Abstract
To determine how binuclear giardia swim, we used video microscopy to observe trophozoites of Giardia intestinalis, which were labeled with an amino-specific Alexa Fluor dye that highlighted the flagella and adherence disc. Giardia swam forward by means of the synchronous beating of anterior, posterolateral, and ventral flagella in the plane of the ventral disc, while caudal flagella swam in a plane perpendicular to the disc. Giardia turned in the plane of the disc by means of a rudder-like motion of its tail, which was constant rather than beating. To determine how giardia divide, we used three-dimensional confocal microscopy, the same surface label, nuclear stains, and antitubulin antibodies. Giardia divided with mirror-image symmetry in the plane of the adherence disc, so that the right nucleus of the mother became the left nucleus of the daughter. Pairs of nuclei were tethered together by microtubules which surrounded nuclei and prevented mother or daughter giardia from receiving two copies of the same nucleus. New adherence discs formed upon a spiral backbone of microtubules, which had a clockwise rotation when viewed from the ventral surface. These dynamic observations of the parasite begin to reveal how giardia swim and divide.
Giardia intestinalis (also known as Giardia lamblia), which was likely first visualized by von Leeuwenhoek, is a protist that causes intestinal malabsorption and diarrhea (3, 21). Although giardia cause an unattractive disease, they are among the most beautiful organisms, as shown by scanning and transmission microscopy (4, 7, 10). Trophozoites of G. intestinalis have two similar-appearing nuclei, which are both transcriptionally active (13). The giardia nuclei are bilaterally symmetric, as are four other microtubule-associated structures: the ventral adherence disc, four pairs of flagella, the median body, and the funis (2, 7, 10). The ventral disc, by which giardia adhere to the surface of intestinal epithelial cells, is composed of α- and β-tubulin and at least three different unique cytoskeletal proteins called giardins (2, 12, 17, 18, 20). Giardia have four pairs of flagella (anterior, posterolateral, ventral, and caudal), which are composed of microtubules in a 9-plus-2 arrangement (7, 10). All four pairs of flagella originate from basal bodies, composed of microtubule triplets, which are located between the two nuclei and are dorsal to the adherence disc. The funis is a set of single microtubules, which run parallel to the caudal flagella from the disc to the tip of the tail (2). The median body, which is a bundle of microtubules bound by a unique protein called the median body protein, is perpendicular to the funis and caudal to the adherence disc (15).
Holberton (10) used phase microscopy to show that the ventral flagella of adherent giardia were constantly beating in a synchronized manner in the plane of the adherence disc. Electron micrographs of adhering giardia to mouse intestines suggested that parasites were drawing the intestinal villi up to the adherence disc. Holberton proposed that the motion of the ventral flagella creates a vacuum under the disc that sucks the intestinal epithelium to the giardia (11). It is likely that lectins on the surface of giardia also bind sugars on the surface of intestinal epithelial cells (5). Because of the limits of phase microscopy, Holberton was unable to determine the motion of the ventral flagella when giardia swim or divide and was unable to determine the motion of the anterior, posterolateral, and caudal flagella. Here we used video microscopy and amino-specific Alexa Fluor dyes, which were recently used to demonstrate the motion of bacterial flagella (23), to determine the motion of each pair of flagella of adherent and swimming giardia.
As giardia are motile and nonadherent when they divide, little is known about how the organisms replicate themselves. For example, early investigators thought that mother and daughter giardia divide along a sagittal plane, so that the mother got two identical copies of one nucleus while the daughter got two identical copies of the other nucleus (6). Others suggested that the daughter giardia slid off the back of the mother, so that the left nuclei of mother and daughter giardia were the same (when viewed from the dorsal surface) and that the right nuclei were the same (13). In contrast, our results here suggest that giardia divide with mirror-image symmetry in the plane of the adherence disc, so that the left nucleus of the mother becomes the right nucleus of the daughter. Further, we used antitubulin antibodies to identify perinuclear tethers, which bind pairs of nuclei together during cell division.
MATERIALS AND METHODS
Labeling surface of giardia.
