Abstract
Objective:
Serial sectioning optical coherence tomography (OCT) enables accurate volumetric reconstruction of several cubic centimeters of human brain samples. We aimed to identify anatomical features of the ex vivo human brain, such as intraparenchymal blood vessels and axonal fiber bundles, from the OCT data in 3D, using intrinsic optical contrast.
Methods:
We developed an automatic processing pipeline to enable characterization of the intraparenchymal microvascular network in human brain samples.
Results:
We demonstrated the automatic extraction of the vessels down to a 20 μ m in diameter using a filtering strategy followed by a graphing representation and characterization of the geometrical properties of microvascular network in 3D. We also showed the ability to extend this processing strategy to extract axonal fiber bundles from the volumetric OCT image.
Conclusion:
This method provides a viable tool for quantitative characterization of volumetric microvascular network as well as the axonal bundle properties in normal and pathological tissues of the ex vivo human brain.
Keywords: Ex vivo human brain, Optical Coherence Tomography, Serial sectioning, Vasculature characterization
I. INTRODUCTION
A comprehensive atlas of blood vessels of the human cerebral cortex is important in understanding the human brain anatomy and functionality [1, 2]. Alterations in the brain vasculature are associated with normal aging [3] as well as neurodegenerative diseases, such as the Alzheimer’s disease (AD) [4–7]. Changes in vascular morphology have been found in the postmortem brain tissue using standard histology and optical microscopy. For example, increased tortuosity has been observed in the white matter vessels that are downstream of the penetrating arterioles as early as in middle age (around 55 years old) [8, 9]. It is also reported that there is an increase in spiraling and looped vessels in several areas of the brain in AD [10]. Several studies also suggest decreased capillary density in older human brains [11, 12]. Those pathological studies were exclusively conducted by traditional histology, which stained and imaged thin tissue slices in 2D planes. Three-dimensional (3D) vascular morphology of the human brain in centimeter scale has not been reported.
Optical coherence tomography (OCT) has emerged as an imaging technique that produces depth-resolved microstructural images of biological tissue, using endogenous sources of optical contrast [13]. Serial sectioning OCT, which integrates an OCT scanner and a tissue slicer, has been substantially utilized to study the postmortem human brain, given its capability of reconstructing large-scale volumetric microstructure and connectivity [14–18]. Previous studies demonstrated the visualizations of cortical layers and neurons in human brain with OCT [19–21], and pathological changes in disease, such as brain tumors [22–24]. Moreover, with polarization sensitive OCT, it is possible to measure the orientation of fiber tracts in both mouse [14, 15, 25] and human [26]. However, studies of the intraparenchymal microvasculature in postmortem human brain tissue using OCT have not been reported.
In this study, we developed a processing pipeline to map the volumetric microvasculature in postmortem human brain tissues with serial sectioning OCT. To extract the microvascular network in 3D, we first imaged three brain sample blocks and reconstructed the volumetric OCT images with a 12 μ m isotropic resolution. We then applied an automated segmentation method to extract the blood vessels of the human brain samples from the OCT intrinsic contrast, which showed up as black tubes in the OCT volume. We demonstrated how we could validate the vessel segments identified by OCT by comparing with histological techniques. We further characterized the geometrical properties of the microvascular network. Lastly, we showed how we could generalize our vessel segmentation pipeline to extract other structures with similar geometry, such as axonal fiber tracts, in the volumetric OCT data.
II. METHODS
A. Tissue Preparation
Three human brain sample blocks were obtained in this work. Two of the samples (I and II) were obtained from the Alzheimer’s Disease Research Center and VA-BU-CLF brain banks, one from a brain donor with neuropathologically confirmed AD and one from a brain donor with neuropathologically confirmed chronic traumatic encephalopathy (CTE), and a third sample (III) was obtained from the Massachusetts General Hospital Autopsy Suite, from a neurologically normal subject without no neuropathological abnormalities. This study and its use of postmortem tissue was approved by the Institutional Review Board of the Massachusetts General Hospital and the VA-BU-CLF brain banks. Sample blocks I and II from the parietal-frontal cortex and sample III was from primary motor cortex. The postmortem intervals did not exceed 24 h. The brain samples were fixed in Periodate-lysine-paraformaldehyde (PLP) and stored at 4 °C for at least two months. All the sample blocks were washed for one month in 0.01M Phosphate Buffer Saline solution (PBS) at room temperature while gently shaking to remove excessive fixation agent in the tissue blocks. After washing, the tissue blocks were embedded in 4.5% agarose and preserved in 0.01M PBS with 1% sodium azide at 4 °C.
B. OCT System Setup
We built an OCT system following the design in [27]. The light source was a swept-source laser (AxsunTech Inc.), with a center wavelength of 1310 nm and a bandwidth of 135 nm. The axial resolution was estimated to be 5.6 μm in tissue (with a refractive index of 1.4). We used a 4x air objective (Olympus, UPLFLN4x, NA 0.13) in the sample arm to obtain a lateral resolution of 5 μm. The sensitivity of the system was 90 dB and the roll-off was 0.46 dB over 1 mm in depth. The detector was connected to a high-speed digitizer (ATS350, AlazarTech Inc.) for data acquisition at an A-line rate of 100 kHz. With a scanning area of 3 × 3 mm2, one volumetric acquisition composed of 1000 A-lines by 1000 B-lines took 16 s. The raw data was resampled in k-space, compensated for dispersion [28], Fourier transformed into the spatial domain following standard procedures [29], and corrected for sensitivity roll-off [30] before uploaded to a cloud server.
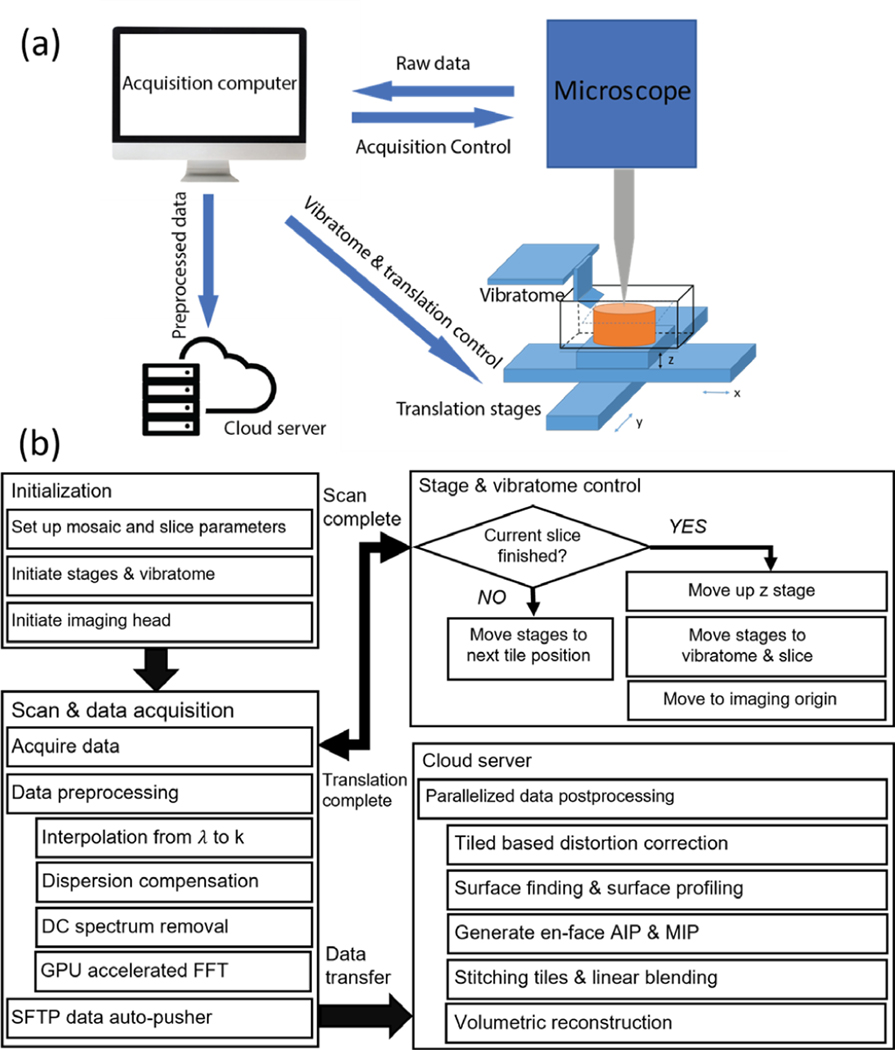
The workflow for serial sectioning OCT acquisition has been previously reported [18] and is summarized in Fig 1. The human brain tissue block was mounted in a water bath filled with PBS during imaging. We used a submerged z spacer attached to the air objective to image the brain block. We custom-made the z spacer: we 3D printed the spacer wall and sealed the wall with a cover glass to protect the objective from water immersion. The z spacer eliminated the effect of high reflection from the water surface. Motorized xyz stages (x and y stages: LTS150, Thorlabs Inc.; z stage: MLJ150, Thorlabs Inc.) were incorporated to translate the samples under the OCT objective to automate image tiling over the entire surface of the tissue. The field of view (FOV) for each image tile was 3 mm x 3 mm and the overlap between adjacent tiles was 15%. A customized vibratome slicer [31] with a 63.5 mm sapphire blade (DDK Inc.) was mounted adjacent to the OCT imaging head to cut off a slice from the top of the tissue block upon completion of the scanning of the sample surface. The sectioning thickness was 150 μm. The blade frequency of the vibratome was 3000 RPM. The slices were preserved for histological validation. Customized stage and vibratome control software were embedded within the image acquisition software for fully automated serial sectioning acquisition. Data transfer and saving were implemented in two relay steps: (1) from local acquisition computer to a local storage server after one image tile and (2) from the storage server to a centralized computational server at the Boston University Shared Computing Cluster (SCC) after the entire serial scan [32]. A parallelized post-processing script written in Matlab was then executed to reconstruct the volumetric sample.
