Abstract
Infectious diseases in livestock industry are major problems for animal health, food safety, and the economy. Zoonotic diseases from farm animals are significant threat to human population as well. These are notifiable diseases listed by the World Organization for Animal Health (OIE). Rapid diagnostic methods can help keep infectious diseases under control in herds. Loop-mediated isothermal amplification (LAMP) is a simple and rapid nucleic acid amplification method that is studied widely for detection of many infectious diseases in the field. LAMP allows biosensing of target DNA or RNA under isothermal conditions with high specificity in a short period of time. An untrained user can analyze results based on color change or turbidity. Here we review LAMP assays to diagnose OIE notifiable ruminant viral diseases in literature highlighting properties of LAMP method considering what is expected from an efficient, field usable diagnostic test.
Keywords: Isothermal nucleic acid amplification, Pen-side testing, Veterinary diagnostics, Infectious diseases
Introduction
Increase in global demand for animal products forces the livestock industry to be highly productive. To increase productivity, animals are bred in crowded lots causing infectious diseases to spread fast in herds with a potential to start an epidemic. Economic losses due to infections can be devastating for the farmer. In addition, some infectious diseases are zoonotic and pose a public health threat [1]. If unchecked, infectious diseases can travel long distances through animal trade. One of the major components of disease control is surveillance. Thus, farm animals need to be monitored closely and tested routinely to control spreading of contagious diseases [2]. The gold standard method for nucleic acid detection in disease diagnosis is quantitative PCR (qPCR). qPCR can detect minute quantities of nucleic acid. However, qPCR tests need to be conducted in a laboratory with a thermal cycler by trained personnel. These requirements make qPCR hard to adopt to field conditions. Serology testing is practical compared to nucleic acid detection, but often it is hard to differentiate acute and chronic infections. Antibody detection tests cannot give positive results until after the immune system is activated which might take couple days in some infectious diseases. During that period infection can spread to the entire herd. On the other hand, long lasting antibodies that remain in circulation even after the infection is cleared, can give false positive test results. Thus, user-friendly, reliable, rapid, pen-side testing is needed in the field to control and minimize the effects of infectious diseases.
World Health Organization (WHO) identified the following properties for an ideal infectious disease diagnostic test that is applicable in field conditions by non-trained users: Affordable, Sensitive, Specific, User-friendly, Rapid, Equipment-free, and Deliverable to the end user, ASSURED criteria in short [3]. These criteria can be considered for pen-side animal disease diagnostics for the livestock industry. Loop-mediated isothermal amplification (LAMP) is a rapid, user friendly isothermal nucleic acid amplification method that has almost all ASSURED properties as discussed below [4, 5]. Thus, LAMP can be considered as a point-of-care/pen-side diagnostic testing method.
LAMP Assay Principle
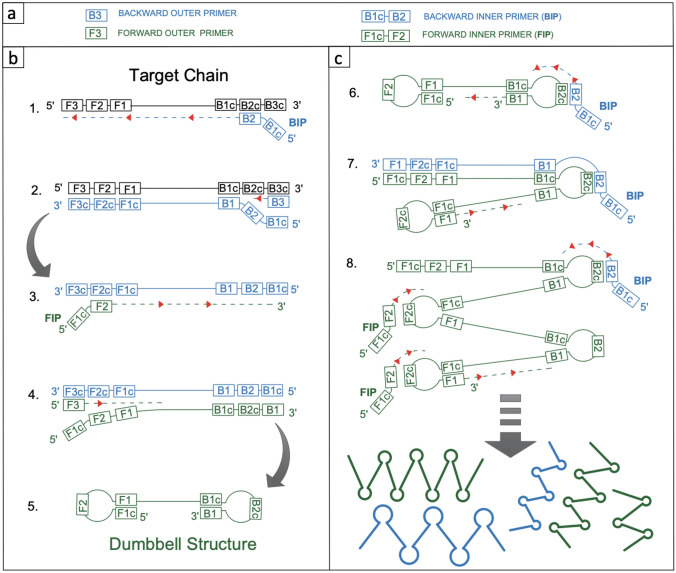
LAMP is a multistep isothermal nucleic acid amplification method that requires 4–6 primers recognizing 6–8 unique sites on the target nucleic acid which makes the assay highly specific for its target. LAMP reaction primers are inner primers (FIP and BIP), outer primers (F3 and B3), and optional loop primers (FL and BL) (Fig. 1a). LAMP reaction can be divided into two parts. First part is formation of the dumbbell structure (Fig. 1b), and the seconds part is exponential amplification (Fig. 1c). Each part of LAMP is composed of multiple steps. For dumbbell structure formation, first backward inner primer (BIP) hybridizes to its complementary sequence on the target DNA and strand displacing DNA polymerase synthesizes the complementary strand. In the second step, outer primer, B3 hybridizes to its complement, B3c at the 3’ end of target DNA. DNA polymerase displaces the strand synthesized in the first step as it synthesizes the new strand. In the third step, forward inner primer (FIP) binds to its complementary region on the strand produced in the first step and DNA polymerase completes DNA synthesis starting from F2 site. Then in the fourth step, forward outer primer (F3) hybridizes to its complementary sequence and DNA polymerase elongates the strand, displacing the strand synthesized in the previous step. The displaced strand in the fourth step has complementary regions (F1-F1c and B1-B1c) at each end. And when these complementary regions hybridize, dumbbell structure is formed (Fig. 1b). Dumbbell-shaped DNA contains many different primer-binding sites, including regions for loop primers. Dumbbell structure serves as a template for further amplification. Exponential amplification of this self-priming template through strand displacement activity of DNA polymerase results in formation of different length DNA products which are called concatemers (Fig. 1c).
Fig. 1.
Principle of LAMP reaction (a) LAMP primers: two inner primers (FIP and BIP), two outer primers (F3 and B3), and two optional loop primers (FL and BL). There are two parts in LAMP reaction: formation of dumbbell structure (b) and exponential nucleic acid amplification (c). See text for a detailed explanation
All these steps can be carried out between 60–70 °C at constant temperature thanks to the strand displacing DNA polymerase which makes thermal cycling unnecessary. Single temperature incubation is possible with simple instruments such as a dry heat block, an incubator, or a water bath. This makes the assay affordable and easier to perform in the field. LAMP reaction is highly specific to its target due to four to six primers (inner primers (FIP, BIP), outer primers (F3, B3), and optional loop primers) recognizing six to eight unique sites on the target nucleic acid (Fig. 1a) [6]. At the end of the reaction, LAMP generates more products than PCR in a shorter time. The presence of LAMP products can be observed with the naked eye. A by-product of nucleic acid synthesis reaction, magnesium pyrophosphate (Mg2P2O7) forms a precipitate making the solution opaque. Increased turbidity due to precipitated Mg2P2O7 can be observed by inspection without any instruments. Colorimetric detection is possible with dyes such as hydroxynaphthol blue (HNB) or fluorescent dyes such as SYBR Green I [7]. It is also possible to identify RNA targets by adding reverse transcriptase into LAMP reaction mixture (RT-LAMP) just like RT-qPCR. In addition to easy set-up and running condition, LAMP reaction is less sensitive to inhibitors than PCR such as residual guanidine from nucleic acid extraction step or complex media from biological samples such as urine or serum [8, 9]. LAMP can be performed with dried reaction components or dried field samples such as dried blood. The results are comparable to reactions conducted with fresh reagents [10]. Drying reaction mixture removes the burden of transferring tests to remote areas in cold chain which is one reason why nucleic acid testing is hard to adopt to field conditions [10, 11]. Multiplexing is complex but possible with LAMP [12]. These properties all point to the fact that LAMP has most of the ASSURED criteria.
In this review article, we aim to highlight how LAMP method can be widely applicable in veterinary disease diagnosis and discuss developed assays for early detection of World Organization for Animal Health (OIE) notifiable ruminant viral diseases (included as a list in Table 1) in animal husbandry.
Table 1.
Published LAMP assay conditions for ruminant viral infections
| Virus | Target sequence in genome | Reaction conditions | Reference |
|---|---|---|---|
| BoHV-1 | virion glycoprotein C (gC) gene | 63℃, 1 h | [18] |
| virion glycoprotein C (gC) gene | 65℃, 1 h | [20] | |
|
virion glycoprotein D (gD) and E (gE) genes |
66℃, 1 h | [21] | |
| CaPV | poly(A) polymerase small subunit | 60℃, 1 h | [28] |
| p32 gene | 65℃, 25 min | [29] | |
| p32 gene | 65℃, 1 h | [24] | |
| inverted terminal repeat (ITR) | 62℃, 45–60 min | [30] | |
| DNA-polymerase gene (DPO) | 63℃, 1 h | [31] | |
| BTV | VP2 gene | 63℃, 1 h | [35] |
| NS1 gene | 62℃, 1 h | [36] | |
| Genome segment 1 | 65–68℃, 30–60 min | [37] | |
| BRV | VP6 gene | 63℃, 1 h | [42] |
| BLV | long terminal repeats (LTR) | 63℃, 1 h | [50] |
| BVDV | 5'UTR | 63℃, 1 h | [58] |
| 5'UTR | 63℃, 1 h | [59] | |
| 5'UTR | 60℃, 40 min | [60] | |
| CAEV |
p25 gene, gag region p25 gene, gag region |
63℃, 1 h 63℃, 30 min |
[63] [67] |
| FMDV | 3D RNA-polymerase gene | 65℃, 22 min | [74] |
| 3D RNA-polymerase gene | 63℃, 1 h | [75] | |
| NS2B gene | 64℃, 45 min | [76] | |
| 3D RNA-polymerase gene | 63℃, 1 h | [77] | |
| 3D RNA-polymerase gene | 62℃, 15 min | [79] | |
| Akabane virus | virion nucleocapsid (N) gene | 63℃, 1 h | [90] |
| PPRV | virion matrix (M) gene | 63℃, 1 h | [96] |
| virion nucleocapsid (N) gene | 58℃, 30 min | [97] | |
| virion nucleocapsid (N) gene | 65℃, 20 min | [98] | |
| virion nucleocapsid (N) gene | 65℃, 20 min | [99] | |
| RVFV | virion large (L) segment | 63℃, 30 min | [104] |
| virion large (L) segment | 61℃, 1 h | [105] | |
| virion S segment | 63℃, 1 h | [106] |
Ruminant Viral Diseases and LAMP Assays for Diagnosis
DNA Viruses
Bovine Herpesvirus-1 (BoHV-1)
Bovine herpesvirus-1 (BoHV-1) is a double-stranded DNA virus that belongs to Varicellovirus genus of the Herpesviridae family [13]. It mainly affects cattle, but interspecies transmission is possible to sheep and goats. Main route of transmission is through nose-to-nose contact with nasal secretions, respiratory droplets, and semen of infected animals [14] BoHV-1 is the causative agent of acute respiratory disease, infectious bovine rhinotracheitis (IBR) and correlated with abortions as well [15]. Since it is responsible for major losses of livestock, several European countries eradicated the disease while others run eradication campaigns against BoHV-1 and especially gE (glycoprotein E)-deleted marker vaccines are being used for surveillance of the disease [16, 17].
