Abstract
Aims:
Macrophages are crucial for the initiation and resolution of an inflammatory response. Non-coding circular RNAs are ubiquitously expressed in mammalian tissue, highly conserved among species, and recently implicated in the regulation of macrophage activation. We sought to determine whether circRNAs modulate monocyte/macrophage biology and function.
Materials and Methods:
We performed circRNA microarray analyses to assess transcriptome changes using RNA isolated from bone marrow derived macrophages polarized to a pro-inflammatory phenotype (INFγ + TNFα) or an anti-inflammatory phenotype (IL-10, IL-4, and TGF-β). Among differentially expressed circRNAs, circ-Cdr1as was chosen for further investigation. Additionally, we performed loss or gain of function studies to investigate if circ-Cdr1as is involved in phenotypic switching. For gain of function, we overexpressed circ-Cdr1as using pc3.1 plasmid with laccase2 flanking regions to promote circularization. For loss of function, we used a lentiviral short hairpin RNA targeting the circ-Cdr1as splicing junction.
Key findings:
Among circRNAs that are highly conserved and differentially expressed in pro- and anti-inflammatory lineages, circ-Cdr1as was one of the most downregulated in pro-inflammatory macrophages and significantly upregulated in anti-inflammatory macrophages in vitro. Overexpression of circ-Cdr1as increased transcription of anti-inflammatory markers and percentage of CD206+ cells in naïve and pro-inflammatory macrophages in vitro. Meanwhile, knockdown decreased transcription of anti-inflammatory markers and increased the percentage of CD86+ cells in naïve and anti-inflammatory macrophages in vitro.
Significance:
This study suggests that circ-Cdr1as plays a key role in regulating anti-inflammatory phenotype of macrophages and may potentially be developed as an anti-inflammatory regulator in tissue inflammation.
Keywords: Circular RNA, Macrophage, Phenotypic switching, Microarray, Circ-Cdr1as
1. Introduction
Exacerbated and chronic inflammatory response is involved in numerous pathological diseases including cancer, cardiovascular disease, type 2 diabetes, and inflammatory bowel disease [1–3]. Macrophages play important roles in the progression of inflammatory diseases controlling the initiation, maintenance, and resolution of the inflammatory response. Bone-marrow and tissue-resident macrophages exhibit the unique characteristic to phenotypically switch between a pro-and anti-inflammatory phenotype producing a sequential spectrum of cytokines, chemokines, and growth factors exerting both pro-and anti-inflammatory effects. Pro-inflammatory macrophages are referred to as classically activated macrophages, induced by IFNy, LPS, or TNFα [4]. When stimulated, they initiate pro-inflammatory responses, and secrete high levels of pro-inflammatory cytokines such as TNFα and interleukins (IL) IL-12, IL23, IL-1, and IL-6. During the wound healing stage, they then undergo dynamic changes to an anti-inflammatory, pro-reparative phenotype and secrete high levels of anti-inflammatory cytokines, including IL-10 and growth factors such as vascular endothelial grow factor (VEGF) [5–7]. However, the mechanisms involved in the regulation of macrophage differentiation, polarization, re-polarization, and activation remains unclear.
Given the importance of macrophages in pathological conditions, there have been attempts to develop therapeutics that reduce/deplete monocytes or impair monocyte function and mobilization. However, the unintended consequences were increased risk of infection and prolonging inflammation due to insufficient clearance of tissue debris and impaired tissue remodeling [5,8,9]. Therefore, targeting macrophage phenotypic switching to promote an anti-inflammatory phenotype could improve tissue remodeling and regeneration without these unintended consequences. Advances in high-throughput RNA sequencing (RNA-seq) allowed the identification of novel non-coding transcripts such as microRNAs (miRNA), long non-coding RNAs (lncRNAs), and circular RNAs (circRNA). Several studies identified circRNAs as regulators of biological processes, disease initiation, and disease progression. They have recently been suggested to play a role in immunity by contributing to the process of macrophage polarization, appropriate activation of macrophages when exposed to LPS, and inhibition of macrophage biogenesis [10–12]. CircRNAs originate from precursor mRNA (pre-mRNA) that is back-spliced to form a circular RNA molecule with a 3′-5′ phosphodiester bond at the junction site [13]. The lack of free terminals makes circRNA resistant to RNase R exoribonuclease degradation and allows for the enrichment of circular RNAs by RNase R treatment [1,14]. Previous literature identified the potential roles for circRNAs in regulating genes encoding inflammatory molecules. One study has shown that the delivery of purified exogenous circRNA stimulates a greater innate immune response than that stimulated by linear RNA with the same sequence [15]. Another investigation cataloged circRNA expression in macrophages and found induction of nearly 2000 circular RNAs following TLR4 stimulation. The authors found that circRNA circRasGEF1b is stably expressed, dependent on NF-kB, and needed for activation of macrophages in response to lipopolysaccharide [10–12]. Although these studies implicate circRNA in the involvement of macrophage activation, the role of circRNAs in macrophage phenotypic switching is still obscure.
In this study, we explored the role of circular RNAs in macrophage phenotypic switching. Overall, we provide a comprehensive profile of differentially expressed circular RNAs between pro–/anti-inflammatory macrophages and identify circular RNA Cdr1as as a key promoter of macrophage polarization to an anti-inflammatory phenotype.
2. Materials and methods
2.1. Bone Marrow cell isolation and Monocyte culture
Bone marrow (BM) monocytes were isolated from bone marrow mononuclear cells of C57BL/6 male (approximately 8–10-week old) mice by density-gradient centrifugation with histopaque-1083 (Sigma) and red blood cells were removed with NH4Cl (Stemcell Technologies Cat #07800) as previously described [16,17]. The monocyte population was cultured in RPMI 1640 1× (Gibco) with 20% FBS, 1% penicillin-streptomycin solution, and 20% of L929-conditioned medium. The media was changed the day after culture, on day 3, and on day 5–7. By day 5–7, high purity of macrophages can be observed as previously noted [18].
The L929 cell conditioned medium was collected as previously described [19]. Briefly, 2 × 106 L929 cells were seeded in T75 flask for 3–4 days until reaching confluency of 90% and conditioned media is collected and centrifuged at 3000 rpm, 4°C. The media is then filtered through a 0.45 μm filter and frozen at −80°C in 50 mL aliquots. All animal experiments complied with approved protocols of Temple University animal care and use guidelines.
2.2. Polarization of BMDM
On day 5–7, change to fresh stimulation media: for pro-inflammatory activation [6], 100 ng/mL IFNγ (R&D systems Cat# 485-MI-100) and 100 ng/mL TNFα (PromoKine Cat# D-63720); for anti-inflammatory activation [20], 10 ng/mL IL-4 (R&D systems Cat# 404-ML-010), 10 ng/mL IL-10 (R&D systems Cat# 217-IL-005), and 20 ng/mL TGF-β (R&D systems Cat#7666-MB-005) for a period of 24 h (Fig. 1A).
Fig. 1.
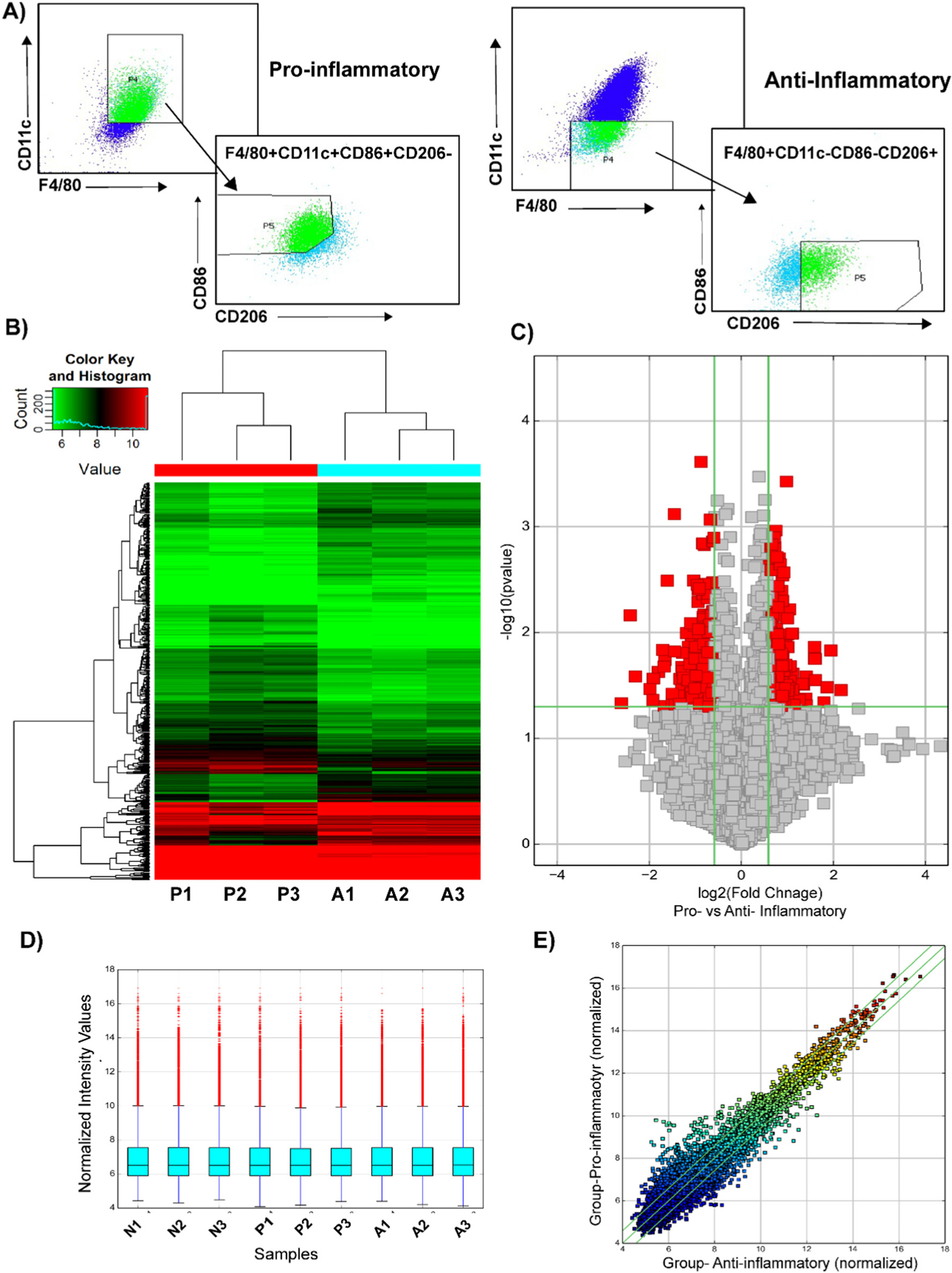
Microarray analysis of differentially expressed circular RNAs in bone marrow derived macrophages. (A) The BMDMs were sorted based on the following cell surface markers: pro-inflammatory- F4/80+, CD86+, CD11c+, CD206−; anti-inflammatory- F4/80+, CD86−, CD11c−, CD206+. Total RNA was isolated. (B) Heat map revealing differential circular RNA expression profiles between pro-inflammatory and anti-inflammatory macrophages. The red color indicates a higher FC value; green indicates a smaller FC value. (C) Volcano plot comparing significantly expressed circular RNA between pro-inflammatory and anti-inflammatory macrophages. The circular RNA expression log 2- transformed FC values (x-axis) were plotted against the log10 P values (on the y-axis). The red dots represent the circular RNAs having an FC value ≥1.5 and P < 0.05 comparing between pro-inflammatory and anti-inflammatory macrophages. (D) Box plot visualizing the distribution of a dataset for the circRNAs profiles. The distributions were nearly the same after normalization. (E) Scatter plot demonstrating the distributions of circular RNAs that are differentially expressed between pro-inflammatory and anti-inflammatory macrophages. The values of the x- and y-axes in the scatter plot were averaged to the normalized signal values of the group. FC, fold change; circRNA, circular RNA; N, naïve; P, pro-inflammatory; A-anti-inflammatory macrophages.
