Abstract
Adenoid cystic carcinoma (ACC) of salivary gland is a slowly growing tumor showing a propensity for delayed recurrence, with decreased survival rates. The identification of poor prognosis patients may help in defining molecular-based targeted strategies in this rare disease orphan of new treatments. Through a gene expression microarray-based approach followed by GSE functional analysis the expression profile of 46 primary untreated ACC samples and of ACC (h-TERT) tumor cells was analyzed. Patients who experienced early relapse showed enrichment in proliferation-related gene sets, including the G2-M checkpoint, E2F and myc targets, and in gene sets related to IFN signaling and aberrant proteostasis (FDR < 0.1), indicating increased mitotic and transcriptional activity in aggressive ACC. Similar functions were enriched in ACC samples classified by immunohistochemical staining as p63-negative, which exhibited increased protein burden and activation of pro-survival stress response pathways compared to p63-positive tumors. Compared to ACC tissues, ACC (h-TERT) cells share transcriptional features of aggressive p63-negative tumors. These data suggest association of specific pathway alterations with histopathological features of ACC, as recapitulated by p63 testing in patient prognostic stratification, anticipating new avenues for therapeutic intervention.
Subject terms: Head and neck cancer, Cancer genomics, Oncology
Introduction
Adenoid Cystic Carcinoma (ACC) is a rare cancer, representing 1% of all Head and Neck Cancers (HNC) and 20% of all Salivary Gland Cancers (SGC)1, with a reported incidence of 4.5 cases/100,000 individuals2. It is generally considered as a long-term poor prognosis tumor with survival rates at 5,10, and 20 years around 68%, 52% and 28% respectively3. Despite advances in the gold standard treatment for the locoregional disease, represented by surgery and radiotherapy (RT), more than 75% of patients with locally advanced disease experience relapse, and from 8 to 60% of patients, according to different studies, develop distant metastasis4,5.
Among the reasons for this poor prognosis is the lack of effective clinical markers, informative of the clinical evolution of the disease, and/or predictive of treatment response, that could pave the way to more tailored therapeutic approaches. Moreover, despite advances in understanding the ACC underlying molecular alterations, tumor biology remains actually largely unknown, and the application of genomic predictors in clinical settings is still limited. The comprehensive genomic analysis of ACC samples has shown few recurrent mutations in known genes and a substantially low mutational burden, confirming the MYB/MYBL1-NFIB fusions as key events in the pathogenesis of ACC and a hallmark of this pathology, being present in more than 50% of cases6,7. Among the prognostic markers and potential therapeutic targets, the NOTCH mutation, reported in 15–20% of tumors, has been associated with advanced stages, solid histology, and shorter Overall Survival (OS), the latter only in univariate analysis8. Recently, a multidimensional analysis integrating mutational, transcriptional and proteomic data, highlighted the existence of two ACC molecular subtypes, ontogenetically distinct, with multiple and unique molecular alterations in each subtype, providing opportunities for combinational therapies9.
While histological and clinical characteristics could not totally describe the behaviour of the disease, a more in-depth molecular and biological analysis of this neoplasm might help both in depicting prognostic information and finding new potential therapeutic targets. The main aim of our study is to evaluate prognostic features and potential targetable pathways using gene expression analysis in a series of curatively treated ACC.
Patients and methods
Patient information
We included all consecutive treatment naïve patients diagnosed and treated with curative intent for ACC at the Unit of Otorhinolaryngology-Head and Neck Surgery, ASST Spedali Civili di Brescia, Italy, from 1996 to 2019. Eligible patients had a histological diagnosis of ACC, they were primarily treated with surgery, they had a histological sample available for tissue analysis and an adequate follow up. Patients could receive postoperative radiation, according to clinical stage and pathological characteristics. Written informed consent was obtained from all patients enrolled as per protocol approved by the Research Review Board-the Ethic Committee- of the ASST Spedali Civili, Brescia, Italy (study reference number: NP4266). The clinical and follow-up information were recorded from patients’ files. Clinical stage was re-assessed according to the 8th edition of the AJCC classification.
Sample collection and histopathological analysis
Paraffin blocks from ACC tissue were generated at the time of surgery, from the primary tumor. All cases were reviewed by two expert head and neck pathologists (A.B. and L.A.) and the diagnosis were confirmed using H&E staining. Histologic grading was obtained by evaluating growth patterns, the percentage of solid areas and the cytological characteristics of the neoplastic cells. Carcinoma mostly tubular with some cribriform patterns, with the absence of solid areas or nuclear pleomorphism and low mitotic activity were classified as grade 1; cases with pure cribriform or mixed tubular/cribriform patterns, solid areas lower than 30% and slightly greater degree of nuclear pleomorphism and mitotic activity were classified as grade 2; cases with solid pattern greater than 30%, with a more significant nuclear pleomorphism, mitotic activity and necrosis were classified as grade 310. Immunohistochemical analysis with p63 monoclonal antibody (DAK-63; Agilent) through Leica Bond III staining platform was performed on serial 5 µm sections of selected FFPE representative tumor blocks. P63 expression was considered positive when uniform or diffuse strong nuclear staining was observed in > 70% of tumor cells, as previously described9.
