Abstract
Aberrant activity of the SUMOylation pathway has been associated with MYC overexpression and poor prognosis in aggressive B-cell lymphoma (BCL) and other malignancies. Recently developed small-molecule inhibitors of SUMOylation (SUMOi) target the heterodimeric E1 SUMO activation complex (SAE1/UBA2). Here, we report that activated MYC signaling is an actionable molecular vulnerability in vitro and in a preclinical murine in vivo model of MYC-driven BCL. While SUMOi conferred direct effects on MYC-driven lymphoma cells, SUMO inhibition also resulted in substantial remodeling of various subsets of the innate and specific immunity in vivo. Specifically, SUMOi increased the number of memory B cells as well as cytotoxic and memory T cells, subsets that are attributed a key role within a coordinated anti-tumor immune response. In summary, our data constitute pharmacologic SUMOi as a powerful therapy in a subset of BCL causing massive remodeling of the normal B-cell and T-cell compartment.
Introduction
The myelocytomatosis oncogene MYC is deregulated in almost half of all human cancers by chromosomal amplification, translocation or mutations in signaling pathways that regulate MYC expression.1-3 MYC belongs to a family of basic helix-loop-helix leucin zipper DNA binding proteins that function as a transcription factor controlling multiple biological processes including cell proliferation, differentiation, apoptosis and metabolism.1 Control of gene transcription is a well-established function of MYC, while MYC also interferes with translation and protein turnover.4,5 Genetic and epigenetic dysregulation of MYC expression accelerates cell proliferation and drives malignant transformation. So far, there are no effective therapies specifically targeting MYC signaling that have been established for clinical use. Uncontrolled cell growth in response to MYC overexpression creates dependencies on MYC-driven pathways to maintain the tumor phenotype.6 Therefore, the idea of targeting these cellular processes has been developed within the concept of “synthetic lethal interactions”.7,8
SUMOylation is a post-translational protein modification that controls localization, function and half-life of target proteins.9,10 It emerged as a crucial regulatory mechanism for fundamental cellular processes like chromatin organization, transcription and cell proliferation.11 The conjugation of SUMO (SUMO1, SUMO2 or SUMO3) to its substrates is controlled by a multi-step cascade involving the E1 SUMO-activating enzyme SAE1/UBA2, the E2 SUMO-conjugating enzyme UBC9, and various E3 SUMO ligases.12 SUMOylation is a fully reversible protein modification. Deconjugation of SUMO from its substrates is catalyzed by SUMO-specific proteases (or deconjugases) of the SENP (sentrin-specific protease) family.13 Disruption of this well-controlled balance contributes to tumor development and progression.14,15 Of note, dysregulation of oncogenes such as MYC activates SUMOylation and hyperSUMOylation often correlates with poor prognosis in cancer.16,17 However, while activated SUMOylation is a key feature of aggressive cancers, the impact of activated SUMOylation on tumor biology is multifaceted.9,10 Besides its effect on cancer biology itself, SUMOylation restrains anti-tumor immunity by repression of interferon signaling.18-20 Since SUMOylation is activated in cancer cells with high MYC levels, targeting MYC-induced SUMOylation as a therapeutic vulnerability seems highly attractive.7,21-23 A selective small-molecule inhibitor of SUMOylation that blocks activation of SUMO by the E1 activation enzyme was developed as ML-792 and further refined as ML-093. The clinically applicable form subasumstat (formerly TAK-981)24,25 is currently investigated in clinical trials (clinicaltrials gov. Identifier: NCT03648372, NCT04074330, NCT04381650).
Here, we investigated SUMOylation as a MYC-induced molecular vulnerability in aggressive BCL. We uncovered that activated MYC signaling confers susceptibility to pharmacologic SUMO inhibition (SUMOi) in MYC-driven BCL. Next to direct effects on MYC-driven lymphoma, SUMOi challenge resulted in pronounced remodeling of the immune cell landscape.
Methods
Chemicals
SUMOi (ML-093 and Tak-981, as specified in the figure legends) was either purchased from MedChemExpress or provided by Millennium Pharmaceuticals, Inc., a wholly owned subsidiary of Takeda Pharmaceutical Company Limited. SUMOi doses and treatment durations are indicated in the figure legends.
Cell culture
Human diffuse large B-cell lymphoma (DLBCL) cell lines were kept in RPMI-1640 (U-2932, NU-DHL-1, SU-DHL-4/5/6/8, DB and Toledo), IMDM (Oci-Ly1) or a-MEM (Oci-Ly19) medium supplemented with 10-20% fetal calf serum (FCS), 1% penicillin streptomycin and 2 mM L-glutamine (Thermo Scientific).
Flow cytometry
Cells were washed in HF2 buffer (ddH2O, 2% FCS, 1% penicillin streptomycin, 1% Hepes, 10% HBSS) and stained on ice for 30 minutes in HF2 (a list of all antibodies is provided in the Online Supplementary Appendix). After washing in HF2 cells were either resuspended in HF2 containing DAPI or propidium iodide (PI) for fluorescence-activated cell sorting (FACS) analysis, stained with the respective antibody combination or fixed with BD Cytofix/Cytoperm for intracellular staining. Data were acquired on Beckman Coulter CytoFLEX S.
Immunoblot analysis
Protein extracts were prepared by solving cell pellets in lysis buffer (150 mM NaCl, 1% NP-40 oder IGEPAL, 0.5% sodium deoxycholate, 0.1% SDS, 50 mM Tris) supplemented with NaF, PMSF and NaVO4 followed by sonification. Protein lysates were fractioned on SDS PAGE gels, transferred to PVDF Transfer Membran (Thermo Scientific) and incubated with primary antibodies overnight. MYC antibody was obtained by Cell signaling (9402S) and b-tubulin by DSHB (E7). HRP-conjugated secondary antibodies allowed signal detection via enhanced chemiluminescence (ECL) reagents (Millipore).
Cell viability assay
Twenty thousand cells per well were seeded and treatments were administered as indicated. After 24, 48 and 72 hours (h) of incubation, CellTiterGlo (Promega, G7572) was added and luminescence was measured and normalized to dimethyl sulfoxide (DMSO) control.
Mice
Wild-type (wt) mice (CD45.1, C57Bl/6J) were obtained from the Jackson Laboratory. Mice were examined twice a week. All animal experiments were performed in accordance to local authorities (Regierung von Oberbayern, Munich, Germany).
Transplantation and in vivo treatment of mice
1x106 Em-myc cells (CD45.2) were transplanted intravenously (i.v.) into C57Bl/6J wt mice. SUMOi or carrier treatment to tumor or wt mice was administered i.v., doses and treatment duration are indicated in the figure legend.
