Abstract
Previous research has suggested that the adhesin encoded by the F18 fimbrial operon in Escherichia coli is either the FedE or FedF protein. In this work, we show that anti-FedF antibodies, unlike anti-FedE serum, were able to inhibit E. coli adhesion to porcine enterocytes. Moreover, specific adhesion to enterocytes was shown with purified FedF-maltose binding protein.
The operons of many fimbrial adhesins of Escherichia coli are well characterized (4). They contain genes coding for the major subunit protein, molecular chaperone and usher proteins, minor subunits, adhesin, and proteins of unknown function (4, 11, 12). The genes involved in the biosynthesis of F18 fimbria have been only partially described (5, 6). The major protein of the F18 fimbria, FedA, is not sufficient for recognizing the F18 receptor (5). Two additional genes from the fed gene cluster, fedE and fedF, have been described as essential for fimbrial adhesion and fimbrial length (6). However, so far it has not been possible to assess F18 adhesion function with regard to either of the two gene products.
In this study, we sequenced the unknown region of the E. coli fed gene cluster and produced and purified FedF and FedE as fusion proteins with maltose binding protein (MBP) for raising antisera for adhesion studies. Furthermore, using indirect immunofluorescence microscopy and adhesion inhibition tests, we have characterized the FedF proteins as the adhesin of F18 fimbriae.
Sequencing of the plasmid pIH120.
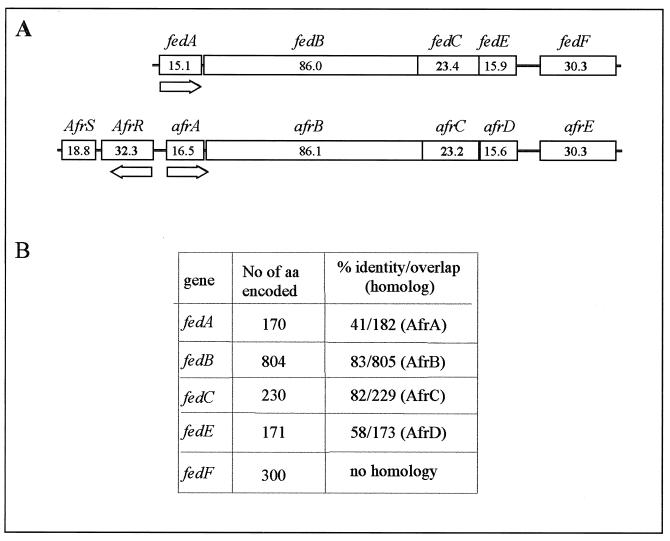
The entire gene cluster encoding E. coli F18 fimbria was sequenced from the plasmid pIH120 (6) with an ABI 310 sequencer according to the manual of the manufacturer (PE Applied Biosystems). pIH120 was transferred into an E. coli HB101 host, resulting in strain ERF2055. Sequence analyses revealed that the fed gene cluster is composed of five genes. The gene coding for the major protein of F18 fimbria (fedA) and the genes encoding two minor proteins (fedE and fedF) were described earlier (5, 6). Two additional open reading frames were found between fedA and fedE and were designated fedB and fedC. FedB showed the highest similarity (83% identity) to the AfrB protein (AAC28316) from E. coli RDEC-1 (Fig. 1) and significant homology to other usher proteins involved in the biosynthesis of microbial pili (3). The second open reading frame (fedC) overlapped the 3′ end of fedB, and its product had high identity (82%) with the periplasmic chaperone AfrC (AAC228317) from E. coli RDEC-1. Both FedB and FedC possess a predicted signal peptide for transmembrane secretion with a putative cleavage site for a signal peptidase between amino acids 23 and 24. The calculated molecular masses of the mature FedB and FedC are 86,001 and 23,418 Da, respectively. The fedF gene was also PCR cloned and sequenced from a Finnish E. coli O141 isolate (data not shown) and found to have 99.6% identity with the fedF derived from pIH120. In addition to the previously reported transcription terminator, located downstream of fedA (5), an inverted repeat (ΔG of −17.3 kcal mol−1) for the putative transcription terminator of the fed gene cluster was found from 11 to 94 nucleotides downstream of the stop codon of fedF.
