ABSTRACT
Posttreatment controllers (PTCs) are rare HIV-infected individuals who can limit viral rebound after antiretroviral therapy interruption (ATI), but the mechanisms of this remain unclear. To investigate these mechanisms, we quantified various HIV RNA transcripts (via reverse transcription droplet digital PCR [RT-ddPCR]) and cellular transcriptomes (via RNA-seq) in blood cells from PTCs and noncontrollers (NCs) before and two time points after ATI. HIV transcription initiation did not significantly increase after ATI in PTCs or in NCs, whereas completed HIV transcripts increased at early ATI in both groups and multiply-spliced HIV transcripts increased only in NCs. Compared to NCs, PTCs showed lower levels of HIV DNA, more cell-associated HIV transcripts per total RNA at all times, no increase in multiply-spliced HIV RNA at early or late ATI, and a reduction in the ratio of completed/elongated HIV RNA after early ATI. NCs expressed higher levels of the IL-7 pathway before ATI and expressed higher levels of multiple cytokine, inflammation, HIV transcription, and cell death pathways after ATI. Compared to the baseline, the NCs upregulated interferon and cytokine (especially TNF) pathways during early and late ATI, whereas PTCs upregulated interferon and p53 pathways only at early ATI and downregulated gene translation during early and late ATI. In NCs, viral rebound after ATI is associated with increases in HIV transcriptional completion and splicing, rather than initiation. Differences in HIV and cellular transcription may contribute to posttreatment control, including an early limitation of spliced HIV RNA, a delayed reduction in completed HIV transcripts, and the differential expression of the IL-7, p53, and TNF pathways.
IMPORTANCE The findings presented here provide new insights into how HIV and cellular gene expression change after stopping ART in both noncontrollers and posttreatment controllers. Posttreatment control is associated with an early ability to limit increases in multiply-spliced HIV RNA, a delayed (and presumably immune-mediated) ability to reverse an initial rise in processive/completed HIV transcripts, and multiple differences in cellular gene expression pathways. These differences may represent correlates or mechanisms of posttreatment control and may provide insight into the development and/or monitoring of therapeutic strategies that are aimed at a functional HIV cure.
KEYWORDS: HIV posttreatment controllers, noncontrollers, transcription, transcriptional completion, splicing, analytic treatment interruption, cell-mediated immune response, viremic suppression, transcriptional profile, HIV DNA, ART, RNA splicing, human immunodeficiency virus, treatment interruption
INTRODUCTION
Several studies have measured the effects of the dynamic interruption of antiretroviral therapy (ART) to control human immunodeficiency virus (HIV) infection (1–4). These studies have led to the identification of a rare subset of people with HIV (PWH), known as posttreatment controllers (PTCs), who comprise <1% of PWH and can control HIV infection for months to years after stopping ART (1–3, 5, 6). However, identifying PTCs has been difficult, as studies of “analytic therapy interruption” (ATI) are infrequently performed (5).
Various studies have identified and classified posttreatment controllers. The VISCONTI study identified 14 PTCs who were initiated on ART during primary infection (5). Compared to natural controllers, these PTCs tended to lack protective HLA alleles and had higher viral loads, lower CD4 counts, lower HIV-specific CD8+ T cell responses, and less T cell activation during primary infection (5). Like spontaneous controllers, PTCs had a latent reservoir and low total levels of HIV DNA, but the HIV DNA was less concentrated in longer-lived naive and central memory cells and declined over time after ATI (5). The CHAMP study identified PTCs who initiated ART during both early and chronic infection, but it showed that posttreatment control was more common with early ART (6). PTCs frequently showed a transient, low level viral rebound after ATI, and this was followed by a variable period of subsequent virologic control, with some PTCs later experiencing virologic rebound (6).
PTCs have lower average levels of total and intact proviruses than do NCs (7), and low provirus levels have served as one predictor of HIV control (8, 9). However, a subset of PTCs can also have relatively high HIV DNA levels (10), suggesting that HIV DNA levels alone are not sufficient in predicting posttreatment control. Lower levels of cell-associated HIV RNA (11, 12) and elevated pre-ATI levels of glutamine have also been associated with posttreatment control (13). However, the exact mechanisms of posttreatment control remain unknown.
A deeper understanding of these mechanisms may inform new therapies aimed at a functional HIV cure. We hypothesized that posttreatment control may be driven by a lower expression of completed and multiply-spliced HIV transcripts and/or the greater destruction of the cells that make these HIV transcripts. To test our hypothesis, we measured the levels of cell-associated initiated, 5′-elongated, mid-elongated/unspliced, completed, and multiply-spliced HIV transcripts and human cellular transcriptomes in PTCs and NCs before and after ATI. Whereas virologic rebound in NCs was associated with increases in HIV transcriptional completion and splicing, PTCs showed an early ability to prevent increases in multiply-spliced HIV RNA after ATI, a delayed response that reduced completed HIV transcripts, and the differential transcription of multiple cellular gene pathways.
RESULTS
PTCs exhibit lower viral loads (VL) after ATI.
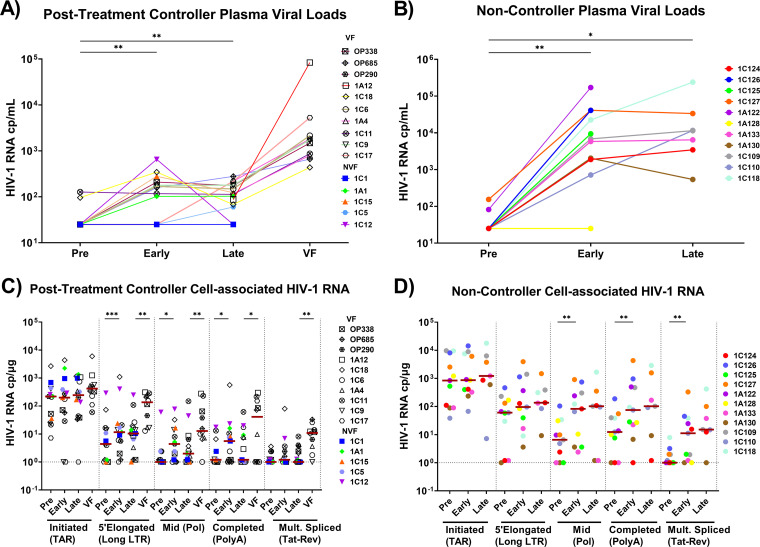
The median ART duration was 4.8 years for PTCs and 3.3 years for NCs (P = 0.5). In addition to other clinical parameters (Table S2), VL were measured to stratify participants as either PTCs or NCs (Fig. S1). While VL increased from pre-ATI to early ATI in both PTCs and NCs (P = 0.01 for both) (Fig. 1A and B), VL were markedly lower in PTCs at both early ATI (medians: 143 versus 6,856 copies/mL; P = 0.0001) and late ATI (medians: 108 versus 11,335 copies/mL; P = 0.0001) (Fig. 2A).
FIG 1.
PTCs and NCs show different temporal changes in plasma and cell-associated HIV RNA after ATI. (A and B) Plasma viral loads were measured by clinical assays in posttreatment controllers (A) and in noncontrollers (B) before ATI (pre), at early and late times after ATI, and at the time of virologic failure (VF). (C and D) Initiated (TAR), 5′-elongated (Long LTR), mid-transcribed (Pol), completed (PolyA), and multiply-spliced (Tat-Rev) cell-associated HIV transcripts were measured at each time point in posttreatment controllers (C) and in noncontrollers (D) using RT-ddPCR. Each participant is represented by a different color or symbol. Open symbols denote PTCs who eventually experienced rebound. Maroon-colored lines indicate medians. Nondetectable transcripts were assigned a value of 1 cp/μg. Statistics were calculated using the Wilcoxon signed-rank test. *, P < 0.05; **, P < 0.01; ***, P < 0.001.