RNA-sequencing and processing of gene expression data
RNA samples were processed and sequenced (paired end, 150 bp/read) by Novogene (Cambridge, UK) on a HiSeq2500 Illumina device with a read depth of >20 M reads. Subsequent quality control, data processing and analysis were performed as previously described.26 RNA-sequencing (RNA-seq) data were uploaded to the European Nucleotide Archive and are available via accession ID: PRJEB53800. Further processing of transcriptomic data including gene set enrichment analysis is described in the Online Supplementary Appendix.
Single-Cell RNA-sequencing analysis
A detailed description of the methods is provided in the Online Supplemental Appendix.
Statistical analysis
Statistical analyses were performed using GraphPad Prism (GraphPad Software). The error bars shown in the figures represent the standard deviation (SD), unless specified otherwise. A P value lower than 0.05 was generally considered significant and all exact P values and tests are indicated in the figures.
Study approval
All animal experiments were performed in accordance with local authorities (Regierung Oberbayern, Munich, Germany and LAGeSo Berlin, Germany).
Results
Activated MYC signaling confers susceptibility to small-molecule SUMO inhibition in diffuse large B-cell lymphoma cell lines
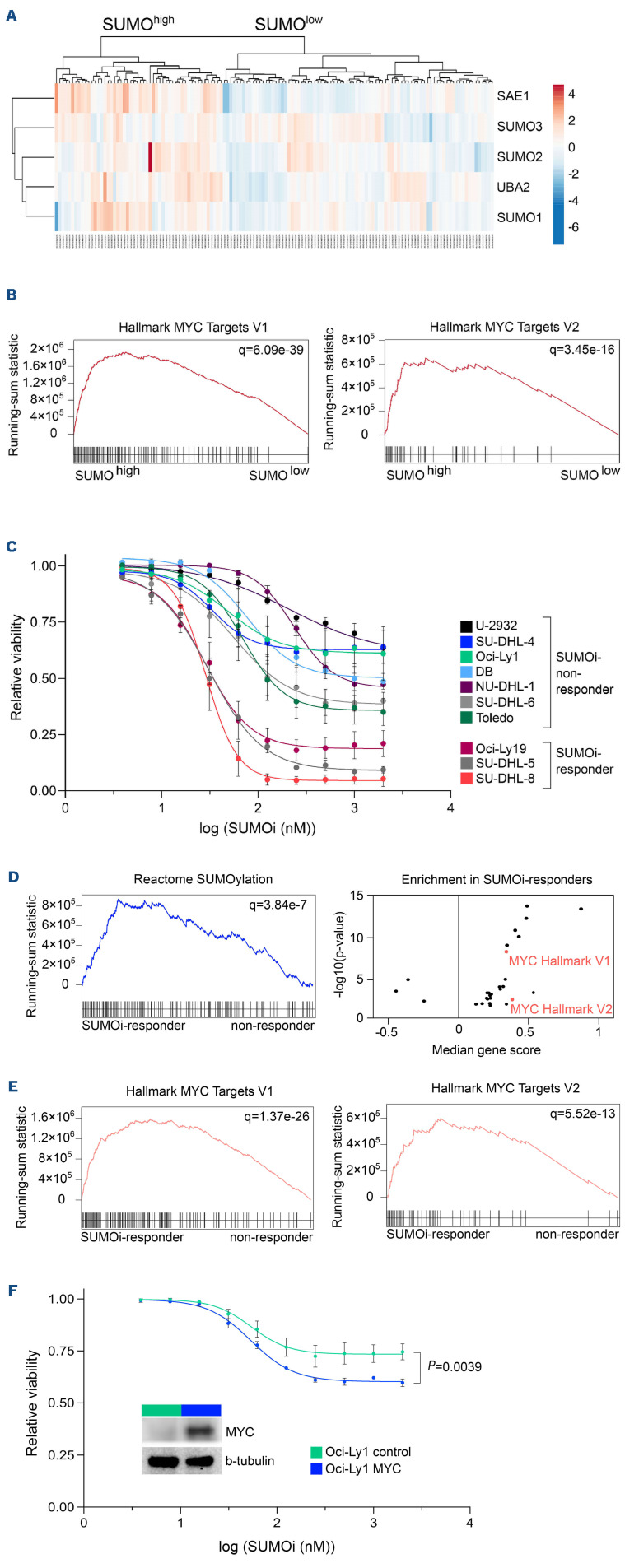
Activation of the oncoprotein MYC is associated with a hyperSUMOylated phenotype in DLBCL (Figure 1A and B) and both MYC overexpression and enhanced SUMO pathway activity are correlated with adverse clinical outcome (Online Supplementary Figure 1SA to C).
Given the role of MYC in DLBCL, we investigated the sensitivity of DLBCL cell lines to pharmacological SUMO inhibition.24,25 Three of ten cell lines (SU-DHL-8, SU-DHL-5 and Oci-Ly19) were responsive to SUMOi with viabilities below 25% at the highest SUMOi concentration (2,000 nM) (Figure 1C, subasumstat; Online Supplementary Figure S1D, ML-093). Importantly, the broad range of response from exquisite sensitivity (SU-DHL-8, SU-DHL5, Oci-Ly19) to very minor response/non-response in the remainder of DLBCL cell lines revealed that the complex genetic and non-genetic background and dependencies of DLBCL likely define the response to SUMOi.
In order to systematically identify potential biomarkers predictive for SUMOi sensitivity, we ranked the cell lines according to their subasumstat 50% growth inhibitory power (GI50) values and classified the lower third of the panel as SUMOi-responders, and the remaining two thirds constituting the SUMOi low sensitivity/non-responder subset. Gene set enrichment analysis identified the SUMO core pathway to be enhanced as well as various SUMOylation signatures to be enriched in the SUMOi-responsive cell lines (Figure 1D; Online Supplementary Figure S1E), whereas baseline growth characteristics and doubling time were not directly correlated with sensitivity to SUMOi (Online Supplementary Figure S1F and G). Moreover, sensitivity to SUMOi treatment was associated with activated MYC signaling and MYC hallmark gene sets scored among the top enriched gene sets in SUMOi-responsive cell lines (Figure 1D and E; Online Supplementary Figure S1H). Of note, the SU-DHL-8 and Oci-Ly19 cell lines, which had the highest activity of MYC signaling (SU-DHL-8 also harbors an IG::MYC translocation), were among the most sensitive cell lines within the investigated panel (Figure 1C; Online Supplementary Figure S1D and G). We thus concluded that activated MYC signaling is an actionable molecular vulnerability and would predict response to SUMOi. In order to experimentally validate these findings and prove a causal relationship, we ectopically expressed MYC in the human Oci-Ly1 cell line, classified as a non-responder (Figure 1C and F; Online Supplementary Figure S1D). Whereas MYC overexpression influenced growth kinetics moderately (Online Supplementary Figure S1I) while not affecting basal apoptotic rate or viability (Online Supplementary Figure S1J and K), ectopic MYC expression sensitized Oci-Ly1 to pharmacological SUMO inhibition (Figure 1F; Online Supplementary Figure S1J to L). Moreover, depletion of SUMO signaling by pharmacological or genetic targeting resulted in impaired MYC pathway activity (Online Supplementary Figure S2; Online Supplementary Table S1).
