ABSTRACT
Rhizopus oryzae lipase (ROL) is one of the most important enzymes used in the food, biofuel, and pharmaceutical industries. However, the highly demanding conditions of industrial processes can reduce its stability and activity. To seek a feasible method to improve both the catalytic activity and the thermostability of this lipase, first, the structure of ROL was divided into catalytic and noncatalytic regions by identifying critical amino acids in the crevice-like binding pocket. Second, a mutant screening library aimed at improvement of ROL catalytic performance by virtual saturation mutagenesis of residues in the catalytic region was constructed based on Rosetta’s Cartesian_ddg protocol. A double mutant, E265V/S267W (with an E-to-V change at residue 265 and an S-to-W change at residue 267), with markedly improved catalytic activity toward diverse chain-length fatty acid esters was identified. Then, computational design of disulfide bonds was conducted for the noncatalytic amino acids of E265V/S267W, and two potential disulfide bonds, S61C-S115C and E190C-E238C, were identified as candidates. Experimental data validated that the variant E265V/S267W/S61C-S115C/E190C-E238C had superior stability, with an increase of 8.5°C in the melting temperature and a half-life of 31.7 min at 60°C, 4.2-fold longer than that of the wild-type enzyme. Moreover, the variant improved the lipase activity toward five 4-nitrophenyl esters by 1.5 to 3.8 times, exhibiting a potential to modify the catalytic efficiency.
IMPORTANCE Rhizopus oryzae lipase (ROL) is very attractive in biotechnology and industry as a safe and environmentally friendly biocatalyst. Functional expression of ROL in Escherichia coli facilitates effective high-throughput screening for positive variants. This work highlights a method to improve both selectivity and thermostability based on a combination of virtual saturation mutagenesis in the substrate pocket and disulfide bond prediction in the noncatalytic region. Using the method, ROL thermostability and activity to diverse 4-nitrophenyl esters could be substantially improved. The strategy of rational introduction of multiple mutations in different functional domains of the enzyme is a great prospect in the modification of biocatalysts.
KEYWORDS: Cartesian_ddg protocol, Rhizopus oryzae lipase, chain-length selectivity, disulfide bond design, thermostability
INTRODUCTION
Microbial lipases (EC 3.1.1.3) are one of the most important biological catalysts applied in various industries, including the food, medical, pulp and paper, bioremediation, and energy industries (1–3). Although there are plenty of lipase resources in nature, their low stability under severe industrial conditions, such as high temperature and the presence of protein denaturants and organic solvents, hampers widespread application (4). It is of great importance to screen lipases with high stability and selectivity (5, 6). Accordingly, protein engineering, as a mainstream strategy, has been used to modify existing lipases into ideal biocatalysts with shortened evolution processes at a minimum cost (4, 7).
As we know, enzyme activity is determined by the interaction between an active pocket and a substrate (8). Numerous studies have demonstrated that manipulating amino acids around the binding pocket by site saturation mutagenesis allows effective improvement of the activities of enzymes (9, 10). Godehard et al. (11) successfully enhanced the acyltransferase activity and broadened the acyl-donor substrate scope of Mycobacterium smegmatis acyltransferase by manipulating residues in the binding site and tunnel. Similarly, through saturation mutagenesis of targeted binding sites along the channel, the substrate specificity of Pseudomonas sp. acidic lipase Lip I.3 shifted to long-chain C18 substrates (12). Additionally, enrichment of long-chain monounsaturated fatty acids (FAs) from Camelina and Crambe oil ethyl ester derivatives was achieved by semirational modification of the tunnel-like structure of Candida antarctica lipase A (13).
Disulfide bonds, closely related to enzyme stability (14–16), can reduce the entropy of an enzyme in the unfolded state by 2.3 to 5.2 kcal/mol per native disulfide bond (17). Scholars have successfully introduced disulfide bonds into enzyme structures to improve stability (18–20). Janssen et al. (21, 22) developed a computational strategy, FRESCO, to design stable disulfide bonds using molecular dynamics simulations and X-ray diffraction. Using this strategy, the apparent melting temperature (Tm) of GH11 xylanase from Neocallimastix patriciarum was increased by 14°C (22). In addition, based on in silico design servers (MODIP, DbD2, and FoldX), disulfide bond mutants with an A-to-C change at residue 211 and S-to-C change at residue 214 (A211C-S214C) of the maltotetraose-forming amylase from Pseudomonas saccharophila STB07 (MFAps) exhibited 2.6-fold improvement in half-life at 60°C (23).
Rhizopus oryzae lipase (ROL) is one of the most important industrial enzymes used in the food, biofuel, and pharmaceutical industries (24, 25). It has been heterologously expressed in Escherichia coli, Saccharomyces cerevisiae, and Pichia pastoris (26, 27). There are three major commercial products available: lipase DF “Amano 15,” supplied by Amano, and two products from Sigma (catalog numbers 62305 and 89445) (26). Zhao et al. (28) introduced a pair of disulfide bonds between positions 190 and 238 using DbD2 and greatly improved the thermal stability of the target enzyme with little change in its catalytic performance. The activity of variant E190C/E238C retained 58.2% after 720 min of incubation at 55°C, at which the half-life of the wild-type (WT) enzyme was only 11.7 min. Ding et al. (29) modified ROL, Rhizomucor miehei lipase (RML), and Thermomyces lanuginosus lipase by rationally engineering the key residues at the crevice-like binding site. The obtained variants exhibited better activity toward 4-nitrophenyl (pNP) esters and the optimum temperature declined by 10 to 20°C. Although multiple bioinformatics tools are available to improve enzyme selectivity and/or thermostability, it remains elusive how to perfect one property without affecting the other (6, 30, 31).
In this study, an efficient strategy was developed to modify ROL for better catalytic activity and thermostability simultaneously. To recognize mutations advantageous for the catalytic efficiency without reducing the thermal stability, residues (except for the highly conservative ones) in the catalytic region were subjected to virtual saturation mutagenesis based on Rosetta’s Cartesian_ddg protocol to screen mutants with reduced binding and folding free energy. To improve the thermostability, potential disulfide bonds in the noncatalytic region were further designed using online prediction servers (MODIP, DbD2, and BridgeD). Combined with the beneficial mutations designed by the aforesaid methods, a most robust variant was obtained with markedly improved thermostability and catalytic performance. In addition, the mechanism underlying the enhanced selectivity and thermostability was investigated.
RESULTS
Identification of hot spot residues.
Based on the crystal structure of ROL in closed conformation (PDB ID 1lgy) and RML in open conformation (PDB ID 4tgl), the open structure of ROL (OROL) was obtained by comparative modeling using RosettaCM (see Fig. S1A in the supplemental material). The structures were ranked by their Rosetta all-atom score, and the top 10 models were visually evaluated for native disulfide bonds. Then, the targeted top-ranked structure (S_0023.pdb) (Fig. 1A) was confirmed by Ramachandran plot analysis using PROCHECK and MolProbity. MolProbity calculation results indicated that the modeled OROL had 97.8% (261/267) of all residues in favored (98%) regions and 100.0% (267/267) of all residues in allowed (>99.8%) regions (Fig. S1B). The PROCHECK analysis revealed that only 0.4% of residues were located in a disallowed region of the Ramachandran plot (Fig. S1C). Therefore, the target model was selected as the open conformation for molecular docking.
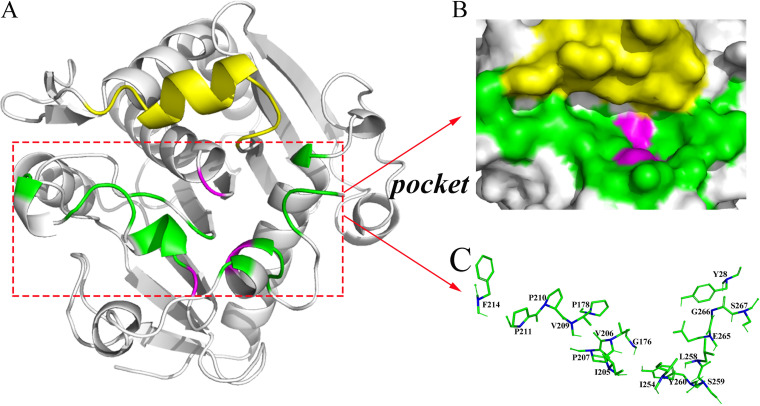
FIG 1.
Three-dimensional structure of the OROL and local structure of the catalytic pocket and mutation sites. (A) Three-dimensional structure of the open conformation of ROL. The structure was constructed by multitemplate modeling using RosettaCM, with the opened lid of RML (PDB ID 4tgl, residues 82 to 95) and the structure of 1lgy without lid region (residues 1 to 82 and 96 to 269) as templates. (B) Catalytic pocket of ROL. (C) Local region of mutation sites in ROL based on Cartesian_ddg. The lid region (residues 83 to 95) is colored yellow, the catalytic triad (S145, D204, H257) is magenta, and the mutation sites in the substrate pocket are green, with alpha-carbon atoms marked in blue.
To better understand the structural basis of chain-length selectivity and ensure the correct orientation of pNP esters in the pocket of OROL, AutoDock Vina was used for molecular docking. Residues within 5 Å from the substrate were considered to make up the catalytic region (Fig. 1B). To ensure normal catalytic function, the highly conservative residues and significant functional amino acids were maintained (3). Therefore, the catalytic triad (S145, D204, H257), oxyanion hole (T83, L146), and lid region (residues 82 to 96) of ROL were excluded. The remaining residues (Y28, G176, P178, I205, V206, P207, V209, P210, P211, F214, I254, L258, S259, Y260, E265, G266, and S267) were considered hot spots that would influence the catalytic activity (Fig. 1C).