Figure 1:
workflow of volumetric reconstruction. (a) The schematic of the serial sectioning OCT system. (b) Flow chart of the acquisition and processing pipeline. The details of preprocessing were explained in section II.B and the details of the post processing were explained in section II.D.
C. Histological Imaging
1). Passive Tissue Clearing & Labeling
After serial sectioning OCT, the sectioned brain slices were cleared and labeled with tomato lectin conjugated to fluorescent dye DyLight 649 (Vector Laboratories) for vasculature validation following the protocol below. The slices were first incubated in a 7:1 mixture of SHIELD-ON buffer and SHIELD-Epoxy solution (LifeCanvas, Cambridge, MA) for 6 hours at 4° C followed by 24 hours at room temperature and washed for 30 min with PBST (PBS with 0.4% TritonX-100) [33]. To delipidate the slices, the slices were then incubated overnight at 37° C in passive clearing buffer (300 mM sodium dodecyl sulfate, 10 mM boric acid, 100 mM sodium sulfite, pH 9, in deionized water) and washed twice with PBST. Slices were photobleached for five days in PBST as previously described [34]. To stain, slices were incubated with 40 μg tomato lectin649 in 1 mL of PBST for 24 hours. Samples were washed three times with PBST and incubated overnight in 4% paraformaldehyde in PBS (Affymetrix, Santa Clara, CA) to prevent dissociation of tomato lectin649. Samples were refraction index-matched in EasyIndex (LifeCanvas, Cambridge, MA) overnight before mounting and imaging.
2). Confocal Imaging of Cleared Tissue
Labeled slices were mounted onto a glass slide (Fisher, Pittsburgh, PA) with EasyIndex (LifeCanvas, Cambridge, MA) mounting media for vasculature validation. Image tiles were acquired with a Nikon Eclipse Ti2-E spinning disk confocal microscope, then digitized and stitched in Nikon Elements software (Nikon, Melville, NY). Tomato lectin 649 was excited using the 640 nm laser line of a Celesta Light Engine (Lumercor, Beaverton, WA), at 75% power with an exposure time of 200 ms. A Plan Apo λ 4x air objective (Nikon, Melville, NY) was used to take the images, resulting in a 0.96 μm lateral resolution. For each image tile, a stack in depth was taken with a 12.5 μm step size. After the image stacks were stitched in x and y, a maximum intensity projection (MIP) was computed for manual vessel labeling in section II.F.
D. Volumetric Reconstruction of Serial Sectioning OCT
1). Tile Based Distortion Correction
To reconstruct the volume, we first correct the distortions of individual tiles, including the field curvature and the shading effect. The field of view of each image tile is curved due to the different path lengths that light travels to each xy position in the image. We calibrated this field curvature by imaging a mirror and corrected the depth location [35]. A non-linear distortion is also observed in xy plane, which was calibrated by imaging a grid target. We used the bUnwarpJ plugin in ImageJ [36, 37] to perform a non-linear registration between the image of the grid target and a synthesized grid pattern. The registration matrix was then applied to each image tile of human brain samples to correct for the xy-plane distortion. Another issue brought by the field curvature is the nonuniform intensity across the FOV. To correct for that, we adopted a shading correction process. We used BaSiC illumination correction method in ImageJ to correct for this shading effect [38]. This method is based on low-rand and decomposition to correct for uneven illumination, and further improves stitching and mosaic image quality. The above procedures significantly reduced the stitching artifacts in the volumetric reconstruction process.
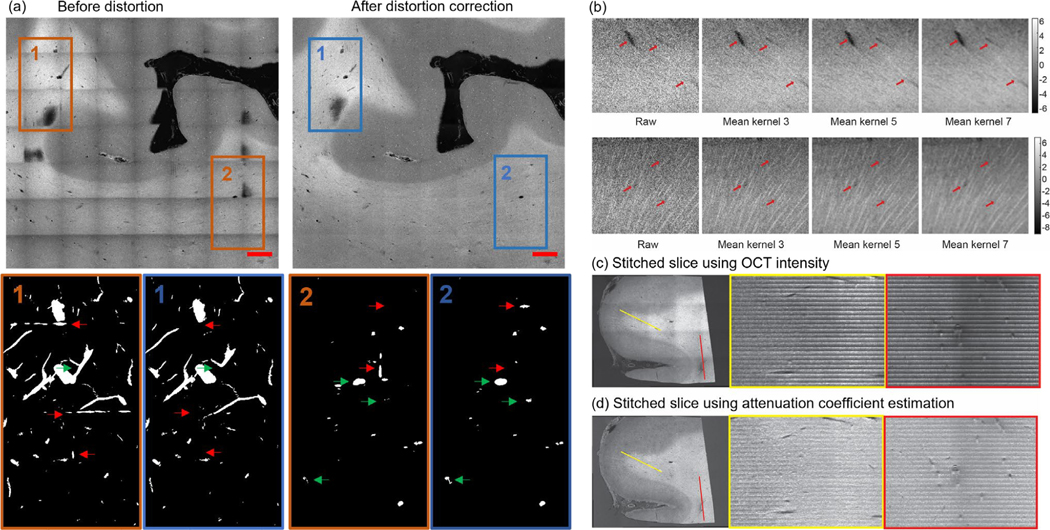
We showed in Fig 2 (a) the improved visualization for a stitched slice after the distortion correction. Moreover, we also observed that the distortion correction procedure improved the vessel segmentation by removing segmentation artifacts and deblurring vessels in the overlapping area of image tiles.
Figure 2:
Distortion correction, volumetric averaging kernel and volumetric stitching. (a) An example XY slice (top row) and the correspondent maximum intensity projection (MIP) of the vessel segmentation of the volume (bottom row) before (left column) and after (right column) the distortion correction. Scale bars: 2 mm. Red arrows: stitching artifacts falsely segmented as vessels. Green arrows: vessels that are not accurately segmented before distortion correction. (b) Kernel averaging of OCT data with various kernel size. Top row: vessels of different size. Bottom row: axonal fiber bundles. (c) The volumetric stitching using OCT intensity. An XY plane is shown at the left side with two lines of interest labeled with yellow and red. The XZ profile at the two lines of interest was plotted on the right side. (d) The volumetric stitching using attenuation coefficient estimation. An XY plane is shown at the left side with two lines of interest labeled with yellow and red. The XZ profile at the two lines of interest was plotted on the right side.
2). Volumetric Stitching
To reconstruct the OCT volume in three dimensions, we first stitched in x and y using a displacement model [39]. We generated the average intensity projection (AIP) of each image tile starting from the tissue surface and over a 250 μm depth range. The tissue surface was determined using an edge detector [40]. Since the laser scanning axes was not parallel to the motorized stage axes, the x and y position of adjacent tiles was shifted with each other. We assumed this distortion could be represented by a tilted angle and an offset distance at both x and y direction, which were obtained by comparing the input positions of the motorized x and y stages and the coordinates calculated from registration based tile stitching [32, 33]. After the initial tile registration, the position of each tile was further adjusted with the Fiji stitching plug-in [41]. To avoid possible local minima in this registration step, we confined the maximum displacement of tiles to be 50 pixels, which was 5% of FOV. After the final x and y coordinates were obtained, we then perform a linear blending to stitch all the tiles together to a mosaic image.
Lastly, we stitched all the tissue slices in z-axis by cropping each tissue slice into 150 μm thickness and stacking them in depth. We applied a uniform volumetric averaging kernel with different sizes (3, 5 and 7 pixels isotropic) to suppress the speckle noise and enhance the image features, as shown in Fig 2 (b). We reconstructed the 3D volumes using both OCT intensity image as well as depth-resolved attenuation coefficient map, which was derived from OCT depth-profile based on the method described in [42]. An example of the reconstructed volume with depth-resolved attenuation coefficient map was shown in Fig. S1. The attenuation coefficient images were used for vessel segmentation later on, since this transformation effectively reduced intensity fluctuation in depth and resulted in a better volumetric representation of the sample blocks, as shown in Fig 2 (c) – (d).