In summary, we here identify pharmacological SUMO inhibition as a vulnerability and rational treatment strategy for MYC-driven BCL.
SUMO inhibition is an effective therapy for MYC-driven B-cell lymphoma in vivo
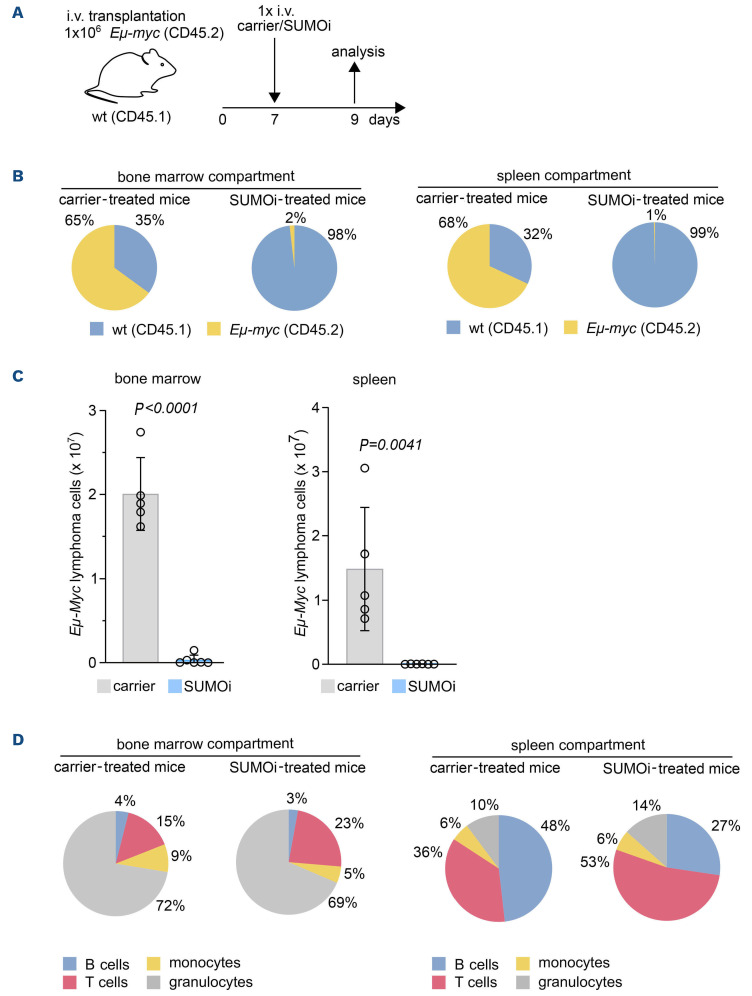
Principal-component analysis of mRNA expression of the SUMO core machinery in wt B cells and MYC-driven BCL from Em-myc mice confirmed enhanced SUMO pathway expression in MYC-driven BCL (GSE7897) (Online Supplementary Figure S3A). In order to test whether inhibition of SUMOylation would offer therapeutic efficacy towards MYC-induced BCL, we treated cell lines derived from three independent primary Em-myc lymphomas with SUMOi and detected striking sensitivity to pharmacological inhibition of SUMOylation (Online Supplementary Figure S3B and C). In order to test therapeutic efficacy in vivo, we transplanted primary Em-myc lymphoma cells into syngeneic wt recipient mice. Seven days after lymphoma cell injection, lymphoma-bearing mice received treatment with SUMOi or carrier solution (Figure 2A). CD45.1 and CD45.2 epitope diversity allowed to discriminate between wt recipient B cells and the syngeneic lymphoma compartment. Strikingly, a single SUMOi therapy resulted in a highly efficient reduction and almost complete eradication of Em-myc-transgenic CD45.2 lymphoma cells in the bone marrow (BM) and spleen (Figure 2B and C; Online Supplementary Figure S4A). Moreover, a single dose of SUMOi treatment significantly reduced spleen weight without causing obvious signs of severe short-term toxicity (Online Supplementary Figure S4B to D). Thus, we here establish activated MYC signaling as an actionable vulnerability and inhibition of SUMOylation as a rational treatment strategy in a preclinical murine model of MYC-driven BCL.
Remarkably, SUMOi therapy not only affected the tumor compartment (CD45.2+), but phenotype analysis hinted towards major changes in the composition of the recipient primary and secondary lymphoid organs. Analysis of the CD45.1 wt compartment in lymphoma-grafted mice revealed a significant increase in the abundance of CD3+ T cells in both BM and spleen after SUMOi treatment that was accompanied by a distinct reduction of the recipient B220+ B-cell compartment, particularly in the spleen (Figure 2D; Online Supplementary Figure S4E and F), revealing that B cells were more sensitive to SUMO inhibition than T cells (Online Supplementary Figure S4G). Besides, the effect on B cells was less pronounced in recipient B cells than in MYC-driven lymphoma B cells (Online Supplementary Figure S4H). The abundance of granulocytes and monocytes mostly remained unaffected by SUMOi treatment (Figure 2D; Online Supplementary Figure S4E and F). Thus, SUMO inhibition leads to killing of MYC-driven B-cell lymphoma in vivo. This fast and most likely direct effect was accompanied by alterations of cellular components of the immune system.