Pawar and colleagues developed the first BoHV-1 LAMP test using a set of six primers to target conserved glycoprotein-C (gC) gene of the genome. LAMP assay was run for 60 min at 63℃ and the results were visualized using SYBR Green I, HNB dye and confirmed with agarose gel electrophoresis. The LOD of the assay was determined as 10 fg DNA/reaction showing LAMP assay was 100-fold more sensitive than conventional PCR. Furthermore, LAMP assay was able to detect 0.2 TCID50 virus particles/reaction showing equal sensitivity as real-time PCR in bovine semen samples. In clinical samples, BoHV-1 LAMP assay had 97% sensitivity and 100% specificity which was again indicated that results were in agreement with real-time PCR [18]. Furthermore, same group developed another LAMP assay to differentiate the wild-type BoHV-1 strain from gE-deleted marker vaccine strain [19]. Another BoHV-1 LAMP test was designed targeting again the glycoprotein-C gene with a set of six primers. The optimum reaction conditions were set as 65℃ and 60 min. Results were analyzed with agarose gel electrophoresis and with naked eye following the color change after the addition of SYBR Green I dye. BoHV-1 LAMP assay showed no cross-reactivity with other tested viruses while having an LOD equal to 1 fg DNA/reaction which was comparable with conventional PCR. When tested in clinical samples, developed test was successful to detect the virus in all nasal swab samples [20]. Socha and colleagues established two BoHV-1 LAMP assays with two sets of six primers; one targeting glycoprotein-D and the other targeting glycoprotein-E gene to differentiate infected and vaccinated animals. Optimum reaction conditions were 66℃ and 60 min; and results were analyzed visually with SYBR Green dye I and confirmed with agarose gel electrophoresis. However, BoHV-1 LAMP glycoprotein-D and glycoprotein-E assays have an LOD equal to 2 × 104 copies and 2 × 105 copies of viral genome, respectively. LOD of these differentiating LAMP assays were high compared to previously developed BoHV-1 LAMP assays [18, 20] and conventional PCR, indicating lower sensitivity. In clinical nasal swab samples, differentiating LAMP assays have lower diagnostic sensitivity and specificity. Neither assay developed by Socha and colleagues showed any cross-reactivity with other alpha herpesviruses, and LAMP glycoprotein-E successfully differentiated gE-deleted vaccine strains from the wild-type strains [21]. LAMP assays’ target region in the viral genome and assay conditions are summarized in Table 1.
Capripoxviruses (CaPV): Sheeppox Virus (SPPV) and Goatpox Virus (GTPV)
Capripoxvirus genus is a member of Poxviridae family. Capripoxviruses are double-stranded DNA viruses and there are three species in this genus: sheeppox virus (SPPV), goatpox virus (GTPV), and lumpy skin disease virus (LSDV). [22]. Cattle are susceptible to LSDV while goatpox and sheeppox affect goats and sheep, respectively. The infection can be easily transmitted via aerosol or direct contact, manifests as widespread skin eruption, non-wool skins, and even internal lesions in lungs and gastrointestinal system which leads to death [23]. Capripoxvirus infection causes great economic losses due to reduced milk production, damaged wool and high rates of abortion [23, 24]. Despite the availability of live-attenuated vaccines, recent outbreaks continue to be seen in many African, Middle Eastern, and European countries. Sheeppox and goatpox viruses are still listed as notifiable diseases by World Organization of Animal Health (OIE) [25–27].
Das and colleagues developed the first LAMP assay to detect capripoxviruses (CaPV) by targeting the conserved poly(A) polymerase small subunit (VP39). LAMP products were generated at 60℃ in 1 h in the presence of six primers including loop primers. Results were monitored by color change using hydroxynaphthol blue (HNB) dye and confirmed with agarose-gel electrophoresis. Developed test has similar limit of detection (6.3 TCID50/ml) with gold-standard real-time qPCR. Analyzed clinical samples showed that there is 90–95% agreement in diagnostic specificity and 89–100% agreement in diagnostic sensitivity between two tests [28]. Similarly, a following study designed a LAMP assay for capripoxviruses based on conserved envelope immunogenic protein, P32 gene using a set of six primers. Rapid amplification was observed at 65℃. Results were analyzed using DNA intercalating Picogreen dye. In addition, LAMP primers were labeled with biotin and fluorescein. At the end of LAMP reaction, dual-labeled products detected by a lateral flow device enabling simple and rapid point-of-care amplification. The CaPV LAMP test showed no cross-reactivity, being able to detect all CaPV strains but none of the other mammalian poxviruses, indicating specificity. Limit of detection of the developed assay was measured as 163 DNA copies/μl, indicating high sensitivity which is equal to gold-standard real-time qPCR [29]. Batra and colleagues also focused on P32 gene to design a rapid and accurate LAMP assay for CaPV identification. They used a set of six primers. Optimized assay results obtained at 65℃, in 1 h in agarose gel electrophoresis, while monitoring color change with Picogreen dye gave results in 30 min. Their limit of detection of this assay has proven to be 100 times more sensitive than gold-standard qPCR technique [24]. Sheeppox and goatpox viruses are specific to ruminants and they are clinically and serologically very similar. Zhao and colleagues designed first LAMP assay to differentiate these two viruses by targeting inverted terminal repeat (ITR) region of the viral genome. They used a combination of 3 set of primers in a single assay; specific to goatpox virus (GTPV), specific to sheeppox virus (SPPV) and universal primers (GSPV) that can detect both viruses. Each set contains four primers, amplification occurred at 62℃, in 45–60 min and results were analyzed in agarose-gel electrophoresis. All sets were highly specific, showing no cross-reactivity with tested viruses. Universal GSPV primer set had the lowest limit of detection (103 copies), being the most sensitive group; SPPV primers were more sensitive (104 copies) than GTPV primers (106 copies). In clinical samples, all primers showed 98.8–100% detection rates, consistent with laboratory results. Combining all three sets of primers in a single assay enabled the quick and efficient detection of GTPV and SPPV [30]. A recent study by Venkatesan and colleagues targeted the conserved DNA polymerase gene (DPO) of CaPV genome, using a set of four primers. The assay took 60 min at 63℃, results were analyzed with agarose gel electrophoresis and color change of SYBR Green I and HNB dyes. This assay’s LOD was 2 TCID50/ml and 10 copies of DNA template/assay indicating that it was more sensitive than P32-based [28, 29] and ITR-based CaPV LAMP assays [30]. This DPO-based CaPV LAMP assay is comparable to gold-standard qPCR with 96.6% sensitivity and 100% specificity in tested clinical samples [31].
RNA Viruses
Bluetongue Virus (BTV)
Bluetongue virus (BTV) genome is composed of ten double-stranded RNA fragments. BTV is from orbivirus genus within the Reoviridae family and classified into 29 confirmed serotypes worldwide [32]. BTV can infect ruminants, cattle, deer, and camelids. Depending on the infection causing serotype the symptoms can vary widely from subclinical infection to fetus malformation in pregnant animals to mortality. Clinical signs of infection are more pronounced in sheep such as severe facial edema, hemorrhages in the nose, lips, and tongue. Bluetongue name is derived from sometimes occurring cyanosis of the infected animal’s tongue. Transmission of BTV occurs mainly through insects, but direct contact and mother to fetus (vertical) transmission was reported recently. Live-attenuated vaccines contribute to control BT outbreaks, but strict surveillance is still needed especially during outbreaks [33, 34]. There are live-attenuated and inactivated vaccines against BTV with various drawbacks, thus alternative BTV vaccine development continues [32].
