Abstract
Motivation
The Division of Cancer Epidemiology and Genetics (DCEG) and the Division of Cancer Prevention (DCP) at the National Cancer Institute (NCI) have recently generated genome-wide association study (GWAS) data for multiple traits in the Prostate, Lung, Colorectal, and Ovarian (PLCO) Genomic Atlas project. The GWAS included 110 000 participants. The dissemination of the genetic association data through a data portal called GWAS Explorer, in a manner that addresses the modern expectations of FAIR reusability by data scientists and engineers, is the main motivation for the development of the open-source JavaScript software development kit (SDK) reported here.
Results
The PLCO GWAS Explorer resource relies on a public stateless HTTP application programming interface (API) deployed as the sole backend service for both the landing page’s web application and third-party analytical workflows. The core PLCOjs SDK is mapped to each of the API methods, and also to each of the reference graphic visualizations in the GWAS Explorer. A few additional visualization methods extend it. As is the norm with web SDKs, no download or installation is needed and modularization supports targeted code injection for web applications, reactive notebooks (Observable) and node-based web services.
Availability and implementation
code at https://github.com/episphere/plco; project page at https://episphere.github.io/plco
1 Introduction
Modern FAIR principles for stewardship of scientific data (Wilkinson et al., 2016) ultimately aim at using the web as a distributed data space (Heath and Bizer, 2011). Pursuing this goal requires stateless application programming interface (API) ecosystems, the narrow middle of data commons (Grossman, 2018), where each data source maintains cross-domain data exchange methods. PLCOjs is a contribution to the operation of such an (API), of the PLCO Atlas project (exploregwas.cancer.gov/plco-atlas), such that interactive graphical representations and analytical workflows can be assembled without requiring download or installations. This ‘beyond the data deluge’ (Bell et al., 2009) approach takes advantage of the ubiquity of the web stack by injecting code (JavaScript) to act on behalf of a user in a client machine where authentication is resolved independently. This is in contrast with the conventional, less scalable, approach of aggregating raw data in a single backend.
Engaging a client-side software development kit (SDK) has the advantages of reducing the computational needs of the remote data backend by allowing the development of serverless ‘Web APIs’ (Almeida et al., 2019). The scalability of this approach is particularly clear when engaging distributed data resources in real time, as illustrated by SDKs developed to track coronavirus disease 2019 data (Almeida et al., 2021).
As demonstrated in this report, the reliance on portable client-side web SDKs is equally compelling for large complex studies such as genome-wide association studies (GWASs) that generate millions of association statistics of traits with genotypes (Machiela and Chanock, 2018), such as the PLCO Genetic Atlas Project. This project generated summary statistics from GWAS analyses for multiple traits on 110 000 subjects participating in the PLCO Screening Trial (cdas.cancer.gov/plco) (Gohagan et al., 2015). Traits included cancer outcomes and a myriad of anthropometric, biochemical, lifestyle and non-cancer medical factors. The changing dynamics of PLCO data generation and dissemination over 30 years of updates in clinical outcomes (Black et al., 2015) are essentially those of a real-time project benefiting from SDKs abstracting variability in interoperability models. As discussed in Almeida et al. (2019), a JavaScript SDK adds the important feature that the code is programmatically injected where the data access is governed—in the low permission sandbox encapsulating the user’s web browser. Accordingly, the software construct reported here tests a web-enabled operational architecture as a platform for distributed GWAS.
2 Materials and methods
The data portal for the GWAS summary data generated from PLCO, the GWAS Explorer (see also Supplementary Material) was the starting point for the SDK development as a modularization effort. A close inspection of the GWAS Explorer web application reveals that all interactions with the data backend are mediated using the same API reported there: no component of the portal is rendered server-side. Instead, the components are dynamically assembled by the resources of the client machine by injected code acting on behalf of the user. The PLCOjs SDK uses this full decoupling between the data layer and the presentation layer to modularize the latter as a series of methods that mediate (i) the operation of the API/plco.api, including downloads; and (ii) the client-side assembly of interactive interfaces,/plco.plot, including the data wrangling workflows needed to populate them. For example, extending PLCOjs SDK to integrate the operation of other GWAS APIs, such as those of NHGRI-EBI Catalog (MacArthur et al., 2021), can be similarly approached by either adding new methods or generalizing existing ones in a shared SDK.