SUMO inhibition remodels immune cell abundance in vivo
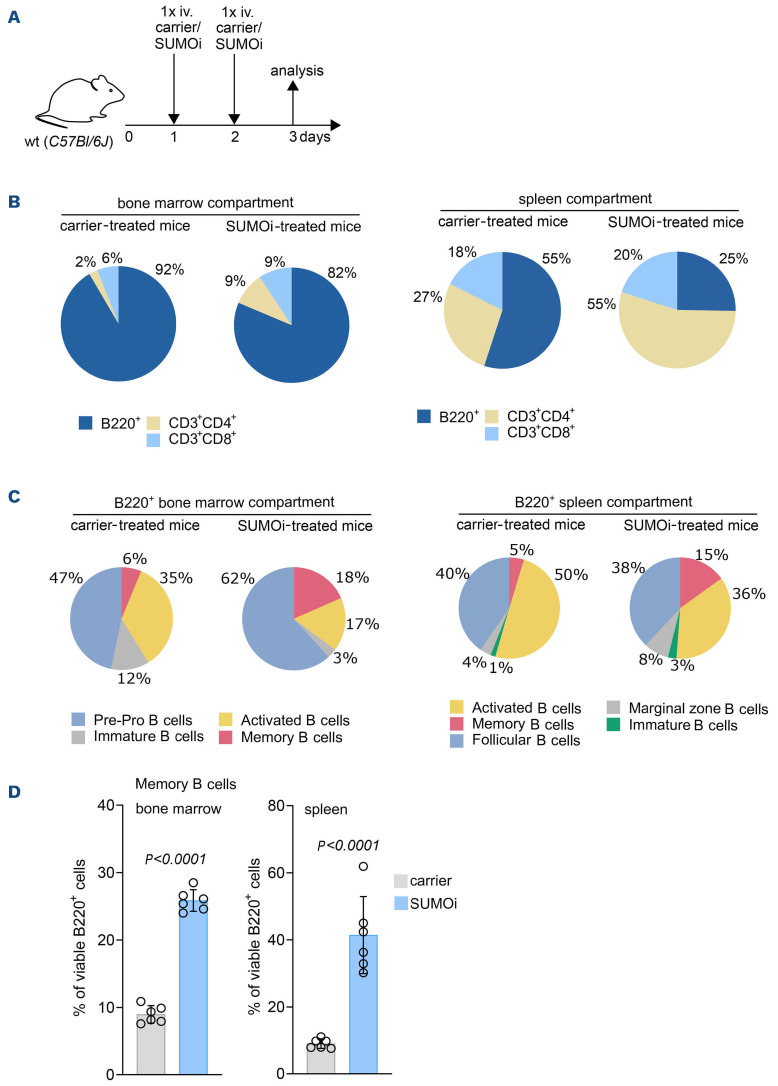
Impaired immune surveillance is involved in lymphoma pathogenesis27 and induction of immune activity contributes to cancer control.28,29 In order to test if immune effects by SUMOi treatment are a general feature independent of the presence of lymphoma, we treated C57Bl/6J mice with either SUMOi or carrier control on 2 consecutive days and analyzed splenocytes and BM cells by multicolor flow cytometry (Figure 3A; Online Supplementary Figure S5A and D). In line with our previous findings in lymphoma bearing mice, SUMOi treatment led to a relative increase of the CD3+CD4+ T-cell compartment (Figure 3B; Online Supplementary Figure S5B, C, E and F). The increase of CD3+CD4+ T cells, a cellular subset holding an important role in anti-tumor immunity,30,31 was most prominent in the spleen. These effects were accompanied by a reduction of the B220+ B-cell compartment (Figure 3B; Online Supplementary Figures S5B, C, E and F; and S6A and D). Shifts in immune cell abundance were detected in both BM and spleen, despite of a decline in overall BM and spleen cell numbers upon SUMOi challenge (Online Supplementary Figure S5A and D).
Figure 1.
Active MYC signaling confers susceptibility to small-molecule SUMO inhibition in diffuse large B-cell lymphoma cell lines. (A) Hierarchical clustering (Euclidean/Ward) of indicated SUMO core components of normalized human diffuse large B-cell lymphoma (DLBCL) transcriptome profiles (GSE98588) revealed SUMOhigh and SUMOlow subgroups as indicated. (B) Gene set enrichment analysis of SUMOhigh and SUMOlow subgroups using Hallmark gene sets from the molecular-signature database (MSigDb). Indicated MYC target gene signatures have been identified using GeneTrail v3 (Kolgomorov-Smirnov-Test). Adjusted P values (q) are indicated. (C) Flow cytometry analysis of relative viability of indicated DLBCL cell lines treated with increasing SUMO inhibitor (SUMOi) concentrations (Tak-981, 0, 3.9, 7.8, 15.6, 31.2, 62.5, 125, 250, 500, 1,000, 2,000 nM) for 72 hours (h) (n=3). (D) Gene set enrichment analysis of SUMOi-responder (Oci-Ly19, SU-DHL-5, SU-DHL-8) vs. SUMOi-non-responder (U-2932, SUDHL-4, DB, NU-DHL-1) on expression profiles accessed via GSE53798. Enrichment plot on SUMOylation signatures, obtained from the Reactome knowledgebase. Adjusted P values (q) are indicated. Volcano plot displays significant gene signatures of the Hallmark gene set (MSigDb) with both MYC Hallmark signatures (V1, V2) highlighted. (E) Enrichment plot on MYC Hallmark signatures, based on the analysis described in (D). Adjusted P values (q) are indicated. (F) Relative viability of Oci-Ly1 cells transduced with a MYC expression plasmid or a control plasmid, treated with increasing SUMOi concentrations (Tak-981, 0, 3.9, 7.8, 15.6, 31.2, 62.5, 125, 250, 500, 1,000, 2,000 nM) for 72 h (n=3). P values refers to 2,000 nM SUMOi and is determined by unpaired t-test. Immunoblot analysis of Oci-Ly1 control and MYC cell lines.
We next substantiated the in vivo effects of pharmacological SUMOi on immune cell distribution in more detail.
Figure 2.
SUMO inhibition is an effective therapy for Myc-driven B-cell lymphoma in vivo. (A) Schematic illustration showing transplantation of Em-myc lymphoma cells (CD45.2) into wild-type (wt) (CD45.1) recipient mice. Mice were treated with carrier or SUMO inhibitor (SUMOi) (ML-093, 50 mg/kg) at day 7 post transplantation. Analysis of bone marrow, spleen and blood was performed at day 9, n=6 per condition. (B) Pie charts representing the frequencies of wt and Em-myc lymphoma cells in the bone marrow (BM) and spleen after carrier vs. SUMOi treatment. (C) Total number of Em-myc lymphoma cells in the BM and spleen following SUMOi treatment compared to carrier. N=6, P values were determined by unpaired t-test. (D) Pie charts representing the frequencies of B cells (B220+), T cells (CD3+), granulocytes (Gr.1+CD11b+) and monocytes (Gr.1- CD11b+) in the BM and spleen after carrier vs. SUMOi treatment. i.v.: intravenously.
Figure 3.
SUMO inhibition induces major changes in the B-cell compartment, favoring a memory B-cell phenotype. (A) Schematic illustration showing treatment of wild-type (wt) mice with carrier or SUMO inhibitor (SUMOi) (ML-093, 50 mg/kg) at day 1 and 2. Analysis of bone marrow (BM) and spleen was performed at day 3, n=6 per condition. (B) Pie charts representing the frequencies of indicated cell populations (B220+ B cells, CD3+CD4+ T cells, CD3+CD8+ T cells) in the BM and spleen after carrier vs. SUMOi treatment. (C) Pie charts representing the frequencies of indicated cell populations in the B220+ BM compartment (B220+IgM- pre-pro B cells, B220+IgM+IgD- immature B cells, B220+CD19+MHC-II+ activated B cells, B220+CD80+CD86+ memory B cells) and B220+ spleen compartment (B220+CD93+ Immature B cells, B220+CD21highCD23low marginal zone B cells, B220+CD21lowCD23high follicular B-cells, B220+CD80+CD86+ memory B cells, B220+CD19+MHC-II+ activated B cells) after carrier vs. SUMOi treatment. (D) Percentage of memory B cells (B220+ CD80+ CD86+) of B220+ BM and spleen cells. N=6, P values were determined by unpaired t-test. i.v.: intravenously.