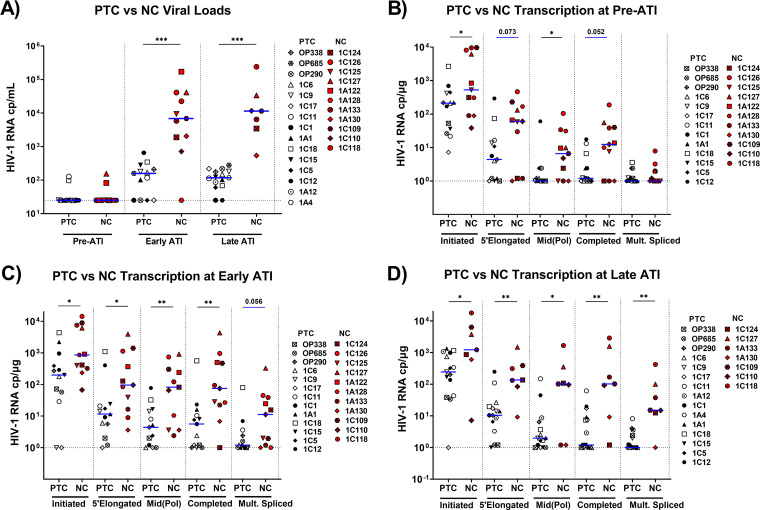
FIG 2.
PTCs have lower levels of measured HIV-1 transcripts at all time points studied. Viral loads (A) and cell-associated HIV RNA levels at pre-ATI (B), early ATI (C), and late ATI (D) time points were compared between posttreatment controllers (PTCs) and noncontrollers (NCs). PTCs are represented by unique white or black symbols. Open symbols denote PTCs who eventually experienced virologic rebound. NCs are represented by unique red symbols. Blue lines indicate median detectable transcripts. All viral loads of <50 copies/mL were assigned a value of 25 copies/mL, and undetected transcripts were assigned a value of 1 cp/μg. Statistics were calculated using the Mann-Whitney U test. *, P < 0.05; **, P < 0.01; ***, P < 0.001. Statistically trending data are denoted using blue bars, with the associated P values being above the data sets.
Unlike NCs, PTCs limit the levels of completed and spliced HIV RNA after ATI.
Initiated (TAR), 5′-elongated (LongLTR), mid-elongated (Pol), completed (PolyA), and multiply-spliced (Tat-Rev) HIV transcripts were quantified using reverse transcription droplet digital PCR (RT-ddPCR). In PTCs, we did not observe any significant increase in the initiated or multiply-spliced HIV transcripts per μg of RNA (approximately 106 cells) early or late after ATI (Fig. 1C). From pre-ATI to early ATI, PTCs showed increases in 5′-elongated, mid-elongated, and completed HIV transcripts (median fold changes: 2.6, 4.4, and 4.7, respectively; P = 0.001, 0.011, and 0.025, respectively). Between early and late ATI, the median levels of the mid-elongated and completed HIV transcripts tended to decrease in PTCs (P = NS). In the subset of PTCs who eventually experienced virologic rebound, the 5′-elongated, mid-elongated, completed, and multiply-spliced HIV RNA all increased from late ATI to virologic failure (P = 0.006, 0.004, 0.032, and 0.006, respectively), whereas no change was observed in the initiated HIV transcripts.
Like PTCs, NCs showed no significant increase in initiated HIV transcripts after ATI. While the median levels of 5′-elongated HIV transcripts tended to increase after ATI, the differences were not significant. Like PTCs, NCs showed increases in mid-elongated and completed HIV transcripts from pre-ATI to early ATI (median fold changes: 12.3 and 17.8, respectively; P = 0.007 and 0.002, respectively), but these were also accompanied by a large increase in multiply-spliced Tat-Rev (fold change of 28.9; P = 0.008) (Fig. 1D). In NCs, the median levels of Pol, PolyA, and Tat-Rev remained stable or increased (P = NS) from early to late ATI, in contrast to those of PTCs.
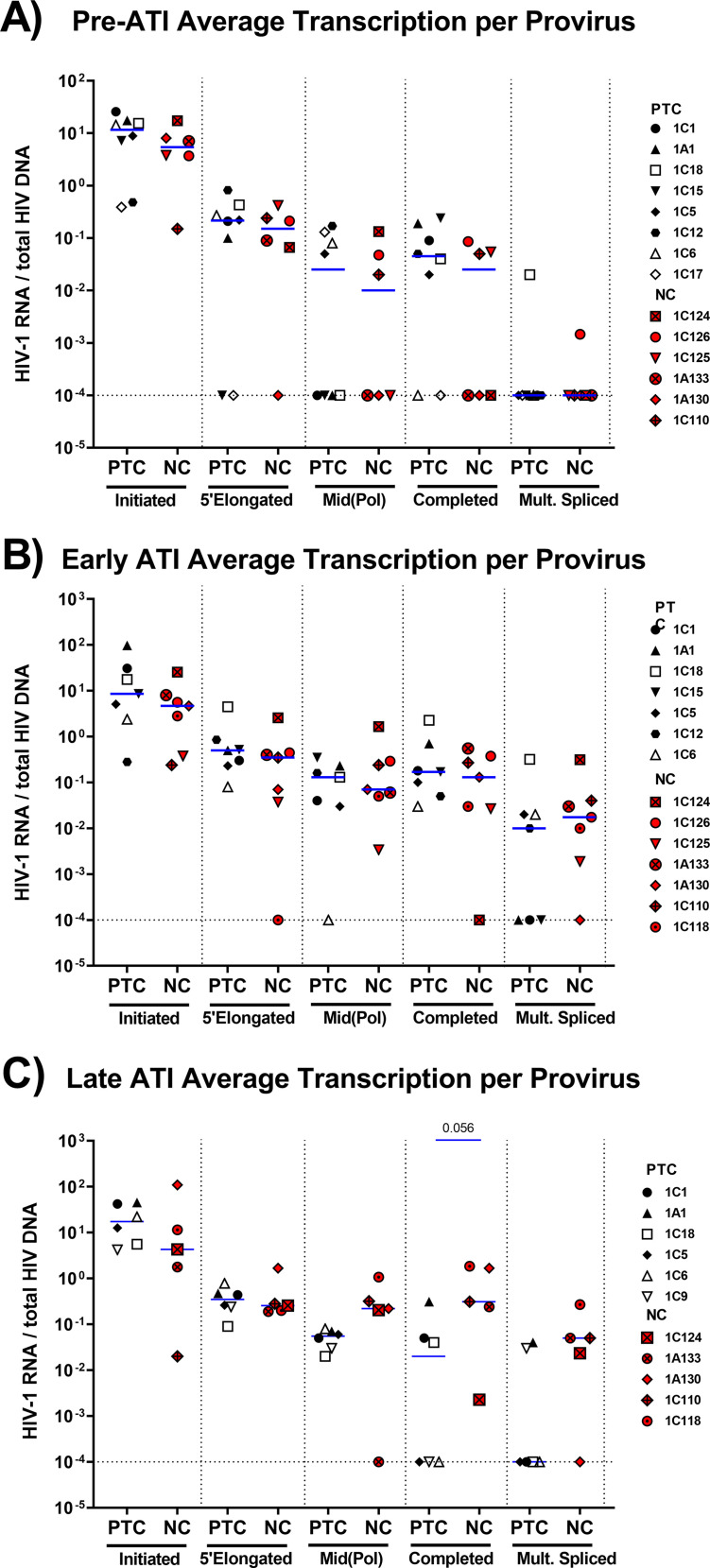
Compared to NCs, PTCs exhibited lower levels of initiated and mid-elongated HIV transcripts before ATI (P = 0.041 for both) and a trend toward lower levels of 5′-elongated and completed transcripts (P = 0.075 and 0.052, respectively) (Fig. 2B). In both groups, multiply-spliced Tat-Rev was usually undetectable before ATI. Early after ATI, PTCs had lower levels of initiated, 5′-elongated, mid-elongated, and completed transcripts than did NCs (P = 0.018, 0.026, 0.008, and 0.007, respectively), and they tended to have lower levels of multiply-spliced Tat-Rev (P = 0.056) (Fig. 2C). By late ATI, all transcripts were lower in PTCs (P = 0.05, 0.004, 0.05, 0.002, and 0.002, respectively) (Fig. 2D). Since the total HIV RNA levels reflect both the infection frequency and the HIV transcription per provirus, we also analyzed the levels of total and intact HIV DNA, as measured in a subset of participants. From pre-ATI to early ATI, both PTCs and NCs showed increases in total HIV DNA (P = 0.02, P = 0.035) (Fig. S2), whereas neither group showed significant changes between early and late ATI. We did not observe significant differences between any time points for the intact HIV DNA, although the power was limited.