2.3. RNA isolation
Total RNA was isolated from cells using miRNeasy Mini Kit (Qiagen). For RNase R treatment, 500 ng was incubated 30 mins at 37°C with or without 2.5 U of Rnase R (Epicentre Technologies Cat# RNR07250).
2.4. Circular RNA microarray
CircRNA expression analysis was performed on total RNA extracted from naïve, pro-inflammatory, and anti-inflammatory macrophages 24 h after polarization (n = 3). Total RNA from each sample was quantified using the NanoDrop ND-1000. The sample preparation and microarray hybridization were performed based on the Arraystar’s standard protocols (Arraystar MD, USA). Briefly, total RNAs were digested with Rnase R (Epicentre, Inc.) to remove linear RNAs and enrich circular RNAs. Then, the enriched circular RNAs samples were then amplified and transcribed into fluorescent cRNA utilizing a random priming method (Arraystar Super RNA Labeling Kit; Arraystar) and hybridized onto the Arraystar Mouse circRNA Array V2 (8×15K, Arraystar). The slides were washed, and the arrays were scanned by the Agilent Scanner G2505C.
2.5. Microarray data analysis
To analyze acquired array images, Agilent Feature Extraction software (version 11.0.1.1) was used. The limma package from the R software package was used for quantile normalization and subsequent data processing. Differentially expressed circRNAs with statistical significance between two groups were identified through Volcano Plot filtering. Hierarchical Clustering was performed to show the distinguishable circRNAs expression pattern among samples. Differentially expressed circRNAs between two samples were identified through Fold Change filtering and differential analysis between two groups was performed by t-test. The cutoffs were FC ≥ 1.5 and p ≤ 0.05. Differentially expressed circular RNAs identified by microarray analysis were first uploaded into Qiagen’s IPA system for core analysis and then overlaid with the global molecular network in the Ingenuity Pathway Knowledge Base (IPKB). IPA was performed to identify canonical pathways, diseases, and gene networks that are most significant to microarray outcomes. The top 10 circular RNA were ranked by fold change and www.circbase.org was used for identification of human circular RNA sequences followed by pairwise alignment between human and murine circular RNA by blast NCBI as indicated in Table 1.
Table 1.
The top 10 upregulated and downregulated conserved circular RNAs in Bone marrow derived macrophages ranked by fold changes.
| circRNA ID | circRNA type | Chromosome | Source Gene | Fold Change | Padj Value | % homology of splicing junction (human vs mouse) | Regulation in BMDM |
|---|---|---|---|---|---|---|---|
|
| |||||||
| mmu_circRNA_005039 | Exonic | Chr3 | Elf2 | 10.4129583 | 0.031236023 | 81.1% | Upregulated in M1 |
| mmu_circRNA_005434 | Antisense | Chr8 | Zfhx3 | 3.4138622 | 0.019556071 | 94.4% | Downregulated in M1 |
| mmu_circRNA_010385 | Exonic | Chr1 | Gigyf2 | 2.7595952 | 0.05860264 | 92.0% | Upregulated in M1 |
| mmu_circRNA_001946 | antisense | ChrX | Cdr1 | 2.4249507 | 0.013344931 | 74.0% | Downregulated in M1 |
| mmu_circRNA_43784 | Exonic | Chr9 | Amotl1 | 1.9046831 | 0.017305584 | 84.0% | Downregulated in M1 |
| mmu_circ_0001136 | Intronic | Chr3 | Rsrc1 | 1.8513597 | 0.013099638 | 84.8% | Upregulated in M1 |
| mmu_circRNA_014660 | Exonic | Chr4 | Bnc2 | 1.7290235 | 0.037834294 | 90.7% | Downregulated in M1 |
| mmu_circ_0000612 | Sense overlapping |
Chr15 | Ptk2 | 1.673395 | 0.037898566 | 45.1% | Downregulated in M1 |
| mmu_circRNA_25157 | Exonic | Chr12 | Mipol1 | 1.5065144 | 0.04025052 | 88.7% | Upregulated in M2 |
| mmu_circ_0001160 | Exonic | Chr3 | Slc30a7 | 1.1896273 | 0.118923222 | 89.5% | Downregulated in M1 |
2.6. Generation of circ-Cdr1as overexpression plasmid
To generate a circ-Cdr1as expression plasmid, the previously described pcDNA3.1 (+) Lacasse2 MCS exon vector (Addgene plasmid # 69893; http://n2t.net/addgene:69893; RRID:Addgene_69893) [21]. This plasmid contains bacterial resistance to ampicillin and G418/neomycin resistance in eukaryotes. Expression of the circRNA is driven by a CMV promoter. Circ-Cdr1as (1263 bp) sequence gene fragment could not be synthesized as a double stranded DNA fragment due to its high complexity and so, it was cloned in pUC57 by EcoRV between EcoRI and HindIII sites (Genescript Project ID U8508GC140-2). The plasmid was then digested, the mature sequence fragment was purified by gel purification and subcloned into the above vector. Fragment insertion was verified by gel electrophoresis and sanger sequencing. The following sequence was inserted between PacI and SacII sites of pcDNA3.1 (+) Lacasse2 MCS exon vector (restriction enzyme sites are in lowercase):
tatgcagaaattaattaaGGTTTCCAGTGGTGCCAGTACCAAGGTCTTCCAACATCTCCAGGTCTTCCAGCAACTGCAAGTCTTCCAACACTGTCAAGGTCTTCCAGACAATCGTGATCTTCCAGAAAATATGTCTTCCAGATGATATATGTCTTCCAACAACCTCAGAGTCTTCCAGTAAATTCAGTCTTCCATGGTATCTAGATCTTCCGTGATATCTAGATCTTCTAGGAAAATCTGTGTCTTCCAGGAAAATCTTTGTCTTCCAAGGGATCCATGTCTTCCAGAAAAAAATCCACGTCTTCCAGAAAAATTCATGTCTTCCAGGAAAATCCATGTCTTCCAGAAAAATCCAAGTCTTCCAGAAGAATCTAAGTCTTCCTGAAAAATCCATGTCTTCCATAACAATCTACATCTTCCAGAATAATCAACGTCTTCCACAAAATTCAGGTCTTCCAGAAAAATATCCAGGTCTTCCAACCAGTCTATGTCTTCCAGAAAAATCCAGGTCTTCCAGCTCATCTATGTCTTCCATCAGTTCAAGTCTTCCAGATACCCATGTCTTCCATTAAATCTAGCCATGTCTTCCTGAAAATCCATGACTTCCCAAAAATCCAAATCTTCCTGAAAAATCCATATCTTCCCAAAAATCCACGACTTCCCAAAAATCCATGTCTTCGTGAATATCCACGTCTTCCTGAAAATCCAAGTCTTCCCAGAAATCCATGTCCTTCCAAAAATCCATGTCTTCCTGAAAATCTATGCCTTCCATAAATCTACATCTTCCTGAAAATCCACATCTTCCTGAAAATCTACATCTTCCTGAAAATCCACATCTTCCATAAATCCCAGTCTTCCCAGAAATCCAAGTCTTCTGGAAAATCCATGTCTTCCTGAAAATCTATGCCTTCCACAAATCCATGTCCTCCTAACAATCCAAGTCTTCCTAAAAATCTACGCCTTCCACAAATCTACATCTTCCTGAAAATCCACATCTTCCCAAAATCCATGTCTTCGGAAAAAATCCATGTCTTCCAAGGAGTACATGTGTCTTCCTTCACCTCCAAGTCTTCCAGCATCTCCAGGGCTTCCAGCATCTGCCTGTCTTCCAACATCTCCACATCTTCCAGCATCTTTATGTCTTCCAACAACTGCGCAGTGTCTCCAGTGTATCGGCGTTTTGACATTCAGGTTTTCTGGTGTCTGCCGTATCCAGccgcggagcgtaagtat.
2.7. BMDM cell and in vitro transfection with overexpression plasmid
BMDMs prior to transfection were polarized for 24 h followed by transiently transfection with 100 nM of control or circ-Cdr1as overexpression plasmid for 24 h. The plasmid was added in 500 uL of Opti-MEM followed by addition of 5 uL of Lipofectamine RNAiMAX in 500 uL of Opti-MEM and incubated for 5 min. The two mixtures were pooled and incubated for 15 min at room temperature (RT). The solution mixture was then added to the plate and incubated for 24 h. The following day, the media was changed to media containing 3 μg/mL of G418 antibiotic. Cells were harvested and changes in the expression or transcription of macrophage markers were measured by FACS or RT-qPCR, respectively.
