Abstract
Chromatin and associated epigenetic marks provide important platforms for gene regulation in response to metabolic changes associated with environmental exposures, including physiological stress, nutritional deprivation, and starvation. Numerous studies have shown that fluctuations of key metabolites can influence chromatin modifications, but their effects on chromatin structure (e.g. chromatin compaction, nucleosome arrangement, and chromatin loops) and how they appropriately deposit specific chemical modification on chromatin are largely unknown. Here, focusing on methionine metabolism, we discuss recent developments of metabolic effects on chromatin modifications and structure, as well as consequences on gene regulation.
Keywords: Metabolism, Chromatin, Epigenetics, Transcription, Methionine
Introduction
Cells metabolize distinctive extracellular nutrients into other products to provide energy and chemical building blocks for their survival. Diverse metabolic pathways are interconnected and tightly regulated to allow cells to respond to changing environmental conditions (e.g. fasting, nutrient deficiency/excess). For example, fasting causes the liver to engage glycogenolysis, gluconeogenesis, and ketogenesis pathways to produce energy [1,2]. Modulation of cellular metabolism (e.g. methionine availability) has been suggested to contribute to mammalian lifespan and to be relevant to cancer pathogenesis [3,4]. How metabolism contributes to transcriptional and other gene regulatory responses is still incompletely known.
Genes encoded in DNA are packaged in the nucleus by wrapping around histone octamer complexes to form nucleosomes. The amino-terminal tails of histones are subject to chemical modifications, which can be recognized by specific binding proteins. Adjacent nucleosomes are linked by DNA and progressively fold into higher-order chromatin structures. The rearrangement of nucleosomes can expose or hide DNA binding sites from DNA-binding proteins. These chromatin features provide diversity in the mechanisms through which genes can be activated or suppressed. Emerging studies suggest that metabolism and diets connect with epigenetic modifications, chromatin compaction, nucleosome arrangement, and long-range chromatin interactions via metabolites, intermediates, metabolic enzymes, histone modifiers, and signal-responsive proteins (Fig. 1). Here, we summarize several major advances in understanding the connections between metabolism and chromatin with a focus on methionine, and also discuss their downstream effects on gene transcription.
Figure 1. Overview of how metabolism influences gene activation via chromatin regulations.
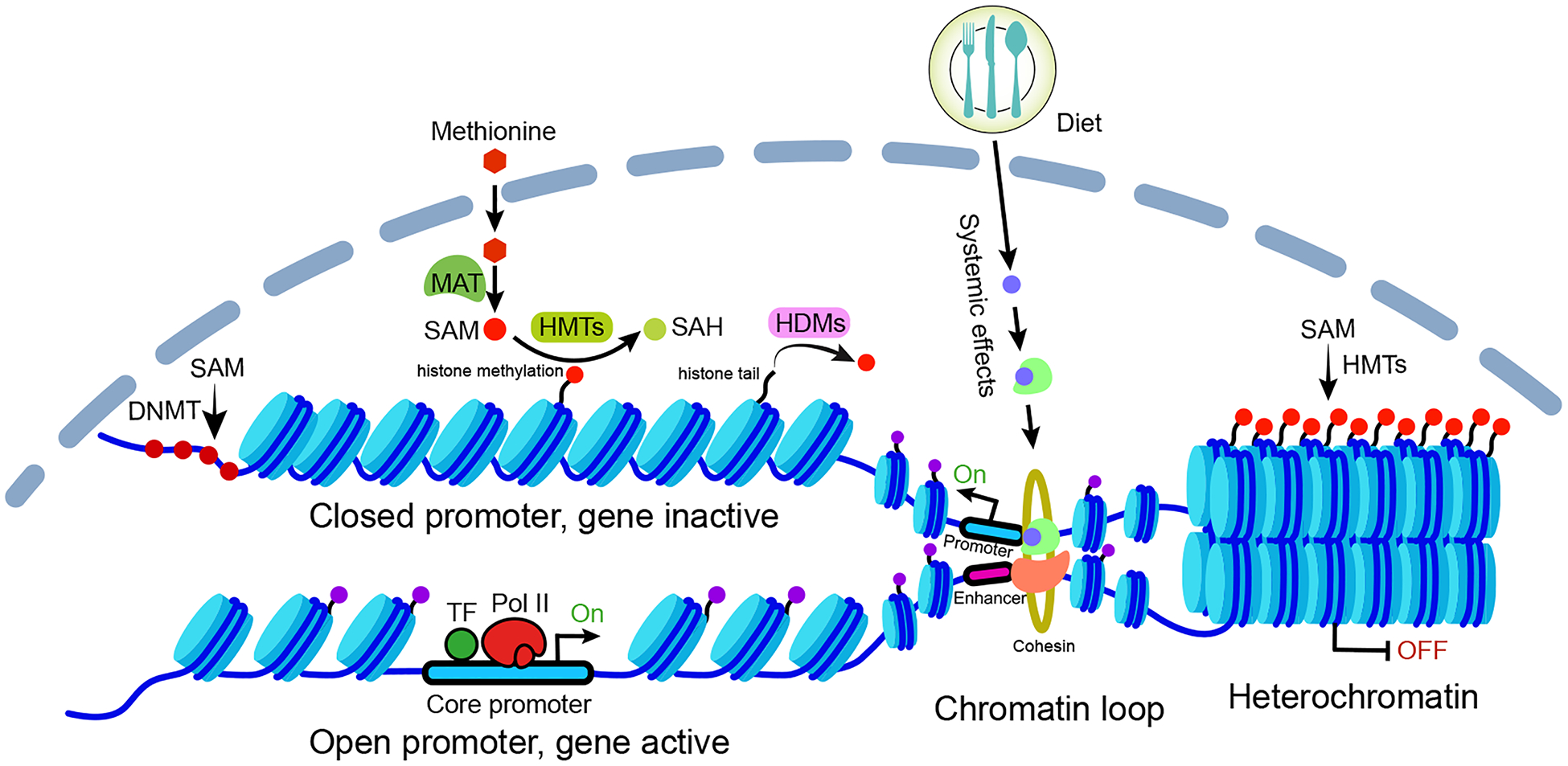
The gene activation and repression are mediated by chromatin structure and modifications, which can be regulated by metabolism through several mechanisms. Metabolic intermediates (e.g. S-adenosylmethionine [SAM]), catalyzed from the primary metabolites (e.g. methionine) by various enzymes (e.g. methionine adenosyltransferase [MAT]), are transported into the nucleus and serve as the substrates for histone and DNA modifications. Enzymes, such as methyltransferases and demethylases, are responsible for the deposition and removal of these modifications. Various modifications can either activate or repress gene transcription by altering DNA accessibility, for example, forming compact heterochromatin structure, exposing DNA regulatory elements to transcription factors (TFs) as well as transcription machinery (Pol II complex) by nucleosome remodeling. Moreover, the systemic effects of diets (e.g. activation of the nuclear receptor) can mediate chromatin looping and drive gene expression by promoting the interactions between enhancers and promoters. SAH: S-adenosylhomocysteine, is a product of methylation reaction involving SAM. DNMT, DNA methyltransferases; HMT, histone methyltransferase; HDM, histone demethylase.