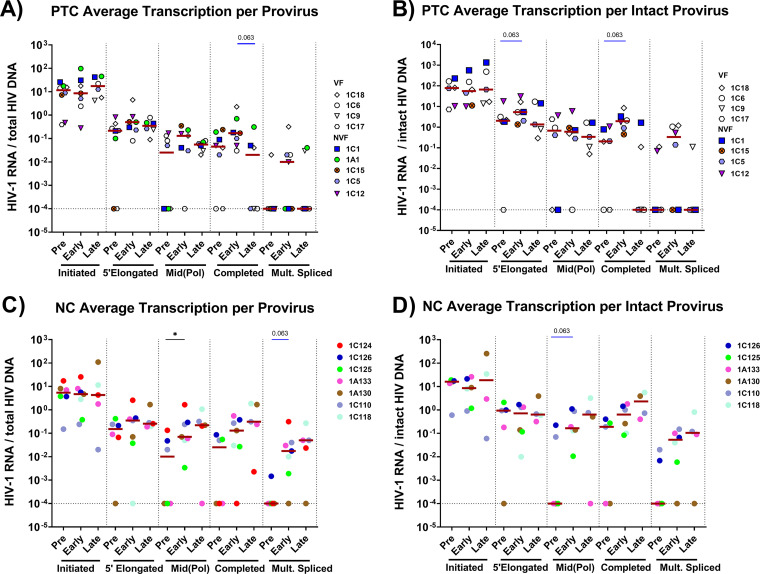
To account for differences in infection frequency, we calculated the ratio of each HIV-1 RNA to the total and intact HIV DNA (where available) to express the average level of each transcript per provirus (Fig. 3). The trends were similar to those observed for the total HIV RNA. From pre-ATI to early ATI, PTCs showed a trending increase in 5′-elongated and completed transcripts per intact HIV DNA (P = 0.063) (Fig. 3B), whereas the levels of completed HIV RNA per total DNA tended to decrease from early to late ATI (P = 0.063) (Fig. 3A).
FIG 3.
PTCs and NCs show different temporal changes in average HIV RNA levels per provirus (HIV RNA/HIV DNA) after ATI. To account for differences in infection frequency, levels of each HIV transcript were normalized to the total HIV DNA (A and C) and the intact HIV DNA (B and D) levels, as measured using the intact proviral DNA assay (IPDA) in a subset of individuals. Each participant is represented by a different color or symbol. Open symbols denote PTCs who eventually experienced viral rebound. Maroon lines indicate ratio medians. Ratios with a value of 0 were assigned a value of 0.0001. Statistics were calculated using the Wilcoxon signed-rank test or the Mann-Whitney U-test. *, P < 0.05. Statistically trending data are denoted using blue bars, with the associated P values being above the data sets.
After ATI, NCs showed no significant increase in initiated or 5′-elongated transcripts per total or intact HIV DNA (Fig. 3C and D). From pre-ATI to early ATI, NCs showed an increase in mid-elongated transcripts per total HIV DNA (P = 0.04), a trend toward an increase in multiply-spliced Tat-Rev per total HIV DNA (P = 0.063) (Fig. 3C), and a trend toward an increase in mid-elongated transcripts per intact HIV (P = 0.063) (Fig. 3D).
Comparing PTCs and NCs, we found no difference in the levels of any HIV RNA per total HIV DNA at the pre-ATI time point, suggesting that the lower total levels of HIV RNA in PTCs before ATI may be driven by lower infection frequencies (Fig. 4). Likewise, we did not detect significant differences between PTCs and NCs at the early ATI time point. However, the lack of HIV DNA data for some participants limits the power with which to detect differences. Moreover, the median levels of mid-elongated, completed, and multiply-spliced HIV RNA per total HIV DNA tended to be higher in NCs than in PTCs by late ATI, although only the difference in completed HIV RNA per DNA approached statistical significance (P = 0.056) (Fig. 4C). By late ATI, PTCs had significantly lower levels of completed transcripts per intact provirus (Fig. S3). These findings suggest that the differences between PTCs and NCs at the late ATI time point are not solely due to differences in HIV infection frequency.
FIG 4.
Average HIV RNA levels per provirus do not differ between NCs and PTCs before ATI, but differences start to appear by late ATI. The average levels of each HIV transcript per provirus (HIV RNA/total HIV DNA) were compared between PTCs and NCs at pre-ATI (A), early ATI (B), and late ATI (C). Blue lines indicate medians. Ratios with a value of 0 were assigned a value of 0.0001. Statistics were calculated via the Mann-Whitney U test. Statistically trending data are denoted using blue bars, with the associated P values being above the data sets.
PTCs show less HIV transcriptional completion and splicing after ATI.
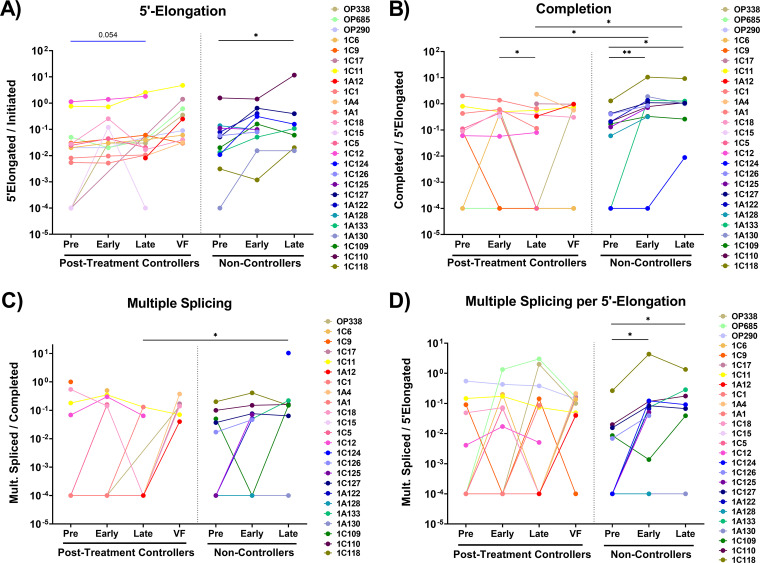
The ratios of one HIV RNA to another were calculated to express the amount of HIV transcriptional elongation (5′-elongated/initiated), completion (completed/elongated), and multiple splicing (multiply-spliced/completed and multiply-spliced/5′-elongated). Of note, these measures are independent of the cell infection frequency and the methods used to normalize HIV RNA levels to cell numbers. From pre-ATI to late ATI, HIV transcriptional elongation tended to increase in PTCs and increased in NCs (P = 0.02) (Fig. 5A). In PTCs, we did not detect an increase in HIV transcriptional completion at early ATI, and completion decreased significantly from early to late ATI (P = 0.04). In contrast, NCs showed an increase in HIV transcriptional completion from pre-ATI to early ATI (P = 0.004) as well as from pre-ATI to late ATI (P = 0.04) (Fig. 5B). At both early and late ATI, completion was higher in NCs than in PTCs.
FIG 5.
PTCs have lower levels of HIV transcriptional completion and splicing after ATI. The ratios of one HIV RNA to another were used to express the extent of progression through various blocks to HIV transcription, including: (A) 5′ elongation (elongated/initiated HIV RNA, (B) completion (completed/elongated HIV RNA), (C) multiple splicing (multiply-spliced/completed HIV RNA), and (D) multiply-spliced as a fraction of elongated (multiply-spliced/elongated HIV RNA). Each participant is represented by a different color. Statistics were calculated via the Wilcoxon signed-rank test for within-group comparisons and the Mann-Whitney U test for between-group comparisons. *, P < 0.05; **, P < 0.01. Statistically trending data are denoted using blue bars, with the associated P values being above the data sets.
By late ATI, multiple splicing (Tat-Rev/PolyA) was lower in PTCs than in NCs (P = 0.015) (Fig. 5C). In addition, the fraction of elongated transcripts that are multiply-spliced (Tat-Rev/Long LTR) increased from pre-ATI to early ATI in NCs (P = 0.008) (Fig. 5D), whereas no such increase was detected in PTCs. Generally, completion and multiple splicing were lower in PTCs by late ATI.
PTCs demonstrated a distinct transcriptome, compared to NCs.