2.8. Lentivirus short hairpin RNA construction
To generate a knockdown circ-Cdr1as plasmid, a custom shRNA plasmid was generated. Vector TR30023 pGFP-C-shLenti containing a TCAAGAG loop targeting the splicing junction sequence of circ-Cdr1as was constructed. This plasmid contains bacterial resistance to chloramphenicol and puromycin in eukaryotes. The shRNA circ-Cdr1as plasmid and sh scrambled control plasmid were expanded and packaged into Lentivirus. All viruses were produced using the third-generation lentivirus system. Briefly, HEK293 cells were plated at 1×107cells/15 cm plate one day prior to transfection. Production of 3rd generation lentivirus was performed using the combined ratio of transfer plasmid (Custom shRNA plasmid from Origene, Inc. TR30023 Lenti GFP-C-sh-9B, TTCCAACAACTGCGCAGTGTCTCCAGTGT), packaging plasmid, Env plasmid and pRSV-Rev plasmid at 2:1:1:1, respectively. Polyethylenimine HCl MAX, MW 40000, Transfection Grade (Polysciences, Inc) was used as the transfection reagent at a ratio of 1:2 μg DNA: μg PEI-Max. Without changing the media on the 15 cm plates, the DNA-PEI reagent was added to the cells drop-wise. At 72 h post transfection, supernatant of 293 T cell cultures were PEG precipitated (1 volume of 40 % PEG to 3 volumes of supernatant) for overnight at 4 °C and the samples were centrifuged at 1600 ×g for 45 min at 4 °C and the pellets were gently re-suspended in 15 mL 1× PBS containing 0.001 % Pluronic F68 and 5 % Glycerol, buffer exchanged two times and concentrated down to 1 mL using Amicon Ultra-15 100 K filter. The purified virus was then recovered and filtered through a 0.45 μm PES filter and quantified using Lenti-X™ qRT-PCR Titration Kit (Takara Bio, USA). The viruses were aliquoted and stored at −80 °C [22–24].
2.9. BMDM cell and in vitro knockdown of circ-Cdr1as
BMDMs were transfected with MOI 15 lentiviral shRNA (titer concentration for shRNA scrambled: 2.9 × 109; shRNA circ-Cdr1as: 3.67 × 10 [9] vp/mL). The cells were transfected with lentivirus shRNA and viral enhancer (ABM Cat# G515) for 48 h. The media was then changed to media containing 3μg/mL of puromycin antibiotic for selection of stably transfected cells. The cells were polarized for 24 h. Cells were harvested and changes in the expression or transcription of macrophage markers were measured by FACS or RT-qPCR, respectively.
2.10. FACS analysis and cell sorting
BMDMs were harvested and washed with PBS staining buffer containing 2 % FBS and 0.05 % EDTA. The cells were pre-incubated with 0.25 μg of TrueStain FcX PLUS (anti-mouse CD16/32) antibody (Biolegend Cat# 156603) in 100 uL volume for 10 mins on ice. The cells were then washed with staining buffer and incubated with the following antibodies at 4 °C for 30 min then washed with staining buffer 3× and resuspended in 1 mL of staining buffer. The BDMD were sorted using BD FACS Aria IIμ based on the following markers: pro-inflammatory- F4/80+, CD86+, CD11c+, CD206−; anti-inflammatory- F4/80+, CD86−, CD11c−, CD206+ (Supplemental Fig. 1A). BD LSR II was used for FACS analysis, 1.5 × 106 cells were collected and the cells were stained with the following markers: F4/80, CD86, and CD206. Gating strategies were based on fluorescence minus one (FMO).
| Antibody | Fluorophore | Final concentration | Company and catalog number |
|---|---|---|---|
|
| |||
| F4/80 | FITC | 0.5 μg/100uL | Invitrogen 11-4801-82 |
| CD11c | PE-Cy7 | 0.5 μg/100uL | Thermofisher 25-0114-82 |
| CD86 | Super Bright 600 | 0.125 μg/100uL | Thermofisher 63-0862-82 |
| CD206 | Brilliant Violet 421 | 5 μg/mL | Biolegend 141717 |
2.11. Real-time quantitative reverse transcription PCR
Reverse Transcription (RT) was performed using High-Capacity cDNA RT kit (Invitrogen Cat# 4368813) and quantitative PCR (qPCR) was performed using SYBR Green master mix (Applied Biosystems). To quantify expression of circRNA transcripts, divergent primers designed to amplify across the backsplicing junction. To quantify linear transcripts, convergent primers were designed to amplify exonic sequences not present in the circular RNA. Amplification was performed using the StepOnePLus Real-Time PCR system (Applied Biosystems 7000 apparatus) and ct thresholds were determined by the software. Expression was quantified using fold change of 2e (delta ct) compared to naïve control, normalized to 18S rRNA used as a reference gene. All primer sequences are listed in Supplementary Table 2.
2.12. Identification of circRNA-miRNA interactions
Circular RNA-miRNA interactions were predicted by Arraystar’s miRNA target prediction software based on TargetScan and miRanda, and the differentially expressed circRNAs within all the comparisons annotated in detail including 2D structure, local AU, position, conservation. The miRNA and top 5 gene targets are illustrated in Fig. 6.
Fig. 6.
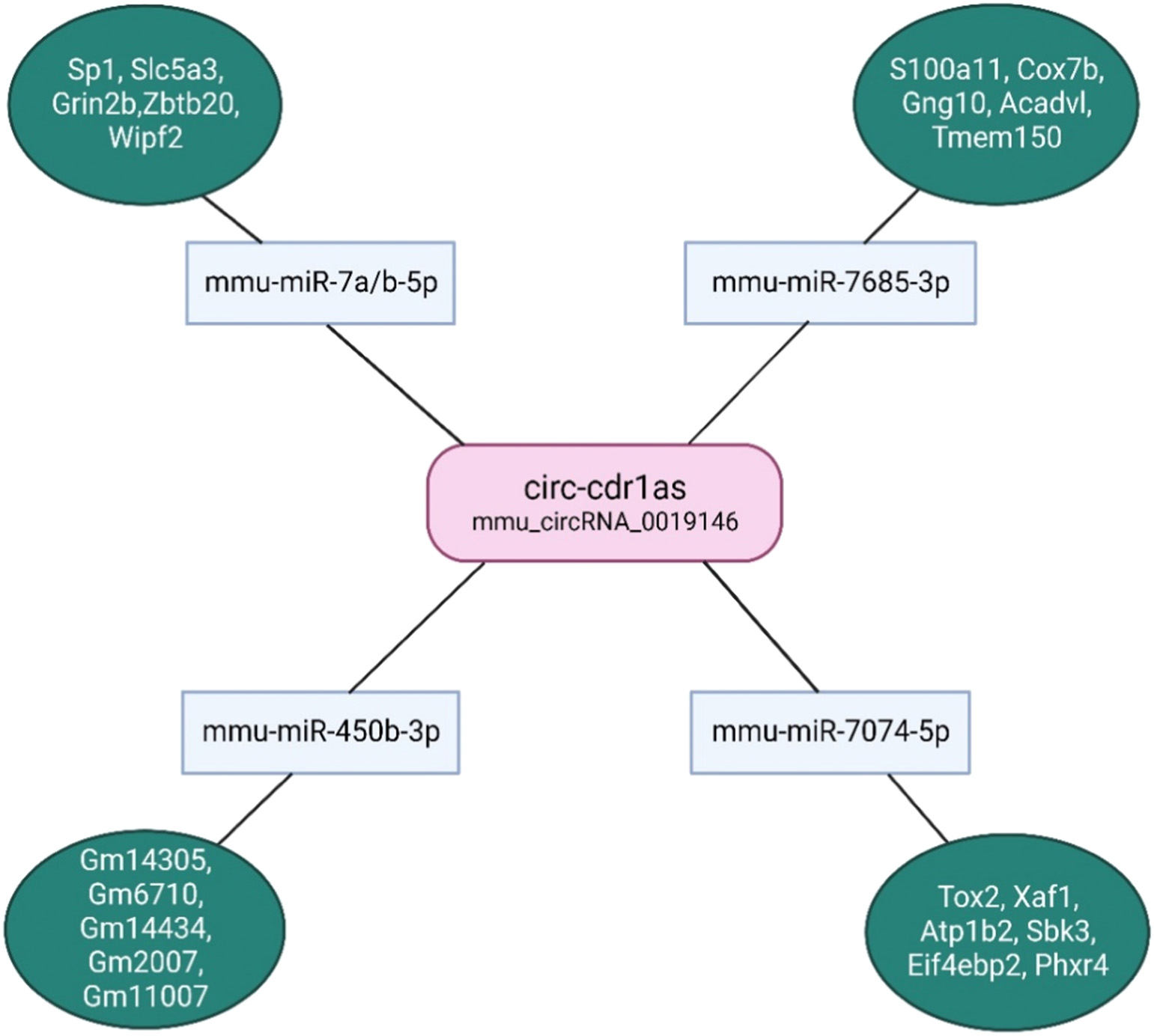
Predicted circ-cdr1as-miRNA-mRNA targets. The microRNA targets were identified using Arraystar’s miRNA target prediction software based on on TargetScan and miRanda from all the differentially expressed circRNAs between naïve, pro-, and anti-inflammatory macrophages. Predicted miRNA targets (only top 5 included) were determined using TargetScan and sorted by cumulative weighted context ++ score that is the sum of the contribution of 14 features, previously described. (Illustration created with Biorender.com) [32]
2.13. TargetScan and gene ontology analysis
The miRNA targets were input into TargetScan software (release 8.0) to determine microRNA targets and presence of conserved 8mer, 7mer, and 6mer sites that match the seed region of each miRNA. The Gene Ontology (GO) enrichment analysis of the miRNA target genes using Consortium’s online tool (http://www.geneontology.org/) found several significantly enriched (False Discovery Rate: FDR, p-value <0.05) GO terms, including associated biological process, molecular function, PANTHER pathways (Supplementary Table S1).
2.14. Statistical analysis
Data are expressed as Mean ± SEM. Analyses were performed using Prism (GraphPad Software Inc). Mean of the groups were compared using unpaired two tail t-test (for two groups) and One-way ANOVA (more than two groups). P values of <0.05 indicate statistical significance. GraphPad Prism version 9.4.0was used for making data visualization and for statistical analysis. Data are expressed as mean ± standard error of the mean (S.E.M.), and P < 0.05 was accepted as statistically significant.
3. Results
3.1. Identification of differentially expressed circular RNAs between pro-inflammatory and anti-inflammatory macrophages
To investigate the circular RNA transcriptome changes between naïve, pro-, and anti- inflammatory macrophages, we performed circRNA microarray expression profile from the RNA isolated from bone marrow derived macrophages (BMDM) polarized to a pro-inflammatory phenotype (stimulated with INFγ and TNFα, 24 h) or an anti-inflammatory phenotype (stimulated with IL-10, IL-4, and TGF-β, 24 h; Supplemental Fig. 1A). The BDMD were sorted based on the following markers: pro-inflammatory- F4/80+, CD86+, CD11c+, CD206−; anti-inflammatory- F4/80+, CD86−, CD11c−, CD206+ (Fig. 1A). Additionally, RT-qPCR analysis was performed for further validation of BMDM polarization based on pro or anti-macrophage markers (Supplemental Fig. 1B, C) [6,25,26]. We found differential expression of circRNAs in all experimental groups as depicted in the heat map (Fig. 1B, Supplemental Fig. 1D, E). Circular RNAs were then selected based on a log fold change of >1.5 and a p-value of <0.05 (Fig. 1C, Supplemental Fig. 1D, E). We found a total of 419 circRNAs differently expressed in pro-inflammatory macrophages compared with anti-inflammatory macrophages. Among these, 218 circRNAs were upregulated and 201 were downregulated (Fig. 1C). The box plot data indicates the distribution of the intensities from all the data sets after normalization and similar distribution in all biological replicates (Fig. 1D). Moreover, the scatter plot demonstrates the aberrantly expressed circRNAs between pro- and anti-inflammatory macrophages (Fig. 1E). these analyses discovered the distinguishable circRNAs expression profiles between pro- and anti-inflammatory macrophages.