Mulholland et. al. developed first RT-LAMP assay specific for serotype 8 of BTV targeting VP2 region of the viral genome. Serotype 8 caused a major epidemic in Europe in 2006. RT-LAMP reaction was conducted at 63℃ and amplification was detected with three different methods: fluorescent dye, UV light, and agarose gel electrophoresis. Specificity of the assay was confirmed with all BTV strains and four strains of closely related epizootic hemorrhagic disease virus (EHDV). System was able to detect more than 5 × 102 copies of BTV-8 RNA specifically after 60 min at 63℃. This is less sensitive than pan-serotype RT-PCR assay that can detect a single copy of BTV-8 RNA [35]. Another study targeting highly conserved NS1 gene of BTV was able to detect seven serotypes with RT-LAMP assay. RT-LAMP reaction was conducted at 62℃ for one hour. The assay results were visualized colorimetrically and by agarose gel electrophoresis. Specificity of the assay was assessed by testing foot and mouth disease, peste-des-petits-ruminants, goatpox, sheeppox, and orf viruses along with seven serotypes of BTV. Sensitivity of the assay was reported to be 0.3 TCID50/reaction which was very similar to RT-PCR test [36]. A 2016 publication by Maan et. al. differentially identified genome segment-1 of eastern and western topotype of BTV strains in circulation in India with RT-LAMP assay. RT-LAMP reaction was conducted at 65–68℃ for 30 min for eastern and 60 min for western topotypes. Results were analyzed colorimetrically and by agarose gel electrophoresis. LOD of RT-LAMP assay for eastern topotype was 10 copies in 30 min which was 1000 times more sensitive than RT-PCR. Whereas LOD of RT-LAMP assay for western topotype was 5000 copies in one hour which was 10 times more sensitive than RT-PCR. Specificity of the assay was confirmed with viruses causing similar clinical symptoms to BTV infection: foot-and-mouth-disease, peste des petits ruminants, capripox, and Orf viruses [37].
Bovine Rotavirus (BRV)
Bovine rotavirus (BRV) is a double-stranded RNA virus that belongs to Rotavirus genus of Reoviridae family [38, 39]. Rotavirus A is one of the 7 serotypes and it is a zoonotic virus causing acute diarrheal disease in livestock and humans [40] Rotavirus transmission occurs through fecal–oral route [41].
The first BRV-based RT-LAMP test was designed targeting the conserved VP6 gene of rotavirus group A. A set of six primers used at 63℃ with 60 min of incubation time and test results were determined with naked-eye monitoring turbidity and color change with SYBR Green I dye. The test detected only BRV among other tested bovine pathogens proving its specificity. BRV-based RT-LAMP test had an LOD equal to 3 copies/tube which was equal to LOD of gold-standard RT-PCR technique. Furthermore, in tested clinical samples novel RT-LAMP test showed 100% agreement with RT-PCR which indicated RT-LAMP assay was highly reliable [42].
Bovine Leukemia Virus (BLV)
Bovine leukemia virus (BLV) is a positive-sense RNA virus that belongs to the Deltaretrovirus genus of the Retroviridae family [43]. It causes enzootic bovine leukemia (EBL) mainly in dairy and beef cattle [44]. Recently, BLV was found to be in circulation in sheep in Colombia [45]. Infection affects B cells in cattle, impairs the immune system, reduces milk yield, shortens lifespan, and causes tumor formation in approximately 1–5% of animals [46, 47]. BLV infection spread to the herd during standard farm practices via infected blood or bodily fluids and vertical transmission is possible to the fetus as well [48]. BLV infection is asymptomatic in 70% of animals, thus its infection rate is quite high which causes great economic losses in livestock industry. The OIE declared BLV as a serious disease affecting animal trade, thus eradication of the infection has become a priority of the European Union. Even though research on vaccine development continues, there is no approved vaccine for BLV yet [49].
Komiyama and colleagues targeted the LTR region of BLV genome using six primers to develop the first BLV LAMP test. The LAMP reaction was incubated at 63 °C for 1 h and the results were confirmed by turbidity and agarose gel electrophoresis. The LAMP assay was able to detect 2 copies of BLV DNA and it was found to be more sensitive compared to conventional PCR and real-time PCR. In the study conducted with field samples, results of the LAMP assay was in agreement with serology test results [50].
Bovine Viral Diarrhea Virus (BVDV)
Bovine viral diarrhea virus (BVDV) is a single stranded, positive-sense RNA virus, and a member of Pestivirus genus of Flaviviridae family [51]. The two strains (BVDV-1 and BVDV-2) mainly infect cattle. Other member of Pestivirus genus border disease virus (BDV) mainly infects ruminants; but it is known that BVDV and BDV are not host-specific viruses. BVDV is able to infect especially sheep and goats that are in close-contact with cattle [52, 53]. Main transmission route of the virus is through bodily fluids and close-contact [54]. BVDV has various clinical manifestation such as enteric and respiratory disease, reduced milk production and high mortality due to immunosuppressive effects. Depending on the time of gestation, fetal infections can result with abortions or birth of persistently infected (PI) calves which secretes the virus throughout their lives and continuously transmit BVDV to the herd [55]. BVDV also reduces the reproductivity of animals causing major economic losses [56]. There are live attenuated, inactivated, and recombinant vaccines available against BVDV. However, it is still a major threat against livestock industry worldwide and diagnosis and isolation of PI lambs are crucial for eradication of the disease [56, 57].
The first BVDV RT-LAMP test was designed to target most conserved region of the genome, 5’UTR with a set of four primers. Optimum reaction conditions were 63℃, 60 min. Test results were analyzed by turbidity and color change of SYBR Green I, and confirmed with agarose gel electrophoresis. The LOD of the BVDV RT-LAMP test was determined as 4.7 × 100 RNA copies/reaction which was a 1000-fold better than RT-PCR, indicating high sensitivity. Also, developed test showed no cross-reactivity with other tested viruses and was able to detect both strains of BVDV. In field samples, BVDV RT-LAMP test results were in agreement with real-time RT-PCR results [58]. Aebischer and colleagues modified the previously designed primer set [58] and added an LF primer to target same 5’UTR region of BVDV with an RT-LAMP test. Optimum reaction conditions were 63℃, 60 min and results were obtained in real-time monitoring ResoLight dye signal. The LOD of the novel RT-LAMP test was 5 × 103 RNA copies/reaction which was low compared to gold-standard RT-qPCR. However, BVDV RT-LAMP test showed high specificity by demonstrating no cross-reactivity with other tested pestiviruses. In clinical samples, BVDV RT-LAMP test was able to detect all BVDV-1 and BVDV-2a infected samples, indicating specificity for these strains only [59]. A recent study targeted the 5’UTR region of the BVDV genome with a primer set called “P25” containing three novel primers (BIP, B3, and BL) and three primers (FIP, F3, and FL) from Aebischer et al. study. Additionally, P25 primer set contained total eight primers, 2 BIP and 2 BL species-specific primers to detect both BVDV-1 and BVDV-2 strains. Reaction conditions were 60℃, 40 min. P25 primers were able to detect both strains with higher sensitivity compared to primers of previous study by Aebischer et al. However, authors reported that P25 primer set weakly cross-reacts with pestivirus D and H. In clinical samples, BVDV RT-LAMP test had comparable results with RT-qPCR. An additional species-specific primer set (P26) designed to differentiate BVDV-1 from BVDV-2 as well [60].
Caprine Arthritis-Encephalitis Virus (CAEV)
Caprine arthritis-encephalitis virus (CAEV) is a single-stranded positive-sense RNA virus that belongs to the genus Lentivirus, Retroviridae family [61, 62]. The virus targets host immune system and causes multi-organ progressive inflammatory diseases such as chronic polyarthritis, cachexia, leukoencephalomyelitis, chronic pneumonia, mastitis, and progressive weight loss in goats [63, 64]. Main route of disease transmission occurs vertically through ingestion of colostrum or milk, but horizontal transmission via aerosol and bodily fluids of infected animals is also possible [65]. Infected animals become life-long carriers of the virus and shed virus throughout their lives [66]. There are no vaccine or approved treatment for CAEV infection. Therefore, CAEV poses a great risk against dairy goat industry with a potential to create significant economic losses [67, 68].
First CAEV RT-LAMP assay was developed targeting highly conserved p25 gene in gag region using a set of six primers. The assay was developed with CAEV DNA and gave positive result within 1 h at 63℃ in a real-time turbidimeter. Furthermore, when whole blood extract was tested with RT-LAMP assay result was positive without separation of PBMCs and nucleic acid extraction processes. The overall test time was reduced from 150 to 80 min. This test was also confirmed to work in field samples [63]. Another RT-LAMP assay was designed targeting the gag region of CAEV genome sequence obtained in Philippines and it was able to give result in 30 min at 63℃. Gag region targeting RT-LAMP assay was two times faster than previously developed test [63]. Assay results were visualized with SYBR Green dye in agarose gel electrophoresis. This assay showed no cross-reactivity with porcine epidemic diarrhea (PED) and foot-mouth disease (FMD), proving its specificity. CAEV RT-LAMP showed comparable sensitivity in field samples against a conventional nested PCR method [67].
Foot and Mouth Disease Virus (FMDV)
Foot and mouth disease virus (FMDV) is a single-stranded positive-sense RNA virus that belongs to Aphthovirus genus of the Picornaviridae family and it has 7 serotypes (A, C, O, Asia 1, South African Territories 1, 2, and 3) [69, 70]. FMDV can affect all members of Artiodacytla; specifically cattle, swine, sheep, goats, and many wildlife species [71]. FMDV is the causative agent of the foot and mouth disease (FMD) which is transmitted through close-contact and aerosol. It is correlated with weight lost and reduced milk production in animals [72]. It is a highly contagious virus that is responsible for many outbreaks throughout the history all around the world. The cost of vaccines and production losses were estimated to be between 5.3–17 billion euros per year to countries in FMDV-endemic areas (in Asia, Sub-Saharan Africa, and South America) [71]. Traditional FMD vaccines have certain drawbacks, especially related to high heterogeneity of the virus, hence, efforts to develop alternative vaccines continue [73].
