein levels of SP1 in BPDE-treated Swan 71 or HTR-8/SVneo cells, with GAPDH as internal standard. The relative intensity of each band was quantified and their of three replicates was shown in Figure S8O. (H–I) SP1 ChIP assay analysis (each ) of the relative enrichment of SP1 in the promoter region of PLD1 gene in untreated or BPDE-treated Swan 71 (H) or HTR-8/SVneo (I) cells. (J–K) SP1 ChIP assay analysis (each ) of the relative enrichment of SP1 in the promoter region of PLD1 gene in BPDE-treated Swan 71 (J) or HTR-8/SVneo (K) cells with overexpression of lnc-HZ09. (L–M) The mRNA stability of PLD1 (each ) in BPDE-treated Swan 71 (L) or HTR-8/SVneo (M) cells with overexpression or knockdown of lnc-HZ09. (N) Representative western blot analysis of the protein levels of PLD1 in BPDE-treated Swan 71 or HTR-8/SVneo cells with overexpression or knockdown of HuR, with as internal standard. The relative intensity of each band was quantified and their of three replicates was shown in Figure S9C. (O) The mRNA stability of PLD1 (each ) in BPDE-treated Swan 71 cells with overexpression or knockdown of HuR. (P) Representative western blot analysis of the protein levels of HuR in BPDE-treated Swan 71 or HTR-8/SVneo cells, with as internal standard. The relative intensity of each band was quantified and their of three replicates was shown in Figure S9H. (Q–R) RIP assay analysis (each ) of the relative levels of PLD1 mRNA that was pulled down by HuR protein in BPDE-treated Swan 71 (Q) or HTR-8/SVneo (R) cells with overexpression of lnc-HZ09. (S) Representative western blot analysis of MSX1 protein levels in BPDE-treated Swan 71 or HTR-8/SVneo cells, with GAPDH as internal standard. The relative intensity of each band was quantified and their of three replicates was shown in Figure S9L. (T) MSX1 ChIP assay analysis (each ) of the relative enrichment of MSX1 in the promoter region of lnc-HZ09 in untreated or BPDE-treated Swan 71 cells. (U) Representative western blot analysis of the protein levels of METTL3 in BPDE-treated Swan 71 or HTR-8/SVneo cells, with GAPDH as internal standard. The relative intensity of each band was quantified and their of three replicates was shown in Figure S9O. (V) MeRIP assay analysis (each ) of the relative levels of m6A RNA methylation on lnc-HZ09 in untreated or BPDE-treated Swan 71 cells. (W) The RNA stability of lnc-HZ09 (each ) in untreated or BPDE-treated Swan 71 cells. (X) The mRNA stability of PLD1 (each ) in untreated or BPDE-treated Swan 71 cells. The summary data of these bar charts and diagram were shown in Excel Table S1. The RNA levels in NC or Vector group were set as “1” in all of mRNA stability assays; the DNA or RNA levels in IgG group were set as “1” in all of ChIP, RIP, and MeRIP assays; and the band intensity in NC or Vector group was set as “100” in all of western blot assays. (A–G,N,P,S,U) show the representative data from three independent experiments. Data in (H–M,O,Q,R,T,V–X) show of three independent experiments. Two-tailed Student’s -test for (H–K, Q–R, T, V); *, **, and ***. Note: BPDE, benzo(a)pyrene-7,8-dihydrodiol-9,10-epoxide; ChIP, chromatin immunoprecipitation; GADPH, glyceraldehyde-3-phosphate dehydrogenase; HuR, overexpression of HuR; HZ09, overexpression of lnc-HZ09; IgG, immunoglobulin G; MeRIP, methylated RNA immunoprecipitation; NC, negative control of siRNA; ns, nonsignificance; PLD1, phospholipase D hydrolyze 1; RIP, RNA immunoprecipitation; SD, standard deviation; si-HuR, knockdown of HuR; si-HZ09, knockdown of lnc-HZ09; Vector, empty vector of pcDNA3.1.
The Effects of lnc-HZ09 on PLD1 mRNA Transcription and mRNA Stability in BPDE-Exposed Human Trophoblast Cells
How lnc-HZ09 regulated PLD1 mRNA transcription was further investigated in BPDE-treated trophoblast cells. The mRNA and protein levels of the transcription factor SP1 were lower in BPDE-treated Swan 71 or HTR-8/SVneo cells (Figure 4G; Figure S8O–P). SP1 ChIP assays showed that the occupancy of SP1 on the promoter region of PLD1 was less after BPDE treatment in both cells (Figure 4H–I). In BPDE-treated Swan 71 or HTR-8/SVneo cells, cells with lnc-HZ09 overexpression had lower SP1 mRNA and protein levels, whereas cells with lnc-HZ09 knockdown had higher SP1 mRNA and protein levels (Figure 4F; Figure S8G–J,Q–R). Furthermore, SP1 ChIP assays showed that in BPDE-treated Swan 71 or HTR-8/SVneo cells, cells with overexpression of lnc-HZ09 had less occupancy of SP1 on the promoter region of PLD1, whereas cells with knockdown of lnc-HZ09 had greater occupancy of SP1 on the promoter region of PLD1 (Figure 4J–K; Figure S8S–T).
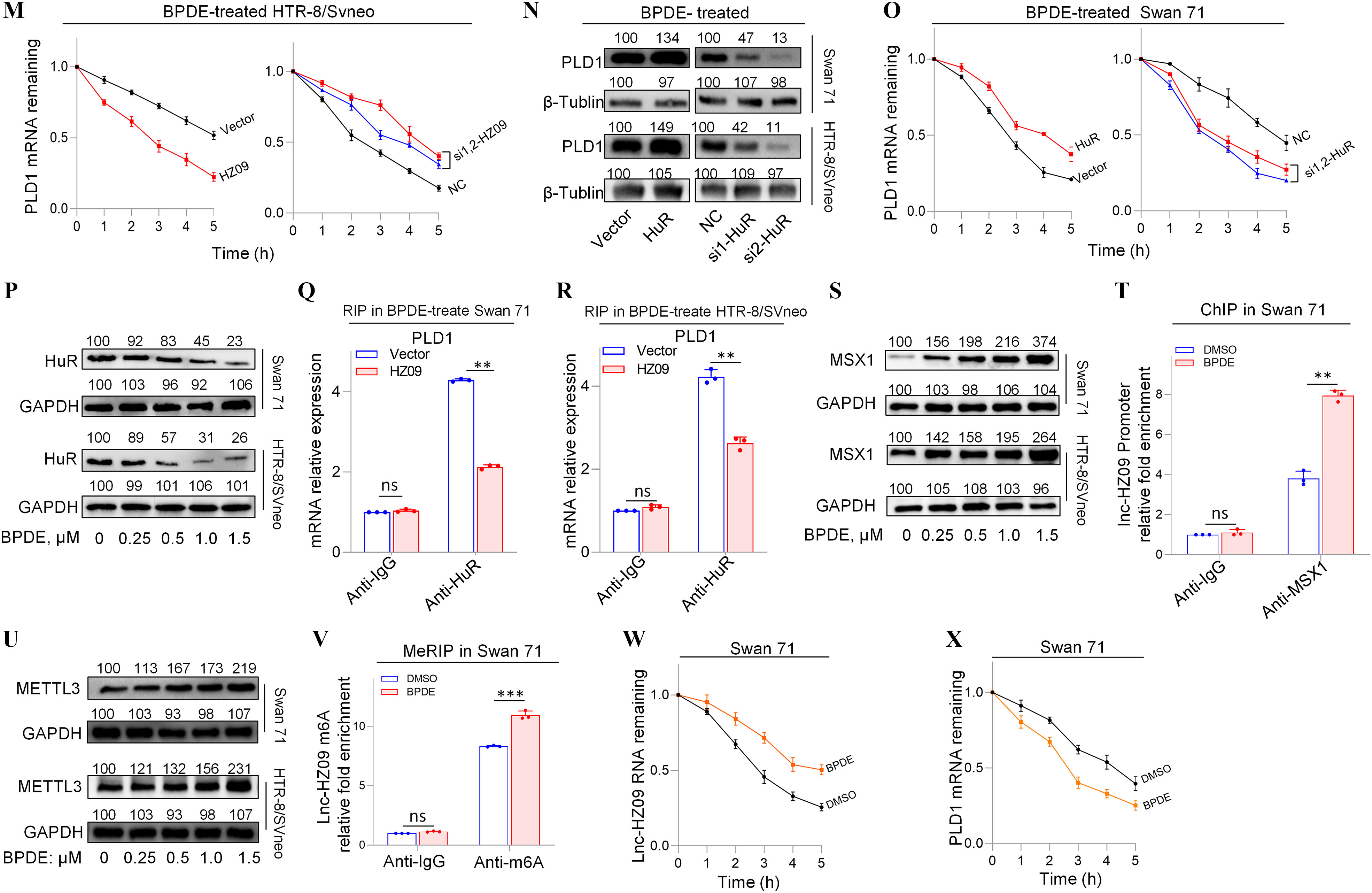