SUMO inhibition resulted in a decline of most B-cell subsets within both primary lymphoid BM tissue and the secondary lymphoid organ spleen (Figure 3C), which is in line with recent reports.19 Besides, we detected a significant increase in B220+ B-memory cells after SUMO inhibition in BM and spleen (Figure 3D; Online Supplementary Figures S6D and S7A), indicating effects on a cellular subset important for a coordinated antibody-dependent immune response.32
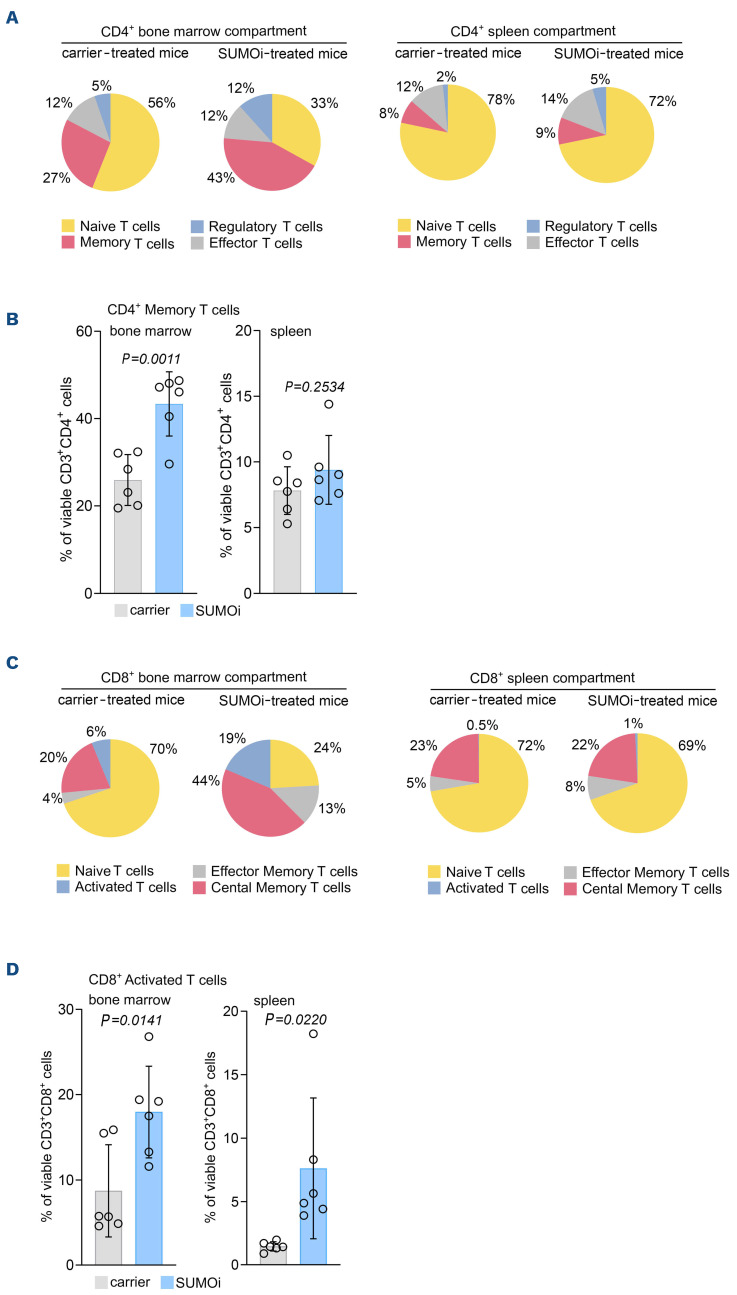
SUMO-directed intervention not only exerted broad effects on the B-cell subsets, but also on the T-cell compartment, as we observed a significant increase in CD4+ memory and regulatory T cells within BM and spleen upon SUMOi (Figure 4A, B; Online Supplementary Figures S6B and E; and S7B). Beyond, the abundance of BM CD8+ effector memory cells was significantly increased (Figure 4C; Online Supplementary Figures S6C and S7C). Both CD4+ and CD8+ T memory cells are antigen-specific long-term persisting cells that arise from naïve T-cells upon encounter to a cognate antigen and are designated to execute a protective immune response upon antigen reencounter.33,34 Of note, SUMO inhibition was accompanied by a substantially higher abundance of activated CD8+ T cells (Figure 4C and D; Online Supplementary Figures S6C and F; and S7D), a population referred to as cytotoxic T cells with well-described features in host defense and tumor cytolysis.35 This finding is in line with current reports linking SUMO inhibition to enhanced T-cell activation.19,20,36
In summary, our analyses revealed a substantial remodeling of immune cell subsets following SUMO-directed intervention, underscoring the important role of the SUMO pathway in the immune system.