The first RT-LAMP assay was developed targeting 3D RNA-polymerase gene of FMDV with a set of six primers. Amplification was done at 65℃ and results were interpreted from color change and real-time measurements with Picogreen dye. RT-LAMP test was proven to have high specificity since it showed no cross-reactivity with viruses that showed similar clinical symptoms. Its LOD was equal to 10 RNA copies detected in 22 min while real-time PCR detected same number of copies in 55 min. In field samples, FMDV RT-LAMP successfully identified 82.3% of all 7 FMDV serotypes [74]. Similarly, another study targeted 3D RNA polymerase gene of FMDV using a set of six primers. Developed RT-LAMP assay was run at 63℃ for an hour. Results were analyzed with naked eye using SYBR Green I and confirmed with agarose gel electrophoresis. The LOD of the assay was determined as 10 DNA copies which was equal to real-time PCR but tenfold higher than RT-PCR. Novel test results were in agreement with RT-PCR and real-time PCR when tested in field samples and 98% of O, A, Asia-1, and C serotypes were detected successfully [75]. In another study, Chen and colleagues designed an RT-LAMP test targeting FMDV 2B region with a set of four primers. Amplification conditions were 64℃, 45 min and results were analyzed with agarose gel electrophoresis. The RT-LAMP assay in question has an LOD equal to 10 RNA copies per reaction which was tenfold higher than RT-PCR. In field samples, FMDV 2B RT-LAMP identified O, A, Asia-1, and C serotypes while showing no cross-reactivity with other tested viruses [76]. Yamazaki and colleagues established a multiplex RT-LAMP test to cover all 7 serotypes of FMDV. For this purpose, they designed 4 novel primer sets targeting 3D RNA polymerase gene of the FMDV and combined those with 2 previously designed primer sets targeting the same region [74, 75]. Reaction conditions were 63℃, 60 min and results were analyzed with real-time turbidimeter. The combination of primer set 82 and a previous set [75] has highest diagnostic sensitivity (98%) and specificity (98.1%) in tested field samples [77]. Lim and colleagues designed a probe-based real-time RT-LAMP assay targeting 3D RNA polymerase gene of FMDV. They modified their earlier primer design [78] and also designed assimilating probes from scratch. Reaction temperature was 62℃ and results were detected within 15 min by measuring real-time fluorescence intensities of FAM-labeled probes. The LOD of the assay was 102 RNA copies/µl which was equal to RT-qPCR sensitivity. Developed test identified all 7 FMDV serotypes while having no cross-reactivity with other tested viruses. In diagnostic samples, RT-LAMP assay had 97.1% agreement with gold-standard RT-qPCR as well [79]. There were other FMDV RT-LAMP assays designed to identify serotypes circulating in Korea [78], India [80], and Middle East [81]. Furthermore, RT-LAMP assays for differentiating serotypes A [82], Asia-1 [83], C [84], and O [85] were reported in literature.
Akabane Virus
Akabane virus is a single-stranded negative-sense RNA virus that belongs to the Orthobunyavirus genus of the Bunyaviridae family. It is the causative agent of Akabane disease which affects cattle and ruminants [86]. Transmission occurs horizontally through blood-sucking insect vectors; and vertically from mother to child [87]. Akabane disease causes abortions, stillbirths, and birth anomalies in the herd and most common symptoms are hydranencephaly and arthrogryposis [88]. A live-attenuated vaccine and inactive vaccines have been applied in Japan and Korea but antigenic differences in vaccine strains create the need for new vaccine development [89].
An RT-LAMP test against Akabane virus was developed targeting the conserved nucleoprotein (N) gene of the virus, using a set of four primers. Optimum conditions were set as 63℃ and 60 min. Test results were visualized with color change using SYBR Green I and confirmed with agarose gel electrophoresis. Developed RT-LAMP test was successful at differentiating the Akabane virus from the other viruses with similar clinical symptoms such as BVDV, Bluetongue virus, Bovine parvovirus, and Bovine herpesvirus-1. Its LOD is 5.0 TCID50 /mL and when it was used to test clinical samples, it showed 97% agreement with well-established semi-nested RT-PCR [90].
Peste des Petits Ruminants Virus (PPRV)
The causative agent of peste de petits ruminants (PPR) disease, peste de petits ruminants virus (PPRV) belongs to Morbillivirus genus of Paramyxoviridae family [91]. It is a single-stranded negative-sense RNA virus [92]. PPRV is highly contagious virus that can be transmitted through close contact. Symptoms associated with PPRV are high fever, nasal and ocular discharge, severe diarrhea and pneumonia in ruminants [93]. PPR disease is responsible for major economic losses and thus, the World Organization of Animal Health (OIE) and the Food and Agriculture Organization (FAO) runs a campaign to globally eradicate PPRV until 2030 [94]. Although live-attenuated vaccines are available against PPRV, they have certain drawbacks and outbreaks continue to be seen in Africa, Asia, and Europe [95].
First PPRV RT-LAMP assay was developed using a set of six primers and targeting matrix protein, M-protein gene of the virus. Reaction was carried out at 63℃, for 1 h. Results were observed by color change using FDR as fluorescent detection agent and confirmed with agarose-gel electrophoresis. The LOD of the test was 1.41 × 10–4 ng total RNA per assay, which was tenfold more sensitive compared to RT-PCR. PPRV RT-LAMP test was highly specific that it showed no cross-reactivity with other tested viruses. Furthermore, it was able to detect 8 strains and results were in agreement with gold-standard RT-PCR in clinical samples [96]. Another RT-LAMP test was developed against PPRV, targeting nucleocapsid N-protein gene of the virus with a set of six primers. Optimum amplification temperature was 58℃ and developed test detected positivity around 30 min. Results were observed in fluorimeter, confirmed with agarose-gel electrophoresis; and addition of SYBR Green dye enabled naked-eye detection by color change. RT-LAMP test has a 10–4 TCID50/ml LOD which indicates tenfold higher sensitivity than gold-standard RT-PCR. Furthermore, analytical sensitivity and specificity of novel RT-LAMP test showed perfect agreement with RT-PCR [97]. Mahapatra and colleagues developed PPRV RT-LAMP assay targeting conserved N-protein gene region of the virus using a set of six primers. Template RNA was detected in 20 min at 65℃ and results were monitored using a fluorimeter. PPRV RT-LAMP test has an LOD of 102 RNA copies which is tenfold lower than PPRV RT-qPCR. On the other hand, both N gene-based RT-LAMP assays had equivalent diagnostic sensitivity. RT-LAMP assay developed by Ashraf and colleagues could only detect two lineages (II and IV) [97], whereas RT-LAMP assay developed by Mahapatra and coworkers was able to detect all four lineages of the virus while showing no cross-reactivity, indicating higher sensitivity[98]. A recent RT-LAMP assay against PPRV, also targeted N-protein gene using a set of six primers. Two primer sets were able to amplify all four-lineages of PPRV without showing any cross-reactivity with other tested viruses. Amplification of the target RNA was observed in less than 20 min at 65℃ in a fluorimeter. Developed assay has an LOD around 0.3–0.8 log10 TCID50/ml. In clinical samples collected from recent PPRV outbreaks and experimentally infected goats, PPRV RT-LAMP test showed 97% diagnostic sensitivity and 100% specificity compared to gold-standard technique RT-PCR [99]. Summary of assay conditions and target region of the viral genome are listed in Table 1.
Rift Valley Fever Virus (RVFV)
Rift valley fever virus (RVFV) is a single-stranded negative-sense RNA virus which is a member of Phlebovirus genus of Bunyaviridae family [100]. RVFV has an economic impact in animal husbandry since it is associated with abortions and high mortality rates of newborns of ruminants. It is a zoonotic virus that can be transmitted through infected mosquito bites to domestic livestock and close contact with sick animals cause infections in humans [101]. Infected humans generally have mild flu-like symptoms but in a small percentage of the patients, it might lead to severe disease [102]. Despite the availability of live and inactivated vaccines for ruminants, there is an urgent need for a human vaccine to prevent spread of RVFV from Africa to non-endemic areas [103].
Peyrefitte and colleagues developed first RVFV RT-LAMP assay by targeting large (L) segment of the virus which encoded the polymerase with a set of six primers. The assay was optimized to give results in less than 30 min at 63℃. Results were obtained in real-time with fluorescence reading using ethidium bromide, also with turbidity and confirmed with agarose gel electrophoresis. Developed assay was able to detect 7 strains containing 4 subtypes of RVFV while it showed no cross-reactivity with other phleboviruses, flaviviruses, and alphavirus, indicating high specificity. RVFV RT-LAMP test has an LOD of 10 RNA copies per assay, the same sensitivity as the gold-standard RT-qPCR [104]. Another study also targeted RNA-dependent RNA polymerase gene in the L segment of RVFV genome with a set of six primers. Optimum amplification was reached at 61℃ in 60 min and results were monitored in real-time turbidimeter and agarose gel electrophoresis. Moreover, fluorescent detection reagent FDR was added to LAMP reactions for naked-eye visualization. This RVFV RT-LAMP assay had high specificity since it was able to detect all RVFV strains collected between 1944 and 2007. Developed test had an LOD of 10 RNA copies per reaction, equal analytical sensitivity with RT-qPCR. Furthermore, RVFV RT-LAMP and RT-qPCR showed same diagnostic sensitivity in clinical samples from humans and animals [105]. Another, recent study established RVFV RT-LAMP assay with a set of six primers targeting S segment of ZH501 strain which encoded nucleocapsid and non-structural proteins. Optimum amplification was achieved at 63℃, around 60 min. Different from previous studies a vertical flow visualization strip was designed to amplify results with sandwich strategy, labeling 5’ end of LF and LB primers with FITC and biotin, respectively. The LOD of the test was found as 1.94 × 100 copies/µl which proved that it was 100 times more sensitive than RT-qPCR. Developed test also showed no cross-reactivity with other viruses causing similar symptoms proving assay’s high specificity [106].