3.2. Gene interaction networks and disease enrichment analysis
Among the circular RNA found to be differentially expressed between pro-inflammatory vs naïve and anti-inflammatory vs naive, 932 circular RNA were present in pro-inflammatory macrophages, 136 circular RNA present in anti-inflammatory macrophages, and 78 circular RNA found in all three phenotypes (Fig. 2A).
Fig. 2.
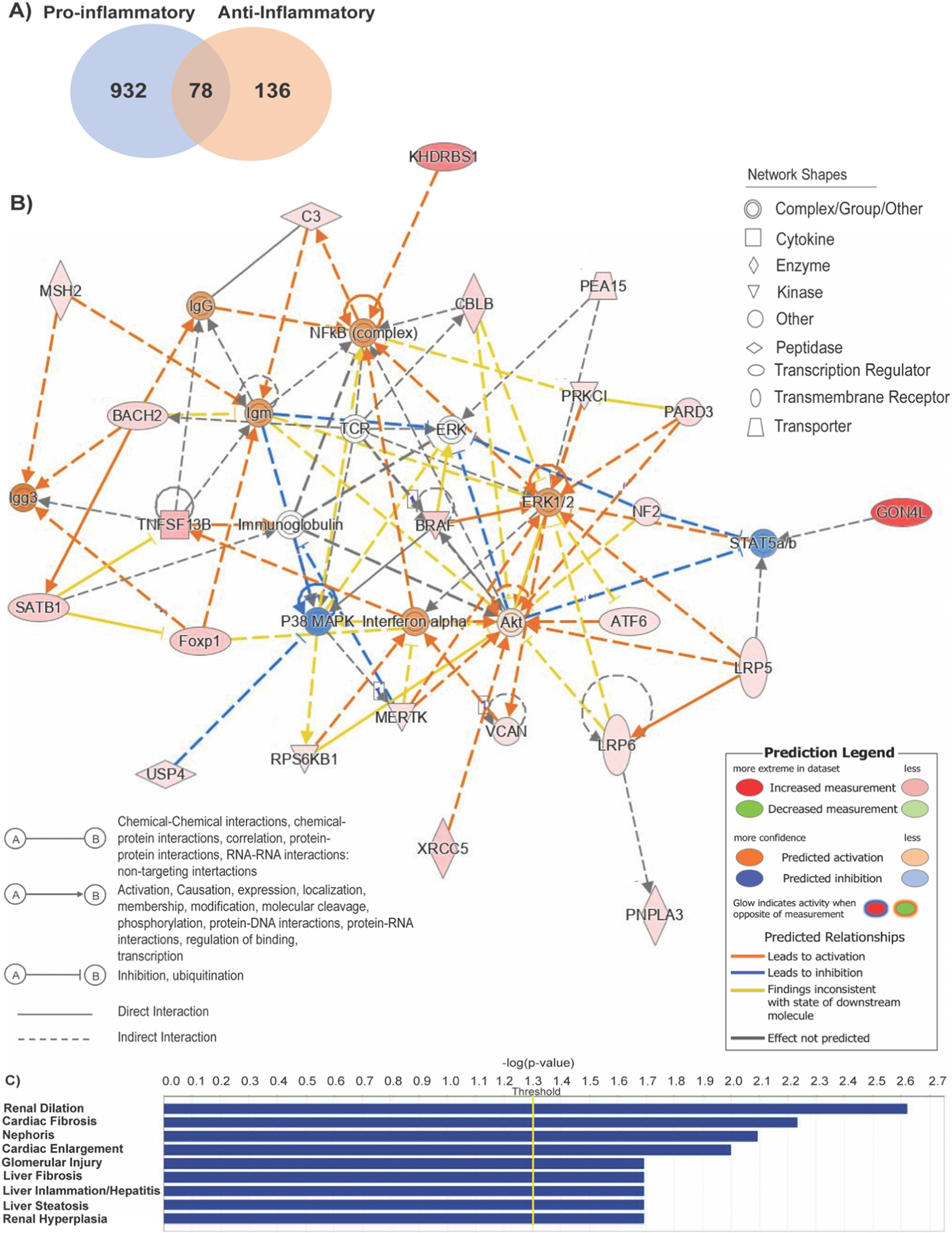
Ingenuity pathway analysis for differentially expressed circular RNAs between pro-inflammatory and anti-inflammatory macrophages. (A) Venn Diagram illustrating the overlap of circular RNA found between pro-inflammatory vs naïve or anti- inflammatory vs. naïve macrophages. (B) Overlapping interaction network analysis regulated by circular RNAs found between all data sets indicating predicted activated and inhibited genes that either directly or indirectly interact. (C) Disease enrichment analysis based on −log(p-value) >4 threshold on the IPA system.
Ingenuity Pathway Analysis (IPA) generated gene interaction networks overlapping all data sets indicating predicted activated and inhibited genes associated with cell death and survival, cell-to cell signaling, cardiovascular system development and function, and cell-mediated immune response. (Fig. 2B, Supplemental Fig. 2B, C). Of note, the most activated predicted genes include crucial inflammatory regulators nuclear factor-κB (NFkB), interferon α (INFα) (Fig. 2B), and IL-10 (Supplemental Fig. 2B). Moreover, the genes involved are linked to the top enriched canonical pathways between all data sets including senescence, and B cell receptor signaling (Supplemental Fig. 2A). Disease enrichment analysis identified top diseases associated with circular RNA differently expressed between pro- and anti-inflammatory macrophages. Among them, the most common diseases are related to the liver in which macrophages are a key cellular component and essentially for tissue homeostasis (Fig. 2C) [27]. Interestingly, cardiac fibrosis and cardiac enlargement are also noted as the top related disease in which macrophages are the predominant leukocyte type after cardiac injury (Fig. 2C) [28].
3.3. Identification of target circ-Cdr1as from the top ten conserved and differentially expressed circular RNAs
Based on the microarray analysis, we chose the top ten differentially expressed circular RNAs based on their conservation between human and mouse ranked by fold change (Table 1). Among them, 6 were downregulated (circ-Zfhx3, circ-Cdr1as, circ-Amotl1, circ-Bnc2, circ-Ptk2, and circ-Slc30a7) and 3 (circ-Elf2, circ-Gigyf2, and circ-Rsrc1) were upregulated in pro-inflammatory (M1) macrophages; and 1 (circ-Mipol1) was upregulated anti-inflammatory (M2) macrophages (Table 1).
RT-qPCR confirmation of profile data based on detection of divergent primers showed circ-Zfhx3, circ-Amotl1, and circ-Bnc2 downregulated in both phenotypes; circ-Elf2, and circ-Mipol1 downregulated in anti-inflammatory phenotypes; circ-Gigyf2, circ-Ptk2, and circ-Slc30a7 were non-significantly expressed between phenotypes (Fig. 3). Interestingly, RT-qPCR validation confirmed circular RNA Cdr1as expression to be significantly downregulated in pro-inflammatory macrophages while significantly upregulated in anti-inflammatory macrophages (Fig. 3). In addition to this, circ-Cdr1as has a 79% sequence similarity with human sequence based on pairwise alignment (Supplemental Fig. 3A). Furthermore, RT-qPCR analysis of convergent primer for linear transcript cerebellar degeneration-related protein 1 (cdr1) indicates no significant change in parental cdr regardless of phenotype (Supplemental Fig. 3B). There are currently no reports on the role of circ-Cdr1as in macrophage phenotypic switching; however, circ-Cdr1as was reported to be involved in the regulation of brain development, diabetes, increase in cardiac infarct size, and cardiomyocyte apoptosis [29,30]. Therefore, we selected circ-Cdr1as as a candidate circRNA for further investigation.
Fig. 3.
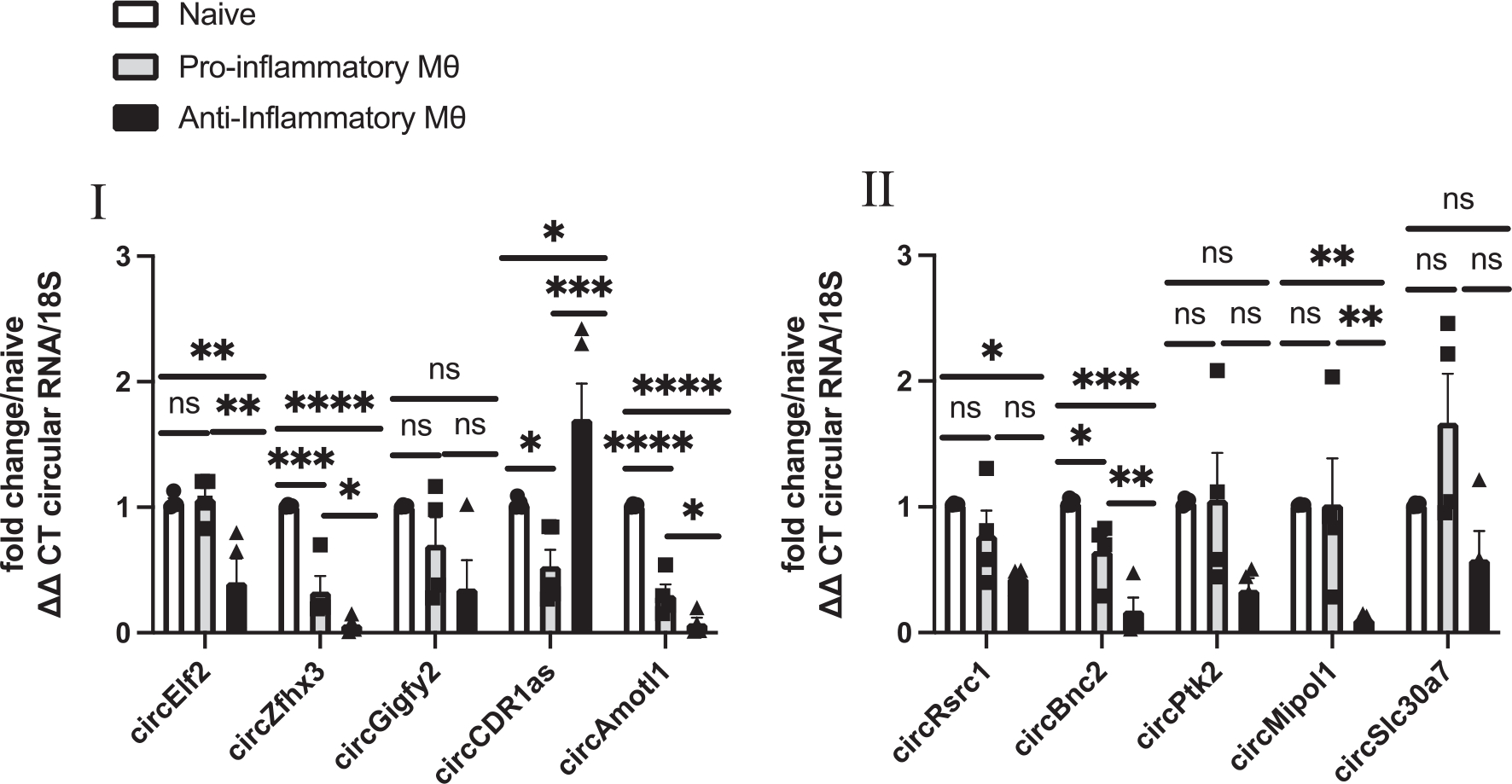
Circular RNAs are differentially expressed between naïve, pro-inflammatory, and anti-inflammatory macrophages. Validation of the top 10 differentially expressed circRNAs by RT-qPCR analysis (CT value <31) of bone marrow derived macrophages polarized to pro-inflammatory or anti-inflammatory phenotype compared to naïve macrophages, normalized to 18S. n = 4–5. Data are Mean ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, ns non-significant vs naïve BMDM (one-way ANOVA).