E. Frangi Filtering for Vessel Enhancement & Segmentation
We adopted a Frangi filtering strategy to enhance the visualization of vessels in OCT volume. Frangi filter performed an eigenvector decomposition on the local second order derivatives of the image intensity to enhance tubular structures in the image [43–47]. Since vessels appeared in different sizes, an adjustable scale parameter was incorporated into the filter. With this filtering process, we constructed a “vesselness” measure (ω) based on the eigenvalues obtained from the Frangi filter to represent the likelihood of pixels being a vessel between 0 and 1. In this work, we adopted a previously constructed vesselness measure proposed by Sato et al. [46]:
Where λ1, λ2 and λ3 are the eigenvalues of the local image patch, γ23 ≥ 0 and γ12 ≥ 0 controls the diameter range selectivity of the filter, and 0≤α≤1 introduces asymmetrical behavior of the filter in the negative and positive regions of λ1. We set initial value of the parameters of Frangi filter based on a previous work in CT vasculature imaging [46] and fine-tuned them by exhaustively searching the neighborhood of the parameter space to obtain the final parameters. In practice, we found γ23 = γ12 = 0.5 and α = 0.25 worked well with the volumetric OCT data. We applied a threshold on the filtered images to obtain the initial vessel segmentation. The threshold of the vesselness filtering was determined by visual inspection, where we overlaid the thresholded image with the original OCT image and determined no visible artifacts was included in the thresholded image. We then removed the unconnected components that were less than 100 voxels in the segmentation using the connectivity analysis function (bwconncomp.m) in Matlab® 2019b since most of them were artifacts from the filtering process.
F. Graph Construction & Geometrical characterization
We computed the graph representation of the vessel segmentation for further geometrical characterization. A graph is defined as a set of nodes and a set of edges that connect the nodes. To obtain the graph of a vessel segmentation, we first skeletonized the segmentation using a Matlab function (bwskel.m), then traced each segment of the skeleton to record the nodes and edges of the vessel segmentation in 3D. The nodes were defined to have at least a 10-pixel Euclidean distance with each other. After that we pruned the graph by removing the spur segments and breaking up self-loops in the graph that were artifacts other than the vessels. All the graphing and pruning process was done using a customized Matlab script (https://github.com/BUNPC/vesGraphValidate).
We further extracted geometrical parameters of vessels from the graph using a customized Matlab script, including the segment length and the vessel tortuosity. The segment length was the length of the segments in the graph. The tortuosity of the segment is defined as the ratio of the segment length and the Euclidean distance between the two end nodes.
G. Validation of OCT Vasculature
To validate the accuracy of the automatic vessel segmentation described in section II.E, we performed manual segmentation of blood vessels in three OCT datasets, constructed the graph representation and extracted the geometrical parameters as described in section II.F. The lumen of the vessels was identified and labeled in the process of manual segmentation.
To validate the vessel identification by OCT, we compared the results with tomato lectin labeled confocal images in 6 slices. We registered the confocal image to the OCT image using a landmark based affine registration [48]. Due to the severe distortion of confocal images introduced during tissue slicing, clearing and staining, the accuracy of registration did not allow us to perform pixel-wise comparison. We used an alternative strategy to compare the results of vessel identification between OCT and confocal images: for each slice, an observer who was naïve to both images randomly selected 3 groups of vessels with different diameter ranges (40 vessels in 20–40 μm, 40 vessels in 40–60 μm, and all vessels in 60–100 μm) on one modality, and then searched the corresponding vessels in the other modality. In 3 out of 6 slices the observer was asked to examine the OCT images first, and for the rest the confocal images first. Based on the searching results, we calculated the true positive (TP) rate (the ratio of vessels found in OCT that were also found in the confocal image), the false negative (FN) rate (the ratio of vessels not found in the OCT but found in the confocal image) and the false positive (FP) rate (the ratio of vessels found in the OCT but not found in the confocal image) for each slice and each vessel diameter group.
H. Fiber Tract Extraction on Serial Sectioning OCT
We extended the usage of the Frangi filter and segmentation to neuronal axonal tracts, which appeared as similar tubular structures but with bright intensity and smaller diameter in the OCT images. The extraction of axonal tracts is more challenging compared to the extraction of blood vessels since the contrast between the axonal tracts and the surrounding tissue is much less than the contrast between the blood vessel wall and the interior of the vessel, which is usually a dark cavity in the post-mortem human brain sample.
The axonal tracts could be enhanced by the Frangi filter with a smaller scaling factor compared to vessels. We fine-tuned the scaling parameter of the Frangi filter as well as the threshold for the binary mask in order to extract the fiber tracts. To obtain these parameters, we first determined the optimal scaling parameter by using a grid search of the parameter space. We went from 0.1 to 3 with a step size of 0.1 and found that θ=0.1, 0.2, and 0.3 were able to capture most fiber tract structure. Once we found the scaling parameter, we tuned the threshold by visual inspection, where we overlaid the thresholded image with the original OCT image and determined no visible artifacts was included in the thresholded image. The γ and α in the vesselness measure related to extracting local tubular structure were unchanged for the fiber tracts extraction. One problem in the segmentation is the discontinuity between voxels along a fiber tract because of the speckle noise and the low contrast-to-noise ratio. To improve the continuity between fragmented tracts, we further applied a marching ellipsoid method [49] on the initial segmentation results of the axonal tracts. More specifically, we constructed the graph representation and utilized randomly selected nodes in the graph as seed points to initialize the marching ellipsoid procedure. The marching ellipsoid procedure fit an ellipsoid centered at the seed point to connect the adjacent tract fragment, as the axonal tracts were ellipsoidal-like structures. Then the central point was moved along the primary direction of the fitted ellipsoid and the fitting of the ellipsoid was repeated. This fitting and moving procedure were stopped if the central point met one of the ending criteria, including a low intensity value, no ellipsoid fitting achieved or close to the boundary of the image. While the marching ellipsoid was running, all the seed points within the fitted ellipsoid were marked as “marched” on the graph. The process was repeated until all seed points were marked as “marched”.
III. RESULTS
A. Volumetric Reconstruction
All three brain samples were volumetrically reconstructed with our postprocessing pipeline. Sample I was consisted of 4 slices and each slice was consisted of 100 tiles (10 tiles in x and 10 tiles in y). Sample II was consisted of 7 slices and each slice was consisted of 100 tiles (10 tiles in x and 10 tiles in y). Sample III was consisted of 8 slices and each slice was consisted of 100 tiles (10 tiles in x and 10 tiles in y).
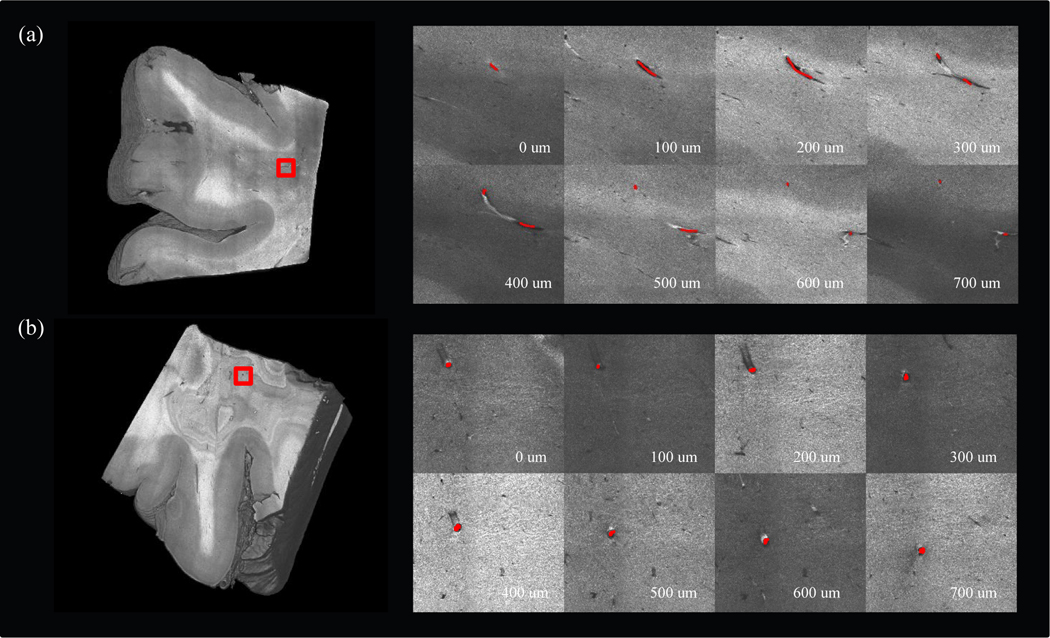
Volumetric rendering of two brain samples after running our reconstruction pipeline is shown in Fig 3. Sample I (a) was 2×2×0.5 cm3 in size and sample II (b) was 2×2×1 cm3 in size. The vessels in OCT appeared as dark cavities confined by highly scattered medium around. The vessel wall appeared bright below the vessel cavity in the OCT images since the light intensity was not attenuated inside the vessel. To demonstrate the trace of vessels in OCT images, we selected a 3×3 mm2 region of interest (ROI) from each sample and manually annotated a vessel in consecutive depths, as shown in the right panel of Fig. 3 (a) – (b).