Virtual saturation mutagenesis based on the Cartesian_ddg protocol.
The binding and folding free energy changes of ROL variants predicted by Cartesian_ddg are presented in Table S1. Forty-one virtual mutants involving nine residues (G176, P178, P211, I254, L258, S259, E265, G266, and S267) displayed reduced binding free energy. To assess the impact of mutation on structural stability, the folding free energy changes were further calculated using Rosetta’s Cartesian_ddg protocol. When analyzing the folding free energy changes (last column of Table S1), the values of 24 variants were found to be positive. Hence, the virtual saturated mutant library was reduced from 41 to 17 and was used for subsequent experimental validations.
Characterization of recombinant ROL with single and double mutations.
The WT and aforesaid 17 mutants were successfully expressed in E. coli and purified. All target proteins produced a single band around 45 kDa in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) (Fig. S2A).
Based on melting temperature changes (ΔTm), the influence of mutations on the stability of ROL was measured by the differential scanning fluorimetry (DSF) method (32). The ΔTm values of the variants are shown in Fig. S2B. Seven out of 17 variants exhibited negative values, one had a value of zero, and nine had positive values, corresponding to baleful, effect-free, and beneficial mutations, respectively. To reduce the workload and considerably improve the screening efficiency, the seven harmful mutations were excluded from subsequent experiments.
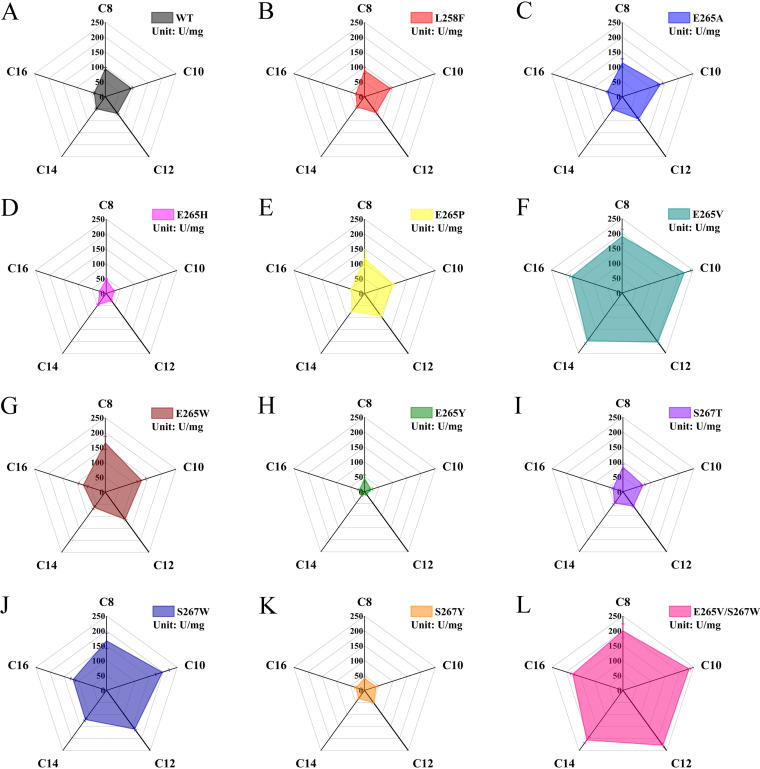
The hydrolysis of five pNP esters varying in alkyl chain length was measured to assess the substrate selectivity of WT and the candidates. In comparison with WT (Fig. 2A), single-site mutants L258F, E265H, E265Y, S267T, and S267Y exhibited decreased specific activity toward all five esters; mutants E265A, E265P, and E265W displayed enhanced activity, albeit not significantly; and mutants 265V and S267W showed superior ability (Fig. 2B to K).The specific activities of mutant E265V toward substrates with carbon chain lengths ranging from 8 to 16 were 2.0, 2.4, 2.9, 3.9, and 4.6 times that of the WT, and those of mutant S267W were 1.8, 2.2, 2.3, 2.4, and 3.0 times that of the WT. Interestingly, the performance of target residues of mutants E265V and S267W was altered from hydrophilic to hydrophobic. A double mutant, E265V/S267W, was constructed to analyze the possible synergistic effect of the mutations. As shown in Fig. 2L, E265V/S267W showed 2.1, 2.6, 3.3, 4.1, and 4.6 times higher hydrolytic activity than that of WT.
FIG 2.
Chain-length substrate specificity of WT and mutant ROL toward p-nitrophenyl esters at 35°C in pH 8.0 50 mM Tris-HCl buffer. (A) WT; (B to K) single mutants (L258F, E265A, E265H, E265P, E265W, E265V, E265Y, S267T, S267W, S267Y); (L) double mutant S265V/S267W.
Disulfide bond prediction using MODIP, DbD2, and BridgeD.
A strategy based on multiple computational methods (DbD2, MODIP, and BridgeD) was used to design possible residue pairs that might form disulfide bonds properly. There were 117 residue pairs predicted by BridgeD, 30 residue pairs satisfying the default criteria of DbD2, and 72 potential pairs rated as A to C grade using the MODIP algorithm. They, along with the corresponding algorithms, are listed in Table S2, with natural disulfide bonds (C29-C268, C40-C43, and C235-C244) shown in bold italics. Amino acid residues involved in the functionally important catalytic region were excluded so as not to sacrifice substrate specificity. Thus, only 63 pairs of residues were retained.
Stability assessment and visual validation of the designed disulfide bonds.
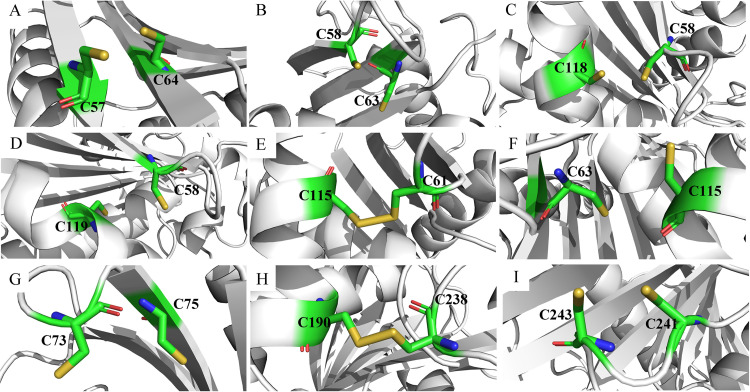
As shown in Table S2, 54 out of the 63 pairs of disulfide bonds had positive folding free energy changes, suggesting that mutation in these sites would be detrimental to the thermal stability of the enzyme. The formation of disulfide bonds in the mutated structures of the other nine variants was visually inspected. As shown in Fig. 3, seven predicted residue pairs were free cysteines, and only S61C-S115C and E190C-E238C could effectively form disulfide bonds. Figure 3E and H show the locations of the newly designed disulfide bonds in three-dimensional (3D) structures. Both S61C-S115C and E190C-E238C are located in the surface and successfully form bridge connections between one flexible loop and one α-helix. Similarly, a combination variant E265V/S267W/S61C-S115C/E190C-E238C (M6) with two pairs of mutated disulfide bonds was constructed to verify the additive effects of disulfide bonds on thermostability, and the structure is displayed in Fig. S3.
FIG 3.
Visual inspection of disulfide bond formation of the mutated structures computed by FoldX. (A) Variant E265V/S267W/T57C-N64C; (B) variant E265V/S267W/S58C-T63C; (C) variant E265V/S267W/S58C-Q118C; (D) variant E265V/S267W/S58C-V119C; (E) variant E265V/S267W/S61C-S115C; (F) variant E265V/S267W/T63C-S115C; (G) variant E265V/S267W/Q73C-T75C; (H) variant E265V/S267W/E190C-E238C; (I) variant E265V/S267W/T241C-D243C.
Determination of the formation of disulfide bonds.
To assess the effect of the introduced disulfide bonds on thermal stability, WT, E265V/S267W, E265V/S267W/S61C-S115C, E265V/S267W/E190C-E238C, and M6 were expressed and purified. SDS-PAGE was used to determine the molecular weights of the proteins. All variants produced a single band around 45 kDa, consistent with the molecular weight of ROL (Fig. S4). Next, the soluble protein (in milligrams) per liter of medium was measured after purification by nickel affinity chromatography. In comparison with WT (60 mg/liter medium), the solubility of mutant M6 (20 mg/liter medium) was decreased. As disulfide bond formation in E. coli is a rate-limiting step for protein expression (33, 34), the introduction of the newly designed thios may have led to an increase in the probability of inclusion body formation and, thus, a decrease in the amount of mutant M6.
As shown in Fig. S3, the six native cysteine residues formed three pairs of disulfide bonds (C40-C43, C29-C268, and C235-C244) in the 3D structure (35). All mutants behaved as expected, and no free sulfhydryl group was detected in any of the five enzymes under normal conditions using the Ellman method (Table S3). After treatment with dithiothreitol (DTT), 6 mol of free sulfhydryl groups was released from E265V/S267W and WT, 8 mol from the two mutants containing one pair of mutated disulfide bonds (E265V/S267W/S61C-S115C and E265V/S267W/E190C-E238C), and 10 mol from the combined mutant M6 (Table S3). Therefore, it can be concluded there are three natural disulfide bonds in E265V/S267W and WT, four in E265V/S267W/S61C-S115C and E265V/S267W/E190C-E238C, and five in M6. The differences in the number of disulfide bonds demonstrated that they were successfully formed in the three variants, as expected.