RNA extraction and gene expression profiling
Total RNA was isolated with the RNeasy DSP FFPE Kit (Qiagen) according to the manufacturer’s instructions. RNA concentration and 260/280 absorbance ratio (A260/280) was measured with the Infinite M200 spectrophotometer (TECAN) and quality control of extracted RNA assessed with RT-qPCR for 18S rRNA. RNA from ACC (h-TERT) cells was kindly gifted by Prof. Sandra Sigala (University of Brescia).
To generate gene expression profiles, total RNA was converted to cRNA and then to cDNA, and hybridized, according to the manufacturer’s guidelines, on the Human Clariom S GeneChip (ThermoFisher Scientific), optimized for FFPE samples and able to accurately measure gene-level expression from > 20,000 well-annotated genes. The Scanner 3000 7G (ThermoFisher Scientific) was used in conjunction with GeneChip Operation Software (ThermoFisher Scientific) to generate one CEL file per hybridized cDNA.
Microarray data processing and analysis
Arrays underwent quality control before being processed with RMA algorithm11. Hierarchical clustering was performed using Euclidean distance metric and Ward’s agglomerative algorithm12. Genes were filtered based on association with DFS evaluated using gene-level multivariable Cox models. Top genes were selected based on unadjusted p-value less than 5%. Differential expression (DE) between groups was modelled using linear models with t-statistic computation based on Empirical Bayesian algorithm (limma)13. Filtered features were further processed to evaluate potential genesets enrichment using Gene Set Enrichment Analysis (GSEA)14,15 and both Hallmark16 and GO geneset lists.
An enrichment score for selected genesets (hereafter global score) for each patient were computed using single sample GSEA (ssGSEA)17 which computes an enrichment score for every subject-geneset based on a non-parametric approach: such score can be interpreted as the level of global expression of a specific geneset for every subject.
Statistical analysis
Data were described using mean (standard deviation) and median (Interquartile range, IQR) for quantitative variables and percentages for categorical.
Association between categorical variables was evaluated using a Chi-squared test with p-value computed using Monte Carlo simulation (B = 2000).
Disease-free survival (DFS) was modelled as a function of clinical variables using Cox models. Follow-up median time was estimated using the Kaplan–Meier method. Optimal cutpoints for continuous variables, as predictors of time-to-event outcomes, were computed using maximally selected log-rank statistics18. All tests were two sided and assumed a significance level of 5%. All analyses were performed using R (version 4.1.1).
Institutional review board statement
The study was conducted according to the guidelines of the Declaration of Helsinki and approved by the Institutional Review Board of ASST Spedali Civili, University of Brescia, Italy (study reference number: NP4266).
Results
Clinical and pathological description of ACC cohort
We identified 83 patients with ACC matching inclusion criteria, whose clinical and pathological characteristics are summarized in Table 1. Mean age at diagnosis was 53.4 years (median 53.5, range [21–85]) and gender was predominantly female (67%). In 74% of cases ACC developed from minor salivary glands. At diagnosis, 77% of tumors were classified as T3–4, 88% had no nodal involvement, and 70% of the patients with nodal involvement presented extranodal extension. Adjuvant radiotherapy was performed in 81% of the patients. During follow-up, 29 patients died (35%) and 42 (51%) had a disease relapse; median follow-up time was 124 months (IQR 61–167), while median time to relapse was 77 months. Overall, 37 patients were excluded from subsequent analysis for either poor quality of FFPE tumor block (N = 5) or poor quality of extracted RNA (N = 32). Characteristics of the 37 patients excluded were compared to those of eligible patients, to investigate possible selection bias. No significant differences were found (Supplementary Table S1).
Table 1.
Patient cohort description.
| Overall ACC cohort n (%) |
Gene expression study ACC cohort n (%) |
|
|---|---|---|
| Number of patients | 83 (100%) | 46 (55%) |
| Gender | ||
| Female | 56 (67) | 35 (76) |
| Male | 27 (33) | 11 (24) |
| Age at presentation (years) | ||
| Median | 53.5 | 51.0 |
| Range | (21–85) | (21–85) |
| pT | ||
| pT1 | 10 (12) | 5 (11) |
| pT2 | 6 (7) | 3 (7) |
| pT3 | 14 (17) | 10 (22) |
| pT4 | 50 (60) | 27 (59) |
| Miss | 3 | 1 |
| pN | ||
| − | 73 (88) | 41(89) |
| + | 10 (12) | 5 (11) |
| ENE | ||
| − | 3 (30) | 2 (40) |
| + | 7 (70) | 3 (60) |
| Grade | ||
| I–II | – | 35 (76) |
| III | – | 11 (24) |
| Site | ||
| Major | 22 (26) | 14 (30) |
| Minor | 61 (74) | 32 (70) |
| Adjuvant RT | ||
| No | 15 (18) | 12 (26) |
| Yes | 67 (81) | 33 (72) |
| Miss | 1 | 1 |
| Recurrence at last follow up | ||
| No | 41 (49) | 19 (41) |
| Yes | 42 (51) | 27 (59) |
| Dead at last follow up | ||
| No | 54 (65) | 29 (63) |
| Yes | 29 (35) | 17 (37) |
ENE extra nodal extension, RT radiotherapy, N lymph node.