The selective effect of methionine metabolism on histone methylation
Histones are modified by a plethora of chemical groups (e.g. methyl, acetyl, crotonyl, lactyl, and serotonyl), which represents a key chromatin feature that is often associated with gene regulation. Histone lysine methylation is a particularly important modification that involves both gene activation (e.g. H3K4me3 and H3K79me2) and repression (e.g. H3K27me3 and H3K9me3). The deposition of methyl groups on histone tails is catalyzed by methyltransferases that use S-adenosylmethionine (SAM) from the methionine cycle as the major donor of the methyl group (Fig. 2). Dietary methionine restriction has been used to elucidate its function on histone methylation and gene expression because of its profound impact on SAM production and promising influence on cancer therapeutic outcomes [3,4]. We and others have found that different types of cells respond to this nutritional variation selectively. For the cancer cells and certain types of lymphocytes such as Th cells, methionine restriction significantly impairs SAM synthesis, leading to loss of H3K4me3 and changes in gene expression relevant to one-carbon metabolism [5,6]. While for CD8+ T cells, low methionine selectively reduces H3K79me2 but not other marks, resulting in low expression of STAT5 and impaired T cell function [7] (Fig. 3a). What causes this disparity of epigenetic response to the same stimulus between different cell types? First, the cellular SAM level is highly dependent on the efficiency of methionine uptake. Two members of the solute carrier family (SLC7A5 and SLC43A2) responsible for methionine transportation are relatively lower expressed in CD8+ T cells than tumor cells, suggesting lower methionine consumption and lower SAM production in CD8+ T cells comparing to tumor cells [7]. Second, adding methyl groups donated by SAM to histone tails is catalyzed by a specific methyltransferase. Both H3K4me3 and H3K79me2 are histone marks of actively transcribed genes but deposited by different methyltransferases. H3K4me3 is catalyzed by the COMPASS-like methyltransferase family which requires a high level of SAM, whereas H3K79me2 is specifically catalyzed by methyltransferase DOT1L [8,9], which has a relatively low Michaelis constant (Km) thus use small amounts of SAM [10,11]. Therefore, both the deficiency of methionine transporters and low requirement of DOT1L to SAM concentration in CD8+ T cells may explain why H3K79me2 is more sensitive than other histone marks to methionine changes. However, tumor cells take in more methionine and generate more SAM for H3K4me3 deposition (Fig. 3a). According to Michaelis-Menten kinetic theory, each of these histone methylations may be highly sensitive to SAM accumulation when SAM concentration is close to the Km value of the corresponding methyltransferase [12]. A recent study reports that hepatocyte nuclear factor 4α (HNF4α) and key metabolic enzymes which mediate sulfur amino acid metabolism dictate the sensitivity of liver cancer to methionine restriction, but the underlying molecular mechanisms and their impacts on histone methylation warrant further investigation [13]. Interestingly, the breadth or length of the H3K4me3 domain, which has previously been suggested to be associated with transcription activity [14] and cell identity [15–18], can also respond to methionine restriction and shows a positive correlation with the differential gene expression [19]. Different methyltransferases are reported to specifically establish broad or narrow H3K4me3 domains [20]. Overall, both SAM availability and highly specific methyltransferases with different susceptibility to the fluctuation of its substrate determine the selective effects of methionine metabolism on histone modification and its breadth.
Figure 2. Methionine cycle provides methyl group for methylation.
