Abstract
The repetitive shed acute-phase antigen (SAPA) from Trypanosoma cruzi was thoroughly mapped by SPOT peptides and phage display strategies, showing that a single SAPA repeat is composed of multiple overlapping B-cell epitopes. We propose that this intricate antigenic structure constitutes an alternative device to repetitiveness in order to improve its immunogenicity.
Several repetitive antigens from parasitic protozoa and pathogenic bacteria were identified through immunoscreening of genomic and/or cDNA libraries with sera from infected individuals (7–9, 14, 15, 20). It is hypothesized that the immunodominant character of these repetitive antigens relies on their high epitope dosage (on a molar ratio basis) and/or on their particular two-dimensional organization (19). However, the existence of additional mechanisms contributing to the immunodominance of repetitive antigens could not be disregarded. To clarify this issue, we performed a detailed antigenic study on the trypomastigote-restricted shed acute-phase antigen (SAPA) from Trypanosoma cruzi, one example of this kind of molecule. It elicits a strong immunoglobulin G response early after infection that is lost with the progression of the disease (1, 17, 18). SAPA is constituted by 12-amino-acid repetitive units (DSSAHSTPSTPA) displayed in tandem on the C terminus of trans-sialidase, a T. cruzi virulence factor (3, 5, 11) which enhances its half-live in blood and is associated with the induction of trans-sialidase-neutralizing antibodies (2).
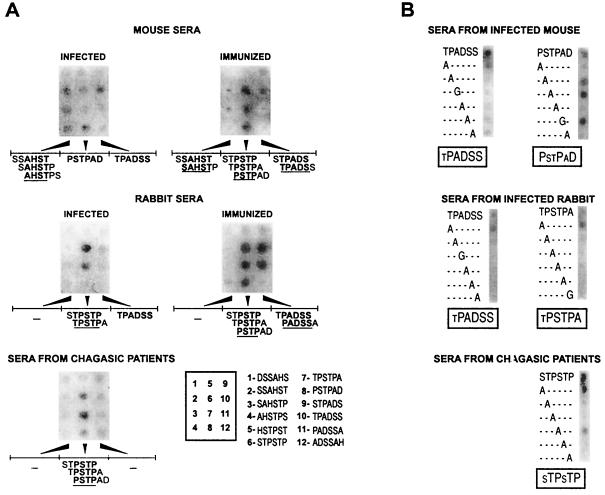
One central and still-unsolved concern regarding the antigenic structure of SAPA is whether this antigen displays a single or several B-cell epitopes per repeat unit. To analyze this issue, 12 peptides covering all possible hexamers in the SAPA repeat sequence were custom synthesized by the SPOT technique (Genosys Biotechnologies, The Woodlands, Tex.) (4) and probed with sera from infected individuals of different species. This peptide length was chosen to minimize the possibility that a single SPOT could span more than one epitope. Sera from six New Zealand white rabbits and eight mice (C3H/HeN, CF1, or BALB/c) infected with different T. cruzi strains (RA, CAI, Y, and Acosta) along with those from 10 acute-phase Chagasic patients were assayed individually, and representative results are shown in Fig. 1A. (Human and animal materials were obtained in accordance with national regulations.) Reactions were developed by the addition of secondary antibodies coupled to alkaline phosphatase (Dako Corporation, Carpinteria, Calif.) followed by 5-bromo-4-chloro-3-indolyl phosphate–3-(4,5-dimethylthiazol-2-yl) 2,5-diphenyltetrazolium bromide (Sigma Chemical Co., St. Louis, Mo.). Sera from both infected rabbits and mice reacted against two sets of peptides spanning the sequences TPSTPAD and TPADSS. These epitopes are partially overlapped, thus defining a major B-cell restricted antigenic spot on the SAPA sequence (spanning the sequence TPSTPADSS). In addition, sera from infected mice recognized a third epitope spanning the sequence SAHSTP (Fig. 1A) that also was strongly recognized by an anti-SAPA mouse monoclonal antibody (MAb) (10) (data not shown). Interestingly, similar sequences for each species were highlighted when sera from mice and rabbits immunized with the recombinant glutathione S-transferase (GST)–SAPA protein (2) were tested (Fig. 1A). Therefore, the two species disclose a multiepitope arrangement within the SAPA repeat. Acute-phase Chagasic human sera reacted with the consensus sequence PSTP, which is similar to one of the major epitopes defined for the other two species (Fig. 1A). In this case, the humoral response seemed to be restricted to this epitope and not spread over different antigenic determinants present on the SAPA sequence.