Furthermore, the effects of lnc-HZ09 on PLD1 mRNA stability were also studied in BPDE-treated trophoblast cells. In BPDE-treated trophoblast cells, cells with overexpression of lnc-HZ09 had lower, whereas cells with knockdown of lnc-HZ09 had greater PLD1 mRNA stability (Figure 4L–M). As control, alteration of lnc-HZ09 did not affect GAPDH mRNA stability in BPDE-treated both cells (Figure S9A,B). In BPDE-treated Swan 71 or HTR-8/SVneo cells, cells with overexpression of HuR had higher PLD1 mRNA and protein levels, whereas cells with knockdown of HuR had lower PLD1 mRNA and protein levels (Figure 4N; Figure S9C–F). Furthermore, BPDE-treated Swan 71 cells overexpressing HuR also had greater PLD1 mRNA stability; BPDE-treated Swan 71 cells with HuR knockdown had less PLD1 mRNA stability (Figure 4O). However, alteration of HuR did not affect GAPDH mRNA stability (Figure S9G). Moreover, cells (both Swan 71 and HTR8/SVneo) treated with BPDE had lower HuR protein expression levels (Figure 4P; Figure S9H). However, HuR levels were not affected by lnc-HZ09 in BPDE-treated trophoblast cells (Figure S9I,J). Furthermore, cells with overexpression of lnc-HZ09 had lower levels of PLD1 mRNA pulled down by HuR protein in BPDE-treated trophoblast cells, whereas those with knockdown of lnc-HZ09 had higher levels of PLD1 mRNA pulled down by HuR protein in BPDE-treated trophoblast cells (Figure 4Q,R; Figure S9K,L), suggesting lnc-HZ09 competed with PLD1 mRNA to bind with HuR.
Examination of lnc-HZ09 Transcription and Stability in BPDE-Exposed Human Trophoblast Cells
Subsequently, lnc-HZ09 transcription was investigated in BPDE-treated trophoblast cells. The mRNA and protein levels of MSX1 were higher in BPDE-treated trophoblast cells (Figure 4S; Figure S9M,N). ChIP assays showed that the occupancy of MSX1 on the promoter region of lnc-HZ09 was greater in BPDE-treated Swan 71 or HTR-8/SVneo cells (Figure 4T; Figure S9O).
Finally, lnc-HZ09 stability was also investigated in BPDE-treated trophoblast cells. The mRNA and protein levels of METTL3 (Figure 4U; Figure S9P,Q), as well as the levels of m6A RNA methylation on lnc-HZ09 (Figure 4V; Figure S9R), were all higher in BPDE-treated trophoblast cells. Lnc-HZ09 RNA stability was greater in BPDE-treated Swan 71 cells, whereas PLD1 mRNA stability was lower in BPDE-treated Swan 71 cells (Figure 4W,X). As control, GAPDH mRNA stability was not affected with BPDE treatment (Figure S9S,T).
The Correlation of lnc-HZ09 with PLD1/RAC1/CDC42 Pathway in RM Villous Tissues
Having known the regulatory roles of lnc-HZ09 in trophoblast cells, its roles in villous tissues were further explored. We collected villous tissue samples from unexplained the RM and the matched healthy control (HC) groups (each ). The parameters, such as body mass index, age, gestational days, did not show significant differences between the RM and HC groups (Table S12). However, the levels of BPDE-DNA adducts were significantly higher in RM group than those in HC group (Table S12; Figure S10A). We also found that lnc-HZ09 was significantly highly expressed in RM group relative to those in HC group (Figure 5A), and this trend was consistent with that observed in BPDE-treated human trophoblast cells, suggesting that lnc-HZ09 might simultaneously regulate BPDE-induced dysfunctions of human trophoblast cells and the occurrence of miscarriage. The mRNA and protein levels of PLD1/RAC1/CDC42 were all lower in RM tissues than those in HC tissues (Figure 5B,C; Figure S10B,C). Correlation analysis showed that the levels of PLD1, RAC1, and CDC42 were all negatively correlated with those of lnc-HZ09 in RM tissues (Figure 5D,E; Figure S10D–G). The locations of most data points in HC and RM groups were relatively separated, manifesting that this pathway was differentially regulated.
Figure 5.
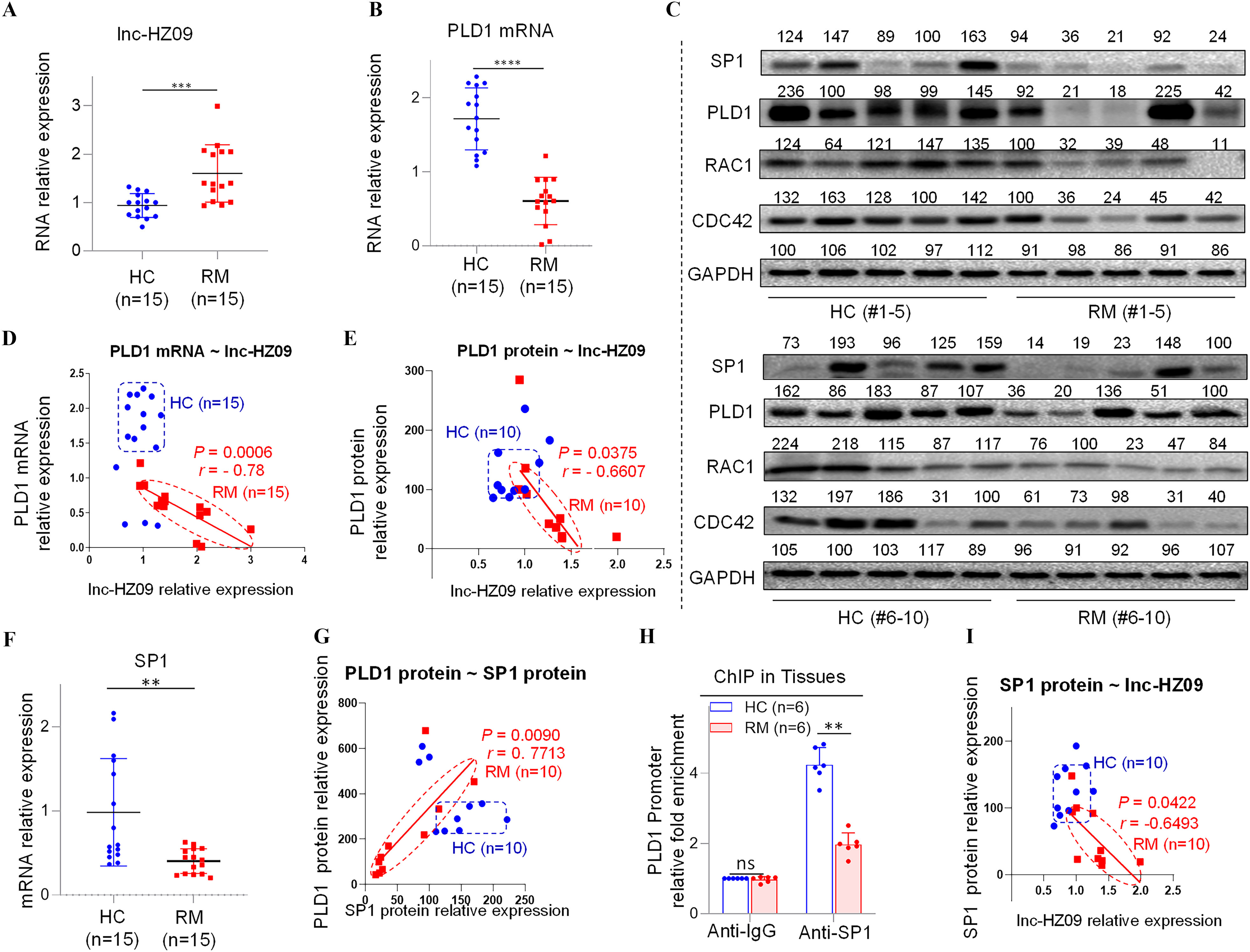
Regulation roles of lnc-HZ09 in human villous tissues. (A–B) RT-qPCR analysis of the levels of lnc-HZ09 (A) or PLD1 mRNA (B) in HC (healthy control, round) and RM (recurrent miscarriage, square) tissues (each ). (C) Western blot analysis of the protein levels of SP1, PLD1, RAC1, and CDC42 in HC and RM tissues (each ), with GAPDH as internal standard. The relative intensity of each band was quantified, and their levels were shown in Figure S10C. (D) The correlation between PLD1 mRNA levels and lnc-HZ09 levels in HC (round) and RM (square) tissues (each ). (E) The correlation between PLD1 protein levels and lnc-HZ09 levels in HC (round) and RM (square) groups (each ). (F) RT-qPCR analysis (each ) of the mRNA levels of SP1 in HC (round) and RM (square) tissues (each ). (G) The correlation between the protein levels of PLD1 and SP1 in HC (round) and RM (square) groups (each ). (H) SP1 ChIP assay analysis of the relative enrichment of SP1 in