Abstract
There is widespread recognition of the need to increase research opportunities in biomedical science for undergraduate students from underrepresented backgrounds. Here, we describe the implementation of team-based science combined with intensive mentoring to conduct a large-scale project examining the evolution of behavior. This system can be widely applied in other areas of STEM to promote research-intensive opportunities in STEM fields and to promote diversity in science.
Keywords: CURE, evolutionary genetics, research, undergraduate education
At many universities, undergraduate access to research in STEM fields is limited, and only a fraction of students are engaged in undergraduate research. Course-based undergraduate research experiences (CURE) have the potential to provide unique exposure to research for students who may otherwise not have opportunities to participate in undergraduate research experiences (Bangera & Brownell, 2014; Quick & Cisneros, 2022). While CURE classes have historically been applied exclusively to provide undergraduates with research experience, there are an increasing number of examples when they are used for research projects that yield publishable results (Sun et al., 2020; Olson et al., 2019). In this issue, Oliva et al. describe results from a CURE course specifically designed to provide students an opportunity to participate in research in a team-based science setting, to provide support networks for students, and to generate publication-quality scientific data (Oliva et al., 2022).
To do so, we developed a hybrid between a CURE course and classic undergraduate research project, allowing students to perform mentored undergraduate research in a classroom setting. Briefly, 10 students supported by an Undergraduate Research Training Initiative for Student Enhancement (URISE) award from the National Institutes of Health were paired in research groups consisting of two undergraduates and a peer mentor (graduate student or postdoctoral fellow). These research teams then functioned as units with a defined research focus on a component of a larger project (Figure 1). This system was designed to foster interactions between undergraduate students and mentors. In doing so, the students developed a support network centered around their interest in the project.
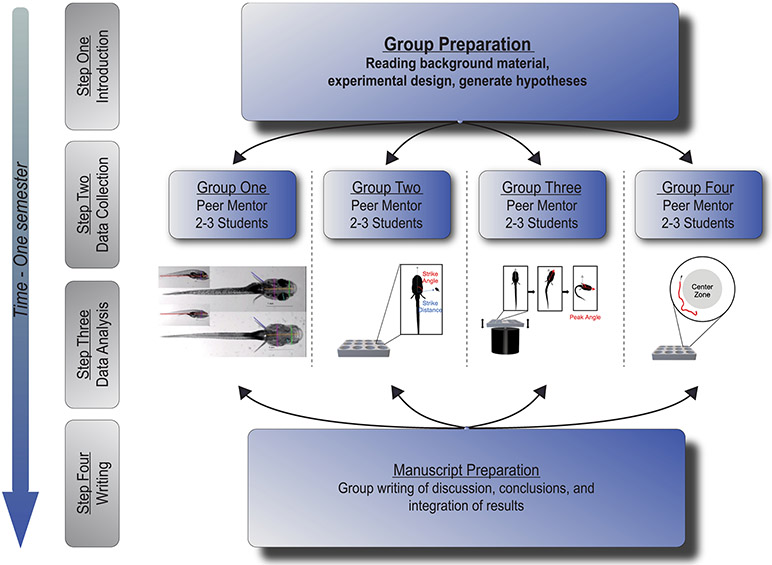
FIGURE 1.
Schematic depicting the course organization. The course took place in four steps, over the course of two semesters. In Step 1, the group met to develop a background in the field and make decisions regarding experimental design. Steps 2 and 3 consisted of small group data collection and analysis with the support of near-peer mentors. Step 4 involved group writing and data presentation.