2.1 Data model and SDK development
Both the data model and a swagger.io-based API sandbox are provided in the PLCO GWAS Explorer landing page under ‘API Access’. The PLCOjs SDK is fully developed in JavaScript (ECMAScript), as an in-browser code injection construct that can be executed with no downloads or installations in any web computing environment such as the web browser and NodeJS services. The underlying libraries are delivered with version control as Github pages. The code base can be explored under ‘docs’ in the Supplementary Information. As its inspection will show, PLOSjs has a single dependency on the popular open-source cross-language graphics library Plotly (plotly.com/javascript): the JSON structures instantiating the graphic representations in JavaScript will generate the same representation by the corresponding command in R, Python, Julia, F# and Matlab (plotly.com/graphing-libraries). For expediency, an in-browser tool (no data transitioning by servers) was developed at episphere.github.io/plot to render graphic representations from such JSON data structures, with the option to locally download the underlying data tables in CSV format.
3 Results and discussion
All graphic representations in the PLCO GWAS Explorer data atlas portal (exploregwas.cancer.gov/plco-atlas) are generated by calling the PLCO APIs directly. Accordingly, the PLCOjs SDK described here is, architecturally, decoupling those graphic components from the interface of the landing page, such that they can be used in different contexts, from reactive notebooks (Perkel, 2021) to web applications in general.
3.1 Libraries and applications
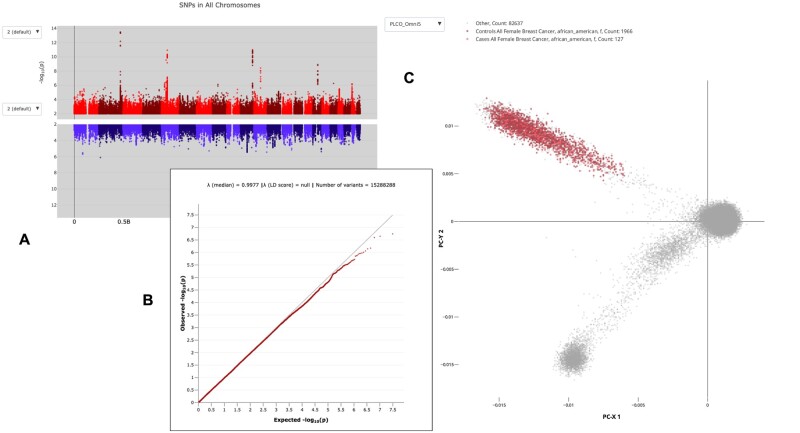
The PLCOjs library (see Availability), and demonstration composite live applications (see Supplementary Information), is assembled by code served from the same GitHub pages, with versioned hosting. This result is illustrated in Figure 1, assembled as a composite of plots independently generated by distinct PLCOjs methods without download/installation.
Fig. 1.
Three examples of graphic representations of PLCO data pulled directly by PLCOjs SDK from this data resource API. The live individual plots, (A) Manhattan plot comparing two populations, (B) quantile–quantile log plot of expected and observed variation frequency, (C) First two principal component representations of case versus control variations for 84 730 individuals. These and other plots can be reproduced, and engaged interactively, in the Observable Notebook Supplementary Material. Plot A also illustrates the portability of calling PLCOjs SDK from within HTML Custom Elements
Conflict of Interest: none declared.
Contributor Information
Eric Ruan, Department of Biomedical Informatics, Stony Brook University, Stony Brook, NY 11794, USA.