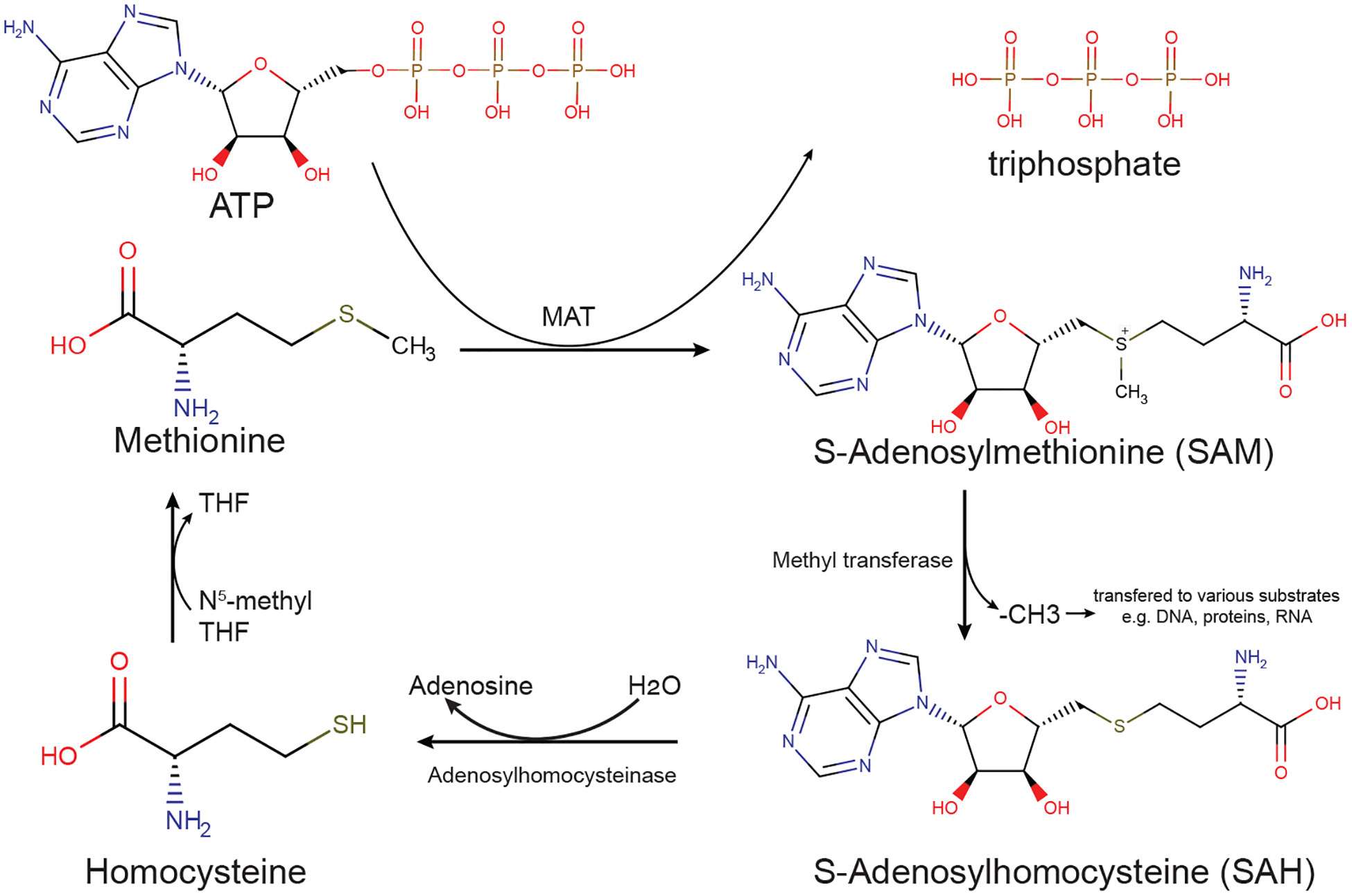
Methionine is converted into SAM by MAT in an ATP-dependent process. Various methyltransferases use SAM as the methyl donor for DNA, RNA, proteins, etc. and produce SAH. SAH is then catalyzed by adenosylhomocysteinase, forming adenosine and homocysteine. 5-methyl-THF donates its methyl group to homocysteine to synthesize methionine and produce tetrahydrofolate (THF). Chemical structures were obtained from the Human Metabolome Database (www.hmdb.ca).
Figure 3. Representative models for methionine metabolism-mediated chromatin regulations.

(a) Selective effect of methionine availability on histone methylation in tumor cell and CD8+ T cell. Cells take in methionine through transporters such as SLC7A5 and SLC43A2, which are highly expressed in the tumor cells than in CD8+ T cells. The lack of methionine transporters in CD8+ T cells leads to lower methionine uptake and less SAM production. The methyltransferase DOTL1 is sensitive to a small amount of SAM and specifically deposit H3K79me2. Although the tumor cells consume much more methionine and produce a large amount of SAM, which is used by COMPASS (Complex of Proteins Associated with Set1) to deposit H3K4me3. Both H3K79me2 and H3K4me3 are associated with active gene expression. (b) Reconfiguration of epigenetic modifications keeps genome stable and sustains DNA repetitive elements repression during metabolic stress. Heterochromatin is a highly condensed chromatin structure and is often marked by H3K9 and H3K27 methylation. Under the S-adenosylmethionine (SAM) limitation conditions, epigenetic modifications are regulated to preserves heterochromatin stability.