Pathological grading was performed on H&E sections of the 46 ACC patients eligible for gene expression profiling. As shown in Table 1, 35 out of 46 patients (76%) were classified as grade 1/2, while 11 cases were classified as grade 3 (24%).
Survival analysis of clinical and pathological risk factors for local–regional recurrence or distant metastasis indicate that T class, histological grade, and adjuvant RT have prognostic impact on DFS, with advanced stage and high grade independently associated with reduced DFS (Table 2).
Table 2.
Disease-free survival analysis for ACC patients (n = 83). Clinical and pathological risk factors for local–regional recurrence or distant metastasis were analyzed using Multivariable Cox’s proportional-hazards models.
| Characteristics | DFS | |||
|---|---|---|---|---|
| Univariate | Multivariate | |||
| HR (95% CI) | p value | HR (95% CI) | p value | |
| Gender | ||||
| Male vs female | 1.72 (0.92–3.24) | 0.09 | 1.49 (0.77–2.88) | 0.293 |
| T stage | ||||
| T3–4 vs T1–2 | 2.64 (0.94–7.41) | 0.065 | 4.69 (1.41–15.6) | 0.012 |
| Histological grade | ||||
| G3 vs G1–2 | 3.92 (1.72–8.94) | *0.001 | 4.92 (1.86–13) | *0.001 |
| Site | ||||
| Major vs Minor | 1.12 (0.56–2.22) | 0.754 | 0.90 (0.43–1.87) | 0.778 |
| Adjuvant RT | ||||
| Yes vs no | 0.75 (0.35–1.63) | 0.472 | 0.30 (0.12–0.78) | 0.013 |
HR hazard ratio, CI confidence interval.
*n = 46.
Different biological processes are associated with prognosis in ACC
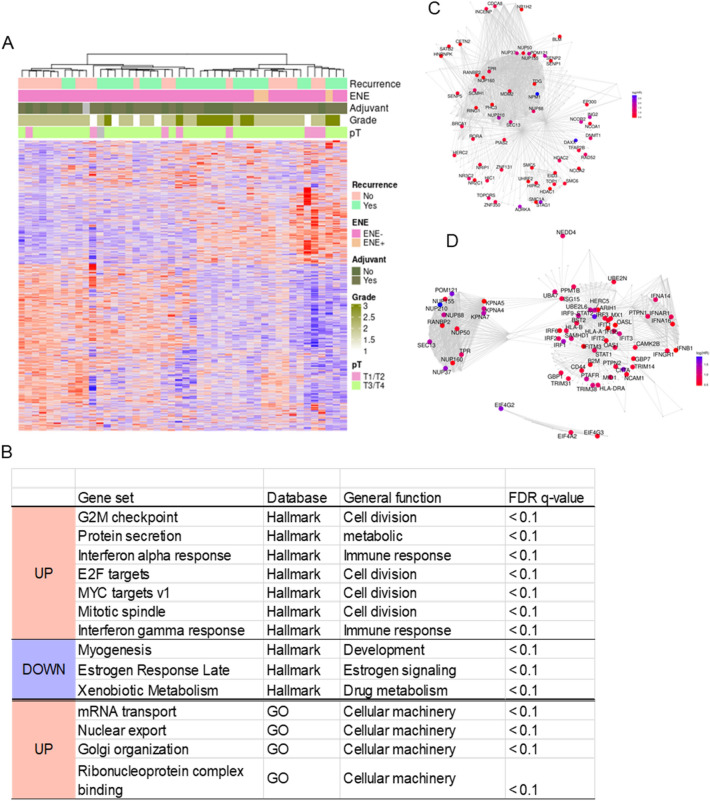
The global gene expression profile of ACC samples was generated by microarray platform, to obtain a comprehensive picture of transcriptomic landscape characterizing relapsing patients. Analysis was performed at gene-level using multivariate Cox models. Of the ~ 20,000 genes present on the chip, we found 650 genes significantly associated with DFS (p-value < 5%). Hierarchical clustering of the selected genes identified two distinct clusters with no evident enrichment for advanced stage tumors, grade or specific subsites nor difference according to adjuvant treatment strategy among the subgroups (Fig. 1A).