Impact of introduced disulfide bonds on thermostability.
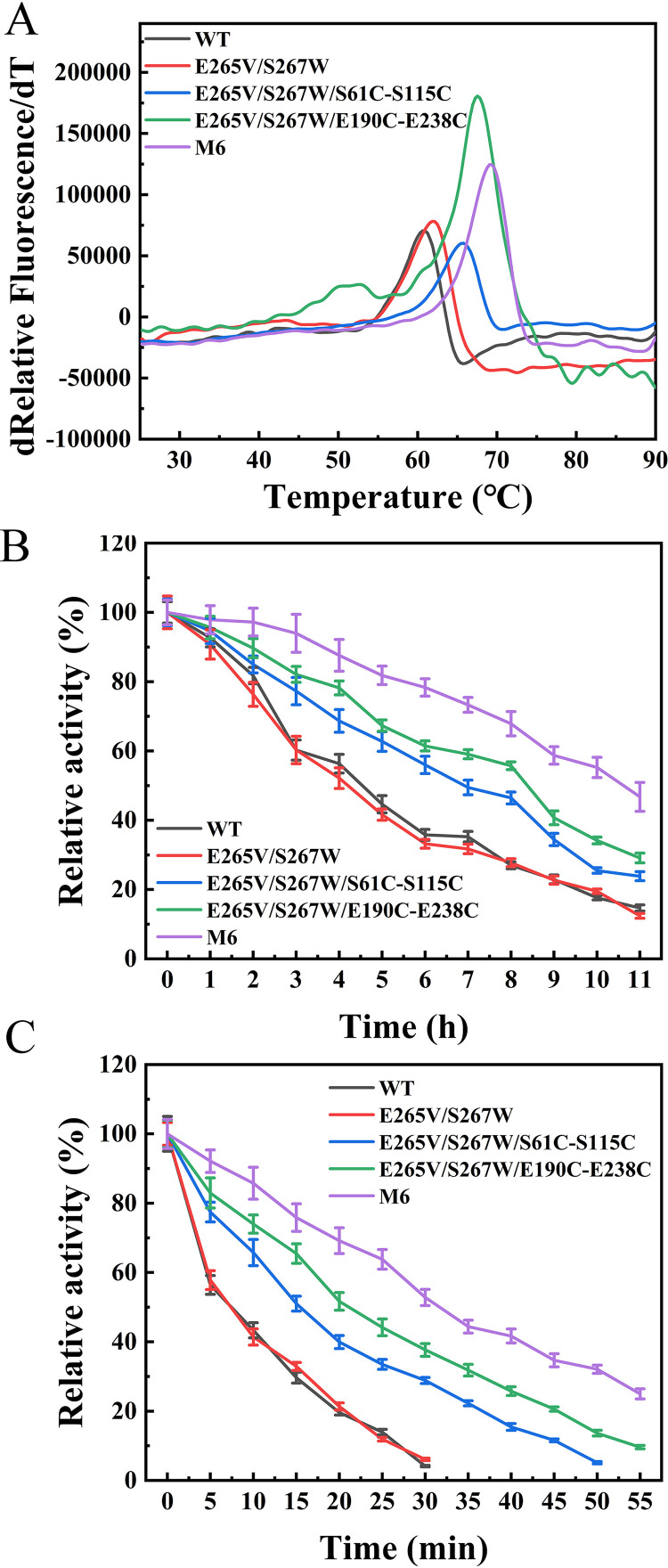
To determine the impact of the introduced disulfide bonds on thermostability, Tm values of purified WT and variant ROL were measured based on the DSF method. The results are presented in Fig. 4A and Table 1. The Tm value of variant M6 increased to 69.1 ± 0.2°C (mean ± standard deviation), which was 8.5°C higher than that of WT. The Tm values of the variants with a single designed disulfide bond (E265V/S267W/S61C-S115C and E265V/S267W/E190C-E238C) increased by 5.0°C and 6.9°C, respectively. Thus, the thermal stability of the enzyme was improved synergistically by introducing two new pairs of disulfide bonds.
FIG 4.
Thermostability profiles of WT and the E265V/S267W, E265V/S267W/S61C-S115C, E265V/S267W/E190C-E238C, and M6 mutants. (A) Apparent melting temperatures of WT and the E265V/S267W, E265V/S267W/S61C-S115C, E265V/S267W/E190C-E238C, and M6 mutants determined by DSF, Tm represents the temperature at which 50% of the protein is unfolded. (B) Half-life at 45°C measured using C8 as a substrate at the respective optimal pH. (C) Half-life at 60°C measured using C8 as a substrate at the respective optimal pH.
TABLE 1.
Thermostability characteristics of WT and mutant ROL based on single or double disulfide bond mutagenesis
| ROL variant | Tm (°C)a | ΔTm (°C) | Topt (°C) | t45 1/2 (h) | t60 1/2 (min) |
|---|---|---|---|---|---|
| WT | 60.6 ± 0.2 | 35 | 4.5 | 7.5 | |
| E265V/S267W | 61.8 ± 0.2 | 1.2 | 35 | 4.2 | 7.3 |
| E265V/S267W/S61C-S115C | 65.7 ± 0.2 | 5.0 | 40 | 7.0 | 15. 5 |
| E265V/S267W/E190C-E238C | 67.5 ± 0.2 | 6. 9 | 40 | 8.4 | 21.0 |
| M6 | 69.1 ± 0.2 | 8.5 | 45 | 10.6 | 31.7 |
Values are means ± standard deviations.
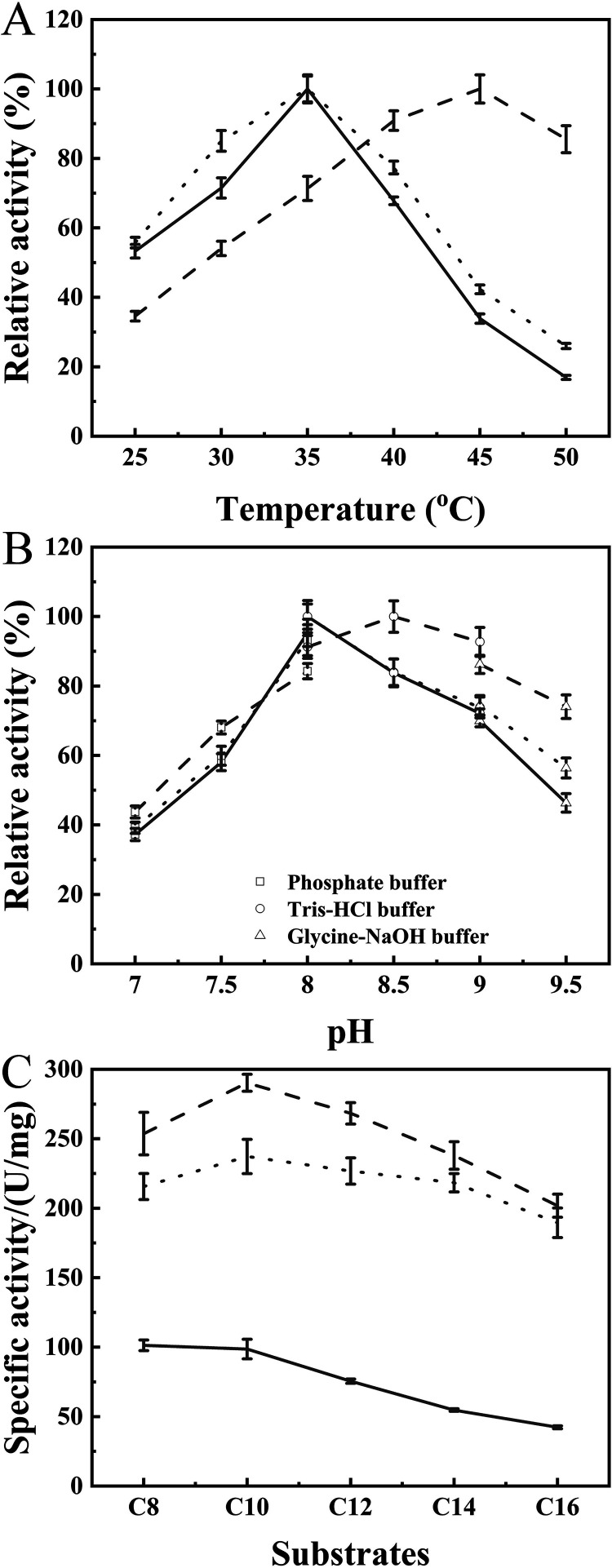
As listed in Table 1, the optimum temperature (Topt) of WT and the E265V/S267W mutant was 35°C, whereas variant M6 was more active at 45°C. The Topt values of mutants E265V/S267W/S61C-S115C, E265V/S267W/E190C-E238C, and M6 were increased by 5°C, 5°C, and 10°C, respectively, compared to that of WT and were close to the Tm.
The thermal inactivation also was measured to assess the kinetic stability of the variants (Fig. 4B and C) except for Tm and Topt. The half-life at 45°C (t45 1/2) of the WT and mutant E265V/S267 were about 4 h, and the values of the mutant E265V/S267W/S61C-S115C, E265V/S267W/E190C-E238C, and M6 were increased to 8.4 h, 7.0 h, and 10.6 h, respectively. When incubated at 60°C, WT and E265V/S267W were deactivated rapidly, with a half-life (t60 1/2) of no more than 7.5 min. The t60 1/2 of variants E265V/S267W/S61C-S115C, E265V/S267W/E190C-E238C, and M6 were 15.5 min, 21.0 min, and 31.70 min, respectively, which were 2 to 4 times higher than that of WT. Hence, the combined variant M6 was selected for further investigation of enzymatic properties.