Conclusion
This review discusses loop-mediated isothermal amplification (LAMP) method-based assays developed to detect eleven OIE notifiable, highly contagious viral disease agents infecting ruminants in the livestock industry. Due to crowded living conditions in industrial animal farms, animals must be closely monitored for infections. Otherwise, infection can spread to the entire herd rapidly and may cause an epidemic. There is an urgent need for rapid, affordable testing method to make disease monitoring easier in the field. LAMP is highly specific, rapid, affordable nucleic acid detection method, and comparable with qPCR in performance. As a result, there is increasing trend in developing LAMP-based diagnostic assays for these infectious diseases.
Despite these advantages, currently there are no commercial animal disease diagnostic test working with LAMP method in the market yet [107]. There are commercial LAMP tests available for food industry and for some human pathogens such as malaria, tuberculosis, and SARS-CoV-2. These LAMP tests are recommended to be used in laboratory setting because most often sample preparation requires nucleic acid isolation before LAMP reaction. There is no efficient, field applicable nucleic acid isolation method in the market yet and it is the bottleneck for field usable isothermal nucleic acid assays. Coronavirus pandemic accelerated sample preparation, nucleic acid isolation methods, however, improvement is still needed especially when separating nucleic acid material from tissue, blood, or serum [108]. Visual detection with color change can be improved with these other dyes in the literature [109–116].
Due to its practicality and high tolerance to presence of inhibitors in sample solution increased use of LAMP-based diagnostics systems in laboratories would pave the way for field usable LAMP tests. As the nucleic acid isolation methods improve, field usable LAMP systems are expected to increase in number.
Acknowledgements
Authors wish to thank the Scientific and Technological Research Council of Turkey (TÜBİTAK) for funding (2210597) for S.A.K. and H.T.S. We would also like to thank Dr. Ahmet Sait from Pendik Veterinary Control Institute, Istanbul, Turkiye for critically reading the manuscript and for helpful discussion.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.McElwain TF, Thumbi SM. Animal pathogens and their impact on animal health, the economy, food security, food safety and public health. Revue scientifique et technique (International Office of Epizootics) 2017;36(2):423–433. doi: 10.20506/RST.36.2.2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holm A, Hill R, Farsang A, Jungbäck C. Diagnostics in the veterinary field: The role in health surveillance and disease identification. Biologicals. 2019;61:80–84. doi: 10.1016/j.biologicals.2019.07.002. [DOI] [PubMed] [Google Scholar]
- 3.Mabey D, Peeling RW, Ustianowski A, Perkins MD. Diagnostics for the developing world. Nature Reviews Microbiology. 2004;2(3):231–240. doi: 10.1038/nrmicro841. [DOI] [PubMed] [Google Scholar]
- 4.Everitt ML, Tillery A, David MG, Singh N, Borison A, White IM. A critical review of point-of-care diagnostic technologies to combat viral pandemics. Analytica Chimica Acta. 2021;1146:184–199. doi: 10.1016/j.aca.2020.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K, Amino N, Hase T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Research. 2000;28(12):e63–e63. doi: 10.1093/NAR/28.12.E63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tomita N, Mori Y, Kanda H, Notomi T. Loop-mediated isothermal amplification (LAMP) of gene sequences and simple visual detection of products. Nature Protocols. 2008;3(5):877–882. doi: 10.1038/nprot.2008.57. [DOI] [PubMed] [Google Scholar]
- 7.Tanner NA, Zhang Y, Evans TC. Visual detection of isothermal nucleic acid amplification using pH-sensitive dyes. BioTechniques. 2015;58(2):59–68. doi: 10.2144/000114253/. [DOI] [PubMed] [Google Scholar]
- 8.Francois P, Tangomo M, Hibbs J, Bonetti EJ, Boehme CC, Notomi T, Schrenzel J. Robustness of a loop-mediated isothermal amplification reaction for diagnostic applications. FEMS Immunology and Medical Microbiology. 2011;62(1):41–48. doi: 10.1111/j.1574-695X.2011.00785.x. [DOI] [PubMed] [Google Scholar]
- 9.Kaneko H, Kawana T, Fukushima E, Suzutani T. Tolerance of loop-mediated isothermal amplification to a culture medium and biological substances. Journal of Biochemical and Biophysical Methods. 2007;70(3):499–501. doi: 10.1016/j.jbbm.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 10.Hayashida K, Kajino K, Hachaambwa L, Namangala B, Sugimoto C. Direct blood dry LAMP: A rapid, stable, and easy diagnostic tool for human African Trypanosomiasis. PLOS Neglected Tropical Diseases. 2015;9(3):e0003578. doi: 10.1371/JOURNAL.PNTD.0003578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yaren O, Alto BW, Bradley KM, Moussatche P, Glushakova L, Benner SA. Multiplexed isothermal amplification based diagnostic platform to detect Zika, Chikungunya, and Dengue 1. Journal of Visualized Experiments. 2018 doi: 10.3791/57051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yaren O, Alto BW, Gangodkar PV, Ranade SR, Patil KN, Bradley KM, Benner SA. Point of sampling detection of Zika virus within a multiplexed kit capable of detecting dengue and chikungunya. BMC Infectious Diseases. 2017;17(1):1–13. doi: 10.1186/s12879-017-2382-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tikoo SK, Campos M, Babiuk LA. Bovine herpesvirus 1 (BHV-1): Biology, pathogenesis, and control. Advances in Virus Research. 1995;45:191–223. doi: 10.1016/S0065-3527(08)60061-5. [DOI] [PubMed] [Google Scholar]
- 14.Biswas S, Bandyopadhyay S, Dimri U, Patra H. Bovine herpesvirus-1 (BHV-1) - a re-emerging concern in livestock: A revisit to its biology, epidemiology, diagnosis, and prophylaxis. The Veterinary Quarterly. 2013;33(2):68–81. doi: 10.1080/01652176.2013.799301. [DOI] [PubMed] [Google Scholar]
- 15.Hostnik P, Černe D, Mrkun J, Starič J, Toplak I. Review of infections with bovine herpesvirus 1 in Slovenia. Frontiers in Veterinary Science. 2021;8:728. doi: 10.3389/FVETS.2021.676549/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Raaperi K, Orro T, Viltrop A. Epidemiology and control of bovine herpesvirus 1 infection in Europe. Veterinary Journal. 2014;201(3):249–256. doi: 10.1016/J.TVJL.2014.05.040. [DOI] [PubMed] [Google Scholar]
- 17.Sayers RG. Associations between exposure to bovine herpesvirus 1 (BoHV-1) and milk production, reproductive performance, and mortality in Irish dairy herds. Journal of Dairy Science. 2017;100(2):1340–1352. doi: 10.3168/JDS.2016-11113. [DOI] [PubMed] [Google Scholar]
- 18.Pawar SS, Meshram CD, Singh NK, Sonwane AA, Saini M, Rautmare SS, Gupta PK. Rapid detection of bovine herpesvirus 1 in bovine semen by loop-mediated isothermal amplification (LAMP) assay. Archives of Virology. 2014;159(4):641–648. doi: 10.1007/S00705-013-1869-2. [DOI] [PubMed] [Google Scholar]
- 19.Pawar SS, Meshram CD, Singh NK, Saini M, Mishra BP, Gupta PK. Loop-mediated isothermal amplification for rapid detection and differentiation of wild-type bovine herpesvirus-1 and glycoprotein E-deleted marker vaccine strain. Animal Biotechnology. 2015;26(4):268–272. doi: 10.1080/10495398.2015.1015680. [DOI] [PubMed] [Google Scholar]
- 20.El-Kholy AA, Abdelrahman K, Soliman H. Rapid detection of BoHV-1 genomic DNA by loop-mediated isothermal amplification assay. Journal of Virological Methods. 2014;204:81–85. doi: 10.1016/J.JVIROMET.2014.04.011. [DOI] [PubMed] [Google Scholar]
- 21.Socha W, Rola J, Urban-Chmiel R, Zmudziński JF. Application of Loop-Mediated Isothermal Amplification (LAMP) assays for the detection of bovine herpesvirus 1. Polish Journal of Veterinary Sciences. 2017;20(3):619–622. doi: 10.1515/pjvs-2017-0078. [DOI] [PubMed] [Google Scholar]
- 22.Babiuk S, Bowden TR, Boyle DB, Wallace DB, Kitching RP. Capripoxviruses: An emerging worldwide threat to sheep, goats and cattle. Transboundary and Emerging Diseases. 2008;55(7):263–272. doi: 10.1111/J.1865-1682.2008.01043.X. [DOI] [PubMed] [Google Scholar]
- 23.TakeleTesgera H, Zhizhong J, Huaijie J, Guohua C, Xiao-Bing H. A review on Sheeppox and Goatpox: Insight of epidemiology, diagnosis, treatment and control measures in Ethiopia. Journal of Infectious Diseases and Epidemiology. 2018 doi: 10.23937/2474-3658/1510057. [DOI] [Google Scholar]
- 24.Batra K, Kumar A, Kumar V, Nanda T, Maan N, Maan S. Development and evaluation of loop-mediated isothermal amplification assay for rapid detection of Capripoxvirus. Veterinary World. 2015;8(11):1286–1292. doi: 10.14202/VETWORLD.2015.1286-1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bhanuprakash V, Indrani BK, Hegde R, Kumar MM, Moorthy ARS. A classical live attenuated vaccine for sheep pox. Tropical Animal Health and Production. 2004;36(4):307–320. doi: 10.1023/B:TROP.0000026661.88631.50. [DOI] [PubMed] [Google Scholar]
- 26.(OIE), O. I. des E. (World H. O. for animals). (2017). Sheep pox and Goat Pox. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (p. Chapter 2.7.13.). Office International des Epizootes (OIE). 10.1145/3132847.3132886