FIG. 1.
Comparison of the fed gene cluster with the AF/R1 pilus operon. (A) Gene organization of the operons. The AF/R1 pilus operon is as described by Cantey et al. (2). Numbers in the boxes are molecular masses (in kilodaltons). (B) Levels of identity of the protein homologs.
Production of fusion proteins.
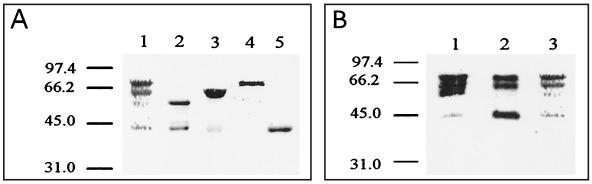
The genes encoding FedC, FedE, and FedF were cloned into E. coli with pMAL-p2 (New England Biolabs) and sequenced. The resulting recombinant strains were designated ERF2021 (for fedE), ERF2022 (for fedF), and ERF2048 (for fedC). All strains were shown to produce derivatives of MBP with predicted sizes of the corresponding full-length fusion protein (Fig. 2A). However, truncated MBP-FedF was seen in addition to the full-length product. The fusion proteins were purified from sonicated crude cell extracts by affinity chromatography according to the manual of the pMAL protein and purification system (New England Biolabs) and used for antibody production in rabbits (Fig. 2A, lanes 1 to 3). In immunoblots, both MBP-FedF and MBP antisera detected the corresponding full-length proteins and the truncated forms of MBP-FedF. The same bands were also seen after staining with Coomassie R-250 dye (Fig. 2B). To obtain antiserum against the pure full-length MBP-FedF protein, its band was excised from sodium dodecyl sulfate (SDS)-polyacrylamide gels, followed by electroelution (Fig. 2A, lane 4) and immunization of mice. Before immunization, the identity and purity of MBP-FedF, excised and eluted from an SDS–9% polyacrylamide gel, were confirmed by peptide mass mapping analysis (9). Ten peptides were analyzed, and all of them could be localized to the amino acid sequences of either MBP or FedF.
FIG. 2.
(A) Fusion proteins purified by affinity chromatography. Lanes 1 to 5, MBP-FedF, MBP-FedE, MBP-FedC, and MBP-FedF purified from SDS-polyacrylamide gels and MBP, respectively. Molecular masses (in kilodaltons) of marker proteins are on the left. The calculated molecular masses of the fusion proteins are 74.1 kDa for MBP-FedF, 58.8 kDa for MBP-FedE, and 66.4 kDa for MBP-FedC. (B) Immunoblots with MBP-FedF antiserum, MBP antiserum, and MBP-FedF stained with Coomassie dye (lanes 1 to 3, respectively).
In vitro adhesion analyses.
Epithelial cells were isolated from the small intestine (segments of ileum and jejunum) of an 8-week-old pig, and phase-contrast microscopic examination of the adherence of F18 fimbrial E. coli to epithelial cells was performed essentially as described by Alwan et al. (1). To obtain a semiquantitative estimation of the level of adhesion, the number of bacteria adhering to 15 randomly chosen epithelial cells was counted. The average numbers of ERF2055 bacteria adhering per ileal or jejunal cell when the bacteria were preincubated with different antisera, which had been raised in rabbits or mice and diluted in phosphate-buffered saline (PBS), are listed in Table 1. Representative pictures are also shown for each adhesion analysis (Fig. 3 and 4). Abolishment of the adhesion capability of ERF2055 cells was observed after preincubation (at 25°C for 2 h) of ERF2055 cells with MBP-FedF-specific antibodies or antibodies directed against the entire F18 fimbria. In contrast, antibodies to MBP-FedE or MBP-FedC were not able to inhibit the adhesion of the ERF2055 cells, even though a minor decrease in the adhesion capability was found.
TABLE 1.