The WB strain of G. intestinalis was grown axenically at 37°C in TYI-S-33 medium supplemented with bile salts (14). Dividing parasites, which are motile and nonadherent, were collected by removing the supernatant of unchilled cultures. Nondividing giardia, which are adherent to the plastic culture flask in logarithmic-phase cultures, were collected by adding ice-cold phosphate-buffered saline (PBS), pH 7, to the remaining parasites. For studies of giardia cysts, organisms were incubated in encystation medium for 24 h, as described earlier in reference 9. Cysts were recognized by their characteristic ovalness, the absence of flagella, and the presence of four nuclei. Giardia were washed in PBS; placed in 100 μl of PBS, pH 8.5, to maximize the number of deproteinated amino groups on their surface; and incubated for 1 h at 37°C with 0.25 mg of Alexa Fluor 488 or Alexa Fluor 584 (Molecular Probes, Eugene, Oreg.). Alexa Fluor dyes are carboxylic acid succinimidyl esters, which react with deproteinated amino groups (23). Alexa Fluor 488, which fluoresces like fluorescein, was predominantly used for confocal microscopy studies. Alexa Fluor 584, which fluoresces like Texas red, was predominantly used for video microscopy, because it tended to bleach less. After labeling, giardia were washed in PBS four times and were then returned to complete culture medium.
Video microscopy.
Surface-labeled giardia were observed with a Nikon Eclipse TE 300 inverted-fluorescence microscope, to which was attached a Hamamatsu digital video camera, a Lambda 10-2 controller (Sutter), and a MetaMorph (Universal Imaging) shutter and image analysis system. Digital recordings were made of swimming giardia using 60× or 100× objectives and the streaming mode, which captured ∼20 images per s. The zoom feature was used to focus and enlarge individual giardia, while individual frames were examined to determine the motion of each pair of flagella. Dozens of hours of observations and recordings were distilled into the video micrographs shown (see Fig. 2 and 3).
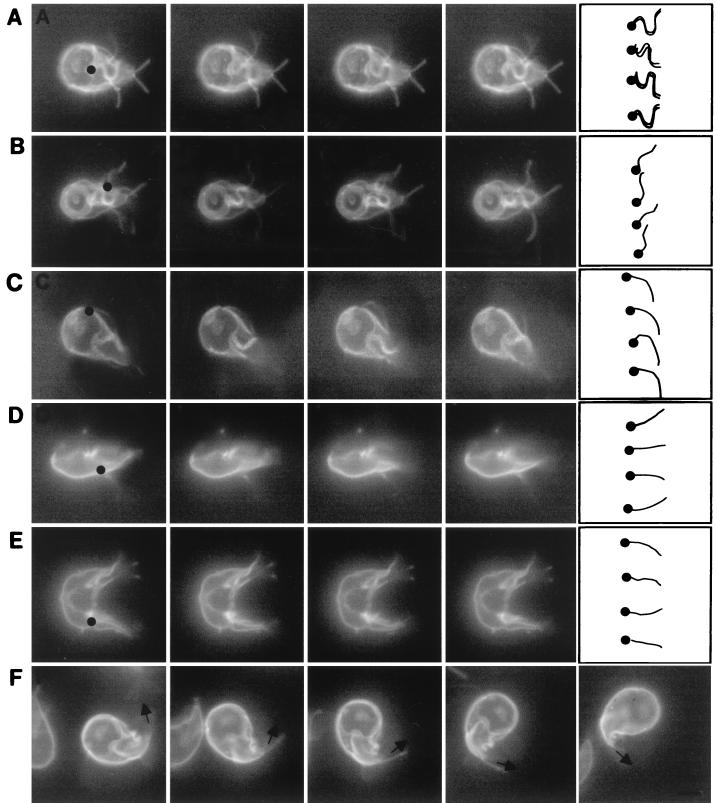
FIG. 2.
Videos of adherent and swimming giardia, each of which was labeled on its surface with Alexa Fluor 584. (A) Ventral flagella of an adherent giardia moved with a series of bends, which originated where the ventral flagella exited the cytosol (black dot) and propagated to the tip (drawing at right of row A). In contrast, the anterior, posterolateral, and caudal flagella of this adherent giardia were still. (B) The posterolateral flagellum of a second adherent giardia moved with a series of bends which originated where the posterolateral flagella exited the cytosol (black dot) and propagated to the tip (drawing at right of row B). The beat of the posterolateral flagella had the same frequency and wavelength but less amplitude than that of the ventral flagella. (C) An anterior flagellum of a swimming giardia moved with a single bend of low amplitude (drawing at right of row C), which originated where the anterior flagellum exited the adherence disc (black dot). More subtly, the beating motion of the caudal flagella in a plane perpendicular to the adherence disc caused the tails of swimming giardia to go in and out of focus. In contrast, the wavelike bending of the caudal flagella (drawings in rows D and E) was easier to see when swimming (D) or dividing (E) giardia were viewed in profile. The bending of the caudal flagella begins within the cytosol of the tail (black dots in rows D and E). (F) The tail of a turning giardia remained curved in the direction of the turn, like the rudder on a boat. Bars in the drawing at the right of row F indicate direction of the caudal flagella. Each set of videos was composed of consecutive frames shot at 20 frames per s. Bar, 5 μm.