- 27.Tuppurainen ESM, Venter EH, Shisler JL, Gari G, Mekonnen GA, Juleff N, Babiuk LA. Review: Capripoxvirus diseases: Current status and opportunities for control. Transboundary and Emerging Diseases. 2017;64(3):729–745. doi: 10.1111/TBED.12444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Das A, Babiuk S, McIntosh M. Development of a loop-mediated isothermal amplification assay for rapid detection of capripoxviruses. Journal of Clinical Microbiology. 2012;50(5):1613–1620. doi: 10.1128/JCM.06796-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Murray L, Edwards L, Tuppurainen E, Bachanek-Bankowska K, Oura C, Mioulet V, King D. Detection of capripoxvirus DNA using a novel loop-mediated isothermal amplification assay. BMC Veterinary Research. 2013 doi: 10.1186/1746-6148-9-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao Z, Fan B, Wu G, Yan X, Li Y, Zhou X, Zhang Q. Development of loop-mediated isothermal amplification assay for specific and rapid detection of differential goat pox virus and sheep pox virus. BMC Microbiology. 2014 doi: 10.1186/1471-2180-14-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Venkatesan G, Balamurugan V, Bhanuprakash V, Singh RK, Pandey AB. Loop-mediated isothermal amplification assay for rapid and sensitive detection of sheep pox and goat pox viruses in clinical samples. Molecular and Cellular Probes. 2016;30(3):174–177. doi: 10.1016/j.mcp.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 32.Jiménez-Cabello L, Utrilla-Trigo S, Calvo-Pinilla E, Moreno S, Nogales A, Ortego J, Marín-López A. Viral vector vaccines against bluetongue virus. Microorganisms. 2020;9(1):1–21. doi: 10.3390/MICROORGANISMS9010042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Van Der Sluijs MTW, De Smit AJ, Moormann RJM. Vector independent transmission of the vector-borne bluetongue virus. Critical Reviews in Microbiology. 2016;42(1):57–64. doi: 10.3109/1040841X.2013.879850. [DOI] [PubMed] [Google Scholar]
- 34.Rojas JM, Rodríguez-Martín D, Martín V, Sevilla N. Diagnosing bluetongue virus in domestic ruminants: current perspectives. Veterinary Medicine (Auckland, NZ) 2019;10:17–27. doi: 10.2147/VMRR.S163804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mulholland C, Hoffmann B, McMenamy MJ, Korthase C, Earley B, Markey B, Welsh MD. The development of an accelerated reverse-transcription loop mediated isothermal amplification for the serotype specific detection of bluetongue virus 8 in clinical samples. Journal of Virological Methods. 2014;202:95–100. doi: 10.1016/j.jviromet.2014.03.004. [DOI] [PubMed] [Google Scholar]
- 36.Mohandas SS, Muthuchelvan D, Pandey AB, Biswas SK, Chand K, Venkatesan G, Mondal B. Development of reverse transcription loop mediated isothermal amplification assay for rapid detection of bluetongue viruses. Journal of Virological Methods. 2015;222:103–105. doi: 10.1016/j.jviromet.2015.06.005. [DOI] [PubMed] [Google Scholar]
- 37.Maan S, Maan NS, Batra K, Kumar A, Gupta A, Rao PP, Mertens PPC. Reverse transcription loop-mediated isothermal amplification assays for rapid identification of eastern and western strains of bluetongue virus in India. Journal of Virological Methods. 2016;234:65–74. doi: 10.1016/j.jviromet.2016.04.002. [DOI] [PubMed] [Google Scholar]
- 38.Maldonado YA, Yolken RH. Rotavirus. Bailliere’s Clinical Gastroenterology. 1990;4(3):609–625. doi: 10.1016/0950-3528(90)90052-I. [DOI] [PubMed] [Google Scholar]
- 39.Estes MK, Palmer EL, Obijeski JF. Rotaviruses: A review. Current Topics in Microbiology and Immunology. 1983;105:123–184. doi: 10.1007/978-3-642-69159-1_3/COVER. [DOI] [PubMed] [Google Scholar]
- 40.Luchs A, Timenetsky M. Group A rotavirus gastroenteritis: Post-vaccine era, genotypes and zoonotic transmission. Einstein. 2016;14(2):278. doi: 10.1590/S1679-45082016RB3582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dennehy PH. Transmission of rotavirus and other enteric pathogens in the home. The Pediatric Infectious Disease Journal. 2000 doi: 10.1097/00006454-200010001-00003. [DOI] [PubMed] [Google Scholar]
- 42.Xie Z, Fan Q, Liu J, Pang Y, Deng X, Xie Z, Khan MI. Reverse transcription loop-mediated isothermal amplification assay for rapid detection of Bovine Rotavirus. BMC Veterinary Research. 2012 doi: 10.1186/1746-6148-8-133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kyei GB, Powderly WG. Retroviruses and retroviral infections. Infectious Diseases. 2017;1:1483–1492. doi: 10.1016/B978-0-7020-6285-8.00174-X. [DOI] [Google Scholar]
- 44.Juliarena MA, Barrios CN, Lützelschwab CM, Esteban EN, Gutiérrez SE. Bovine leukemia virus: Current perspectives. Virus Adaptation and Treatment. 2017;9:13–26. doi: 10.2147/VAAT.S113947. [DOI] [Google Scholar]
- 45.Olaya-Galán NN, Corredor-Figueroa AP, Velandia-Álvarez S, Vargas-Bermudez DS, Fonseca-Ahumada N, Nuñez K, Gutiérrez MF. Evidence of bovine leukemia virus circulating in sheep and buffaloes in Colombia: Insights into multispecies infection. Archives of Virology. 2022;167(3):807–817. doi: 10.1007/s00705-021-05285-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kuczewski A, Orsel K, Barkema HW, Mason S, Erskine R, van der Meer F. Invited review: Bovine leukemia virus-transmission, control, and eradication. Journal of Dairy Science. 2021;104(6):6358–6375. doi: 10.3168/JDS.2020-18925. [DOI] [PubMed] [Google Scholar]
- 47.Marawan MA, Alouffi A, El Tokhy S, Badawy S, Shirani I, Dawood A, Selim A. Bovine leukaemia virus: Current epidemiological circumstance and future prospective. Viruses. 2021;2021(13):2167. doi: 10.3390/V13112167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hopkins SG, DiGiacomo RF. Natural transmission of bovine leukemia virus in dairy and beef cattle. The Veterinary Clinics of North America: Food Animal Practice. 1997;13(1):107–128. doi: 10.1016/S0749-0720(15)30367-4. [DOI] [PubMed] [Google Scholar]
- 49.Bartlett PC, Ruggiero VJ, Hutchinson HC, Droscha CJ, Norby B, Sporer KRB, Taxis TM. Current developments in the epidemiology and control of enzootic bovine leukosis as caused by bovine leukemia virus. Pathogens. 2020;9(12):1–13. doi: 10.3390/PATHOGENS9121058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Komiyama C, Suzuki K, Miura Y, Sentsui H. Development of loop-mediated isothermal amplification method for diagnosis of bovine leukemia virus infection. Journal of Virological Methods. 2009;157(2):175–179. doi: 10.1016/J.JVIROMET.2008.12.015. [DOI] [PubMed] [Google Scholar]
- 51.Nettleton PF, Entrican G. Ruminant pestiviruses. The British Veterinary Journal. 1995;151(6):615–642. doi: 10.1016/S0007-1935(95)80145-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kalaycioglu AT. Bovine viral diarrhoea virus (BVDV) diversity and vaccination. A review. The Veterinary Quarterly. 2007;29(2):60–67. doi: 10.1080/01652176.2007.9695228. [DOI] [PubMed] [Google Scholar]
- 53.Ridpath JF. BVDV genotypes and biotypes: Practical implications for diagnosis and control. Biologicals: Journal of the International Association of Biological Standardization. 2003;31(2):127–131. doi: 10.1016/S1045-1056(03)00028-9. [DOI] [PubMed] [Google Scholar]
- 54.Qi L, Beaunée G, Arnoux S, Dutta BL, Joly A, Vergu E, Ezanno P. Neighbourhood contacts and trade movements drive the regional spread of bovine viral diarrhoea virus (BVDV) Veterinary Research. 2019;50(1):1–15. doi: 10.1186/S13567-019-0647-X/FIGURES/8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.(OIE), O. I. des E. (World H. O. for animals) Bovine Viral Diarrhoea. Manual of Diagnostic Tests and Vaccines for Terrestrial Animals, Chapter. 2018;3(4):7. [Google Scholar]
- 56.Pinior B, Firth CL, Richter V, Lebl K, Trauffler M, Dzieciol M, Käsbohrer A. A systematic review of financial and economic assessments of bovine viral diarrhea virus (BVDV) prevention and mitigation activities worldwide. Preventive Veterinary Medicine. 2017;137:77–92. doi: 10.1016/J.PREVETMED.2016.12.014. [DOI] [PubMed] [Google Scholar]
- 57.Al-Kubati AAG, Hussen J, Kandeel M, Al-Mubarak AIA, Hemida MG. Recent advances on the bovine viral diarrhea virus molecular pathogenesis, immune response, and vaccines development. Frontiers in Veterinary Science. 2021;8:475. doi: 10.3389/FVETS.2021.665128/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Fan Q, Xie Z, Xie L, Liu J, Pang Y, Deng X, Wang X. A reverse transcription loop-mediated isothermal amplification method for rapid detection of bovine viral diarrhea virus. Journal of Virological Methods. 2012;186(1–2):43–48. doi: 10.1016/J.JVIROMET.2012.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Aebischer A, Wernike K, Hoffmann B, Beer M. Rapid genome detection of Schmallenberg virus and bovine viral diarrhea virus by use of isothermal amplification methods and high-speed real-time reverse transcriptase PCR. Journal of Clinical Microbiology. 2014;52(6):1883. doi: 10.1128/JCM.00167-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mungthong K, Khaing ST, Otsubo T, Hatanaka C, Yoneyama S, Hisamatsu S, Tsukamoto K. Broad detection and quick differentiation of bovine viral diarrhea viruses 1and 2 by a reverse transcription loop-mediated isothermal amplificationtest. The Journal of Veterinary Medical Science. 2021;83(8):1321. doi: 10.1292/JVMS.20-0742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cork LC, Hadlow WJ, Crawford TB, Gorham JR, Piper RC. Infectious leukoencephalomyelitis of young goats. Journal of Infectious Diseases. 1974;129(2):134–141. doi: 10.1093/infdis/129.2.134. [DOI] [PubMed] [Google Scholar]
- 62.Robinson WF, Ellis TM. Caprine arthritis-encephalitis virus infection: From recognition to eradication. Australian Veterinary Journal. 1986;63(8):237–241. doi: 10.1111/J.1751-0813.1986.TB02983.X. [DOI] [PubMed] [Google Scholar]
- 63.Huang J, Sun Y, Liu Y, Xiao H, Zhuang S. Development of a loop-mediated isothermal amplification method for rapid detection of caprine arthritis-encephalitis virus proviral DNA. Archives of Virology. 2012;157(8):1463–1469. doi: 10.1007/S00705-012-1322-Y. [DOI] [PubMed] [Google Scholar]
- 64.Mdurvwa EG, Ogunbiyi PO, Gakou HS, Reddy PG. PATHOGENIC MECHANISMS OF CAPRINE ARTHRITIS-ENCEPHALITIS VIRUS. 1994;18:483–490. doi: 10.1007/BF01839425. [DOI] [PubMed] [Google Scholar]
- 65.(OIE), O. I. des E. (World H. O. for animals). (2017). Caprine arthritis-encephalitis and maedi- visna. In Manual of Diagnostic Tests and Vaccines for Terrestrial Animals (pp. 1–10). Office International des Epizootes (OIE).