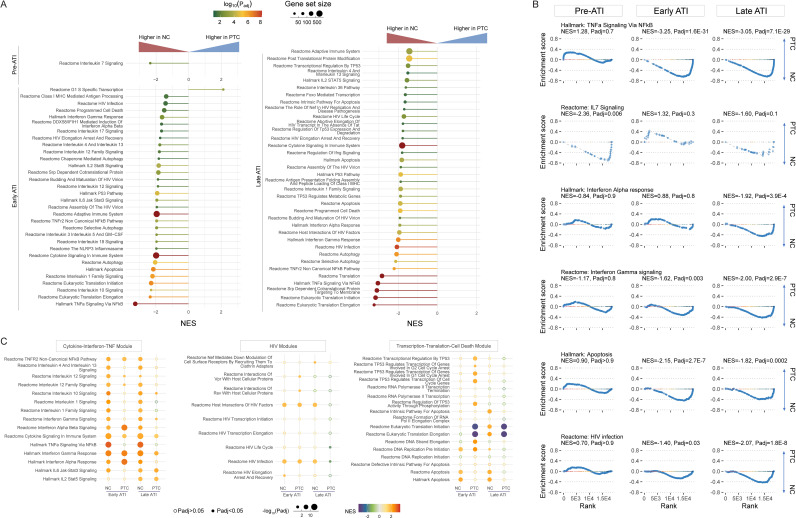
To understand the mechanisms behind posttreatment control and the higher levels of transcriptional completion in NCs, we performed a gene set enrichment analysis (GSEA) on bulk CD4+ T cell RNA sequencing (RNA-seq) data (14). Three PTCs without virologic failure and three NCs with longitudinal RNA-seq data were included in this analysis (sampling schema shown in Fig. S4A). Before ATI, only a limited number of pathways from the curated gene sets differed between PTCs and NCs, including the higher expression of the interleukin-7 (IL-7) pathway in NCs (Fig. 6A and B). However, during both early and late ATI, NCs upregulated multiple pathways related to inflammation, interferon type I and II activity, the HIV life cycle, and cell death (Fig. 6A and B). One representative inflammatory response pathway that was persistently enriched in NCs was the tumor necrosis factor (TNF) pathway, which is tightly linked to inflammation, HIV transcription, and cell death (Fig. 6A and B) (15). A longitudinal within-group comparison revealed the differential upregulation of certain pathways in PTCs versus NCs (Fig. 6C). NCs demonstrated persistent global activation of cytokine, interferon, and TNF pathways during early and late ATI, whereas PTCs only showed selected, non-durable upregulation of some pathways, especially the interferon pathways, early in the ATI (Fig. 6C, Cytokine-Interferon-TNF Module). Both PTCs and NCs upregulated host proteins that interacted with HIV in early ATI, whereas PTCs downregulated certain HIV-dependent factors in late ATI (Fig. 6C, HIV Module). Finally, PTCs upregulated p53-related cell cycle arrest pathways and downregulated gene translation during early ATI (Fig. 6C, Transcription-Translation-Cell Death Module). When evaluating potential mechanisms behind complete HIV RNA transcription (PolyA level) during ATI, we discovered that the high PolyA level was associated with elevated inflammatory response (TNF, interferons, IL-2) and apoptosis pathways as well as with downregulated mRNA decay/deadenylation, global gene transcription, and translation (Fig. S4B).
FIG 6.
PTCs had distinct transcriptomic features from NCs during ATI. PTCs had lower levels of inflammation, cell death, and HIV-dependent factors during ATI. (A) Normalized Enrichment Scores (NES) for the curated pathways, highlighting the enrichment of inflammatory, cell death, and HIV-dependent pathways in NCs, compared to PTCs, during pre-ATI, early ATI, and late ATI. (B) Gene set enrichment in selected pathways at different time points. NES, normalized enrichment score. (C) Longitudinal NES for different pathways in PTCs and NCs. The NES were derived from a within-group comparison, namely, the transcriptome obtained during ATI versus that obtained in pre-ATI in PTCs and in NCs, respectively.
DISCUSSION
We analyzed longitudinal changes in HIV DNA and various HIV transcripts before and after ATI in 15 PTCs and 11 NCs. Whereas previous studies have measured one region of HIV RNA, the results presented here provide greater insight into how the mechanisms governing HIV expression change from ART suppression to virologic rebound in PTCs and NCs.
We detected no change in HIV transcriptional initiation after ATI in either NCs or PTCs, even at the time of virologic failure (Fig. 2B–D). This result seems surprising, since one would expect that the principal effect of stopping ART is to allow for the infection of new cells, which should increase HIV transcriptional initiation. It is possible that many of the observed increases in completed and/or multiply-spliced HIV RNA (and perhaps virus production) occur in cells that were infected prior to ATI and/or that many of the newly infected cells are in tissues, migrate to tissues, or are cleared by immune responses. However, HIV DNA increased from pre-ATI to early ATI in both PTCs and NCs, suggesting the infection of new cells, although other explanations (proliferation of previously infected cells) might contribute. Alternatively, it is possible that the newly infected cells are rare and that their levels of HIV transcriptional initiation are not much greater than the much larger population of preinfected cells, such that they do not perceptibly change initiated transcripts, but they do have much higher rates of transcriptional completion and splicing. Finally, the increase in HIV transcription in newly infected cells could be matched by a decrease in transcription from previously infected cells.
It is also possible that the analyzed cells could harbor cell-bound virions that contain genomic HIV RNA, especially during viremia in NCs, and that these could create the appearance of increased HIV transcriptional elongation and completion. However, virion HIV RNA should not contain multiply-spliced Tat-Rev, and it should be detected as 2 copies of HIV TAR for every one copy of LongLTR, Pol, and U3-PolyA RNA. The effect of bound virions would be to disproportionately increase HIV transcriptional initiation and not increase the ratios of Tat-Rev/PolyA or Tat-Rev/LongLTR. The fact that we observed the opposite in NCs suggests that our results cannot be explained by bound virions. Instead, our data suggest that the transition from a non-productive to a productive HIV infection is mostly associated with increases in HIV transcriptional processivity and splicing, which may occur in newly infected cells. These findings accord with prior studies that suggest that reversible blocks to HIV transcriptional elongation, completion, and splicing are the main mechanisms of viral latency in the blood of ART-suppressed humans (16), whereas blocks to elongation and splicing also operate in primary cell latency models (17). Future studies should investigate whether the levels of completed and multiply-spliced HIV transcripts are early harbingers of a rebound after ATI.
PTCs and NCs differed in three major aspects of HIV transcription. First, PTCs had lower total levels of initiated HIV transcripts and most other HIV transcripts per million cell equivalents at all time points. Before ATI, the differences between PTCs and NCs in HIV RNA levels were not apparent after normalizing to HIV DNA or another HIV RNA, suggesting that these differences were largely driven by the lower infection frequency in PTCs. The differences in infection frequency may also contribute to some of the differences between PTCs and NCs in HIV RNA levels after ATI. However, other key findings cannot be explained by differences in infection frequency. For example, the different types of HIV transcripts showed different temporal changes in NCs and PTCs. Moreover, even at early ATI, PTCs and NCs started to show different changes in the ratios of completed and multiply-spliced to elongated HIV transcripts, which are measured from the same infected cells. Third, by late ATI, we started to see differences between PTCs and NCs in levels of some HIV transcripts per provirus, which are also independent of infection frequency.
A second major difference between PTCs and NCs is that PTCs are able to limit increases in multiply-spliced HIV Tat-Rev RNA at both early and late ATI. Multiply-spliced Tat-Rev is used to encode Tat and Rev proteins. Tat creates a positive feedback loop that increases HIV transcriptional elongation and has been deemed necessary for productive infection, whereas Rev facilitates the nuclear export of unspliced genomic HIV RNA (18). Whereas NCs had increases in multiply-spliced Tat-Rev and in the ratio of multiply-spliced/elongated HIV RNA, even at early ATI, and though these became more pronounced at late ATI, PTCs exhibited no such increases at early or late ATI, even though the unspliced and completed HIV transcripts had increased at early ATI. The ability of PTCs to limit multiply-spliced HIV RNA may be due to lower production (higher frequency of proviruses with mutations affecting splicing or Tat/Rev regions, differential expression of cellular factors that govern HIV splicing [17], fewer newly infected cells), increased destruction (immune mediated responses that rapidly kill and clear infected cells with Tat-Rev), and/or cell shifts (migration or homing of cells with Tat-Rev RNA). However, 10 of the 15 PTCs eventually had a viral rebound, at which time 9 of the 10 showed increases in Tat-Rev, indicating that the mechanisms that limit multiply-spliced HIV RNA in these individuals are not sustained or can be overcome.