the promoter region of PLD1 gene in HC and RM tissues (each ). (I) The correlation between SP1 protein levels and lnc-HZ09 levels in HC (round) and RM (square) groups (each ). (J) RT-qPCR analysis of the mRNA levels of MSX1 in HC and RM tissues (each ). (K) Western blot analysis of the protein levels of HuR, MSX1, and METTL3 in HC and RM tissues (each ), with GAPDH as internal standard. The relative intensity of each band was quantified, and their levels were shown in Figure S10H,J,K. (L) MSX1 ChIP assay analysis of the relative enrichment of MSX1 in the promoter region of lnc-HZ09 in HC and RM tissues (each ). (M) The correlation between the levels of lnc-HZ09 and the protein levels of MSX1 in HC (round) and RM (square) groups (each ). (N) RT-qPCR analysis (each ) of the mRNA levels of METTL3 in HC and RM tissues (each ). (O) MeRIP assay analysis (each ) of the levels of m6A RNA methylation on lnc-HZ09 in HC and RM tissues (each ). The summary data of these bar charts and scatter plots were shown in Excel Table S1. The DNA or RNA level in IgG group was set as “1” in all of ChIP and MeRIP assays and the middle intensity value was set to “100” in all of western blot assays. (C–E, G, I, K, M) shows representative data from three independent experiments. Data in (H, L, O) show of six independent experiments. Two-tailed Student’s -test for (A–B,F,H,J,L,N–O); Pearson analysis for (D,E,G,I,M). *, **, and ***. Note: ChIP, chromatin immunoprecipitation; GADPH; glyceraldehyde-3-phosphate dehydrogenase; HC, healthy control group; IgG, immunoglobulin G; MeRIP, methylated RNA immunoprecipitation; n, the number of biologically independent samples; ns, nonsignificance; PLD1, phospholipase D hydrolyze 1; RM, recurrent miscarriage group.
The Effects of lnc-HZ09 on PLD1 mRNA Transcription and mRNA Stability in RM Villous Tissues
Subsequently, the effects of lnc-HZ09 on PLD1 mRNA transcription were studied in villous tissues. The mRNA and protein levels of PLD1 and its transcription factor SP1 were all expressed at a low level in RM tissues relative to those in HC tissues (Figure 5B,C,F; Figure S10C). The levels of PLD1 and SP1 were positively correlated in RM tissues (Figure 5G). SP1 ChIP assays showed that the occupancy of SP1 on the promoter region of PLD1 was lower in RM tissues relative to that in HC tissues (Figure 5H), suggesting that SP1-mediated PLD1 transcription was suppressed in RM tissues. Furthermore, the levels of lnc-HZ09 were negatively correlated with the levels of SP1 in RM tissues (Figure 5I).
Then, the effects of lnc-HZ09 on PLD1 mRNA stability were also studied in villous tissues. The levels of HuR were lower in RM tissues in comparison with those in HC tissues (Figure 5K; Figure S10H). The protein levels of HuR and PLD1 were positively correlated in RM tissues (Figure S10I).
Measurement of lnc-HZ09 Transcription and RNA Stability in RM Tissues
In tissues, the mRNA and protein levels of MSX1 were higher in RM tissues relative to HC tissues (Figure 5J,K; Figure S10J). MSX1 ChIP assays showed that the occupancy of MSX1 on the promoter region of lnc-HZ09 was greater in RM tissues relative to HC tissues (Figure 5L). The levels of MSX1 were positively correlated with the levels of lnc-HZ09 in RM tissues (Figure 5M).
Lnc-HZ09 stability was also studied in villous tissues. The mRNA and protein levels of METTL3 (Figure 5K,N; Figure S10K), as well as the levels of m6A RNA methylation on lnc-HZ09 (Figure 5O), were all higher in RM tissues relative to those in HC tissues. The levels of METTL3 were positively correlated with the levels of lnc-HZ09 in RM tissues (Figure S10L).
Evaluation of Sp1-Mediated Pld1 mRNA Transcription in Placental Tissues of Mice with B(a)P-Induced Miscarriage
To investigate the mechanism by which B(a)P induced miscarriage in vivo, we constructed a mouse model by treating pregnant mice with 0, 0.05, or B(a)P to induce miscarriage, as described previously.28,29,31 Considering that B(a)P is metabolized into BPDE in organisms, we treated mice with B(a)P directly. After sequence alignment, we found that PLD1, RAC1, CDC42, and SP1 genes are evolutionarily conservative in human, rhesus, elephants, dogs, and mice (Figure S11A–D; Table S13), implying that this migration and invasion pathway might be conserved among these species. However, lnc-HZ09 sequence was conserved in only human and rhesus, but not in mice (Figure S11E). Using this miscarriage model, we found that the mRNA and protein levels of murine Pld1, Rac1 and Cdc42 were all lower in placental tissues with increasing B(a)P concentrations (Figure 6A–C,E; Figure S11F). This change trend was consistent with that found in BPDE-treated human trophoblast cells and in RM tissues.
Figure 6.