Figure 3:
Tracing vessels in volumetric OCT data. (a) 3D rendering of sample I (left). A region of interest was labeled in red rectangle to show the branching of the manually labeled vessel in different depths (right). (b) 3D rendering of sample II (left). A region of interest was labeled in red rectangle to show the manually labeled vessel in different depth (right).
B. Automatic Vessel Segmentation & Graph Construction
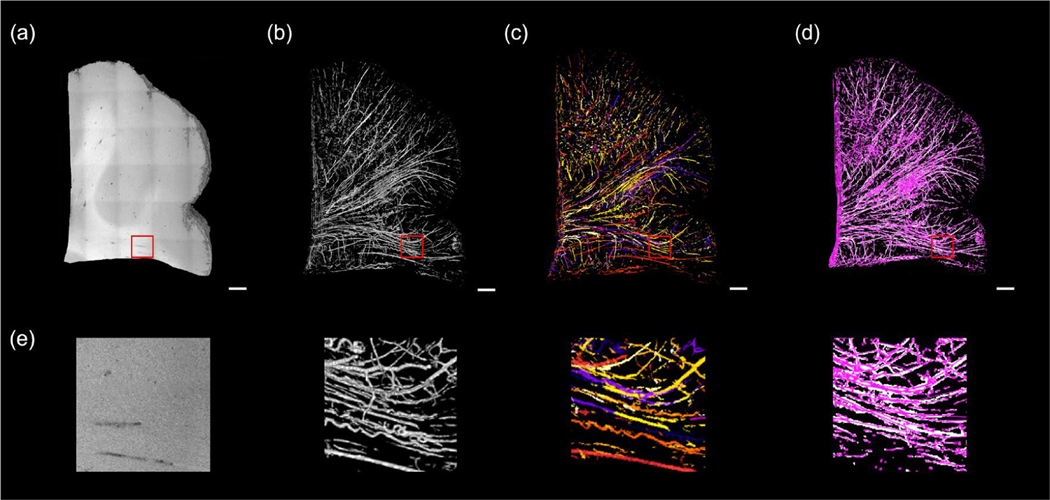
In Fig 4 (a) – (d), we show a graph representation of the vessel network generated from the volumetric OCT data in individual steps. Since the data size is too large for downstream processing (~1 GB for each cubic centimeter), the original OCT volume was first downsampled 4x to 12 μm isotropic voxel size. In this case, the minimal diameter of a resolvable vessel was 24 μm (2 pixels). To identify blood vessels with a wide diameter range, we applied the Frangi filter with different scales for the Hessian matrices. Our study focused on vessels with a diameter from 20 to 500 μm in diameter. Therefore, we tuned the size of the Hessian matrix by setting the scale variable θ=1,2,3 that are mostly sensitive to vessels within this diameter range. After the Frangi filtering process, the intensity of the vesselness enhanced volume was normalized between 0 to 1 and a threshold of a=0.18 was applied to binarize the volume and extract the vessel segments, as shown in Fig. 4 (b). The vessel segmentation was further skeletonized (c) and converted into a graph (d) following the procedure mentioned in section II.F.
Figure 4:
Frangi filtering and graph reconstruction of volumetric OCT. Scale bars: 1 mm. (a) 3D rendering of the OCT volume. (b) 3D rendering of the Frangi segments. (c) Skeletonization and tracing each segment from the Frangi segments. End-point voxels are displayed in blue, slab voxels in orange and junction voxels in purple. (d) The final graph representation of the vessel network of the OCT volume after pruning. (e) The zoom-in view of the red rectangles in (a) – (d).
C. Validation by Comparing with Manual Annotation
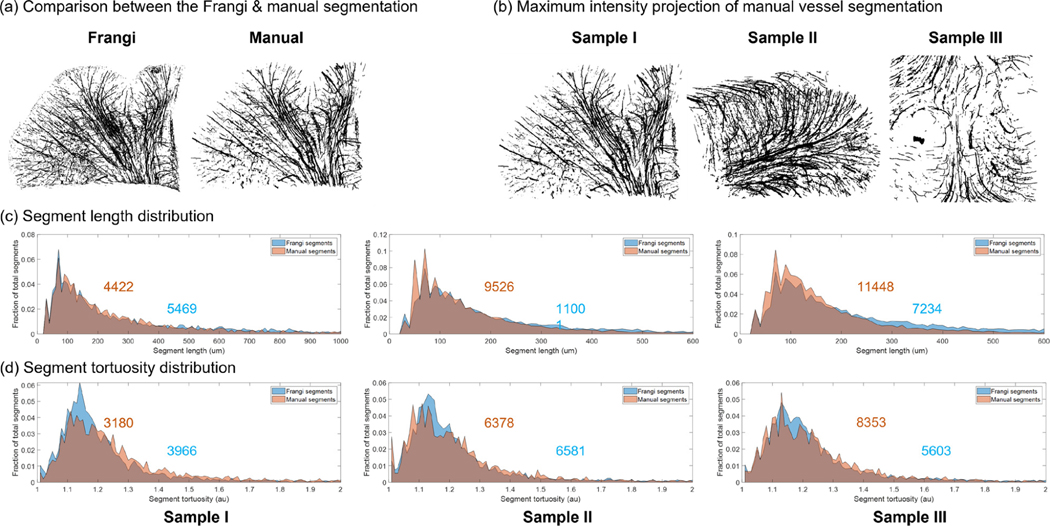
To validate that the automatic segmentation enabled accurate quantification of the geometrical properties of the vasculature network, we performed manual vessel annotations on the 3 OCT volumes and compared the derived geometrical parameters with those obtained from the automatic segmentation. The maximum intensity projection (MIP) of the Frangi vessel segmentations of each OCT volume is shown in Fig. 5 (a). The distribution of the segment length and the segment tortuosity of both the Frangi segmentations and the manual segmentations were plotted in Fig. 5 (b) and (c). The distributions were normalized by the total number of segments, which were indicated in each panel. To better visualize the tortuosity distribution, we excluded segments that are shorter than 100 μm in Fig 5. (c), as they appeared straight with a tortuosity very close to 1. This resulted in the difference of the total number of segments between Fig. 5 (b) and (c). We observed that the distributions of the Frangi segmentations and the manual segmentations were in agreement with each other. To further examine the difference between the two set of segmentations, we performed an F test in the distributions of the segment length between the Frangi segmentations and the manual segmentations and found there was no statistically significant difference (p-values of 0.604, 0.5467 and 0.4501, respectively). In addition, we calculated the structural similarity index (s) between the manual segmentations and the Frangi segmentations, defined as:
Figure 5:
Validation of the Frangi vessel segments by comparing with the manual segments. (a) An example comparison of the maximum intensity projection of the Frangi segmentation and the manual segmentation. (b) The MIP of the manually labeled vessel segments of each dataset. (c) The normalized segment length distribution of each dataset. The Frangi segments were plotted in blue whereas the manual segments were plotted in orange. The gray regions in the histogram are the overlapping of the two distributions. The total number of segments was labeled on the distribution. (d) The normalized segment tortuosity distribution of each dataset. The Frangi segments were plotted in blue whereas the manual segments were plotted in orange. The gray regions in the histogram are the overlapping of the two distributions. The total number of segments was labeled on the distribution.
Where μx and μy are the average of x and y, and are the variance of x and y, σxy is the covariance of x and y, and c1 and c2 are the variables to stabilize the division with weak denominator. We found a high structural similarity index between the two segmentations for all the dataset (s=0.9237 For sample I, s=0.9634 For sample II and s=0.9741 For sample III), which suggested that the Frangi segmentation allowed for accurate extraction of the vessel network properties from volumetric OCT data.
