Abstract
A major obstacle in the treatment of acute myeloid leukemia (AML) is refractory disease or relapse after achieving remission. The latter arises from a few therapy-resistant cells within minimal residual disease (MRD). Resistant cells with long-term self-renewal capacity that drive clonal outgrowth are referred to as leukemic stem cells (LSC). The cancer stem cell concept considers LSC as relapse-initiating cells residing at the top of each genetically defined AML subclone forming epigenetically controlled downstream hierarchies. LSC display significant phenotypic and epigenetic plasticity, particularly in response to therapy stress, which results in various mechanisms mediating treatment resistance. Given the inherent chemotherapy resistance of LSC, targeted strategies must be incorporated into first-line regimens to prevent LSC-mediated AML relapse. The combination of venetoclax and azacitidine is a promising current strategy for the treatment of AML LSC. Nevertheless, the selection of patients who would benefit either from standard chemotherapy or venetoclax + azacitidine treatment in first-line therapy has yet to be established and the mechanisms of resistance still need to be discovered and overcome. Clinical trials are currently underway that investigate LSC susceptibility to first-line therapies. The era of single-cell multi-omics has begun to uncover the complex clonal and cellular architectures and associated biological networks. This should lead to a better understanding of the highly heterogeneous AML at the inter- and intra-patient level and identify resistance mechanisms by longitudinal analysis of patients’ samples. This review discusses LSC biology and associated resistance mechanisms, potential therapeutic LSC vulnerabilities and current clinical trial activities.
Introduction
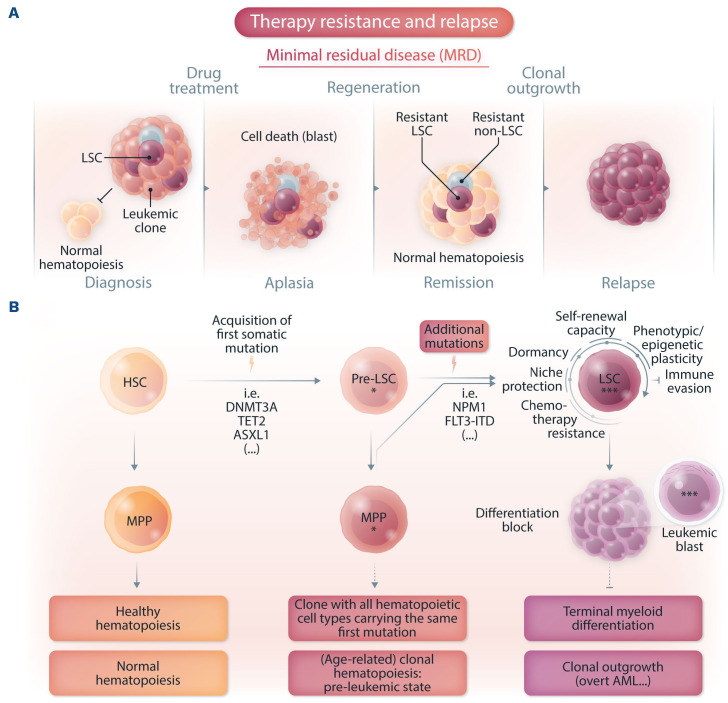
Acute myeloid leukemia (AML) is a heterogeneous disease with a complex cytogenetic and molecular landscape.1 Following conventional induction chemotherapy, patients are assigned to risk-adapted post-remission consolidative therapies.2 Although the majority of AML patients respond to induction chemotherapy, refractory disease is common and relapse is a major challenge.2 Currently, risk stratification is based on cytogenetic diagnostics for recurrent structural genomic abnormalities and targeted sequencing-based diagnostics for recurrent gene mutations.3 However, the origin of relapse has been traced back to therapy-resistant leukemia cells referred to as minimal residual disease (MRD), containing leukemic stem cells (LSC).4 The cancer stem cell concept attributes the origin of relapse to these therapy-resistant leukemia cells exhibiting specific gene expression signatures related to stemness properties.5 The early detection of these drug-tolerant persister cells enables allocation of patients to salvage therapies or enrollment in clinical trials prior to overt AML relapse (Figure 1A).
The combination of venetoclax and the hypomethylating agent azacitidine has become the standard of care for patients with newly diagnosed AML ≥75 years of age or those who have comorbidities that preclude the use of standard intensive chemotherapy, as a phase III clinical trial demonstrated durable remissions.6 Furthermore, there is a role for venetoclax + azacitidine in refractory/relapsed AML patients to induce remission prior to allogeneic stem cell transplantation and in newly diagnosed patients in whom intensive chemotherapy is not justifiable (given leukemic organ infiltration or a serious infectious complication in neutropenia).7
Since MRD assessment is routinely performed either by cytogenetics, targeted sequencing detecting specific molecular alterations present at diagnosis, such as NPM1 mutations, or by multiparameter flow cytometry, AML evolution trajectories and dynamic properties of therapy-resistant AML cells have not so far been captured. The era of single-cell multi-omics provides precious insights into clonal dynamics and enables tracing of distinct subclones and the detection of therapy-resistant leukemia cells during the course of AML therapy. Here, we discuss LSC biology and emphasize resistance mechanisms and therapeutic vulnerabilities thereby highlighting a role for single-cell multi-omics to detect MRD, to uncover clonal dynamics of AML and to identify new therapeutic targets aiming to prevent the re-emergence of AML.
Leukemic stem cells, clonal evolution dynamics and tumor heterogeneity
Mature blood and immune cells exhibit tremendous diversity in cell morphology and function. The majority of these cells are derived from multipotent hematopoietic stem cells (HSC), at the top of the hierarchy within the hematopoietic organization.8 The hallmark of these HSC is their capacity to self-renew maintaining the resident HSC population and generating various progenitors that proliferate and differentiate into mature blood cells and immune cells. By contrast, committed progenitors have limited and steadily decreasing self-renewal capacity, are exposed to lineage fate and are exhausted within a certain time.9 At steady state, most HSC are inactive or in a long-term quiescent, but reversible G0 phase of the cell cycle to maintain their long-term function, a state termed dormancy.10,11 Classically, HSC have been considered as a discrete homogeneous population. However, more recent studies showed significant HSC heterogeneity including early lineage priming and the presence of lineage-biased HSC within the HSC compartment.8,12,13 HSC reside in a highly specialized, hypoxic bone marrow microenvironment referred to as a niche. The niche concept was proposed in 1978 and is now viewed as a complex network that provides molecular mechanisms and physical interactions that are essential for HSC localization, maintenance and differentiation.14,15
Figure 1.
Evolution and relapse of acute myeloid leukemia. (A) Illustration of minimal residual disease (MRD) and leukemic stem cell (LSC)-mediated therapy resistance in acute myeloid leukemia (AML). Drug-tolerant persister cells, persisting over treatment and fueling relapse, are illustrated (LSC and non-LSC). (B) Scheme of AML evolution illustrating normal hematopoiesis, clonal hematopoiesis (an age-dependent pre-leukemic state) and clonal outgrowth (overt AML). Pre-LSC, in contrast to LSC, maintain their differentiation ability capable of giving rise to mature blood and immune cells. These mutation-bearing progenitors favor an inflammatory environment, thereby contributing to cardiovascular disease and probably also to clonal expansion. Additional mutations in pre-LSC or mutated multipotent progenitor cells then result in LSC fueling clonal outgrowth. HSC: hematopoietic stem cell, MPP: multipotent progenitor cell, pre-LSC: pre-leukemic stem cell.
