Abstract
Summary
Amidst the continuing spread of coronavirus disease-19 (COVID-19), real-time data analysis and visualization remain critical the general public to track the pandemic’s impact and to inform policy making by officials. Multiple metrics permit the evaluation of the spread, infection and mortality of infectious diseases. For example, numbers of new cases and deaths provide easily interpretable measures of absolute impact within a given population and time frame, while the effective reproduction rate provides an epidemiological measure of the rate of spread. By evaluating multiple metrics concurrently, users can leverage complementary insights into the impact and current state of the pandemic when formulating prevention and safety plans for oneself and others. We describe COVID-19 Spread Mapper, a unified framework for estimating and quantifying the uncertainty in the smoothed daily effective reproduction number, case rate and death rate in a region using log-linear models. We apply this framework to characterize COVID-19 impact at multiple geographic resolutions, including by US county and state as well as by country, demonstrating the variation across resolutions and the need for harmonized efforts to control the pandemic. We provide an open-source online dashboard for real-time analysis and visualization of multiple key metrics, which are critical to evaluate the impact of COVID-19 and make informed policy decisions.
Availability and implementation
Our model and tool are publicly available as implemented in R and hosted at https://metrics.covid19-analysis.org/. The source code is freely available from https://github.com/lin-lab/COVID19-Rt and https://github.com/lin-lab/COVID19-Viz.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
Data-driven efforts for pandemic tracking have remained essential as the disease has continued to spread and resurge in previously affected regions due to persistent circulation of variants and emergence of new variants, to the detriment of the healthcare system, economy and general welfare. Vaccination efforts have been broadening to control the pandemic’s spread; however, they may not be sufficient alone amidst the emergence of new variants. Significant efforts including non-pharmaceutical interventions (NPIs) are still needed to mitigate continued risks and contain the coronavirus disease-19 (COVID-19) pandemic (Fontanet and Cauchemez, 2020; Kim et al., 2021; Moore et al., 2021). There are many metrics for assessing COVID-19 spread that are valuable for allowing the public and authorities to examine trends and for policymakers to guide containment efforts; multiple metrics must be evaluated concurrently to provide complementary insights. Resources have tracked counts or rates of cases and deaths to measure absolute impact within a given population and time frame, but these do not fully characterize the pandemic’s spread due to difficulty in attributing deaths specifically to COVID-19, lack of reliability or lag in counts. Models including compartment and AI approaches have been developed to track transmission and metrics (Ahmad et al., 2021; Pham et al., 2021; Tang et al., 2020). However, compartmental approaches do not provide continuous Rt measures and these models are limited by stringent assumptions, computational tractability for real-time modeling, applicability only to certain locales and intervention-based specification (Supplementary Material). A key, dynamic measure of the pandemic’s spread is the effective reproduction number, Rt. It is a time-dependent measure of how fast the pandemic is spreading and is defined as the average number of people who become infected from an infectious person (Inglesby, 2020). Some existing and former tools incorporated Rt (Abbott et al., 2020; Systrom and Krieger, 2020; Worden et al., 2020) but were limited to a small set of geographic locations. Thus resources have provided location- and metric-specific insight but consequently do not completely capture the manifold, heterogeneous impact of COVID-19. We developed the COVID-19 Spread Mapper approach and site to provide the quantification, visualization and comparison of a set of metrics on COVID-19 spread across multiple geographic resolutions.
2 Implementation
We developed COVID-19 Spread Mapper, a unified framework and online dashboard for estimating, quantifying uncertainty in and visualizing the daily effective reproduction number Rt, case rate and death rate in a region. Our open-source website Visualizing COVID-19 Spread Metrics (https://metrics.covid19-analysis.org) uses a flexible non-parametric log-linear model to estimate and display epidemiological measures leveraging a data feed from multiple reporting sources that have been aggregated and retrieved daily. It allows for real time reporting of COVID-19 metrics in an interactive dashboard easily used by the general public, accounting for aspects such as case and death reporting delays and differences by weekday. It is essential to provide regularly updated reporting in order to allow for corresponding action by authorities, while accounting for the fact that there can be case and death reporting delays and differences by weekday. Specifically, our framework estimates smooth curves of time-varying metrics, including Rt, case rate and death rate curves, using a B-spline of time by accounting for overdispersion on aggregated case and death data in a region, and offers visualization of their trajectories (Supplementary Material). As Rt is an imperative measure to reduce, real-time reporting of Rt facilitates continual assessment of disease transmission, impact of NPIs and vaccination programs, and managing resources such as hospital beds (Inglesby, 2020). In contrast to existing tools, our framework and dashboard allows for the quantification, visualization and comparison of a comprehensive set of real-time essential metrics on COVID-19 spread for multiple geographic resolutions.