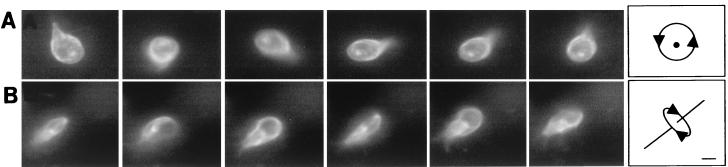
FIG. 3.
Videos of rotating giardia, each of which was labeled on its surface with Alexa Fluor 584. (A) Head-on view of a giardia rotating clockwise around its axis at ∼3 revolutions per s. (B) Side view of another giardia rotating around its long axis at ∼3 revolution per s. Each set of videos was composed of consecutive frames shot at 20 frames per s. Bar, 5 μm.
Labeling cytoskeleton and nuclei of giardia.
WB giardia, some of which were labeled on their surface with Alexa Fluor 488, were fixed in 2% formaldehyde at 4°C for 10 min, washed in PBS, and permeabilized with 0.1% Triton X-100 for 1 to 2 min. To highlight different structures in dividing giardia, parasites were stained with three different antibodies to α-tubulin. Some giardia were incubated for 1 h at 37°C with a mouse monoclonal antibody to bovine α-tubulin (Molecular Probes), which was diluted 1:100 in PBS with 2 mg of bovine serum albumin/ml. The giardia were washed four times in PBS and were then incubated for 1 h at room temperature with a rhodamine-conjugated goat anti-mouse antibody. Alternatively, some giardia were incubated with a polyclonal rabbit antibody to α-tubulin (Sigma), diluted 1:100, which was detected with a Texas red-conjugated goat anti-rabbit antibody (20). Finally, some giardia were incubated with a mouse monoclonal anti-α-tubulin antibody, diluted 1:100, which was also detected with a rhodamine-conjugated goat anti-rabbit antibody. In addition, some giardia were incubated with 1 μg of propidium iodide or Sytox green per ml to label the nuclei.
Three-dimensional confocal laser scanning microscopy.
Immunolabeled giardia were placed in mounting media, coverslipped, and examined using a Leica TCS NT confocal laser scanning microscope (Leica Inc., Exton, Pa.). A band-pass (530 + 30 nm) filter was used to select light emitted from the nucleus-specific probe (Sytox green), and a long-pass 590-nm filter was used to detect the rhodamine- and Texas red-conjugated antibodies under multiple-channel fluorescent mode. Serial sections were collected from the apical surfaces of giardia with step increments of 0.5 to 1 μm in the z axis. Three-dimensional reconstructions which simultaneously showed features on the inside and outside of giardia were created using VoxelView (Vital Images, Fairfield, Iowa). Projections were electronically repositioned to view the opposite side of these organisms.
Scanning electron microscopy.
Giardia were fixed in 2% glutaraldehyde in PBS, postfixed in 1% osmium tetroxide, dehydrated in a graded ethanol series to absolute followed by hexamethyldisilazine, and vacuum desiccated. Dry specimens were gold sputtered and imaged using an XL30 ESEM-FEG (Fei Co., Hillsborough, Oreg.).
FISH.
WB parasites transfected with a pGEM-based plasmid (p5n-Pac), which contains a bacterial puromycin-N-acetyltransferase gene under a short G. intestinalis promoter, were a generous gift from Steven Singer of Georgetown University (19). Transfected WB, which had been cloned once by limiting dilution, was recloned on soft agar, as described earlier (8), and was maintained in 100 μM puromycin. For fluorescence in situ hybridization (FISH), giardia, which were adherent to poly-Lys-coated slides by means of the ventral disc, were swollen in 70 mM KCl, fixed in 3:1 methanol:acetic acid, and denatured with 70% formamide at 70°C (16). Slide contents were incubated overnight with biotin-labeled pGEM (20 μg/ml) in 50% formamide and 2× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) at 37°C and were washed in 2× SSC and in phosphate buffer plus detergent (PDB; Oncor). Biotin-labeled probes were detected with fluorescein isothiocyanate-avidin and were amplified with antiavidin antibody and a second incubation with fluorescein isothiocyanate-avidin. Parasites were treated with RNase and counterstained with propidium iodide. Slides were examined with a Cytovision imaging system, which is designed for FISH, or a Leica confocal microscope.
RESULTS
Anatomy of giardia labeled on surface with amino-specific Alexa Fluor dyes.