3.4. Overexpression of circ-Cdr1as increases transcription of anti-inflammatory markers and percentage of CD206 positive cells
Based on expression data indicating circ-Cdr1as upregulation in anti-inflammatory macrophages, we sought to determine if overexpression promotes phenotypic switching to an anti-inflammatory phenotype. We generated circ-Cdr1as overexpression plasmid containing laccasse2 intronic regions to promote circularization (Supplemental Fig. 4A). The overexpression plasmid was then transiently transfected into naïve or pro-inflammatory BMDMs (Supplemental Fig. 4B). Overall, overexpression did not significantly change transcription of linear cdr (Supplemental Fig. 4C) or macrophage markers cd11b and F4/80 (Supplemental Fig. 4E, F). Meanwhile, overexpression increased circ-Cdr1as levels by >100-fold compared to scrambled plasmid control and un-transfected naïve controls, confirming successful transfection of plasmid. in all phenotypes (Supplemental Fig. 4D). FACS analysis showed an increase in anti-inflammatory macrophage marker CD206 in cdr1as overexpressed naïve and pro-inflammatory macrophages compared to scrambled control (Fig. 4A, C). Additionally, density plots demonstrated a shift in the distribution of cells compared to scrambled control (Supplemental Fig. 4F). The mRNA levels of reparative anti-inflammatory macrophage markers such as chitinase-3-like protein 1 (CHI3I3), CD68, arginase 1 (Arg-1), CD206, and CD163 were also increased in treated naïve and pro-inflammatory macrophages compared to scrambled naïve control. The IL-10 and TGFβ mRNA levels were also increased in both phenotypes (Fig. 4B, D). These data suggest that overexpression of circ-cdr1as not only drives naïve macrophages to M2/anti-inflammatory phenotype but more interestingly has similar effect in macrophages that are stimulated towards M1/pro-inflammatory phenotype.
Fig. 4.
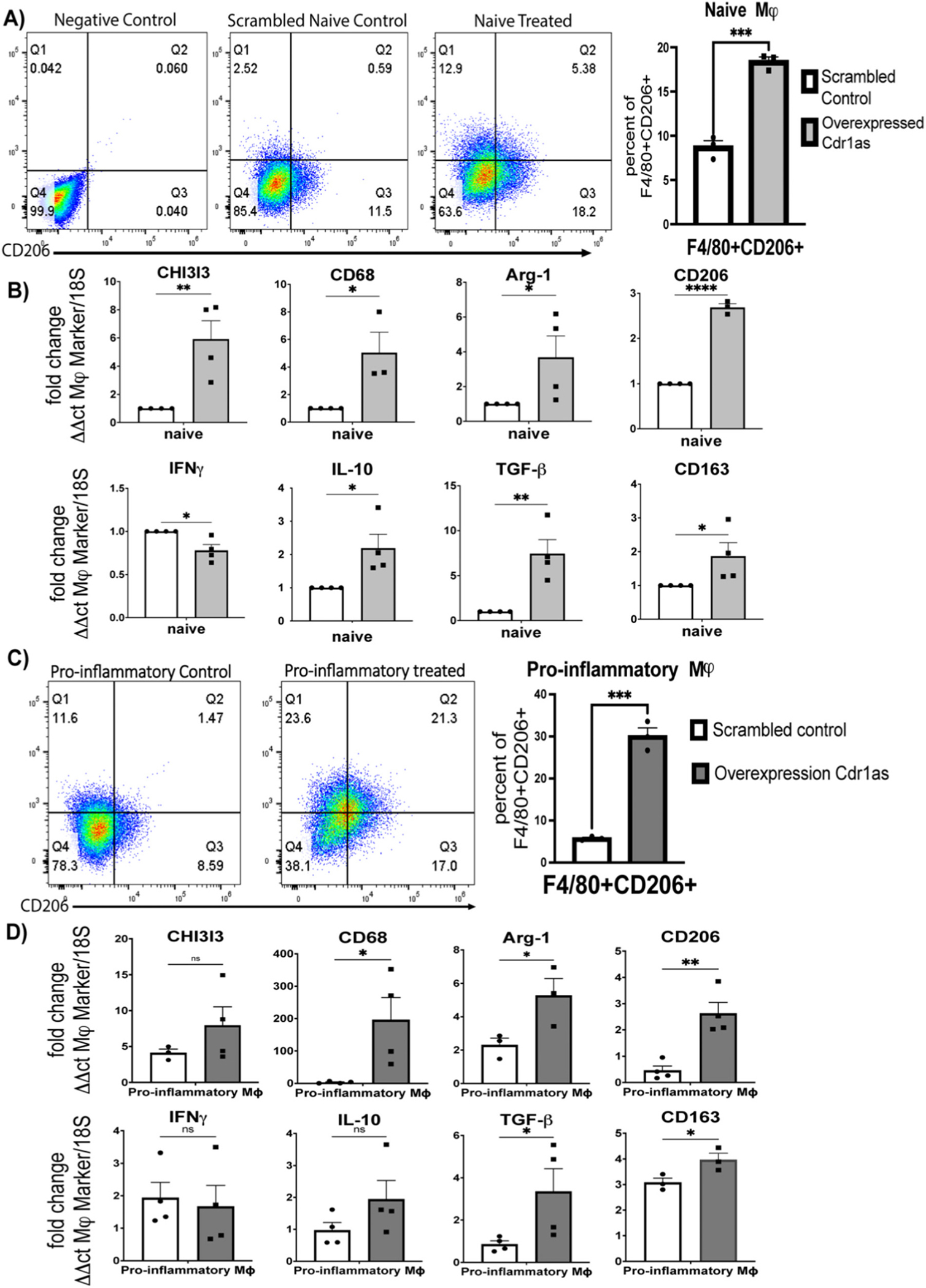
Overexpression of circular cdr1as upregulate expression of anti-inflammatory macrophage markers in naïve macrophages and pro-inflammatory macrophages stimulated with pro-inflammatory cytokines. (A, C) FACS analysis of F4/80 + CD206+ cells in naïve (A) or pro-inflammatory (C) macrophages treated with overexpression plasmid compared to scrambled control, normalized to 18S. (B, D) qRT-qPCR analysis of transcriptional changes of macrophage markers in naïve (B) or pro-inflammatory (D) macrophages compared to scrambled control, normalized to 18S. n = 3–4. Data are Mean ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, ns (non-significant) vs control (unpaired two tail t-test). CHI3I3, chitinase-3-like protein 1; Arg-1, arginase 1; INFy, Interferon y; IL-10, Interleukin 10; TGFb-Tumor grow factor-b.
3.5. Knockdown of circ-Cdr1as decreases transcription of anti-inflammatory markers and increases percentage of CD86 positive cells
To investigate if downregulation of circ-Cdr1as has the opposite effect of gain in function experiments noted above, knockdown of circ-Cdr1as was done by lentiviral small hairpin RNA (shRNA) targeting the splicing junction specific for circular RNA (Supplemental Fig. 5A) and stably transduced into naïve BMDMs prior to polarization to an anti-inflammatory phenotype (Supplemental Fig. 5B). Importantly, circ-cdr1as knockdown did not significantly change the expression of linear cdr1 (Supplemental Fig. 5C) or macrophage markers cd11b and F4/80 (Supplemental Fig. 5E, F). Meanwhile, knockdown significantly decreased circ-Cdr1as transcription compared to scrambled control in all phenotypes (Supplemental Fig. 5D). FACS analysis showed an increase in pro-inflammatory macrophage marker CD86 in treated naïve and anti-inflammatory macrophages compared to scrambled naïve control (Fig. 5A, C). Additionally, density plots demonstrated a shift in the distribution of cells compared to scrambled control (Supplemental Fig. 5F). The mRNA levels of anti-inflammatory macrophage lineage markers Arg-1, CD206, and CD163 were decreased in shRNA treated naïve and anti-inflammatory macrophages compared to control. The IL-10 and TGFβ mRNA levels were also decreased in both phenotypes (Fig. 5B, D). These findings suggest that circ-Cdr1as participates in promoting phenotypic switching to an anti-inflammatory phenotype.
Fig. 5.