3 Application
To assess the pandemic’s spread and impact, we first estimated metrics globally at subnational and national levels. The COVID-19 Spread Mapper tool demonstrates the dynamic, heterogeneous nature of the metrics, such as fluctuations in Rt by season as captured in snapshots from our tool (Fig. 1). We observe the trajectory of spread from Asia to Europe, Africa and the Americas; this has been evidenced by genetic evolution of COVID-19. These snapshots demonstrate that many continents were impacted in Spring 2020, with higher rates concentrating in Europe, Asia and Africa in Summer 2020.
Fig. 1.
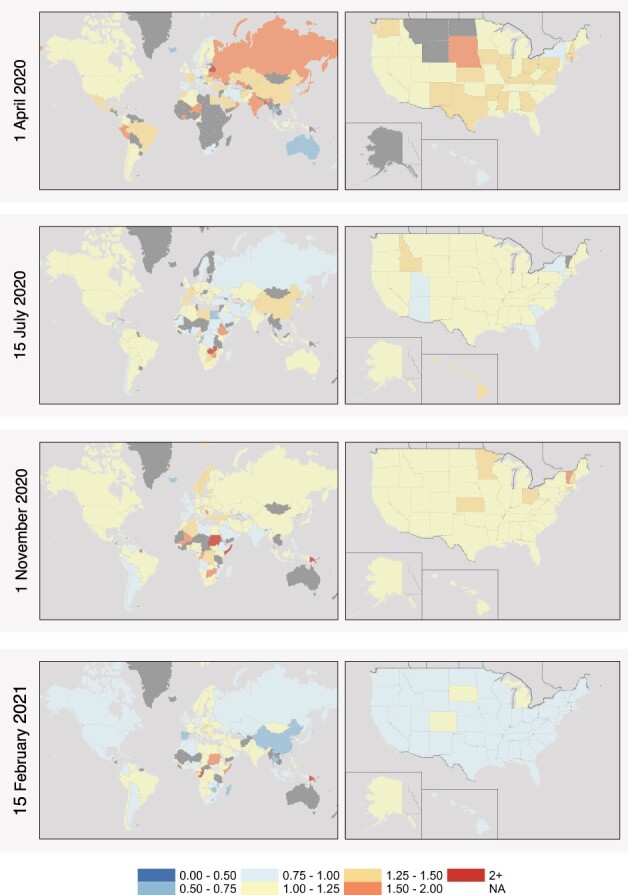
Effective reproductive number map over time. Seasonal maps of effective reproductive number Rt estimated on April 1, 2020, July 15, 2020, November 1, 2020 and February 15, 2021, by both country and US state for locations with sufficient data
We further investigate the spread at two levels in the United States: states (Fig. 1, Supplementary Figs S1 and S2) and counties (Supplementary Fig. S3). Given differences throughout the country in aspects such as demographics, health care access, population density and pandemic response, the pandemic’s impact has varied over time in the United States. States initially impacted in Spring 2020 demonstrated lower rates in Summer 2020, and most states experienced decreasing spread from Fall 2020 into Winter 2021. There is significant spatial variation in estimated metrics across geographic units, where it is evident in the United States that state-level measures obscure more local trends at country and regional levels in cases and spread. County-level measures in New York (Supplementary Fig. S3) further illustrates heterogeneity in pandemic spread, even with a state. Given that strategies and resources are provisioned from multiple levels of governance and more generally that geographic units do not function in isolation, this demonstrates that accurate tracking and targeted interventions require study at multiple geographic resolutions. Subnational units are provided for additional countries on the web resource. Given that the impact of the pandemic is heterogeneous across finer geographic levels, it is critical to track at multiple resolutions and target responses and interventions accordingly.