SUMO inhibition substantially alters the normal B-cell landscape
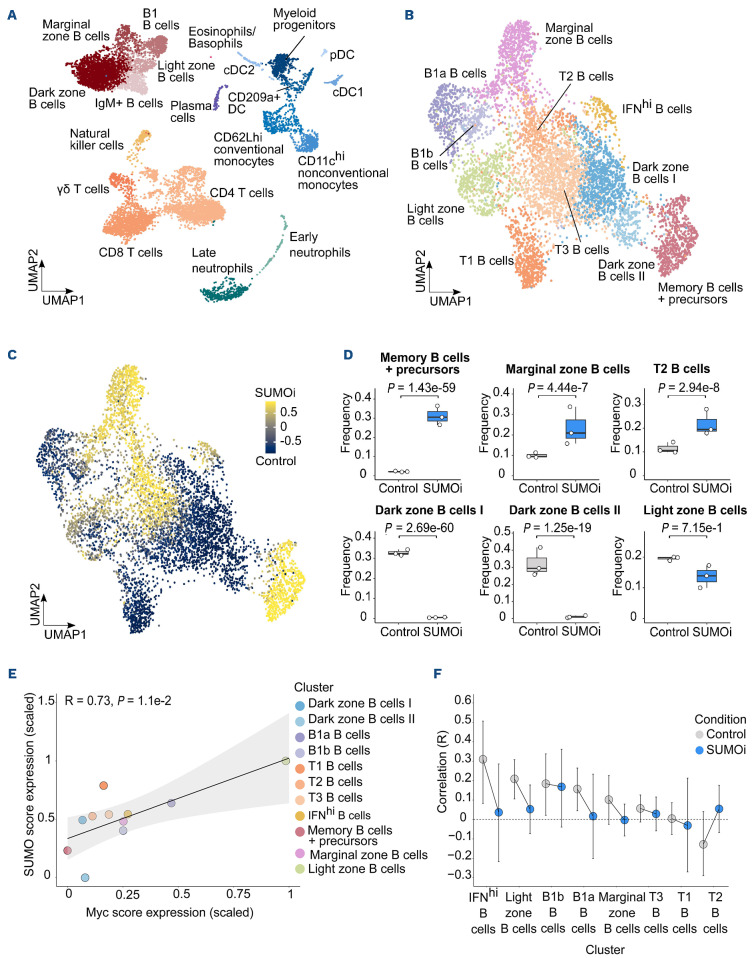
Our preceding analyses revealed pharmacologic inhibition of SUMOylation as a vulnerability in MYC-driven lymphoma that was associated with pronounced changes in the immune cell compartment. In order to decipher the effects of short term highly specific small-molecule SUMOi on B-cell subsets in more detail, we next opted for Cellular Indexing of Transcriptomes and Epitopes by Sequencing (CITE-seq) analysis (dataset GSE193359).20 CITE-seq combines the measurements of surface protein levels and transcriptome analysis to determine cellular states and their alterations at single-cell level.37 In this experiment wt mice were challenged with a lower dose of the SUMO inhibitor subasumstat in line with a recent publication reporting enhanced anti-tumor T-cell capacity upon SUMOi.36 CITE-seq analysis was conducted on spleens from three control and three SUMOi-treated mice. Prior to analysis cells were marked with oligo-conjugated antibodies allowing discrimination between B- and T-cell subsets (Figure 5A). Transcriptomics and surface proteomics were conducted on the 10x Genomics platform. In total 18,361 cells (9,878 cell from control mice and 8,483 from SUMOi-treated mice) were analyzed. Using expression data of B-cell surface markers and B-cell specific marker genes, we annotated 11 different murine B-cell populations (Figure 5B; Online Supplementary Figure S8). SUMOi treatment resulted in decreased abundance of the early and more immature splenic B-cell subset T1 and T3 (Online Supplementary Figure S9A). The abundance of memory and marginal zone B cells was substantially higher in SUMOi-treated mice (Figure 5C and D). Moreover, we detected a striking decline in dark zone and light zone B cells (Figure 5C and D). Of note, MYC expression is induced in light-zone B cells in direct proportion to antigen capture during the dark-zone to light-zone germinal center transition to coordinate residence time in the dark-zone.38 Accordingly, we observed high expression of the MYC core machinery in light-zone B cells that was associated with enhanced expression of the SUMO core machinery (Figure 5E). Remarkably, SUMO inhibition abrogated this effect (Figure 5F; Online Supplementary Figure S9B). Assessing the question of whether cellular proliferation needs to be considered as a confounder effect when evaluating the changes in abundance between treatment conditions, we performed proliferation score analysis on B and T cells within the CITE-seq dataset (Online Supplementary Figure S10A, B and D to F). Differences in proliferation among the different cell populations were overall moderate and not significantly altered after SUMO inhibition among all subsets analyzed (Online Supplementary Figure S10C).
In summary, these data revealed the complexity of a SUMO-directed therapeutic intervention on immune cell abundance and specifically on the B-cell landscape. Furthermore, we could link MYC expression to expression of the SUMO core machinery on a single-cell level, highlighting MYC signaling as an actionable vulnerability targeted by SUMO inhibition.
Discussion
Here, we unraveled activation of SUMOylation as a striking vulnerability in MYC-driven BCL. Next to direct effects of specific pharmacological SUMO inhibition on MYC-induced lymphoma, our investigations depicted a substantial remodeling of components of the innate and specific immunity in vivo by SUMOi.
Defining strategies to tackle MYC-driven tumors is of huge relevance based on the well-established activation of MYC in the vast majority of cancers, however, direct MYC targeting remains challenging.39 Although MYC targeting via a dominant-negative peptide OMOMYC showed promising preclinical results and is currently tested in a phase I/II clinical trial (clinicaltrials gov. Identifier: NCT04808362),40 no direct MYC inhibitor has reached clinical practice.8 These unmet challenges favor the idea of targeting deregulated cellular pathways in MYC-overexpressing tumors to translate the concept of “synthetic lethality”7, 8 into clinical cancer therapy. We here identified aberrant activity of the SUMO core pathway in aggressive BCL with activated MYC signaling and identified activated MYC signaling as an actionable molecular vulnerability in vitro and in a preclinical murine model of MYC-driven lymphoma in vivo. Notably, SUMO inhibition alone or administered as combination therapy with rituximab, a standard therapy for BCL patients, showed remarkable efficacy in preclinical DLBCL xenograft models.25,41 Besides, SUMO inhibition is currently tested in various clinical trials (clinicaltrials gov. Identifier: NCT03648372, NCT04074330, NCT04381650), emphasizing the relevance of our findings for biomarker-informed clinical applications. Our data is in line with an unbiased synthetic lethality screen in MYC-driven breast cancer that uncovered a role for the SUMO activation complex SAE1/2 in MYC-driven tumorigenesis.21 Furthermore, a previous loss-of-function study in BCL-linked silencing of SAE2 to tumor regression in vivo.22 The MYC-SUMO connection and MYC-related sensitivity to SUMOi was also identified in aggressive pancreatic cancer.23 We here provide first data indicating that highly specific small-molecule SUMO inhibition is an effective therapy for MYC-driven BCL in vivo and demonstrate a key role for activated MYC signaling in conferring susceptibility to SUMO inhibition in B cells, while non-cancer syngeneic B cells showed remarkably lower killing rates as compared to the MYC-driven lymphoma population. This MYC-dependent effect of SUMO inhibition was confirmed by ectopic expression of MYC in the human DLBCL cells. Moreover, following in vivo low dose SUMOi challenge, CITE-seq analysis showed diminished abundance of wt light-zone B cells that are characterized by high MYC expression and concurrent high expression of the SUMO core machinery. Beyond depicting MYC signaling as a vulnerability, we would like to emphasize that uncovering additional biomarkers predicting SUMOi sensitivity in cancer therapy is an area worthy of further investigations.
Figure 4.
SUMO inhibition remodels the T-cell compartment towards a memory and activated T-cell phenotype. (A) Experimental setup as outlined in Figure 3A. Pie charts representing the frequencies of the indicated cell populations in the CD3+CD4+ bone marrow (BM) and spleen compartment (CD3+CD4+CD44lowCD62L+ naïve T cells, CD3+CD4+CD44highCD62L- memory T cells, CD3+CD4+CD44lowCD62L- effector T cells, CD3+CD4+CD25+CD69+FoxP3+ regulatory T cells). (B) Percentage of memory T cells (CD3+CD4+CD44highCD62L-) of the CD3+CD4+ BM and spleen cells. N=6, P values were determined by unpaired t-test. (C) Pie charts representing the frequencies of the indicated cell populations in the CD3+CD8+ BM and spleen compartment (CD3+CD8+CD44lowCD62L+ naïve T cells, CD3+CD8+ CD44highCD62L- effector memory T cells, CD3+CD8+CD44highCD62L+ central memory T cells, CD3+CD8+CD25+CD69+ activated T cells) after carrier vs. SUMOi treatment. (D) Percentage of activated T cells (CD3+CD8+CD25+CD69+) of the CD3+CD8+ BM and spleen cells. N=6, P values were determined by unpaired t-test.