D. Validation by Vessel Labeling & Confocal Imaging
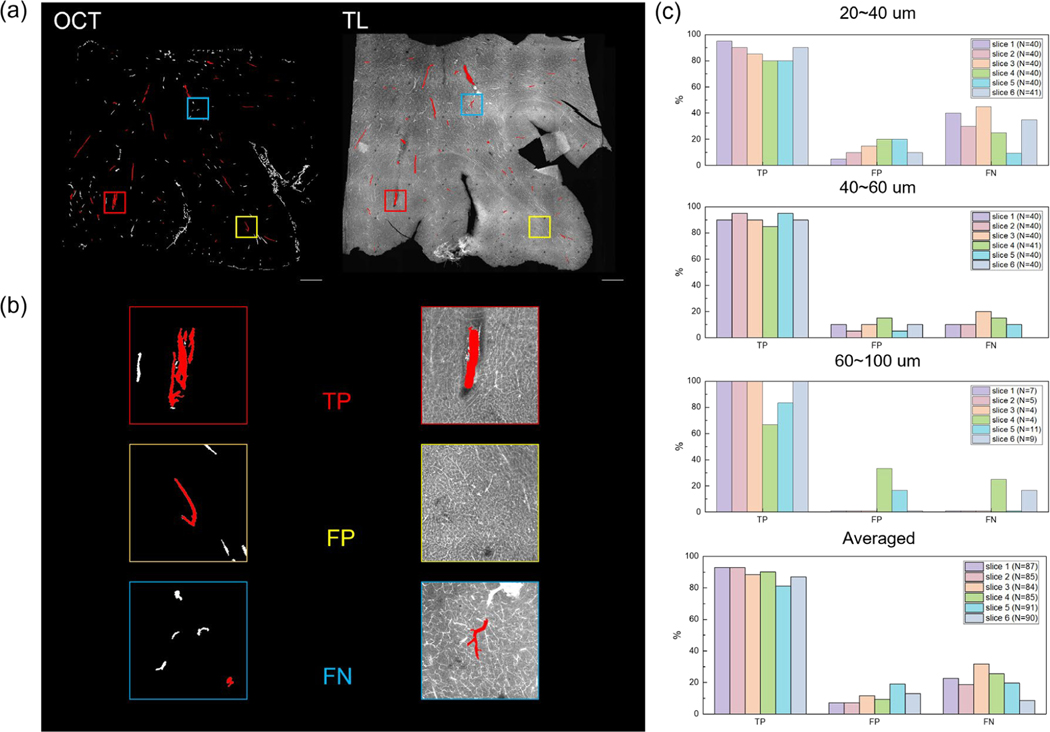
We imaged 6 tomato-lectin labeled slices that were taken from serial sectioning OCT by a confocal microscopy. A comparison between OCT vessel segmentation and the labeled fluorescent image on the same slice is shown in Fig. 6 (a). The total number of vessels selected for the six slices was 87, 85, 84, 85, 90, and 91, respectively. For vessels that were 20–40 μm in diameter, the average true positive rate in OCT images was 86.7% across the slices. The average false positive and false negative rate were 13.3% and 30.8%. For vessels that were 40–60 μm in diameter, the average true positive rate was 90.8% across the slices. The average false positive and false negative rate were 9.2% and 12.5%. For vessels that were 60–100 μm, the average true positive rate was 91.7%. The average false positive and false negative rate were 8.3% and 7.0%. While combining all the vessel diameter ranges, the average true positive rate in OCT images was 88.7%. The average false positive and false negative rate were 11.3% and 21.13%. Moreover, we also calculated the average true positive, false positive and false negative rate of the slices based on which modality the human grader was first exposed. For slices that the grader looked the OCT images first, the average true positive rate is 87.5%. The average false positive and false negative rate was 12.5% and 24.7%. For slices that the grader looked the confocal images first, the average true positive rate is 90.0%. The average false positive and false negative rate was 10.0% and 17.6%. The quantitative assessment for each slice is shown in Fig. 6 (b) and (c). In general, we showed high accuracy in identifying the vessels with a diameter range of 20–100 μm from volumetric OCT. The accuracy was slightly higher in vessels with diameter greater than 40 μm.
Figure 6:
Validation of OCT imaging vasculature by comparing with tomato lectin staining. (a) An example of slice-by-slice comparison between the vessel segments from volumetric OCT and the tomato lection stained image. The vessels selected by the independent observer were labeled in red. (b) An example of a true positive (TP), a false positive (FP) and a false negative (FN) case in the example slice in (a). (c) Bar plot of the true positive (TP), false positive (FP) and false negative (FN) rate for the selected vessels of different diameters in each slice, as well as averaged across all slices. The total number of vessels selected was labeled in caption.
E. Fiber Tract Extraction
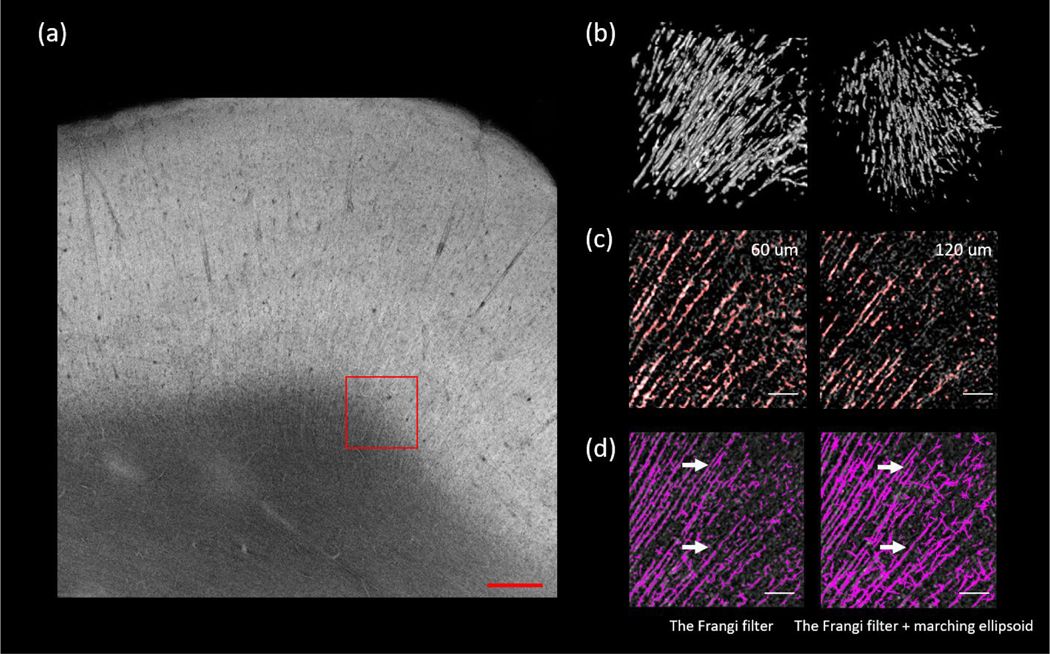
We further expanded our Frangi filtering based approach to extract the axonal fiber bundles from volumetric OCT data. In this case we examined the axonal fiber bundles coming from the white matter projecting into the cortex (as shown in Fig 7 (a), ROI size: 500 μm x 500 μm x 500 μm), which usually have diameters up to 16 μm. We tuned the scale of the Frangi filter to have the most sensitivity to structures within this diameter range. We applied a marching ellipsoid method on the segmented image to obtain a more continuous axonal tract segmentation, as shown in Fig. 7 (b). A visual inspection on the overlaid segmentation and the original volume at two imaging depths revealed that the automatic segmentation reliably identified the majority of the fiber tracts, as shown in Fig.7 (c). We then calculated the total number of segments from the Frangi filtered segmentation and the marching ellipsoid segmentation. We found that of the Frangi filtered segmentation produced 808 tracts while the marching ellipsoid processing reduced this number to 637 by connecting the fragmented tracts in the original Frangi segmentation, as shown in Fig. 7 (d). It was also noticed that the marching ellipsoid method could introduce some artificially connected fibers and pruning strategies need to be investigated for a more robust fiber tracing. To quantify the results we obtained with the marching ellipsoid method, we adopted the percentage missing segments and the percentage false segments as measures from a previous study [50]. The percentage missing segments was defined as the ratio of the number of fiber tract segments not extracted to the total number of segments seen in the original OCT images, whereas the percentage false segments was defined as the ratio of the number of false connections to the total number of segments seen in the original OCT images. We asked a human grader who is naïve to the dataset to manually inspected side by side the segmentation and the original image to count the total number of false connections in this ROI. The human grader reported 66 missing segments and 21 false connections out of the total of 837 segments of the ROI, which corresponded to a percentage missing segments of 7.9% and a percentage false segments of 2.5%. This is much better compared with a previous study by Kulkarni et al. where they reported the percentage missing segments and the percentage false segments using marching ellipsoid method to be 20.8% and 9.0%, respectively, in tracing astrocyte in confocal images.
Figure 7:
Extending the Frangi filtering strategy to extract axonal fiber tracts. (a) The AIP of a selected slice. The region of interest is labeled with red rectangle. Scale bar: 500 μm. (b) 3D rendering of the axonal fiber tracts segmentation in two different perspectives. (c) The overlay of the segmentation and the original image. Two planes of depth (60 μm and 120 μm) were shown. Red is the binary segmentation, and the gray is original image. Scale bars: 100 μm. (d) Comparison of the Frangi filtering (left) and the Frangi filtering plus marching ellipsoid (right). White arrows point to the segments that were connected after the marching ellipsoid. Scale bars: 100 μm.