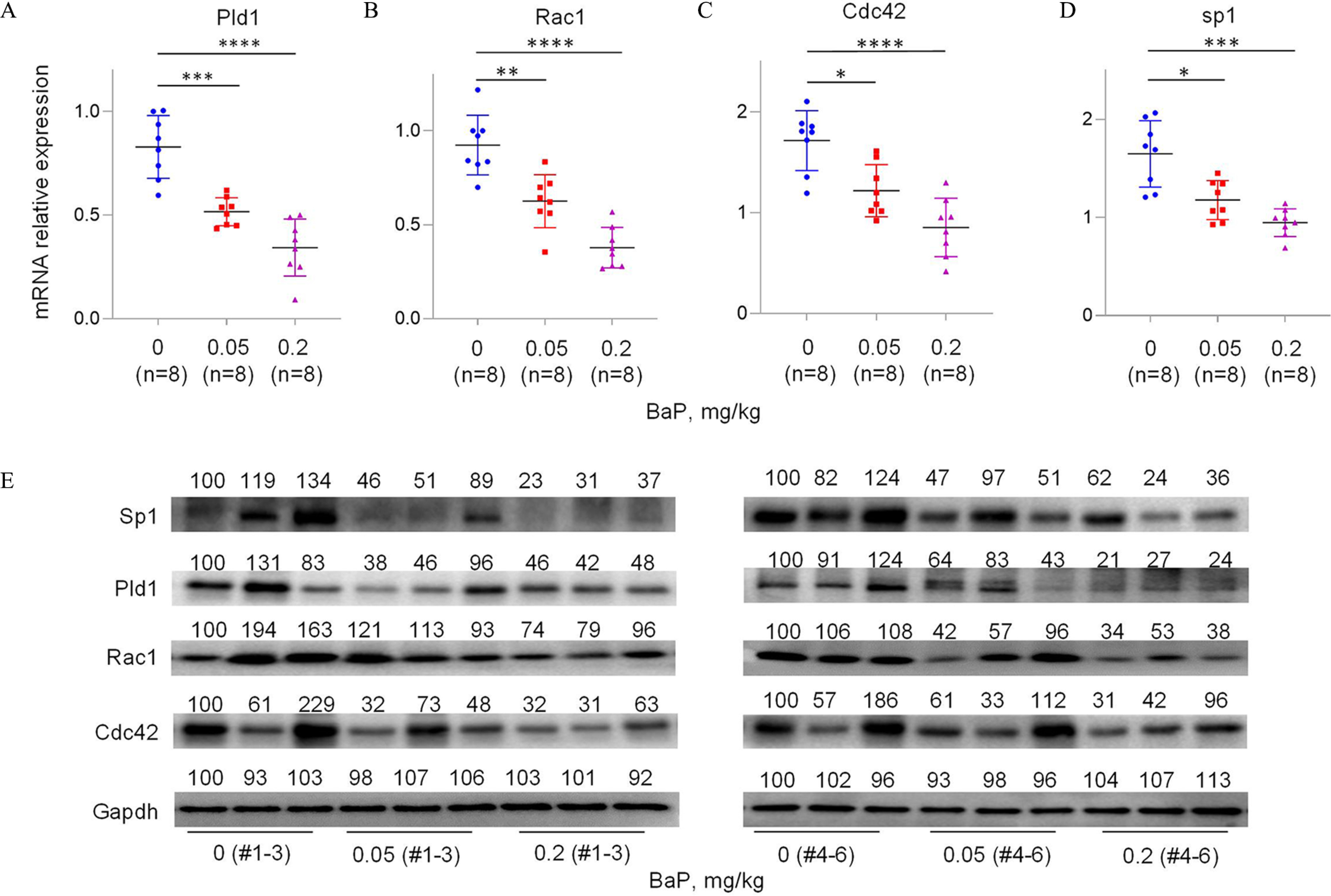
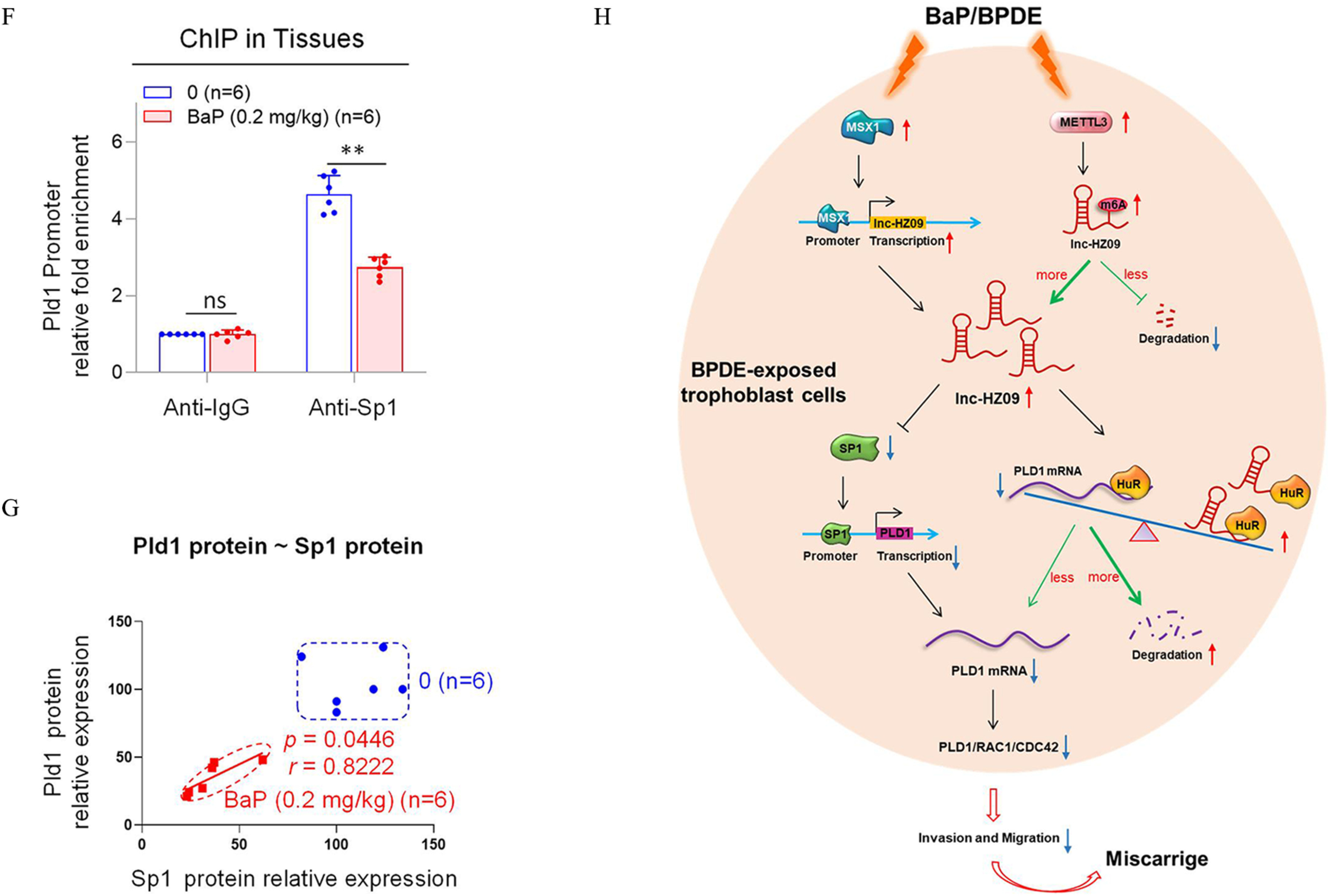
Pld1/Rac1/Cdc42 pathway in placental tissues of mice with BaP-induced miscarriage. (A–D) RT-qPCR analysis of the mRNA levels of Pld1 (A), Rac1 (B), Cdc42 (C), and Sp1 (D) in each B(a)P-treated mouse group (each ). (E) Western blot analysis of the protein levels of Sp1, Pld1, Rac1, and Cdc42 in each B(a)P-treated mouse group (each ), with GAPDH as internal standard. The relative intensity of each band was quantified and their levels were shown in Figure S11F. (F) Sp1 ChIP assay analysis of the relative enrichment of Sp1 in the promoter region of Pld1 gene in control and B(a)P-treated mouse groups (each ). (G) The correlation between the protein levels of Pld1 and Sp1 in control (round) and B(a)P-treated (square) groups (each ). (H) The proposed regulation mechanism of lnc-HZ09. Lnc-HZ09 suppressed SP1-mediated PLD1 mRNA transcription and also reduced PLD1 mRNA stability by competitively restraining the binding of PLD1 mRNA with HuR, a protein that could stabilize PLD1 mRNA. BPDE exposure promoted MSX1-mediated lnc-HZ09 transcription and also enhanced lnc-HZ09 RNA stability by up-regulating its m6A methylation modification. Thus, BPDE exposure might up-regulate lnc-HZ09 expression level, suppress PLD1/RAC1/CDC42 pathway, inhibit the migration and invasion, and might further induce miscarriage. The summary data of these bar charts and scatter plots were shown in Excel Table S1. The DNA level in IgG group was set as “1” in ChIP assay, and the middle intensity value was set as “100” in western blot assay. (A–E,G) shows representative data from three independent experiments. Data in (F) show of six independent experiments. Two-tailed Student’s -test for (F); one-way ANOVA analysis for (A–D), Pearson analysis for (G). (H) was generated by Microsoft Office PowerPoint. *, **, and ***. Note: B(a)P, benzo(a)pyrene; BPDE, benzo(a)pyrene-7,8-dihydrodiol-9,10-epoxide; ChIP, chromatin immunoprecipitation; GADPH, glyceraldehyde-3-phosphate dehydrogenase; IgG, immunoglobulin G; n, the number of biologically independent samples; ns, nonsignificance; RT-qPCR, quantitative reverse transcription polymerase chain reaction; SD, standard deviation.