Histone methylation connects metabolism to chromatin compaction
Chromatin exists in two broadly distinct states: densely arranged heterochromatin and less condensed euchromatin. Compared with euchromatin, heterochromatin is transcriptionally inactivated or silenced because its tightly packaged conformation precludes regulatory elements and transcription factors (TFs) to access the chromatin. It is further categorized into facultative and constitutive heterochromatin based on their histone marks. Facultative heterochromatin is marked by H3 lysine 27 tri-methylation (H3K27me3) and can switch to more open, transcriptionally active conformation at a specific cell differential phase [21–23], whereas the constitutive heterochromatin is enriched with H3K9 methylation and is generally associated with the stable maintenance of gene silencing [24,25]. These modifications can be recognized by chromatin “readers” containing specific domains (e.g. HP1) and regulate chromatin compaction [26]. Glutamine deficiency, isocitrate dehydrogenase 1 mutation, and inhibition of H3K27 demethylases can regulate H3K27me3/H3K9me3 and contribute to gene regulation by affecting the production of α-ketoglutarate (α-KG), a key cofactor for the Jumonji-domain-containing histone demethylases (JHDMs) and 2-hydroxyglutarate (2HG), an inhibitor of α-KG [27–30]. Decondensation of heterochromatin in the absence of H3K27me2/3 is found to become more interactive with other euchromatin regions [31,32]. H3K9me3-decorated heterochromatin loss at protein-coding genes is concomitant with cell type-specific gene expression [33]. Therefore, these studies suggest the critical role of histone methylation in maintaining the compact chromatin structure. Heterochromatin also represents a compensatory system to maintain genome stability and suppress the activity of transposable elements to prevent their potentially harmful effects [34]. Recently, two studies reveal that the reconfiguration of epigenetic modifications compensates for the losses of histone/DNA methylation due to the insufficiency of the methyl-group donor SAM, allowing cells to preserve a compacted state and maintain the repressed state of repetitive DNA elements that enriched in heterochromatic regions (Fig. 3b). Haws et al. demonstrate that cells prefer mono methylation of H3K9 at the expense of broad losses in histone di- and tri-methylation at repetitive and transposable loci under SAM-depleted conditions [35]. Deblois et al. report that impairment of methionine metabolism significantly reduces the SAM level in breast cancer cells, leading to a global decrease in DNA methylation and accompanying with the reallocation of H3K27me3 modifications into regions enriched in transposable elements [36]. As we described in preceding paragraph, these different epigenetic changes may reflect the kinetic properties of various methylation-modifying enzymes and their sensitivities to fluctuations of cellular SAM. However, the detailed mechanisms of how heterochromatin organization is established and maintained remains a mystery. Thus, how methionine metabolism affects chromatin structure remains an emerging area of inquiry. Recent studies suggest that the formation of heterochromatin is mediated by phase separation [37,38], whereas the role of epigenetic modifications or in this process is still not answered.
Metabolic influences on DNA accessibility through nucleosome remodeling
The accessibility of DNA at promoters and enhancers to regulatory factors and transcriptional machinery is an important parameter of regulating transcription initiation. Both the chromatin compactness (as described in the preceding paragraph) and the nucleosome arrangement are thought to determine DNA accessibility. ATP-dependent chromatin remodeling complexes, such as INO80 and SWI/SNF, alter nucleosome position and occupancy (e.g. nucleosome sliding or eviction) to adjust the space between adjacent nucleosomes and contribute to nucleosome-free region that required for TFs binding [39,40]. Interestingly, recent studies suggest that these complexes regulate metabolic gene expression in response to changing nutrient environments [41–46]. A study by Gowans et al. reveals that INO80 complex rapidly and reversibly changes DNA accessibility of periodical genes crucial for coordinating cell division with cellular respiration in the yeast metabolic cycle [45]. Each remodeling complex possesses the same core ATPase subunit but different bromodomains that can read specific histone modifications [47]. For example, the mammalian SWI/SNF complex preferentially targets to specific enhancers and interact with p300 (acetyltransferase) to modulate H3K27ac [48]. The modifications, such as acetylation and succinylation, may reduce the affinity between DNA and histone core by altering the charge of histone tail [49,50]. H3K27ac is well recognized as a marker for active promoters and enhancers, and its distribution on chromatin is tied to metabolism because the substrate of histone acetyltransferase (i.e. acetyl-CoA) is produced from metabolic processes (e.g. glucose, fatty acid, and amino acid catabolism) that are highly dependent on nutrient availability. Recent studies show that under stress conditions, for example, glucose restriction, hypoxia, and mitochondrial stress, cells regulate the histone acetylation and DNA accessibility by controlling the pool of acetyl-CoA generated from available nutrients [51–53]. Nutritional supplements can increase the accumulation of cytoplasmic acetyl-CoA and facilitates the transportation of acetyl-CoA into the nucleus for histone acetylation, where significant changes in DNA accessibility could be detected by ATAC-seq at the activated genes along with the increasing of H3K27ac. Non-enzymatic covalent modifications (e.g. glycation) also connect the metabolic state with DNA accessibility by disrupting the assembly and stability of nucleosomes [54]. These and other studies suggest that regulation of gene transcription by rapidly altering DNA accessibility promotes metabolic homeostasis and affects cell differentiation [55–58]. How methionine and histone methylation might influence DNA accessibility warrants future study. Further investigation is required to investigate the precise mechanism of how metabolic status regulates nucleosome organization.