The sequential acquisition of somatic mutations contributing to subsequent clonal evolution constitutes a basic principle in cancer biology.16 This sequence was introduced in studies investigating mutations across different stages of colorectal cancer establishing that genetic alterations cause phenotypic manifestations.17 Since the acquisition of somatic mutations in HSC results in a mutated progeny that is endowed with a Darwinian fitness advantage, these cells are empowered to clonal expansion and will dominate the site in which they originate.16 Additional mutations have the potential to strengthen the growth advantage, resulting in different subclones contributing to independent phylogenetic lineage trees within a tumor reminiscent of a branching evolution.16 Thus, many different subclones are conceivable alongside the dominant clone at diagnosis and these do not contribute significantly to the tumor bulk population. This view shows clearly that tumors are not a collection of homogeneous cells with equal capacity for proliferation but rather a heterogeneous assembly consisting of differently functioning cells working together to maintain tumor growth as a pathophysiological organ.16 The LSC phenotype and its plasticity are shaped by distinct mechanisms including gene mutations, epigenetic modifications that result in specific gene expression programs and the metabolic states that shape a patient’s unique leukemic cell heterogeneity. Complexity is further enhanced through the crosstalk between leukemic cells and non-tumor elements, referred to as the tumor microenvironment.18
Leukemic stem cells re-initiate leukemia
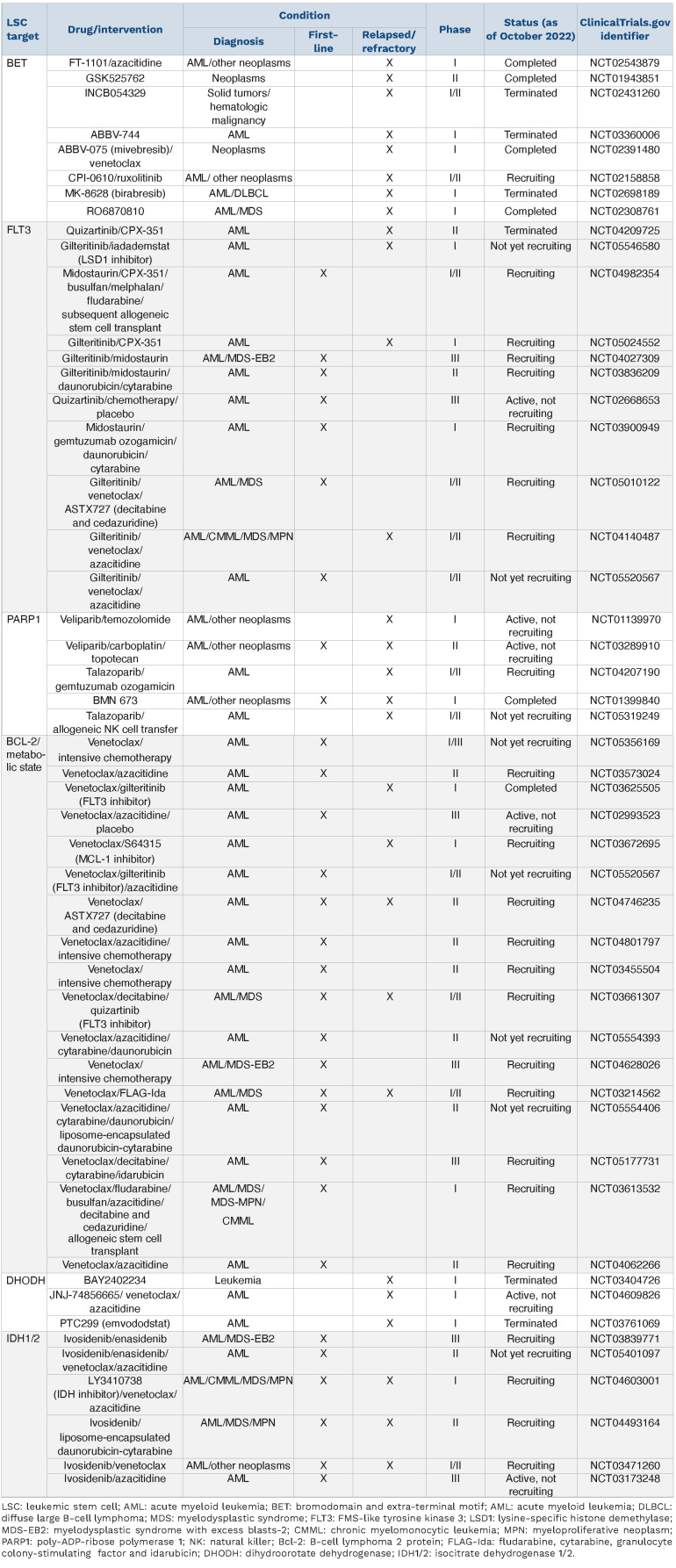
Stem cells can be functionally identified by testing self-renewal in clonal serial in vivo repopulation assays.16 AML is a prime example in which the capacity to self-renew is tested in xenotransplantation assays where LSC engraft in immune-deficient recipient mice giving rise to leukemia.19 This was first achieved 30 years ago, when it was possible to engraft normal human hematopoietic cells and leukemic cells in mice.20-22 Engraftment and the potential to initiate leukemia was restricted to the flow-sorted CD34+CD38– fraction, establishing that AML is organized as a hierarchy with CD34+CD38– leukemia-initiating cells sitting at its top.21,23 The ability of xenografts to capture even rare relapse-relevant LSC enables comprehensive investigations of therapy resistance and therapeutic approaches. The origin of leukemic cells in relapse samples can be traced back by using specific mutations as lineage tracking marks. Therefore, cells within the diagnostic and relapse samples sharing the same mutational profile will most likely originate from the same founder LSC. Individual variant allele frequencies can then be used to follow the evolution of LSC clones from diagnosis to relapse.24 Leukemic cells capable of engrafting in NSG mice have been demonstrated to be transiently quiescent in the G0 phase of the cell cycle. Following serial transplantation, a rare quiescent long-term leukemia-initiating cell population with extensive self-renewal capacity and an extremely low proliferation rate was identified.25 These data suggest that only LSC subsets drive relapse. Studies with paired diagnostic/relapse samples provide evidence that relapse arises from re-emergence or clonal evolution of a pre-existing and chemotherapy-resistant clone generated before treatment.26-28 Thus, a role for LSC in AML relapse has been demonstrated by combining sequencing of purified AML subpopulations and xenotransplantation assays from paired diagnostic/relapse samples identifying the presence of genetically diverse LSC at diagnosis and two distinct patterns of relapse based on the cell type from which relapse originates.28 In the relapse origin-primitive (ROP) group, a rare population of cells with an HSC-like phenotype already present at diagnosis generates bulk blasts that exhibit extensive myeloid differentiation. By contrast, in the relapse origin-committed group (ROC), relapse originates from cells with an immunophenotype of a more committed progenitor. In both groups of patients relapse is linked to stem cell properties, manifested either as a primitive LSC population giving rise to relapse or as stemness transcriptional programs that are retained in the more differentiated bulk population.28 These findings have considerable implications for cancer biology as well as for how AML should be monitored and treated. The identification of distinct relapse patterns emphasizes that improved methods (including single-cell multi-omics as discussed below) tracking the complex evolutionary history of AML within individual patients are inevitable in the design of further clinical trials as an attempt to overcome LSC-mediated therapy resistance and relapse. Furthermore, the shared functional and transcriptional stemness properties that underlie both cellular origins of relapse emphasize the importance of integrating new therapeutic approaches targeting stemness properties to prevent AML relapse (Table 1).
Clonal hematopoiesis: an age-related pre-leukemic state
LSC can give rise to leukemic blasts that carry leukemia-related mutations and are characterized by a differentiation block. By contrast, pre-leukemic stem cells (pre-LSC) harbor recurrent pre-leukemic variants and maintain differentiation and maturation abilities capable of giving rise to mature functional progenitor cells bearing the same variants.29
Clonal hematopoiesis (CH), also called CHIP (clonal hematopoiesis of indeterminate potential), is an age-related condition defined as the presence of myeloid malignancy-associated somatic driver mutations in the peripheral blood without diagnostic criteria for hematologic malignancies.30,31 CH is associated with an increased risk of leukemia and increased mortality largely mediated by cardiovascular disease.32,33 The latter is considered to be caused by a hyperinflammatory phenotype mediated by monocytes and macrophages bearing CH mutations that show increased production of pro-inflammatory cytokines, such as interleukin-1β and interleukin-6, in mice.34,35 DNA methyltransferase 3A (DNMT3A) mutations are the most common driver of this state and most variants exhibit reduced protein stability correlating with strengthened clonal expansion and AML development.36 The tet methylcytosine dioxygenase 2 (TET2) gene has a functionally opposite effect on DNA methylation and is also recurrently mutated in myeloid malignancies and CH.37-3 9 These CH mutations confer a selection advantage to the mutated cell resulting in clonal expansion.40 Thus, DNMT3A mutation-bearing HSC in AML remission samples, without coincident NPM1 mutations present in AML blasts, have a competitive multilineage repopulation advantage over non-mutated HSC in xenografts, thereby establishing their identity as pre-leukemic HSC.29 These early mutations in pre-LSC precede leukemic transformation and define a pre-leukemic state capable of generating the entire hematopoietic hierarchy (Figure 1B). AML can evolve from such a clonally expanded pre-LSC pool detected in remission samples, indicating that pre-LSC survive chemotherapy and might serve as a reservoir for clonal evolution leading to recurrent disease.29 This was shown by performing deep targeted sequencing of commonly mutated leukemia genes, which revealed that DNMT3A mutations occur in an ancestral cell that gives rise to both T cells and the dominant AML clone present at diagnosis.29 Xenograft repopulation assays then demonstrated that phenotypically defined DNMT3A-mutated HSC were functional pre-LSC endowed with a competitive repopulation advantage.29 These data also showed that mutations in healthy HSC or at least multi-potent progenitors can serve as the cell-of-origin for myeloid leukemias in humans.