4 Discussion
Dashboards and epidemiological metrics have provided an integral resource for the general public and policymakers during the pandemic. Our framework and tool, COVID-19 Spread Mapper, critically provides a comprehensive and real time suite of tracking and analyses at multiple geographical resolutions. We provide multiple metrics from case counts to Rt, allowing individuals from all backgrounds to be informed regarding the pandemic’s impact at multiple levels and to encourage and expedite the consideration of complementary measures when facilitating pandemic surveillance and making informed decisions regarding interventions, behaviors and vaccination. The multiple levels and comparisons provided allow for users to target their regions of interest and understand the pandemic’s impact in their location and broadly. While there is an interplay of many factors contributing to the pandemic spread, tracking trajectories of spread, cases and deaths allows for understanding of and response to the pandemic’s impact on communities.
Data Availability
The data underlying this article are from the COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (Dong, 2020) and are available online at https://github.com/CSSEGISandData/COVID-19.
Funding
This work was supported by a grant from the Partners in Health.
Conflict of Interest: none declared.
Supplementary Material
Contributor Information
Andy Shi, Department of Biostatistics, Harvard TH Chan School of Public Health, Boston, MA 02115, USA.
Sheila M Gaynor, Department of Biostatistics, Harvard TH Chan School of Public Health, Boston, MA 02115, USA.
Rounak Dey, Department of Biostatistics, Harvard TH Chan School of Public Health, Boston, MA 02115, USA.
Haoyu Zhang, Department of Biostatistics, Harvard TH Chan School of Public Health, Boston, MA 02115, USA.
Corbin Quick, Department of Biostatistics, Harvard TH Chan School of Public Health, Boston, MA 02115, USA.
Xihong Lin, Department of Biostatistics, Harvard TH Chan School of Public Health, Boston, MA 02115, USA; Department of Statistics, Harvard University, Cambridge, MA 02138, USA.
References
- Abbott S. et al. ; CMMID COVID Modelling Group. (2020) Estimating the time-varying reproduction number of SARS-CoV-2 using national and subnational case counts. Wellcome Open Res., 5, 112. [Google Scholar]
- Ahmad A. et al. (2021) The number of confirmed cases of COVID-19 by using machine learning: methods and challenges. Arch. Comput. Methods Eng., 28, 2645–2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong,E. et al. (2020) An interactive web-based dashboard to track COVID-19 in real time. Lancet Infect. Dis., 20, 533–534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontanet A., Cauchemez S. (2020) COVID-19 herd immunity: where are we? Nat. Rev. Immunol., 20, 583–584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inglesby T.V. (2020) Public health measures and the reproduction number of SARS-CoV-2. J. Am. Med. Assoc., 323, 2186–2187. [DOI] [PubMed] [Google Scholar]
- Kim J.H. et al. (2021) Looking beyond COVID-19 vaccine phase 3 trials. Nat. Med., 27, 205–207. [DOI] [PubMed] [Google Scholar]
- Moore S. et al. (2021) Vaccination and non-pharmaceutical interventions for COVID-19: a mathematical modelling study. Lancet Infect. Dis., 21, 793–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pham Q.-V. et al. (2021) Artificial intelligence (AI) and big data for coronavirus (COVID-19) pandemic: a survey on the state-of-the-arts. IEEE access, 8, 130820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Systrom K., Krieger M. (2020) Rt.live. https://rt.live (1 December 2020, date last accessed).
- Tang L. et al. (2020) A review of multi-compartment infectious disease models. Int. Stat. Rev., 88, 462–513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Worden L. et al. (2020) Estimation of COVID-19 transmission rates in California and the US with reporting delays. medRxiv. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data underlying this article are from the COVID-19 Data Repository by the Center for Systems Science and Engineering (CSSE) at Johns Hopkins University (Dong, 2020) and are available online at https://github.com/CSSEGISandData/COVID-19.