Erika Nemeth, Department of Biomedical Informatics, Stony Brook University, Stony Brook, NY 11794, USA.
Richard Moffitt, Department of Biomedical Informatics, Stony Brook University, Stony Brook, NY 11794, USA.
Lorena Sandoval, Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, Rockville, MD 20850, USA.
Mitchell J Machiela, Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, Rockville, MD 20850, USA.
Neal D Freedman, Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, Rockville, MD 20850, USA.
Wen-Yi Huang, Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, Rockville, MD 20850, USA.
Wendy Wong, Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, Rockville, MD 20850, USA.
Kai-Ling Chen, Center for Biomedical Informatics and Information Technology (CBIIT), National Cancer Institute, Rockville, MD 20850, USA.
Brian Park, Center for Biomedical Informatics and Information Technology (CBIIT), National Cancer Institute, Rockville, MD 20850, USA.
Kevin Jiang, Center for Biomedical Informatics and Information Technology (CBIIT), National Cancer Institute, Rockville, MD 20850, USA.
Belynda Hicks, Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, Rockville, MD 20850, USA.
Jia Liu, Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, Rockville, MD 20850, USA.
Daniel Russ, Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, Rockville, MD 20850, USA.
Lori Minasian, Division of Cancer Prevention, National Cancer Institute, Rockville, MD 20850, USA.
Paul Pinsky, Division of Cancer Prevention, National Cancer Institute, Rockville, MD 20850, USA.
Stephen J Chanock, Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, Rockville, MD 20850, USA.
Montserrat Garcia-Closas, Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, Rockville, MD 20850, USA.
Jonas S Almeida, Division of Cancer Epidemiology and Genetics (DCEG), National Cancer Institute, Rockville, MD 20850, USA.
Data Availability
Reference data at https://exploregwas-dev.cancer.gov/plco-atlas.
Tutorial at https://youtu.be/87dXT9YtbfY (17 min).
References
- Almeida J.S. et al. (2019) Serverless OpenHealth at data commons scale—traversing the 20 million patient records of New York’s SPARCS dataset in real-time. PeerJ, 7, e6230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Almeida J.S. et al. (2021) Mortality tracker: the COVID-19 case for real time web APIs as epidemiology commons. Bioinformatics, 37, 2073–2074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. et al. (2009) Computer science. Beyond the data deluge. Science, 323, 1297–98. [DOI] [PubMed] [Google Scholar]
- Black A. et al. (2015) PLCO: evolution of an epidemiologic resource and opportunities for future studies. Rev. Recent Clin. Trials, 10, 238–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohagan J.K. et al. (2015) The PLCO cancer screening trial: background, goals, organization, operations, results. Rev. Recent Clin. Trials, 10, 173–180. [DOI] [PubMed] [Google Scholar]
- Grossman R.L. (2018) Progress toward cancer data ecosystems. Cancer J., 24, 126–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath T., Bizer C. (2011). Linked Data: Evolving the Web into a Global Data Space. Morgan & Claypool Publishers, San Rafael, CA, USA. [Google Scholar]
- MacArthur J.A.L. et al. (2021) Workshop proceedings: GWAS summary statistics standards and sharing. Cell Genomics, 1, 100004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Machiela M.J., Chanock S.J. (2018) LDassoc: an online tool for interactively exploring genome-wide association study results and prioritizing variants for functional investigation. Bioinformatics, 34, 887–889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perkel J.M. (2021) Reactive, reproducible, collaborative: computational notebooks evolve. Nature, 593, 156–157. [DOI] [PubMed] [Google Scholar]
- Wilkinson M.D. et al. (2016) The FAIR guiding principles for scientific data management and stewardship. Sci. Data, 3, 160018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
Reference data at https://exploregwas-dev.cancer.gov/plco-atlas.
Tutorial at https://youtu.be/87dXT9YtbfY (17 min).