Thus, the accumulation of mutated mature blood cells arising from CH clones/pre-LSC can have an impact on atherosclerosis via monocytes/macrophages but also contribute to clonal expansion and in some rare cases give rise to frank leukemia.37 Within individuals with CH, those with a high risk of developing AML can be identified in predictive models, thereby distinguishing between benign CH and the pre-leukemic state.41
In contrast, other leukemias are considered to be related to specific translocations that can be detected in these cases. AML with chromosomal rearrangements inv(16)(p13q22) or t(16;16)(p13;q22) – collectively referred to as inv(16) – and t(8;21)(q22;q22) are classified as core-binding factor (CBF) leukemias and result in the oncogenic fusion proteins CBFB-MYH11 and RUNX1-RUNX1T1 (AML1/ETO), respectively.42 Unique translocations are likely not sufficient to drive leukemogenesis alone and additional mutational events are needed for leukemic evolution and relapse.42-44 However, in those AML cases, pre-LSC/LSC might not evolve from a pre-existent CH clone but structural variations may spontaneously occur in stem and progenitor cells or after exposure to genotoxic agents including chemotherapy.
The mechanisms of clonal fitness in CH constitute a field of highly competitive research and there is evidence that a pro-inflammatory phenotype contributes to cardiovascular disease. This suggests that clonal expansion is also strengthened through inflammatory pathways; although this needs to be explored further, recent data demonstrate that an enhanced inflammatory response in TET2-mutated mice correlates with progression of myeloid neoplasms.45,46
Leukemic stem cell vulnerabilities and mechanisms driving drug-resistance
Chemotherapy resistance
LSC are considered to harbor inherent resistance to anti-proliferative therapies. This is linked to their capacity to acquire transient quiescence, dormancy and senescence states and thought to be mediated by several mechanisms including resistance to DNA damage.47-49 Furthermore, it has been suggested that DNMT3A mutations in pre-LSC drive AML chemoresistance.50,51 However, this view was challenged by a recent study demonstrating in a patient-derived xenograft model that resistant AML cells were neither enriched in immature quiescent cells nor in LSC after treatment with cytarabine, thereby showing that cytarabine similarly depleted quiescent G0 AML cells and proliferating cells (blasts).52 Another study revealed a unique and transient molecular state of leukemia-regenerating cells responsible for re-outgrowth of leukemia distinct from therapy-naïve LSC.53 Furthermore, recent research identified a senescence-like resilience phenotype conferred with superior engraftment potential through which AML cells can survive and repopulate leukemia.49 The authors demonstrated that this transient phenotype of AML cells occurs regardless of their stem cell status and that these cells give rise to relapsed AML with increased stem cell potential.49 Together, these data show that the LSC landscape is shaped by chemotherapy, indicating transient LSC stages with dynamic therapy resistance properties during the course of AML therapy. Hence, targeting distinct LSC states is difficult as they are subject to plasticity, likely patient-specific, and there might be specific situations in which LSC undergo phenotypic plasticity, which may also affect cell surface marker expression. These stages are expected to be dynamic, transient and likely reversible. Thus, targeting LSC by surface markers would still have an impact on eliminating the LSC clone.
Table 1.
Selection of clinical trials, registered with clinicaltrials.gov, which include strategies targeting leukemic stem cell vulnerabilities in patients with newly diagnosed or relapsed/refractory acute myeloid leukemia.
MYC is an essential transcription factor regulating metabolic properties including the balance between dormancy and proliferation of stem cells comprising HSC.11,54,55 Recently it has been shown that a distantly located MYC enhancer cluster (BENC) controls these properties and its activity in human LSC is linked to chemosensitivity.56
The chemotherapy-resistant LSC phenotype is shaped by different mechanisms including recurrent genotypes, epigenetic modifications and resulting gene expression programs and also the metabolic state, which are not mutually exclusive. Efforts to identify differentially expressed surface markers distinguishing LSC and HSC in AML patients have spawned specific surface markers enriched in the LSC compartment, including CD34,21 CD123,57 CLEC12A (CLL-1),58 GPR56,59 CD44,60 CD47,61 and CD96.62
HSC reside in a highly specialized bone marrow environment referred to as a niche. In these hypoxic niches HIF1α regulates quiescence by HIF1α-dependent gene expression including CXCR4, which is also upregulated on the membrane of LSC.63 There is evidence that LSC within their niches may be protected from chemotherapy.48 The area of niche-related potential therapeutic targets and LSC niche-mediated drug resistance mechanisms is not discussed here and is reviewed elsewhere.64
Leukemic stem cell gene signatures and therapeutic targets
Differential gene expression analyses identified altered gene expression programs in LSC that predict clinical parameters including overall survival.65,66 These programs are regulated by the chromatin state (accessibility for transcription factors), epigenetic mechanisms contributing to transcriptional output and LSC plasticity via activation or repression of gene expression.
Gene expression analysis of functionally defined LSC revealed that these cells harbor a transcriptional profile related to HSC and that stemness-related gene expression programs are highly predictive of response to standard AML therapy.65,66 A subset of genes within this transcriptional program of stemness (17-gene signature) yielded a LSC17 score that can serve as a predictor of clinical parameters.66 Another study proposed an RNA-sequencing-based risk stratification model capable of recovering all relevant chromosomal translocations and inversions.67
A recent study investigating HSC-derived AML marked by high expression of the oncogenic transcription factor EVI1 showed that p53 protein expression is influenced in an EVI1-dependent manner.68 The authors demonstrated that the cell-of-origin of leukemia initiation dictates therapeutic sensitivity to inhibitors of LSD1, a histone demethylase implicated in DNA damage responses and in p53 pathways, and that drug resistance could be overcome in HSC-derived leukemias by combining LSD1 inhibition with venetoclax.68
Bromodomain and extra-terminal motif (BET) proteins that modify MYC expression and Brd4, a BET family protein, represent another potential new target for AML therapy. However, BET inhibitor resistance emerges from LSC, is related to transcriptional plasticity and a role for the Wnt pathway has been described.69,70 Interestingly, LSD1 inhibition re-sensitizes AML cells that are resistant to BET inhibition.71
Approaches using preclinical models, including drugs targeting the epigenetic and metabolic state or a specific immunophenotype, exhibit the potential to eradicate relapse-relevant LSC. For example, it was shown that inhibition of miR-126, a microRNA controlling the PI3K-Akt-mTOR pathway, attenuates LSC activity.72 Furthermore, a recent study developed a combinatorial approach linking the LSC concept to immune evasion.73 AML cells that express natural killer group 2D ligands (NKG2DL) are cleared by natural killer (NK) cells, whereas NKG2DL-negative LSC escape killing by NK cells (Figure 2). Poly-ADP-ribose polymerase 1 (PARP1) is an enzyme involved in several cellular processes, such as DNA repair and gene regulation, which uses NAD+ to transfer ADP-ribose to other proteins. PARP1 represses NKG2DL expression and pharmacological inhibition of PARP1 (by talazoparib) induces NKG2DL re-expression on the LSC surface, rendering these cells amenable to NK cell control in vivo.73 This concept is being translated into the latest clinical research; upcoming trials will have to prove the clinical efficacy of PARP1 inhibition with subsequent transfer of alloreactive NK cells (clinicaltrials.gov identifier NCT05319249) (Table 1).
Targeting the metabolic state
While HSC adapt their metabolic program depending on their state of activation, LSC are considered to be rather metabolically inflexible, uniquely reliant on mitochondrial oxidative phosphorylation (OXPHOS) for ATP production despite the necessity to retain low levels of reactive oxygen species.74,7 5 Due to the decreased glycolytic activity of LSC, one of the three metabolic fuels for the mitochondrial tricarboxylic acid cycle is unavailable and LSC must rely on amino acids and/or fatty acids to fuel OXPHOS.75,76 By contrast, quiescent HSC rely on anaerobic glycolysis metabolizing pyruvate to lactate in their low-oxygen environment (hypoxic niche) reserving OXPHOS to meet increased energy requirements during expansion and differentiation.75,77 This unique reliance has drawn attention to the pharmacological inhibition of OXPHOS in LSC.74 ,76 ,78 ,7 9
Figure 2.