Enzymatic properties of WT, E265V/S267W, and M6.
The Topt value of M6 was 45°C, which was 10°C higher than that of WT and E265V/S267W (Fig. 5A and Table S4). As shown in Fig. 5B and Table S4, the optimum pH of E265V/S267W was the same as that of WT (8.0), and the optimum pH of M6 shifted to 8.5. The relative activity of M6 was no less than 65% under alkaline conditions (pH 7.5 to 9.5), exhibiting greater stability than WT. In addition, the chain length specificity at optimum conditions was tested under optimal reaction conditions (Fig. 5C). E265V/S267W was more active than WT, which was in agreement with the result shown in Fig. 2L. Interestingly, the specific activities of E265V/S267W and M6 were not significantly different, demonstrating that the newly introduced disulfide bonds markedly improved thermostability, without sacrificing selectivity.
FIG 5.
Enzymatic properties of WT (solid line) and the E265V/S267W (dotted line) and M6 (dashed line) mutants. (A) Optimal reaction temperature measured using C8 as a substrate at pH 8.0. (B) Optimal pH for hydrolytic reaction measured at respective optimal temperatures. (C) Specific activity toward five pNP esters of various alkyl chain lengths at optimum temperatures and pH.
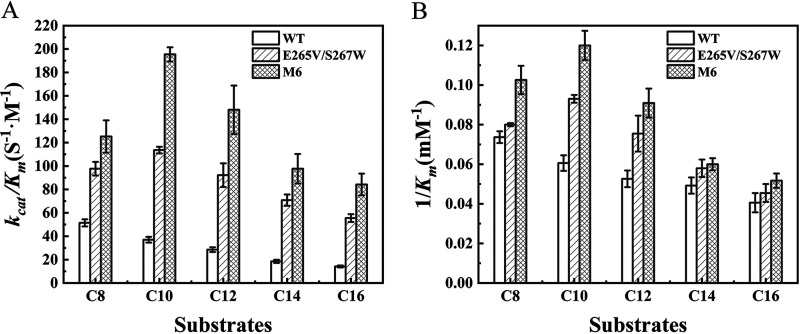
Kinetic parameters of WT, E265V/S267W, and M6 were tested using five pNP esters of varying alkyl chain lengths with different concentrations at respective optimum temperatures and pH of three enzymes (Fig. 6). For the WT, the catalytic efficiency (kcat/Km) and 1/Km values decreased as the carbon chain length increased; the catalytic efficiency toward C8 (4-nitrophenyl octanoate) was 2.6-fold higher than that toward C16 (4-nitrophenyl palmitate), which was consistent with the specific activity of WT toward C8 being higher than that toward others (Fig. 5C). Whereas mutants E265V/S267W and M6 exhibited a substrate specificity shift from C8 to C10 (4-nitrophenyl myristate) and improvement in kcat/Km and 1/Km toward all five pNP esters compared with those of WT, especially against the longer-length chain. The catalytic efficiency of mutant E265V/S267W toward C8 was 0.9-fold higher than that of WT, while the catalytic efficiency of mutant E265V/S267W toward C16 was 2.9-fold higher. Moreover, mutant M6 had a catalytic efficiency superior to that of WT and mutant E265V/S267W, indicating that the extra introduced disulfide bonds also influenced the kinetic parameters of the enzyme.
FIG 6.
Determination of the kinetic parameters of WT and the E265V/S267W and M6 mutants using five pNP esters of various alkyl chain lengths as substrates under optimum reaction conditions of each enzyme. (A) Catalytic efficiency (kcat/Km); (B) 1/Km.
Fatty acid production from different chain length natural fatty acid esters using ROL variants.
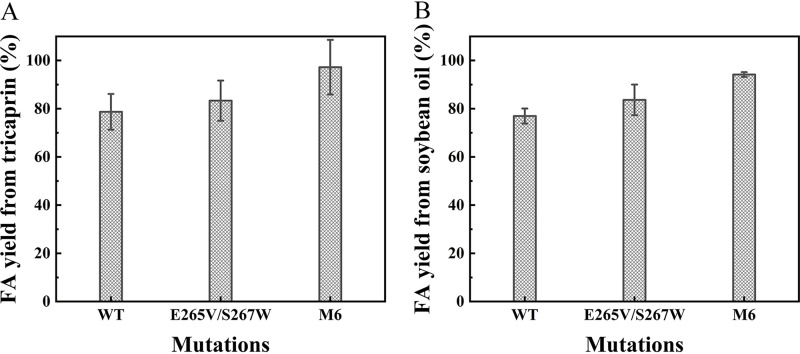
The abilities of the mutants to produce fatty acids (FAs) from tricaprin and soybean oil at the same enzyme protein concentrations were investigated. As shown in Fig. 7A, WT and mutant E265V/S267W exhibited relatively poor performance, achieving only a 78.7% and 83.3% hydrolysis rate after 12 h toward tricaprin. In comparison, the hydrolysis rate of mutant M6 reached 97.2% during the same period, demonstrating that an increase in thermostability could affect catalytic efficiency (36, 37). Likewise, mutant M6 achieved 94.2% FA yield toward soybean oil after 24 h and exhibited the best performance (Fig. 7B), while WT and mutant E265V/S267W yields reached 76.9% and 83.6% FA yields, respectively.
FIG 7.
Fatty acid production of WT, E265V/S267W, and M6 at 45°C. (A) Enzymatic hydrolysis was performed on 600 μL tricaprin for 12 h. (B) Enzymatic hydrolysis was performed on 2 mL soybean oil for 24 h.
DISCUSSION
To promote the industrial application of fungal lipase, studies on the improvement of the catalytic activity and/or thermostability of lipases via protein engineering have become a hot spot (Table S5). Through structure-guided rational design of lipase from Aspergillus oryzae, mutant V269D exhibited 6-fold-higher hydrolysis activity compared to the WT, while the thermostability remained unchanged (38). In combinations of multiple sequence alignment and disulfide bond design, the Topt of the quadruple variant V209L/D262G/E190C/E238C of ROL shifted from 40°C to 55°C, but the substrate affinity and catalytic efficiency were negatively affected (28). Similarly, in our previous work, based on integrated rational design methods, the mutant T18K/T22I/E230I/S56C-N63C/V189C-D238C of RML exhibited 15°C improvement in Topt with only a 0.4-fold increase in catalytic activity (34). In this study, we first found the positive impact of mutations E265V/S267W on the specific activity without hampering thermostability by virtual saturation mutagenesis in the catalytic region (with reduced binding and folding free energy changes) (Table S1) and experimental determinations (Fig. 2 and Fig. S2B). After that, disulfide bonds (S61C-S115C/E190C-E238C) were identified using online prediction servers (MODIP, DbD2, and BridgeD), stability evaluation (based on folding free energy changes by FoldX) (Table S2), and experimental analyses (Fig. 4 and Table 1). Finally, by integrating the beneficial mutations, the variant M6 showing better improved specific activity toward pNP esters (1.5- to 3.8-fold increases), and heat resistance (10°C increase in Topt and 3.2-fold increase in t60 1/2) was obtained. With the best performance in overcoming the reported activity-stability trade-offs (Table S5), mutant M6 could be a favorable competitor in industrial applications, such as the low-cost production of fatty acids and biodiesel.
Substrates enter into the active pocket and interact with amino acids along the tunnel, positioning the substrate in a favorable conformation to complete reactions by the catalytic residues (7, 39, 40). Therefore, the residues in the catalytic region play an important role in substrate binding and enzymatic reactions (20, 41, 42). As can be seen from the anatomy of the ROL pocket, there is a hydrophobic, crevice-like binding site located near the surface (Fig. 1B) (43). By estimating the binding and folding free energy change and ΔTm values, 10 mutants in 3 sites (258, 265, 267) were predicted to exhibit potential enhancement of catalytic activity (Table S1 and Fig. S2B). Thereinto, double mutant E265V/S267W displayed synergistically improved hydrolytic activity, 2.1 to 4.6 times higher than that of WT (Fig. 2). The phenomenon was probably due to the altered hydrophobic and hydrophilic performances of mutation sites in the positive variants that lead to an increased affinity for hydrophobic substrates (C10 and C12 [4-nitrophenyl decanoate and 4-nitrophenyl dodecanoate]) (44, 45). The interactions between residues in the activity pocket of E265V/S267W and the five bound ligands are graphically represented in Fig. S5 and indicate that the enhanced activity was attributed to the additional hydrophobic contacts generated by introducing hydrophobic amino acids at positions 265 and 267.