- 66.Clements JE, Christine Zink M. Molecular biology and pathogenesis of animal lentivirus infections. Clinical Microbiology Reviews. 1996;9(1):100–117. doi: 10.1128/CMR.9.1.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Balbin MM, Belotindos LP, Abes NS, Mingala CN. Caprine arthritis encephalitis virus detection in blood by loop-mediated isothermal amplification (LAMP) assay targeting the proviral gag region. Diagnostic Microbiology and Infectious Disease. 2014;79(1):37–42. doi: 10.1016/j.diagmicrobio.2013.12.012. [DOI] [PubMed] [Google Scholar]
- 68.Leitner G, Krifucks O, Weisblit L, Lavi Y, Bernstein S, Merin U. The effect of caprine arthritis encephalitis virus infection on production in goats. The Veterinary Journal. 2010;183(3):328–331. doi: 10.1016/j.tvjl.2008.12.001. [DOI] [PubMed] [Google Scholar]
- 69.Domingo E, Baranowski E, Escarmís C, Sobrino F. Foot-and-mouth disease virus. Comparative Immunology, Microbiology and Infectious Diseases. 2002;25(5–6):297–308. doi: 10.1016/S0147-9571(02)00027-9. [DOI] [PubMed] [Google Scholar]
- 70.Jamal, S. M., & Belsham, G. J. (2013). Foot-and-mouth disease: Past, present and future (pp. 1–14). [DOI] [PMC free article] [PubMed]
- 71.Poonsuk K, Giménez-Lirola L, Zimmerman JJ. A review of foot-and-mouth disease virus (FMDV) testing in livestock with an emphasis on the use of alternative diagnostic specimens. Animal Health Research Reviews. 2018;19(2):100–112. doi: 10.1017/S1466252318000063. [DOI] [PubMed] [Google Scholar]
- 72.Grubman MJ, Baxt B. Foot-and-mouth disease. Clinical Microbiology Reviews. 2004;17(2):465–493. doi: 10.1128/CMR.17.2.465-493.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Hardham JM, Krug P, Pacheco JM, Thompson J, Dominowski P, Moulin V, Rieder E. Novel foot-and-mouth disease vaccine platform: Formulations for safe and DIVA-compatible FMD vaccines with improved potency. Frontiers in Veterinary Science. 2020;7:659. doi: 10.3389/FVETS.2020.554305/BIBTEX. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Dukes JP, King DP, Alexandersen S. Novel reverse transcription loop-mediated isothermal amplification for rapid detection of foot-and-mouth disease virus. Archives of Virology. 2006;151(6):1093–1106. doi: 10.1007/S00705-005-0708-5. [DOI] [PubMed] [Google Scholar]
- 75.Shao J-J, Chang H-Y, Zhou G-Q, Cong G-Z. Rapid Detection of Foot-and-Mouth Disease Virus by Reverse Transcription Loop-mediated Isothermal Amplification (RT-LAMP) Journal of Applied Research in Veterinary Medicine. 2010;8:133–142. [Google Scholar]
- 76.Chen, H., Zhang, J., Liu, Y., & Liu, X. (2011). Detection of foot-and-mouth disease virus rna by reverse transcription loop-mediated isothermal amplification (pp. 2–5). [DOI] [PMC free article] [PubMed]
- 77.Yamazaki W, Mioulet V, Murray L, Madi M, Haga T, Misawa N, King DP. Development and evaluation of multiplex RT-LAMP assays for rapid and sensitive detection of foot-and-mouth disease virus. Journal of Virological Methods. 2013;192(1–2):18–24. doi: 10.1016/j.jviromet.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 78.Lim D-R, Park Y-R, Park S-Y, Kim H-R, Park M-J, Ku B-K, Park C-K. Pan-serotype reverse transcription loop-mediated isothermal amplification (RT-LAMP) for the rapid detection of foot-and-mouth disease virus. Korean Journal of Veterinary Service. 2018;41(1):29–39. doi: 10.7853/KJVS.2018.41.1.29. [DOI] [Google Scholar]
- 79.Lim DR, Kim HR, Chae HG, Ku BK, Nah JJ, Ryoo S, Park CK. Probe-based real-time reverse transcription loop-mediated isothermal amplification (RRT-LAMP) assay for rapid and specific detection of foot-and-mouth disease virus. Transboundary and Emerging Diseases. 2020;67(6):2936–2945. doi: 10.1111/tbed.13669. [DOI] [PubMed] [Google Scholar]
- 80.Madhanmohan M, Nagendrakumar SB, Manikumar K, Yuvaraj S, Parida S, Srinivasan VA. Development and evaluation of a real-time reverse transcription-loop-mediated isothermal amplification assay for rapid serotyping of foot-and-mouth disease virus. Journal of Virological Methods. 2013;187(1):195–202. doi: 10.1016/j.jviromet.2012.08.015. [DOI] [PubMed] [Google Scholar]
- 81.Reid SM, Mioulet V, Knowles NJ, Shirazi N, Belsham GJ, King DP. Development of tailored real-time RT-PCR assays for the detection and differentiation of serotype O, A and Asia-1 foot-and-mouth disease virus lineages circulating in the Middle East. Journal of Virological Methods. 2014;207:146–153. doi: 10.1016/J.JVIROMET.2014.07.002. [DOI] [PubMed] [Google Scholar]
- 82.Lim DR, Kim HR, Park MJ, Chae HG, Ku BK, Nah JJ, Park CK. A tailored reverse transcription loop-mediated isothermal amplification for sensitive and specific detection of serotype A foot-and-mouth disease virus circulating in pool 1 region countries. Transboundary and Emerging Diseases. 2018;65(6):1898–1908. doi: 10.1111/tbed.12971. [DOI] [PubMed] [Google Scholar]
- 83.Chen H, Zhang J, Liu Y, Liu X. Rapid typing of foot-and-mouth disease serotype Asia 1 by reverse transcription loop-mediated isothermal amplification. Virology journal. 2011;8(1):489. doi: 10.1186/1743-422X-8-489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Ding Y, Zhou J, Ma L, Qi Y, Wei G, Zhang J, Zhang Y. A reverse transcription loop-mediated isothermal amplification assay to rapidly diagnose foot-and-mouth disease virus C. Journal of Veterinary Science. 2014;15(3):423. doi: 10.4142/JVS.2014.15.3.423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Lim DR, Kim HR, Park MJ, Chae HG, Ku BK, Nah JJ, Park CK. An improved reverse transcription loop-mediated isothermal amplification assay for sensitive and specific detection of serotype O foot-and-mouth disease virus. Journal of Virological Methods. 2018;260:6–13. doi: 10.1016/j.jviromet.2018.06.017. [DOI] [PubMed] [Google Scholar]
- 86.Charles JA. Akabane virus. The Veterinary Clinics of North America: Food Animal Practice. 1994;10(3):525–546. doi: 10.1016/S0749-0720(15)30537-5. [DOI] [PubMed] [Google Scholar]
- 87.Kirkland RD. Akabane virus infection. Revue scientifique et technique (International Office of Epizootics) 2015;34(2):403–410. doi: 10.20506/RST.34.2.2366. [DOI] [PubMed] [Google Scholar]
- 88.Wang J, Blasdell KR, Yin H, Walker PJ. A large-scale serological survey of Akabane virus infection in cattle, yak, sheep and goats in China. Veterinary Microbiology. 2017;207:7–12. doi: 10.1016/J.VETMIC.2017.05.014. [DOI] [PubMed] [Google Scholar]
- 89.Spickler, A. R. (2018). Akabane Disease.