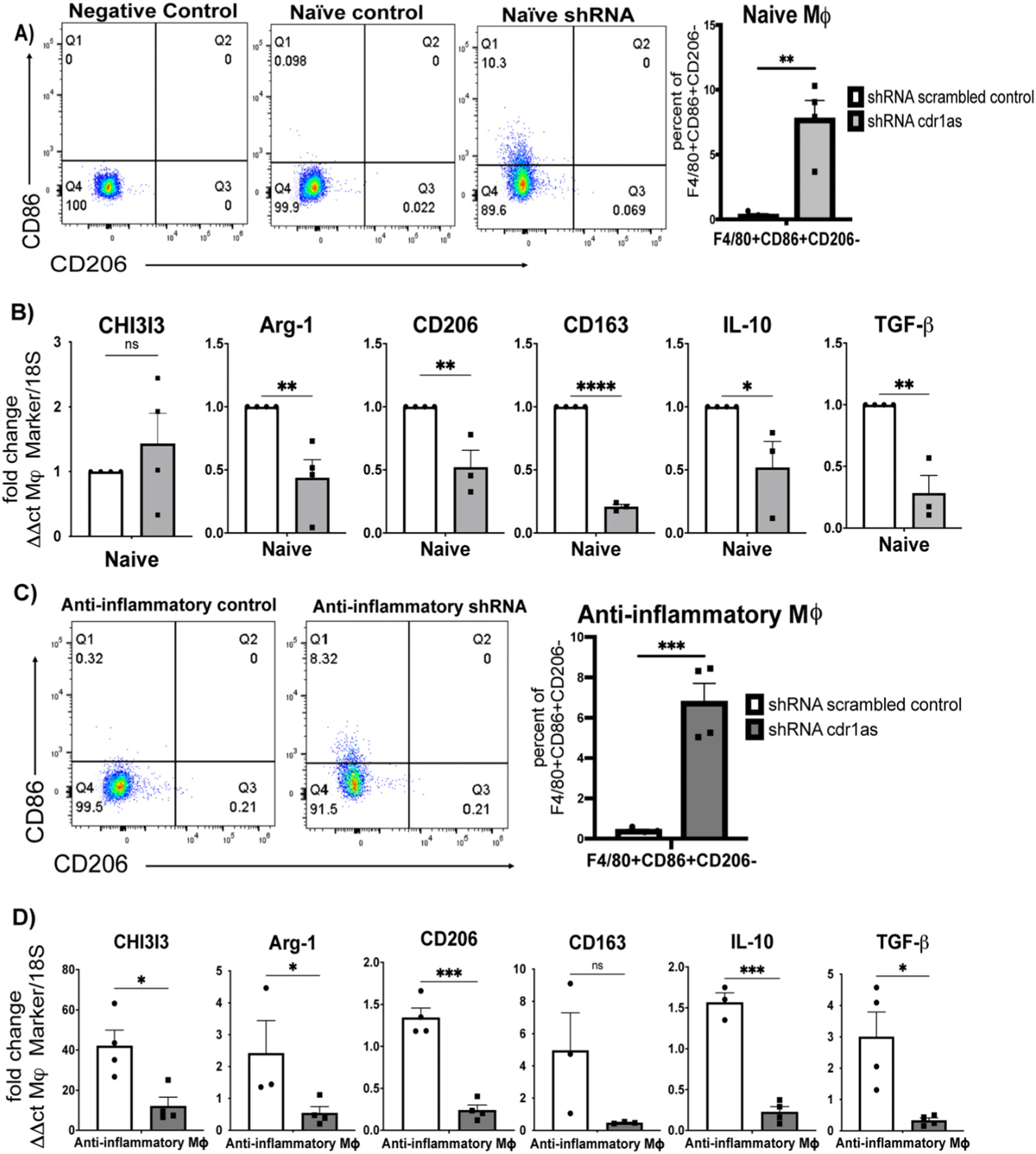
Knockdown of circular cdr1as downregulates expression of anti-inflammatory macrophage markers in naïve macrophages or anti-inflammatory macrophages stimulated with anti-inflammatory cytokines. (A, C) FACS analysis of F4/80 + CD86 + CD206− cells in naïve (A) or anti-inflammatory (C) macrophages treated with lentiviral shRNA cdr1as compared to lentivirus scrambled control, normalized to 18S. (B, D) qRT-qPCR analysis of transcriptional changes of cytokines and macrophage markers in naïve (B) or anti-inflammatory (D) macrophages compared to lentivirus scrambled control, normalized to 18S. n = 3–4. Data are Mean ± SEM. * p < 0.05, ** p < 0.01, *** p < 0.001, ns (non-significant) vs control (unpaired two tail t-test). CHI3I3, chitinase-3-like protein 1; Arg-1, arginase 1; IL-10, Interleukin 10; TGFb-Tumor grow factor-b.
3.6. Circular RNA cdr1as-miRNA-mRNA network
Recent evidences have demonstrated that circular RNAs play a crucial role in fine tuning the level of miRNA mediated regulation of gene expression by sequestering the target miRNAs [1,13,31]. Therefore, to identify possible miRNA targets specific binding patterns between circRNAs and miRNA were predicted with Arrays miRNA target prediction software based on TargetScan and miRanda according to the complementary miRNA matching sequence. A total of 4 miRNAs (miR-7a/b-5p, miR-450b-3p, miR-7685-3p, miR-7074-5p) and corresponding target mRNAs were predicted to have an interaction with circ-cdr1as (Fig. 6). The highest conserved miRNA based on TargetScan and miRanda analysis is miR-7-5p (Supplementary Table S1). To find the functional significance of the downstream target genes in the circRNA-miRNA-mRNA regulatory network, we performed GO enrichment analysis for the biological process, molecular function, and pathways involved (Supplementary Table S1).
4. Discussion
Macrophages mediate tissue damage and inflammatory responses in both physical and pathological conditions [6,27]. Macrophages are key cellular elements of chronic inflammatory responses associated with the most common causes of morbidity such as cardiovascular disease and cancer [2,26,28]. These leukocytes are remarkably dynamic cells that can switch phenotypes depending on environmental stimuli and signals. Two major subpopulations include classically activated pro-inflammatory macrophages or anti-inflammatory macrophages [4,7,20,33]. Deciphering the mechanisms by which macrophage polarization is regulated may provide insights for the development of new therapies to manipulate the balance between pro- and anti-inflammatory phenotypes to help or dampen an inflammatory response without the unintended consequences of therapies that impair monocyte function and recruitment [9].
Recent studies with the use of high-throughput RNA sequencing have identified a new class of non-coding RNAs called circular RNAs. Circular RNAs form covalently closed transcripts generated when the pre-mRNA splicing machinery back splices to join a downstream 5′ splice site to an upstream 3′ splice site [34]. They are derived from exonic, intronic sequences, or both. Overall, circRNAs are highly conserved among species and have a high degree of stability in mammalian cells. These properties provide circRNAs with the potential therapeutic targets [14,35,36].
Whether circRNAs are involved in macrophage polarity and phenotype is not yet established. This study investigated expression patterns of circRNAs between pro- and anti- inflammatory macrophages. We found 419 circular RNAs significantly different between these two phenotypes with 218 circRNAs upregulated and 201 circRNAs downregulated in pro-inflammatory macrophages. Additionally, 78 circRNAs were present in naive, pro-, and anti-inflammatory macrophages. Furthermore, it should be noted that macrophage activation deregulate expression of other potential circular RNAs that are not annotated by microarray chip.
Several studies have implicated circRNAs to regulate physiological conditions and disease progression [37]. Numerous investigators have shown that circRNAs are involved in brain-related disorders such as Alzheimer’s disease [38,39], Parkinson’s disease [40], and glioblastoma [41]. Secondly, circRNAs are also highly expressed in the heart. Werfel et al. [14], found that in mouse, rat, and human failing hearts, there is re-expression of fetal genes in cardiomyocytes along with an overall increase in circRNA expression arising from the titin (Ttn) gene across all species. Additionally, circular RNAs are reported to play a role in cardiac remodeling [42,43], immune tolerance [44], fibrosis [45–47], and endothelial to mesenchymal transition [48].
Ingenuity Pathway analysis of differentially expressed circular RNA between pro- and anti- inflammatory macrophages generated gene interaction networks associated with cell death and survival, cell-to cell signaling, cardiovascular system development and function, and cell-mediated immune response. Among these networks, master inflammatory regulators NF-κB, INFα, and IL-10 were predicted to be activated. These master regulators are vital to many of the roles that macrophages play in orchestrating an inflammatory response. Signaling of these regulators is modulated based on shifting thresholds of activation and regulation of transcription [ 33,49,50]. How we may utilize circRNAs to modulate signaling is an important field of future research.
Disease enrichment analysis identified top diseases associated with circular RNA differently expressed between pro- and anti-inflammatory macrophages. The top two targeted organs were the liver and the heart, in which macrophages play a major role in the initiation and resolution of inflammation after injury [27,28]. Currently, circular RNAs have been implicated to play a role in cardiovascular diseases such as cardiac hypertrophy [43], coronary artery disease [51], cardiac fibrosis [45–47], rheumatic heart disease [52], and myocardial infarction [42]. One of the most characterized circular RNA, heart-related circular RNA (HRCR), is repressed in hypertrophic and failing hearts. HRCR sequesters miR-223, a miRNA that induces cardiac hypertrophy by inhibition of the protein apoptosis inhibitor with a CARD domain. Enforced expression of HRCR attenuated cardiomyocyte hypertrophy in vitro and in vivo [43]. Functional experiments demonstrated CircRNA_010567 silencing upregulated miR-141 and downregulated TGF-ß resulting in suppression of fibrosis-associated protein expression of cardiac fibroblast [47]. Comparative analysis of circular RNA expression profiles of atrial tissues from patients with persistent atrial fibrillation (AF) that had rheumatic heart disease and non-AF healthy patients found 108 circular RNA to be differentially expressed. Lastly, circular RNA circFndc3b modulates cardiac repair after MI. This circular RNA is down regulated in post-MI murine hearts and in ischemic cardiomyopathy patients. CircFndc3b interacts with RNA binding protein fused in sarcoma (FUS) to regulate VEFG expression. Overexpression of circFndc3b reduces endothelial apoptosis, cardiomyocyte apoptosis, and promotes angiogenesis after cardiac injury highlighting the potential of therapeutically targeting circRNA promote cardiac function and remodeling post MI [53]. Therefore, our future studies will focus on translational functionality of circ-Cdr1as following myocardial injury.
Based on the microarray analysis, we selected the top 10 differentially expressed and highly conserved between human and mouse. Validation by RT-qPCR demonstrated discrepancies between the microarray data and RT-qPCR analyses indicating validation is an essential step of screening for circular targets. We chose circ-Cdr1as for further investigation as both analyses indicated a downregulation in pro-inflammatory macrophages and RT-qPCR analysis showed a significant upregulation in anti-inflammatory macrophages.
Although there are limited studies on the role of circ-Cdr1as in immunity, there are circular RNAs previously found to be involved in T cell and macrophage biology. Circular RNA expression profile of human hematopoietic cells and differentiated lymphoid and myeloid cells found that during hematopoietic differentiation, the expression levels of circRNA of lymphocytes were the highest. Moreover, circ-FNDC3B had the highest expression in natural killer cells, while circ-ELK4, circ-MYBL1, and circ-SLFN12L showed the highest expression in T cells and natural killer cells [54]. A bioinformatics study on circ-Cdr1as analyzing 868 cancer samples applied Gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG), gene set enrichment analysis (GSEA), CIBERSORT, Estimating the Proportion of Immune and Cancer cells (EPIC), and the Malignant Tumors using Expression data (ESTIMATE) algorithms to identify potential functions of Cdr1as in cancer. The results indicated that they may be involved in immune and stromal cell infiltration in tumor tissue, especially those of CD8+ T cells, activated NK cells, anti-inflammatory macrophages, cancer-associated fibroblasts (CAFs) and endothelial cells [55].