The project was structured around a scientific question and developed with two factors that were critical to ensuring the success over the course time frame. First, the scientific question was descriptive, so that finite experiments could be planned from the start of the project. Second, the project was divided into four distinct yet complementary parts, so that each group could be independently responsible for each portion of the analysis. Third, each group of students was assigned a near-peer mentor who was a senior graduate student or postdoctoral fellow with extensive expertise related to the project. In Oliva et al. (2022), the project examined how evolutionarily derived traits segregate in the Mexican cavefish, a commonly used model to study convergent evolution (McGaugh et al., 2020). River-dwelling surface fish were crossed with fish from a cavefish population of Astyanax mexicanus to generate cave-surface F2 hybrids. The project sought to perform a detailed analysis of morphological and behavioral traits in these hybrid fish to determine whether traits are regulated by shared, or independent genetic architecture. This is a question that had not been explored for many of the traits that were investigated, providing novelty and the potential of discovery for the students.
Each group of students focused on a distinct set of traits: morphological traits, prey capture, startle response, and locomotor activity, all of which have been shown to differ between surface fish and cavefish. This required the undergraduates to not only learn the assays but also perform quantitative analysis of the data over the course of a single semester. Pairing students with an individual mentor who ran experiments along with them, and assisted with data analysis, gave students the intensive feedback needed to gain expertise over a short period of time. All the students involved in the project met on a weekly basis to discuss their progress on the project, as well as to participate in a journal club aimed at developing a background in the field of evolutionary genetics that would allow them to interpret the findings.
Within one semester students had developed a sufficient understanding of the field to discuss primary literature, mastered their assays and analysis, and collected the majority of data needed for the project. The second semester was largely focused on data analysis, presentation, and manuscript preparation. This involved each group writing the introduction and methods associated with their portion of the project and receiving intensive feedback, followed by group discussion. This feedback included a discussion of how results are interpreted, data presentation, the order of the manuscript, and writing style. In all cases, feedback came from the PIs and mentors and was discussed with the entire group. This specifically served the purpose of guiding students through the process of manuscript preparation. At the conclusion of the second semester, students had taken an idea from conception to the generation of a fully formed manuscript.
Students developed several unique skills that are not only essential to cutting-edge science but also ones that will train them as they move into graduate programs. First, students learned how to identify a scientific question and devise hypotheses that could be tested to address that question. A critical aspect leading to the success of this goal was that the experimental design was formulated in a group with the students. Second, students learned how to collaborate with one another, and how to work in groups. Lastly, and most importantly, students learned how to perform cutting-edge research as well as the statistical analyses that followed. Because the results were submitted and published, students learned about manuscript preparation and submission, as well as the peer-review process. These skills are those that are not always obtained at the undergraduate level and undoubtedly gave a preview into the world of science that undergraduate students may otherwise not get.
As part of the URISE program, following the completion of this course and manuscript submission, all students were then placed in research labs where they are currently engaged in independent projects. These projects and interests were in many ways shaped by the course, and several students took on projects evaluating different aspects of behavior. Other students were particularly attracted to the data analysis side, and are currently working in biocomputational labs doing cutting-edge research on molecule modeling. This course was also instrumental to the development of students' resumes, underscored by one student being the first recipient of a Barry Goldwater award at her host institution. Moreover, all students received summer internships at prestigious universities. Thus, this course, both in the concepts that it taught and the manuscript that was produced was an important factor in students reaching these goals.
This approach has a number of aspects that can be expanded upon in the future. First, the mentored, team-based approach could be expanded to include formal training on mentoring for the graduate student and postdoctoral fellows who participate in experiences such as these. While it is well recognized that being an effective mentor has a number of positive impacts, many academic scientists receive little training in mentoring (Hund et al., 2018). Thus, pairing a program with multiple participating mentors provide an opportunity for formally training mentors in best mentoring practices (Stelter et al., 2021). Further, in settings where a year-long program is not practical, alternative ways to obtain publishable results could be used. For example, there are now journals that publish short manuscripts on novel findings that have been effectively used to publish undergraduate research (McCullough et al., 2021). With the increasing implementation of CUREs, there is the opportunity to design these courses such that the publishable unit(s) at the end of the course are smaller, and thus fit more effectively into a semester-long course setting.