FIG. 1.
Mapping of SAPA B-cell epitopes through SPOT peptides. (A) Filters containing 12 overlapping hexapeptides synthesized in a defined array (enclosed box) were probed with sera from infected (1:100 dilution) or immunized (1:200) animals and humans. Peptides that gave positive reactions are indicated below the corresponding filter, and the consensus sequences of the defined epitopes are underlined. (B) Series of alanine or glycine-substituted analogs synthesized on the basis of the parent peptide shown above each panel were probed with the indicated serum and processed as described above. Critical residues for antibody recognition, as judged by peptide reactivity, are indicated by uppercase letters below each panel.
To define critical residues involved in the antibody recognition for each identified epitope, a series of alanine-substituted analogs were synthesized by the SPOT method and probed as indicated before. Five sequences were deemed core epitope motifs (TPADSS and PSTPAD for mice, TPADSS and TPSTPA for rabbits, and STPSTP for humans) based on their degree of antibody recognition in the SPOT assay (Fig. 1A). The recognition of the TPADSS peptide by antibodies from infected rabbits and mice involved almost the entire sequence (Fig. 1B). Alanine replacements at any position resulted in a significant reduction in binding except for the N-terminal threonine residue. On the other hand, the critical residues from the other major SAPA epitope seemed to be species specific. Mouse sera recognized the sequence PXXPXD, whereas rabbit sera required the sequence XPSTPA and human sera recognized the sequence XTPXTP. It is noteworthy that in all cases the replacement of either proline residue completely impaired the binding of the antibody. Thus, the epitope defined by the three species contains the minimal PXXP sequence, known as the polyproline II motif (22), that has been identified in a variety of surface proteins and might be related to adhesive and/or antigenic properties (13).
As a complementary technique to SPOT peptides, phage display was chosen to search for SAPA-restricted B-cell epitopes (12). This technique allows the detection of longer and/or discontinuous epitopes, thus overcoming two of the major drawbacks of the SPOT hexapeptides assayed before. Two libraries made up of random nonapeptides were screened. In one of them, peptides are flanked by cysteine residues, allowing the location of the epitopes in a solvent-exposed loop on phage pVIII protein (12). For these screenings, both the MAb anti-SAPA (10) and purified immunoglobulin G from GST-SAPA-immunized rabbits were used as described previously (16). Screening of the libraries with the MAb anti-SAPA yielded five positive phages (group I) with the consensus sequence SAH(A/V)XP(S/T), where X denotes a permissive position (Fig. 2A). This mimotope-based sequence is very similar to one of the epitopes (SAHSTP) previously defined (Fig. 1A). When SAPA-immunized-rabbit antibodies were assayed, 11 phages were obtained that could be arranged in three separate groups (Fig. 2B). The consensus defined for group II (n = 4) was STPSXXP(A/G), showing a stretch of four residues (STPS) that perfectly matches the SAPA sequence. In addition, three of these phages retained a second proline residue at a fixed position that may correspond to the other proline present on SAPA, though all of these mimotopes accommodated an extra hydrophobic residue between the prolines. As a result, the consensus of this second group of phages seems to mimic TPSTP, one of the major SAPA epitopes highlighted in the SPOT experiments (Fig. 1A).
FIG. 2.
Sequence analysis of phage-derived mimotopes. Mimotopes expressed by phages cloned through library screening with the MAb anti-SAPA (group I) or rabbit polyclonal anti-SAPA antibodies (groups II and III) are aligned with the SAPA sequence. Hyphens are introduced for better alignment. Another four phages bearing the sequences KVRNWRESM (phage 1), CRWPDVLLWC (phage 2), CQKAGGVAL (phage 6), and NSHAARDAW (phage 9) without obvious homology to SAPA were also obtained by rabbit antibody screening.