Inhibition of adhesion of E. coli strain ERF2055 to porcine ileal or jejunal epithelial cells
| Epithelial cell type | Antiserum (dilution in PBS) | No. of adherent E. coli cells/epithelial cell |
|---|---|---|
| Ileal | Rabbit anti-MBP-FedF (1/100) | <1 |
| Rabbit anti-MBP-FedE (1/100) | 30 | |
| Rabbit anti-MBP-FedC (1/100) | 34 | |
| Rabbit anti-F18 fimbria (1/100) | 3 | |
| None (PBS control)a | >50 | |
| Jejunal | Mouse anti-MBP-FedF (1/10) | 6 |
| Mouse anti-MBP-FedF (1/30) | 14 | |
| Mouse anti-MBP (1/10) | >50 |
E. coli HB101 and PBS were used as a negative control and yielded <1 adherent bacterial cell per epithelial cell.
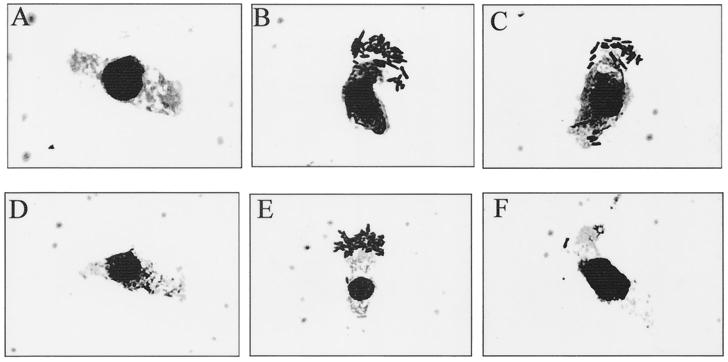
FIG. 3.
Adhesion of E. coli ERF2055 to porcine ileal epithelial cells after preincubation with rabbit antisera raised against MBP-FedF (A), MBP-FedE (B), MBP-FedC (C), or F18 fimbriae (D) or preincubated with PBS as a positive adhesion control (E). (F) Strain HB101 was used as a negative adhesion control.
FIG. 4.
Adhesion of E. coli ERF2055 to jejunal epithelial cells after preincubation with mouse MBP-FedF or MBP antiserum. ERF2055 cells were preincubated with antiserum raised against MBP-FedF (A) or MBP (B) and diluted 1/10 in PBS or preincubated with PBS as a positive control (C).
These results confirmed that from the antisera directed against Fed subunits, only MBP-FedF antibodies were able to efficiently inhibit the adherence of the F18 fimbria-expressing strain (ERF2055). As expected, a distinct reduction in the adhesion capability of ERF2055 cells, when preincubated with antibodies raised against whole F18 fimbriae, could also be demonstrated. The protective function of antibodies raised against F18 fimbriae has been described (7, 13). Despite promising results with antibodies directed against whole F18 fimbriae, antibodies raised directly against the adhesin inhibited bacterial attachment more efficiently. Adhesins attached to similar receptor moieties possess a high degree of antigenic conservation and could protect a wider range of bacteria, whereas the major immunodominant component of pilus fibers is often antigenically variable (8). No significant agglutination of E. coli cells was observed with any of the antisera under the test conditions used.
Indirect immunofluorescence microscopy.