Disulfide bonds are not only closely related to protein folding but also of great significance in maintaining the stability of the advanced structure, as their bonding energy is substantially greater than that of other secondary bonds (19). The introduction of disulfide bonds effectively reduces the conformation entropy of the protein backbone in the unfolded state and improves the rigidity of enzyme molecules, which has become a widely used stability-enhancing method presently (18). The precise construction of disulfide bonds also depends on the strict geometry of the structure, and not all randomly designed disulfide bonds are effective. Unfavorable strains generated by disulfide bonds in improper mutation positions may offset or weaken the stabilization effect (46, 47). Recent reports have verified that the online servers (MODIP, DBD2, and SSBOND) show great potential to ensure the suitability of mutation sites for disulfide bonds (23, 34, 48). Although as many as possible disulfide bonds were predicted with respect to ROL based on combinations of the three methods (Table S2), only two new pairs of mutations (S61C-S115C and E190C-E238C) were capable of forming covalent disulfide bridges (Fig. 3). Moreover, the irreversible intramolecular cross-linking of the designed disulfide bond between the α-helix and loop region can support the scaffold of the enzyme robustly, resulting in enhanced thermotolerance (34, 49). Mutations of amino acids in the flexible surface region avoid the formation of inappropriate interactions with internal residues and minimize the entropy effect, and thus they are more likely to generate a positive mutation (50). Therefore, the newly added disulfide bonds may mainly account for the enhanced stability (t1/2 and Tm in Fig. 4) of the mutant M6 by constraining the motion of loop 58–62 and loop 236–244 on the surface (Fig. S3). Interestingly, a slight increase from 8.0 to 8.5 in the optimum pH was observed with the addition of the disulfide bonds, which was in line with findings in our previous study (34). Catherine et al. (36) also reported a similar phenomenon, and they deduced that intermolecular interactions were strengthened with the increasing number of salt bridges, resulting in enhanced alkaline resistance. The improved affinity (1/Km) and catalytic efficiency (kcat/Km) of M6 indicated that modification of thermostability away from the catalytic region is not necessarily at the expense of enzyme affinity and activity (41). Further, on account of the improved catalytic and thermostability of ROL, we evaluated the effects of the mutations on the production of FAs from tricaprin and soybean oil. The FA yields of WT and mutant E265V/S267W were more obviously lower than that of the mutant M6 during the same reaction time, owing both to the improved reaction rates and to the shortened reaction times by the higher thermostability (37, 51).
In conclusion, we effectively overcame the trade-off between enzyme activity and stability through a combination strategy using the Rosetta’s Cartesian_ddg protocol and disulfide bond prediction servers. Using ROL as a model, hydrophobic amino acids in the catalytic region were mutated and disulfide bond mutations were introduced in the noncatalytic region. This study not only strikes an appropriate balance between catalytic activity and thermostability, but also it provides a feasible and targeting strategy to tailor the catalytic properties of biocatalysts in the future.
MATERIALS AND METHODS
Gene, plasmids, and strains.
The WT gene encoding ROL and containing a prosequence (ProROL; GenBank accession number JN689988.1) came from our laboratory (52). To ensure that the translated protein sequence corresponded with the resolved crystal structure in closed conformation (PDB ID 1lgy), histidine (H) at site 113 was exchanged for asparagine (N). The plasmid pET-28a(+) with an N-terminal His6 was employed as the vector. E. coli DH5α was used as a host for gene cloning and E. coli BL21(DE3) was used for ROL expression.
Construction of variants.
All variants in this study were constructed in the same manner. Variants were prepared using PrimeSTAR HS DNA polymerase (TaKaRa Biomedical Technology, Beijing, China) by whole-plasmid PCR amplification using plasmid with the WT ProROL gene as a template. Fifty microliters of PCR product was digested with 1 μL of DpnI (TaKaRa Biomedical Technology) at 37°C for 1 h to remove the template. The digestion product was transformed into DH5α cells after efficient cyclization by homologous recombination in vitro using the ClonExpress II one-step cloning kit (Nanjing Vazyme Biotech). Positive transformants were verified by sequencing, and correctly cloned plasmids were transformed into BL21(DE3) cells to obtain the target recombinant strains. The primers used to construct the mutants are listed in Table 2.
TABLE 2.
Primers used in this study
| Primer | Sequence (5′→3′)a |
|---|---|
| P211W-F | GATATCGTTCCTCACGTTCCTtggCAATCCTTCGGATTCCTTCAT |
| P211W-R | ATGAAGGAATCCGAAGGATTGccaAGGAACGTGAGGAACGATATC |
| L258F-F | CACCTCTATCCTTGATCACttcAGTTACTTTGATATCAACG |
| L258F-R | CGTTGATATCAAAGTAACTgaaGTGATCAAGGATAGAGGTG |
| L258Y-F | CTTTCACCTCTATCCTTGATCACtacAGTTACTTTGATATCAACGAAGG |
| L258Y-R | CCTTCGTTGATATCAAAGTAACTgtaGTGATCAAGGATAGAGGTGAAAG |
| E265A-F | TGAGTTACTTTGATATCAACgctGGAAGCTGTTTGGCGGCC |
| E265A-R | GGCCGCCAAACAGCTTCCagcGTTGATATCAAAGTAACTCA |
| E265D-F | AGTTACTTTGATATCAACgatGGAAGCTGTTTGGCGGC |
| E265D-R | GCCGCCAAACAGCTTCCatcGTTGATATCAAAGTAACT |
| E265N-F | TGAGTTACTTTGATATCAACaacGGAAGCTGTTTGGCGGCC |
| E265N-R | GGCCGCCAAACAGCTTCCgttGTTGATATCAAAGTAACTCA |
| E265C-F | CACTTGAGTTACTTTGATATCAACtgtGGAAGCTGTTTGGCGGCCGC |
| E265C-R | GCGGCCGCCAAACAGCTTCCacaGTTGATATCAAAGTAACTCAAGTG |
| E265H-F | TGAGTTACTTTGATATCAACcacGGAAGCTGTTTGGCGGCC |
| E265H-R | GGCCGCCAAACAGCTTCCgtgGTTGATATCAAAGTAACTCA |
| E265F-F | CACTTGAGTTACTTTGATATCAACtttGGAAGCTGTTTGGCGGCCGC |
| E265F-R | GCGGCCGCCAAACAGCTTCCaaaGTTGATATCAAAGTAACTCAAGTG |
| E265P-F | CTTGAGTTACTTTGATATCAACcctGGAAGCTGTTTGGCGGCCG |
| E265P-R | CGGCCGCCAAACAGCTTCCaggGTTGATATCAAAGTAACTCAAG |
| E265W-F | CACTTGAGTTACTTTGATATCAACtggGGAAGCTGTTTGGCGGCCGC |
| E265W-R | GCGGCCGCCAAACAGCTTCCccaGTTGATATCAAAGTAACTCAAGTG |
| E265V-F | TGAGTTACTTTGATATCAACgttGGAAGCTGTTTGGCGGCC |
| E265V-R | GGCCGCCAAACAGCTTCCaacGTTGATATCAAAGTAACTCA |
| E265Y-F | AGTTACTTTGATATCAACcaaGGAAGCTGTTTGGCGG |
| E265Y-R | CCGCCAAACAGCTTCCttgGTTGATATCAAAGTAACT |
| S267L-F | CTTGAGTTACTTTGATATCAACGAAGGAttgTGTTTGGCGGCCGCAC |
| S267L-R | GTGCGGCCGCCAAACAcaaTCCTTCGTTGATATCAAAGTAACTCAAG |
| S267T-F | ACTTTGATATCAACGAAGGAaccTGTTTGGCGGCC |
| S267T-R | GGCCGCCAAACAggtTCCTTCGTTGATATCAAAGT |
| S267W-F | AGTTACTTTGATATCAACGAAGGAtggTGTTTGGCGGCCGC |
| S267W-R | GCGGCCGCCAAACAccaTCCTTCGTTGATATCAAAGTAACT |
| S267Y-F | AGTTACTTTGATATCAACGAAGGAtacTGTTTGGCGGCCGC |
| S267Y-R | GCGGCCGCCAAACAgtaTCCTTCGTTGATATCAAAGTAACT |
| E265V/S267W -F | CACTTGAGTTACTTTGATATCAACgttGGAtggTGTTTGGCGGCCGCACT |
| E265V/S267W-R | AGTGCGGCCGCCAAACAccaTCCaacGTTGATATCAAAGTAACTCAAGTG |
| S61C-F | TCCTTGCTTtgtGACACAAATG |
| S61C-R | CATTTGTGTCacaAAGCAAGGA |
| S115C-F | TGGTTTCCTTTCCtgtTATGAGCAAGTTG |
| S115C-R | CAACTTGCTCATAacaGGAAAGGAAACCA |
| E190C-F | GCTTACTATGTTtgtTCTACCGGTA |
| E190C-R | TACCGGTAGAacaAACATAGTAAGC |
| E238C-F | TCTGTACTTCTtgtATTGAAACCAAG |
| E238C-R | CTTGGTTTCAATacaAGAAGTACAGA |
Mutation sites are shown in italic lowercase.
ROL expression in E. coli BL21(DE3) and purification.
For WT and mutant protein production, the recombinant strains were cultured in 800 mL of Luria-Bertani medium containing 50 μg/mL kanamycin at 37°C. When the cells reached an optical density at 600 nm of 0.6 to 0.8, they were treated with isopropyl β-d-1-thiogalactopyranoside at a final concentration of 0.1 mM at 16°C for 16 h. The cells were harvested by centrifugation at 5,000 × g for 10 min and eluted with 0.9% NaCl twice to remove the medium. The purified proteins were harvested by ultrasonic fragmentation in 25 mL of lysis buffer (50 mM Tris, 0.5 mM EDTA, 50 mM NaCl) and Ni-Sepharose purification using a gradient elution buffer containing different concentrations of imidazole (2 mM, 4 mM, 10 mM, and 30 mM). Finally, the proteins were dialyzed using 50 mM pH 8.0 phosphate buffer overnight to remove imidazole and Tris, which can interfere with subsequent determinations of protein concentration, enzymatic activity, and melting temperature (34).
Comparative modeling of the open conformation using RosettaCM.