The consensus of group III of the mimotopes (n = 3) was defined as RSNPAHMA(E/W) (Fig. 2B). The alignment of this consensus sequence with SAPA is not so obvious, but it might resemble the sequence of the other previously defined epitope (TPADSS). Alternatively, these phages may bear conformational epitopes. The remaining four isolated phages (listed in the legend to Fig. 2) displayed mimotopes that could not be assigned to any given SAPA sequence, a frequent finding when these libraries are used (12). Altogether, these results reinforce those obtained by the SPOT method and further support the multiepitope composition of a single SAPA repeat unit.
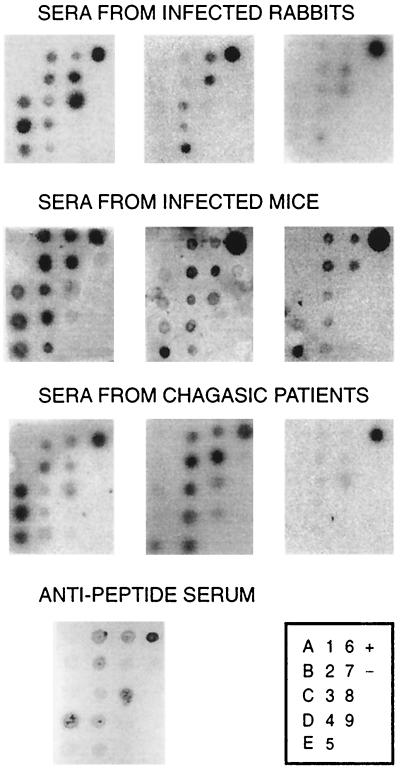
The specificity of the identified mimotopes was evaluated by phage-dot assay. As depicted in Fig. 3, phages belonging to different defined groups were recognized by a rabbit antiserum raised against a synthetic 24-mer peptide spanning two SAPA repeats (21), strongly suggesting that they are in fact mimicking SAPA epitopes. In addition, sera derived from infected rabbits (5 of 6), infected mice (11 of 12), and Chagasic patients (7 of 8) but none of the normal sera used were able to recognize several phages (Fig. 3 and data not shown). In these assays, reactivity was developed by alkaline phosphatase, 125I-protein A, or a secondary antibody coupled to horseradish peroxidase (Dako) followed by SuperSignal substrate (Pierce, Rockford, Ill.). As expected, the recognition profile for these epitopes showed subtle individual differences that might be attributed to parasite and/or host polymorphism. More important, phage-dot experiments suggest the existence of multiple B-cell epitopes even for human antibodies whose recognition profile was shifted toward a unique SAPA sequence in the SPOT assays (PSTP) (Fig. 1A). These results might be ascribed to differences in the sensitivity of both methods or, alternatively, to a constraint imposed on the repertoire of human antibody recognition by the length of the SPOT peptides (hexamer). A similar effect was observed for infected rabbit sera that recognized mimotopes corresponding to the SAHSTP epitope in the phage-dot assays (Fig. 3) but not the epitope in the SPOT experiments (Fig. 1A).
FIG. 3.
Recognition of phage-displayed mimotopes by sera from T. cruzi infection. Polyethylene glycol-purified phages were spotted (108 PFU) onto nitrocellulose membranes in a defined array (box). Filters were tested with the indicated sera at a 1:100 dilution. Results for the rabbit anti-SAPA peptide serum are also displayed. Negative (−) and positive (+) controls were M13K07 helper phage (108 PFU; Amersham-Pharmacia Biotech, Uppsala, Sweden) and purified GST-SAPA protein (20 ng), respectively.
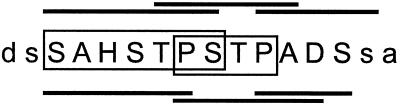
Overall, our data revealed that SAPA displays a complex antigenic array, made up of multiple linear overlapping epitopes per repeat unit (schematized in Fig. 4). Different species showed subtle variations in the sequence and length of their defined epitopes, but they all seem to elicit multiple B-cell immune responses against SAPA. In addition to the linear B-cell epitopes demonstrated within the repeat unit, a further discontinuous epitope(s) can be proposed (Fig. 2). The complexity of this scenario would be further increased by the presence of a weak T-cell epitope(s) (rendering stimulation indexes of about 2 to 4) within SAPA as detected by us (data not shown) and others (6). Such an intricate antigenic arrangement provides an alternative mechanism contributing to explain the overall immunogenicity of SAPA during T. cruzi infections. More important, these data give us a new perspective on the molecular structure and the interplay with the immune system of repetitive antigens, a common theme among infectious agents.