IV. DISCUSSION
Integrating serial sectioning with OCT enabled volumetric imaging of large tissue blocks up to several centimeters [16, 18]. In this study, we implemented a data processing pipeline to advance the serial sectioning OCT techniques. We removed the stitching artifacts with a distortion correction algorithm and created the depth-resolved attenuation coefficient maps to stitch the slices in 3D. This resulted in a better visualization of the volume, as the discontinuity between slices was significantly suppressed compared to the OCT intensity images, and the quality of feature visualization was improved (Fig. 2). The residual discontinuity was caused by a bias of elevated attenuation coefficient estimation at deeper regions. The attenuation coefficient was approximated based the assumption of infinite pixels in depth [42]. However, due to the heavy burden of data streaming during acquisition, each image tile was trimmed to 200 pixels in depth, which was insufficient for OCT signals to decay to the noise floor.
The slice thickness of 150 μm was chosen based on the optimal performance of OCT. The OCT objective has a confocal parameter of ~150 μm and the effective imaging depth in ex vivo human brain tissue covers 150 μm. Therefore, we cut a 150 μm slice from tissue blockface to remove the portion that has been imaged and allow OCT to image deeper regions of the tissue block.
With the volumetric OCT data, we identified the vessel network in the human brain samples. The signal we are detecting is the “black tubes” in OCT which corresponds to the lumen of the vessel. Since the lumen is empty and hollow, its scattering coefficient is very distinct from surrounding tissue and would not be affected by the diameter or orientation of the vessel. We utilized a Frangi filter to automatically segment the vessels in 3D. This algorithm was widely used to extract vessels in different imaging techniques such as cortical vasculature from magnetic resonance imaging (MRI) and magnetic resonance angiography (MRA), bronchi from a chest CT, and portal veins in the liver from an abdominal CT [45, 46].
With a histological validation, we showed that the Frangi filter-based segmentation had a high accuracy in extracting vessels that are larger than 20 microns in diameter in the human brain samples imaged with serial sectioning OCT. Converting the vessel segmentation into a graph enabled us to characterize the geometrical properties of the vessel network, such as the vessel segment length and tortuosity. By comparing the geometrical parameters extracted by automatic vessel segmentation with manual segmentation, we confirmed that the Frangi filtering strategy could accurately reveal the network properties of the vasculature. This segmentation and graph-building framework can be applied to study the pathology of diseased brains. For example, neurodegenerative disease such as Alzheimer’s disease and chronic traumatic encephalopathy are known to be accompanied by vascular abnormities, such as increasing number of spiraling vessels and sparser vessel network [4, 10]. This could be reflected by the vessel density change and the increased tortuosity of vessels in the network, which can be readily quantified within our processing framework. Additionally, since OCT is a label-free imaging technique, it does not suffer from the variations introduced during molecular labeling and tissue sectioning thickness as in traditional histological methods [1]. Therefore, OCT-based techniques enable a quantitative comparison across subjects and large-scale sample size.
One big advantage of OCT-based technique is that it removes the challenge of tissue distortions and tears that are inevitable in histological sections of human tissue. As traditional histological imaging is performed on thin slices, a significant distortion is introduced during tissue slicing and mounting thin slice on glass slide [51, 52], which results in inevitable warping and tears in each slice. Those slice-specific and nonlinear distortions are difficult to correct in volumetric registration across slices. In serial sectioning OCT, imaging is performed on the block face of the tissue and prior to tissue sectioning. Therefore, it preserves the spatial information of the tissue block and eliminates the distortion across slices in volumetric registration. The distortion we described in the OCT was introduced by the optics system. This type of distortion can be well characterized and corrected by the method described in section II.D. Tissue deformation introduced during fixation steps is another type of distortion that is common for all ex vivo studies [53, 54].
We used tomato-lectin labeled confocal imaging to validate the OCT vasculature. Lectins are small-molecule carbohydrate-binding proteins, distinct from antibodies, enzymes, and transport proteins, which makes them easily tagged fluorescently or with biotin [55]. They have been used to label vascular elements in fixed tissue sections, through their binding to carbohydrate components of the endothelial plasmalemma [56–58]. Results reported thus far indicate wide variation in the efficacy of vascular labeling of different lectins, and that tomato lectin may provide the best labeling of central nervous system vasculature [57, 59–61]. Previous studies have widely used confocal microscopy in imaging histological slices stained with tomato lectin [62, 63], which is one of the most available imaging methods that identifies vasculature down to the single capillary level [64].
There are a few limitations in the vessel segmentation approach. First, previous studies suggested that it is possible for the vessels to collapse during tissue preparation/fixation steps [65, 66]. As a matter of fact, it is a common limitation for ex vivo studies and it could affect our detection technique. Specifically, if the lumen was partially collapsed, the diameter of the vessel could be inaccurately estimated. If the lumen was completely collapsed, the vessel would not be detectable by our technique. Second, since the Frangi filter involving eigendecomposition was computational expensive, we downsampled the large 3D volume before applying it for vessel segmentation. This limited the smallest vessels we were able to detect. In this work, vessel segmentations were performed on volumetric OCT data with 12 micron resolution, and thus all microvessels smaller than 2 pixels were neglected in our framework. From the confocal images we observed that the gray matter was more vascularized than the white matter due to a dense capillary network. However, in our OCT based vasculature map, the capillary bed was excluded, leading to an incomplete microvascular network, as shown in Fig. S2. An alternative strategy is to retain the original resolution while extracting vessels from a smaller region. Moreover, the sensitivity of the Frangi filter was not remarkably high, which resulted in missing vessel identification after the filtering process. This could be reflected by the smaller total number of segments from the Frangi filter than the manual segmentation, as shown in Fig 5 (b) and (c).
We imaged 3 human brain samples to prove the reliability of our approach; however, the geometrical properties of the vasculature were not comparable to evaluate the variability across the samples. Since the sample volume as well as the anatomical region were different, the difference in the total number of segments across the samples was probably attributed to those factors rather than pathological conditions.
Extracting axonal bundles is more challenging compared to the vessel identification, given that the contrast between the fibers and the surrounding medium is much weaker compared to the that between the lumen and the vessel walls. We demonstrated the ability of axonal bundle segmentation in volumetric OCT using the Frangi filtering at a different scale. The axonal bundle information is useful in studying brain connectivity. Future work could improve the quality of segmentation by incorporating deep learning based methods, such as convolutional neural networks [67, 68].
V. CONCLUSION
A novel framework of automatically constructing large-scale OCT volumes and extracting microvasculature was demonstrated. The OCT signal was distortion-corrected and transformed into an attenuation coefficient for better volumetric visualization. The extraction of vasculature in 3D was performed with a Frangi filtering strategy. We validated that this segmentation scheme was able to capture the geometrical characteristics of the intraparenchymal microvascular network. The Frangi filtering strategy could be further extended to extract other tubular structures such as axonal tracts with proper scale parameters. This framework enables us to quantitatively characterize the 3D microvascular networks in ex vivo human brain.
Acknowledgments
This work was submitted for review on January 12, 2021. This work was supported by NIH Brain Initiative U01MH117023, , the National Institute for Biomedical Imaging and Bioengineering (P41EB015896, 1R01EB023281, R01EB006758, R21EB018907, R01EB019956, P41EB030006, R00EB023993), the National Institute on Aging (1R56AG064027, 1R01AG064027, 5R01AG008122, R01AG016495, 1R01AG070988), the National Institute of Mental Health (R01 MH123195, R01MH121885, 1RF1MH123195), the National Institute for Neurological Disorders and Stroke (R01NS0525851, R21NS072652, R01NS070963, R01NS083534, 5U01NS086625, 5U24NS10059103, R01NS105820), the Eunice Kennedy Shriver National Institute of Child Health and Human Development (R21HD106038), the Chan-Zuckerberg Initiative DAF (2019-198101), and was made possible by the resources provided by Shared Instrumentation Grants 1S10RR023401, 1S10RR019307, and 1S10RR023043. Additional support was provided by the NIH Blueprint for Neuroscience Research (5U01-MH093765), part of the multi-institutional Human Connectome Project. In addition, BF has a financial interest in CorticoMetrics, a company whose medical pursuits focus on brain imaging and measurement technologies. BF’s interests were reviewed and are managed by Massachusetts General Hospital and Partners HealthCare in accordance with their conflict of interest policies.
Contributor Information
Jiarui Yang, Department of Biomedical Engineering, Boston University, USA..
Shuaibin Chang, Department of Electrical and Computer Engineering, Boston University, USA..
Ichun Anderson Chen, Department of Biomedical Engineering, Boston University, USA..
Sreekanth Kura, Department of Biomedical Engineering, Boston University, USA..
Grace A. Rosen, Department of Neurology, Boston University Medical Center, USA.
Nicole A. Saltiel, Department of Neurology, Boston University Medical Center, USA.
Bertrand R. Huber, Department of Neurology, Boston University Medical Center, USA.
Divya Varadarajan, Department of Radiology, Massachusetts General Hospital, USA..
Yael Balbastre, Department of Radiology, Massachusetts General Hospital, USA..