The resolved crystal structure of ROL (PDB ID 1lgy) in the RCSB PDB database is in the closed conformation (35, 53). A sufficiently open conformation is required for substrate binding modeling. To construct an OROL, the opened lid of RML (PDB ID 4tgl, residues 82 to 95) and the 1lgy structure without lid region (residues 1 to 82 and 97 to 269) were used as templates, based on the RosettaCM protocol (54). Multiple sequence alignments were conducted using the PROMALS3D server (http://prodata.swmed.edu/promals3d/promals3d.php) (55) and visualized using Jalview (56), with colors based on percentage identity. The X-ray crystal structures of ROL in closed conformation (PDB ID 1lgy) and RML in open conformation (PDB ID 4tgl) were downloaded from the RCSB PDB database. One thousand models were generated, and the topmost model without disrupted native disulfide bonds was selected based on the Rosetta all-atom score. The reliability of the targeted model was evaluated based on a Ramachandran plot using PROCHECK (57) and MolProbity (58). When the proportion of residues in the disallowed region of the selected model structure is below 5%, it can be used for molecular docking (57, 58).
Semiflexible molecular docking using AutoDock Vina.
All amino acids located in the hydrophobic tunnel leading to the active site of OROL were assessed with AutoDock Vina using pNP esters of various lengths as the substrate (59). All parameters were set to the standard values. All crystallographic bound waters were removed, and hydrogen atoms were added to the protein prior to docking. The area within 5 Å of the substrate of the modeled enzyme-substrate complex structure was defined as the catalytic region, using PyMOL (-select cata_reg, byres substrate around 5).
Virtual saturation mutagenesis using the Cartesian_ddg protocol.
Rosetta’s Cartesian_ddg protocol is a fast and accurate free energy prediction method that can be used to calculate the difference of free energy after site mutation in a single protein or a small protein-ligand complex (60). The changes in activity toward pNP esters of diverse chain lengths (C8 to C16) were predicted from the binding free energy based on virtual saturation mutagenesis using Rosetta’s Cartesian_ddg protocol. To shrink the virtual saturated mutant library, the folding free energy change of ROL (PDB ID 1lgy) was calculated for the residues in the catalytic region using the Rosetta’s Cartesian_ddg protocol. Notably, highly conservative residues, such as the catalytic triad, lid, and oxyanion hole, which would have allowed further shrinking of the mutant library, were not manipulated.
The following command lines were used for Rosetta’s Cartesian_ddg: Cartesian_ddg.linuxgccrelease -s $pdbfile -ddg:mut_file all.mutfile -ddg:iterations 3 -ddg::cartesian -ddg::dump_pdbs False -bbnbrs 1 -fa_max_dis 9.0 -score:weights ref2015_cart -relax:cartesian -relax:min_type lbfgs_armijo_nonmonotone -ex1 -ex2 -use_input_sc -flip_HNQ -crystal_refine -interface_ddg 1 -extra_res_fa LIG.params -score:extra_improper_file LIG.tors -optimization:default_max_cycles 1000.
Rational design of potential disulfide bonds using MODIP, DbD2, and BridgeD.
Three computational tools, Disulfide by Design 2.0 (DbD2, http://cptweb.cpt.wayne.edu/DbD2/) (61), MODIP (http://caps.ncbs.res.in/iws/modip.html) (62), and BridgeD (http://biodev.cea.fr/bridged/) (63), were used to analyze selected mutagenic residue pairs and introduce more potential disulfide bonds, aiming to further improve the thermostability of above-mentioned variants. It has been reported that when disulfides are designed 6 to 8 Å from the active site, favorable disulfide bonds are more likely to be formed (64). Therefore, only predicted residues not involved in the catalytic region or in original pairs of disulfide bonds (residues 29, 40, 43, 235, 244, and 268) were considered.
Stability and visual validation of designed disulfide bonds.
FoldX was used to evaluate the impact of each disulfide bond mutation on protein stability (65). The RepairPDB module was used to identify and repair the bad torsion angles or conflicting van der Waals interactions. The output file was submitted to the BuildModel module to optimize the conformation structures and calculate the fold free energy change with default parameters. The running number was set to 5 for better average results. A negative free energy change indicated that the newly formed disulfide bond had a positive influence on protein stability. The optimized structures from FoldX were visually inspected using PyMOL to validate disulfide bond formation to reduce the number of candidates.
Determination of disulfide bond formation using the Ellman method.
Protein samples were treated with 6 M guanidine hydrochloride for 15 min to expose the buried cysteines. Then, the samples were divided into two equal parts for different posttreatment methods, including the Ellman method (33). One portion was treated with DTT at a final concentration of 0.1 M to break the S-S bonds and expose the sulfhydryl group. The other portion was treated with 50 mM pH 8.0 sodium phosphate buffer. Both the reduced and the nonreduced samples were dialyzed overnight in 50 mM pH 8.0 sodium phosphate buffer to remove excess reducing agent. Finally, the samples were mixed with 2 mM 5,5′-dithio-bis-(2-nitrobenzoic acid) solution at a 1:1 ratio and incubated at 37°C for 10 min. The number of sulfhydryl groups was calculated based on the absorbance at 412 nm. The experiment was repeated three times in parallel.
Determination of melting temperatures.
DSF is a strategy to assess protein thermostability based on the melting temperature (Tm) (32). Samples were mixed with SYPRO orange dye (5,000×; Life Technologies, Carlsbad, CA, USA) and slowly heated from 25°C to 90°C at a rate of 0.3°C per min in a StepOnePlus real-time PCR system (Applied Biosystems, Waltham, MA, USA). Tm values were determined as the maximum rate of fluorescence change relative to temperature (dRFU/dT).
Standard colorimetric determination of lipase activity.
The determination of lipase activity was performed on emulsified pNP esters (Sigma-Aldrich, St. Louis, MO, USA) with different chain lengths, in accordance with methods reported in a previous study (34). The 1-mL reaction system was composed of 940 μL of Tris-HCl buffer (50 mM), 40 μL of absolute ethyl alcohol, 10 μL of diluted enzyme solution, and 10 μL of 50 mM pNP esters (dissolved in acetonitrile). As a control, the same volume of Tris-HCl instead of enzyme was used. One unit of lipase activity was defined as the amount of enzyme consumed to release 1 μmol 4-nitrophenol per min (34).
Thermal inactivation analysis.
The thermostability of WT and mutant ROL was investigated by testing residual activities of samples withdrawn under the heat treatment at 45°C and 60°C at specified intervals against the optimal substrate and pH.
Determination of enzymatic properties.
(i) Optimum temperature. To analyze the effect of temperature on the activity of WT and mutant ROL, the hydrolysis reaction was conducted at temperatures ranging from 25°C to 50°C with 5°C intervals and pH 8.0 in 50 mM Tris-HCl buffer, using C8 as the substrate. For each enzyme, the specific activity in optimum temperature was considered 100%.
(ii) Optimum pH. The optimum pH of WT and mutant ROL were determined by examining the enzyme activity in phosphate buffer (pH 7.0 to 8.0), Tris-HCl buffer (pH 8.0 to 9.0), and glycine-NaOH buffer (pH 9.0 to 9.5) at 35°C, using C8 as the substrate. For each enzyme, the specific activity in optimum pH was considered 100%.
(iii) Analysis of chain length selectivity of WT and mutant ROL. The catalytic activity of WT and mutant ROL toward five pNP esters of varying alkyl chain lengths were investigated using the standard colorimetric assay. WT, single-site mutants, and double mutant E265V/S267W were reacted in 50 mM Tris-HCl buffer at 35°C, pH 8.0. Variant M6 was reacted in 50 mM Tris-HCl buffer at 45°C, pH 8.5.
(iv) Determination of kinetic parameters. Kinetic parameters (kcat, Km, and kcat/Km) of WT and mutant ROL were determined by measuring the initial reaction rate in systematic substrate concentrations ranging from 10 mM to 100 mM at 10 mM intervals. WT and variant E265V/S267W were reacted with pNP esters of different chain lengths in pH 8.0 50 mM Tris-HCl buffer at 35°C, whereas variant M6 was reacted in 50 mM Tris-HCl buffer at 45°C, pH 8.5. The Michaelis-Menten constant (Km) and maximum reaction velocity (Vmax) values were calculated using the Lineweaver-Burk method (34). The catalytic turnover rate (kcat) was calculated based on Vmax and the enzyme concentration.
(v) Enzymatic hydrolysis reactions of ROL variants using tricaprin and soybean oil as substrates. The enzymatic hydrolysis of tricaprin was measured by following the method of Wang et al. (42). The reaction system consisted of 600 μL of tricaprin, 5 mL 50 mM Tris-HCl buffer, and an appropriate amount of ROL incubated at 45°C for 12 h. For the hydrolysis of soybean oil, the 6-mL reaction volume contained 2 mL of oil, 3 mL of 50 mM Tris-HCl buffer, and 1 mL of purified enzyme solution mixed in a 50-mL triangular conical flask at 45°C for 24 h. All lipids were first emulsified by sonication, and the reactions were finally terminated by adding 10 mL of absolute ethanol. The released fatty acid was neutralized with 50 mM NaOH, and phenolphthalein was used as an indicator. FA yield (percent) was calculated as the ratio of FA quality produced in a period to the initial quality of substrate at a defined reaction time.
In addition, all enzymatic activity experiments were performed using a standard colorimetric method and conducted in triplicate. Buffers with various pH values were prepared at the same temperatures they were used in the experiments.
ACKNOWLEDGMENTS
This work was financially supported by the National Natural Science Foundation of China (grant 31971206), Fundamental Research Funds for HUST (2014NY007, 2017KFYXJJ212, 2017KFXKJC010, and 2017KFTSZZ001), and the Fundamental Research Funds for the Central Universities, China (S202210487445).