Furthermore, the transcription of Pld1 was also explored in B(a)P-treated mouse model. The mRNA and protein levels of murine transcription factor Sp1 were lower with increasing B(a)P concentrations (Figure 6D,E). Moreover, Sp1 ChIP assays showed that the occupancy of Sp1 on the promoter region of Pld1 was lower in B(a)P-treated mouse group relative to those in the untreated group (Figure 6F). The levels of Sp1 were positively correlated with the levels of Pld1 in group (Figure 6G). The locations of most data points in these groups were relatively separated.
Discussion
Increasing attention has been paid to environmental carcinogens and human reproductive health. B(a)P, one of the most widely spread and unavoidably environmental carcinogens, could induce various adverse pregnancy outcomes.50 Emerging studies have revealed that lncRNAs may regulate the occurrence of miscarriage.26,51 In our recent works, we identified several novel lncRNAs that regulated trophoblast cell proliferation, apoptosis, and other cell functions by different pathways.28,29 However, evidence that lncRNAs regulate trophoblast invasion and migration under B(a)P exposure conditions and thus affect miscarriage is still lacking. In this work, we performed in vitro cellular experiments, human tissue experiments, and mouse model experiments, and we found that lnc-HZ09 was highly expressed in BPDE-treated human trophoblast cells and in RM tissues relative to HC tissues (Figure 6H). The mRNA and protein levels of members of the PLD1/RAC1/CDC42 pathway were lower in BPDE-treated human trophoblast cells, in RM relative to HC tissues, and in the placental tissues of B(a)P-treated mice. All these results suggest that lnc-HZ09 suppressed the invasion and migration of trophoblast cells by down-regulating the PLD1/RAC1/CDC42 pathway in BPDE-exposed human trophoblast cells and in RM tissues. However, by searching the NCBI database, we did not find the murine counterpart of lnc-HZ09 in mouse systems, implying that lnc-HZ09 might have specific epigenetic regulation roles in human system.
Regulation Mechanisms of lnc-HZ09
The regulation mechanisms of lnc-HZ09 in human trophoblast cells were proposed (Figure 6H). Based on these data, we hypothesize that MSX1, a transcription factor of lnc-HZ09, promoted lnc-HZ09 transcription. Furthermore, we suggest that METTL3 promoted m6A methylation modification on lnc-HZ09 and enhanced its RNA stability. Thus, both MSX1 and METTL3 may positively regulate lnc-HZ09 expression level. Subsequently, we hypothesize that lnc-HZ09 suppressed SP1 expression, which was a transcription factor of PLD1 and thus inhibited SP1-mediated PLD1 transcription and reduced PLD1 expression level. In addition, lnc-HZ09 and PLD1 mRNA may competitively bind with HuR, which is an RNA binding protein and could maintain RNA stability. We posit that lnc-HZ09 impaired the binding of PLD1 mRNA with HuR and thus reduced PLD1 mRNA stability and its expression level. Therefore, we suggest that lnc-HZ09 finally down-regulated PLD1 expression level, which further suppressed the PLD1/RAC1/CDC42 pathway and inhibited the migration and invasion of human trophoblast cells. Once trophoblast cells are exposed to environmental B(a)P or BPDE, the expression levels of MSX1 and METTL3 would be increased, which subsequently increased the lnc-HZ09 level, down-regulated the PLD1/RAC1/CDC42 pathway, suppressed the migration and invasion of trophoblast cells, and might further induce miscarriage.
Roles of lnc-HZ09 in Regulation of PLD1 Expression
In general, PLD1 is abnormally up-regulated in various cancers and is related to tumor malignancy, maintenance of self-renewal of cancer stem cells and resistance to radiotherapy and chemotherapy.52 PLD1 was shown to promote the invasion, migration, and proliferation of glioblastoma cell lines.53 In glioblastoma, abnormally elevated transcription factor SP1 enhanced PLD1 transcription and increased PLD1 expression level, which further enhanced the drug resistance of glioblastoma tumor cells.53 It has been proposed that early embryonic development and tumor metastasis might have similar biological manifestations.54 In our work, we found that lnc-HZ09 suppressed SP1-mediated PLD1 transcription in human RM tissues and in BPDE-treated human trophoblast cells, which might possibly be similar in cancer cell lines. Moreover, lnc-HZ09 also restrained the binding of PLD1 mRNA with HuR and reduced PLD1 mRNA stability. Finally, due to the regulatory roles of lnc-HZ09, PLD1 transcription was inhibited and its degradation was increased, and thus the expression level of PLD1 was ultimately decreased. However, the actual regulatory mechanisms of lnc-HZ09 on PLD1 expression should be further explored in a specific cancer cell line.
The Roles of m6A Modification in Regulation of Miscarriage
M6A modification regulates various physiological and pathological processes.55 It has been reported that linc1281 played an important role in proper differentiation of embryonic stem cells by acting as a ceRNA to attenuate the functions of let-7 miRNAs.56 The m6A enrichment on linc1281 was required for this linc1281-mediated ceRNA model. In our recent work, we found that m6A modification on lnc-HZ01 enhanced lnc-HZ01 RNA stability, further regulated trophoblast cell proliferation, and was associated with the occurrence of miscarriage.29 In this work, we found that lnc-HZ09 also contained m6A modification, which was associated with greater lnc-HZ09 stability and higher expression level. In BPDE-treated trophoblast cells and RM tissues, the levels of m6A modification on lnc-HZ09 are higher, which was associated with higher lnc-HZ09 expression level, and less migration and invasion of trophoblast cells.