However, there are no reports on the role of circ-Cdr1as in the regulation of macrophage biology and function. Our data, for the first time, reports that overexpression of circ-Cdr1as promotes phenotypic switching to an anti-inflammatory phenotype in naïve and pro-inflammatory macrophages. Additionally, knockdown of circ-Cdr1as has the opposite effect, promoting a more pro-inflammatory phenotype in naïve and anti-inflammatory macrophages. Meanwhile, loss or gain of function experiments did not significantly alter the linear counterpart.
Recent studies focusing on the role of circ-Cdr1as identified that it acts to sequester miRNAs and thereby, regulate miRNA-mediated regulation of gene expression in neurological and cardiovascular disease [1,13,29]. This present study identified 4 miRNA and subsequent mRNA targets that provide us with another research strategy to explore the mechanism of circ-cdr1as in the regulation of associated miRNA-mRNA interactions. Among them, circ-Cdr1as contains >70 binding sites for miR-7 acting as a miRNA sponge to inhibit miRNA-7 function [30,56]. Previously, functional studies demonstrated that overexpression of CDR1as induced development defects in the midbrain of embryonic zebrafish and overexpression of miR-7 reduced the effects of CDR1as [57]. In a cardiac injury context, one study studied circ-Cdr1as 24 h post MI and found upregulation of this circular RNA associated with increased cardiac infarct size. They further investigated the role of this circRNA under hypoxic conditions in primary cardiomyocytes and mouse cardiac myocytes cell line demonstrating that CDR1as promoted cardiomyocyte apoptosis by acting as a miR-7 sponge to prevent miRNA inhibition of pro-apoptotic binding targets, poly (ADP-ribose) polymerase (PARP), and member of the SP/KLF family of transcription factors SP1 [29].
Overall, co-expression with miR-7 and circ-Cdr1as was reported to reduce inhibition of miR-7 to its target by competing with target gene for binding with miR-7 and thereby, leading to increased expression of miR-7 target [31]. Although there are some reports describing dysregulation of miR-7 is associated with aberrant NFkB activation in cancer and respiratory diseases, there is currently limited research on whether circ-Cdr1as indirectly target key inflammatory regulators via miR-7 inhibition [58,59]. Additionally, mRNA targets for miR-7 have previously been shown to play a role in inflammation. For example, Sp1 binding element as an important determinant of macrophage inflammatory protein 2 (MIP-2) activity [60], Slc5a3 overexpression associated with inflammatory infiltration in sporadic inclusion body myositis [61], and Zbtb20 linked to inflammatory responses of oxidized LDL induced macrophages in atherosclerosis [62]. Overall, this current study identified miR-7 as a target for circ-cdr1as in BMDMs and GO enrichment analysis provided insight into the possible biological process, molecular functions, and pathways involved including inflammatory pathways such as interferon-gamma signaling and interleukin signaling pathways.
Therefore, future studies are needed on elucidating the regulatory relationship between circ-cdr1as and miRNAs in association with phenotypic switching.
5. Conclusion
In conclusion, the present study provides the first investigation the expression profile of circRNA in naïve, pro-, and anti-inflammatory macrophages and validated top 10 circRNA that are conserved and differentially expressed between pro- and anti-inflammatory macrophages. Our studies identified circ-Cdr1as to be significantly downregulated in pro-inflammatory macrophages and upregulated in anti-inflammatory macrophages. The functional importance of circ-crd1as was investigated by gain and loss of function experiments which established that overexpression of circ-cdr1as promotes phenotypic switching to an anti-inflammatory phenotype and knockdown has the opposite effect. In the future, it is necessary to explore the detailed molecular mechanism by which circ-Cdr1as functions to regulate macrophage polarization. Given the importance of circ-Cdr1as in cardiovascular disease and our IPA disease enrichment analysis further work is needed to determine whether circ-Cdr1as may modulate monocyte/macrophage biology during cardiac injury and therapeutically regulate post-injury cardiac inflammation. These studies are currently in the planning stage.
Supplementary Material
Funding
This work was in part funded by the National Institutes of Health grants HL091983, HL143892, HL134608 and HL147841 and F31 Pre-doctoral fellowship HL162543-01.
Footnotes
Declaration of competing interest
The authors declare no competing interests.
Credit authorship contribution statement
Carolina Gonzalez: Conceptualization, Methodology, Software, Formal analysis, Data curation, Writing – original draft, Visualization, Funding acquisition. Maria Cimini, Zhongjian Cheng: Methodology, Resources, Visualization. Cindy Benedict: Investigation, Methodology, Resources. Chunlin Wang, May Trungcao: Investigation. Vandana Mallaredy: Visualization. Sudarsan Rajan: Resources. Venkata Naga Srikanth Garikipati: Conceptualization, methodology. Raj Kishore: Conceptualization, Methodology, Validation, Formal analysis, Investigation, Data curation, Writing – review & editing, Supervision, Funding acquisition.
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.lfs.2022.121003.
Data availability
All source data that support the findings of this study and any material used are available upon request from the corresponding author.
References
- [1].Kishore R, Garikipati VNS, Gonzalez C, Role of circular RNAs in cardiovascular disease, J. Cardiovasc. Pharmacol. 76 (2020) 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Parisi L, Gini E, Baci D, Tremolati M, Fanuli M, Bassani B, Farronato G, Bruno A, Mortara L, Macrophage polarization in chronic inflammatory diseases: killers or builders? J. Immunol. Res. 2018 (2018), 8917804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Steinbach EC, Plevy SE, The role of macrophages and dendritic cells in the initiation of inflammation in IBD, Inflamm. Bowel Dis. 20 (2014) 166–175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Das A, Sinha M, Datta S, Abas M, Chaffee S, Sen CK, Roy S, Monocyte and macrophage plasticity in tissue repair and regeneration, Am. J. Pathol. 185 (2015) 2596–2606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].O’Rourke SA, Dunne A, Monaghan MG, The role of macrophages in the infarcted myocardium: orchestrators of ECM remodeling, Front. Cardiovasc. Med. 6 (2019) 101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, Sahebkar A, Macrophage plasticity, polarization, and function in health and disease, J. Cell. Physiol. 233 (2018) 6425–6440. [DOI] [PubMed] [Google Scholar]
- [7].Mantovani A, Biswas SK, Galdiero MR, Sica A, Locati M, Macrophage plasticity and polarization in tissue repair and remodelling, J. Pathol. 229 (2013) 176–185. [DOI] [PubMed] [Google Scholar]
- [8].Mann DL, Innate immunity and the failing heart: the cytokine hypothesis revisited, Circ. Res. 116 (2015) 1254–1268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Ponzoni M, Pastorino F, Di Paolo D, Perri P, Brignole C, Targeting macrophages as a potential therapeutic intervention: impact on inflammatory diseases and cancer, Int. J. Mol. Sci. 19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Holdt LM, Stahringer A, Sass K, Pichler G, Kulak NA, Wilfert W, Kohlmaier A, Herbst A, Northoff BH, Nicolaou A, Gäbel G, Beutner F, Scholz M, Thiery J, Musunuru K, Krohn K, Mann M, Teupser D, Circular non-coding RNA ANRIL modulates ribosomal RNA maturation and atherosclerosis in humans, Nat. Commun. 7 (2016) 12429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Ng WL, Marinov GK, Chin YM, Lim YY, Ea CK, Transcriptomic analysis of the role of RasGEF1B circular RNA in the TLR4/LPS pathway, Sci. Rep. 7 (2017) 12227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ng WL, Marinov GK, Liau ES, Lam YL, Lim YY, Ea CK, Inducible RasGEF1B circular RNA is a positive regulator of ICAM-1 in the TLR4/LPS pathway, RNA Biol. 13 (2016) 861–871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Chen LL, The biogenesis and emerging roles of circular RNAs, Nat. Rev. Mol. Cell Biol. 17 (2016) 205–211. [DOI] [PubMed] [Google Scholar]
- [14].Werfel S, Nothjunge S, Schwarzmayr T, Strom TM, Meitinger T, Engelhardt S, Characterization of circular RNAs in human, mouse and rat hearts, J. Mol. Cell. Cardiol. 98 (2016) 103–107. [DOI] [PubMed] [Google Scholar]
- [15].Chen YG, Kim MV, Chen X, Batista PJ, Aoyama S, Wilusz JE, Iwasaki A, Chang HY, Sensing self and foreign circular RNAs by intron identity, Mol. Cell 67 (2017) 228–238 e5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Cimini M, Garikipati VNS, de Lucia C, Cheng Z, Wang C, Truongcao MM, Lucchese AM, Roy R, Benedict C, Goukassian DA, Koch WJ, Kishore R, Podoplanin neutralization improves cardiac remodeling and function after acute myocardial infarction, JCIInsight 5 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Krishnamurthy P, Thal M, Verma S, Hoxha E, Lambers E, Ramirez V, Qin G, Losordo D, Kishore R, Interleukin-10 deficiency impairs bone marrow-derived endothelial progenitor cell survival and function in ischemic myocardium, Circ. Res. 109 (2011) 1280–1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Ying W, Cheruku PS, Bazer FW, Safe SH, Zhou B, Investigation of macrophage polarization using bone marrow derived macrophages, J. Vis. Exp. (76) (2013) 50323, 10.3791/50323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Bender AT, Ostenson CL, Giordano D, Beavo JA, Differentiation of human monocytes in vitro with granulocyte-macrophage colony-stimulating factor and macrophage colony-stimulating factor produces distinct changes in cGMP phosphodiesterase expression, Cell. Signal. 16 (2004) 365–374. [DOI] [PubMed] [Google Scholar]
- [20].Martinez FO, Sica A, Mantovani A, Locati M, Macrophage activation and polarization, Front. Biosci. 13 (2008) 453–461. [DOI] [PubMed] [Google Scholar]