A third major difference between PTCs and NCs is that PTCs are able to reduce the levels of completed HIV RNA after an initial increase at early ATI. Both PTCs and NCs showed significant increases in Pol and completed HIV RNA from pre-ATI to early ATI, and these trends seemed similar even after normalization to the total HIV DNA, although only NCs showed an increase in the ratio of completed/elongated HIV RNA at early ATI. However, between early and late ATI, Pol and completed HIV RNA tended to decrease in PTCs only, and the ratio of completed/elongated RNA decreased significantly in PTCs, whereas no such decreases were observed in NCs. The limitation of completed transcripts in PTCs could be due to mechanisms affecting RNA production and/or destruction (described above for multiply-spliced HIV RNA), as reflected in the link between high PolyA levels and downregulated mRNA decay/deadenylation pathways via transcriptomic analysis. However, the reduced activity of other global gene transcription and translation pathways suggest that the downregulation of the host transcription/translational machinery could also be a consequence of high level viral replication and of an elevated inflammatory state. In addition, the increase and subsequent decrease in completed HIV transcripts suggests a delayed but sensed response that begins after early ATI, such as an HIV-specific immune response.
PTCs may have cell intrinsic or extrinsic mechanisms that limit HIV transcriptional processivity (after an initial delay) or selectively kill the HIV-infected cells that transcribe polyadenylated HIV transcripts, which are also more likely to be exported and translated into HIV proteins. Previous studies have indicated the preservation of HIV-specific CD4+ and CD8+ cells among PTCs (19), and in addition to their cytolytic activities, CD8+ T cells have noncytolytic mechanisms that can reduce HIV transcription (20–24). Moreover, auxiliary inflammatory responses from natural killer cells in PTCs have shown increased production of IFN-γ (25).
In our transcriptomic analysis, we identified a collection of cellular transcriptional pathways that were persistently enriched in NCs but not in PTCs. These include pathways linked to inflammation, interferon responses, the HIV life cycle, and cell death. One particularly prominent example was the TNF pathway. TNF triggers a series of intracellular signaling pathways, including those mediated by NF-κB, JNK, and the p38 MAP kinase (15). In HIV-infected cells, TNF stimulates HIV transcriptional activity by potentiating the translocation of NF-κB into the nucleus as well as by enhancing the binding of the NF-κB and AP-1 complexes to their binding sites on the HIV LTR (26–28). In addition, NCs demonstrated persistent activation of the IL-1 signaling pathway. IL-1 signaling is related to pyroptosis and inflammasome activation, which can preferentially affect “bystander” CD4+ T cells that undergo abortive HIV infection (29). Moreover, NCs showed persistent activation of interferon signaling, which has been previously linked to HIV reservoir expansion and persistent immune activation (30). In contrast, PTCs showed a transient upregulation in the interferon and p53 pathways after ATI as well as a sustained downregulation in the gene translation pathways. Besides the TNF and proinflammatory pathways, we also noted the upregulation of the gene sets related to the HIV life cycle, HIV transcription, and virion assembly in NCs.
Additional studies are needed to determine how PTCs and NCs may differ in intrinsic and extrinsic immune responses as well as how these sense different HIV transcripts or antigens. If PTCs have a mechanism by which to kill cells expressing completed HIV RNA, it does not result in a detectable decrease in HIV DNA between early and late ATI, although our power to detect such a decrease was more limited, and it could be that only a minority of HIV-infected cells in PTCs transcribe completed HIV RNA after ATI. Moreover, completed HIV transcripts rose after late ATI in most PTCs who experienced virologic failure, suggesting that in these individuals, the mechanisms that limit completed HIV transcripts are not sustained or can be overcome.
It would be interesting to know how the 5 PTCs who did not experience virologic rebound differed from the 10 who eventually rebounded. We did not detect any significant differences between these two groups in HIV DNA or RNA levels or ratios at any time point, but the power to detect such differences was limited.
The proposed mechanisms of control exhibited by PTCs largely differ from those observed in HIV elite controllers. The model of elite control suggests provirus integration into transcriptionally silent regions of the host genome, which leads to a state of deep latency, as well as the killing of provirus-containing cells via host immune responses, such as CD8+ T cell responses, that are associated with favorable HLA alleles (31, 32). We propose that posttreatment control is not driven by preferential integration into noncoding host genomic regions, which should result in lower HIV transcription per provirus before ATI, but rather by cell intrinsic or extrinsic immune mechanisms after the expression of completed and spliced transcripts. Indeed, the RNA-seq data suggest that PTCs and NCs have differentially regulated pathways involving inflammation, cell death, gene translation, and mRNA decay during ATI.
There are multiple limitations of this study. Due to the rare occurrence of posttreatment control, we were only able to study 15 PTCs. The definition of PTCs varies between studies (1, 6, 33–35), and our PTCs may differ from those in other cohorts (3, 34, 36–40) and may be heterogeneous. We did not have access to tissue samples, and there was some variation in the timing of blood collection, both between and within PTC and NC groups, due in part to participant availability and also to differences in the timings of rebounds. Some PWH were unable to participate for all of the times of sample collection, and the IPDA data were not available for all participants. Whereas ratios of our HIV RNA targets can be used to measure the proportion of HIV transcripts that are incomplete (and therefore non-infectious) and nonpolyadenylated (and therefore less translation-competent), our assays cannot distinguish which proportion of each RNA (including PolyA) may have been transcribed from intact versus defective proviruses. Furthermore, there can be variation and imprecision in measuring HIV levels, due to low copy numbers or primer/probe sequence mismatches, and this error gets multiplied when expressing ratios of one HIV target to another. Combined with some undetectable levels of HIV transcripts (especially Tat-Rev) and intact proviruses, this limits the power with which to detect statistically significant differences. Finally, we were only able to perform bulk RNA-seq, and most of the CD4+ T cells were not infected with HIV. Nonetheless, we were able to find key differences between PTCs and NCs, and these results provide insights into the CD4+ T cellular milieu that fosters posttreatment control.
The findings presented here provide new observations into the mechanisms of HIV transcriptional control by PTCs, including an early limitation of multiply-spliced HIV RNA and a later, presumably immune-mediated limitation of more processive/completed HIV transcripts after ATI. More studies into these mechanisms will provide better understanding and insight into the development of therapeutic strategies that are aimed at a functional cure. These strategies will likely need to limit increases in completed and multiply-spliced HIV RNA after ATI through mechanisms that either limit HIV expression, block new infection, and/or kill infected cells.
MATERIALS AND METHODS
Study participants.
Peripheral blood mononuclear cells (PBMC) were obtained from participants enrolled in AIDS Clinical Trials Group (ACTG) studies A371 (39), A5024 (41), A5068 (42), A5170 (43), and A5178 (44) as well as in the University of California, San Francisco OPTIONS cohort (45). Samples were isolated before ATI (pre-ATI) and at early and later time points after ATI. Sample collection schedules were dictated by the original studies (39, 41–45). PTCs and NCs were identified based on published criteria (6). Samples were available from 11 NCs and 15 PTCs. For some participants, no late ATI time point was available. In 10 of the 15 PTCs, the viral loads (VL) increased beyond defined thresholds after the late ATI time point, such that an additional viremic failure (VF) time point was collected. The samples were analyzed at three time points: pre-ATI, early post-ATI, and late post-ATI. For the NCs, the early post-ATI time point was a median of 6 (Q1, Q3: 4, 8) weeks after treatment interruption, and the late time point was a median of 17 (Q1, Q3: 16, 20) weeks after treatment interruption. For the PTCs, the early and late post-ATI time points were medians of 12 (Q1, Q3: 12, 13) and 60 (Q1, Q3: 43, 96) weeks after treatment interruption, respectively. The slightly delayed timing for the early post-ATI time points in the PTCs was due to a desire to assess the PTCs after any transient early post-ATI viral rebound, which was seen in a subset of the PTCs prior to viral control and was described in the original description of the CHAMP study (6). For safety reasons, NCs were restarted on ART after a high-level viral rebound, and this generally occurred within 24 weeks of treatment interruption. All of the study participants provided written informed consent.
Nucleic acid isolation.
Cryovials containing 5 to 10 × 106 PBMC were thawed, transferred to Eppendorf DNA LoBind Microcentrifuge Tubes, and centrifuged at 1,500 rpm for 5 min to pellet the cells. The DNA and RNA were extracted using a Qiagen AllPrep DNA/RNA/miRNA Universal Kit with an on-column DNase I treatment of the RNA and the manufacturer’s modification to enhance the recovery of short transcripts. Nucleic acids were quantified using a NanoDrop One spectrophotometer (Thermo Scientific).
Reverse transcription (RT).