Figure 1.
Transcriptional profiling to predict prognosis of ACC patients. (A) Heatmap for hierarchical clustering of relevant genes selected based on DFS hazard ratio (p value < 0.05). Clinicopathologic ACC features shown in the upper panels. (B) Pathways enriched in patients with short DFS by GSEA with Hallmark and GO genesets. (C,D) REACTOME pathways for SUMOylation and Interferon Signaling. Color band displays the log (HR) value for the leading-edge genes in each pathways.
Using Hallmark collection, GSEA shows that the top genesets positively associated with shorter DFS are G2-M checkpoint genes, mitotic spindle, E2F and myc targets (all FDR < 0.1) These are largely recapitulated by GO functional enrichment analysis, which indicates cell cycle regulation, protein synthesis and ribosome genesis pathways significantly up-regulated in patients with early relapse (all FDR < 0.1) and collectively suggest that ACC progression is marked by both an increased mitotic activity and rates of protein synthesis. Moreover, the up-regulation of Interferon alpha and gamma response genesets in poor-prognosis patients indicates increased expression of target genes in response to IFN stimulation (Fig. 1B, Supplementary Table S2). The top down-regulated genesets include xenobiotic metabolism, estrogen response and myogenesis (all FDR < 0.1), the latter highlighting the loss of myoepithelial component in aggressive tumors (Fig. 1B). REACTOME analysis shows a similarity of enriched pathways in high-risk ACC, confirming a dysregulation of cell cycle and IFN signaling, and an increase in SUMOylation, a key post-translational modification associated with chromatin remodeling which play stress-mitigation function under strong oncogenic stimuli (FDR < 0.05, Fig. 1C,D, Supplementary Table S2).
Defining ACC molecular subgroups with different prognosis
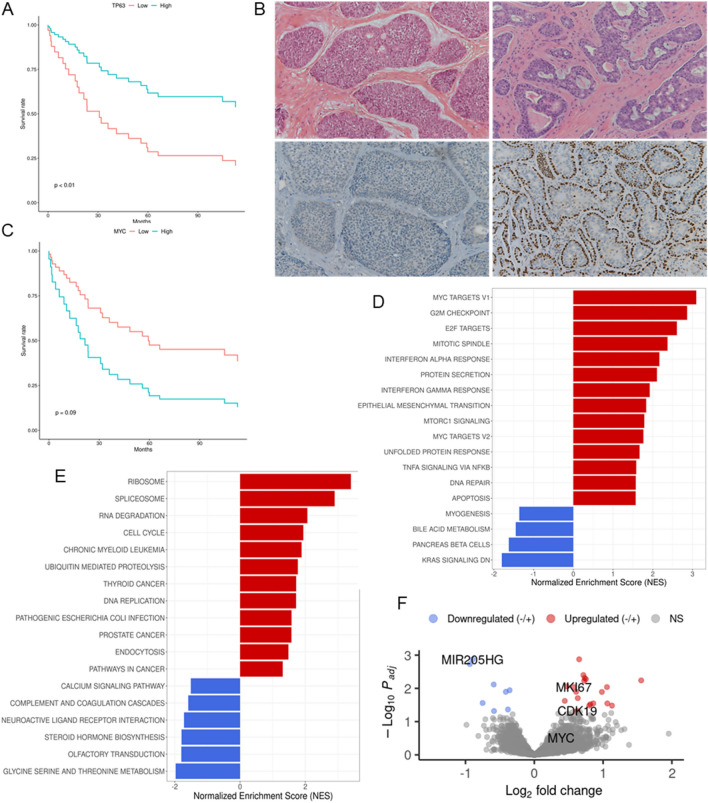
A molecular subtyping of ACC centered on TP63 expression has been proposed as opposed to constitutive activation of MYC induced by NOTCH mutation/pathway alteration, both indicated as drivers of unique regulatory programs in ACC, with direct prognostic implications9. Following this concept, we evaluated patients' survival as a function of TP63 mRNA expression level, whose optimal cut-off value was computed using maximally selected statistics, and we observed a significantly reduced DFS in patients with low TP63 expression (HR: 0.34, CI95% 0.15–0.77, p = 0.01, Fig. 2A). This prognostic relevance is independent of tumor stage and adjuvant treatment but strongly correlated to histological grade. Indeed, immunohistochemistry confirmed negative p63 expression in almost all grade 3 cases (9/11, 82%), with only two high grade cases revealing strong and diffuse expression of p63 in solid areas, whereas all grade 1–2 ACC showed positive p63 expression with a strong nuclear staining in > 30% of tumor cells (100% positive in grade I and II vs 18% in grade III, Chi-squared test p-value < 0.01) (Supplementary Table S3, Fig. 2B). At functional level, these data are consistent with the down-regulation of myogenesis pathway observed in early relapsing patients, and indicate p63 loss as a predictor of unfavourable outcome.