Figure 5.
SUMO inhibition substantially alters the normal B-cell landscape. (A) UMAP visualization of spleen scRNA-sequencing data from control and SUMO inhibitor (SUMOi)-treated mice (subasumstat 7.5 mg/kg on day 1 and 4, spleen cell analysis day 5; dataset GSE193359). (B) B-cell populations identified in (A) were subsetted and reclustered. The UMAP visualization represents B cells from both conditions. (C) Differentially abundant B-cell populations in control and SUMOi-treated mice identified with DA-seq. Cells are colored by DA-seq measure. Yellow = more abundant after SUMOi challenge. Dark blue = more abundant after control treatment. (D) Differential abundance testing on mouse-wise pseudobulks (indicated by white dots, n=3). Bar plots represent the subpopulation frequencies stratified by condition. The median is indicated by the center line of the box plot. The box extends from the 25th to 75th percentiles, whisker length reaches from minimum to maximum. Significance is determined by a Negative Binomial Generalized Linear Model. (E) SUMO and MYC score correlation analysis in control mice. Scaled mean expression values are plotted against each other for each cell population identified in (B). R indicates the Pearson correlation coefficient. The regression line is shown in black. The grey area indicates the 95% confidence interval. (F) SUMO and MYC score correlation analysis in control and SUMOi-treated mice. For each population the Pearson correlation coefficient (R) is plotted. Black bars indicate the range of the 95% confidence interval. The dashed black line at R=0 is a reference line and plotted as a visualization aid.
Apart from driving cancer progression through tumor cell intrinsic acquisition of cancer hallmarks, MYC restrains the anti-cancer immune response and dysregulates the tumor microenvironment.27,42 We show substantial alteration of the B-cell landscape upon SUMO inhibition in a wt mouse model and report higher abundance of memory B cells after pharmacological SUMO inhibition, accompanied by a decline in immature subsets, which is in line with recent publications.19 B-memory cells are accounted with a key role in sustaining a long-term immune response.32,43 SUMO inhibition not only altered B-cell abundance, but also strikingly increased the abundance of CD8+ cytotoxic T cells as well as CD4+ and CD8+ memory T cells and particularly T-regulatory cells. CD8+ effector T cells exert direct anti-tumor cytotoxicity and CD4+ T cells enhance cytolytic efficacy of CTL by antigen cross-presentation and mediate plasma cell differentiation.30,31,35 T-regulatory cells are accounted with a double-edged role in anti-tumor immunity.44 They suppress anti-cancer immunity, but also drive and control the T-cell immune responses against tumor neoantigens.45 So far, only enhanced CD8+ T-cell activation and augmented anti-tumor sensitivity by SUMOimediated reactivation of type I interferon signaling have been reported before.19,20,36 We here identified a novel function of SUMO inhibition in remodeling the immune cell landscape by enhancing abundance of cellular subsets involved in the innate and specific immune response.
In summary, we here depict activated MYC signaling as an actionable molecular vulnerability for SUMO inhibition in vitro and in a preclinical murine model of MYC-driven BCL in vivo. Next to direct effects on MYC-driven lymphoma, SUMO inhibition substantially remodels the immune cell landscape in vivo independent of a tumor microenvironment. Our findings thus identify SUMO inhibition as a powerful therapy in a subset of MYC-driven B-cell lymphomas and suggest SUMO-targeted therapies as a potential therapeutic strategy for cancer immunotherapy.
Supplementary Material
Acknowledgments
The authors would like to thank Millennium Pharmaceuticals, Inc., a wholly owned subsidiary of Takeda Pharmaceutical Company Limited, for providing ML-093 and subasumstat/Tak-981.
Funding Statement
Funding: This work was supported by Deutsche Forschungsgemeinschaft (DFG, SFB824/C3 and SFB1335/P3 to UK; grant KE 222/10-1 to UK and SM, DFG grants MU 1764/7-1 (#494535244) and MU 1764/7-1 to to SM), Deutsche Krebshilfe (grants 70114425 and 70114724 to UK, grant 70114823 to SM), Stiftung Charité (to UK), and Wilhelm-Sander Foundation (2017.048.2 to UK). UD is a participant in the BIH-Charité Junior Clinician Scientist program funded by the Charité - Universitätsmedizin Berlin and the Berlin Institute of Health.
References
- 1.Dang CV. MYC on the path to cancer. Cell. 2012;149(1):22-35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schick M, Habringer S, Nilsson JA, Keller U. Pathogenesis and therapeutic targeting of aberrant MYC expression in haematological cancers. Br J Haematol. 2017;179(5):724-738. [DOI] [PubMed] [Google Scholar]
- 3.Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. 2018;3:5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Singh K, Lin J, Zhong Y, et al. c-MYC regulates mRNA translation efficiency and start-site selection in lymphoma. J Exp Med. 2019;216(7):1509-1524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ruggero D. The role of Myc-induced protein synthesis in cancer. Cancer Res. 2009;69(23):8839-8843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Stine ZE, Walton ZE, Altman BJ, Hsieh AL, Dang CV. MYC, metabolism, and cancer. Cancer Discov. 2015;5(10):1024-1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kaelin WG. The concept of synthetic lethality in the context of anticancer therapy. Nat Rev Cancer. 2005;5(9):689-698. [DOI] [PubMed] [Google Scholar]
- 8.Dhanasekaran R, Deutzmann A, Mahauad-Fernandez WD, Hansen AS, Gouw AM, Felsher DW. The MYC oncogene - the grand orchestrator of cancer growth and immune evasion. Nat Rev Clin Oncol. 2022;19(1):23-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Seeler JS, Dejean A. SUMO and the robustness of cancer. Nat Rev Cancer. 2017;17(3):184-197. [DOI] [PubMed] [Google Scholar]
- 10.Hendriks IA, Vertegaal AC. A comprehensive compilation of SUMO proteomics. Nat Rev Mol Cell Biol. 2016;17(9):581-595. [DOI] [PubMed] [Google Scholar]
- 11.Flotho A, Melchior F. Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem. 2013;82:357-385. [DOI] [PubMed] [Google Scholar]
- 12.Cappadocia L, Lima CD. Ubiquitin-like protein conjugation: structures, chemistry, and mechanism. Chem Rev. 2018;118(3):889-918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunz K, Piller T, Muller S. SUMO-specific proteases and isopeptidases of the SENP family at a glance. J Cell Sci. 2018;131(6);jcs211904. [DOI] [PubMed] [Google Scholar]