- 90.Qiao J, Wang J, Meng Q, Wang G, Liu Y, He Z, Chen C. Rapid detection of Akabane virus by a novel reverse transcription loop-mediated isothermal amplification assay (RT-LAMP) Virology Journal. 2013 doi: 10.1186/1743-422X-10-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Gibbbs EPJ, Taylor WP, Lawman MJP, Bryant J. Classification of peste des petits ruminants virus as the fourth member of the genus morbillivirus. Intervirology. 1979;11(5):268–274. doi: 10.1159/000149044. [DOI] [PubMed] [Google Scholar]
- 92.Parida S, Muniraju M, Mahapatra M, Muthuchelvan D, Buczkowski H, Banyard AC. Peste des petits ruminants. Veterinary Microbiology. 2015;181(1–2):90. doi: 10.1016/J.VETMIC.2015.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Bailey D, Banyard A, Dash P, Ozkul A, Barrett T. Full genome sequence of peste des petits ruminants virus, a member of the Morbillivirus genus. Virus Research. 2005;110(1–2):119–124. doi: 10.1016/j.virusres.2005.01.013. [DOI] [PubMed] [Google Scholar]
- 94.Altan E, Parida S, Mahapatra M, Turan N, Yilmaz H. Molecular characterization of Peste des petits ruminants viruses in the Marmara Region of Turkey. Transboundary and emerging diseases. 2019;66(2):865–872. doi: 10.1111/TBED.13095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rojas JM, Sevilla N, Martín V. A new look at vaccine strategies against PPRV focused on adenoviral candidates. Frontiers in Veterinary Science. 2021;8:1–14. doi: 10.3389/fvets.2021.729879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li L, Bao J, Wu X, Wang Z, Wang J, Gong M, Li J. Rapid detection of peste des petits ruminants virus by a reverse transcription loop-mediated isothermal amplification assay. Journal of Virological Methods. 2010;170(1–2):37–41. doi: 10.1016/j.jviromet.2010.08.016. [DOI] [PubMed] [Google Scholar]
- 97.Ashraf W, Unger H, Haris S, Mobeen A, Farooq M, Asif M, Khan QM. Genetic detection of peste des petits ruminants virus under field conditions: A step forward towards disease eradication. BMC Veterinary Research. 2017;13(1):1–13. doi: 10.1186/S12917-016-0940-0/TABLES/5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Mahapatra M, Howson E, Fowler V, Batten C, Flannery J, Selvaraj M, Parida S. Rapid Detection of Peste des Petits Ruminants Virus (PPRV) Nucleic Acid Using a Novel Low-Cost Reverse Transcription Loop-Mediated Isothermal Amplification (RT-LAMP) Assay for Future Use in Nascent PPR Eradication Programme. Viruses. 2019;11:8. doi: 10.3390/V11080699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rajko-Nenow P, Flannery J, Arnold H, Howson E, Darpel K, Stedman A, Batten C. A rapid RT-LAMP assay for the detection of all four lineages of Peste des Petits Ruminants Virus. Journal of Virological Methods. 2019 doi: 10.1016/J.JVIROMET.2019.113730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Flick R, Bouloy M. Rift valley fever virus. Current Molecular Medicine. 2005;5:827–834. doi: 10.2174/156652405774962263. [DOI] [PubMed] [Google Scholar]
- 101.Wright D, Kortekaas J, Bowden TA, Warimwe GM. Rift Valley fever: Biology and epidemiology. The Journal of General Virology. 2019;100(8):1187–1199. doi: 10.1099/JGV.0.001296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Quellec J, Salinas S, Simonin Y, Cêtre-Sossah C. Rift Valley fever virus infection : Physiopathology and pathogenesis. Virologie (Montrouge, France) 2021;25(5):263–279. doi: 10.1684/VIR.2021.0919. [DOI] [PubMed] [Google Scholar]
- 103.Mansfield KL, Banyard AC, McElhinney L, Johnson N, Horton DL, Hernández-Triana LM, Fooks AR. Rift Valley fever virus: A review of diagnosis and vaccination, and implications for emergence in Europe. Vaccine. 2015;33(42):5520–5531. doi: 10.1016/J.VACCINE.2015.08.020. [DOI] [PubMed] [Google Scholar]
- 104.Peyrefitte C, Boubis L, Coudrier D, Bouloy M, Grandadam M, Tolou H, Plumet S. Real-time reverse-transcription loop-mediated isothermal amplification for rapid detection of rift valley Fever virus. Journal of Clinical Microbiology. 2008;46(11):3653–3659. doi: 10.1128/JCM.01188-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Le Roux C, Kubo T, Grobbelaar A, van Vuren P, Weyer J, Nel L, Paweska J. Development and evaluation of a real-time reverse transcription-loop-mediated isothermal amplification assay for rapid detection of Rift Valley fever virus in clinical specimens. Journal of Clinical Microbiology. 2009;47(3):645–651. doi: 10.1128/JCM.01412-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Han Q, Zhang S, Liu D, Yan F, Wang H, Huang P, Xia X. Development of a visible reverse transcription-loop-mediated isothermal amplification assay for the detection of rift valley fever virus. Frontiers in Microbiology. 2020 doi: 10.3389/fmicb.2020.590732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Cullinane A, Garvey M. A review of diagnostic tests recommended by the World Organisation for Animal Health Manual of Diagnostic Tests and Vaccines for Terrestrial Animals. OIE Revue Scientifique et Technique. 2021;40(1):75–89. doi: 10.20506/rst.40.1.3209. [DOI] [PubMed] [Google Scholar]
- 108.Paul R, Ostermann E, Wei Q. Advances in point-of-care nucleic acid extraction technologies for rapid diagnosis of human and plant diseases. Biosensors and Bioelectronics. 2020;169:112592. doi: 10.1016/J.BIOS.2020.112592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Mahdi MA, Yousefi SR, Jasim LS, Salavati-Niasari M. Green synthesis of DyBa2Fe3O7.988/DyFeO3 nanocomposites using almond extract with dual eco-friendly applications: Photocatalytic and antibacterial activities. International Journal of Hydrogen Energy. 2022;47(31):14319–14330. doi: 10.1016/J.IJHYDENE.2022.02.175. [DOI] [Google Scholar]
- 110.Yousefi SR, Alshamsi HA, Amiri O, Salavati-Niasari M. Synthesis, characterization and application of Co/Co3O4 nanocomposites as an effective photocatalyst for discoloration of organic dye contaminants in wastewater and antibacterial properties. Journal of Molecular Liquids. 2021;337:116405. doi: 10.1016/J.MOLLIQ.2021.116405. [DOI] [Google Scholar]
- 111.Yousefi SR, Ghanbari D, Salavati-Niasari M, Hassanpour M. Photo-degradation of organic dyes: Simple chemical synthesis of Ni(OH)2 nanoparticles, Ni/Ni(OH)2 and Ni/NiO magnetic nanocomposites. Journal of Materials Science: Materials in Electronics. 2016;27(2):1244–1253. doi: 10.1007/S10854-015-3882-6/FIGURES/16. [DOI] [Google Scholar]
- 112.Yousefi SR, Ghanbari M, Amiri O, Marzhoseyni Z, Mehdizadeh P, Hajizadeh-Oghaz M, Salavati-Niasari M. Dy2BaCuO5/Ba4DyCu3O9.09 S-scheme heterojunction nanocomposite with enhanced photocatalytic and antibacterial activities. Journal of the American Ceramic Society. 2021;104(7):2952–2965. doi: 10.1111/JACE.17696. [DOI] [Google Scholar]
- 113.Yousefi SR, Sobhani A, Alshamsi HA, Salavati-Niasari M. Green sonochemical synthesis of BaDy2NiO5/Dy2O3 and BaDy2NiO5/NiO nanocomposites in the presence of core almond as a capping agent and their application as photocatalysts for the removal of organic dyes in water. RSC Advances. 2021;11(19):11500–11512. doi: 10.1039/D0RA10288A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Yousefi SR, Amiri O, Salavati-Niasari M. Control sonochemical parameter to prepare pure Zn0.35Fe2.65O4 nanostructures and study their photocatalytic activity. Ultrasonics Sonochemistry. 2019;58:104619. doi: 10.1016/J.ULTSONCH.2019.104619. [DOI] [PubMed] [Google Scholar]
- 115.Yousefi SR, Ghanbari D, Salavati-Niasari M. Hydrothermal synthesis of nickel hydroxide nanostructures and flame retardant poly vinyl alcohol and cellulose acetate nanocomposites. Journal of Nanostructures. 2016;6(1):80–85. doi: 10.7508/JNS.2016.01.013. [DOI] [Google Scholar]
- 116.Yousefi SR, Sobhani A, Salavati-Niasari M. A new nanocomposite superionic system (CdHgI4/HgI2): Synthesis, characterization and experimental investigation. Advanced Powder Technology. 2017;28(4):1258–1262. doi: 10.1016/J.APT.2017.02.013. [DOI] [Google Scholar]