FIG. 4.
Multiple overlapping B-cell epitopes in SAPA. A single SAPA unit (capital letters) and two residues from the flanking units are shown. Epitopes defined by sera from infected rabbits (upper lines), mice (lower lines), and humans (boxes) are indicated.
Acknowledgments
This work was supported by grants from the Agencia Nacional de Promoción Científica y Tecnológica, the Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET) from Argentina, and the World Bank/UNDP/WHO Special Program for Research and Training in Tropical Diseases (TDR). P.A., C.A.B., and T.A.P. are Fellows and M.S.L. and O.C. are Researchers from CONICET.
We thank F. Felici (Istituto di Ricerche di Biologia Molecolare P. Angeletti, Rome, Italy) for the phage-displayed libraries, G. Russomando (Instituto de Investigaciones en Ciencias de la Salud, Universidad Nacional de Asunción, Asuncións, Paraguay) for the human sera, and A. C. C. Frasch for critical reading of the manuscript.
REFERENCES
- 1.Affranchino J L, Ibáñez C F, Luquetti A O, Rassi A, Reyes M B, Macina R A, Aslund L, Pettersson U, Frasch A C. Identification of a Trypanosoma cruzi antigen that is shed during the acute phase of Chagas' disease. Mol Biochem Parasitol. 1989;34:221–228. doi: 10.1016/0166-6851(89)90050-9. [DOI] [PubMed] [Google Scholar]
- 2.Buscaglia C A, Campetella O, Leguizamón M S, Frasch A C. The repetitive domain of Trypanosoma cruzi trans-sialidase enhances the immune response against the catalytic domain. J Infect Dis. 1998;177:431–436. doi: 10.1086/514199. [DOI] [PubMed] [Google Scholar]
- 3.Chuenkova M, Pereira M E. Trypanosoma cruzi trans-sialidase: enhancement of virulence in a murine model of Chagas' disease. J Exp Med. 1995;181:1693–1703. doi: 10.1084/jem.181.5.1693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Frank R, Doring R. Simultaneous multiple peptide synthesis under continuous flow conditions on cellulose paper discs as segmental solid synthesis. Tetrahedron. 1988;44:6031–6040. [Google Scholar]
- 5.Gao W, Pereira M A. Trypanosoma cruzi trans-sialidase potentiates T cell activation through antigen-presenting cells: role of IL-6 and Bruton's tyrosine kinase. Eur J Immunol. 2001;31:1503–1512. doi: 10.1002/1521-4141(200105)31:5<1503::AID-IMMU1503>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 6.Gironès N, Rodriguez C I, Carrasco-Marin E, Hernaez R F, de Rego J L, Fresno M. Dominant T- and B-cell epitopes in an autoantigen linked to Chagas' disease. J Clin Invest. 2001;107:985–993. doi: 10.1172/JCI10734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gravekamp C, Horensky D S, Michel J L, Madoff L C. Variation in repeat number within the alpha C protein of group B streptococci alters antigenicity and protective epitopes. Infect Immun. 1996;64:3576–3583. doi: 10.1128/iai.64.9.3576-3583.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibáñez C F, Affranchino J L, Macina R A, Reyes M B, Leguizamón S, Camargo M E, Aslund L, Pettersson U, Frasch A C. Multiple Trypanosoma cruzi antigens containing tandemly repeated amino acid sequence motifs. Mol Biochem Parasitol. 1988;30:27–33. doi: 10.1016/0166-6851(88)90129-6. [DOI] [PubMed] [Google Scholar]
- 9.Kemp D J, Coppel R L, Anders R F. Repetitive proteins and genes of malaria. Annu Rev Microbiol. 1987;41:181–208. doi: 10.1146/annurev.mi.41.100187.001145. [DOI] [PubMed] [Google Scholar]