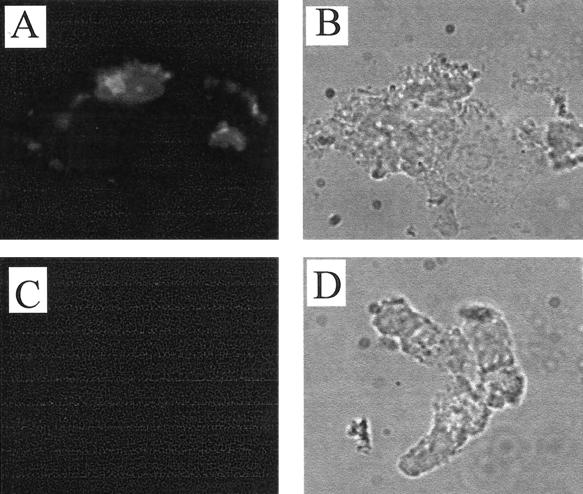
Adhesion of 0.8 mg of fusion proteins/ml to 106 epithelial cells/ml (incubation for 1 h at 37°C) was detected with fluorescence microscopy after incubation with rabbit anti-MBP antiserum and fluorescein isothiocyanate-labeled anti-rabbit antibodies. In the adhesion studies, 1% bovine serum albumin was used as a blocking reagent. The epithelial cells incubated in the presence of MBP-FedF exhibited bright fluorescence (Fig. 5A), whereas epithelial cells incubated with equal amounts of MBP-FedE (Fig. 5C) or MBP (data not shown) did not show any fluorescence. These data further confirmed that the FedF protein represents the adhesin of the fed gene cluster encoding the F18 fimbria and also indicated that the FedF, produced as a fusion protein with MBP but without its chaperone, retained its capacity to bind to epithelial cells. Similar results have also been reported for the F17 fimbria (10), where a truncated adhesin, produced in the absence of the chaperone, was still able to bind to its receptor in vitro.
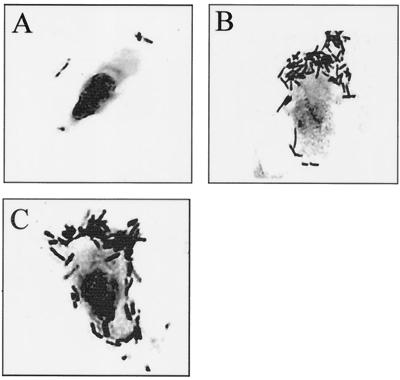
FIG. 5.
Indirect immunofluorescence of porcine ileal epithelial cells after incubation with purified MBP-FedF (A and B) and MBP-FedE (C and D) fusion proteins. (A and C) Adhesion viewed by fluorescence microscopy with a fluorescein isothiocyanate filter; (B and D) corresponding fields viewed by phase-contrast microscopy.
Alignment analyses of Fed proteins and the AF/R1 pilus subunits, with BLAST and the SIM program (BLOSUM62), revealed that all subunits from the F18 fimbria found their corresponding homologue from the AF/R1 pilus, with exception of FedF, which exhibited no homology with AfrE, reported as one of the putative adhesive subunits on the AF/R1 pilus (2). These adhesion results together with alignment analyses suggest that FedF is the adhesin of the F18 fimbria. A number of in-frame deletions within the fedF gene were also shown to inhibit F18-mediated attachment (A. Smeds et al., unpublished data), further confirming this interpretation. Although the fed gene cluster is very similar to the gene cluster encoding AF/R1 pilus, it seems that the expression of F18 fimbriae is regulated by a different mechanism. No genes encoding transcriptional activators like afrR, located immediately upstream of the AF/R1 gene cluster, could be found on the upstream region of the fed gene cluster.
Nucleotide sequence accession number.
The DNA sequences of the fedB and fedC gene regions have been deposited in the EMBL sequence data bank under accession number AF222806.
Acknowledgments
This work was supported by the Academy of Finland and the Faculty of Veterinary Medicine, University of Helsinki.
We are grateful to Sinikka Ahonen, Ulla Viitanen, and Esa Pohjolainen for excellent technical assistance. Ilkka Palva is acknowledged for valuable discussions and critical reading of the manuscript. We also thank Tarja Pohjanvirta for providing E. coli ERF2014 and Nisse Kalkkinen for performing the peptide mass-mapping.