We declare no conflicts of interest.
J.H., L.X., and Y.Y. conceived the project and wrote the manuscript. J.H. designed the experimental scheme and performed the majority of calculations, experiments, and analyses. S.D. helped with the experiments. X.C. helped with the computational data analysis. All authors reviewed the manuscript and agreed to the published version of the manuscript.
Footnotes
Supplemental material is available online only.
Contributor Information
Li Xu, Email: xuli@hust.edu.cn.
Yunjun Yan, Email: yanyunjun@hust.edu.cn.
Nicole R. Buan, University of Nebraska—Lincoln
REFERENCES
- 1.Adlercreutz P. 2013. Immobilisation and application of lipases in organic media. Chem Soc Rev 42:6406–6436. 10.1039/c3cs35446f. [DOI] [PubMed] [Google Scholar]
- 2.Palomo JM. 2020. Synthetic complexity created by lipases. Nat Catal 3:335–336. 10.1038/s41929-020-0453-x. [DOI] [Google Scholar]
- 3.Casas-Godoy L, Gasteazoro F, Duquesne S, Bordes F, Marty A, Sandoval G. 2018. Lipases: an overview. Methods Mol Biol 1835:3–38. 10.1007/978-1-4939-8672-9_1. [DOI] [PubMed] [Google Scholar]
- 4.Liu Q, Xun GH, Feng Y. 2019. The state-of-the-art strategies of protein engineering for enzyme stabilization. Biotechnol Adv 37:530–537. 10.1016/j.biotechadv.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 5.Qu G, Li AT, Acevedo-Rocha CG, Sun ZT, Reetz MT. 2020. The crucial role of methodology development in directed evolution of selective enzymes. Angew Chem Int Ed Engl 59:13204–13231. 10.1002/anie.201901491. [DOI] [PubMed] [Google Scholar]
- 6.Sun ZT, Liu Q, Qu G, Feng Y, Reetz MT. 2019. Utility of B-factors in protein science: interpreting rigidity, flexibility, and internal motion and engineering thermostability. Chem Rev 119:1626–1665. 10.1021/acs.chemrev.8b00290. [DOI] [PubMed] [Google Scholar]
- 7.Wu LJ, Qin L, Nie Y, Xu Y, Zhao YL. 2022. Computer-aided understanding and engineering of enzymatic selectivity. Biotechnol Adv 54:107793. 10.1016/j.biotechadv.2021.107793. [DOI] [PubMed] [Google Scholar]
- 8.Yang B, Wang HJ, Song W, Chen XL, Liu J, Luo QL, Liu LM. 2017. Engineering of the conformational dynamics of lipase to increase enantioselectivity. ACS Catal 7:7593–7599. 10.1021/acscatal.7b02404. [DOI] [Google Scholar]
- 9.Reetz MT. 2011. Laboratory evolution of stereoselective enzymes: a prolific source of catalysts for asymmetric reactions. Angew Chem Int Ed Engl 50:138–174. 10.1002/anie.201000826. [DOI] [PubMed] [Google Scholar]
- 10.Chen K, Arnold FH. 2020. Engineering new catalytic activities in enzymes. Nat Catal 3:203–213. 10.1038/s41929-019-0385-5. [DOI] [Google Scholar]
- 11.Godehard SP, Badenhorst CPS, Müller H, Bornscheuer UT. 2020. Protein engineering for enhanced acyltransferase activity, substrate scope, and selectivity of the Mycobacterium smegmatis acyltransferase MsAcT. ACS Catal 10:7552–7562. 10.1021/acscatal.0c01767. [DOI] [Google Scholar]
- 12.Panizza P, Cesarini S, Diaz P, Rodríguez Giordano S. 2015. Saturation mutagenesis in selected amino acids to shift Pseudomonas sp. acidic lipase Lip I.3 substrate specificity and activity. Chem Commun (Camb) 51:1330–1333. 10.1039/c4cc08477b. [DOI] [PubMed] [Google Scholar]
- 13.Zorn K, Oroz-Guinea I, Brundiek H, Dörr M, Bornscheuer UT. 2018. Alteration of chain length selectivity of Candida antarctica lipase a by semi-rational design for the enrichment of erucic and gondoic fatty acids. Adv Synth Catal 360:4115–4131. 10.1002/adsc.201800889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Banaś AM, Bocian-Ostrzycka KM, Jagusztyn-Krynicka EK. 2019. Engineering of the Dsb (disulfide bond) proteins: contribution towards understanding their mechanism of action and their applications in biotechnology and medicine. Crit Rev Microbiol 45:433–450. 10.1080/1040841X.2019.1622509. [DOI] [PubMed] [Google Scholar]
- 15.Kuan SL, Bergamini FRG, Weil T. 2018. Functional protein nanostructures: a chemical toolbox. Chem Soc Rev 47:9069–9105. 10.1039/c8cs00590g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matsumura M, Signor G, Matthews BW. 1989. Substantial increase of protein stability by multiple disulphide bonds. Nature 342:291–293. 10.1038/342291a0. [DOI] [PubMed] [Google Scholar]
- 17.Dombkowski AA, Sultana KZ, Craig DB. 2014. Protein disulfide engineering. FEBS Lett 588:206–212. 10.1016/j.febslet.2013.11.024. [DOI] [PubMed] [Google Scholar]
- 18.Xu Z, Cen YK, Zou SP, Xue YP, Zheng YG. 2020. Recent advances in the improvement of enzyme thermostability by structure modification. Crit Rev Biotechnol 40:83–98. 10.1080/07388551.2019.1682963. [DOI] [PubMed] [Google Scholar]
- 19.Wu H, Chen QM, Zhang WL, Mu WM. 2021. Overview of strategies for developing high thermostability industrial enzymes: discovery, mechanism, modification and challenges. Crit Rev Food Sci Nutr 1–18. 10.1080/10408398.2021.1970508. [DOI] [PubMed] [Google Scholar]
- 20.Teufl M, Zajc CU, Traxlmayr MW. 2022. Engineering strategies to overcome the stability-function trade-off in proteins. ACS Synth Biol 11:1030–1039. 10.1021/acssynbio.1c00512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wijma HJ, Fürst MJLJ, Janssen DB. 2018. A computational library design protocol for rapid improvement of protein stability: FRESCO. Methods Mol Biol 1685:69–85. 10.1007/978-1-4939-7366-8_5. [DOI] [PubMed] [Google Scholar]
- 22.Bu YF, Cui YL, Peng Y, Hu MR, Tian YE, Tao Y, Wu B. 2018. Engineering improved thermostability of the GH11 xylanase from Neocallimastix patriciarum via computational library design. Appl Microbiol Biotechnol 102:3675–3685. 10.1007/s00253-018-8872-1. [DOI] [PubMed] [Google Scholar]
- 23.Wang YL, Li CM, Ban XF, Gu ZB, Hong Y, Cheng L, Li ZF. 2022. Disulfide bond engineering for enhancing the thermostability of the maltotetraose-forming amylase from Pseudomonas saccharophila STB07. Foods 11:1207. 10.3390/foods11091207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Di Lorenzo M, Hidalgo A, Haas M, Bornscheuer UT. 2005. Heterologous production of functional forms of Rhizopus oryzae lipase in Escherichia coli. Appl Environ Microbiol 71:8974–8977. 10.1128/AEM.71.12.8974-8977.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Melani NB, Tambourgi EB, Silveira E. 2020. Lipases: from production to applications. Sep Purif Rev 49:143–158. 10.1080/15422119.2018.1564328. [DOI] [Google Scholar]
- 26.López-Fernández J, Benaiges MD, Valero F. 2020. Rhizopus oryzae lipase, a promising industrial enzyme: biochemical characteristics, production and biocatalytic applications. Catalysts 10:1277. 10.3390/catal10111277. [DOI] [Google Scholar]
- 27.Yu XW, Xu Y, Xiao R. 2016. Lipases from the genus Rhizopus: characteristics, expression, protein engineering and application. Prog Lipid Res 64:57–68. 10.1016/j.plipres.2016.08.001. [DOI] [PubMed] [Google Scholar]
- 28.Zhao JF, Wang Z, Gao FL, Lin JP, Yang LR, Wu MB. 2018. Enhancing the thermostability of Rhizopus oryzae lipase by combined mutation of hot-spots and engineering a disulfide bond. RSC Adv 8:41247–41254. 10.1039/c8ra07767c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ding X, Tang XL, Zheng RC, Zheng YG. 2019. Identification and engineering of the key residues at the crevice-like binding site of lipases responsible for activity and substrate specificity. Biotechnol Lett 41:137–146. 10.1007/s10529-018-2620-6. [DOI] [PubMed] [Google Scholar]
- 30.Fang JW. 2020. A critical review of five machine learning-based algorithms for predicting protein stability changes upon mutation. Brief Bioinform 21:1285–1292. 10.1093/bib/bbz071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sequeiros-Borja CE, Surpeta B, Brezovsky J. 2021. Recent advances in user-friendly computational tools to engineer protein function. Brief Bioinform 22:bbaa150. 10.1093/bib/bbaa150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vedadi M, Niesen FH, Allali-Hassani A, Fedorov OY, Finerty PJ, Wasney GA, Yeung R, Arrowsmith C, Ball LJ, Berglund H, Hui R, Marsden BD, Nordlund P, Sundstrom M, Weigelt J, Edwards AM. 2006. Chemical screening methods to identify ligands that promote protein stability, protein crystallization, and structure determination. Proc Natl Acad Sci USA 103:15835–15840. 10.1073/pnas.0605224103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mamathambika BS, Bardwell JC. 2008. Disulfide-linked protein folding pathway. Annu Rev Cell Dev Biol 24:211–235. 10.1146/annurev.cellbio.24.110707.175333. [DOI] [PubMed] [Google Scholar]