Caroline Magnain, Department of Radiology, Massachusetts General Hospital, USA..
Shih-chi Chen, Department of Mechanical Engineering, the Chinese University of Hong Kong, Hong Kong..
Bruce Fischl, Department of Radiology, Massachusetts General Hospital, USA; Health Science & Technology/Computer Science & Artificial Intelligence Laboratory, Massachusetts Institute of Technology, USA..
Ann C. McKee, Departments of Neurology and Pathology, Boston University School of Medicine, USA.
David A. Boas, Department of Biomedical Engineering, Boston University, USA.
Hui Wang, Department of Radiology, Massachusetts General Hospital, Boston, MA 02114 USA.
REFERENCES
- [1].Duvernoy HM, Delon S, and Vannson J, “Cortical blood vessels of the human brain,” Brain research bulletin, vol. 7, no. 5, pp. 519–579, 1981. [DOI] [PubMed] [Google Scholar]
- [2].Cassot F, Lauwers F, Lorthois S, Puwanarajah P, Cances-Lauwers V, and Duvernoy H, “Branching patterns for arterioles and venules of the human cerebral cortex,” Brain research, vol. 1313, pp. 62–78, 2010. [DOI] [PubMed] [Google Scholar]
- [3].Diaz-Otero JM, Garver H, Fink GD, Jackson WF, and Dorrance AM, “Aging is associated with changes to the biomechanical properties of the posterior cerebral artery and parenchymal arterioles,” American Journal of Physiology-Heart and Circulatory Physiology, vol. 310, no. 3, pp. H365–H375, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Attems J.and Jellinger KA, “The overlap between vascular disease and Alzheimer’s disease-lessons from pathology,” BMC medicine, vol. 12, no. 1, pp. 1–12, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].BRETELER MM, “Vascular involvement in cognitive decline and dementia: epidemiologic evidence from the Rotterdam Study and the Rotterdam Scan Study,” Annals of the New York Academy of Sciences, vol. 903, no. 1, pp. 457–465, 2000. [DOI] [PubMed] [Google Scholar]
- [6].Hofman A.et al. , “Atherosclerosis, apolipoprotein E, and prevalence of dementia and Alzheimer’s disease in the Rotterdam Study,” The Lancet, vol. 349, no. 9046, pp. 151–154, 1997. [DOI] [PubMed] [Google Scholar]
- [7].Farkas E, De Vos RA, Steur ENJ, and Luiten PG, “Are Alzheimer’s disease, hypertension, and cerebrocapillary damage related?☆,” Neurobiology of aging, vol. 21, no. 2, pp. 235–243, 2000. [DOI] [PubMed] [Google Scholar]
- [8].Bullitt E.et al. , “The effects of healthy aging on intracerebral blood vessels visualized by magnetic resonance angiography,” Neurobiology of aging, vol. 31, no. 2, pp. 290–300, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Thore CR, Anstrom JA, Moody DM, Challa VR, Marion MC, and Brown WR, “Morphometric analysis of arteriolar tortuosity in human cerebral white matter of preterm, young, and aged subjects,” Journal of neuropathology and experimental neurology, vol. 66, no. 5, pp. 337–345, 2007. [DOI] [PubMed] [Google Scholar]
- [10].Fischer V, Siddiqi A, and Yusufaly Y, “Altered angioarchitecture in selected areas of brains with Alzheimer’s disease,” Acta neuropathologica, vol. 79, no. 6, pp. 672–679, 1990. [DOI] [PubMed] [Google Scholar]
- [11].Ambrose CT, “Neuroangiogenesis: a vascular basis for Alzheimer’s disease and cognitive decline during aging,” Journal of Alzheimer’s Disease, vol. 32, no. 3, pp. 773–788, 2012. [DOI] [PubMed] [Google Scholar]
- [12].Farkas E.and Luiten PG, “Cerebral microvascular pathology in aging and Alzheimer’s disease,” Progress in neurobiology, vol. 64, no. 6, pp. 575–611, 2001. [DOI] [PubMed] [Google Scholar]
- [13].Huang D.et al. , “Optical coherence tomography,” science, vol. 254, no. 5035, pp. 1178–1181, 1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Wang H.et al. , “Polarization sensitive optical coherence microscopy for brain imaging,” Optics letters, vol. 41, no. 10, pp. 2213–2216, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Wang H.et al. , “Reconstructing micrometer-scale fiber pathways in the brain: multi-contrast optical coherence tomography based tractography,” Neuroimage, vol. 58, no. 4, pp. 984–992, 2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Wang H, Zhu J, and Akkin T, “Serial optical coherence scanner for large-scale brain imaging at microscopic resolution,” Neuroimage, vol. 84, pp. 1007–1017, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu CJ, Williams KE, Orr HT, and Akkin T, “Visualizing and mapping the cerebellum with serial optical coherence scanner,” Neurophotonics, vol. 4, no. 1, p. 011006, 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Wang H.et al. , “as-PSOCT: Volumetric microscopic imaging of human brain architecture and connectivity,” NeuroImage, vol. 165, pp. 56–68, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Srinivasan VJ, Radhakrishnan H, Jiang JY, Barry S, and Cable AE, “Optical coherence microscopy for deep tissue imaging of the cerebral cortex with intrinsic contrast,” Optics express, vol. 20, no. 3, pp. 2220–2239, 2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Magnain C.et al. , “Optical coherence tomography visualizes neurons in human entorhinal cortex,” Neurophotonics, vol. 2, no. 1, p. 015004, 2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Magnain C.et al. , “Blockface histology with optical coherence tomography: a comparison with Nissl staining,” NeuroImage, vol. 84, pp. 524–533, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Böhringer H, Lankenau E, Stellmacher F, Reusche E, Hüttmann G, and Giese A, “Imaging of human brain tumor tissue by near-infrared laser coherence tomography,” Acta neurochirurgica, vol. 151, no. 5, pp. 507–517, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Böhringer H.et al. , “Time‐domain and spectral‐domain optical coherence tomography in the analysis of brain tumor tissue,” Lasers in Surgery and Medicine: The Official Journal of the American Society for Laser Medicine and Surgery, vol. 38, no. 6, pp. 588–597, 2006. [DOI] [PubMed] [Google Scholar]
- [24].Assayag O.et al. , “Imaging of non-tumorous and tumorous human brain tissues with full-field optical coherence tomography,” NeuroImage: clinical, vol. 2, pp. 549–557, 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Nakaji H, Kouyama N, Muragaki Y, Kawakami Y, and Iseki H, “Localization of nerve fiber bundles by polarization-sensitive optical coherence tomography,” Journal of neuroscience methods, vol. 174, no. 1, pp. 82–90, 2008. [DOI] [PubMed] [Google Scholar]
- [26].Wang H.et al. , “Cross-validation of serial optical coherence scanning and diffusion tensor imaging: a study on neural fiber maps in human medulla oblongata,” Neuroimage, vol. 100, pp. 395–404, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Golde J.et al. , “Detection of carious lesions utilizing depolarization imaging by polarization sensitive optical coherence tomography,” Journal of biomedical optics, vol. 23, no. 7, p. 071203, 2018. [DOI] [PubMed] [Google Scholar]
- [28].Wojtkowski M, Srinivasan VJ, Ko TH, Fujimoto JG, Kowalczyk A, and Duker JS, “Ultrahigh-resolution, high-speed, Fourier domain optical coherence tomography and methods for dispersion compensation,” Optics express, vol. 12, no. 11, pp. 2404–2422, 2004. [DOI] [PubMed] [Google Scholar]
- [29].De Boer JF, Cense B, Park BH, Pierce MC, Tearney GJ, and Bouma BE, “Improved signal-to-noise ratio in spectral-domain compared with time-domain optical coherence tomography,” Optics letters, vol. 28, no. 21, pp. 2067–2069, 2003. [DOI] [PubMed] [Google Scholar]
- [30].Yang J.et al. , “Improving the characterization of ex vivo human brain optical properties using high numerical aperture optical coherence tomography by spatially constraining the confocal parameters,” Neurophotonics, vol. 7, no. 4, p. 045005, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Wang J, Li C, and Chen S-C, “Sectioning soft materials with an oscillating blade,” Precision Engineering, vol. 56, pp. 96–100, 2019. [Google Scholar]
- [32].Costantini I.et al. , “A multimodal imaging and analysis pipeline for creating a cellular census of the human cerebral cortex,” bioRxiv, 2021. [Google Scholar]