The Upstream Effects of Environment Carcinogens on Trophoblast-Related Adverse Pregnancy Outcomes
To study the possible causes and mechanism of unexplained RM, we collected villous tissue samples from RM and HC groups, with exclusion of the known causes of miscarriage. We found that the levels of BPDE-DNA adducts were significantly higher in the RM group than those in the HC group. This effect is possibly because B(a)P is a ubiquitous environmental carcinogen,57,58 and some women may inevitably intake more B(a)P. It has been reported that female smokers have significantly higher levels of BaP () in their follicular fluid in comparison with their nonsmoking counterparts ().13 The levels of BPDE-DNA adducts in RM and HC villous tissues detected in this study also agreed with a previous case–control study of miscarriage, in which 2.2-fold more BPDE-DNA adducts were detected in maternal blood of the miscarriage group relative to the health control group.14 In this work, our data showed that BPDE exposure might up-regulate the lnc-HZ09 level, inhibit the PLD1/RAC1/CDC42 pathway, suppress migration and invasion, and finally induce miscarriage. Notably, it would likely be the suppressed invasion and migration of trophoblast cells rather than BPDE or B(a)P exposure that directly induces miscarriage. When the PLD1/RAC1/CDC42 pathway is inhibited, the upstream BPDE exposure might become less vital for the ultimate miscarriage. In a larger sense, not only BPDE or B(a)P but other environmental factors or pathways might also suppress trophoblast cell invasion and migration. For example, bisphenol A and para-nonylphenol suppressed the migration and invasion of HTR-8/SVneo cells.59 Heavy metal cadmium (Cd)-induced apoptosis and inhibited migration and invasion of HTR-8/SVneo cells in a dose-dependent manner.60 Besides of miscarriage, dysfunctions of trophoblast cells might also induce other trophoblast-related adverse pregnancy outcomes, such as eclampsia, preeclampsia, intrauterine growth restriction, and gestational diabetes. Epidemiological studies have shown that PAHs exposure might be correlated with these adverse pregnancy outcomes.15,62 BPDE inhibited the invasion and migration of trophoblast cells, which implies that BPDE might be one possible compound that induces these adverse pregnancy outcomes. In a larger sense, other environmental carcinogens, such as air pollutants,63 toxic metals (Cd and palladium),64,65 or trihalomethanes in drinking water,63 may also lead to human trophoblast cell dysfunctions, which suggests that these environmental carcinogens might also induce these trophoblast-related adverse pregnancy outcomes. It is a long stream line from the upstream environmental carcinogen exposure to the ultimate adverse pregnancy outcomes, providing multiple potential biotargets or biomarkers for diagnosis and treatment against these trophoblast-related adverse pregnancy outcomes.
The Limitation of This Work
In this work, we collected villous tissues () to verify the cellular results. More samples should be collected to exclude the possible demographic differences. Because lnc-HZ09 counterpart was not identified in mouse system, only the Pld1/Rac1/Cdc42 pathway was studied in the mouse model. However, the epigenetic regulation on this pathway in mouse models should be further explored. In these tissue samples, we evaluated the internal exposure level of B(a)P by detecting BPDE-DNA adducts in genomic DNA in RM and HC villous tissues, which might reflect the total external exposures of B(a)P. However, we did not consider a single external environmental exposure, such as smoking, in this work. Although the B(a)P level was detected to be higher in the RM group relative to the HC group, we cannot exclude the possibility that other carcinogens or genetic differences might also induce miscarriage. Other carcinogens that might induce miscarriage should be further explored.
Conclusion
RM is a global problem that produces severe challenges to socioeconomic development. Although many studies have indicated that various risk factors may induce miscarriage in the first trimester, about half of RMs have unknown clinical causes.66 In this work, we have identified a novel lnc-HZ09, which suppressed the migration and invasion of human trophoblast cells and affected the occurrence of miscarriage. In mechanism, this lnc-HZ09 suppressed SP1-mediated PLD1 mRNA transcription and reduced PLD1 mRNA stability. BPDE exposure promoted MSX1-mediated lnc-HZ09 transcription and also increased lnc-HZ09 stability by up-regulating its m6A methylation modification. Thus, BPDE exposure up-regulated the lnc-HZ09 level, suppressed the PLD1/RAC1/CDC42 pathway, inhibited migration and invasion, and might further induce miscarriage. In summary, this work provided novel insights in the roles of lnc-HZ09 in regulation of BPDE-induced dysfunctions of human trophoblast cells and the occurrence of unexplained miscarriage.
Supplementary Material
Acknowledgments
The authors acknowledge K. Inman, Science Editor of Environmental Health Perspectives, for careful and comprehensive revision of this manuscript. The authors acknowledge financial support from the Natural Science Foundation of China (NSFC No. 82073589), the Fundamental Research Funds for the Central Universities, Shenzhen Science and Technology Program (No. JCYJ20210324114814038) and Futian Health Care Research Project (No. FTWS2021009). The authors also acknowledge the facility supports by Central Laboratory of the Eighth Affiliated Hospital at Sun Yat-sen University.
M.D., W.H., and H.Z. designed this study. M.D. and W.H. performed most of the experiments. M.D., W.H., and H.Z. wrote the draft manuscript. X.H. and C.M. were responsible for animal experiments. R.W. and P.T. contributed to the tissue preparation. W.C., Y.Z., and C.M. conducted partial experiments.
References
- 1.Jurkovic D, Overton C, Bender-Atik R. 2013. Diagnosis and management of first trimester miscarriage. BMJ 346:f3676, PMID: , 10.1136/bmj.f3676. [DOI] [PubMed] [Google Scholar]
- 2.Ammon Avalos L, Galindo C, Li DK. 2012. A systematic review to calculate background miscarriage rates using life table analysis. Birth Defects Res A Clin Mol Teratol 94(6):417–423, PMID: , 10.1002/bdra.23014. [DOI] [PubMed] [Google Scholar]
- 3.Savitz DA, Hertz-Picciotto I, Poole C, Olshan AF. 2002. Epidemiologic measures of the course and outcome of pregnancy. Epidemiol Rev 24(2):91–101, PMID: , 10.1093/epirev/mxf006. [DOI] [PubMed] [Google Scholar]
- 4.Pereza N, Ostojic S, Kapovic M, Peterlin B. 2017. Systematic review and meta-analysis of genetic association studies in idiopathic recurrent spontaneous abortion. Fertil Steril 107(1):150–159.e2, PMID: , 10.1016/j.fertnstert.2016.10.007. [DOI] [PubMed] [Google Scholar]
- 5.Vaiman D. 2015. Genetic regulation of recurrent spontaneous abortion in humans. Biomed J 38(1):11–24, PMID: , 10.4103/2319-4170.133777. [DOI] [PubMed] [Google Scholar]
- 6.Chan YY, Jayaprakasan K, Tan A, Thornton JG, Coomarasamy A, Raine-Fenning NJ. 2011. Reproductive outcomes in women with congenital uterine anomalies: a systematic review. Ultrasound Obstet Gynecol 38(4):371–382, PMID: , 10.1002/uog.10056. [DOI] [PubMed] [Google Scholar]
- 7.Temple R, Aldridge V, Greenwood R, Heyburn P, Sampson M, Stanley K. 2002. Association between outcome of pregnancy and glycaemic control in early pregnancy in type 1 diabetes: population based study. BMJ 325(7375):1275–1276, PMID: , 10.1136/bmj.325.7375.1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.ESHRE Capri Workshop Group. 2008. Genetic aspects of female reproduction. Hum Reprod Update 14:293–307, PMID: , 10.1093/humupd/dmn009. [DOI] [PubMed] [Google Scholar]