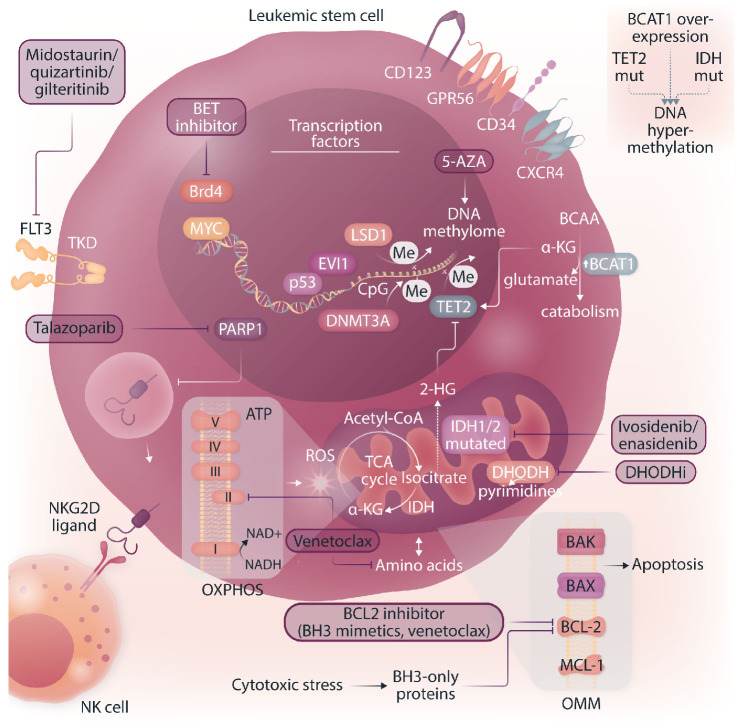
Leukemic stem cell vulnerabilities and targeted therapeutic approaches. The figure illustrates a leukemic stem cell (LSC) and highlights phenotypic characteristics, vulnerabilities and potential therapeutic approaches. LSC are considered metabolically inflexible and uniquely reliant on amino acids and fatty acids to fuel oxidative phosphorylation. BCL-2 and MCL-1 are anti-apoptotic members of the BCL-2 family residing in the outer mitochondrial membrane (OMM). BAX and BAK are pore-forming proteins and the BH3-only proteins are pro-apoptotic. All BCL-2 family proteins interact to maintain the integrity of the OMM. Upon cellular stress, the cell is committed to induce apoptosis via upregulation of the pro-apoptotic BH3-only proteins and downregulation of BCL-2/MCL-1. A shift in the BCL-2 family interactome releases the effector proteins BAX/BAK and promotes homo-oligomerization to form cytotoxic pores in the OMM.80 Hence, BCL-2 inhibitors (called BH3-mimetics) induce apoptosis. Dihydroorotate dehydrogenase localized at the outer layer of the inner mitochondrial membrane is crucial for de novo pyrimidine synthesis thereby providing substrates for nucleic acid synthesis. DNMT3A and TET2 have opposite effects on DNA methylation. DNMT3A catalyzes de novo methylation of cytosine residues (CpG dinucleotide), while TET2 catalyzes the conversion of 5-methylcytosine to 5-hydroxymethylcytosine, the initial step of DNA demethylation. FLT3: FMS-like tyrosine kinase 3; TKD: tyrosine kinase domain; Brd4: member of the BET (bromodomain and extra-terminal motif) family; MYC/p53/EVI1: transcription factors; LSD1: lysine-specific histone demethylase; PARP1: poly-ADP-ribose polymerase 1, me: methylation; DNMT3A: DNA methyltransferase 3A; TET2: tet methylcytosine dioxygenase 2; BCAA: branched-chain amino acids, BCAT1: BCAA transaminase 1; α-KG: alpha-ketoglutarate; 2-HG: 2-hydroxyglutarate; ROS: reactive oxygen species; OXPHOS: oxidative phosphorylation; TCA: tricarboxylic acid cycle; DHODHi: dihydroorotate dehydrogenase inhibitors; 5-AZA: 5-azacitidine; NK cell: natural killer cell; NKG2D ligand: natural killer group 2D ligand; IDH: isocitrate dehydrogenase.
BCL-2 and MCL-1 are anti-apoptotic members of the BCL-2 family present in the outer mitochondrial membrane. Upon cellular stress, the cell is commited to induce apoptosis via upregulation of the pro-apoptotic BH3-only proteins and downregulation of BCL-2/MCL-1. This results in a release of the BAX and BAK proteins, forming cytotoxic pores in the outer mitochondrial membrane.80 De novo LSC seem to be dependent on OXPHOS fueled by amino acids and thus rely on amino acid metabolism to provide substrates for the tricarboxylic acid cycle.76 The combination of BCL-2 inhibition (by venetoclax) and the hypomethylating agent azacitidine significantly decreases OXPHOS through amino acid depletion and ETC complex II inhibition thereby selectively targeting LSC.78,80 Interestingly, in relapsed/refractory AML LSC exhibit metabolic plasticity allowing for compensation via upregulation of fatty acid metabolism, becoming resistant to BCL-2 inhibition, and can be re-sensitized to azacitidine/venetoclax by targeting fatty acid transport.76,81 The β-oxidation of fatty acids results in acetyl-CoA producing NADH and FADH2 and fueling OXPHOS to generate ATP. The targeting of fatty acid oxidation and thereby its role in fueling OXPHOS is an exciting new direction in overcoming LSC-mediated therapy resistance in AML.75
In phenotypically monocytic AML, resistance has been mechanistically linked to a distinct transcriptomic profile and a physiological switch from BCL-2 to an MCL-1-mediated pro-survival program. This leads to a loss of BCL-2 expression and dependency and thus mediates insensitivity of such monocytic blasts to venetoclax; it remains unclear whether LSC from these more differentiated AML also behave similarly.82 MCL-1 inhibitors are currently in clinical evaluation and combination therapies could be an efficient approach with side effects that remain manageable.80,82 Finally, recent data indicate that acquired BAX mutations represent another mechanism of adaptive resistance to venetoclax-based AML therapy.83
Maintenance of low levels of reactive oxygen species as well as mitochondrial function are required for LSC stemness.75 LSC use different mechanisms to avoid oxidative stress and maintain low reactive oxygen species levels, including juxtaposition to hypoxic niches,84,85 activation of FOXO transcription factors,86 generation of more glutathione and the removal of damaged mitochondria via mitophagy.87 These highly reactive byproducts of aerobic metabolism contribute to stem cell aging, force cells out of quiescence and compromise their ability to maintain the LSC population.75
Another role in the interplay of LSC metabolic function and therapeutic resistance has emerged for branched-chain amino acids (BCAA) produced by BCAA transaminase 1 (BCAT1). BCAT1 is overexpressed in a subset of LSC resulting in a survival advantage by depleting α-ketoglutarate, a critical co-factor for TET2, thus mimicking the effects of IDH and TET2 mutations.88 Since amino acid metabolism is crucial for ATP production in LSC, BCAA metabolism constitutes a potential pharmacological target to compromise LSC function selectively (Figure 2).
Dihydroorotate dehydrogenase (DHODH) is an enzyme localized in the inner mitochondrial membrane which catalyzes the fourth step of de novo pyrimidine synthesis. The inhibition of DHODH reduced leukemic burden and decreased levels of leukemia-initiating cells highlighting that pyrimidine synthesis constitutes another metabolic vulnerability.89 Blunting glutamine metabolism and pyrimidine synthesis has been shown to inhibit residual leukemiainitiating cells and such treatment schemes improved survival in leukemia mouse models and patient-derived xenografts.90 Recent data show that the novel DHODH inhibitor AG636 leads to inhibition of the protein translation machinery and confirm that LSC are dependent on de novo pyrimidine synthesis.91 Interestingly, by performing a CRISPR-Cas9 knockout screen using a focused library of epigenetic regulators, CDK5 was identified as a sensitizer to DHODH inhibition, thereby raising the possibility of simultaneously targeting different mitochondrial processes.