- 10.Leguizamón M S, Campetella O E, Reyes M B, Ibáñez C F, Basombrio M A, Rincon J, Örn A, Frasch A C C. Bloodstream Trypanosoma cruzi parasites from mice simultaneously express antigens that are markers of acute and chronic human Chagas' disease. Parasitology. 1991;102:379–385. doi: 10.1017/s0031182000064337. [DOI] [PubMed] [Google Scholar]
- 11.Leguizamón M S, Mocetti E, Garcia Rivello H, Argibay P, Campetella O. Trans-sialidase from Trypanosoma cruzi induces apoptosis in cells from the immune system in vivo. J Infect Dis. 1999;180:1398–1402. doi: 10.1086/315001. [DOI] [PubMed] [Google Scholar]
- 12.Luzzago A, Felici F, Tramontano A, Pessi A, Cortese R. Mimicking of discontinuous epitopes by phage-displayed peptides. I. Epitope mapping of human H ferritin using a phage library of constrained peptides. Gene. 1993;128:51–57. doi: 10.1016/0378-1119(93)90152-s. [DOI] [PubMed] [Google Scholar]
- 13.Mathiesen M J, Holm A, Christiansen M, Blom J, Hansen K, Ostergaard S, Theisen M. The dominant epitope of Borrelia garinii outer surface protein C recognized by sera from patients with neuroborreliosis has a surface-exposed conserved structural motif. Infect Immun. 1998;66:4073–4079. doi: 10.1128/iai.66.9.4073-4079.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McKean P G, Trenholme K R, Rangarajan D, Keen J K, Smith D F. Diversity in repeat-containing surface proteins of Leishmania major. Mol Biochem Parasitol. 1997;86:225–235. doi: 10.1016/s0166-6851(97)00035-2. [DOI] [PubMed] [Google Scholar]
- 15.Peterson D S, Wrightsman R A, Manning J E. Cloning of a major surface-antigen gene of Trypanosoma cruzi and identification of a nonapeptide repeat. Nature. 1986;322:566–568. doi: 10.1038/322566a0. [DOI] [PubMed] [Google Scholar]
- 16.Pitcovsky T A, Mucci J, Alvarez P, Leguizamon M S, Burrone O, Alzari P M, Campetella O. Epitope mapping of trans-sialidase from Trypanosoma cruzi reveals the presence of several cross-reactive determinants. Infect Immun. 2001;69:1869–1875. doi: 10.1128/IAI.69.3.1869-1875.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Prioli R P, Ortega-Barria E, Mejía J S, Pereira M E A. Mapping of a B-cell epitope in the neuraminidase of Trypanosoma cruzi. Mol Biochem Parasitol. 1992;52:85–96. doi: 10.1016/0166-6851(92)90038-l. [DOI] [PubMed] [Google Scholar]
- 18.Reyes M B, Lorca M, Muñoz P, Frasch A C C. Fetal IgG specificities against Trypanosoma cruzi antigens in infected newborns. Proc Natl Acad Sci USA. 1990;87:2846–2850. doi: 10.1073/pnas.87.7.2846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schofield L. On the function of repetitive domains in protein antigens of Plasmodium and other eukaryotic parasites. Parasitol Today. 1991;7:99–105. doi: 10.1016/0169-4758(91)90166-l. [DOI] [PubMed] [Google Scholar]
- 20.Singh K K, Zhang X, Patibandla A S, Chien P, Jr, Laal S. Antigens of Mycobacterium tuberculosis expressed during preclinical tuberculosis: serological immunodominance of proteins with repetitive amino acid sequences. Infect Immun. 2001;69:4185–4191. doi: 10.1128/IAI.69.6.4185-4191.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vergara U, Lorca M, Veloso C, Gonzalez A, Engstrom A, Aslund L, Petterson U, Frasch A C C. Assay for detection of Trypanosoma cruzi antibodies in human sera based on reaction with synthetic peptides. J Clin Microbiol. 1991;29:2034–2037. doi: 10.1128/jcm.29.9.2034-2037.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Williamson M P. The structure and function of proline-rich regions in proteins. Biochem J. 1994;297:249–260. doi: 10.1042/bj2970249. [DOI] [PMC free article] [PubMed] [Google Scholar]