Diet and metabolism contribute to chromatin looping
The human genome contains hundreds of thousands of regulatory elements, such as enhancers that contain binding sites for TFs, allowing them to act together to control gene activation. Enhancer-associated histone marks (e.g. H3K4me1 and H3K27ac) can be regulated by methyltransferases and concentration of metabolites (e.g. glucose), then consequently influence the enhancer activity [59–61]. Enhancer elements can be located as much as a million base pairs away from target promoters, where the chromatin looping can bring them into very close spatial proximity (<200 nm) by “looping factors” (e.g. CTCF and cohesin), thereby facilitating enhancer-promoter interactions [62,63]. This mechanism allows for gene regulation. The DNA and histone methylation states can regulate the CTCF binding and recruitment of the cohesin, which, in turn, control chromatin loops at specific DNA sites and thus impact transcriptome diversity [64,65]. In such a way, T cells translate αKG-sensitive metabolic changes into context-dependent gene expression [65]. Studies have suggested that several proteins induced by chronic metabolic stress contribute to chromatin looping. Manuel et. al [66] have reported that during fasting, elevated glucocorticoids can regulate lipid metabolism through the glucocorticoid receptor (GR). Once stimulated by glucocorticoid, GR is translocated into the nucleus and interacts with SETDB2 which works as a signal responsive protein other than methyltransferase to facilitate long-range chromatin looping. The formation of chromatin loops consequently activates the target genes relevant to the metabolic stress of fasting in the liver, such as INSIG2. INSIG2 protein suppresses SREBP (sterol regulatory-element binding proteins) accumulation in the liver and inhibits lipogenesis. Another study shows that during β-adrenergic stimulation, phosphorylated JMJD1A, an H3K9 demethylase, can function as a cAMP-responsive protein that interacts with the SWI/SNF chromatin remodeling complex and DNA-bound PPARγ to induce long-range chromatin interactions which influence metabolic gene expression in brown adipocytes [67]. These cases highlight the diverse roles of histone modification proteins in response to environmental signals. With the development of chromatin conformation assays [68,69], more chromatin interactions involved in gene regulation to respond to nutrient availability are beginning to be identified. Using promoter capture Hi-C, Qin et. al. have detected more long-range enhancer-promoter interactions in the liver cells during metabolic adaptation to lipid-rich and carbohydrate-rich diets [70]. Depending on the given diet, the chromatin interactions are regulated either by activating pre-established chromatin loops or by forming new loops. Hnf4α is suggested to be activated by its ligand and can bind with other regulatory factors to activate chromatin loops under a lipid-rich diet. These chromatin loops upregulate genes involved in fatty acid oxidation and downregulate genes associated with de novo lipogenesis. Further studies are required to determine how such systemic effects on chromatin are induced, and which proteins are required to form new chromatin loops to adapt to the carbohydrate-rich diet. The extent to which metabolism itself such as methionine metabolism, aside from hormonal regulation influences three-dimensional structure is currently unclear. Nevertheless, these early studies support a model describing that the regulation of chromatin loops can be mediated by the cooperative action of stimulus-responsive molecules and proteins, which ensures gene activation during chronic metabolic stress. Chromatin looping often drives interactions between distal regulatory elements and target gene promoters in a cell-type-specific manner [71], it is thus interesting to investigate whether the chromatin interactions vary across different cell types under different metabolic stresses, and what metabolic consequences are associated with this variation.
Conclusions
We have discussed that various nutrients or metabolic stimuli elicit diverse changes in epigenetic modifications and chromatin structure, which enable cells to alter their gene expression programs. The selective impact of methionine metabolism on epigenetics is involved in cell proliferation and anti-tumor immunity, thus which have implications for tumor therapy. Because both tumor cells and T cells existing in the same tumor microenvironments compete for the limited nutrients, disrupting the entry of methionine to tumor cells while allowing T cell to take in more methionine may suppress the proliferation of tumor cells and restore T cell immunity [7]. The reconfiguration of histone methylation makes chromatin robust to tolerate metabolic alterations although the role of these modifications in organizing the compact chromatin structure needs to be clarified. Metabolism can also influence chromatin remodelers and histone modifications to affect DNA accessibility by altering nucleosome mobility, allowing a rapid and reversible way to control gene transcription. Finally, we highlight that chromatin loops induced by diets enable gene activation by promoting enhancer-promoter communications. Such mechanisms may allow for adaptation to long-term dietary changes as they might directly influence chromatin by metabolism through diet itself. Much remains to be discovered about how metabolites, metabolic enzymes, chromatin remodelers, and chromatin modifiers coordinately translate environmental cues into chromatin architecture.
Acknowledgments
J.W.L. gratefully acknowledges funding supports from National Institutes of Health (R01CA193256) and American Cancer Society (RSG-16-214-01-TBE). Z.X. thanks Yudong Sun and Dr. Ziwei Dai for suggestions.
Footnotes
Declaration of competing interest
J.W.L. serves advisory roles in Nanocare Technologies, Raphael Pharmaceuticals, and Restoration Foodworks. Z.X. declare no competing interest.
References
- 1.Goldstein I, Hager GL: Transcriptional and Chromatin Regulation during Fasting - The Genomic Era. Trends Endocrinol Metab 2015, 26:699–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Geisler CE, Hepler C, Higgins MR, Renquist BJ: Hepatic adaptations to maintain metabolic homeostasis in response to fasting and refeeding in mice. Nutr Metab (Lond) 2016, 13:62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sanderson SM, Gao X, Dai Z, Locasale JW: Methionine metabolism in health and cancer: a nexus of diet and precision medicine. Nat Rev Cancer 2019, 19:625–637. [DOI] [PubMed] [Google Scholar]
- 4.Gao X, Sanderson SM, Dai Z, Reid MA, Cooper DE, Lu M, Richie JP Jr., Ciccarella A, Calcagnotto A, Mikhael PG, et al. : Dietary methionine influences therapy in mouse cancer models and alters human metabolism. Nature 2019, 572:397–401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Mentch SJ, Mehrmohamadi M, Huang L, Liu X, Gupta D, Mattocks D, Gomez Padilla P, Ables G, Bamman MM, Thalacker-Mercer AE, et al. : Histone Methylation Dynamics and Gene Regulation Occur through the Sensing of One-Carbon Metabolism. Cell Metab 2015, 22:861–873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roy DG, Chen J, Mamane V, Ma EH, Muhire BM, Sheldon RD, Shorstova T, Koning R, Johnson RM, Esaulova E, et al. : Methionine Metabolism Shapes T Helper Cell Responses through Regulation of Epigenetic Reprogramming. Cell Metab 2020, 31:250–266. [DOI] [PubMed] [Google Scholar]
- 7.Bian Y, Li W, Kremer DM, Sajjakulnukit P, Li S, Crespo J, Nwosu ZC, Zhang L, Czerwonka A, Pawlowska A, et al. : Cancer SLC43A2 alters T cell methionine metabolism and histone methylation. Nature 2020, 585:277–282. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates that the consumption of methionine through transporter SLC43A2 in CD8+ T cells selectively influences H3K79me2 which controls STAT5 expression and affects T cell immunity.