Figure 2.
Distinct transcriptional features are associated with p63-negative ACC. (A–C) Kaplan–Meier curves showing the DFS as a function of TP63 or MYC mRNA expression. p-value computed from Likelihood Ratio Test (LRT). (B) Representative cases of ACC showing either diffuse strong nuclear p63 immunostaining in more than 70% of tumor cells (low right panel) or completely negative p63 expression (low left panel). Corresponding H&E sections are shown in the upper panel (10 × magnification). (D,E) Functional enrichment analysis depicting biological processes up-regulated in p63neg ACC group compared to p63pos tumors, considering both Hallmark and KEGG genesets. Each distribution represents the t-statistic associated to the core-enrichment genes of each pathway. (F) Volcano plot of genes with increased expression in p63neg (red dots) and p63pos (blue dots) ACCs.
We then performed survival analysis according to MYC mRNA expression. Although with marginal statistical significance, evident survival differences were observed between patients in high and low MYC score (HR: 2.60, CI95% 0.94–7.15, p = 0.065, Fig. 2C). Of interest, exploring MYC expression in our cohort of patients classified as p63-positive (p63pos) or p63-negative (p63neg) according to IHC staining, we found an increase of myc mean expression in p63neg samples (FC = 1.311, p = 0.008, Supplementary Table S3). GSEA based on Hallmark collection, show that cell-cycle and proliferation related genesets, such as MYC targets, G2M checkpoint and E2F targets were significantly enriched in the p63neg tumors, along with protein secretion and unfolded protein response (UPR), a network of signaling pathways that respond to stress in the endoplasmic reticulum (ER), and referred to as important mediators in maintaining cell survival in stressed conditions (all FDR < 0.05). MTORC1 signaling and DNA damage, two closely related pathways that communicate to face metabolic and xenotoxic stresses, were also enriched in the p63neg group, together with the immune-related pathways interferon gamma/alpha responses. The RAS signaling, composed of genes that are downregulated when the KRAS signaling pathway is activated, is among the most significantly enriched pathways in p63pos ACCs (Fig. 2D). KEGG pathway analysis19–21 shows consistent enrichment in cell cycle, ribosome and spliceosome functions and increase ubiquitin-mediated proteolysis in p63neg, overall suggestive of a compromised metabolic environment (Fig. 2E).
Among genes with the highest differential expression between p63pos and p63neg ACC we shortlisted the proliferation marker MKI67 and the cyclin dependent kinase CDK19, both highly upregulated in p63neg tumor (FC > 1.2, FDR < 0.05, p < 0.001). MIR205HG, a long non-coding RNA enriched in the basal layer of epithelia and whose expression is regulated by p63, was among the highest up-regulated genes among the p63pos (FC > 1.2, FDR < 0.05, p < 0.001) (Fig. 2F, Supplementary Table S4).
The transcriptional profile of aggressive ACC is recapitulated in cell model of the disease
We exploited microarray data to link ACC transcriptomics with pharmacological information previously obtained in ACC (h-TERT) tumor cells22, a well-established experimental model of ACC with epi-myoepithelial derivation, lacking p63 expression23. By comparing the expression profile of ACC patients to cells, we found that the transcriptional profile of ACC (h-TERT) resembles that of ACC recurrent patients and indirectly links to myc expressing (Supplementary Fig. S1), suggesting that this molecular profile is maintained also when cancer cells are cultured in vitro, and collectively indicating ACC (h-TERT) as a cell model of aggressive ACC.