Resistance to targeted therapies for acute myeloid leukemia
While venetoclax targets a distinct metabolic state, other targeted therapeutic approaches inhibit specific oncogenic proteins such as mutant FLT3 or IDH1/2.75 RAS mutations are common mechanisms of resistance to FLT3- and IDH-inhibitors and also to BCL-2-inhibitor-based therapies.2 One of the most commonly mutated genes in AML is FLT3, which encodes a receptor tyrosine kinase. The most common type of FLT3 mutation is an internal tandem duplication (FLT3-ITD), consisting of an in-frame amino acid insertion in the juxtamembrane domain of the receptor, which results in constitutive kinase activity.92 While there is also a role for FLT3-tyrosine kinase domain (FLT3-TKD) mutations, in particular, are associated with increased risk of relapse and inferior survival which is influenced by both co-mutations and the ratio of FLT3-ITD to wildtype FLT3 alleles.2,93 Three FLT3 inhibitors (midostaurin, quizartinib and gilteritinib) have been demonstrated to improve overall survival compared with conventional chemotherapy (for gilteritinib, 9.3 months vs. 5.6 months) in randomized phase III trials.94-96 However, secondary mutations of the FLT3 gene frequently lead to therapy resistance.2 Next-generation sequencing studies using primary cells from AML patients have established that FLT3-ITD mutations occur relatively late in leukemogenesis.97 In contrast, competitive transplantation experiments in mice indicated that the mutated FLT3 is expressed on HSC.98 However, another study using single-cell mRNA-sequencing found essentially the opposite.92,99 A recent study shed new light on the subclonal architecture of FLT3-ITD-mutant AML providing evidence that FLT3-ITD mutations may also occur early in leukemic precursor cells and that CD99 may serve as a therapeutic target.100 Furthermore, the combination of single-cell RNA-sequencing and genotyping from bone marrow samples of 16 AML patients demonstrated that FLT3-ITD-mutated cells were enriched in the cell populations with undifferentiated HSC/progenitor-like cell signatures, suggesting that FLT3-ITD confers a strong differentiation block.101 The expression of FLT3-ITD in the MUTZ-3AML cell line and examination of resultant cellular phenotypes by flow cytometry demonstrated that FLT3 expression increased the percent of primitive CD34+ MUTZ-3 cells and that this effect was most pronounced with the FLT3-ITD construct.101 These results help to understand how FLT3-ITD mutations may be associated with HSC and progenitor-like cells. Although the role of FLT3 inhibitors in the clinic is emerging, with demonstration of improved outcomes, their effect on eliminating LSC remains enigmatic.
IDH1 and IDH2 catalyze the oxidative decarboxylation of isocitrate to produce α-ketoglutarate (Figure 2). Mutant IDH1/2 acquire neomorphic catalytic activity and produce 2-hydroxyglutarate,102 which competitively inhibits α-ketoglutarate-dependent enzymes such as TET2.103 TET2 is an epigenetic regulator mediating active DNA demethylation.39 Consequently, IDH1/2 and TET2 mutations result in a state of genomic hypermethylation.104 There is a strong rationale for combining IDH1/2 inhibitors with hypomethylating agents and also for the combination with BCL-2 inhibitors (venetoclax), as the accumulation of 2-hydroxyglutarate caused by IDH1/2 mutations mimics an oxygen-depriving state, thereby decreasing the mitochondrial threshold for induction of apoptosis.105 Oral inhibitors of both mutant IDH1 (ivosidenib) and IDH2 (enasidenib) have shown efficacy in patients with the corresponding mutations.106,107 The combination of ivosidenib + azacitidine has shown superiority compared to azacitidine alone in patients with newly diagnosed IDH1-mutated AML, who were ineligible for intensive induction chemotherapy (clinicaltrials.gov identifier NCT03173248) (Table 1)108 and the combination of ivosidenib + venetoclax in IDH-mutated patients is currently being tested in a clinical trial (clinicaltrials.gov identifier NCT03471260) (Table 1). In a recent study, genomic analyses of longitudinally collected AML samples indicated that stemness is a major driver of primary IDH inhibitor resistance.109 Since IDH inhibitors induce differentiation of leukemic blasts, this seems mechanistically plausible. However, the mechanisms driving stemness in IDH-mutant AML and the role of LSC in this regard remains poorly understood.
Methodological improvements and their significance for translational research in acute myeloid leukemia
Although LSC remain difficult to isolate because of their scarcity, their pronounced similarity to healthy HSC and their phenotypic plasticity, novel technologies now allow the identification of complex heterogeneous cell mixtures at single-cell resolution. Moreover, more sophisticated multi-omics single-cell approaches are now available to capture surface proteins next to the transcriptomes (CITEseq; cellular indexing of transcriptomes and epitopes by sequencing),110,111 chromatin accessibility (ATAC-seq; assay for transposase-accessible chromatin with sequencing) and importantly can also integrate mutational profiling (single nucleotide variations and structural variants)112 and/or tracking of clonal dynamics based on mitochondrial marker mutations (TARGET-seq,113 GoT [genotyping of transcriptomes]114 and MutaSeq115). Although it is becoming increasingly evident that dynamic changes in metabolism play critical roles in LSC function and treatment resistance, approaches based on mass spectrometry of bulk samples and metabolic flux analysis both require large numbers of cells. These are often not available from patients, in particular if smaller subpopulations such as LSC need to be analyzed and thus the development of better, high-resolution, single-cell technologies is much wanted in this area. Further technical advances, each with its inherent merits and limitations, will pave the way towards a more comprehensive understanding of clonal dynamics and the distinct (transient) single-cell states responding to AML therapy driving the continued AML evolution. The integration of single-cell genotyping adds an additional layer of information, thus allowing the capture of even rare clones and the comparison of the networks active in various (pre-)leukemic subclones and wildtype cells within the same patient. This technical progress also offers new opportunities to analyze rare CH clones in the pre-leukemic state and to capture and characterize residual, therapy-resilient relapse-initiating leukemia cells including LSC present in patients’ MRD.
Conclusion
AML is a highly heterogeneous disease characterized by a complex network of genetically distinct subclones arising in a branching evolution alongside the predominant clone. The cells within each genetically identical subclone show their own clone-specific molecular features and develop a specific non-genetically driven hierarchy of cellular differentiation. There is overwhelming evidence that cancer stem cells and stemness properties are clinically relevant, in particular for AML. In this review we have discussed that overcoming therapy resistance in AML requires not only eradication of bulk tumor cells but also the capture and efficient targeting of therapy-resistant leukemia cells, including LSC. The presence of genetically diverse LSC at diagnosis highlights a major limitation of therapies that target only the specific properties of the dominant clone. The identification of specific patterns of AML relapse demonstrated that these will require different therapies given their distinct stem cell biology.28 Since LSC harbor inherent resistance mechanisms including phenotypic plasticity, dormancy and senescence, conventional chemotherapy is increasingly being added to or replaced by targeted therapeutic strategies to act specifically on LSC properties. Given the chemotherapy resistance of LSC, targeted strategies need to be integrated into first-line regimens to prevent LSC-mediated AML relapse. Venetoclax + azacitidine is a promising approach which is currently reserved for relapsed/refractory patients and newly diagnosed patients of older age or with comorbidities. This combination targets at least some LSC, but the molecular basis of treatment refractoriness and resistance still needs to be better explored and overcome in this setting as well. Nevertheless, BH3 mimetics are among the most promising strategies to treat AML, including LSC. Importantly, with the success of venetoclax + azacitidine and availability of this combination for first-line therapy, the selection of patients who would benefit from either standard chemotherapy or upfront venetoclax + azacitidine treatment is a challenge. Biomarkers need to be developed to stratify patients and clinical trials to monitor LSC-targeting efficacy in AML first-line regimens need to be implemented and used to study, understand and overcome LSC-mediated therapy resistance. Furthermore, new methods of disease monitoring have to be established to track LSC subclones and improve future clinical trials. Despite impressive rates of response to venetoclax + azacitidine, the combination is not curative since LSC exhibit molecular and metabolic plasticity becoming resistant to BCL-2 inhibition (i.e. high expression of MCL-1 or BCL-xL). Importantly, while venetoclax + azacitidine efficiently targets at least some LSC, it may not target all of them in both intra- and inter-patient settings, and the surviving LSC are the drivers of relapse. Thus, many different clinical trials investigating combinatorial therapeutic approaches to target LSC vulnerabilities and thereby attempt to eradicate relapse-initiating cells through different mechanisms are currently being explored in clinical settings (Table 1).