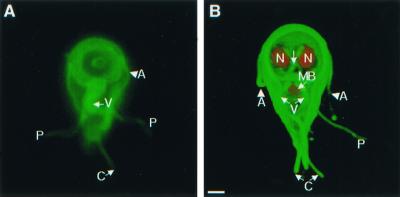
Alexa fluor dyes, which have recently been used to study the motion of bacterial flagella during running and tumbling (23), highlighted four pairs of giardia flagella with no apparent effect on viability or cell division (Fig. 1A). The relationship between surface labels and underlying cytoskeletal structures was shown by three-dimensional confocal microscopy (Fig. 1B). Anterior flagella cross the midline, exit the cytoplasm at the lateral borders of the adherence disc, and extend 8 μm. Ventral flagella, which look thicker than the other flagella, exit the cytoplasm at the posterior border of the disc and extend 12 μm. Posterolateral flagella, which are contained within the caudal cytoplasm for about half their length, exit at the side of the tail and extend 8 μm. Caudal flagella, which are for the most part contained within the cytoplasm, exit at the end of the tail and extend just 4 μm.
FIG. 1.
Important cytoskeletal structures of nondividing Giardia trophozoites demonstrated by video microscopy (A) or confocal microscopy (B). (A) Ventral view of a giardia labeled green on its surface with Alexa Fluor 488, with highlighted anterior (A), posterolateral (P), caudal (C), and ventral (V) flagella. (B) A three-dimensional confocal micrograph of the ventral surface of another giardia. Its surface was labeled green with Alexa Fluor 488, the nuclei (N) were stained red with propidium iodide, and the median body (MB) was stained red with monoclonal antibodies to bovine tubulin. Bar, 2 μm.
Anterior, posterolateral, and ventral flagella of giardia beat with a similar frequency in the plane of the adherence disc.
Giardia prefer to adhere to a surface rather than swim freely in the medium. For the most part, anterior, posterolateral, and caudal flagella of adherent giardia were still (Fig. 2A and 2B). In contrast, the ventral flagella of adherent giardia were constantly beating together in the plane of the adherence disc. The waveform of the flagellar beating was bilaterally symmetric and advanced from the caudal edge of the adherence disc to the tips of the flagella. The waveform had an amplitude of 1.7 μm, a wavelength of 5.8 μm, and a frequency of 5 to 10 beats/s, depending upon the temperature of the medium and the freshness of the parasite preparation. When giardia swam in the direction of the adherence disc, ventral flagella moved with the same bilaterally symmetric, pseudosinusoidal waveform as that of adherent giardia (Fig. 2C). Posterolateral flagella beat in the plane of the adherence disc with a frequency and wavelength similar to those of ventral flagella, but the amplitude was smaller (1.1 μm) (Fig. 2B). The anterior flagella beat in nearly the same plane and with the same frequency as ventral and posterior flagella, but the bending of the anterior flagella was asymmetric (Fig. 2C). Movies of swimming giardia are available for viewing at http://www.hsph.harvard.edu/physiology /projects/bilavi/.
Two motions (beating and turning) of tails of swimming giardia.
For the most part, the tail of adherent giardia was still, with the distal tips of the caudal flagella projecting like two short antennae (Fig. 1A and B). In contrast, the tail of swimming giardia beat in the plane perpendicular to the adherence disc with the same frequency as the beating of ventral, anterior, and posterolateral flagella. When swimming giardia were viewed from the dorsal or ventral surfaces, their tails moved in and out of the plane of focus (Fig. 2C). In contrast, when swimming or dividing giardia were seen in profile, their tails moved up and down (Fig. 2D and E, respectively). Giardia swam straight in the plane of the adherence disc (Fig. 2C) and/or rotated clockwise or counterclockwise along their long axis (Fig. 3A and B). Alternatively, giardia turned in the plane of the adherence disc by curving their tails like the rudder of a boat (Fig. 2F). When giardia turned, the caudal flagella, which were not beating, were parallel to each other. Giardia maintained their curved shape for a number of seconds, so that the parasites turned in multiple circles.
Giardia replicate with mirror-image symmetry in plane of adherence disc.