- 8.Min J, Feng Q, Li Z, Zhang Y, Xu RM: Structure of the catalytic domain of human DOT1L, a non-SET domain nucleosomal histone methyltransferase. Cell 2003, 112:711–723. [DOI] [PubMed] [Google Scholar]
- 9.Nguyen AT, Zhang Y: The diverse functions of Dot1 and H3K79 methylation. Genes Dev 2011, 25:1345–1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horiuchi KY, Eason MM, Ferry JJ, Planck JL, Walsh CP, Smith RF, Howitz KT, Ma H: Assay development for histone methyltransferases. Assay Drug Dev Technol 2013, 11:227–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Richon VM, Johnston D, Sneeringer CJ, Jin L, Majer CR, Elliston K, Jerva LF, Scott MP, Copeland RA: Chemogenetic analysis of human protein methyltransferases. Chem Biol Drug Des 2011, 78:199–210. [DOI] [PubMed] [Google Scholar]
- 12.Reid MA, Dai Z, Locasale JW: The impact of cellular metabolism on chromatin dynamics and epigenetics. Nat Cell Biol 2017, 19:1298–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Q, Li Y, Gao X, Kang K, Williams JG, Tong L, Liu J, Ji M, Deterding LJ, Tong X, et al. : HNF4alpha regulates sulfur amino acid metabolism and confers sensitivity to methionine restriction in liver cancer. Nat Commun 2020, 11:3978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen K, Chen Z, Wu D, Zhang L, Lin X, Su J, Rodriguez B, Xi Y, Xia Z, Chen X, et al. : Broad H3K4me3 is associated with increased transcription elongation and enhancer activity at tumor-suppressor genes. Nat Genet 2015, 47:1149–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Benayoun BA, Pollina EA, Ucar D, Mahmoudi S, Karra K, Wong ED, Devarajan K, Daugherty AC, Kundaje AB, Mancini E, et al. : H3K4me3 breadth is linked to cell identity and transcriptional consistency. Cell 2014, 158:673–688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Xia B, Zhao D, Wang G, Zhang M, Lv J, Tomoiaga AS, Li Y, Wang X, Meng S, Cooke JP, et al. : Machine learning uncovers cell identity regulator by histone code. Nat Commun 2020, 11:2696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dahl JA, Jung I, Aanes H, Greggains GD, Manaf A, Lerdrup M, Li G, Kuan S, Li B, Lee AY, et al. : Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature 2016, 537:548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang B, Zheng H, Huang B, Li W, Xiang Y, Peng X, Ming J, Wu X, Zhang Y, Xu Q, et al. : Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature 2016, 537:553–557. [DOI] [PubMed] [Google Scholar]
- 19.Dai Z, Mentch SJ, Gao X, Nichenametla SN, Locasale JW: Methionine metabolism influences genomic architecture and gene expression through H3K4me3 peak width. Nat Commun 2018, 9:1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sze CC, Ozark PA, Cao K, Ugarenko M, Das S, Wang L, Marshall SA, Rendleman EJ, Ryan CA, Zha D, et al. : Coordinated regulation of cellular identity-associated H3K4me3 breadth by the COMPASS family. Sci Adv 2020, 6:eaaz4764. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study identified different members of the COMPASS family of methyltransferases that responsible for establishing broad and narrow H3K4me3 peaks.
- 21.Klar AJS, Ishikawa K, Moore S: A Unique DNA Recombination Mechanism of the Mating/Cell-type Switching of Fission Yeasts: a Review. Microbiol Spectr 2014, 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jia S, Yamada T, Grewal SI: Heterochromatin regulates cell type-specific long-range chromatin interactions essential for directed recombination. Cell 2004, 119:469–480. [DOI] [PubMed] [Google Scholar]
- 23.Soufi A, Donahue G, Zaret KS: Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 2012, 151:994–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noma K, Allis CD, Grewal SI: Transitions in distinct histone H3 methylation patterns at the heterochromatin domain boundaries. Science 2001, 293:1150–1155. [DOI] [PubMed] [Google Scholar]
- 25.Wang J, Jia ST, Jia S: New Insights into the Regulation of Heterochromatin. Trends Genet 2016, 32:284–294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allshire RC, Madhani HD: Ten principles of heterochromatin formation and function. Nat Rev Mol Cell Biol 2018, 19:229–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pan M, Reid MA, Lowman XH, Kulkarni RP, Tran TQ, Liu X, Yang Y, Hernandez-Davies JE, Rosales KK, Li H, et al. : Regional glutamine deficiency in tumours promotes dedifferentiation through inhibition of histone demethylation. Nat Cell Biol 2016, 18:1090–1101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Carey BW, Finley LW, Cross JR, Allis CD, Thompson CB: Intracellular alpha-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 2015, 518:413–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lu C, Ward PS, Kapoor GS, Rohle D, Turcan S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et al. : IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature 2012, 483:474–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cribbs AP, Terlecki-Zaniewicz S, Philpott M, Baardman J, Ahern D, Lindow M, Obad S, Oerum H, Sampey B, Mander PK, et al. : Histone H3K27me3 demethylases regulate human Th17 cell development and effector functions by impacting on metabolism. Proc Natl Acad Sci U S A 2020, 117:6056–6066. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that inhibition of demethylases KDM6A/B increases H3K27me3 levels and leads to downregulation of the key transcription factors that reduce mitochondrial biogenesis and regulate Th17 cell function.