The era of single-cell multi-omics provides unprecedented opportunities to characterize relapse-initiating cell populations and allows tracking of individual clonal architectures and underlying biological networks. These technical innovations may offer new ways to trace the drivers of relapse, including LSC, within upcoming clinical trials and to identify and target therapy-resistant cells with resilience phenotypes that repopulate leukemia. Many of the cited methods have recently been applied to address basic and translational research questions and offered novel, critical insights into AML biology. However, it seems very difficult that these can be included in clinical routine diagnostics, at least at present, as they are not easily scalable. Nevertheless, these methods will be crucial to characterize the few relapse-initiating cells and to translate the understanding of LSC biology into novel therapeutic strategies for AML therapy. Results obtained from these newer, low-throughput and expensive technologies need to be translated into scalable tools that can, after clinical validation, be implemented in routine clinical practice. Such tools need to fulfill clinical standards regarding specificity, feasibility and cost-efficiency and have to be validated in larger cohorts of patients. Overall, there is increasing evidence for patient-specific approaches that address individual therapeutic vulnerabilities. An inevitable strategy to prevent AML recurrence and improve clinical outcome in the future is the integration of LSC-targeting agents into first-line treatments which may lead to a decrease in relapse frequency and an increase of cure rates of AML patients.
Funding Statement
Funding: PS is funded by a fellowship of the DKFZ Clinician Scientist Program, supported by the Dieter Morszeck Foundation.
References
- 1.Papaemmanuil E, Gerstung M, Bullinger L, et al. Genomic classification and prognosis in acute myeloid leukemia. N Engl J Med. 2016;374(23):2209-2221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Short NJ, Konopleva M, Kadia TM, et al. Advances in the treatment of acute myeloid leukemia: new drugs and new challenges. Cancer Discov. 2020;10(4):506-525. [DOI] [PubMed] [Google Scholar]
- 3.Dohner H, Estey E, Grimwade D, et al. Diagnosis and management of AML in adults: 2017 ELN recommendations from an international expert panel. Blood. 2017;129(4):424-447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jongen-Lavrencic M, Grob T, Hanekamp D, et al. Molecular minimal residual disease in acute myeloid leukemia. N Engl J Med. 2018;378(13):1189-1199. [DOI] [PubMed] [Google Scholar]
- 5.Trumpp A, Haas S. Cancer stem cells: the adventurous journey from hematopoietic to leukemic stem cells. Cell. 2022;185(8):1266-1270. [DOI] [PubMed] [Google Scholar]
- 6.DiNardo CD, Jonas BA, Pullarkat V, et al. Azacitidine and venetoclax in previously untreated acute myeloid leukemia. N Engl J Med. 2020;383(7):617-629. [DOI] [PubMed] [Google Scholar]
- 7.Stahl M, Menghrajani K, Derkach A, et al. Clinical and molecular predictors of response and survival following venetoclax therapy in relapsed/refractory AML. Blood Adv. 2021;5(5):1552-1564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Haas S, Trumpp A, Milsom MD. Causes and consequences of hematopoietic stem cell heterogeneity. Cell Stem Cell. 2018;22(5):627-638. [DOI] [PubMed] [Google Scholar]
- 9.Laurenti E, Gottgens B. From haematopoietic stem cells to complex differentiation landscapes. Nature. 2018;553(7689):418-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wilson A, Laurenti E, Oser G, et al. Hematopoietic stem cells reversibly switch from dormancy to self-renewal during homeostasis and repair. Cell. 2008;135(6):1118-1129. [DOI] [PubMed] [Google Scholar]
- 11.Cabezas-Wallscheid N, Buettner F, Sommerkamp P, et al. Vitamin A-retinoic acid signaling regulates hematopoietic stem cell dormancy. Cell. 2017;169(5):807-823. [DOI] [PubMed] [Google Scholar]
- 12.Yamamoto R, Morita Y, Ooehara J, et al. Clonal analysis unveils self-renewing lineage-restricted progenitors generated directly from hematopoietic stem cells. Cell. 2013;154(5):1112-1126. [DOI] [PubMed] [Google Scholar]
- 13.Notta F, Zandi S, Takayama N, et al. Distinct routes of lineage development reshape the human blood hierarchy across ontogeny. Science. 2016;351(6269):aab2116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pinho S, Frenette PS. Haematopoietic stem cell activity and interactions with the niche. Nat Rev Mol Cell Biol. 2019;20(5):303-320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schofield R. The relationship between the spleen colony-forming cell and the haemopoietic stem cell. Blood Cells. 1978;4(1-2):7-25. [PubMed] [Google Scholar]
- 16.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14(3):275-291. [DOI] [PubMed] [Google Scholar]
- 17.Vogelstein B, Fearon ER, Hamilton SR, Feinberg AP. Use of restriction fragment length polymorphisms to determine the clonal origin of human tumors. Science. 1985;227(4687):642-645. [DOI] [PubMed] [Google Scholar]
- 18.Hanahan D, Coussens LM. Accessories to the crime: functions of cells recruited to the tumor microenvironment. Cancer Cell. 2012;21(3):309-322. [DOI] [PubMed] [Google Scholar]
- 19.Doulatov S, Notta F, Laurenti E, Dick JE. Hematopoiesis: a human perspective. Cell Stem Cell. 2012;10(2):120-136. [DOI] [PubMed] [Google Scholar]
- 20.Lapidot T, Pflumio F, Doedens M, Murdoch B, Williams DE, Dick JE. Cytokine stimulation of multilineage hematopoiesis from immature human cells engrafted in SCID mice. Science. 1992;255(5048):1137-1141. [DOI] [PubMed] [Google Scholar]
- 21.Lapidot T, Sirard C, Vormoor J, et al. A cell initiating human acute myeloid leukaemia after transplantation into SCID mice. Nature. 1994;367(6464):645-648. [DOI] [PubMed] [Google Scholar]
- 22.Kamel-Reid S, Dick JE. Engraftment of immune-deficient mice with human hematopoietic stem cells. Science. 1988;242(4886):1706-1709. [DOI] [PubMed] [Google Scholar]
- 23.Bonnet D, Dick JE. Human acute myeloid leukemia is organized as a hierarchy that originates from a primitive hematopoietic cell. Nat Med. 1997;3(7):730-737. [DOI] [PubMed] [Google Scholar]
- 24.Bahr C, Correia NC, Trumpp A. Stem cells make leukemia grow again. EMBO J. 2017;36(18):2667-2669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hope KJ, Jin L, Dick JE. Acute myeloid leukemia originates from a hierarchy of leukemic stem cell classes that differ in self-renewal capacity. Nat Immunol. 2004;5(7):738-743. [DOI] [PubMed] [Google Scholar]
- 26.Ding L, Ley TJ, Larson DE, et al. Clonal evolution in relapsed acute myeloid leukaemia revealed by whole-genome sequencing. Nature. 2012;481(7382):506-510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Parkin B, Ouillette P, Li Y, et al. Clonal evolution and devolution after chemotherapy in adult acute myelogenous leukemia. Blood. 2013;121(2):369-377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shlush LI, Mitchell A, Heisler L, et al. Tracing the origins of relapse in acute myeloid leukaemia to stem cells. Nature. 2017;547(7661):104-108. [DOI] [PubMed] [Google Scholar]
- 29.Shlush LI, Zandi S, Mitchell A, et al. Identification of pre-leukaemic haematopoietic stem cells in acute leukaemia. Nature. 2014;506(7488):328-333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Steensma DP, Bejar R, Jaiswal S, et al. Clonal hematopoiesis of indeterminate potential and its distinction from myelodysplastic syndromes. Blood. 2015;126(1):9-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Genovese G, Kahler AK, Handsaker RE, et al. Clonal hematopoiesis and blood-cancer risk inferred from blood DNA sequence. N Engl J Med. 2014;371(26):2477-2487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaiswal S, Fontanillas P, Flannick J, et al. Age-related clonal hematopoiesis associated with adverse outcomes. N Engl J Med. 2014;371(26):2488-2498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jaiswal S, Natarajan P, Silver AJ, et al. Clonal hematopoiesis and risk of atherosclerotic cardiovascular disease. N Engl J Med. 2017;377(2):111-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fuster JJ, MacLauchlan S, Zuriaga MA, et al. Clonal hematopoiesis associated with TET2 deficiency accelerates atherosclerosis development in mice. Science. 