- 34.Li GL, Fang XR, Su F, Chen Y, Xu L, Yan YJ. 2018. Enhancing the thermostability of Rhizomucor miehei lipase with a limited screening library by rational-design point mutations and disulfide bonds. Appl Environ Microbiol 84:e02129-17. 10.1128/AEM.02129-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kohno M, Funatsu J, Mikami B, Kugimiya W, Matsuo T, Morita Y. 1996. The crystal structure of lipase II from Rhizopus niveus at 2.2 Å resolution. J Biochem 120:505–510. 10.1093/oxfordjournals.jbchem.a021442. [DOI] [PubMed] [Google Scholar]
- 36.Catherine M, Jenny P, Aurélie M, Pierre V, Isabelle H, Johan W. 2010. Structural insights into the acidophilic pH adaptation of a novel endo-1,4-b-xylanase from Scytalidium acidophilum. Biochimie 92:1407–1415. 10.1016/j.biochi.2010.07.003. [DOI] [PubMed] [Google Scholar]
- 37.Rigoldi F, Donini S, Redaelli A, Parisini E, Gautieri A. 2018. Review: engineering of thermostable enzymes for industrial applications. APL Bioeng 2:e011501. 10.1063/1.4997367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lan DM, Zhao G, Holzmann N, Yuan SG, Wang J, Wang YH. 2021. Structure-guided rational design of a mono- and diacylglycerol lipase from Aspergillus oryzae: a single residue mutant increases the hydrolysis ability. J Agric Food Chem 69:5344–5352. 10.1021/acs.jafc.1c00913. [DOI] [PubMed] [Google Scholar]
- 39.Gautieri A, Rigoldi F, Torretta A, Redaelli A, Parisini E. 2022. In silico engineering of enzyme access tunnels. Methods Mol Biol 2397:203–225. 10.1007/978-1-0716-1826-4_11. [DOI] [PubMed] [Google Scholar]
- 40.Kokkonen P, Bednar D, Pinto G, Prokop Z, Damborsky J. 2019. Engineering enzyme access tunnels. Biotechnol Adv 37:107386. 10.1016/j.biotechadv.2019.04.008. [DOI] [PubMed] [Google Scholar]
- 41.Wang XL, Du JH, Zhao BC, Wang HY, Rao SQ, Du GC, Zhou JW, Chen J, Liu S. 2021. Significantly improving the thermostability and catalytic efficiency of Streptomyces mobaraenesis transglutaminase through combined rational design. J Agric Food Chem 69:15268–15278. 10.1021/acs.jafc.1c05256. [DOI] [PubMed] [Google Scholar]
- 42.Wang S, Xu Y, Yu XW. 2021. Propeptide in Rhizopus chinensis lipase: new insights into its mechanism of activity and substrate selectivity by computational design. J Agric Food Chem 69:4263–4275. 10.1021/acs.jafc.1c00721. [DOI] [PubMed] [Google Scholar]
- 43.Pleiss J, Fischer M, Schmid RD. 1998. Anatomy of lipase binding sites: the scissile fatty acid binding site. Chem Phys Lipids 93:67–80. 10.1016/s0009-3084(98)00030-9. [DOI] [PubMed] [Google Scholar]
- 44.Müller H, Becker AK, Palm GJ, Berndt L, Badenhorst CPS, Godehard SP, Reisky L, Lammers M, Bornscheuer UT. 2020. Sequence-based prediction of promiscuous acyltransferase activity in hydrolases. Angew Chem Int Ed Engl 59:11607–11612. 10.1002/anie.202003635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Yang KS, Sung BH, Park MK, Lee JH, Lim KJ, Park SC, Kim SL, Kim HK, Sohn JH, Kim HM, Kim SC. 2015. Recombinant lipase engineered with amphipathic and coiled-coil peptides. ACS Catal 5:5016–5025. 10.1021/cs502079g. [DOI] [Google Scholar]
- 46.Wang R, Wang S, Xu Y, Yu X. 2020. Enhancing the thermostability of Rhizopus chinensis lipase by rational design and MD simulations. Int J Biol Macromol 160:1189–1200. 10.1016/j.ijbiomac.2020.05.243. [DOI] [PubMed] [Google Scholar]
- 47.Pijning AE, Hogg P. 2019. Classification of protein disulphide bonds. Methods Mol Biol 1967:1–8. 10.1007/978-1-4939-9187-7_1. [DOI] [PubMed] [Google Scholar]
- 48.Craig DB, Dombkowski AA. 2018. Stabilization of peptides and proteins by engineered disulfide bonds, p 381–398. In Feige MJ (ed), Oxidative folding of proteins: basic principles, cellular regulation and engineering. RSC Publishing, Cambridge, United Kingdom. [Google Scholar]
- 49.Gihaz S, Bash Y, Rush I, Shahar A, Pazy Y, Fishman A. 2020. Bridges to stability: engineering disulfide bonds towards enhanced lipase biodiesel synthesis. Chemcatchem 12:181–192. 10.1002/cctc.201901369. [DOI] [Google Scholar]
- 50.Zhang HT, Sang JC, Zhang Y, Sun TW, Liu H, Yue R, Zhang J, Wang HK, Dai YJ, Lu FJ, Liu FF. 2019. Rational design of a Yarrowia lipolytica derived lipase for improved thermostability. Int J Biol Macromol 137:1190–1198. 10.1016/j.ijbiomac.2019.07.070. [DOI] [PubMed] [Google Scholar]
- 51.Qu PP, Li DD, Lazim R, Xu R, Xiao DC, Wang F, Li X, Zhang Y. 2022. Improved thermostability of Thermomyces lanuginosus lipase by molecular dynamics simulation and in silico mutation prediction and its application in biodiesel production. Fuel 327:125039. 10.1016/j.fuel.2022.125039. [DOI] [Google Scholar]
- 52.Jiao LC, Zhou QH, Su ZX, Xu L, Yan YJ. 2018. High-level extracellular production of Rhizopus oryzae lipase in Pichia pastoris via a strategy combining optimization of gene-copy number with co-expression of ERAD-related proteins. Protein Expr Purif 147:1–12. 10.1016/j.pep.2018.02.005. [DOI] [PubMed] [Google Scholar]
- 53.Berman HM, Westbrook J, Feng ZK, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. 2000. The protein data bank. Nucleic Acids Res 28:235–242. 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Song YF, DiMaio F, Wang RY, Kim D, Miles C, Brunette TJ, Thompson J, Baker D. 2013. High-resolution comparative modeling with RosettaCM. Structure 21:1735–1742. 10.1016/j.str.2013.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Pei JM, Kim BH, Grishin NV. 2008. PROMALS3D: a tool for multiple protein sequence and structure alignments. Nucleic Acids Res 36:2295–2300. 10.1093/nar/gkn072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. 2009. Jalview version 2, a multiple sequence alignment editor and analysis workbench. Bioinformatics 25:1189–1191. 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Laskowski RA, Rullmann JAC, MacArthur MW, Kaptein R, Thornton JM. 1996. AQUA and PROCHECK-NMR: programs for checking the quality of protein structures solved by NMR. J Biomol NMR 8:477–486. 10.1007/BF00228148. [DOI] [PubMed] [Google Scholar]
- 58.Chen VB, Arendall WB, III, Headd JJ, Keedy DA, Immormino RM, Kapral GJ, Murray LW, Richardson JS, Richardson DC. 2010. MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr D Biol Crystallogr 66:12–21. 10.1107/S0907444909042073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Eberhardt J, Santos-Martins D, Tillack AF, Forli S. 2021. AutoDock Vina 1.2.0: new docking methods, expanded force field, and python bindings. J Chem Inf Model 61:3891–3898. 10.1021/acs.jcim.1c00203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Park H, Bradley P, Greisen P, Jr, Liu Y, Mulligan VK, Kim DE, Baker D, DiMaio F. 2016. Simultaneous optimization of biomolecular energy functions on features from small molecules and macromolecules. J Chem Theory Comput 12:6201–6212. 10.1021/acs.jctc.6b00819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Craig DB, Dombkowski AA. 2013. Disulfide by Design 2.0: a web-based tool for disulfide engineering in proteins. BMC Bioinformatics 14:346. 10.1186/1471-2105-14-346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Dani VS, Ramakrishnan C, Varadarajan R. 2003. MODIP revisited: re-evaluation and refinement of an automated procedure for modeling of disulfide bonds in proteins. Protein Eng 16:187–193. 10.1093/proeng/gzg024. [DOI] [PubMed] [Google Scholar]
- 63.Pellequer JL, Chen SW. 2006. Multi-template approach to modeling engineered disulfide bonds. Proteins 65:192–202. 10.1002/prot.21059. [DOI] [PubMed] [Google Scholar]
- 64.de Bakker PI, Hünenberger PH, McCammon JA. 1999. Molecular dynamics simulations of the hyperthermophilic protein sac7d from Sulfolobus acidocaldarius: contribution of salt bridges to thermostability. J Mol Biol 285:1811–1830. 10.1006/jmbi.1998.2397. [DOI] [PubMed] [Google Scholar]
- 65.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. 2005. The FoldX web server: an online force field. Nucleic Acids Res 33:W382–W388. 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig. S1 to S5 and Tables S1 to S5. Download aem.01878-22-s0001.pdf, PDF file, 1.2 MB (1.2MB, pdf)