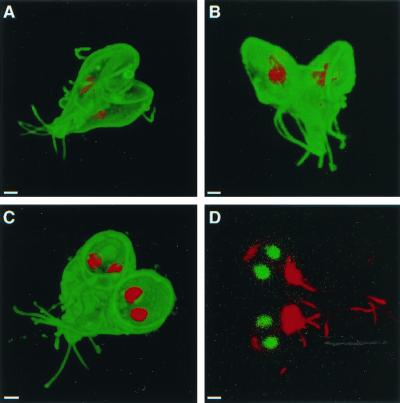
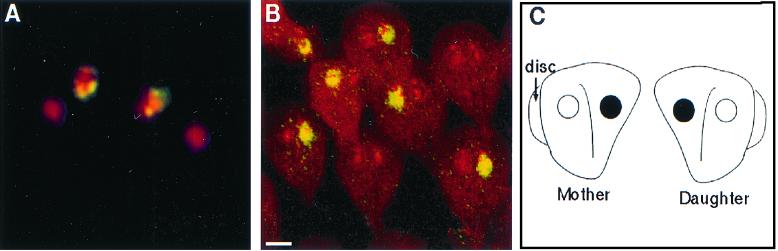
Dividing giardia had a mirror-image symmetry in the plane of the disc, so that it was impossible to distinguish the mother from the daughter. Mirror-image symmetry was present whether discs faced toward, away, or in the same plane as the others (Fig. 4A to C). Giardia also showed mirror-image symmetry when the cytoskeleton rather than the surface was visualized with antibodies to acetylated tubulin (Fig. 4D). A prediction of the mirror-image symmetry hypothesis is that the left nucleus of the mother should become the right nucleus of the daughter giardia. This was shown by introducing a foreign episome into a single nucleus of transfected giardia and localizing the episome using FISH (16, 19). A quadrinucleate, mitotic organism had episomal plasmids in the right nucleus of one pair of nuclei and the left nucleus of the other pair of nuclei (Fig. 5A). The episome-associated nucleus could be either left or right when viewed from the dorsal surface of giardia, which adhered to poly-Lys-coated slides prior to fixation (Fig. 5B). The ∼50:50 mixture of left- and right-labeled nuclei was maintained after transfected WB was recloned on soft agar (8). A summary of the FISH results is shown in Fig. 5C.
FIG. 4.
Bilateral symmetry of dividing giardia, labeled on their surface with Alexa Fluor 488 (A to C) and viewed with three-dimensional confocal microscopy. (A) Dividing giardia, which were also labeled with monoclonal anti-bovine α-tubulin antibodies to demonstrate the median body (red), were bilaterally symmetric with incompletely formed discs facing each other. Two other giardia, the nuclei of which were labeled red with propidium iodide, were bilaterally symmetric with discs facing away from each other (B) or in a plane (C). Bilateral symmetry was also present in dividing giardia labeled with Sytox green and antibodies to acetylated tubulin (red) (D). Bars, 2 μm.
FIG. 5.
FISH with a probe to a foreign plasmid in transfected giardia. (A) Fluorescence micrograph of a quadrinucleate, dividing giardia which has plasmids (yellow) on the right side of one pair of nuclei and on the left side of the other pair. (B) Confocal micrographs of transfected and cloned giardia, which show that the episomal plasmids (yellow) are present in just one nucleus, which may be left and right. Bars, 2 μm. (C) Cartoon shows that giardia divide with mirror-image symmetry, so that episomal plasmids in the right nucleus of the mother are present in the left nucleus of the daughter.
Pairs of nuclei are tethered together when giardia replicate.
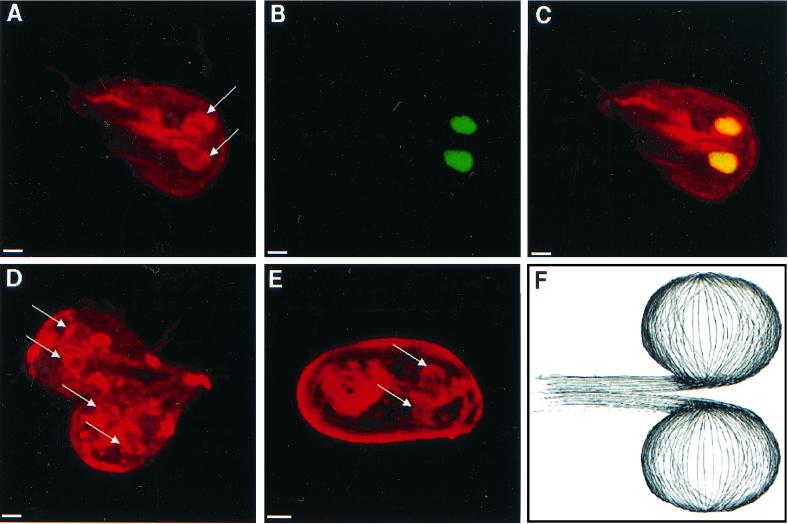
Nuclei of nondividing giardia were surrounded by perinuclear tethers, which were composed of microtubules that were highlighted by a polyclonal rabbit anti-α-tubulin antibody (Fig. 6A to C). Microtubules within the perinuclear tethers did not appear to be acetylated, because they were poorly visualized with antibodies to acetylated tubulin (Fig. 4D and 7B and C) (20). During cell division, both pairs of nuclei were surrounded by perinuclear tethers of microtubules (Fig. 6D). These perinuclear tethers also surrounded the nuclei of giardia cysts (Fig. 6E). Three-dimensional reconstructions of confocal images suggested that perinuclear tethers were developed from a set of microtubules, which ran along the central axis of giardia (Fig. 6F).