In conclusion, CURE classes have the potential to expand undergraduate research opportunities, and have an enormous impact on student trajectory. The positive outcomes for students of this program suggest that it should be replicated in the future. In particular, we advocate for a year-long set of courses that will allow students time to both participate in research and write a manuscript in class-based settings, something that would have been difficult to achieve in a single semester. Expanding research opportunities for undergraduates through CURES or alternative mechanisms will be critical to the diversity of STEM fields.
DATA AVAILABILITY STATEMENT
The data that support the findings of this study are openly available in re3data at https://www.re3data.org.
REFERENCES
- Bangera G, & Brownell SE (2014). Course-based undergraduate research experiences can make scientific research more inclusive. CBE—Life Sciences Education, 13(4), 573–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Call GB, Olson JM, Chen J, Villarasa N, Ngo KT, Yabroff AM, Cokus S, Pellegrini M, Bibikova E, Bui C, Cespedes A, Chan C, Chan S, Cheema AK, Chhabra A, Chitsazzadeh V, Do M-T, Fang QA, Folick A, … Goodstein GL Genetics education innovations in teaching and learning genetics genomewide clonal analysis of lethal mutations in the Drosophila melanogaster eye: Comparison of the X chromosome and autosomes. Genetics, 177(2), 689–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hund AK, Churchill AC, Faist AM, Havrilla CA, Stowell SML, McCreery HF, Ng G, Pinzone CA, & Scordato ESC (2018). Transforming mentorship in STEM by training scientists to be better leaders. Ecology and Evolution, 8(20), 9962–9974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough J, Fey P, Rahman RJ, Wallace M, Morey S, Sahlberg M, McGonagle E, Hess D, Hatfield C, Sarmiento M-R, Velasquez J, & Gomer RH (2021). Annotating putative D. discoideum proteins using I-TASSER. MicroPublication Biology, 2021. 10.17912/micropub.biology.000420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGaugh SE, Kowalko JE, Duboué E, Lewis P, Franz-Odendaal TA, Rohner N, Gross JB, & Keene AC (2020). Dark world rises: The emergence of cavefish as a model for the study of evolution, development, behavior, and disease. Journal of Experimental Zoology Part B: Molecular and Developmental Evolution, 334, 397–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliva C, Hinz NK, Robinson W, Barrett Thompson AM, Booth J, Crisostomo LM, Zanineli S, Tanner M, Lloyd E, O'Gorman M, McDole B, Paz A, Kozol R, Brown EB, Kowalko JE, Fily Y, Duboue ER, & Keene AC (2022). Characterizing the genetic basis of trait evolution in the Mexican cavefish. Evolution & Development, 1–14. 10.1111/ede.12412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson JM, Evans CJ, Ngo KT, Jong Kim H, Duy Nguyen J, Gurley KGH, Ta T, Patel V, Han L, Truong-N KT, Liang L, Chu MK, Lam H, Ahn HG, Banerjee AK, Choi IY, Kelley RG, Moridzadeh N, Khan AM, … Johnson EB (2019). Expression-based cell lineage analysis in Drosophila through a course-based research experience for early undergraduates. G3, 9(11), 3791–3800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quick CM, & Cisneros MR (2022). Vertically-integrated course-based undergraduate research experiences (CUREs) structure a biomedical research certificate program that promotes inclusivity. The FASEB Journal, 36(Suppl 1). [Google Scholar]
- Stelter RL, Kupersmidt JB, & Stump KN (2021). Establishing effective STEM mentoring relationships through mentor training. Annals of the New York Academy of Sciences, 1483, 224–243. [DOI] [PubMed] [Google Scholar]
- Sun E, Graves ML, & Oliver DC (2020). Propelling a course-based undergraduate research experience using an open-access online undergraduate research journal. Hypothesis and Theory, 11, 589025. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The data that support the findings of this study are openly available in re3data at https://www.re3data.org.