- [33].Park Y-G et al. , “Protection of tissue physicochemical properties using polyfunctional crosslinkers,” Nature biotechnology, vol. 37, no. 1, pp. 73–83, 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Ku T.et al. , “Elasticizing tissues for reversible shape transformation and accelerated molecular labeling,” Nature Methods, pp. 1–5, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Graf BW, Adie SG, and Boppart SA, “Correction of coherence gate curvature in high numerical aperture optical coherence imaging,” Optics letters, vol. 35, no. 18, pp. 3120–3122, 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Arganda-Carreras I, Sorzano CO, Marabini R, Carazo JM, Ortiz-de-Solorzano C, and Kybic J, “Consistent and elastic registration of histological sections using vector-spline regularization,” in International Workshop on Computer Vision Approaches to Medical Image Analysis, 2006, pp. 85–95: Springer. [Google Scholar]
- [37].Sorzano COS, Thévenaz P, and Unser M, “Elastic registration of biological images using vector-spline regularization,” IEEE Transactions on Biomedical Engineering, vol. 52, no. 4, pp. 652–663, 2005. [DOI] [PubMed] [Google Scholar]
- [38].Peng T.et al. , “A BaSiC tool for background and shading correction of optical microscopy images,” Nature communications, vol. 8, no. 1, pp. 1–7, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Lefebvre J, Castonguay A, Pouliot P, Descoteaux M, and Lesage F, “Whole mouse brain imaging using optical coherence tomography: reconstruction, normalization, segmentation, and comparison with diffusion MRI,” Neurophotonics, vol. 4, no. 4, p. 041501, 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Canny J, “A computational approach to edge detection,” IEEE Transactions on pattern analysis and machine intelligence, no. 6, pp. 679–698, 1986. [PubMed] [Google Scholar]
- [41].Preibisch S, Saalfeld S, and Tomancak P, “Globally optimal stitching of tiled 3D microscopic image acquisitions,” Bioinformatics, vol. 25, no. 11, pp. 1463–1465, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Vermeer K, Mo J, Weda J, Lemij H, and De Boer J, “Depth-resolved model-based reconstruction of attenuation coefficients in optical coherence tomography,” Biomedical optics express, vol. 5, no. 1, pp. 322–337, 2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Haralick RM, “Ridges and valleys on digital images,” Computer Vision, Graphics, and Image Processing, vol. 22, no. 1, pp. 28–38, 1983. [Google Scholar]
- [44].Koller TM, Gerig G, Szekely G, and Dettwiler D, “Multiscale detection of curvilinear structures in 2-D and 3-D image data,” in Proceedings of IEEE International Conference on Computer Vision, 1995, pp. 864–869: IEEE. [Google Scholar]
- [45].Sato Y.et al. , “3D multi-scale line filter for segmentation and visualization of curvilinear structures in medical images,” in CVRMed-MRCAS’97, 1997, pp. 213–222: Springer. [DOI] [PubMed] [Google Scholar]
- [46].Sato Y.et al. , “Three-dimensional multi-scale line filter for segmentation and visualization of curvilinear structures in medical images,” Medical image analysis, vol. 2, no. 2, pp. 143–168, 1998. [DOI] [PubMed] [Google Scholar]
- [47].Lorenz C, Carlsen I-C, Buzug TM, Fassnacht C, and Weese J, “Multi-scale line segmentation with automatic estimation of width, contrast and tangential direction in 2D and 3D medical images,” in CVRMed-MRCAS’97, 1997, pp. 233–242: Springer. [Google Scholar]
- [48].Lowe DG, “Distinctive image features from scale-invariant keypoints,” International journal of computer vision, vol. 60, no. 2, pp. 91–110, 2004. [Google Scholar]
- [49].Mukherjee A, Methods for Automated Analysis of Curvilinear Structures in Three-Dimensional and Spatio-temporal Microscopy Images using Orientation Distribution Functions. Rensselaer Polytechnic Institute, 2011. [Google Scholar]
- [50].Kulkarni PM et al. , “Quantitative 3-D analysis of GFAP labeled astrocytes from fluorescence confocal images,” Journal of neuroscience methods, vol. 246, pp. 38–51, 2015. [DOI] [PubMed] [Google Scholar]
- [51].Ju T.et al. , “3D volume reconstruction of a mouse brain from histological sections using warp filtering,” Journal of Neuroscience Methods, vol. 156, no. 1–2, pp. 84–100, 2006. [DOI] [PubMed] [Google Scholar]
- [52].Lowder ML, Li S, Carnell PH, and Vito RP, “Correction of distortion of histologic sections of arteries,” Journal of Biomechanics, vol. 40, no. 2, pp. 445–450, 2007. [DOI] [PubMed] [Google Scholar]
- [53].Doughty M, Bergmanson J, and Blocker Y, “Shrinkage and distortion of the rabbit corneal endothelial cell mosaic caused by a high osmolality glutaraldehyde-formaldehyde fixative compared to glutaraldehyde,” Tissue and Cell, vol. 29, no. 5, pp. 533–547, 1997. [DOI] [PubMed] [Google Scholar]
- [54].Weisbecker V, “Distortion in formalin-fixed brains: using geometric morphometrics to quantify the worst-case scenario in mice,” Brain Structure and Function, vol. 217, no. 2, pp. 677–685, 2012. [DOI] [PubMed] [Google Scholar]
- [55].Robertson RT et al. , “Use of labeled tomato lectin for imaging vasculature structures,” Histochemistry and cell biology, vol. 143, no. 2, pp. 225–234, 2015. [DOI] [PubMed] [Google Scholar]
- [56].Nag S, “Ultrastructural localization of lectin receptors on cerebral endothelium,” Acta Neuropathologica, vol. 66, no. 2, pp. 105–110, 1985. [DOI] [PubMed] [Google Scholar]
- [57].Jilani SM, Murphy TJ, Thai SN, Eichmann A, Alva JA, and Iruela-Arispe ML, “Selective binding of lectins to embryonic chicken vasculature,” Journal of Histochemistry & Cytochemistry, vol. 51, no. 5, pp. 597–604, 2003. [DOI] [PubMed] [Google Scholar]
- [58].Baratta JL, Ngo A, Lopez B, Kasabwalla N, Longmuir KJ, and Robertson RT, “Cellular organization of normal mouse liver: a histological, quantitative immunocytochemical, and fine structural analysis,” Histochemistry and cell biology, vol. 131, no. 6, pp. 713–726, 2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Debbage PL, Griebel J, Ried M, Gneiting T, DeVries A, and Hutzler P, “Lectin intravital perfusion studies in tumor-bearing mice: micrometer-resolution, wide-area mapping of microvascular labeling, distinguishing efficiently and inefficiently perfused microregions in the tumor,” Journal of Histochemistry & Cytochemistry, vol. 46, no. 5, pp. 627–639, 1998. [DOI] [PubMed] [Google Scholar]
- [60].Lokmic Z.and Mitchell GM, “Visualisation and stereological assessment of blood and lymphatic vessels,” Histology ang histopathology, Vol. 26, nº6 (2011), 2011. [DOI] [PubMed] [Google Scholar]
- [61].Smolkova O, Zavadka A, Bankston P, and Lutsyk A, “Cellular heterogeneity of rat vascular endothelium as detected by HPA and GSI lectin-gold probes,” Medical Science Monitor, vol. 7, no. 4, pp. BR659-BR668, 2001. [PubMed] [Google Scholar]
- [62].Dickie R.et al. , “Three-dimensional visualization of microvessel architecture of whole-mount tissue by confocal microscopy,” Microvascular research, vol. 72, no. 1–2, pp. 20–26, 2006. [DOI] [PubMed] [Google Scholar]
- [63].Jiao C, Adler K, Liu X, Sun W, Mullins RF, and Sohn EH, “Visualization of mouse choroidal and retinal vasculature using fluorescent tomato lectin perfusion,” Translational vision science & technology, vol. 9, no. 1, pp. 1–1, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Amos WB, White J, and Fordham M, “Use of confocal imaging in the study of biological structures,” Applied optics, vol. 26, no. 16, pp. 3239–3243, 1987. [DOI] [PubMed] [Google Scholar]
- [65].Feneley M.and Burton G, “Villous composition and membrane thickness in the human placenta at term: a stereological study using unbiased estimators and optimal fixation techniques,” Placenta, vol. 12, no. 2, pp. 131–142, 1991. [DOI] [PubMed] [Google Scholar]
- [66].Schwarzmaiera SM, Knarr MR, Hu S, Ertürk A, Hellal F, and Plesnila N, “Perfusion pressure determines vascular integrity and histomorphological quality following perfusion fixation of the brain,” Journal of Neuroscience Methods, p. 109493, 2022. [DOI] [PubMed] [Google Scholar]
- [67].Zaimi A, Wabartha M, Herman V, Antonsanti P-L, Perone CS, and Cohen-Adad J, “AxonDeepSeg: automatic axon and myelin segmentation from microscopy data using convolutional neural networks,” Scientific reports, vol. 8, no. 1, pp. 1–11, 2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Gupta V, Thomopoulos SI, Corbin CK, Rashid F, and Thompson PM, “Fibernet 2.0: an automatic neural network based tool for clustering white matter fibers in the brain,” in 2018 IEEE 15th International Symposium on Biomedical Imaging (ISBI 2018), 2018, pp. 708–711: IEEE. [Google Scholar]