- 31.Klocko AD, Ormsby T, Galazka JM, Leggett NA, Uesaka M, Honda S, Freitag M, Selker EU: Normal chromosome conformation depends on subtelomeric facultative heterochromatin in Neurospora crassa. Proc Natl Acad Sci U S A 2016, 113:15048–15053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xiong J, Zhang Z, Zhu B: Polycomb “polypacks” the chromatin. Proc Natl Acad Sci U S A 2016, 113:14878–14880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicetto D, Donahue G, Jain T, Peng T, Sidoli S, Sheng L, Montavon T, Becker JS, Grindheim JM, Blahnik K, et al. : H3K9me3-heterochromatin loss at protein-coding genes enables developmental lineage specification. Science 2019, 363:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Slotkin RK, Martienssen R: Transposable elements and the epigenetic regulation of the genome. Nat Rev Genet 2007, 8:272–285. [DOI] [PubMed] [Google Scholar]
- 35.Haws SA, Yu D, Ye C, Wille CK, Nguyen LC, Krautkramer KA, Tomasiewicz JL, Yang SE, Miller BR, Liu WH, et al. : Methyl-Metabolite Depletion Elicits Adaptive Responses to Support Heterochromatin Stability and Epigenetic Persistence. Mol Cell 2020, 78:210–223. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that H3K9 mono-methylation compensates the broad losses in H3K9 di- and tri-methylation under S-adenosylmethionine depleted condition, which enables cells to keep heterochromatin structure stable.
- 36.Deblois G, Tonekaboni SAM, Grillo G, Martinez C, Kao YI, Tai F, Ettayebi I, Fortier AM, Savage P, Fedor AN, et al. : Epigenetic Switch-Induced Viral Mimicry Evasion in Chemotherapy-Resistant Breast Cancer. Cancer Discov 2020, 10:1312–1329. [DOI] [PubMed] [Google Scholar]; This study shows that the redistribution of H3K27me3 compensates the loss of DNA methylation to sustain transposable elements repression, allowing cells to adapt the drug-induced metabolic stress.
- 37.Gibson BA, Doolittle LK, Schneider MWG, Jensen LE, Gamarra N, Henry L, Gerlich DW, Redding S, Rosen MK: Organization of Chromatin by Intrinsic and Regulated Phase Separation. Cell 2019, 179:470–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sanulli S, Trnka MJ, Dharmarajan V, Tibble RW, Pascal BD, Burlingame AL, Griffin PR, Gross JD, Narlikar GJ: HP1 reshapes nucleosome core to promote phase separation of heterochromatin. Nature 2019, 575:390–394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jiang C, Pugh BF: Nucleosome positioning and gene regulation: advances through genomics. Nat Rev Genet 2009, 10:161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Clapier CR, Verma N, Parnell TJ, Cairns BR: Cancer-Associated Gain-of-Function Mutations Activate a SWI/SNF-Family Regulatory Hub. Molecular Cell 2020, 80:712–725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pan D, Kobayashi A, Jiang P, Ferrari de Andrade L, Tay RE, Luoma AM, Tsoucas D, Qiu X, Lim K, Rao P, et al. : A major chromatin regulator determines resistance of tumor cells to T cell-mediated killing. Science 2018, 359:770–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Porter EG, Dhiman A, Chowdhury B, Carter BC, Lin H, Stewart JC, Kazemian M, Wendt MK, Dykhuizen EC: PBRM1 Regulates Stress Response in Epithelial Cells. iScience 2019, 15:196–210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yao W, King DA, Beckwith SL, Gowans GJ, Yen K, Zhou C, Morrison AJ: The INO80 Complex Requires the Arp5-Ies6 Subcomplex for Chromatin Remodeling and Metabolic Regulation. Mol Cell Biol 2016, 36:979–991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Beckwith SL, Schwartz EK, Garcia-Nieto PE, King DA, Gowans GJ, Wong KM, Eckley TL, Paraschuk AP, Peltan EL, Lee LR, et al. : The INO80 chromatin remodeler sustains metabolic stability by promoting TOR signaling and regulating histone acetylation. PLoS Genet 2018, 14:e1007216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gowans GJ, Schep AN, Wong KM, King DA, Greenleaf WJ, Morrison AJ: INO80 Chromatin Remodeling Coordinates Metabolic Homeostasis with Cell Division. Cell Rep 2018, 22:611–623. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study demonstrates the chromatin remodeling complex plays an important role in controlling the accessibility of periodic gene promoters in YMC. This mechanism ensures the coordination between cell division and cellular respiration.
- 46.Morrison AJ: Chromatin-remodeling links metabolic signaling to gene expression. Mol Metab 2020, 38:100973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dann GP, Liszczak GP, Bagert JD, Muller MM, Nguyen UTT, Wojcik F, Brown ZZ, Bos J, Panchenko T, Pihl R, et al. : ISWI chromatin remodellers sense nucleosome modifications to determine substrate preference. Nature 2017, 548:607–611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Alver BH, Kim KH, Lu P, Wang X, Manchester HE, Wang W, Haswell JR, Park PJ, Roberts CW: The SWI/SNF chromatin remodelling complex is required for maintenance of lineage specific enhancers. Nat Commun 2017, 8:14648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Fenley AT, Anandakrishnan R, Kidane YH, Onufriev AV: Modulation of nucleosomal DNA accessibility via charge-altering post-translational modifications in histone core. Epigenetics Chromatin 2018, 11:11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jing Y, Ding D, Tian G, Kwan KCJ, Liu Z, Ishibashi T, Li XD: Semisynthesis of site-specifically succinylated histone reveals that succinylation regulates nucleosome unwrapping rate and DNA accessibility. Nucleic Acids Res 2020, 48:9538–9549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Qiu J, Villa M, Sanin DE, Buck MD, O’Sullivan D, Ching R, Matsushita M, Grzes KM, Winkler F, Chang CH, et al. : Acetate Promotes T Cell Effector Function during Glucose Restriction. Cell Rep 2019, 27:2063–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Li Y, Gruber JJ, Litzenburger UM, Zhou Y, Miao YR, LaGory EL, Li AM, Hu Z, Yip M, Hart LS, et al. : Acetate supplementation restores chromatin accessibility and promotes tumor cell differentiation under hypoxia. Cell Death Dis 2020, 11:102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhu D, Wu X, Zhou J, Li X, Huang X, Li J, Wu J, Bian Q, Wang Y, Tian Y: NuRD mediates mitochondrial stress-induced longevity via chromatin remodeling in response to acetyl-CoA level. Sci Adv 2020, 6:eabb2529. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study shows that acetyl-CoA is a critical metabolic signal to regulate aging through histone deacetylases complex (NuRd) and chromatin remodeling during mitochondrial stress.