2017;355(6327):842-847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cai Z, Kotzin JJ, Ramdas B, et al. Inhibition of inflammatory signaling in Tet2 mutant preleukemic cells mitigates stress-induced abnormalities and clonal hematopoiesis. Cell Stem Cell. 2018;23(6):833-849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang YH, Chen CW, Sundaramurthy V, et al. Systematic profiling of DNMT3A variants reveals protein instability mediated by the DCAF8 E3 ubiquitin ligase adaptor. Cancer Discov. 2022;12(1):220-235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Challen GA, Goodell MA. Clonal hematopoiesis: mechanisms driving dominance of stem cell clones. Blood. 2020;136(14):1590-1598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang L, Rau R, Goodell MA. DNMT3A in haematological malignancies. Nat Rev Cancer. 2015;15(3):152-165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rasmussen KD, Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016;30(7):733-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowman RL, Busque L, Levine RL. Clonal hematopoiesis and evolution to hematopoietic malignancies. Cell Stem Cell. 2018;22(2):157-170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Abelson S, Collord G, Ng SWK, et al. Prediction of acute myeloid leukaemia risk in healthy individuals. Nature. 2018;559(7714):400-404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sood R, Hansen NF, Donovan FX, et al. Somatic mutational landscape of AML with inv(16) or t(8;21) identifies patterns of clonal evolution in relapse leukemia. Leukemia. 2016;30(2):501-504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Borthakur G, Kantarjian H. Core binding factor acute myelogenous leukemia-2021 treatment algorithm. Blood Cancer J. 2021;11(6):114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Faber ZJ, Chen X, Gedman AL, et al. The genomic landscape of core-binding factor acute myeloid leukemias. Nat Genet. 2016;48(12):1551-1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jaiswal S, Ebert BL. Clonal hematopoiesis in human aging and disease. Science. 2019;366(6465):eaan4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yeaton A, Cayanan G, Loghavi S, et al. The impact of inflammation-induced tumor plasticity during myeloid transformation. Cancer Discov. 2022;12(10):2392-2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walter D, Lier A, Geiselhart A, et al. Exit from dormancy provokes DNA-damage-induced attrition in haematopoietic stem cells. Nature. 2015;520(7548):549-552. [DOI] [PubMed] [Google Scholar]
- 48.Ishikawa F, Yoshida S, Saito Y, et al. Chemotherapy-resistant human AML stem cells home to and engraft within the bone-marrow endosteal region. Nat Biotechnol. 2007;25(11):1315-1321. [DOI] [PubMed] [Google Scholar]
- 49.Duy C, Li M, Teater M, et al. Chemotherapy induces senescence-like resilient cells capable of initiating AML recurrence. Cancer Discov. 2021;11(6):1542-1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ho TC, LaMere M, Stevens BM, et al. Evolution of acute myelogenous leukemia stem cell properties after treatment and progression. Blood. 2016;128(13):1671-1678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Guryanova OA, Shank K, Spitzer B, et al. DNMT3A mutations promote anthracycline resistance in acute myeloid leukemia via impaired nucleosome remodeling. Nat Med. 2016;22(12):1488-1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Farge T, Saland E, de Toni F, et al. Chemotherapy-resistant human acute myeloid leukemia cells are not enriched for leukemic stem cells but require oxidative metabolism. Cancer Discov. 2017;7(7):716-735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boyd AL, Aslostovar L, Reid J, et al. Identification of chemotherapy-induced leukemic-regenerating cells reveals a transient vulnerability of human AML recurrence. Cancer Cell. 2018;34(3):483-498. [DOI] [PubMed] [Google Scholar]
- 54.Wilson A, Murphy MJ, Oskarsson T, et al. c-Myc controls the balance between hematopoietic stem cell self-renewal and differentiation. Genes Dev. 2004;18(22):2747-2763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scognamiglio R, Cabezas-Wallscheid N, Thier MC, et al. Myc depletion induces a pluripotent dormant state mimicking diapause. Cell. 2016;164(4):668-680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bahr C, von Paleske L, Uslu VV, et al. A Myc enhancer cluster regulates normal and leukaemic haematopoietic stem cell hierarchies. Nature. 2018;553(7689):515-520. [DOI] [PubMed] [Google Scholar]
- 57.Jordan CT, Upchurch D, Szilvassy SJ, et al. The interleukin-3 receptor alpha chain is a unique marker for human acute myelogenous leukemia stem cells. Leukemia. 2000;14(10):1777-1784. [DOI] [PubMed] [Google Scholar]
- 58.van Rhenen A, van Dongen GA, Kelder A, et al. The novel AML stem cell associated antigen CLL-1 aids in discrimination between normal and leukemic stem cells. Blood. 2007;110(7):2659-2666. [DOI] [PubMed] [Google Scholar]
- 59.Pabst C, Bergeron A, Lavallee VP, et al. GPR56 identifies primary human acute myeloid leukemia cells with high repopulating potential in vivo. Blood. 2016;127(16):2018-2027. [DOI] [PubMed] [Google Scholar]
- 60.Jin L, Hope KJ, Zhai Q, Smadja-Joffe F, Dick JE. Targeting of CD44 eradicates human acute myeloid leukemic stem cells. Nat Med. 2006;12(10):1167-1174. [DOI] [PubMed] [Google Scholar]
- 61.Majeti R, Chao MP, Alizadeh AA, et al. CD47 is an adverse prognostic factor and therapeutic antibody target on human acute myeloid leukemia stem cells. Cell. 2009;138(2):286-299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Hosen N, Park CY, Tatsumi N, et al. CD96 is a leukemic stem cell-specific marker in human acute myeloid leukemia. Proc Natl Acad Sci U S A. 2007;104(26):11008-11013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mohle R, Bautz F, Rafii S, Moore MA, Brugger W, Kanz L. The chemokine receptor CXCR-4 is expressed on CD34+ hematopoietic progenitors and leukemic cells and mediates transendothelial migration induced by stromal cell-derived factor-1. Blood. 1998;91(12):4523-4530. [PubMed] [Google Scholar]
- 64.Schepers K, Campbell TB, Passegue E. Normal and leukemic stem cell niches: insights and therapeutic opportunities. Cell Stem Cell. 2015;16(3):254-267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Eppert K, Takenaka K, Lechman ER, et al. Stem cell gene expression programs influence clinical outcome in human leukemia. Nat Med. 2011;17(9):1086-1093. [DOI] [PubMed] [Google Scholar]
- 66.Ng SW, Mitchell A, Kennedy JA, et al. A 17-gene stemness score for rapid determination of risk in acute leukaemia. Nature. 2016;540(7633):433-437. [DOI] [PubMed] [Google Scholar]
- 67.Docking TR, Parker JDK, Jadersten M, et al. A clinical transcriptome approach to patient stratification and therapy selection in acute myeloid leukemia. Nat Commun. 2021;12(1):2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Cai SF, Chu SH, Goldberg AD, et al. Leukemia cell of origin influences apoptotic priming and sensitivity to LSD1 inhibition. Cancer Discov. 2020;10(10):1500-1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Fong CY, Gilan O, Lam EY, et al. BET inhibitor resistance emerges from leukaemia stem cells. Nature. 2015;525(7570):538-542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Rathert P, Roth M, Neumann T, et al. Transcriptional plasticity promotes primary and acquired resistance to BET inhibition. Nature. 2015;525(7570):543-547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Bell CC, Fennell KA, Chan YC, et al. Targeting enhancer switching overcomes non-genetic drug resistance in acute myeloid leukaemia. Nat Commun. 2019;10(1):2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Lechman ER, Gentner B, Ng SW, et al. miR-126 regulates distinct self-renewal outcomes in normal and malignant hematopoietic stem cells. Cancer Cell. 2016;29(2):214-228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Paczulla AM, Rothfelder K, Raffel S, et al. Absence of NKG2D ligands defines leukaemia stem cells and mediates their immune evasion. Nature. 2019;572(7768):254-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Lagadinou ED, Sach A, Callahan K, et al. BCL-2 inhibition targets oxidative phosphorylation and selectively eradicates quiescent human leukemia stem cells. Cell Stem Cell. 2013;12(3):329-341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Culp-Hill R, D'Alessandro A, Pietras EM. Extinguishing the embers: targeting AML metabolism. Trends Mol Med. 2021;27(4):332-344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Jones CL, Stevens BM, D'Alessandro A, et al. Inhibition of amino acid metabolism selectively targets human leukemia stem cells. Cancer Cell. 2018;34(5):724-740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Suda T, Takubo K, Semenza GL. Metabolic regulation of hematopoietic stem cells in the hypoxic niche. Cell Stem Cell. 2011;9(4):298-310. [DOI] [PubMed] [Google Scholar]