FIG. 6.
Three-dimensional confocal micrographs highlighting microtubules which form perinuclear tethers. (A to C) Micrographs of a giardia which was stained with polyclonal antibodies to α-tubulin (red in panels A and C) and Sytox green (green in panel B and yellow in panel C) show tethers of microtubules (arrows) that surround both nuclei. A dividing giardia (D) and an encysted giardia (E) stained with the same antitubulin antibodies have prominent perinuclear tethers (arrows). Note that the wall of the cyst is nonspecifically stained with the antibodies. Bars, 2 μm. (F) Cartoon of perinuclear tethers of microtubules which connect to microtubules in the central axis of the parasite.
FIG. 7.
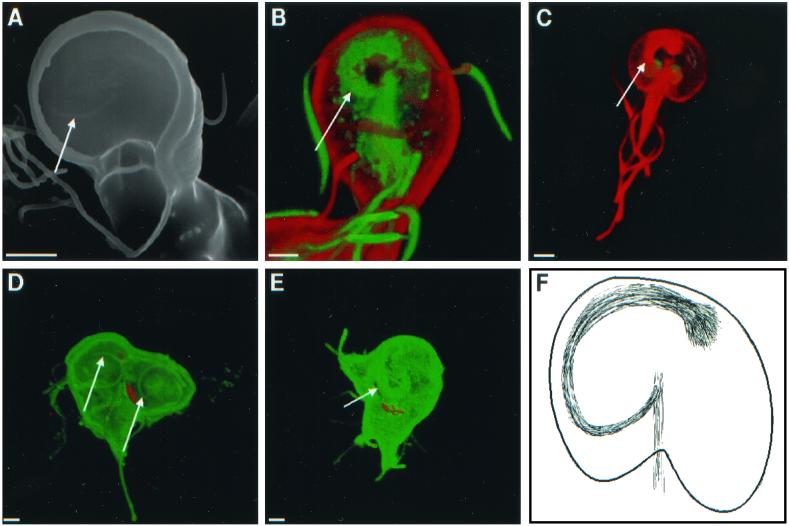
Scanning (A) and confocal (B to E) micrographs highlight spirals of microtubules (arrows), which underlie adherence discs. (B) Giardia labeled red on its surface with Alexa Fluor 584 and stained green with antibodies to acetylated tubulin. (C) Giardia stained red with antibodies to acetylated tubulin. (D and E) Dividing giardia labeled green on their surface with Alexa Fluor 488 and with median bodies labeled red with monoclonal antibodies to bovine tubulin. Bars, 2 μm. (F) Cartoon shows the spiral of microtubules, which underlies the disc and attaches to the axis of microtubules.
Spiral symmetry of new adherence discs.
The ventral adherence disc of nondividing giardia contained a clockwise spiral marking, which was visible with scanning electron microscopy (Fig. 7A). This spiral is made up in part by acetylated tubulin, as shown by confocal microscopy of giardia, which were also labeled on their surface with Alexa Fluor (Fig. 7B) or on their nuclei with Sytox green (Fig. 7C). During cell division, the adherence disc of the mother disassembled, so that dividing giardia often appeared as ovals with eight pairs of projecting flagella (not shown). New adherence discs developed from a spiral backbone of microtubules, which ran clockwise when viewed from the ventral surface of the parasites. (Fig. 7D and E). Three-dimensional reconstructions of confocal images suggested that the spirals developed from a set of microtubules which ran along the central axis of giardia (Fig. 7F). The adherence discs and flagella were all well formed before mother and daughter giardia separated from each other.
DISCUSSION
A more complete description of motion of flagella of adherent and swimming giardia.