- 9.Regan L, Rai R. 2000. Epidemiology and the medical causes of miscarriage. Baillieres Best Pract Res Clin Obstet Gynaecol 14(5):839–854, PMID: , 10.1053/beog.2000.0123. [DOI] [PubMed] [Google Scholar]
- 10.Bostrom CE, Gerde P, Hanberg A, Jernström B, Johansson C, Kyrklund T, et al. 2002. Cancer risk assessment, indicators, and guidelines for polycyclic aromatic hydrocarbons in the ambient air. Environ Health Perspectives 110(suppl 3):451–488, PMID: , 10.1289/ehp.110-1241197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Alegbeleye OO, Opeolu BO, Jackson VA. 2017. Polycyclic aromatic hydrocarbons: a critical review of environmental occurrence and bioremediation. Environ Manage 60(4):758–783, PMID: , 10.1007/s00267-017-0896-2. [DOI] [PubMed] [Google Scholar]
- 12.Sun K, Song Y, He F, Jing M, Tang J, Liu R. 2021. A review of human and animals exposure to polycyclic aromatic hydrocarbons: health risk and adverse effects, photo-induced toxicity and regulating effect of microplastics. Sci Total Environ 773:145403, PMID: , 10.1016/j.scitotenv.2021.145403. [DOI] [PubMed] [Google Scholar]
- 13.Neal MS, Zhu J, Holloway AC, Foster WG. 2007. Follicle growth is inhibited by benzo-[a]-pyrene, at concentrations representative of human exposure, in an isolated rat follicle culture assay. Hum Reprod 22(4):961–967, PMID: , 10.1093/humrep/del487. [DOI] [PubMed] [Google Scholar]
- 14.Ptashekas J, Ciuniene E, Barkiene M, Zurlyte I, Jonauskas G, Sliachtic N, et al. 1996. Environmental and health monitoring in Lithuanian cities: exposure to heavy metals and benz(a)pyrene in Vilnius and Siauliai residents. J Environ Pathol Toxicol Oncol 15(2–4):135–141, PMID: . [PubMed] [Google Scholar]
- 15.Wu J, Hou H, Ritz B, Chen Y. 2010. Exposure to polycyclic aromatic hydrocarbons and missed abortion in early pregnancy in a Chinese population. Sci Total Environ 408(11):2312–2318, PMID: , 10.1016/j.scitotenv.2010.02.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao Y, Chen X, Liu X, Ding Y, Gao R, Qiu Y, et al. 2014. Exposure of mice to benzo(a)pyrene impairs endometrial receptivity and reduces the number of implantation sites during early pregnancy. Food Chem Toxicol 69:244–251, PMID: , 10.1016/j.fct.2014.04.021. [DOI] [PubMed] [Google Scholar]
- 17.Li X, Shen C, Liu X, He J, Ding Y, Gao R, et al. 2017. Exposure to benzo[a]pyrene impairs decidualization and decidual angiogenesis in mice during early pregnancy. Environ Pollut 222:523–531, PMID: , 10.1016/j.envpol.2016.11.029. [DOI] [PubMed] [Google Scholar]
- 18.Ji L, Brkić J, Liu M, Fu G, Peng C, Wang Y-L. 2013. Placental trophoblast cell differentiation: physiological regulation and pathological relevance to preeclampsia. Mol Aspects Med 34(5):981–1023, PMID: , 10.1016/j.mam.2012.12.008. [DOI] [PubMed] [Google Scholar]
- 19.Ball E, Bulmer JN, Ayis S, Lyall F, Robson SC. 2006. Late sporadic miscarriage is associated with abnormalities in spiral artery transformation and trophoblast invasion. J Pathol 208(4):535–542, PMID: , 10.1002/path.1927. [DOI] [PubMed] [Google Scholar]
- 20.Zong S, Li C, Luo C, Zhao X, Liu C, Wang K, et al. 2016. Dysregulated expression of IDO may cause unexplained recurrent spontaneous abortion through suppression of trophoblast cell proliferation and migration. Sci Rep 6:19916, PMID: , 10.1038/srep19916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ye Y, Jiang S, Zhang C, Cheng Y, Zhong H, Du T, et al. 2020. Environmental pollutant benzo[a]pyrene induces recurrent pregnancy loss through promoting apoptosis and suppressing migration of extravillous trophoblast. BioMed Res Int 2020:8983494, PMID: , 10.1155/2020/8983494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jenkins GM, Frohman MA. 2005. Phospholipase D: a lipid centric review. Cell Mol Life Sci 62(19–20):2305–2316, PMID: , 10.1007/s00018-005-5195-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burridge K, Wennerberg K. 2004. Rho and Rac take center stage. Cell 116(2):167–179, PMID: , 10.1016/s0092-8674(04)00003-0. [DOI] [PubMed] [Google Scholar]
- 24.Powner DJ, Hodgkin MN, Wakelam MJ. 2002. Antigen-stimulated activation of phospholipase D1b by Rac1, ARF6, and PKCalpha in RBL-2H3 cells. Mol Biol Cell 13(4):1252–1262, PMID: , 10.1091/mbc.01-05-0235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Costa FF. 2010. Non-coding RNAs: meet thy masters. Bioessays 32(7):599–608, PMID: , 10.1002/bies.200900112. [DOI] [PubMed] [Google Scholar]
- 26.Zhu Y, Liu Q, Liao M, Diao L, Wu T, Liao W, et al. 2019. Overexpression of lncRNA EPB41L4A-AS1 induces metabolic reprogramming in trophoblast cells and placenta tissue of miscarriage. Mol Ther Nucleic Acids 18:518–532, PMID: , 10.1016/j.omtn.2019.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Huang Z, Du G, Huang X, Han L, Han X, Xu B, et al. 2018. The enhancer RNA lnc-SLC4A1-1 epigenetically regulates unexplained recurrent pregnancy loss (URPL) by activating CXCL8 and NF-kB pathway. EBioMedicine 38:162–170, PMID: , 10.1016/j.ebiom.2018.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liang T, Xie J, Zhao J, Huang W, Xu Z, Tian P, et al. 2021. Novel lnc-HZ03 and miR-hz03 promote BPDE-induced human trophoblastic cell apoptosis and induce miscarriage by upregulating p53/SAT1 pathway. Cell Biol Toxicol 37(6):951–970, PMID: , 10.1007/s10565-021-09583-3. [DOI] [PubMed] [Google Scholar]
- 29.Xu Z, Tian P, Guo J, Mi C, Liang T, Xie J, et al. 2021. Lnc-HZ01 with m6A RNA methylation inhibits human trophoblast cell proliferation and induces miscarriage by up-regulating BPDE-activated lnc-HZ01/MXD1 positive feedback loop. Sci Total Environ 776:145950, PMID: , 10.1016/j.scitotenv.2021.145950. [DOI] [PubMed] [Google Scholar]
- 30.Huang W, Dai M, Qiu T, Liang T, Xie J, Mi C, et al. 2021. Novel lncRNA-HZ04 promotes BPDE-induced human trophoblast cell apoptosis and miscarriage by upregulating IP3 R1/CaMKII/SGCB pathway by competitively binding with miR-hz04. FASEB J 35(9):e21789, PMID: , 10.1096/fj.202100376RR. [DOI] [PubMed] [Google Scholar]
- 31.Xie J, Liang T, Zhao J, Xu Z, Tian P, Wang R, et al. 2022. Lnc-HZ08 regulates BPDE-induced trophoblast cell dysfunctions by promoting PI3K ubiquitin degradation and is associated with miscarriage. Cell Biol Toxicol 38(2):291–310, PMID: , 10.1007/s10565-021-09606-z. [DOI] [PubMed] [Google Scholar]
- 32.Straszewski-Chavez SL, Abrahams VM, Alvero AB, Aldo PB, Ma Y, Guller S, et al. 2009. The isolation and characterization of a novel telomerase immortalized first trimester trophoblast cell line, Swan 71. Placenta 30(11):939–948, PMID: , 10.1016/j.placenta.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, et al. 2000. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25(1):25–29, PMID: , 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gene Ontology Consortium. 2021. The Gene Ontology resource: enriching a GOld mine. Nucleic Acids Res 49:D325–D334, PMID: , 10.1093/nar/gkaa1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Tian F-J, He X-Y, Wang J, Li X, Ma X-L, Wu F, et al. 2018. Elevated tristetraprolin impairs trophoblast invasion in women with recurrent miscarriage by destabilization of HOTAIR. Mol Ther Nucleic Acids 12:600–609, PMID: , 10.1016/j.omtn.2018.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 36.Wang L, Park HJ, Dasari S, Wang S, Kocher J-P, Li W. 2013. CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res 41(6):e74, PMID: , 10.1093/nar/gkt006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mistry J, Chuguransky S, Williams L, Qureshi M, Salazar GA, Sonnhammer ELL, et al. 2021. Pfam: the protein families database in 2021. Nucleic Acids Res 49(D1):D412–D419, PMID: , 10.1093/nar/gkaa913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Farre D. 2003. Identification of patterns in biological sequences at the ALGGEN server: PROMO and MALGEN. Nucleic Acids Res 31(13):3651–3653, PMID: , 10.1093/nar/gkg605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Messeguer X, Escudero R, Farré D, Núñez O, Martínez J, Albà MM. 2002. PROMO: detection of known transcription regulatory elements using species-tailored searches. Bioinformatics 18(2):333–334, PMID: , 10.1093/bioinformatics/18.2.333. [DOI] [PubMed] [Google Scholar]