- 78.Pollyea DA, Stevens BM, Jones CL, et al. Venetoclax with azacitidine disrupts energy metabolism and targets leukemia stem cells in patients with acute myeloid leukemia. Nat Med. 2018;24(12):1859-1866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Jones CL, Stevens BM, D'Alessandro A, et al. Cysteine depletion targets leukemia stem cells through inhibition of electron transport complex II. Blood. 2019;134(4):389-394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Widden H, Placzek WJ. The multiple mechanisms of MCL1 in the regulation of cell fate. Commun Biol. 2021;4(1):1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Stevens BM, Jones CL, Pollyea DA, et al. Fatty acid metabolism underlies venetoclax resistance in acute myeloid leukemia stem cells. Nat Cancer. 2020;1(12):1176-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pei S, Pollyea DA, Gustafson A, et al. Monocytic subclones confer resistance to venetoclax-based therapy in patients with acute myeloid leukemia. Cancer Discov. 2020;10(4):536-551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Moujalled DM, Brown FC, Chua CC, et al. Acquired mutations in BAX confer resistance to BH3-mimetic therapy in acute myeloid leukemia. Blood. 2022. Oct 11. [Epub ahead of print] [DOI] [PubMed] [Google Scholar]
- 84.Parmar K, Mauch P, Vergilio JA, Sackstein R, Down JD. Distribution of hematopoietic stem cells in the bone marrow according to regional hypoxia. Proc Natl Acad Sci U S A. 2007;104(13):5431-5436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Mohyeldin A, Garzon-Muvdi T, Quinones-Hinojosa A. Oxygen in stem cell biology: a critical component of the stem cell niche. Cell Stem Cell. 2010;7(2):150-161. [DOI] [PubMed] [Google Scholar]
- 86.Tothova Z, Gilliland DG. FoxO transcription factors and stem cell homeostasis: insights from the hematopoietic system. Cell Stem Cell. 2007;1(2):140-152. [DOI] [PubMed] [Google Scholar]
- 87.Pei S, Minhajuddin M, Adane B, et al. AMPK/FIS1-mediated mitophagy is required for self-renewal of human AML stem cells. Cell Stem Cell. 2018;23(1):86-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Raffel S, Falcone M, Kneisel N, et al. BCAT1 restricts alphaKG levels in AML stem cells leading to IDHmut-like DNA hypermethylation. Nature. 2017;551(7680):384-388. [DOI] [PubMed] [Google Scholar]
- 89.Sykes DB, Kfoury YS, Mercier FE, et al. Inhibition of dihydroorotate dehydrogenase overcomes differentiation blockade in acute myeloid leukemia. Cell. 2016;167(1):171-186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.van Gastel N, Spinelli JB, Sharda A, et al. Induction of a timed metabolic collapse to overcome cancer chemoresistance. Cell Metab. 2020;32(3):391-403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.So J, Lewis AC, Smith LK, et al. Inhibition of pyrimidine biosynthesis targets protein translation in acute myeloid leukemia. EMBO Mol Med. 2022;14(7):e15203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Levis M. FLT3 dancing on the stem cell. J Exp Med. 2017;214(7):1857-1859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schlenk RF, Kayser S, Bullinger L, et al. Differential impact of allelic ratio and insertion site in FLT3-ITD-positive AML with respect to allogeneic transplantation. Blood. 2014;124(23):3441-3449. [DOI] [PubMed] [Google Scholar]
- 94.Stone RM, Mandrekar SJ, Sanford BL, et al. Midostaurin plus chemotherapy for acute myeloid leukemia with a FLT3 mutation. N Engl J Med. 2017;377(5):454-464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Perl AE, Martinelli G, Cortes JE, et al. Gilteritinib or chemotherapy for relapsed or refractory FLT3-mutated AML. N Engl J Med. 2019;381(18):1728-1740. [DOI] [PubMed] [Google Scholar]
- 96.Cortes JE, Khaled S, Martinelli G, et al. Quizartinib versus salvage chemotherapy in relapsed or refractory FLT3-ITD acute myeloid leukaemia (QuANTUM-R): a multicentre, randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2019;20(7):984-997. [DOI] [PubMed] [Google Scholar]
- 97.Welch JS, Ley TJ, Link DC, et al. The origin and evolution of mutations in acute myeloid leukemia. Cell. 2012;150(2):264-278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Chu SH, Heiser D, Li L, et al. FLT3-ITD knockin impairs hematopoietic stem cell quiescence/homeostasis, leading to myeloproliferative neoplasm. Cell Stem Cell. 2012;11(3):346-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Mead AJ, Neo WH, Barkas N, et al. Niche-mediated depletion of the normal hematopoietic stem cell reservoir by Flt3-ITD-induced myeloproliferation. J Exp Med. 2017;214(7):2005-2021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Travaglini S, Angelini DF, Alfonso V, et al. Characterization of FLT3-ITD(mut) acute myeloid leukemia: molecular profiling of leukemic precursor cells. Blood Cancer J. 2020;10(8):85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.van Galen P, Hovestadt V, Wadsworth Ii MH, et al. Single-cell RNA-seq reveals AML hierarchies relevant to disease progression and immunity. Cell. 2019;176(6):1265-1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ward PS, Patel J, Wise DR, et al. The common feature of leukemia-associated IDH1 and IDH2 mutations is a neomorphic enzyme activity converting alpha-ketoglutarate to 2-hydroxyglutarate. Cancer Cell. 2010;17(3):225-234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of alpha-ketoglutarate-dependent dioxygenases. Cancer Cell. 2011;19(1):17-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 mutations result in a hypermethylation phenotype, disrupt TET2 function, and impair hematopoietic differentiation. Cancer Cell. 2010;18(6):553-567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Chan SM, Thomas D, Corces-Zimmerman MR, et al. Isocitrate dehydrogenase 1 and 2 mutations induce BCL-2 dependence in acute myeloid leukemia. Nat Med. 2015;21(2):178-184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.DiNardo CD, Schuh AC, Stein EM, et al. Enasidenib plus azacitidine versus azacitidine alone in patients with newly diagnosed, mutant-IDH2 acute myeloid leukaemia (AG221-AML-005): a single-arm, phase 1b and randomised, phase 2 trial. Lancet Oncol. 2021;22(11):1597-1608. [DOI] [PubMed] [Google Scholar]
- 107.Roboz GJ, DiNardo CD, Stein EM, et al. Ivosidenib induces deep durable remissions in patients with newly diagnosed IDH1-mutant acute myeloid leukemia. Blood. 2020;135(7):463-471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Montesinos P, Recher C, Vives S, et al. Ivosidenib and azacitidine in IDH1-mutated acute myeloid leukemia. N Engl J Med. 2022;386(16):1519-1531. [DOI] [PubMed] [Google Scholar]
- 109.Wang F, Morita K, DiNardo CD, et al. Leukemia stemness and co-occurring mutations drive resistance to IDH inhibitors in acute myeloid leukemia. Nat Commun. 2021;12(1):2607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Stoeckius M, Hafemeister C, Stephenson W, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017;14(9):865-868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Miles LA, Bowman RL, Merlinsky TR, et al. Single-cell mutation analysis of clonal evolution in myeloid malignancies. Nature. 2020;587(7834):477-482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Jeong H, Grimes K, Bruch P-M, et al. Haplotype-aware singlecell multiomics uncovers functional effects of somatic structural variation. bioRxiv. 2021. Nov 13. doi: 10.1101/2021.11.11.468039 [Preprint, not peer-reviewed] [DOI] [Google Scholar]
- 113.Rodriguez-Meira A, Buck G, Clark SA, et al. Unravelling intratumoral heterogeneity through high-sensitivity single-cell mutational analysis and parallel RNA sequencing. Mol Cell. 2019;73(6):1292-1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Nam AS, Kim KT, Chaligne R, et al. Somatic mutations and cell identity linked by genotyping of transcriptomes. Nature. 2019;571(7765):355-360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Velten L, Story BA, Hernandez-Malmierca P, et al. Identification of leukemic and pre-leukemic stem cells by clonal tracking from single-cell transcriptomics. Nat Commun. 2021;12(1):1366. [DOI] [PMC free article] [PubMed] [Google Scholar]