The wavelike beating of the anterior, posterolateral, and caudal flagella was dramatically revealed when the surfaces of giardia were labeled with the Alexa Fluor dyes (23). As previously described (10), the ventral flagella had a pseudosinusoidal beat, which was symmetric in the plane of the adherence disc. Consistent with Holberton's vacuum model of giardia adherence (10, 11), fluorescent beads were sucked from the side of the parasite up under the adherence disc and were shot to its caudal end. In addition, giardia adhered to but slid along the surface of silenized slides. Three unexpected findings follow: first, all four pairs of flagella of swimming giardia appeared to beat at about the same rate, even though the shape and/or the plane of the beat was different for each. How the beating is “centrally controlled” remains to be determined. Second, the tails of giardia moved up and down in a plane perpendicular to the plane of the adherence disc. It is likely that this movement was caused by the caudal flagella beating synchronously together, because caudal flagella are present throughout the entire length of the tail and because the beating of the tail was at the same rate as the beating of the other three pairs of flagella. Third, giardia turned by using their tail as a rudder rather than by adjusting the stroke of the flagella. This is in contrast to Chlamydomonas, which turns by changing the symmetry of the beat of its two flagella, or Paramecia, which turns by changing the stroke of its many cilia, or bacteria, which turn by tumbling (1, 23). Because turning giardia maintain their curved shape for a long time, it is likely that the median body and/or funis microtubules are responsible for turning rather than the caudal flagella.
Mirror-image hypothesis for cell division by giardia.
How giardia replicate their nuclei and distribute them to mother and daughter has been debated for a long time (6, 13). The surface and cytoskeletal labels used here strongly suggest that giardia are replicating with mirror-image symmetry in the plane of the adherence disc. The mirror-image hypothesis was confirmed by FISH, which showed that an episomal plasmid was present in the right nucleus of a transfected mother giardia and the left nucleus of the daughter, while a set of cloned giardia contained a 50:50 mixture of parasites with episomes in the left and right nuclei. The previous models of cytokinesis by giardia are incorrect because they predict that both nuclei or no nuclei will contain the episomes (6) or predict that each giardia clone will have all foreign episomes in the left nucleus or the right nucleus but not in a 50:50 mixture of left and right nuclei (13).
Two new cytoskeletal structures which are likely involved in sorting nuclei of dividing giardia and building the disc.
One copy of each nucleus is appropriately distributed to mother and daughter giardia, because at least one pair of nuclei is bound at all times by the perinuclear tethers, which are composed of nonacetylated microtubules. In the absence of these tethering microtubules, 50% of the mother and daughter giardia would be homonucleate, which might dramatically reduce the genetic diversity of this asexual parasite (22). The adherence discs, which are composed of microtubules and ribbons of giardins (2, 12, 17, 18), appear to develop on a spiral backbone of acetylated microtubules. Although giardia generally divide with mirror-image symmetry in the plane of the adherence disc, the spirals always turn clockwise when discs are viewed from the ventral surface. While perinuclear tethers and spirals appear to originate from a central axis of microtubules, which joins the funis at its caudal end, the relationship of tethers and spirals to each other was difficult to determine. It is likely that future studies will reveal specific microtubule-associated proteins which assist in the assembly of tethers and spirals in the same way that dineins, giardins, and median body proteins direct microtubules in flagella, the adherence disc, and the median body, respectively (12, 15, 17, 18).
ACKNOWLEDGMENTS
This work was supported in part by NIH grants AI-33492 (to J.S.) and HL-33009 and 2HL-43510 (to R.R.). This work made use of the MRSEC Shared Experimental Facilities supported by the National Science Foundation under award number DMR98-08941.
We acknowledge the expert technical support of Jean Lai for confocal microscopy. We thank Barbara Burleigh for help with the video microscopy and constructive comments on the manuscript. We thank Jennifer E. Rogers for drawings in Fig. 6F and 7F.
REFERENCES
- 1.Bray D. Cell movements. New York, N.Y: Garland Publishing; 1993. [Google Scholar]