- 40.Szklarczyk D, Gable AL, Nastou KC, Lyon D, Kirsch R, Pyysalo S, et al. 2021. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res 49(D1):D605–D612, PMID: , 10.1093/nar/gkaa1074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhou Y, Zeng P, Li YH, Zhang Z, Cui Q. 2016. SRAMP: prediction of mammalian N6-methyladenosine (m6A) sites based on sequence-derived features. Nucleic Acids Res 44(10):e91, PMID: , 10.1093/nar/gkw104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Finn RD, Coggill P, Eberhardt RY, Eddy SR, Mistry J, Mitchell AL, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res 44(D1):D279–D285, PMID: , 10.1093/nar/gkv1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gao S, Murakami M, Ito H, Furuhata A, Yoshida K, Tagawa Y, et al. 2009. Mutated ras induced PLD1 gene expression through increased Sp1 transcription factor. Nagoya J Med Sci 71(3–4):127–136, PMID: . [PMC free article] [PubMed] [Google Scholar]
- 44.Guo J, Li R, Xu Z, Tian P, Wang R, Li Y, et al. 2021. Upregulated lnc-HZ02 and miR-hz02 inhibited migration and invasion by downregulating the FAK/SRC/PI3K/AKT pathway in BPDE-treated trophoblast cells. J Biochem Mol Toxicol 35(6):1–13, 10.1002/jbt.22757. [DOI] [PubMed] [Google Scholar]
- 45.Yang Y, Zhu X, Jia X, Hou W, Zhou G, Ma Z, et al. 2020. Phosphorylation of Msx1 promotes cell proliferation through the Fgf9/18-MAPK signaling pathway during embryonic limb development. Nucleic Acids Res 48(20):11452–11467, PMID: , 10.1093/nar/gkaa905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gu C, Wang Z, Zhou N, Li G, Kou Y, Luo Y, et al. 2019. Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N6-methyladenosine of Notch1. Mol Cancer 18(1):168, PMID: , 10.1186/s12943-019-1084-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Patil DP, Chen C-K, Pickering BF, Chow A, Jackson C, Guttman M, et al. 2016. m(6)a RNA methylation promotes XIST-mediated transcriptional repression. Nature 537(7620):369–373, PMID: , 10.1038/nature19342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu J, Yue Y, Han D, Wang X, Fu Y, Zhang L, et al. 2014. A METTL3-METTL14 complex mediates mammalian nuclear RNA N6-adenosine methylation. Nat Chem Biol 10(2):93–95, PMID: , 10.1038/nchembio.1432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fustin J-M, Doi M, Yamaguchi Y, Hida H, Nishimura S, Yoshida M, et al. 2013. RNA-methylation-dependent RNA processing controls the speed of the circadian clock. Cell 155(4):793–806, PMID: , 10.1016/j.cell.2013.10.026. [DOI] [PubMed] [Google Scholar]
- 50.da Silva Moreira S, de Lima Inocêncio LC, Jorge BC, Reis ACC, Hisano H, Arena AC. 2021. Effects of benzo(a)pyrene at environmentally relevant doses on embryo-fetal development in rats. Environ Toxicol 36(5):831–839, PMID: , 10.1002/tox.23085. [DOI] [PubMed] [Google Scholar]
- 51.Che D, Huang W, Fang Z, Li L, Wu H, Pi L, et al. 2019. The lncRNA CCAT2 rs6983267 G allele is associated with decreased susceptibility to recurrent miscarriage. J Cell Physiol 234(11):20577–20583, PMID: , 10.1002/jcp.28661. [DOI] [PubMed] [Google Scholar]
- 52.Brown HA, Thomas PG, Lindsley CW. 2017. Targeting phospholipase D in cancer, infection and neurodegenerative disorders. Nat Rev Drug Discov 16(5):351–367, PMID: , 10.1038/nrd.2016.252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kang DW, Hwang WC, Noh YN, Kang Y, Jang Y, Kim J-A, et al. 2021. Phospholipase D1 is upregulated by vorinostat and confers resistance to vorinostat in glioblastoma. J Cell Physiol 236(1):549–560, PMID: , 10.1002/jcp.29882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Murray MJ, Lessey BA. 1999. Embryo implantation and tumor metastasis: common pathways of invasion and angiogenesis. Semin Reprod Endocrinol 17(3):275–290, PMID: , 10.1055/s-2007-1016235. [DOI] [PubMed] [Google Scholar]
- 55.Jiang X, Liu B, Nie Z, Duan L, Xiong Q, Jin Z, et al. 2021. The role of m6A modification in the biological functions and diseases. Signal Transduct Target Ther 6(1):74, PMID: , 10.1038/s41392-020-00450-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yang D, Qiao J, Wang G, Lan Y, Li G, Guo X, et al. 2018. N6-Methyladenosine modification of lincRNA 1281 is critically required for mESC differentiation potential. Nucleic Acids Res 46(8):3906–3920, PMID: , 10.1093/nar/gky130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Reizer E, Csizmadia IG, Palotas AB, Viskolcz B, Fiser B. 2019. Formation mechanism of benzo(a)pyrene: one of the most carcinogenic polycyclic aromatic hydrocarbons (PAH). Molecules 24(6):1040, PMID: , 10.3390/molecules24061040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi Q, Godschalk RWL, van Schooten FJ. 2017. Inflammation and the chemical carcinogen benzo[a]pyrene: Partners in crime. Mutat Res Rev Mutat Res 774:12–24, PMID: , 10.1016/j.mrrev.2017.08.003. [DOI] [PubMed] [Google Scholar]
- 59.Spagnoletti A, Paulesu L, Mannelli C, Ermini L, Romagnoli R, Cintorino M, et al. 2015. Low concentrations of bisphenol a and para-Nonylphenol affect extravillous pathway of human trophoblast cells. Mol Cell Endocrinol 412:56–64, PMID: , 10.1016/j.mce.2015.05.023. [DOI] [PubMed] [Google Scholar]
- 60.Liao Y, Zheng H, Wu L, He L, Wang Y, Ou Y, et al. 2021. Cadmium cytotoxicity and possible mechanisms in human trophoblast HTR-8/SVneo cells. Environ Toxicol 36(6):1111–1124, PMID: , 10.1002/tox.23110. [DOI] [PubMed] [Google Scholar]
- 61.Robert S, Gicquel T, Bodin A, Lagente V, Boichot E. 2016. Characterization of the MMP/TIMP imbalance and collagen production induced by IL-1β or TNF-α release from human hepatic stellate cells. PLoS One 11(4):e0153118, PMID: , 10.1371/journal.pone.0153118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wu X, Li M, Chen SQ, Li S, Guo F. 2018. Pin1 facilitates isoproterenol-induced cardiac fibrosis and collagen deposition by promoting oxidative stress and activating the MEK1/2ERK1/2 signal transduction pathway in rats. Int J Mol Med 41(3):1573–1583, PMID: , 10.3892/ijmm.2017.3354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Manka SW, Bihan D, Farndale RW. 2019. Structural studies of the MMP-3 interaction with triple-helical collagen introduce new roles for the enzyme in tissue remodelling. Sci Rep 9:18785, PMID: , 10.1038/s41598-019-55266-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gardner DK, Lane M. 2003. Blastocyst transfer. Clin Obstet Gynecol 46(2):231–238, PMID: , 10.1097/00003081-200306000-00005. [DOI] [PubMed] [Google Scholar]
- 65.Zigrino P, Brinckmann J, Niehoff A, Lu Y, Giebeler N, Eckes B, et al. 2016. Fibroblast-derived MMP-14 regulates collagen homeostasis in adult skin. J Invest Dermatol 136(8):1575–1583, PMID: , 10.1016/j.jid.2016.03.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Muyayalo KP, Li ZH, Mor G, Liao AH. 2018. Modulatory effect of intravenous immunoglobulin on Th17/Treg cell balance in women with unexplained recurrent spontaneous abortion. Am J Reprod Immunol 80(4):e13018, PMID: , 10.1111/aji.13018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