To quantify the total initiated (TAR) HIV transcripts, an initial polyadenylation step was performed to enhance the RT of short transcripts (16), except that the polyadenylation reaction was reduced proportionally to 15.5 μL and the subsequent RT reaction was performed in 30 μL. For the quantification of the 5′-elongated (R-U5/pre-Gag; “LongLTR”), mid-elongated (Pol; unspliced), U3-Poly(A) (“PolyA”), and multiply-spliced (Tat-Rev) transcripts, a separate RT reaction was performed (16), and the reactions were modified proportionally for 30 μL.
Droplet digital PCR.
TAR HIV transcripts were quantified in single-plex (16). The other HIV transcripts were detected using duplex combinations: LongLTR [FAM]+Pol [VIC] and PolyA [FAM]+Tat-Rev [VIC]. The duplex assay sensitivities were comparable to the single-plex sensitivities (Table S1). PCR amplification was performed on a Bio-Rad C1000 Touch Thermal Cycler, using a previously described protocol (16). Droplets were read using a QX100 or QX200 (Bio-Rad) and were analyzed using Quantasoft (Bio-Rad).
Intact proviral DNA assay (IPDA).
In a subset of participants who were enrolled in another study, the levels of total and intact HIV DNA were measured at Accelevir using the IPDA (46).
RNA sequencing (RNA-seq).
PTCs were selected for RNA-seq, primarily based on stored sample availability and on their ability to maintain viral suppression without evidence of virologic failure. PBMCs were first enriched for CD4+ T cells using an EasySep Human CD4+ T Cell Enrichment Kit (StemCell). RNA was extracted using an AllPrep DNA/RNA Kit (Qiagen) with subsequent rRNA depletion. RNA was further reverse transcribed to a cDNA library and underwent sequencing via NovaSeq (Illumina). The sequencing results were processed using the VIPER pipeline (47) for alignment, counting, and quality control. Next, we used the raw gene counts for a further differentially expressed gene (DEG) analysis using the DESeq2 package (48) and a gene set enrichment analysis (GSEA) using the fgsea package with the adaptive, multilevel, splitting Monte Carlo approach (the n for the simple fgsea in the preliminary estimation of the P values was 10,000 [49, 50]).
Statistics.
The HIV RNA levels were normalized by the input of cellular RNA (via NanoDrop) to determine the HIV RNA copies/μg RNA (approximately 106 cells). When the levels of total or intact HIV DNA were available, we calculated the ratio of each HIV RNA to the HIV DNA. In addition, the ratios of one HIV RNA to another HIV RNA were calculated to express the degree of progression through the different blocks to HIV transcription. The HIV DNA and RNA levels and ratios were compared between time points using the Wilcoxon signed-rank test, and the comparisons between PTCs and NCs were evaluated using the Mann-Whitney Test in GraphPad Prism (Version 9).
Study approval.
Deidentified samples were obtained from participants enrolled in AIDS Clinical Trials Group (ACTG) studies A371 (39), A5024 (41), A5068 (42), A5170 (43), and A5178 (44) as well as in the University of California, San Francisco OPTIONS cohort (45). These studies were approved by their local Institutional Review Boards. All participants provided written informed consent.
Data availability.
The tabulated data for the HIV RNA and DNA levels are included in an Excel file in the supplemental material (Table S3). The RNA-seq data have been submitted to the NCBI database under BioProject number PRJNA859199.
ACKNOWLEDGMENTS
We thank the study participants for their generous donations of samples, the site staff and investigators of the AIDS Clinical Trial Group, and the SCOPE project staff at Zuckerberg San Francisco General Hospital. In addition, we thank Zach Herbert and his group from the Molecular Biology Core Facilities (MBCF) at the Dana-Farber Cancer Institute (DFCI) and the Advanced Lab Technologies Core from the Harvard University Center for AIDS Research in Boston, MA for their assistance with the RNA-seq. This work was supported by the National Institute of Allergy and Infectious Diseases (R01150396 [to J.Z.L. and S.A.Y.], R01AI132128 [to S.A.Y. and J.K.W.], UM1AI068634 [to AIDS Clinical Trials Group (ACTG)], UM1AI068636 [to ACTG], UM1AI106701 [to ACTG and J.Z.L.], and 1P01AI169606 [to S.A.Y.]) as well as the National Institute of Diabetes and Digestive and Kidney Diseases (R01DK108349 [to S.A.Y.] and R01DK120387 [to S.A.Y.]). S.T. is supported by a CFAR Mentored Scientist in HIV Award (grant number P30 7AI027763, award number A120163, PI: P.V.) as well as the California HIV/AIDS Research Program (award number BB19-SF-009/A135087). Y.L was supported by an NIH T32 training grant (5T32AI007387-32).
Findings from the manuscript were previously presented at CROI 2022.
The authors have declared that no conflict of interest exists.
S.A.Y. and J.Z.L. designed the study. E.C., J.M.J., D.M.M., D.S., P.V., F.H., and S.D. provided the samples. A.W., H.A.M., Y.L., S.T., G.N.K., M.M., and B.E. designed the experiments. A.W., H.A.M., Y.L., S.T., G.N.K., M.M., and B.E. conducted the experiments. A.W., H.A.M., Y.L., S.T., M.M., B.E., J.Z.L., and S.A.Y. analyzed the data. A.W., H.A.M., Y.L., S.T., and S.A.Y. wrote the original draft. All authors reviewed and edited the manuscript. J.Z.L., S.A.Y., S.T., and J.K.W. provided the supervision and the funding. Shared author positions were determined alphabetically by first name.
Footnotes
Supplemental material is available online only.
Contributor Information
Jonathan Z. Li, Email: jli@bwh.harvard.edu.
Steven A. Yukl, Email: Steven.Yukl@ucsf.edu.
Viviana Simon, Icahn School of Medicine at Mount Sinai.
REFERENCES
- 1.Hocqueloux L, Prazuck T, Avettand-Fenoel V, Lafeuillade A, Cardon B, Viard JP, Rouzioux C. 2010. Long-term immunovirologic control following antiretroviral therapy interruption in patients treated at the time of primary HIV-1 infection. AIDS 24:1598–1601. 10.1097/qad.0b013e32833b61ba. [DOI] [PubMed] [Google Scholar]
- 2.Salgado M, Rabi SA, O'Connell KA, Buckheit RW, 3rd, Bailey JR, Chaudhry AA, Breaud AR, Marzinke MA, Clarke W, Margolick JB, Siliciano RF, Blankson JN. 2011. Prolonged control of replication-competent dual- tropic human immunodeficiency virus-1 following cessation of highly active antiretroviral therapy. Retrovirology 8:97. 10.1186/1742-4690-8-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goujard C, Girault I, Rouzioux C, Lecuroux C, Deveau C, Chaix ML, Jacomet C, Talamali A, Delfraissy JF, Venet A, Meyer L, Sinet M, Group ACPS . 2012. HIV-1 control after transient antiretroviral treatment initiated in primary infection: role of patient characteristics and effect of therapy. Antivir Ther 17:1001–1009. 10.3851/IMP2273. [DOI] [PubMed] [Google Scholar]