REFERENCES
- 1.Alwan A, Deignan T, O'Sullivan M, Kelly J, O'Farrelly C. Quantitative assay of Salmonella adherence to intestinal epithelial cells: a new method for assessing novel intervention products. J Microbiol Methods. 1998;33:163–170. [Google Scholar]
- 2.Cantey J R, Blake R K, Williford J R, Moseley S L. Characterization of the Escherichia coli AF/R1 pilus operon: novel genes necessary for transcriptional regulation and for pilus-mediated adherence. Infect Immun. 1999;67:2292–2298. doi: 10.1128/iai.67.5.2292-2298.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dodson K W, Jacob-Dubuisson F, Striker R T, Hultgren S J. Outer-membrane PapC molecular usher discriminately recognizes periplasmic chaperone-pilus subunit complexes. Proc Natl Acad Sci USA. 1993;90:3670–3674. doi: 10.1073/pnas.90.8.3670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaastra W, Svennerholm A-M. Colonization factors of human enterotoxigenic Escherichia coli (ETEC) Trends Microbiol. 1996;4:444–452. doi: 10.1016/0966-842x(96)10068-8. [DOI] [PubMed] [Google Scholar]
- 5.Imberechts H, De Greve H, Schlicker C, Bouchet H, Pohl P, Charlier G, Bertschinger H U, Wild P, Vandekerckhove J, Van Damme J, Van Montagu M, Lintermans P. Characterization of F107 fimbriae of Escherichia coli 107/86, which causes edema disease in pigs, and nucleotide sequence of the F107 major fimbrial subunit gene, fedA. Infect Immun. 1992;60:1963–1971. doi: 10.1128/iai.60.5.1963-1971.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Imberechts H, Wild P, Charlier G, De Greve H, Lintermans P, Pohl P. Characterization of F18 fimbrial genes fedE and fedF involved in adhesion and length of enterotoxemic Escherichia coli strain 107/86. Microb Pathog. 1996;21:183–192. doi: 10.1006/mpat.1996.0053. [DOI] [PubMed] [Google Scholar]
- 7.Imberechts H, Deprez P, Van Driessche E, Pohl P. Chicken egg yolk antibodies against F18ab fimbriae of Escherichia coli inhibit shedding of F18 positive E. coli by experimentally infected pigs. Vet Microbiol. 1997;54:329–341. doi: 10.1016/s0378-1135(96)01293-x. [DOI] [PubMed] [Google Scholar]
- 8.Langermann S, Palaszynski S, Barnhart M, Auguste G, Pinkner J S, Burlein J, Barren P, Koenig S, Leath S, Jones C H, Hultgren S J. Prevention of mucosal Escherichia coli infection by FimH-adhesin-based systemic vaccination. Science. 1997;276:607–611. doi: 10.1126/science.276.5312.607. [DOI] [PubMed] [Google Scholar]
- 9.Nyman T A, Matikainen S, Sareneva T, Julkunen I, Kalkkinen N. Proteome analysis reveals ubiquitin-conjugating enzymes to be a new family of interferon-α-regulated genes. Eur J Biochem. 2000;267:4011–4019. doi: 10.1046/j.1432-1327.2000.01433.x. [DOI] [PubMed] [Google Scholar]
- 10.Saarela S, Taira S, Nurmiaho-Lassila E-L, Makkonen A, Rhen M. The Escherichia coli G-fimbrial lectin protein participates both in fimbrial biogenesis and in recognition of the receptor N-acetyl-d-glucosamine. J Bacteriol. 1995;177:1477–1484. doi: 10.1128/jb.177.6.1477-1484.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Smyth C J, Marron M B, Twohig J M G J, Smith S G J. Fimbrial adhesins: similarities and variations in structure and biogenesis. FEMS Immunol Med Microbiol. 1996;16:127–139. doi: 10.1111/j.1574-695X.1996.tb00129.x. [DOI] [PubMed] [Google Scholar]
- 12.Soto G E, Hultgren S J. Bacterial adhesins: common themes and variations in architecture and assembly. J Bacteriol. 1999;181:1059–1071. doi: 10.1128/jb.181.4.1059-1071.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yokoyama H, Hashi T, Umeda K, Kuroki M, Ikemori Y, Kodama Y. Effect of oral egg antibody in experimental F18+ Escherichia coli infection in weaned pigs. J Vet Med Sci. 1997;59:917–921. doi: 10.1292/jvms.59.917. [DOI] [PubMed] [Google Scholar]