Discussion
The clinical behavior of ACC is not straightforward, as many patients with locally advanced disease show a relapse, but with a different timing of recurrence, disease aggressiveness, type of relapse and response to treatments. This unpredictable clinical course is mirrored by the lack of strong pathological and molecular data, which do not fully capture the extent of heterogeneity present in this tumor. Herein, we exploited microarray-generated gene expression data to identify functional profiles associated with poor disease-free survival in curatively treated ACC, in the attempt to clarify the molecular mechanism driving its propensity to recur and to provide additional information to current prognostic assessment. By integrating transcriptomic alteration with patient clinical and pathological information, we demonstrated the prognostic relevance of multiple signaling pathways altered in ACC, extending the vision on the molecular processes mediating ACC biological aggressiveness beyond mutational mechanism24–26. These include the downregulation of the myogenesis pathway and the enrichment of proliferation and cell-cycle related processes such as myc targets activation and increased expression of Ki-67 mRNA, as well as increased protein burden and signs of energetic stress. Of note, functional analysis of samples clustered based on p63 immunostaining revealed highly consistent pathway alterations in p63-negative tumors and in early relapsing patients, suggesting a link between molecular events underlying the pathway p63 negative (mostly solid tumor and with high Ki67) and ACC adverse outcome. Together these results are in agreement with Ferrarotto and colleagues9 and functionally reflect the good prognosis of p63-positive ACC type II and the poor-prognosis of myc-driven ACC type I. An increased level of UPR signaling observed in p63-negative ACCs marks the occurrence of ER stress in this tumor subgroup and provides further connection with myc oncogenic activation27. This pro-survival stress response pathway, which helps cells to adapt to the higher demand of protein synthesis to sustain oncogenic-driven unrestricted cell proliferation, has also been implicated in chemotherapy resistance in several tumors, including breast and colorectal cancers28,29 through a mechanism that remains poorly understood, but that at the same time could pave the way towards new therapeutic application for chemo and radiotherapy refractory patients. For instance, small-molecule UPR inhibitors that directly target UPR sensor, such as IRE1alpha, have demonstrated antitumor activity and enhanced response to chemotherapy in preclinical model of triple negative breast cancer and pancreatic neuroendocrine tumors30,31. Along with the global increase of transcriptional activity, ER stress was also reported to enhance chromatin accessibility in activated genes32, suggesting the occurrence of epigenetic deregulation during ACC progression and potentially anticipating new avenues for therapeutic intervention.
Beside key alterations involving cell cycle related functions, pathway analysis indicates an activation of endogenous IFN signaling in aggressive ACC tumors. At odds with the well-recognized pro-apoptotic, pro-inflammatory, antitumor role of IFNs several lines of evidence raised in support of the pro-tumoral properties of this cytokine33,34 and the activation of IFN signaling in tumor cells has been linked to an unfavorable response to DNA damage intervention, including radiation35. In this regard, specific signatures of IFN-stimulated genes were traced in breast cancer patients and correlated with radiotherapy resistance and poor prognosis36.
Of note, the transcriptional profile of aggressive ACC is recapitulated in the cell model of the disease. Indeed, when compared with tumors, ACC (h-TERT) cells reveal the molecular features of aggressive ACC and look transcriptionally representative of patients with poor prognosis. As recently published by Savarese and colleagues testing ACC (h-TERT) cells with targeted agents show poor efficacy for either multikinase inhibitor lenvatinib and PI3-K inhibitor everolimus22. This finding is in line with the downregulation of RAS signaling pathways observed in poor prognosis patients and consistent with the upregulation of receptor tyrosine kinases genes in p63-positive ACC type II reported by Ferrarotto9, highlighting the importance of molecular screening for combination treatments, that currently does not represent a selection criterion for tailored treatments. Besides, our data strengthen the usefulness of p63 assessment for predicting biological behavior as p63 might represent a more reliable and reproducible marker than histopathological grading, which is collinear with other clinical pathological factors and suffers from interobserver variation37. Further studies in a large series of ACCs are warranted to confirm the potential of p63 as a prognostic marker as well as molecular marker for patient stratification in clinical trials.
From the technical standpoint, we proved the feasibility of a microarray-based transcriptional approach on FFPE samples, yet with a moderate successful rate as less than 40% of the retrospective cases turned out to be analyzable for poor quality of purified RNA. However, in our cohort, this appears not related to the archival time of clinical samples but rather to specific protocols required for routine diagnostic practice which may have a detrimental impact on nucleic acid quality.
A limitation of this study is represented by the lack of mutational data, specifically regarding the NOTCH status. Furthermore, we critically recognize that the lack of validation on an independent external cohort limits the generalizability of our findings. Still, results from pathway analysis are highly concordant to that recently published on fresh-frozen primary ACC, where a myc overexpressing subtype enriched in NOTCH activating mutations and associated with worse prognosis emerged from clustering by RNAseq and protein expression analysis9. Our work adds to this context by suggesting an association between specific transcriptional alterations and histopathological features of ACC, providing biological information relevant to the survival of ACC patients and new therapeutic perspectives.
Supplementary Information
Acknowledgements
We sincerely thank the family of Claudia Massoni, who generously and enthusiastically supported this work in memory of their loved daughter.
Author contributions
C.R., S.C., P.B. conceptualised the study, interpreted data and wrote the original draft; C.R., A.B., E.B., L.A., M.B. performed the experiments; S.C. performed data analysis; L.L., M.T., D.T., M.R., C.G. provided human samples and patients' clinical information; A.R., S.B., D.L., D.M., C.P. reviewed and edited the manuscript; P.B. supervised the research; all authors read and approved the final manuscript.
Data availability
Gene expression data are deposited at GEO DataSets with ID GSE214969 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214969). The access is restricted to reviewers using the following token: cxgroeyandmbhwd. No other information is needed for accessing the data. The data will be made readily made publicly accessible once the paper is accepted for publication.
Competing interests
The authors declare no competing interests.
Footnotes
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
The online version contains supplementary material available at 10.1038/s41598-023-28901-9.