- 2.Crossley R, Marshall J, Clark J T, Holberton D V. Immunocytochemical differentiation of microtubules in the cytoskeleton of Giardia lamblia using monoclonal antibodies to alpha-tubulin and polyclonal antibodies to associated low molecular weight proteins. J Cell Sci. 1986;80:233–252. doi: 10.1242/jcs.80.1.233. [DOI] [PubMed] [Google Scholar]
- 3.Dobell C A. The discovery of the intestinal protozoa of man. Proc R Soc Med. 1920;13:1–15. doi: 10.1177/003591572001301601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Erlandsen S L, Macechko P T, van Keulen H, Jarroll E L. Formation of the Giardia cyst wall: studies on extracellular assembly using immunogold labeling and high resolution field emission SEM. J Eukaryot Microbiol. 1996;43:416–429. doi: 10.1111/j.1550-7408.1996.tb05053.x. [DOI] [PubMed] [Google Scholar]
- 5.Farthing M J G, Periera M E A, Keusch G T. Description and characterization of a surface lectin from Giardia lamblia. Infect Immun. 1986;51:661–667. doi: 10.1128/iai.51.2.661-667.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Filice F P. Studies on the cytology and life history of a Giardia from the laboratory rat. Univ Calif Publ Zool. 1952;57:53–143. [Google Scholar]
- 7.Friend D S. The fine structure of Giardia muris. J Cell Biol. 1966;29:317–332. doi: 10.1083/jcb.29.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gillin F D, Diamond L S. Clonal growth of Giardia lamblia trophozoites in a semisolid agarose medium. J Parasitol. 1980;66:350–352. [PubMed] [Google Scholar]
- 9.Gillin F D, Reiner D S, Boucher S E. Small-intestinal factors promote encystation of Giardia lamblia in vitro. Infect Immun. 1988;56:705–707. doi: 10.1128/iai.56.3.705-707.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Holberton D V. Mechanism of attachment of Giardia to the wall of the small intestine. Trans R Soc Trop Med Hyg. 1973;67:29–30. doi: 10.1016/0035-9203(73)90299-x. [DOI] [PubMed] [Google Scholar]
- 11.Holberton D V. Attachment of Giardia—a hydrodynamic model based on flagellar activity. J Exp Biol. 1974;60:207–221. doi: 10.1242/jeb.60.1.207. [DOI] [PubMed] [Google Scholar]
- 12.Holberton D, Baker D A, Marshall J. Segmented alpha-helical coiled-coil structure of the protein giardin from the Giardia cytoskeleton. J Mol Biol. 1988;204:789–795. doi: 10.1016/0022-2836(88)90370-1. [DOI] [PubMed] [Google Scholar]
- 13.Kabnick K S, Peattie D A. In situ analyses reveal that the two nuclei of Giardia lamblia are equivalent. J Cell Sci. 1990;95:353–360. doi: 10.1242/jcs.95.3.353. [DOI] [PubMed] [Google Scholar]
- 14.Keister D B. Axenic culture of Giardia lamblia in TYI-S-33 medium supplemented with bile. Trans R Soc Trop Med Hyg. 1983;77:487–488. doi: 10.1016/0035-9203(83)90120-7. [DOI] [PubMed] [Google Scholar]
- 15.Marshall J, Holberton D V. Sequence and structure of a new coiled coil protein from a microtubule bundle in Giardia. J Mol Biol. 1993;231:521–530. doi: 10.1006/jmbi.1993.1303. [DOI] [PubMed] [Google Scholar]
- 16.McNeil J A, Johnson C V, Carter K C, Singer R H, Lawrence J B. Localizing DNA and RNA within nuclei and chromosomes by fluorescence in situ hybridization. Genet Anal Tech Appl. 1991;8:41–58. doi: 10.1016/1050-3862(91)90049-w. [DOI] [PubMed] [Google Scholar]
- 17.Nohria A, Alonso R A, Peattie D A. Identification and characterization of gamma-giardin and the gamma-giardin gene from Giardia lamblia. Mol Biochem Parasitol. 1992;56:27–37. doi: 10.1016/0166-6851(92)90151-9. [DOI] [PubMed] [Google Scholar]
- 18.Peattie D A, Alonso R A, Hein A, Caulfield J P. Ultrastructural localization of giardins to the edges of disk microribbons of Giardia lamblia and the nucleotide and deduced protein sequence of alpha giardin. J Cell Biol. 1989;109:2323–2335. doi: 10.1083/jcb.109.5.2323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Singer S M, Yee J, Nash T E. Episomal and integrated maintenance of foreign DNA in Giardia lamblia. Mol Biochem Parasitol. 1998;92:59–69. doi: 10.1016/s0166-6851(97)00225-9. [DOI] [PubMed] [Google Scholar]
- 20.Soltys B J, Gupta R S. Immunoelectron microscopy of Giardia lamblia cytoskeleton using antibody to acetylated alpha-tubulin. J Eukaryot Microbiol. 1994;41:625–632. doi: 10.1111/j.1550-7408.1994.tb01524.x. [DOI] [PubMed] [Google Scholar]
- 21.Steiner T S, Thielman N M, Guerrant R L. Protozoal agents: what are the dangers for the public water supply? Annu Rev Med. 1997;48:329–340. doi: 10.1146/annurev.med.48.1.329. [DOI] [PubMed] [Google Scholar]
- 22.Tibayrenc M, Kjellberg F, Arnaud J, Oury B, Breniere S F, Darde M-L, Ayala F J. Are eukaryotic microorganisms clonal or sexual? A population genetics vantage. Proc Natl Acad Sci USA. 1991;88:5129–5133. doi: 10.1073/pnas.88.12.5129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Turner L, Ryu W S, Berg H C. Real-time imaging of fluorescent flagellar filaments. J Bacteriol. 2000;182:2793–2801. doi: 10.1128/jb.182.10.2793-2801.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]