- 4.Fidler S, Porter K, Ewings F, Frater J, Ramjee G, Cooper D, Rees H, Fisher M, Schechter M, Kaleebu P, Tambussi G, Kinloch S, Miro JM, Kelleher A, McClure M, Kaye S, Gabriel M, Phillips R, Weber J, Babiker A, SPARTAC Trial Investigators . 2013. Short-course antiretroviral therapy in primary HIV infection. N Engl J Med 368:207–217. 10.1056/NEJMoa1110039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Saez-Cirion A, Bacchus C, Hocqueloux L, Avettand-Fenoel V, Girault I, Lecuroux C, Potard V, Versmisse P, Melard A, Prazuck T, Descours B, Guergnon J, Viard JP, Boufassa F, Lambotte O, Goujard C, Meyer L, Costagliola D, Venet A, Pancino G, Autran B, Rouzioux C, Group AVS, ANRS VISCONTI Study Group . 2013. Post-treatment HIV-1 controllers with a long-term virological remission after the interruption of early initiated antiretroviral therapy ANRS VISCONTI Study. PLoS Pathog 9:e1003211. 10.1371/journal.ppat.1003211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Namazi G, Fajnzylber JM, Aga E, Bosch RJ, Acosta EP, Sharaf R, Hartogensis W, Jacobson JM, Connick E, Volberding P, Skiest D, Margolis D, Sneller MC, Little SJ, Gianella S, Smith DM, Kuritzkes DR, Gulick RM, Mellors JW, Mehraj V, Gandhi RT, Mitsuyasu R, Schooley RT, Henry K, Tebas P, Deeks SG, Chun TW, Collier AC, Routy JP, Hecht FM, Walker BD, Li JZ. 2018. The Control of HIV after Antiretroviral Medication Pause (CHAMP) Study: posttreatment controllers identified from 14 clinical studies. J Infect Dis 218:1954–1963. 10.1093/infdis/jiy479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sharaf R, Lee GQ, Sun X, Etemad B, Aboukhater LM, Hu Z, Brumme ZL, Aga E, Bosch RJ, Wen Y, Namazi G, Gao C, Acosta EP, Gandhi RT, Jacobson JM, Skiest D, Margolis DM, Mitsuyasu R, Volberding P, Connick E, Kuritzkes DR, Lederman MM, Yu XG, Lichterfeld M, Li JZ. 2018. HIV-1 proviral landscapes distinguish posttreatment controllers from noncontrollers. J Clin Invest 128:4074–4085. 10.1172/JCI120549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Williams JP, Hurst J, Stöhr W, Robinson N, Brown H, Fisher M, Kinloch S, Cooper D, Schechter M, Tambussi G, Fidler S, Carrington M, Babiker A, Weber J, Koelsch KK, Kelleher AD, Phillips RE, Frater J, Investigators SP . 2014. HIV-1 DNA predicts disease progression and post-treatment virological control. Elife 3:e03821. 10.7554/eLife.03821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Assoumou L, Weiss L, Piketty C, Burgard M, Melard A, Girard P-M, Rouzioux C, Costagliola D, ANRS 116 SALTO study group . 2015. A low HIV-DNA level in peripheral blood mononuclear cells at antiretroviral treatment interruption predicts a higher probability of maintaining viral control. AIDS 29:2003–2007. 10.1097/QAD.0000000000000734. [DOI] [PubMed] [Google Scholar]
- 10.Maggiolo F, Di Filippo E, Comi L, Callegaro A. 2018. Post-treatment controllers after treatment interruption in chronically HIV-infected patients. AIDS 32:623–628. 10.1097/QAD.0000000000001743. [DOI] [PubMed] [Google Scholar]
- 11.Li JZ, Etemad B, Ahmed H, Aga E, Bosch RJ, Mellors JW, Kuritzkes DR, Lederman MM, Para M, Gandhi RT. 2016. The size of the expressed HIV reservoir predicts timing of viral rebound after treatment interruption. Aids 30:343–353. 10.1097/QAD.0000000000000953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sneller MC, Justement JS, Gittens KR, Petrone ME, Clarridge KE, Proschan MA, Kwan R, Shi V, Blazkova J, Refsland EW, Morris DE, Cohen KW, McElrath MJ, Xu R, Egan Michael A, Eldridge JH, Benko E, Kovacs C, Moir S, Chun T-W, Fauci AS. 2017. A randomized controlled safety/efficacy trial of therapeutic vaccination in HIV-infected individuals who initiated antiretroviral therapy early in infection. Sci Transl Med 9:eaan8848. 10.1126/scitranslmed.aan8848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giron LB, Palmer CS, Liu Q, Yin X, Papasavvas E, Sharaf R, Etemad B, Damra M, Goldman AR, Tang H-Y, Johnston R, Mounzer K, Kostman JR, Tebas P, Landay A, Montaner LJ, Jacobson JM, Li JZ, Abdel-Mohsen M. 2021. Non-invasive plasma glycomic and metabolic biomarkers of post-treatment control of HIV. Nat Commun 12:3922. 10.1038/s41467-021-24077-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, Paulovich A, Pomeroy SL, Golub TR, Lander ES, Mesirov JP. 2005. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA 102:15545–15550. 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pasquereau S, Kumar A, Herbein G. 2017. Targeting TNF and TNF receptor pathway in HIV-1 infection: from immune activation to viral reservoirs. Viruses 9:64. 10.3390/v9040064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yukl SA, Kaiser P, Kim P, Telwatte S, Joshi SK, Vu M, Lampiris H, Wong JK. 2018. HIV latency in isolated patient CD4(+) T cells may be due to blocks in HIV transcriptional elongation, completion, and splicing. Sci Transl Med 10. 10.1126/scitranslmed.aap9927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moron-Lopez S, Telwatte S, Sarabia I, Battivelli E, Montano M, Macedo AB, Aran D, Butte AJ, Jones RB, Bosque A, Verdin E, Greene WC, Wong JK, Yukl SA. 2020. Human splice factors contribute to latent HIV infection in primary cell models and blood CD4+ T cells from ART-treated individuals. PLoS Pathog 16:e1009060. 10.1371/journal.ppat.1009060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Karn J, Stoltzfus CM. 2012. Transcriptional and posttranscriptional regulation of HIV-1 gene expression. Cold Spring Harb Perspect Med 2:a006916. 10.1101/cshperspect.a006916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Samri A, Bacchus-Souffan C, Hocqueloux L, Avettand-Fenoel V, Descours B, Theodorou I, Larsen M, Saez-Cirion A, Rouzioux C, Autran B, Group A, ANRS VISCONTI study group . 2016. Polyfunctional HIV-specific T cells in post-treatment controllers. AIDS 30:2299–2302. 10.1097/QAD.0000000000001195. [DOI] [PubMed] [Google Scholar]
- 20.Walker CM, Levy JA. 1989. A diffusible lymphokine produced by CD8+ T lymphocytes suppresses HIV replication. Immunology 66:628–630. [PMC free article] [PubMed] [Google Scholar]
- 21.Walker CM, Erickson AL, Hsueh FC, Levy JA. 1991. Inhibition of human immunodeficiency virus replication in acutely infected CD4+ cells by CD8+ cells involves a noncytotoxic mechanism. J Virol 65:5921–5927. 10.1128/JVI.65.11.5921-5927.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mackewicz CE, Blackbourn DJ, Levy JA. 1995. CD8+ T cells suppress human immunodeficiency virus replication by inhibiting viral transcription. Proc Natl Acad Sci USA 92:2308–2312. 10.1073/pnas.92.6.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klatt NR, Shudo E, Ortiz AM, Engram JC, Paiardini M, Lawson B, Miller MD, Else J, Pandrea I, Estes JD, Apetrei C, Schmitz JE, Ribeiro RM, Perelson AS, Silvestri G. 2010. CD8+ lymphocytes control viral replication in SIVmac239-infected rhesus macaques without decreasing the lifespan of productively infected cells. PLoS Pathog 6:e1000747. 10.1371/journal.ppat.1000747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McBrien JB, Kumar NA, Silvestri G. 2018. Mechanisms of CD8(+) T cell-mediated suppression of HIV/SIV replication. Eur J Immunol 48:898–914. 10.1002/eji.201747172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Scott-Algara D, Didier C, Arnold V, Cummings J-S, Boufassa F, Lambotte O, Hocqueloux L, Sáez-Cirión A, Rouzioux C. 2015. Post-treatment controllers have particular NK cells with high anti-HIV capacity: VISCONTI Study, abstr CROI, Seattle, Washington. [Google Scholar]
- 26.Duh EJ, Maury WJ, Folks TM, Fauci AS, Rabson AB. 1989. Tumor necrosis factor alpha activates human immunodeficiency virus type 1 through induction of nuclear factor binding to the NF-kappa B sites in the long terminal repeat. Proc Natl Acad Sci USA 86:5974–5978. 10.1073/pnas.86.15.5974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto T, Matsuyama T, Mori S, Hamamoto Y, Kobayashi N, Yamamoto N, Josephs SF, Wong-Staal F, Shimotohno K. 1989. Augmentation of human immunodeficiency virus type 1 gene expression by tumor necrosis factor alpha. AIDS Res Hum Retroviruses 5:131–138. 10.1089/aid.1989.5.131. [DOI] [PubMed] [Google Scholar]