- [21].Kramer MC, Liang D, Tatomer DC, Gold B, March ZM, Cherry S, Wilusz JE, Combinatorial control of drosophila circular RNA expression by intronic repeats, hnRNPs, and SR proteins, Genes Dev. 29 (2015) 2168–2182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Benskey MJ, Manfredsson FP, Lentivirus production and purification, Methods Mol. Biol. 1382 (2016) 107–114. [DOI] [PubMed] [Google Scholar]
- [23].Kutner RH, Zhang XY, Reiser J, Production, concentration and titration of pseudotyped HIV-1-based lentiviral vectors, Nat. Protoc. 4 (2009) 495–505. [DOI] [PubMed] [Google Scholar]
- [24].Lo HL, Yee JK, Production of vesicular stomatitis virus G glycoprotein (VSV-G) pseudotyped retroviral vectors, Curr. Protoc. Hum. Genet. (2007), 10.1002/0471142905.hg1207s52. Chapter 12:Unit 12.7. [DOI] [PubMed] [Google Scholar]
- [25].Gombozhapova A, Rogovskaya Y, Shurupov V, Rebenkova M, Kzhyshkowska J, Popov SV, Karpov RS, Ryabov V, Macrophage activation and polarization in post-infarction cardiac remodeling, J. Biomed. Sci. 24 (2017) 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Sica A, Erreni M, Allavena P, Porta C, Macrophage polarization in pathology, Cell. Mol. Life Sci. 72 (2015) 4111–4126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Krenkel O, Tacke F, Liver macrophages in tissue homeostasis and disease, Nat. Rev. Immunol. 17 (2017) 306–321. [DOI] [PubMed] [Google Scholar]
- [28].Duncan SE, Gao S, Sarhene M, Coffie JW, Linhua D, Bao X, Jing Z, Li S, Guo R, Su J, Fan G, Macrophage activities in myocardial infarction and heart failure, Cardiol. Res. Pract. 2020 (2020), 4375127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Geng HH, Li R, Su YM, Xiao J, Pan M, Cai XX, Ji XP, The circular RNA Cdr1as promotes myocardial infarction by mediating the regulation of miR-7a on its target genes expression, PLoS One 11 (2016), e0151753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Xu H, Guo S, Li W, Yu P, The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells, Sci. Rep. 5 (2015) 12453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J, Natural RNA circles function as efficient microRNA sponges, Nature 495 (2013) 384–388. [DOI] [PubMed] [Google Scholar]
- [32].Agarwal V, Bell GW, Nam JW, Bartel DP, Predicting effective microRNA target sites in mammalian mRNAs, elife 4 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Orecchioni M, Ghosheh Y, Pramod AB, Ley K, Macrophage polarization: different gene signatures in M1(LPS+) vs.classically and M2(LPS-) vs. alternatively activated macrophages, Front. Immunol. 10 (2019) 1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Panda AC, Gorospe M, Detection and analysis of circular RNAs by RT-PCR, Bio Protoc. 8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Jeck WR, Sorrentino JA, Wang K, Slevin MK, Burd CE, Liu J, Marzluff WF, Sharpless NE, Circular RNAs are abundant, conserved, and associated with ALU repeats, RNA 19 (2013) 141–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Tan WL, Lim BT, Anene-Nzelu CG, Ackers-Johnson M, Dashi A, See K, Tiang Z, Lee DP, Chua WW, Luu TD, Li PY, Richards AM, Foo RS, A landscape of circular RNA expression in the human heart, Cardiovasc. Res. 113 (2017) 298–309. [DOI] [PubMed] [Google Scholar]
- [37].Qu S, Yang X, Li X, Wang J, Gao Y, Shang R, Sun W, Dou K, Li H, Circular RNA: a new star of noncoding RNAs, Cancer Lett. 365 (2015) 141–148. [DOI] [PubMed] [Google Scholar]
- [38].Dube U, Del-Aguila JL, Li Z, Budde JP, Jiang S, Hsu S, Ibanez L, Fernandez MV, Farias F, Norton J, Gentsch J, Wang F, Dominantly Inherited Alzheimer N, Salloway S, Masters CL, Lee JH, Graff-Radford NR, Chhatwal JP, Bateman RJ, Morris JC, Karch CM, Harari O, Cruchaga C, An atlas of cortical circular RNA expression in Alzheimer disease brains demonstrates clinical and pathological associations, Nat. Neurosci. 22 (2019) 1903–1912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Ma N, Pan J, Ye X, Yu B, Zhang W, Wan J, Whole-transcriptome analysis of APP/PS1 mouse brain and identification of circRNA-miRNA-mRNA networks to investigate AD pathogenesis, Mol. Ther. Nucleic Acids 18 (2019) 1049–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Jia E, Zhou Y, Liu Z, Wang L, Ouyang T, Pan M, Bai Y, Ge Q, Transcriptomic profiling of circular RNA in different brain regions of Parkinson’s disease in a mouse model, Int. J. Mol. Sci. 21 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Guo X, Piao H, Research progress of circRNAs in glioblastoma, Front. Cell Dev. Biol. 9 (2021), 791892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Kishore R, Garikipati VNS, Gonzalez C, Role of circular RNAs in cardiovascular disease, J. Cardiovasc. Pharmacol. 76 (2020) 128–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Wang K, Long B, Liu F, Wang JX, Liu CY, Zhao B, Zhou LY, Sun T, Wang M, Yu T, Gong Y, Liu J, Dong YH, Li N, Li PF, A circular RNA protects the heart from pathological hypertrophy and heart failure by targeting miR-223, Eur. Heart J. 37 (2016) 2602–2611. [DOI] [PubMed] [Google Scholar]
- [44].Zhang Y, Zhang G, Liu Y, Chen R, Zhao D, McAlister V, Mele T, Liu K, Zheng X, GDF15 regulates Malat-1 circular RNA and inactivates NFkappaB signaling leading to immune tolerogenic DCs for preventing alloimmune rejection in heart transplantation, Front. Immunol. 9 (2018) 2407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Zhou B, Yu JW, A novel identified circular RNA, circRNA_010567, promotes myocardial fibrosis via suppressing miR-141 by targeting TGF-beta1, Biochem. Biophys. Res. Commun. 487 (2017) 769–775. [DOI] [PubMed] [Google Scholar]
- [46].Li H, Xu JD, Fang XH, Zhu JN, Yang J, Pan R, Yuan SJ, Zeng N, Yang ZZ, Yang H, Wang XP, Duan JZ, Wang S, Luo JF, Wu SL, Shan ZX, Circular RNA circRNA_000203 aggravates cardiac hypertrophy via suppressing miR26b-5p and miR-140–3p binding to Gata4, Cardiovasc. Res. 116 (7) (2019) 1323–1334, 10.1093/cvr/cvz215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Tang CM, Zhang M, Huang L, Hu ZQ, Zhu JN, Xiao Z, Zhang Z, Lin QX, Zheng XL, Yang M, Wu SL, Cheng JD, Shan ZX, CircRNA_000203 enhances the expression of fibrosis-associated genes by derepressing targets of miR-26b-5p, Col1a2 and CTGF, in cardiac fibroblasts, Sci. Rep. 7 (2017) 40342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hulshoff MS, Xu X, Krenning G, Zeisberg EM, Epigenetic regulation of endothelial-to-mesenchymal transition in chronic heart disease, Arterioscler. Thromb. Vasc. Biol. 38 (2018) 1986–1996. [DOI] [PubMed] [Google Scholar]
- [49].Dorrington MG, Fraser IDC, NF-kappaB signaling in macrophages: dynamics, crosstalk, and signal integration, Front. Immunol. 10 (2019) 705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Chuang Y, Hung ME, Cangelose BK, Leonard JN, Regulation of the IL-10-driven macrophage phenotype under incoherent stimuli, Innate Immun. 22 (2016) 647–657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Zhang S, Wang W, Wu X, Zhou X, Regulatory roles of circular RNAs in coronary artery disease, Mol. Ther. Nucleic Acids 21 (2020) 172–179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hu M, Wei X, Li M, Tao L, Wei L, Zhang M, Cheng H, Yuan Y, Circular RNA expression profiles of persistent atrial fibrillation in patients with rheumatic heart disease, Anatol. J. Cardiol. 21 (2019) 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Garikipati VNS, Verma SK, Cheng Z, Liang D, Truongcao MM, Cimini M, Yue Y, Huang G, Wang C, Benedict C, Tang Y, Mallaredy V, Ibetti J, Grisanti L, Schumacher SM, Gao E, Rajan S, Wilusz JE, Goukassian D, Houser SR, Koch WJ, Kishore R, Circular RNA CircFndc3b modulates cardiac repair after myocardial infarction via FUS/VEGF-A axis, Nat. Commun. 10 (2019) 4317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Nicolet BP, Engels S, Aglialoro F, van den Akker E, von Lindern M, Wolkers MC, Circular RNA expression in human hematopoietic cells is widespread and cell-type specific, Nucleic Acids Res. 46 (2018) 8168–8180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Zou Y, Zheng S, Deng X, Yang A, Xie X, Tang H, Xie X, The role of circular RNA CDR1as/ciRS-7 in regulating tumor microenvironment: a pan-cancer analysis, Biomolecules 9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Guo JU, Agarwal V, Guo H, Bartel DP, Expanded identification and characterization of mammalian circular RNAs, Genome Biol. 15 (2014) 409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M, Loewer A, Ziebold U, Landthaler M, Kocks C, le Noble F, Rajewsky N, Circular RNAs are a large class of animal RNAs with regulatory potency, Nature 495 (2013) 333–338. [DOI] [PubMed] [Google Scholar]
- [58].Azizoglu M, Ayaz L, Bayrak G, Yilmaz BC, Birbicer H, Doruk N, Evaluation of miRNAs related with nuclear factor kappa B pathway in lipopolysaccharide induced acute respiratory distress syndrome, Int. J. Mol. Cell Med. 9 (2020) 130–139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Ye T, Yang M, Huang D, Wang X, Xue B, Tian N, Xu X, Bao L, Hu H, Lv T, Huang Y, MicroRNA-7 as a potential therapeutic target for aberrant NF-kappaB-driven distant metastasis of gastric cancer, J. Exp. Clin. Cancer Res. 38 (2019) 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Lee KW, Lee Y, Kwon HJ, Kim DS, Sp1-associated activation of macrophage inflammatory protein-2 promoter by CpG-oligodeoxynucleotide and lipopolysaccharide, Cell. Mol. Life Sci. 62 (2005) 188–198. [DOI] [PubMed] [Google Scholar]
- [61].De Paepe B, Merckx C, Jarosova J, Cannizzaro M, De Bleecker JL, Myo-inositol transporter SLC5A3 associates with degenerative changes and inflammation in sporadic inclusion body myositis, Biomolecules 10 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Tao J, Qiu J, Lu L, Zhang L, Fu Y, Wang M, Han J, Shi M, Li L, Zhao Z, Wei F, Wang C, Zhang H, Liang S, Zheng J, ZBTB20 positively regulates oxidative stress, mitochondrial fission, and inflammatory responses of ox-LDL-induced macrophages in atherosclerosis, Oxidative Med. Cell. Longev. 2021 (2021), 5590855. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All source data that support the findings of this study and any material used are available upon request from the corresponding author.