References
- 1.Coca-Pelaz A, Rodrigo JP, Bradley PJ, Vander Poorten V, Triantafyllou A, Hunt JL, et al. Adenoid cystic carcinoma of the head and neck—An update. Oral Oncol. 2015;51(7):652–661. doi: 10.1016/j.oraloncology.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 2.Ellington CL, Goodman M, Kono SA, Grist W, Wadsworth T, Chen AY, et al. Adenoid cystic carcinoma of the head and neck: Incidence and survival trends based on 1973–2007 Surveillance, Epidemiology, and End Results data. Cancer. 2012;118(18):4444–4451. doi: 10.1002/cncr.27408. [DOI] [PubMed] [Google Scholar]
- 3.van Weert S, Bloemena E, van der Waal I, de Bree R, Rietveld DHF, Kuik JD, et al. Adenoid cystic carcinoma of the head and neck: A single-center analysis of 105 consecutive cases over a 30-year period. Oral Oncol. 2013;49(8):824–829. doi: 10.1016/j.oraloncology.2013.05.004. [DOI] [PubMed] [Google Scholar]
- 4.Terhaard CHJ, Lubsen H, Van der Tweel I, Hilgers FJM, Eijkenboom WMH, Marres HAM, et al. Salivary gland carcinoma: Independent prognostic factors for locoregional control, distant metastases, and overall survival: Results of the Dutch head and neck oncology cooperative group. Head Neck. 2004;26(8):681–693. doi: 10.1002/hed.10400. [DOI] [PubMed] [Google Scholar]
- 5.van der Wal JE, Becking AG, Snow GB, van der Waal I. Distant metastases of adenoid cystic carcinoma of the salivary glands and the value of diagnostic examinations during follow-up. Head Neck. 2002;24(8):779–783. doi: 10.1002/hed.10126. [DOI] [PubMed] [Google Scholar]
- 6.Drier Y, Cotton MJ, Williamson KE, Gillespie SM, Ryan RJH, Kluk MJ, et al. An oncogenic MYB feedback loop drives alternate cell fates in adenoid cystic carcinoma. Nat. Genet. 2016;48(3):265–272. doi: 10.1038/ng.3502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ross JS, Gay LM, Wang K, Vergilio JA, Suh J, Ramkissoon S, et al. Comprehensive genomic profiles of metastatic and relapsed salivary gland carcinomas are associated with tumor type and reveal new routes to targeted therapies. Ann. Oncol. 2017;28(10):2539–2546. doi: 10.1093/annonc/mdx399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ferrarotto R, Mitani Y, Diao L, Guijarro I, Wang J, Zweidler-McKay P, et al. Activating NOTCH1 mutations define a distinct subgroup of patients with adenoid cystic carcinoma who have poor prognosis, propensity to bone and liver metastasis, and potential responsiveness to Notch1 inhibitors. J. Clin. Oncol. 2017;35(3):352–360. doi: 10.1200/JCO.2016.67.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ferrarotto R, Mitani Y, McGrail DJ, Li K, Karpinets TV, Bell D, et al. Proteogenomic analysis of salivary adenoid cystic carcinomas defines molecular subtypes and identifies therapeutic targets. Clin. Cancer Res. 2021;27(3):852–864. doi: 10.1158/1078-0432.CCR-20-1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Perzin KH, Gullane P, Clairmont AC. Adenoid cystic carcinomas arising in salivary glands: A correlation of histologic features and clinical course. Cancer. 1978;42(1):265–282. doi: 10.1002/1097-0142(197807)42:1. [DOI] [PubMed] [Google Scholar]
- 11.Irizarry RA. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4(2):249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 12.Murtagh F, Legendre P. Ward’s hierarchical agglomerative clustering method: Which algorithms implement ward’s criterion? J. Classif. 2014;31(3):274–295. doi: 10.1007/s00357-014-9161-z. [DOI] [Google Scholar]
- 13.Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47–e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 2005;102:15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Subramanian, A., Tamayo, P. & Castanza, A. GSEA: Gene Set Enrichment Analysis (GSEA). R package version 1.2. http://gsea-msigdb.org (2019).