- 28.Osborn L, Kunkel S, Nabel GJ. 1989. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc Natl Acad Sci USA 86:2336–2340. 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Doitsh G, Galloway NL, Geng X, Yang Z, Monroe KM, Zepeda O, Hunt PW, Hatano H, Sowinski S, Munoz-Arias I, Greene WC. 2014. Cell death by pyroptosis drives CD4 T-cell depletion in HIV-1 infection. Nature 505:509–514. 10.1038/nature12940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cheng L, Ma J, Li J, Li D, Li G, Li F, Zhang Q, Yu H, Yasui F, Ye C, Tsao LC, Hu Z, Su L, Zhang L. 2017. Blocking type I interferon signaling enhances T cell recovery and reduces HIV-1 reservoirs. J Clin Invest 127:269–279. 10.1172/JCI90745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hartana CA, Yu XG. 2021. Immunological effector mechanisms in HIV-1 elite controllers. Curr Opin HIV AIDS 16:243–248. 10.1097/COH.0000000000000693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lunardi LW, Bragatte M, Vieira GF. 2021. The influence of HLA/HIV genetics on the occurrence of elite controllers and a need for therapeutics geotargeting view. Braz J Infect Dis 25:101619. 10.1016/j.bjid.2021.101619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maenza J, Tapia K, Holte S, Stekler JD, Stevens CE, Mullins JI, Collier AC. 2015. How often does treatment of primary HIV lead to post-treatment control? Antivir Ther 20:855–863. 10.3851/IMP2963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Martin GE, Gossez M, Williams JP, Stöhr W, Meyerowitz J, Leitman EM, Goulder P, Porter K, Fidler S, Frater J, the SPARTAC Trial Investigators . 2017. Post-treatment control or treated controllers? Viral remission in treated and untreated primary HIV infection. AIDS 31:477–484. 10.1097/QAD.0000000000001382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fajnzylber J, Sharaf R, Hutchinson JN, Aga E, Bosch RJ, Hartogensis W, Jacobson JM, Connick E, Volberding P, Skiest DJ, Margolis D, Sneller MC, Little SJ, Gulick RM, Mellors JW, Gandhi RT, Schooley RT, Henry K, Tebas P, Deeks S, Chun T-W, Collier AC, Hecht FM, Li JZ, team ftCs . 2021. Frequency of post treatment control varies by antiretroviral therapy restart and viral load criteria. Aids 35:2225–2227. 10.1097/QAD.0000000000002978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lampe FC, Porter K, Fau- Kaldor J, Kaldor J, Fau- Law M, Law M, Fau- Kinloch-de Loes S, Kinloch-de Loes S, Fau- Phillips AN, Phillips AN. 2007. Effect of transient antiretroviral treatment during acute HIV infection: comparison of the Quest trial results with CASCADE natural history study. [PubMed]
- 37.Kinloch– d, Loes S, Hoen B, Smith DE, Autran B, Lampe FC, Phillips AN, Goh L-E, Andersson J, Tsoukas C, Sonnerborg A, Tambussi G, Girard P-M, Bloch M, Battegay M, Carter N, El Habib R, Theofan G, Cooper DA, Perrin L, QUEST Study Group . 2005. Impact of therapeutic immunization on HIV-1 viremia after discontinuation of antiretroviral therapy initiated during acute infection. The J Infectious Diseases 192:607–617. 10.1086/432002. [DOI] [PubMed] [Google Scholar]
- 38.Rosenberg ES, Altfeld M, Poon SH, Phillips MN, Wilkes BM, Eldridge RL, Robbins GK, D'Aquila RT, Goulder PJ, Walker BD. 2000. Immune control of HIV-1 after early treatment of acute infection. Nature 407:523–526. 10.1038/35035103. [DOI] [PubMed] [Google Scholar]
- 39.Volberding P, Demeter L, Bosch RJ, Aga E, Pettinelli C, Hirsch M, Vogler M, Martinez A, Little S, Connick E, ACTG 371 Team . 2009. Antiretroviral therapy in acute and recent HIV infection: a prospective multicenter stratified trial of intentionally interrupted treatment. AIDS 23:1987–1995. 10.1097/QAD.0b013e32832eb285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kaufmann DE, Lichterfeld M, Altfeld M, Addo MM, Johnston MN, Lee PK, Wagner BS, Kalife ET, Strick D, Rosenberg ES, Walker BD. 2004. Limited durability of viral control following treated acute HIV infection. PLoS Med 1:e36. 10.1371/journal.pmed.0010036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kilby JM, Bucy RP, Mildvan D, Fischl M, Santana-Bagur J, Lennox J, Pilcher C, Zolopa A, Lawrence J, Pollard RB, Habib RE, Sahner D, Fox L, Aga E, Bosch RJ, Mitsuyasu R, Adult AIDS Clinical Trials Group A5024 Protocol Team . 2006. A randomized, partially blinded phase 2 trial of antiretroviral therapy, HIV-specific immunizations, and interleukin-2 cycles to promote efficient control of viral replication (ACTG A5024). J Infect Dis 194:1672–1676. 10.1086/509508. [DOI] [PubMed] [Google Scholar]
- 42.Jacobson JM, Pat Bucy R, Spritzler J, Saag MS, Eron JJ, Coombs RW, Wang R, Fox L, Johnson VA, Cu-Uvin S, Cohn SE, Mildvan D, O'Neill D, Janik J, Purdue L, O'Connor DK, Vita CD, Frank I, Allergy NIo, Team IDACTGP . 2006. Evidence that intermittent structured treatment interruption, but not immunization with ALVAC-HIV vCP1452, promotes host control of HIV replication: the results of AIDS clinical trials group 5068. J Infect Dis 194:623–632. 10.1086/506364. [DOI] [PubMed] [Google Scholar]
- 43.Skiest DJ, Su Z, Havlir DV, Robertson KR, Coombs RW, Cain P, Peterson T, Krambrink A, Jahed N, McMahon D, Margolis DM, Team ACTGS . 2007. Interruption of antiretroviral treatment in HIV-infected patients with preserved immune function is associated with a low rate of clinical progression: a prospective study by AIDS Clinical Trials Group 5170. J Infect Dis 195:1426–1436. 10.1086/512681. [DOI] [PubMed] [Google Scholar]
- 44.Kemmer N, Hua L, Andersen JW, Chung RT, Butt AA, Sherman KE, Team AS, ACTG 5178 Study Team . 2012. Health-related quality of life in subjects with HCV/HIV coinfection: results from ACTG 5178 study. J Viral Hepatitis 19:792–800. 10.1111/j.1365-2893.2012.01609.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hecht FM, Busch MP, Rawal B, Webb M, Rosenberg E, Swanson M, Chesney M, Anderson J, Levy J, Kahn JO. 2002. Use of laboratory tests and clinical symptoms for identification of primary HIV infection. AIDS 16:1119–1129. 10.1097/00002030-200205240-00005. [DOI] [PubMed] [Google Scholar]
- 46.Bruner KM, Wang Z, Simonetti FR, Bender AM, Kwon KJ, Sengupta S, Fray EJ, Beg SA, Antar AAR, Jenike KM, Bertagnolli LN, Capoferri AA, Kufera JT, Timmons A, Nobles C, Gregg J, Wada N, Ho YC, Zhang H, Margolick JB, Blankson JN, Deeks SG, Bushman FD, Siliciano JD, Laird GM, Siliciano RF. 2019. A quantitative approach for measuring the reservoir of latent HIV-1 proviruses. Nature 566:120–125. 10.1038/s41586-019-0898-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cornwell M, Vangala M, Taing L, Herbert Z, Koster J, Li B, Sun H, Li T, Zhang J, Qiu X, Pun M, Jeselsohn R, Brown M, Liu XS, Long HW. 2018. VIPER: visualization Pipeline for RNA-seq, a Snakemake workflow for efficient and complete RNA-seq analysis. BMC Bioinformatics 19:135. 10.1186/s12859-018-2139-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Love MI, Huber W, Anders S. 2014. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 15:550. 10.1186/s13059-014-0550-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Alexeev N, Isomurodov J, Sukhov V, Korotkevich G, Sergushichev A. 2020. Markov chain Monte Carlo for active module identification problem. BMC Bioinformatics 21:261. 10.1186/s12859-020-03572-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov M, Sergushichev A. 2021. Fast gene set enrichment analysis. bioRxiv. 10.1101/060012. [DOI]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Tables S1 and S2 and Fig. S1 to S4. Download jvi.01254-22-s0001.pdf, PDF file, 0.8 MB (813.5KB, pdf)
Table S3. Download jvi.01254-22-s0002.xlsx, XLSX file, 0.02 MB (20.8KB, xlsx)
Data Availability Statement
The tabulated data for the HIV RNA and DNA levels are included in an Excel file in the supplemental material (Table S3). The RNA-seq data have been submitted to the NCBI database under BioProject number PRJNA859199.