- 14.Bawa-Khalfe T, Yeh ET. SUMO losing balance: SUMO proteases disrupt SUMO homeostasis to facilitate cancer development and progression. Genes Cancer. 2010;1(7):748-752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schick M, Zhang L, Maurer S, et al. Genetic alterations of the SUMO isopeptidase SENP6 drive lymphomagenesis and genetic instability in diffuse large B-cell lymphoma. Nat Commun. 2022;13(1):281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Driscoll JJ, Pelluru D, Lefkimmiatis K, et al. The sumoylation pathway is dysregulated in multiple myeloma and is associated with adverse patient outcome. Blood. 2010;115(14):2827-2834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun WC, Hsu PI, Yu HC, et al. The compliance of doctors with viral hepatitis B screening and antiviral prophylaxis in cancer patients receiving cytotoxic chemotherapy using a hospitalbased screening reminder system. PLoS One. 2015;10(2):e0116978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Decque A, Joffre O, Magalhaes JG, et al. Sumoylation coordinates the repression of inflammatory and anti-viral gene-expression programs during innate sensing. Nat Immunol. 2016;17(2):140-149. [DOI] [PubMed] [Google Scholar]
- 19.Kumar S, Schoonderwoerd MJA, Kroonen JS, et al. Targeting pancreatic cancer by TAK-981: a SUMOylation inhibitor that activates the immune system and blocks cancer cell cycle progression in a preclinical model. Gut. 2022;71(11):2266-2283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Demel UM, Boger M, Yousefian S, et al. Activated SUMOylation restricts MHC class I antigen presentation to confer immune evasion in cancer. J Clin Invest. 2022;132(9):e152383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kessler JD, Kahle KT, Sun T, et al. A SUMOylation-dependent transcriptional subprogram is required for Myc-driven tumorigenesis. Science. 2012;335(6066):348-353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoellein A, Fallahi M, Schoeffmann S, et al. Myc-induced SUMOylation is a therapeutic vulnerability for B-cell lymphoma. Blood. 2014;124(13):2081-2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Biederstadt A, Hassan Z, Schneeweis C, et al. SUMO pathway inhibition targets an aggressive pancreatic cancer subtype. Gut. 2020;69(8):1472-1482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He X, Riceberg J, Soucy T, et al. Probing the roles of SUMOylation in cancer cell biology by using a selective SAE inhibitor. Nat Chem Biol. 2017;13(11):1164-1171. [DOI] [PubMed] [Google Scholar]
- 25.Langston SP, Grossman S, England D, et al. Discovery of TAK-981, a first-in-class inhibitor of SUMO-activating enzyme for the treatment of cancer. J Med Chem. 2021;64(5):2501-2520. [DOI] [PubMed] [Google Scholar]
- 26.Doffo J, Bamopoulos SA, Kose H, et al. NOXA expression drives synthetic lethality to RUNX1 inhibition in pancreatic cancer. Proc Natl Acad Sci U S A. 2022;119(9):e2105691119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Casey SC, Baylot V, Felsher DW. The MYC oncogene is a global regulator of the immune response. Blood. 2018;131(18):2007-2015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kubli SP, Berger T, Araujo DV, Siu LL, Mak TW. Beyond immune checkpoint blockade: emerging immunological strategies. Nat Rev Drug Discov. 2021;20(12):899-919. [DOI] [PubMed] [Google Scholar]
- 29.Restifo NP, Dudley ME, Rosenberg SA. Adoptive immunotherapy for cancer: harnessing the T cell response. Nat Rev Immunol. 2012;12(4):269-281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Borst J, Ahrends T, Babala N, Melief CJM, Kastenmuller W. CD4(+) T cell help in cancer immunology and immunotherapy. Nat Rev Immunol. 2018;18(10):635-647. [DOI] [PubMed] [Google Scholar]
- 31.Tay RE, Richardson EK, Toh HC. Revisiting the role of CD4(+) T cells in cancer immunotherapy-new insights into old paradigms. Cancer Gene Ther. 2021;28(1-2):5-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sautes-Fridman C, Petitprez F, Calderaro J, Fridman WH. Tertiary lymphoid structures in the era of cancer immunotherapy. Nat Rev Cancer. 2019;19(6):307-325. [DOI] [PubMed] [Google Scholar]
- 33.Martin MD, Badovinac VP. Defining memory CD8 T cell. Front Immunol. 2018;9:2692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stockinger B, Bourgeois C, Kassiotis G. CD4+ memory T cells: functional differentiation and homeostasis. Immunol Rev. 2006;211:39-48. [DOI] [PubMed] [Google Scholar]
- 35.St Paul M, Ohashi PS. The roles of CD8(+) T cell subsets in antitumor immunity. Trends Cell Biol. 2020;30(9):695-704. [DOI] [PubMed] [Google Scholar]
- 36.Lightcap ES, Yu P, Grossman S, et al. A small-molecule SUMOylation inhibitor activates antitumor immune responses and potentiates immune therapies in preclinical models. Sci Transl Med. 2021;13(611):eaba7791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stoeckius M, Hafemeister C, Stephenson W, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14(9):865-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Finkin S, Hartweger H, Oliveira TY, Kara EE, Nussenzweig MC. Protein amounts of the MYC transcription factor determine germinal center B cell division capacity. Immunity. 2019;51(2):324-336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schneider G, Wirth M, Keller U, Saur D. Rationale for MYC imaging and targeting in pancreatic cancer. EJNMMI Res. 2021;11(1):104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Llombart V, Mansour MR. Therapeutic targeting of "undruggable" MYC. EBioMedicine. 2022;75:103756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakamura A, Grossman S, Song K, et al. The SUMOylation inhibitor subasumstat potentiates rituximab activity by IFN1-dependent macrophage and NK cell stimulation. Blood. 2022;139(18):2770-2781. [DOI] [PubMed] [Google Scholar]
- 42.Seton-Rogers S. Oncogenes: driving immune evasion. Nat Rev Cancer. 2018;18(2):67. [DOI] [PubMed] [Google Scholar]
- 43.Sharonov GV, Serebrovskaya EO, Yuzhakova DV, Britanova OV, Chudakov DM. B cells, plasma cells and antibody repertoires in the tumour microenvironment. Nat Rev Immunol. 2020;20(5):294-307. [DOI] [PubMed] [Google Scholar]
- 44.Togashi Y, Shitara K, Nishikawa H. Regulatory T cells in cancer immunosuppression - implications for anticancer therapy. Nat Rev Clin Oncol. 2019;16(6):356-371. [DOI] [PubMed] [Google Scholar]
- 45.Pace L, Tempez A, Arnold-Schrauf C, et al. Regulatory T cells increase the avidity of primary CD8+ T cell responses and promote memory. Science. 2012;338(6106):532-536. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.