- 54.Zheng Q, Omans ND, Leicher R, Osunsade A, Agustinus AS, Finkin-Groner E, D’Ambrosio H, Liu B, Chandarlapaty S, Liu S, et al. : Reversible histone glycation is associated with disease-related changes in chromatin architecture. Nat Commun 2019, 10:1289. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study suggests that histone glycation causes changes in gene transcription by increasing the accessibility of certain genes.
- 55.Yucel N, Wang YX, Mai T, Porpiglia E, Lund PJ, Markov G, Garcia BA, Bendall SC, Angelo M, Blau HM: Glucose Metabolism Drives Histone Acetylation Landscape Transitions that Dictate Muscle Stem Cell Function. Cell Rep 2019, 27:3939–3955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Johnson MO, Wolf MM, Madden MZ, Andrejeva G, Sugiura A, Contreras DC, Maseda D, Liberti MV, Paz K, Kishton RJ, et al. : Distinct Regulation of Th17 and Th1 Cell Differentiation by Glutaminase-Dependent Metabolism. Cell 2018, 175:1780–1795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Leung A, Parks BW, Du J, Trac C, Setten R, Chen Y, Brown K, Lusis AJ, Natarajan R, Schones DE: Open chromatin profiling in mice livers reveals unique chromatin variations induced by high fat diet. J Biol Chem 2014, 289:23557–23567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gowans GJ, Bridgers JB, Zhang J, Dronamraju R, Burnetti A, King DA, Thiengmany AV, Shinsky SA, Bhanu NV, Garcia BA, et al. : Recognition of Histone Crotonylation by Taf14 Links Metabolic State to Gene Expression. Mol Cell 2019, 76:909–921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Yan J, Chen SA, Local A, Liu T, Qiu Y, Dorighi KM, Preissl S, Rivera CM, Wang C, Ye Z, et al. : Histone H3 lysine 4 monomethylation modulates long-range chromatin interactions at enhancers. Cell Res 2018, 28:204–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Loubiere V, Papadopoulos GL, Szabo Q, Martinez AM, Cavalli G: Widespread activation of developmental gene expression characterized by PRC1-dependent chromatin looping. Sci Adv 2020, 6:eaax4001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Miguel-Escalada I, Bonas-Guarch S, Cebola I, Ponsa-Cobas J, Mendieta-Esteban J, Atla G, Javierre BM, Rolando DMY, Farabella I, Morgan CC, et al. : Human pancreatic islet three-dimensional chromatin architecture provides insights into the genetics of type 2 diabetes. Nat Genet 2019, 51:1137–1148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Schoenfelder S, Fraser P: Long-range enhancer-promoter contacts in gene expression control. Nat Rev Genet 2019, 20:437–455. [DOI] [PubMed] [Google Scholar]
- 63.Robson MI, Ringel AR, Mundlos S: Regulatory Landscaping: How Enhancer-Promoter Communication Is Sculpted in 3D. Mol Cell 2019, 74:1110–1122. [DOI] [PubMed] [Google Scholar]
- 64.Nanavaty V, Abrash EW, Hong C, Park S, Fink EE, Li Z, Sweet TJ, Bhasin JM, Singuri S, Lee BH, et al. : DNA Methylation Regulates Alternative Polyadenylation via CTCF and the Cohesin Complex. Mol Cell 2020, 78:752–764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Chisolm DA, Savic D, Moore AJ, Ballesteros-Tato A, Leon B, Crossman DK, Murre C, Myers RM, Weinmann AS: CCCTC-Binding Factor Translates Interleukin 2- and alpha-Ketoglutarate-Sensitive Metabolic Changes in T Cells into Context-Dependent Gene Programs. Immunity 2017, 47:251–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Roqueta-Rivera M, Esquejo RM, Phelan PE, Sandor K, Daniel B, Foufelle F, Ding J, Li X, Khorasanizadeh S, Osborne TF: SETDB2 Links Glucocorticoid to Lipid Metabolism through Insig2a Regulation. Cell Metab 2016, 24:474–484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Abe Y, Rozqie R, Matsumura Y, Kawamura T, Nakaki R, Tsurutani Y, Tanimura-Inagaki K, Shiono A, Magoori K, Nakamura K, et al. : JMJD1A is a signal-sensing scaffold that regulates acute chromatin dynamics via SWI/SNF association for thermogenesis. Nat Commun 2015, 6:7052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Yu M, Ren B: The Three-Dimensional Organization of Mammalian Genomes. Annu Rev Cell Dev Biol 2017, 33:265–289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lieberman-Aiden E, van Berkum NL, Williams L, Imakaev M, Ragoczy T, Telling A, Amit I, Lajoie BR, Sabo PJ, Dorschner MO, et al. : Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 2009, 326:289–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Qin Y, Grimm SA, Roberts JD, Chrysovergis K, Wade PA: Alterations in promoter interaction landscape and transcriptional network underlying metabolic adaptation to diet. Nat Commun 2020, 11:962. [DOI] [PMC free article] [PubMed] [Google Scholar]; This study suggests that the liver cells can adapt to different extreme diets by either activating pre-existing chromatin loops or forming new chromatin loops.
- 71.Diehl AG, Ouyang N, Boyle AP: Transposable elements contribute to cell and species-specific chromatin looping and gene regulation in mammalian genomes. Nat Commun 2020, 11:1796. [DOI] [PMC free article] [PubMed] [Google Scholar]