- 16.Liberzon A, Birger C, Thorvaldsdóttir H, Ghandi M, Mesirov JP, Tamayo P. The molecular signatures database hallmark gene set collection. Cell Syst. 2015;1(6):417–425. doi: 10.1016/j.cels.2015.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Barbie DA, Tamayo P, Boehm JS, Kim SY, Moody SE, Dunn IF, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hothorn T, Zeileis A. Generalized maximally selected statistics. Biometrics. 2008;64(4):1263–1269. doi: 10.1111/j.1541-0420.2008.00995.x. [DOI] [PubMed] [Google Scholar]
- 19.Kanehisa M, Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28(1):27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–1951. doi: 10.1002/pro.3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kanehisa M, Furumichi M, Sato Y, Kawashima M, Ishiguro-Watanabe M. KEGG for taxonomy-based analysis of pathways and genomes. Nucleic Acids Res. 2023;51(D1):D587–D592.22. doi: 10.1093/nar/gkac963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Savarese T, Abate A, Basnet RM, Lorini L, Gurizzan C, Tomasoni M, et al. Cytotoxic effects of targeted agent alone or with chemotherapy in the treatment of adenoid cystic carcinoma: A preclinical study. Sci. Rep. 2022;12(1):9951. doi: 10.1038/s41598-022-14197-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li J, Perlaky L, Rao P, Weber RS, El-Naggar AK. Development and characterization of salivary adenoid cystic carcinoma cell line. Oral Oncol. 2014;50(10):991–999. doi: 10.1016/j.oraloncology.2014.06.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ho AS, Kannan K, Roy DM, Morris LGT, Ganly I, Katabi N, et al. The mutational landscape of adenoid cystic carcinoma. Nat. Genet. 2013;45(7):791–798. doi: 10.1038/ng.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stephens PJ, Davies HR, Mitani Y, Van Loo P, Shlien A, Tarpey PS, et al. Whole exome sequencing of adenoid cystic carcinoma. J. Clin. Investig. 2013;123(7):2965–2968. doi: 10.1172/JCI67201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ho AS, Ochoa A, Jayakumaran G, Zehir A, Valero Mayor C, Tepe J, et al. Genetic hallmarks of recurrent/metastatic adenoid cystic carcinoma. J. Clin. Investig. 2019;129(10):4276–4289. doi: 10.1172/JCI128227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Oakes SA. Endoplasmic reticulum stress signaling in cancer cells. Am. J. Pathol. 2020;190(5):934–946. doi: 10.1016/j.ajpath.2020.01.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Avril T, Vauléon E, Chevet E. Endoplasmic reticulum stress signaling and chemotherapy resistance in solid cancers. Oncogenesis. 2017;6(8):e373. doi: 10.1038/oncsis.2017.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Khaled J, Kopsida M, Lennernäs H, Heindryckx F. Drug resistance and endoplasmic reticulum stress in hepatocellular carcinoma. Cells. 2022;11(4):632. doi: 10.3390/cells11040632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Logue SE, McGrath EP, Cleary P, Greene S, Mnich K, Almanza A, et al. Inhibition of IRE1 RNase activity modulates the tumor cell secretome and enhances response to chemotherapy. Nat. Commun. 2018;9(1):3267. doi: 10.1038/s41467-018-05763-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Moore PC, Qi JY, Thamsen M, Ghosh R, Peng J, Gliedt MJ, et al. Parallel signaling through IRE1α and PERK regulates pancreatic neuroendocrine tumor growth and survival. Cancer Res. 2019;79(24):6190–6203. doi: 10.1158/0008-5472.CAN-19-1116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mueller B, Mieczkowski J, Kundu S, Wang P, Sadreyev R, Tolstorukov MY, et al. Widespread changes in nucleosome accessibility without changes in nucleosome occupancy during a rapid transcriptional induction. Genes Dev. 2017;31(5):451–462. doi: 10.1101/gad.293118.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Musella M, Galassi C, Manduca N, Sistigu A. The Yin and Yang of type I IFNs in cancer promotion and immune activation. Biology. 2021;10(9):856. doi: 10.3390/biology10090856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Padariya M, Sznarkowska A, Kote S, Gómez-Herranz M, Mikac S, Pilch M, et al. Functional interfaces, biological pathways, and regulations of interferon-related DNA damage resistance signature (IRDS) genes. Biomolecules. 2021;11(5):622. doi: 10.3390/biom11050622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cheon H, Borden EC, Stark GR. Interferons and their stimulated genes in the tumor microenvironment. Semin. Oncol. 2014;41(2):156–173. doi: 10.1053/j.seminoncol.2014.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Post AEM, Smid M, Nagelkerke A, Martens JWM, Bussink J, Sweep FCGJ, et al. Interferon-stimulated genes are involved in cross-resistance to radiotherapy in tamoxifen-resistant breast cancer. Clin. Cancer Res. 2018;24(14):3397–3408. doi: 10.1158/1078-0432.CCR-17-2551. [DOI] [PubMed] [Google Scholar]
- 37.Vander Poorten V, Meulemans J, Delaere P, Nuyts S, Clement P. Molecular markers and chemotherapy for advanced salivary cancer. Curr. Otorhinolaryngol. Rep. 2014;2(2):85–96. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Gene expression data are deposited at GEO DataSets with ID GSE214969 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE214969). The access is restricted to reviewers using the following token: cxgroeyandmbhwd. No other information is needed for accessing the data. The data will be made readily made publicly accessible once the paper is accepted for publication.